Abstract
The scientific community has joined forces to mitigate the scope of the current COVID-19 pandemic. The early identification of the disease, as well as the evaluation of its evolution is a primary task for the timely application of medical protocols. The use of medical images of the chest provides valuable information to specialists. Specifically, chest X-ray images have been the focus of many investigations that apply artificial intelligence techniques for the automatic classification of this disease. The results achieved to date on the subject are promising. However, some results of these investigations contain errors that must be corrected to obtain appropriate models for clinical use. This research discusses some of the problems found in the current scientific literature on the application of artificial intelligence techniques in the automatic classification of COVID-19. It is evident that in most of the reviewed works an incorrect evaluation protocol is applied, which leads to overestimating the results.
Keywords: COVID-19, Chest X-rays, Artificial intelligence, Deep learning
Introduction
COVID-19 is a new member of the family of coronaviruses belonging to the Acute Respiratory Syndromes (SARS-CoV) and has been called SARS-CoV-2 [1]. This coronavirus outbreak appeared in China at the end of 2019 and was notified to the world on December 31 of that year, since then to date millions of people have been infected with the disease.1 The main symptoms of the virus are: fever, sore throat, dry cough, muscle ache, and acute respiratory distress [2].
The rapid spread of the coronavirus and the serious effects it causes in humans, make an early diagnosis of the disease imperative [3]. To this day, the gold standard for detecting the presence of the virus is from the Reverse Transcription Polymerase Chain Reaction (RT-PCR). This test was designed by the Nobel laureate in Chemistry, Kary Mullis in the 1980s, which allows making a small amount of DNA millions of copies, so that there is enough to analyze it. Very high variability is introduced into the test sampling process, depending on the site where it is taken, the personnel taking it and the person's viral load at that time [4]. Furthermore, the procedure for PCR testing is a time-consuming process, around 6 to 9 h to confirm infection [5]. On the other hand, the tests have a sensitivity of between 60 and 70% depending of the stage of the disease [6].
One of the variants for the detection of positive patients may be based on the analysis of medical images [7]. The typical characteristics of the images and their evolution play an important role in the detection and management of the disease. The specialists rely on radiological studies, either by chest X-Rays (CXR) or computed tomography (CT) to follow the evolution of the disease. In a CT image, the overlapping structures are removed among slices, improving image contrast and making the internal anatomy more apparent. Studies confirm visible abnormalities in radiographic images, making this an important decision-making tool for human specialists [8]. However, 50% of patients have a normal CT scan within the first two days after symptoms of COVID-19 appear [8]. It is important to note that there are patients who present positive PCR, but do not develop signs or symptoms of the disease. These patients have normal radiographic studies. Therefore, they cannot be detected as positive using an image of their lungs.
The use of CT as a diagnostic method for COVID-19 has several drawbacks. In many hospitals the necessary equipment to acquire the image is not available and the cost of a tomographic study is not cheap. The dose of ionizing radiation delivered to the patient in this equipment is relatively high. The disinfection time among patients for the CT equipment and the room is approximately 15 min. On the other hand, CXR images have some advantages compared to CT, which make this modality a more extended way to patients. For example, this technology is available in most health care facilities. There is a portable modality that prevents the patient to move, minimizing the possibility of spreading the virus and exposing the patient to a lower dose of ionizing radiation and it is cheaper than a CT scan.
In both cases, the main role of diagnosis lies in the presence of radiologists for image analysis. However, the COVID-19 findings are in many cases very subtle. Expert radiologists are able to identify only 65% of positive patients [9]. One way to mitigate this drawback would be the application of Artificial Intelligence (AI) techniques. In this way, clinicians can be equipped with an X-ray imaging-based early warning tool for the detection of COVID-19.
Following this idea, a large number of researchers have been working on the issue of automatic classification of COVID-19 from CXR images [10, 11, 12, 13, 14, 15, 16, 17,18]. These studies report systems with high performance rates. In fact, these results are well above those obtained by experienced radiologists [9]. This issue must be handled with care in order not to generate false expectations in the area [19]. Therefore, this research critically analyzes the main methodologies and results achieved in the works published to date on the subject. Both, studies published in refereed journals and in digital repositories have been taken into account. The aim of the research is to present to the scientific community a summary of the work developed on this topic worldwide in the year 2020. In addition, to make a critical presentation, in the opinion of the authors of this work, of why most of this research leads to unreliable results. This is the main difference of our research with other review studies like [20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30] that analyze automatic classification of COVID-19 using CXR images, because none of them address the problems related to the lack of generalization reported in several papers [31]– [34].
Use of AI in CXR image classification
Computer vision (CV) tasks in recent years have been dominated by deep learning (DL) techniques, implemented by the deep neural networks (DNN) [35]. Compared with traditional neural networks, DNN have the ability to extract hidden and sophisticated structures (both, linear and non-linear features) contained in the raw data. Such ability is intrinsically related, on the one hand, to the capacity to model their own internal representation and, on the other hand, to their ability for generalizing any kind of knowledge. Also, they are extremely flexible in the types of data they can support. Moreover, their learning procedure can be adapted to a great variety of learning strategies, from unsupervised to supervised techniques, going through intermediate strategies. Specifically, convolutional neural networks (CNN) have been used, which specialize in the classification of images. DL has been favored due to three fundamental factors. The first is related to the increase in existing data in the present digital age, as there are large data sets used in the training of these algorithms. The second is related to the increase in computing capacities, with the use of specialized processors such as GPUs (graphic process unit) and TPUs (tensor process unit), implementing advanced processing techniques such as batch partition, in particular on parallel and distributed architectures, allowing DNN models to scale better when dealing with large amounts of data. Finally, there are the high-performance rates achieved in complicated applications that are difficult to explain for humans [36]. Among the technological applications of DL are: audio processing, text analysis, natural language processing and image recognition, among others [37].
These potentialities achieved by DL suggest that it could be an ideal candidate to support radiologists in their diagnosis. In fact, one of the tasks addressed has been the automatic classification of CXR images. Sets of this type of images are available,2 on which many researchers have proposed novel solutions that improve the visual analysis that could be done a priori of the different pathologies. In addition, works have been done to identify the different types of pneumonia from these images [38]. The results of using CNN to diagnose disease have been promising, but X-ray trained models from one hospital or group of hospitals have not yet been shown to work equally well in different hospitals [39]. Among the existing limitations there are the biases that the image sets may contain [40, 41]. For example, in the works [39, 42] there are discrepancies in terms of the results achieved when training and evaluating the DL algorithms on sets that do not come from the same source. Specifically, in the work [42], there were four sets A, B, C and D. It was observed that when training and evaluating on set A (using appropriately techniques to divide the sets) the results are higher than, when trains using sets B, C, and D, and evaluate using set A.
