Abstract
Small, noncoding sequences of ribonucleic acid called microRNAs (miRNAs, miR) are functioning as posttranscriptional regulators of gene expression. As they draw increasing attention of rheumatologists, there is a growing body of evidence concerning specific molecules that may affect the long-term care of patients with inflammatory arthritides. Findings involving children with juvenile idiopathic arthritis (JIA) are still limited though. The aim of the study was to browse the available data on microRNAs which may be utilized as potential biomarkers helpful in diagnosing and monitoring JIA patients. The review contains a brief summary on the most studied sequences: miR-16, miR-125a-5p, miR-146a, miR-155, and miR-223. It is complemented with other miRNAs possibly relevant for JIA (miR-145, miR-23b, miR-27a, and miR-204) and discussion on challenges for using miRNAs in pediatric rheumatology (particularly, issues regarding specificity of biomarkers and measurements involving synovial fluid).
1. Introduction
Small, noncoding sequences of microRNA (miRNA, miR) have already established their position as an innovative part of the diagnostic process in the bulk of disorders, with particular emphasis placed on neoplastic diseases. miRNAs should be considered as negative posttranscriptional regulators affecting the expression of genes involved in both innate and adaptive immune responses [1]. Karami et al. stated that nearly 60% of human genes contain at least one binding site for miRNA [2]. By direct base-pairing with the 3′ untranslated region (3′ UTR) of target mRNAs [3], miRNAs can suppress or impair gene translation leading to the inhibition of target protein synthesis [4]. A brief summary of miRNA biogenesis and its functioning is depicted in Figure 1.
Figure 1.
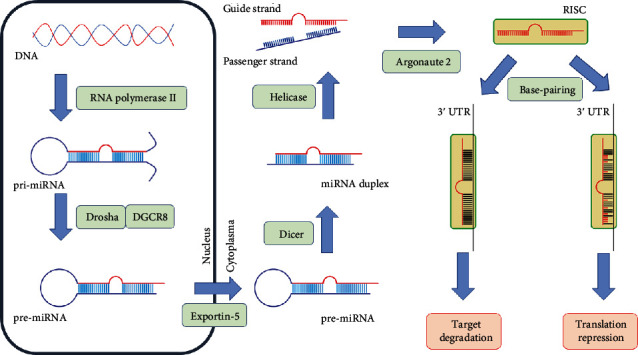
Biogenesis and functioning of miRNA. The figure illustrates the following steps: (a) endonuclear synthesis of the primary double-stranded miRNA transcript (pri-miRNA) of hundreds to thousands of nucleotides in length, mostly by RNA polymerase II; (b) cleavage of pri-miRNA by Drosha endonuclease accompanied by DiGeorge syndrome critical region protein 8 (DGCR8), resulting in formation of 60 to 70 nucleotide long pre-miRNA; (c) export of pre-miRNA from the nucleus into the cytoplasm by Exportin-5 coacting with GTP-binding nuclear protein Ran (Ran-GTP); (d) cleavage of pre-miRNA by Dicer into an asymmetric miRNA duplex containing guide and passenger strand of about 22 nucleotides in length; (e) unwinding the duplex by helicase and release of the passenger miRNA with its subsequent degradation; (f) loading the guide strand by Argonaute 2 into the RNA-induced silencing complex (RISC); (g) binding RISC (containing mature miRNA) to 3′ untranslated region (3′ UTR) of target mRNA; (h) translational repression or target degradation depending on the partial or full complementarity of base-pairing. Based on [60–62].
According to Ormseth et al. [5], miRNAs are not limited to intracellular activity, but they can also be transported to recipient cells through plasma, where they are protected from degradation by several mechanisms, involving exosomes, microvesicles, lipoproteins, and RNA-binding proteins. High robustness of cell-free miRNAs in body fluids (e.g., plasma and serum) supports their potential role as reliable biomarkers [6, 7]. However, concentrations of particular miRNAs may differ from each other depending on the source of the examined material. Interestingly, miR-132, which has been observed to be significantly decreased in serum of RA patients, turned out to be overexpressed in synovial PBMCs [8].
Growing popularity and availability of measuring these molecules reflects from PubMed statistics: only in 2019, the number of the registered papers regarding miRNA exceeded 15,000 articles. Of these, just 121 referred to adult patients with rheumatoid arthritis (RA), whereas juvenile idiopathic arthritis (JIA) was involved in merely eight studies. The first study which postulated the role of miRNA in the pathogenesis of RA dates back to 2007, when Bhanji et al. reported that sera of RA patients include antibodies against Argonaute 2 (a protein required for miRNA-mediated gene silencing) [9]. In the following years, Romo-Garcia et al. speculated that the transcriptional arrest present at the early stages of RA may be related to overexpression of miRNAs and subsequent intensified inhibition of translation [7]. The initial evidence regarding miRNA in JIA, specifically miR-155, was reported in 2014 [10]. A thorough literature review involving the PubMed database provided at least 55 miRNA sequences (listed and briefly described in Supplementary Table 1) which may be applicable to rheumatic diseases. They may affect the pathogenesis of JIA through several mechanisms which include, among others, repressing the expression of genes that are crucial for the TNF/IL-1 pathway [11] or dysregulating the macrophage polarization [12]. Within this paper, we will summarize the available data on the most studied miRNAs in RA and JIA (miR-16, miR-125a-5p, miR-146a, miR-155, and miR-223). Then, we will shed a light on several less known sequences which bear a potential for the future diagnostic process and monitoring disease activity in JIA patients.