That is, CNN performance estimates, based on test data from CXR systems used for model training, may exaggerate their likely performance in actual clinical routine. For example, it was shown that the site of acquisition, both with respect to the CXR system used and the specific department within a hospital, can be predicted with very high precision [39]. This feature should be taken into account when training models of this type, as the network can learn the source of the images rather than the pathology being identified. On the other hand, normally, the greater the amount of data (images) used to train the algorithm, the greater the power of generalization it must have [43]. However, this is not entirely true in these cases, due to possible biases related to imbalances in the amounts of positive and negative images used for training, most of the time with different origin, as well as the different characteristics of the images in each set, due to different mAs, kVp, detection geometry, image size, pixel intensity, artifacts and labels, among others, which if not handled properly, can lead to erroneous results, as will be discussed in the following sections.
CXR and CT in AI models for COVID-19 classification
Diagnosing COVID-19 from CXR images is a complicated task for radiologists. They must identify typical patterns of the disease that are often shared with other types of viral pneumonia, which leads to errors in their diagnosis. A more accurate alternative for disease detection is CT imaging. This technique is considered the most accurate in identifying typical findings in the lungs of COVID-19 [44] and plays a fundamental role in the diagnosis and evaluation of COVID-19 pneumonia [45]. Note that ground glass opacities in the periphery of the right lower lobe on CT, which is one of the typical findings of the disease, are often not visible on CXR [46].
Contrary to what has been explained, the results reported to date seem to be more favorable for CXR than for CT. For example: a comprehensive review of the main sets of images, methods and performance indices achieved in automatic classifications is presented in papers [23, 26]. For example, in [26] a total of 80 articles published between February 21 to June 20, 2020 are reviewed. Of these works, 52 use CXR images, 30 use CT and 2 use both types of images. Taking into account the performance indices reported in the studies consulted, it is observed that automatic classifications using CXR achieve better results than when using CT. Note that the average accuracy (Acc) for CT is 90% and for CXR 96%. These results coincide with those reported in the works of [23, 27] where it is also reported that the performance indices of the models were higher using CXR images than when using CT images. In [22], works [47, 48, 49, 50, 51, 52] were reviewed and it was observed that they were based on small and poorly balanced data sets, with questionable evaluation procedures and without a plan for their inclusion in the flows clinical work.
Several are the advances reported in the scientific literature related to the automatic classification of CXR and CT images for the detection of COVID-19 [20, 21, 23]– [28]. These revision works constitute a starting point since they systematize the main knowledge achieved so far. The main objective of reviewing these works was to learn from the successes and errors of previous research, and to learn about aspects that have been overlooked or slightly studied.
The first published work that reviews the progress made using X-ray images to detect COVID-19 was [20]. This research also explains the role of AI in the prognosis of outbreaks of the disease. As one of the existing challenges to achieve a correct classification using CXR images, the need for large quantities of quality images is raised, which, in general, are not available in international databases. The studies analyzed were [10, 53, 54, 55, 56]. In these investigations, the number of positive images used in the training was less than 100, which greatly limits the generalization power of the models, under the CNN paradigm. In previous studies, binary classification (COVID-19 vs Normal) was performed. It is known that since COVID-19 is a type of pneumonia, a more challenging task is to identify, among the different types of pneumonia, those caused by coronavirus.
The medical imaging scientific community has been assisted by AI in managing COVID-19, an issue reflected in [21]. There is a need to use segmentation methods for the identification of COVID-19, which must be applied in two directions. The first to determine the region of the lungs and the second to fix the lesions that appear within them. However, segmentation in CXR images is a more challenging task compared to CT. In CT, each slice removes the amount of information that is above and below it, improving image contrast. On the other hand, in CXR images the ribs and soft tissues are projected in 2D, thus producing an overlap of information that affects the image contrast. According to what was reviewed in [21], until now, there was no method developed to segment CXR images specific for COVID-19. In fact, the investigations that review the work [18, 47, 54, 56] do not use segmentation methods to locate the region of the lungs, nor to locate the lesions on these. It should be mentioned that due to the dissimilar manifestations of the disease, it is difficult to select regions of interest with useful findings for classification, since they can appear in almost all regions of the lungs. Note that the disease has to be diagnosed only using an image that contains the region of the lungs, which means its bounding box. According to these studies, the COVID-19 positive CXR images used in the experimentation came mostly from the set collected by Cohen [57], which contained 70 images of positive patients. The works [23, 26, 28] confirm this set of images available on GitHub3 as the most used, followed by the sets available on Kaggle2,.4
In [25], works published in reliable databases such as IEEE explore, Web of Science, Science Direct, PubMed and Scopus are analyzed. The study resulted in the review of 11 articles of which only 6 are based on CXR to identify COVID-19, these were [15–17], [58–60]. It was confirmed that the quality and size of the existing images for the task differs greatly from one set to another, as well as the limited number of images that exist for experimentation. Among the proposed alternatives is the increase of the data and the segmentation of regions of interest (ROI). One of the important aspects in obtaining reliable models, according to the authors, is the selection and pre-processing of image sets.
There is a consensus among all these studies that the results obtained in the diagnosis of the disease, based on medical images of CT and CXR are encouraging. Likewise, there is a criticism regarding the limited number of positive images for the correct evaluation of the robustness of the methods, or to obtain models with the power of generalization to be used in clinical settings. Due to this lack of images, the approaches used do not take into account the patients' disease, important information that physicians must handle. In [61] it is stated that the most common causes of risk of bias in diagnostic models based on medical images are, the lack of information to evaluate the selection bias and the lack of a clear report of the image annotation procedures and quality control.
Due to the high complexity of the DNN where a lot of parameters needing to be determined or tuned, a large number of training samples are usually required for deep learning methods. However, previous work agrees that insufficient imaging for training has led research to advance with small sets of images available and apply data augmentation techniques when possible. Even though, the research does not discuss the limitations of the approaches used for the automatic classification of COVID-19. The high performances achieved by the methods used are not questioned either. It should be taken into account that the results obtained by human specialists from the CXR technique are far below of those obtained using AI techniques. Furthermore, the CT technique is considered the most accurate in identifying typical COVID-19 findings, however, the best results using AI techniques are obtained when using CXR.
Biases in used CXR images sets
One of the fundamental aspects to achieve a significant contribution of AI in the battle against the coronavirus, is the compilation of an adequate set of images in terms of quality and quantity. Despite the high number of patients with COVID-19 worldwide, there is no a free set of CXR images with the necessary quality for the construction of a diagnostic system with clinical value for the detection and follow-up of this disease with the use of AI. Radiologists have expressed concern about the limited availability of images to train AI-based models and the possible bias in these models [61], mainly related to the origin place of the positive images to COVID-19.
On the other hand, it is the right of the patient to decide when, how, and to what extent, others can access their medical information. Therefore, the informed consent of the patient must be obtained when their data is used for scientific research purposes. In this case, a process is carried out that includes anonymizing the data. In our view, this is the main reason for the relative low availability of data at present. Hospitals generally protect their patients' confidential information, as improper handling of data over networks can lead to legal problems.
From the publication by Cohen et al. [57] where a set of COVID-19 positive images is freely placed at the service of the international scientific community, a large number of works have been carried out that apply AI techniques for automatic classification of the illness. That is, to this day, this is the main source of COVID-19 positive images freely available worldwide. The formula used by most of the investigations to increase the number of negative images (that do not present COVID-19) has been adding images from sets available from other sources, which have different origin. This way of generating the sets introduces serious problems, which affect the results of the algorithms. For example, if there is any bias in the data set, such as corner labels, typical characteristics of a medical device, or other factors such as similar age of patients, same sex, etc., the classification model learns to recognizing these biases in the data set, rather than focusing on the findings they are trying to determine. In fact, the images contain little or no metadata on age, gender, pathologies present in the subjects, or other necessary information to detect this type of bias.
Another aspect that can introduce biases in the sets is the acquisition parameters such as mAs and kVp, something that the deep model could learn to discriminate. That is, a model can group images according to the scan tool used for the exam; if some scan configurations correspond to all the pneumonia examples, they will generate a false correlation, which the model can exploit to produce apparently favorable classification accuracy. Another example is given by the textual labeling in the images, if all negative examples contain similar markings, the deep model could learn to recognize this characteristic instead of focusing on the lung content, etc. In addition, these sets of images do not represent the severity of the disease in the same amount, with the majority of patients in an advanced stage of the disease, where the signs are more pronounced [62].
Due to the above, it is suspected that the high-performance values obtained so far by AI techniques are mainly due to the fact that the images can present marked differences that make the learning task an easy process for the algorithm. In [31] the current assessment protocols for the identification of COVID-19 from CXR images are strongly criticized. Mainly, the use of the complete image without selecting the region of the lungs and keeping the labels on the images and especially, the non-use of an evaluation set that does not come from any of the sources used in the training. In this study, it is tested how the CNN used was able to classify images that did not contain the region of the lungs. This was replaced by a black square, and even so, the classification was successful, with an Acc greater than 95%. It was demonstrated that the classification algorithms are learning patterns from the set of images, which do not correlate with the presence of the disease to be detected. The heterogeneity of the images makes the CNN learn characteristics that do not belong in themselves to COVID-19 [31, 33, 34]. Due to the existing limit in terms of pages allowed in writing, it was limited to creating Table 1 with the works published in peer-reviewed journals that make use of this methodology of selecting images from different sources to create their sets of images. This way of evaluating the algorithms does not guarantee their generalizability as will be discussed in later sections. Note that the number of images by classes presented in the table refers to the number used at the time of publication of the cited study. Therefore, these amounts may have varied from then to date.
Table 1.
Main papers published in peer-reviewed journals for COVID-19 detection using CXR
| Ref | Available code | Algorithms | Performance Index | Sets of images | Number of images per class |
|---|---|---|---|---|---|
| [10] | no |
-VGG19 -MobileNetv2 -Inception -Xception -Inception ResNet v2 |
Acc=96.78% Se=98.66% Sp=96.46% |
-(Cohen3,RSNA2, Radiopediaa, SIRMb)c -NIH14 |
224 COVID-19 / 700 bacterial pneumonia / 504 normal 224 COVID-19 / 400 bacterial pneumonia y 314 viral pneumonia / 504 normal |
| [12] | no |
-MobileNetv2, -SqueezeNet -ResNet18 -ResNet101 -DenseNet201 -CheXNet, -Inceptionv3 -VGG19 |
Acc=99.7% Pr=99.7% Se=99.7% Sp=99.55% |
-Cohen3,RSNA2, Radiopediaa, SIRMb | 423 COVID-19 / 1485 viral pneumonia / 1579 normal |
| [64] | no | FrMEM, manta-ray Foraging Optimization, Knn |
Acc=96.09% Pr=98.75% Acc=98.09% Pr=98.91% |
Dataset 1 -Cohen3, Kaggled Dataset 2 -same set of images used in [12] |
216 COVID-19 / 1675 negatives 219 COVID-19 / 1341 negatives |
| [13] | no | CNN-LSTM combinada |
Acc=99.4% AUC=99.9 Se=99.3% Sp=99.2% F1score=98.9% |
-(Cohen3, Agchunge,f, Radiopediaa, TCIAg, SIRMb) -Kaggled -NIHh |
613 COVID-19 / 1525 pneumonias / 1525 normal |
| [65] | no |
Resne50 Resnet101 |
Acc=97.77% | Cohen3, Kaggled | 440 COVID-19 / 480 viral pneumonia / 457 bacterial pneumonia / 455 normal |
| [5] | no |
SVM RF BPN ANFIS CNN VGGNet ResNet50 Alexnet GoogleNet Inception V3 Xception modificada |
Acc=97.4% Fmeausre=96.96% Se=97.09% Sp=97.29% Kappa=97.19% |
Same set of images used in [16] | |
| [14] | no |
CNN+Knn CNN+DT CNN+SVM |
Acc=98.97% Se=89.39% Sp=99.75 Fscore=96.72% |
Same set of images used in [12] | 219 COVID-19 / 1345 viral pneumonia / 1341 normal |
| [15] | no | Ensemble Resnet18 |
Acc=88.9% Pr=83.4% Recall=85.9% F1score=84.4% Sp=96.4% Acc=88.9% Pr=83.4% Recall=85.9% F1score=84.4% Sp=96.4% |
Dataset 1 [Cohen3, CoronaHacki, NLC(MC)j, JSRTk] Dataset 2 COVIDxp |
180 COVID-19 / 54 bacterial pneumonia / 20 viral pneumonia / 57 tuberculosis 191 normal 180 COVID-19 / 6012 pneumonias / 8851 normal |
| [16] | yes | DarkCovidNet |
Acc=87.02% Se=85.35% Sp=92.18% Pr=89.96% F1score=87.37 |
Cohen3, ChestX-ray8l | 127 COVID-19 / 500 pneumonias / 500 normal |
| [66] | no | nCOVnet |
Acc=88.09% Se=97.62% Sp=78.57% |
Cohen3, Fig. 1 Actuale Kaggle4 |
192 COVID-19 / 5863 negatives |
| [17] | yes |
Feature Extraction LBP, EQP, LDN, LETRIST, BSIF, LPQ, oBIFs, Inception-V3 Classifiers Knn, SVM, MLP, DT, RF |
F1score=88.89% |
RYDLS-20 [Cohen3, Radiopediaa, Chest X-ray14m] |
180 COVID-19 / 20 MERS / 22 SARS / 20 Varicella / 24 Streptococcus / 22 Pneumocystis / 2000 normal |
| [33] | no | COVID-SDNet | Acc=97.37% | COVIDGR-1.0n | 377 COVID-19 / 377 negatives |
| [59] | yes |
MobileNetV2 SqueezeNet SVM |
Acc=99.27% | Cohen3, Radiopediaa, Kaggle6 | 295 COVID-19 / 98 pneumonias / 65 normal |
| [67] | yes | Inception V3 |
Binary Acc=100% Se=99.0% Sp=100% AUC=100% Ternary Acc=85% Se=94% Sp=92.7% AUC=96% Quaternary Acc=76% Se=93% Sp=91.8% AUC=93% |
Cohen3, RSNA2,Kaggled, Kermanyo | 122 COVID-19 / 150 bacterial pneumonias / 150 viral pneumonias / 150 normal |
| [58] | no | COVIDiagnosis-Net based on SqueezeNet with Bayesian optimization |
Acc=98.3% Spe=99.1% F1score=98.3% MCC=97.4% |
COVIDxu | 76 COVID-19, 4290 pneumonias / 1583 normal |
| [68] | yes |
VGG-19 ResNet-50 COVID-Net |
Acc=93.3% Se=91% |
COVIDxu (Cohen3, Fig. 1 COVID-19j, ActualMed COVID-19k, RSNA2, COVID-19 radiography database5) |
190 COVID-19, 8614 Pneumonia, 8066 normal |
a https://radiopaedia.org/articles/pneumonia