2. miR-16
miR-16 seems to be one of the crucial factors for the pathogenesis of RA and JIA, as through targeting the silencing mediator for retinoid and thyroid hormone receptor (SMRT), it regulates the expression of proinflammatory cytokines, particularly interleukin- (IL-) 1α, IL-6, and IL-8 [13]. Zhou et al. [14] hypothesized the role of miR-16 as a key regulator of Toll-like receptor-signaled inflammatory response. According to Murata et al. [15], miR-16 levels detected in RA patients were markedly higher than in osteoarthritis patients. Furthermore, Demir et al. [16] reported elevated miR-16 concentrations in JIA patients when compared to healthy controls. Nevertheless, the plasma level of this molecule fluctuates in a disease activity-dependent manner which may lead to contradictory findings varying on the measurement protocol. Hence, miR-16 was significantly upregulated in children at the onset of both poly- and oligoarticular JIA [17], but it inversely correlated with Disease Activity Score 28-Joint Count (DAS28) in patients with the average RA duration of 10.4 years [15]. Interestingly, Filková et al. [18] observed that miR-16 is higher in patients with established RA than in the early stage of the disease. Moreover, they hypothesized that this molecule may be utilized as a biomarker of RA outcome because its pretherapy level correlated with disease activity drop after 9 months of treatment. Correlation between high disease activity and miR-16 concentrations was also observed in inflammatory bowel diseases [19] and in ankylosing spondylitis [20]. Nonetheless, children with juvenile spondyloarthropathy had lower miR-16 concentrations than JIA patients [17]. Regarding JIA, miR-16 tends to have higher values in poly- rather than in oligoarthritis [17]. However, the discrepancy observed between subtypes was insufficient to differentiate patients basing only on that parameter.
3. miR-125a-5p
Research in rheumatology on miR-125a-5p involved mainly patients with systemic onset of JIA (SoJIA), which is characterized by autoinflammatory rather than autoimmune response. Monocytes isolated from children in the active phase of SoJIA abundantly overexpressed miR-125a-5p when compared to patients in clinical remission or with active polyarthritis [21]. Moreover, Do et al. [22] noted that miR-125a-5p markedly restricted activation of macrophages' anti-inflammatory phenotype involving expression of CD163. Furthermore, Leong et al. [23] observed a positive correlation between miR-125a-5p and indicators of systemic inflammation (particularly ferritin) with no parallel relation to joint involvement. However, due to the distinct pathogenesis of SoJIA, findings from this subtype cannot be directly extrapolated to all JIA patients. Evangelatos et al. [24] reported increased miR-125a-5p levels in early RA patients, regardless of the results of “classic” serological markers (rheumatoid factor and anticitrullinated protein antibodies). It is also worth interjecting here that miR-125b, which shares the seed sequence and target specificity with miR-125a-5p [25], was the first microRNA recognized as a predictor of treatment outcome in RA patients (specifically, response to rituximab) [26]. Therefore, the final position of measuring miR-125a-5p in JIA patients with active joint inflammation merits further investigation.
4. miR-146a
The pathogenetic role of miR-146a in developing inflammatory arthritis is expressed through several pathways of insufficient negative feedback [27]. By targeting inhibin beta A (INHBA), it regulates differentiation of macrophages [28] towards M2 type [29]. It also targets IL-1 receptor-associated kinase 1 and 2 (IRAK1, IRAK2) as well as tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) [30], which enables suppression of NFkB signaling [3] and modulation of TNF production [31]. Furthermore, miR-146a promotes the expression of proinflammatory cytokines (including, among others, IL-2 and IL-12) by its upregulation in Th1 [32] and Th17 lymphocytes [24] with concurrent downregulation in Th2 cells [33]. Despite being decreased in early RA when compared to the patients with established diagnosis [34], miR-146a was found to be elevated in oligo-, polyarticular, and SoJIA [17, 28]. Besides, the single-nucleotide polymorphism (SNP) of miR-146a sequence was postulated to be associated with susceptibility to develop enthesitis-related arthritis (JIA-ERA) [35] and ankylosing spondylitis [36]. Concerning disease activity, miR-146a positively correlated with DAS28 [15] and Juvenile Arthritis Disease Activity Score 27-Joint Count (JADAS27) [17]. Moreover, Li et al. [28] found miR-146a levels to be associated with the systemic features. Additionally, the molecule has also been assessed as a good predictor of response to methotrexate therapy (AUC 0.760 in the study conducted by Singh et al. [37]).