bhttps://www.sirm.org/en/category/articles/covid-19-database/
chttps://www.kaggle.com/andrewmvd/convid19-X-rays
dhttps://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
ehttps://github.com/agchung/Figure1-COVID-chestxray-dataset
fhttps://github.com/agchung/Actualmed-COVID-chestxray-dataset
ghttps://www.cancerimagingarchive.net/
hhttps://www.kaggle.com/nih-chest-xrays/data?select=Data_Entry_2017.csv
ihttps://www.kaggle.com/praveengovi/coronahack-chest-xraydataset
jhttp://archive.nlm.nih.gov/repos/chestImages.php
lhttps://www.cc.10.nih.gov/drd/summers.html
mhttps://nihcc.app.box.com/v/ChestXray-NIHCC
nhttps://github.com/ari-dasci/OD-covidgr/releases/tag/1.0
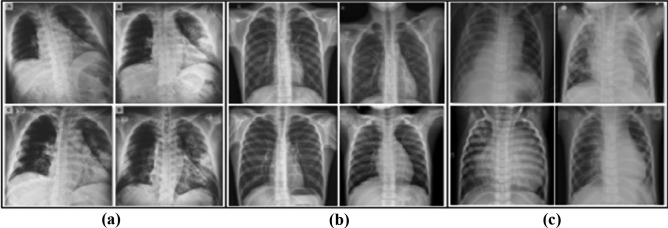
Another important aspect that works against the good performance and reliability of the systems that have been proposed is the large number of artifacts that the images contain. Many of the positive images for COVID-19 present intubated patients, with electrodes and their cables, pacemakers, bras (in women), zippers, among others. This aspect can be another considerable source of bias, since when images acquired under other conditions are classified, not taking into account these characteristics could lead to false negatives. A detailed description of the characteristics of the image sets used in COVID identification studies appears in [63]. This research highlights the biases that exist in each of these sets that can confuse the algorithms. In [59] three sets of public access images are combined. The positive images were obtained from the combination of the images available on GitHub3 and Kaggle,5 76 and 219 respectively. The normal class contains 65 images and the pneumonia class contains 98 images. The image set used is available from Kaggle.6 Figure 1 shows a selection of these images. There are marked differences among the groups of images, perceptible to a not trained human eye; that are not related to differences produced by the diseases they contain. For example, notice in (a) at the top left how a light-colored label always appears. Also, in (a) the black background cannot be seen in the rest of the images. On the other hand, in (c) pulmonary structures are observed totally different from the rest, since they belong to children.
Fig. 1.
Representation of three groups of images. In (a) images positive for COVID-19, in (b) normal images and in (c) images with pneumonia of another type. Taken from [59]
There is no doubt that these sets of images are important for COVID-19 identification studies. However, great attention must be paid to how to use them. Most of the investigations that use sets obtained in a similar way to that explained above, obtain very high-performance indices. Note that the sensitivity of human specialists is around 65% [9]. All of the above suggests that it is necessary to investigate and work on the digital pre-processing of the images to be used to train and validate the systems, so that it is aimed at eliminating the origin biases that the data have, which are generating an overfitting of the algorithms and little or no level of generalizability for their clinical use.
Pre-processing and data augmentation
Medical images can be affected by various sources of distortion and artifacts. As a consequence, the visual evaluation of these images by human specialists, or by AI algorithms, becomes a difficult task. Therefore, one of the initial tasks to obtain better results is the pre-processing of the image. In DL environments, large amounts of images are required to perform training properly and to avoid algorithms overfitting. These large amounts of images are generally not available in medical settings, which involve a variety of techniques. One of the variants used to avoid overfitting of the DL algorithms has been the increase of the set of images [69]. This technique is called data augmentation and consists of applying transformations on the images, with the aim of increasing the set to be used. The main modifications made to the image set as part of its pre-processing, as well as to increase its quantity are discussed below.
One techniques used in the training process to increase the set of images have been, moving the image a number of pixels by rows and / or columns, flipping horizontally and / or vertically, as well as rotating in all directions [70]. In addition, other variants have been applied such as modification of the intensity of the pixels [58, 71] and types of filtering [72].
Although CXR images are grayscale, some studies have used techniques to recolorize them. In [73] four pre-processing and data augmentation schemes are tested in the image set. These were: using the original image without performing any pre-processing, using the CLAHE technique [74], complementing the image and finally combining these modifications in each of the channels. Another alternative has been to use diffuse color techniques as presented in [59]. New images have also been generated based on generative adversarial nets (GAN) technique [75]. In [33] a variant of the GAN technique is used to generate two images per class, which are not interpretable for humans, but help to improve the performance of the algorithms from 77% effectiveness up to 81%.
Another of the applied techniques is the modification of the intensity of the pixels, from the adjustment of the contrast, or simply increasing or decreasing the intensity by a certain amount. In [15], the histogram equalization is carried out as a pre-processing stage, then a gamma correction of its intensity with γ = 0.5 to increase the contrast in the darker regions, which belong to lung, followed by a resizing to 256 × 256 pixels. This results in the intensities of the pixels for the heart and lungs having similar distributions in their histograms in different sets of images. This step should compensate for biases due to differences in the mAs and kVp acquisition parameters among the different image sets.
In the COVID-19 detection environment from CXR, several pre-processing methods have been applied to extract its characteristics or use them directly as input to CNNs. Due to the heterogeneity of images in terms of their dimensions, one of the first steps is to resize them, generally to 224 × 224x3 or 229 × 229x3 pixels. This is because most pre-trained CNNs use these fixed sizes as input. Image normalization has also been applied, using the mean and standard deviation obtained from the ImageNet image set [76]. However, better results have been reported, when training from scratch in the identification of pneumonia and after that apply transfer learning technique [12]. The CNN most used in this task has been ResNet, with different amounts of layers. Its use has been reported in a total of 27 articles [26]. In other cases, the image is resized depending on the input size of the proposed network architecture. For example, in [67] it is resized to 512 × 512 pixels. Something similar is done in [77], using images with three channels (RGB). In [18] it is resized to 480 × 480x3 pixels and in [78] to 200 × 200 pixels. The reduction of the dimensions of the images leads to lightening the computational cost of CNN training. Note that the CNN-based algorithms used in these tasks sometimes have more than 14 million parameters [16, 66].
One problem to attend to when images are resized is that algorithms generally work with square images, but the images used are not always square, which implies modifying the aspect ratio of the image to achieve this. One of the alternatives is reported in the work of [10], where they are scaled in a ratio of 1: 1.5, leaving 200 × 266 pixels. Those images that did not fit this scale were filled with zeros. This step can introduce a bias in the learning of the network. This is because if the images that come from a data set have similar dimensions to those that do not come from that set, they will be marked when they are completed with zeros.
In order to balance the training set, the data augmentation is performed before training [14, 58]. The combination of increasing the data and the balance of the classes improves the performance of the algorithms, reaching approximately 98% Acc in both investigations. However, it is not correct to also increase the test set, as is done in [58], since images that do not belong to a real set are being evaluated. Therefore, these reported results do not guarantee reliability in the final model.
The preprocessing stage corrects the intensity of the pixels to avoid appreciable differences between the different groups of images that make up these sets. However, many of the investigations do not take into account the elimination of the marks that in the images that can help the network to determine which class it belongs to, without this being related to the disease to be classified. One of the alternatives to alleviate this weakness is to use only the region that delimits the lungs. This requires applying a segmentation method. The advantages of performing this step are discussed in the next section.
Segmentation of the lung region
Among the alternatives used to eliminate biases from the data sets related to the labels of the images, it is proposed to work only with an image that contains the region of the lungs. The segmentation technique separates the image into different regions. Each of these regions is made up of a set of pixels that share certain common characteristics. The use of this technique in image processing allows simplifying the representation of the image into something more useful and easier to use. Segmentation can aid in more reliable detection of COVID-19 by extracting the region of the lungs. In this way, areas that do not belong to the region of interest (ROI) are left out of the analysis. Studies are reported that correctly use these methods to extract the region of the lungs and then perform the learning as seen in the works of [15], [32–34], [79–82].
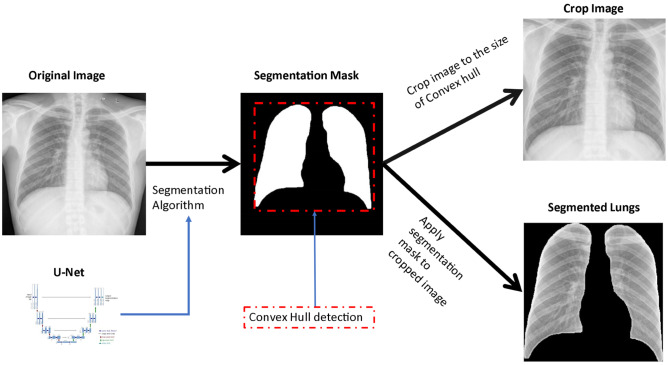
Segmentation can be done manually by human specialists, but it is a time-consuming task. In [17] the images used are manually cropped to avoid these biases. However, there are currently segmentation algorithms capable of doing this automatically. Some DL algorithms have shown good results in segmentation tasks. In the work [15] the algorithms FC-DenceNet67, FC-Dencenet103 and U-Net are compared to segment the region of the lungs in CXR images. It was evidenced that between the last two techniques there are no significant differences in their behavior. In fact, most studies that segment the lungs use U-Net, or some of its variants [26]. Figure 2 shows one of the variants used by the researchers, where we start from a complete CXR image and arrive at a cropped image, which contains only the region of the lungs.
Fig. 2.
Process of extraction of the region of the lungs. U-Net is applied as a segmentation method and a cropped image is obtained
In [81] a new strategy based on CNN ensembles is successfully applied. It is shown that applying transfer learning over a similar domain, as well as iteratively pruning the layers of the CNNs that do not activate, and finally, combining the algorithms, yielded good results in the identification of COVID-19. To remove irrelevant information from the image and ensure reliable DL models, U-Net was applied as a segmentation method. The images used belong to four repositories available online, these were: Pediatric CXR21 [83], RSNA2 [84] which contains images from Chestx-ray8 [85], Twitter COVID-197 and GitHub3. A split was performed at the patient level using 90% for training and 10% for testing.
The study [80] proposed a cascade model to assist doctors in the diagnosis of COVID-19. First, a SEME-ResNet50 architecture is used to classify into three classes: normal, bacterial pneumonia, and viral pneumonia. In the second stage, SEME-DenseNet161 was used to distinguish if viral pneumonia is COVID-19 or not. To exclude the influence of non-pathological features, the images are pre-processed using U-Net in the second stage. The results show an accuracy of 85.6% in the first stage, to determine the type of pneumonia and 97.1% in the second stage, for the identification of COVID-19.
In [32] the effect of performing lung segmentation by applying CNN on CXR images to identify COVID-19 is evaluated. U-Net was used for image segmentation and three popular CNN models like Inception, ResNet and VGG were used for classification. Two explainable artificial intelligence methods were used to visualize the areas on which the models were based to perform the classification. Furthermore, the impact of constructing sets of images from different sources as well as the generalizability of the models was evaluated. However, only the positive images for COVID-19 came from different sources since the negative images came only from RSNA2. It was shown that the main findings that networks use to perform classification using the whole image mainly appear outside the region of the lungs and it is related to marks that the images present. In addition, an experiment was conducted to determine whether the network could classify the database it came from. The result was an F1-Score of 0.92 using the complete images and 0.7 using the segmented images. This shows that segmentation helps to eliminate the bias of algorithms learning to identify the source of provenance related to the labels. However, these results show that even applying the segmentation of the lung region, the network was able to identify its origin set.
These results suggest that CNNs are learning patterns that are not directly related to pathologies associated with images. By using the full images, the networks learn characteristics outside the region of the lungs. It is needed to apply an adequate evaluation protocol to determine the generalizability of the methods.
External set for evaluation of trained models
In previous studies, the use of an external set that did not come from any of the sources used in the training stage was not taken into account for the evaluation of the algorithms. Therefore, the generalizability of the model to new images that do not come from any of the sets used in training is unknown. The investigations that, following the previous approach, have used their own images to evaluate the proposed systems are presented below. In these cases, the results do not correspond to the high-performance values obtained in the majority of investigations that use an evaluation set that is a subset of the training set.
In [82] a cascade architecture to identify COVID-19 was presented. In the first stage, the segmentation of the lungs is carried out. This eliminates unnecessary information that is contained in the images for the purposes of classification of COVID 19 or another disease. U-Net was used to predict the segmentation mask. To prevent the system to learn inconsistent characteristics, it is identified if there is any indication of pneumonia in the region of the lungs. To do this, a binary classification is performed in "Normal" or "Pneumonia" using DenseNet-121 as CNN incrementally. In the next stage, an attempt is made to classify whether the pneumonia is due to COVID-19 or another type of cause. The public repositories used were, Padchest [86], RSNA2 and GitHub3. In addition, three other sets of images called NTUH, TMUH and NHIA were used, from hospitals in Taiwan, which are not available internationally. The training and testing process were carried out independently in the public and private sets. The results showed that, when using the images of the public sets in training and validating and testing on a partition of the same set, the results were very good. The same did not happen when the evaluation was carried out on private groups, where the results were considerably lower. The sensitivity and specificity, using the public repository as a test set, were 85.26% and 85.86% respectively. While, when using the private repository, the sensitivity decreased to 50% and the specificity to 40%, results that demonstrate a random classification. To improve the results, the sets were mixed, adding images of the private set in the training of the models. This time similar values were obtained in both test sets. Sensitivity and specificity were 91.43% and 99.44%, respectively, for the test set, composed of images from public repositories. In the case of the test set of the images from the private repository, values of 100% sensitivity and 75% specificity were obtained. This last evaluation variant does not seem to be adequate, since there is no external evaluation set, but rather the same training and evaluation protocol is followed with images that come from equal sets, and it has been shown that this variant overestimate the results.
In [33] the high sensitivity reached by most models for classification of COVID-19 is demystified. A new set of images called COVIDGR-1.0 was used that contains 754 images distributed in 377 positives and 377 negatives. All images were obtained on the same CXR equipment and using the same settings. All belong to the postero-anterior view (PA). The positive images were divided according to their severity into 76 normal, 80 mild, 145 moderate and 76 severe. This stratification in the positive class allowed to carry out an analysis of the behavior of the models according to the severity of the disease in the patients. The behavior of two of the best performing models was evaluated, these were COVIDNet [18] and COVID-CAPS [87], both trained in the COVIDx set [18]. The experiments show that these models are unable to determine the presence of COVID-19 in the COVIDGR-1.0 set since the Acc reported is approximately 50%. The COVIDNet, COVID-CAPS and ResN-50 models were re-trained using the new set and the results were slightly higher with an Acc of 65%, 61% and 72% respectively. The new proposal presented, called COVID-SDNet, surpassed the performance of the previous models, reaching 77% of Acc. An analysis was carried out by level of severity, and it showed that the model is capable of detecting with an effectiveness of 88% and 97% to moderate and severe cases respectively. However, the images with mild severity and the normal ones reached only 66% and 38%, respectively, of correct classification. This is because images that do not contain marked disease findings are difficult for systems to detect as well. In another experiment, those that were PCR positive with normal radiographs were removed from the set of images. The results showed an increase in performance indexes. The study shows that most of the models proposed to date, trained and evaluated on sets of heterogeneous images, lack the capacity for generalization. However, the study did not evaluate the proposed model on an external validation set. Therefore, there is no evidence of its generalizability power.
The studies developed in [34] appear along this same line. A new set is used for the evaluation called CORDA, obtained in Italy, which contains 447 images from 386 patients. Extensive experimentation was done in the study by combining different sets of images in training and testing. Two of the models with the best reported performances, COVID-Net [18] and ResNet-18, were evaluated. It was evidenced that not even performing the equalization of the histogram and then the segmentation of the region of the lungs, in order to try to eliminate the biases from the sets of images, it was possible to train models with the capacity of generalization. An AUC of 0.55 and 0.61 was obtained for COVID-Net and ResNet-18 respectively when evaluating on the CORDA set. These results demonstrate that algorithms learn characteristics related to the source data set, rather than the disease being classified. Therefore, an appropriate evaluation strategy in this environment is essential to build reliable models. One way to achieve a more reliable evaluation protocol is to separate the training and test images so that the images that belong to the test have a different origin than the images that were used in the training.
Conclusions
There is internationally a limited set of COVID-19 positive CXR images freely available on the internet for the use of the scientific community. Most of the studies complete the data with negative images from other data sources. These images have marked differences among different sets. This leads to very good results in the automatic classification of COVID-19 when evaluating using a subset of images from the set used. However, several studies report little or no power of generalization, when evaluating the trained models in their own sets. Even the models that were trained using pre-processing techniques, which tried to eliminate the biases belonging to the data sets, showed limited results. Therefore, most of the results achieved so far, which are reported in the scientific literature, present models that learn characteristics of the sets where they were trained. The absence of an adequate evaluation protocol means that most of the models developed still present little value in clinical settings.
Declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Footnotes
This article is part of the COVID-19 Health Technology: Design, Regulation, Management, Assessment
Contributor Information
José Daniel López-Cabrera, Email: josedaniellc@uclv.cu.
Rubén Orozco-Morales, Email: rorozco@uclv.cu.
Jorge Armando Portal-Diaz, Email: jportal@uclv.cu.
Orlando Lovelle-Enríquez, Email: lovelle@infomed.sld.cu.
Marlén Pérez-Díaz, Email: mperez@uclv.cu.
References
- 1.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salman S, Salem ML. Routine childhood immunization may protect against COVID-19. Med Hypotheses. 2020;140:109689. doi: 10.1016/j.mehy.2020.109689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie M, Chen Q. Insight into 2019 novel coronavirus — An updated interim review and lessons from SARS-CoV and MERS-CoV. Int J Infect Dis. 2020;94:119–124. doi: 10.1016/j.ijid.2020.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu R, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172–175. doi: 10.1016/j.cca.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narayan Das N, Kumar N, Kaur M, Kumar V, Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. IRBM, 2020. 10.1016/j.irbm.2020.07.001. [DOI] [PMC free article] [PubMed]
- 6.Ai T, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong D, et al. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng, pp. 1–1, 2020. 10.1109/RBME.2020.2990959. [DOI] [PubMed]
- 8.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH. Essentials for radiologists on COVID-19: an update—radiology scientific expert panel. Radiology. 2020;296(2):E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castiglioni I, et al. Artificial intelligence applied on chest X-ray can aid in the diagnosis of COVID-19 infection: a first experience from Lombardy, Italy. MedRxiv, 2020.
- 10.Apostolopoulos ID, Mpesiana TA. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020 doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asif S, Wenhui Y, Jin H, Tao Y, Jinhai S. Classification of COVID-19 from chest X-ray images using deep convolutional neural networks. MedRxiv. 2020. 10.1101/2020.05.01.20088211.
- 12.Chowdhury MEH, et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- 13.Islam MdZ, Islam MdM, Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked. 2020;20:100412. doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nour M, Cömert Z, Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl Soft Comput, p. 106580, 2020. 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed]
- 15.Oh Y, Park S, Ye JC. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans Med Imaging, 2020. 10.1109/TMI.2020.2993291. [DOI] [PubMed]
- 16.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med, vol. 121, p. 103792, 2020. 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed]
- 17.Pereira RM, Bertolini D, Teixeira LO, Silla CN, Costa YMG. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Programs Biomed, p. 105532, 2020. 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed]
- 18.Wang L, Liu ZQ, Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. 2020. http://arxiv.org/abs/2003.08971v4. [DOI] [PMC free article] [PubMed]
- 19.Laghi A. Cautions about radiologic diagnosis of COVID-19 infection driven by artificial intelligence. Lancet Digit Health. 2020;2(5):e225. doi: 10.1016/S2589-7500(20)30079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilyas M, Rehman H, Nait-ali A. Detection of Covid-19 from chest X-ray images using artificial intelligence: an early review. ArXiv Prepr. ArXiv200405436, 2020.
- 21.Shi F, et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev Biomed Eng, pp. 1–1, 2020, 10.1109/RBME.2020.2987975. [DOI] [PubMed]
- 22.Bullock J, Luccioni A, Pham KH, Lam CSN, Luengo-Oroz M. Mapping the landscape of artificial intelligence applications against COVID-19. ArXiv200311336 Cs, 2020, Accessed 25 Aug 2020. [Online]. Available: http://arxiv.org/abs/2003.11336.
- 23.Ulhaq A, Khan A, Gomes D, Paul M. Computer vision for COVID-19 control: a survey. http://arxiv.org/abs/2004.09420. Accessed 11 Jun 2020 [DOI] [PMC free article] [PubMed]
- 24.Nguyen TT. Artificial intelligence in the battle against coronavirus (COVID-19): a survey and future research directions. Prepr. DOI, vol. 10, 2020.
- 25.Albahri OS, et al. Systematic review of artificial intelligence techniques in the detection and classification of COVID-19 medical images in terms of evaluation and benchmarking: Taxonomy analysis, challenges, future solutions and methodological aspects. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah FM, et al. A comprehensive survey of COVID-19 detection using medical images. 2020. https://engrxiv.org/9fdyp/download/?format=pdf. [DOI] [PMC free article] [PubMed]
- 27.Shoeibi A, et al. Automated detection and forecasting of COVID-19 using deep learning techniques: a review. 2020. http://arxiv.org/abs/2007.10785. Accessed 14 Aug 2020.
- 28.Farhat H, Sakr GE, Kilany R. Deep learning applications in pulmonary medical imaging: recent updates and insights on COVID-19. Mach Vis Appl, vol. 31, no. 6, 2020. 10.1007/s00138-020-01101-5. [DOI] [PMC free article] [PubMed]
- 29.Islam MM, Karray F, Alhajj R, Zeng J. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19). 2020. http://arxiv.org/abs/2008.04815. Accessed 28 Aug 2020. [DOI] [PMC free article] [PubMed]
- 30.Chen Y, et al. A Survey on Artificial Intelligence in Chest Imaging of COVID-19. BIO Integr. 2020;1(3):137–146. doi: 10.15212/bioi-2020-0015. [DOI] [Google Scholar]
- 31.Maguolo G, Nanni L. A critic evaluation of methods for COVID-19 automatic detection from X-ray images. 2020. http://arxiv.org/abs/2004.12823. Accessed 21 May 2020. [DOI] [PMC free article] [PubMed]
- 32.Teixeira LO, Pereira RM, Bertolini D, Oliveira LS, Nanni L, Costa YMG. Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest X-ray images. 2020. http://arxiv.org/abs/2009.09780. Accessed 29 Sep 2020. [DOI] [PMC free article] [PubMed]
- 33.Tabik S, et al. COVIDGR dataset and COVID-SDNet methodology for predicting COVID-19 based on chest X-ray images. IEEE J Biomed Health Inform. 2020;24(12):3595–3605. doi: 10.1109/JBHI.2020.3037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tartaglione E, Barbano CA, Berzovini C, Calandri M, Grangetto M. Unveiling COVID-19 from chest X-ray with deep learning: a hurdles race with small data 2020. http://arxiv.org/abs/2004.05405. Accessed 16 Aug 2020. [DOI] [PMC free article] [PubMed]
- 35.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In Adv Neural Inform Process Syst 25, F. Pereira, C. J. C. Burges, L. Bottou, and K. Q. Weinberger, Eds. Curran Associates, Inc., 2012. pp. 1097–1105.
- 36.Goodfellow I, Bengio Y, Courville A, Bengio Y. Deep learning, vol. 1. MIT press Cambridge, 2016.
- 37.Hatcher WG, Yu W. A survey of deep learning: Platforms, applications and emerging research trends. IEEE Access. 2018;6:24411–24432. doi: 10.1109/ACCESS.2018.2830661. [DOI] [Google Scholar]
- 38.Baltruschat IM, Nickisch H, Grass M, Knopp T, Saalbach A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci Rep, vol. 9, no. 1, Art. no. 1, 2019. 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed]
- 39.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLOS Med. 2018;15(11):e1002683. doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao L, Prosky J, Covington B, Lyman K. A strong baseline for domain adaptation and generalization in medical imaging. 2019. http://arxiv.org/abs/1904.01638. Accessed 26 Aug 2020.
- 41.Prevedello LM, et al. Challenges related to artificial intelligence research in medical imaging and the importance of image analysis competitions. Radiol Artif Intell. 2019;1(1):e180031. doi: 10.1148/ryai.2019180031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen JP, Hashir M, Brooks R, Bertrand H. On the limits of cross-domain generalization in automated X-ray prediction. 2020. http://arxiv.org/abs/2002.02497. Accessed 05 Aug 2020.
- 43.Aggarwal CC. Neural networks and deep learning. Springer, 2018.
- 44.Aljondi R, Alghamdi S. Diagnostic value of imaging modalities for COVID-19: Scoping review. J Med Internet Res. 2020;22(8):e19673. doi: 10.2196/19673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poggiali E, et al. Can lung US help critical care clinicians in the early diagnosis of novel coronavirus (COVID-19) pneumonia? Radiology. 2020;295(3):E6–E6. doi: 10.1148/radiol.2020200847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng M-Y, et al. Imaging profile of the COVID-19 infection: Radiologic findings and literature review. Radiol Cardiothorac Imaging. 2020;2(1):e200034. doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghoshal B, Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. 2020. http://arxiv.org/abs/2003.10769. Accessed 29 Jul 2020.
- 48.Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. 2020. [DOI] [PMC free article] [PubMed]
- 49.Bukhari SUK, Bukhari SSK, Syed A, Shah SSH. The diagnostic evaluation of Convolutional Neural Network (CNN) for the assessment of chest X-ray of patients infected with COVID-19. 2020. 10.1101/2020.03.26.20044610.
- 50.Hammoudi K, et al. Deep Learning on chest X-ray images to detect and evaluate pneumonia cases at the era of COVID-19. 2020. http://arxiv.org/abs/2004.03399. Accessed 09 Sep 2020. [DOI] [PMC free article] [PubMed]
- 51.Karim MR, Döhmen T, Rebholz-Schuhmann D, Decker S, Cochez M, Beyan O. DeepCOVIDExplainer: Explainable COVID-19 diagnosis based on chest X-ray images. 2020. https://arxiv.org/abs/2004.04582v3. Accessed 10 Jul 2020.
- 52.Li X, Li C, Zhu D. COVID-MobileXpert: On-device COVID-19 screening using snapshots of chest X-ray. 2020.
- 53.Hemdan EED, Shouman MA, Karar ME. COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. 2020. http://arxiv.org/abs/2003.11055. Accessed 07 Aug 2020.
- 54.Narin A, Kaya C, Pamuk, Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. 2020. [DOI] [PMC free article] [PubMed]
- 55.Sethy PK, Behera SK, Ratha RK, Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. 2020. https://www.preprints.org/manuscript/202003.0300/v2. Accessed 16 Sep 2020.
- 56.Zhang J, Xie Y, Li Y, Shen C, Xia Y. Covid-19 screening on chest X-ray images using deep learning based anomaly detection. 2020.
- 57.Cohen JP, Morrison P, Dao L. COVID-19 image data collection. 2020.
- 58.Ucar F, Korkmaz D. COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med Hypotheses. 2020;140:109761. doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toğaçar M, Ergen B, Cömert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdel-Basset M, Mohamed R, Elhoseny M, Chakrabortty RK, Ryan M. A Hybrid COVID-19 detection model using an improved marine predators algorithm and a ranking-based diversity reduction strategy. IEEE Access. 2020;8:79521–79540. doi: 10.1109/ACCESS.2020.2990893. [DOI] [Google Scholar]
- 61.Naudé W. Artificial intelligence vs COVID-19: limitations, constraints and pitfalls. Ai Soc, p. 1, 2020. 10.1007/s00146-020-00978-0. [DOI] [PMC free article] [PubMed]
- 62.Kundu S, Elhalawani H, Gichoya JW, Kahn CE. How might AI and chest imaging help unravel COVID-19’s mysteries? Radiol Artif Intell. 2020;2(3):e200053. doi: 10.1148/ryai.2020200053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia Santa Cruz B, Sölter J, Nicolas Bossa M, Dominik Husch A. On the composition and limitations of publicly available COVID-19 X-ray imaging datasets. 2020. http://arxiv.org/abs/2008.11572. Accessed 21 Sep 2020.
- 64.Elaziz MA, Hosny KM, Salah A, Darwish MM, Lu S, Sahlol AT. New machine learning method for image-based diagnosis of COVID-19. PLoS One. 2020;15(6):e0235187. doi: 10.1371/journal.pone.0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jain G, Mittal D, Thakur D, Mittal MK. A deep learning approach to detect Covid-19 coronavirus with X-ray images. Biocybern Biomed Eng. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Panwar H, Gupta PK, Siddiqui MK, Morales-Menendez R, Singh V. Application of deep learning for fast detection of COVID-19 in X-rays using nCOVnet. Chaos Solitons Fract, p. 109944, 2020. 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed]
- 67.Tsiknakis N, et al. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp Ther Med. 2020;20(2):727–735. doi: 10.3892/etm.2020.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang L, Lin ZQ, Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep, vol. 10, no. 1, Art. no. 1, 2020. 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed]
- 69.Wong SC, Gatt A, Stamatescu V, McDonnell MD. Understanding data augmentation for classification: When to warp? In 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Gold Coast, Australia, 2016, pp. 1–6. 10.1109/DICTA.2016.7797091.
- 70.Luz EJ, Silva PL, Silva R, Silva LP, Moreira GJ, Menotti D. Towards an effective and efficient deep learning model for COVID-19 patterns detection in X-ray images. 2020. https://arxiv.org/pdf/2004.05717.pdf.
- 71.Farooq M, Hafeez A. COVID-resnet: A deep learning framework for screening of COVID19 from radiographs. 2020. http://arxiv.org/abs/2003.14395. Accessed 07 Aug 20.
- 72.Hassanien AE, Mahdy LN, Ezzat KA, Elmousalami HH, Ella HA. Automatic X-ray COVID-19 lung image classification system based on multi-level thresholding and support vector machine. 2020. 10.1101/2020.03.30.20047787.
- 73.Tahir A, et al. Coronavirus: Comparing COVID-19, SARS and MERS in the eyes of AI. 2020. http://arxiv.org/abs/2005.11524. Accessed 14 Aug 2020.
- 74.Pizer SM, et al. Adaptive histogram equalization and its variations. Comput Vis Graph Image Process. 1987;39(3):355–368. doi: 10.1016/S0734-189X(87)80186-X. [DOI] [Google Scholar]
- 75.Goodfellow I, et al. Generative adversarial nets. 2014. pp. 2672–2680. http://papers.nips.cc/paper/5423-generative-adversarial-nets. Accessed 06 Sep 2020.
- 76.Russakovsky O, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 77.Goodwin BD, Jaskolski C, Zhong C, Asmani H. Intra-model variability in COVID-19 classification using chest X-ray images. 2020. http://arxiv.org/abs/2005.02167. Accessed 11 Jun 2020.
- 78.Apostolopoulos I, Aznaouridis S, Tzani M. Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. 2020. [DOI] [PMC free article] [PubMed]
- 79.Alom MZ, Rahman MMS, Nasrin MS, Taha TM, Asari VK. COVID_mtnet: COVID-19 detection with multi-task deep learning approaches. 2020. http://arxiv.org/abs/2004.03747. Accessed 16 Aug 2020.
- 80.Lv D, Qi W, Li Y, Sun L, Wang Y. A cascade network for detecting COVID-19 using chest X-rays. 2020. http://arxiv.org/abs/2005.01468. Accessed 14 Aug 2020.
- 81.Rajaraman S, Siegelman J, Alderson PO, Folio LS, Folio LR, Antani SK. Iteratively pruned deep learning ensembles for COVID-19 detection in chest X-rays. 2020. http://arxiv.org/abs/2004.08379. Accessed 14 Aug 2020. [DOI] [PMC free article] [PubMed]
- 82.Yeh CF, et al. A cascaded learning strategy for robust COVID-19 pneumonia chest X-ray screening. 2020. http://arxiv.org/abs/2004.12786. Accessed 14 Aug 2020.
- 83.Kermany DS, et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 84.Shih G, et al. Augmenting the national institutes of health chest radiograph dataset with expert annotations of possible pneumonia. Radiol Artif Intell. 2019;1(1):e180041. doi: 10.1148/ryai.2019180041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 2017. pp. 3462–3471, 10.1109/CVPR.2017.369.
- 86.Bustos A, Pertusa A, Salinas J-M, de la Iglesia-Vayá M. PadChest: a large chest X-ray image dataset with multi-label annotated reports. Med Image Anal. 2020;66:101797. doi: 10.1016/j.media.2020.101797. [DOI] [PubMed] [Google Scholar]
- 87.Afshar P, Heidarian S, Naderkhani F, Oikonomou A, Plataniotis KN, Mohammadi A. COVID-CAPS: a capsule network-based framework for identification of COVID-19 cases from X-ray images. 2020. [DOI] [PMC free article] [PubMed]