5. miR-155
Through targeting the suppressor of cytokine signaling 1 (SOCS1), miR-155 acts as a key promoter of M1 macrophages [38]. At the same time, it directly affects IL-13 receptor alpha 1 (IL-13Rα1) which results in the inhibition of M2 activation [39]. Besides, Kurowska-Stolarska et al. found the significant expression of miR-155 in CD68+ macrophages within the lining layer of the synovial membrane in RA patients [40]. Furthermore, miR-155 enhances the production of IL-2 by peripheral blood mononuclear cells (PBMCs) [10]. Additionally, Vigorito et al. [41] reported the impact of miR-155 on B cell maturation into class-switched plasma cells releasing immunoglobulin G1. Interestingly, transcriptional alterations detected in neutrophils isolated from plasma of children with JIA seem to perpetuate regardless of the disease activity [42]. Demir et al. [16] found that upregulation of miR-155 was even more significant in JIA patients with the improvement in disease activity after 6 months of therapy. However, in adults with ankylosing spondylitis, the elevated miR-155 levels correlated with an increase in disease activity index [43]. miR-155 concentrations tended to be higher in children with polyarthritis, but the difference did not reach statistical significance [16]. Moreover, Ma et al. [17] obtained similar miR-155 values in patients diagnosed with JIA or juvenile spondyloarthropathy which further diminished its specificity. As summarized by Su et al. [44], overexpression of miR-155 in RA patients correlated with the inflammatory markers, DAS28 as well as with TNF and IL-1β levels. Similar to miR-146a, miR-155 appeared to be a promising prognostic marker of methotrexate effectiveness (AUC 0.728 in the same study [37]).
6. miR-223
The upregulation of miR-223 in peripheral naïve CD4+ T lymphocytes [45] plays a pivotal role in the development of local joint inflammation. Given that miR-223 is involved in the activation of M2 macrophages through targeting PBX/Knotted 1 Homeobox 1 (Pknox 1) [46] and in the suppression of nucleotide-binding oligomerization domain-like receptor protein 3 (NLRP3) inflammasome activity [47], its detected overexpression may be hypothesized as an indicator of malfunctioning inflammatory response. Intriguingly, miR-223 applies not only to the articular manifestations, as the elevated serum levels of the molecule were found also in children with SoJIA [6]. Moreover, miR-223 concentrations were associated with CRP levels [18] and titer of rheumatoid factor (RF) [48] in RA patients. It also appeared to be a reliable predictor of change in disease activity within a 3- and 12-month observation [18]. Furthermore, elevated miR-223 values before treatment initiation were directly correlated with better response to methotrexate and inversely correlated with anti-TNF efficacy [24]. miR-223 seems to be the first epigenetic factor that was speculated as a novel RA therapeutic target by Li et al. [49], who observed that miR-223 inhibition reduced the disease severity of murine collagen-induced arthritis.
7. Other miRNAs Possibly Relevant for JIA
None of the aforementioned, widely studied miRNA sequences were characterized as being specific to oligoarticular JIA, which is considered as the most common subtype of the disease. Nevertheless, Sun et al. [50] noted that miR-145 is upregulated in oligoarthritis comparing to polyarthritis and SoJIA. Besides, miR-23b appeared to be overexpressed in RA patients with the relevant titer of antinuclear antibodies (ANA) [51]. Given that the variability of gene expression may be correlated with a child's age at disease onset [52], such findings may contribute to the task of defining early onset ANA-positive arthritis which is planned to be discerned in the upcoming reclassification of JIA [53].
Current approach to JIA therapy includes prompt administration of biological agents in patients who did not respond to the first-line treatment, namely, methotrexate [54]. Interestingly, miR-27a may serve as an efficient predictor of whether a patient is likely to improve on anti-TNF therapy. Increased initial miR-27a levels in adults with early RA were associated with better response to a combination therapy of methotrexate with adalimumab, particularly when the concentrations of the molecule decreased within the first 3 months of such treatment [55]. On the other hand, suspending methotrexate in JIA children that achieved clinical remission on medication is another frequent concern of pediatric rheumatologists. Nonetheless, miR-204 turned out to be markedly decreased in patients with inactive disease who continue methotrexate in monotherapy [16]; therefore, it may become a biomarker helpful in therapeutic decisions.
8. Challenges for miRNA in Pediatric Rheumatology
JIA is the most common arthropathy in childhood, though it is not characterized by homogenous natural history of the disease. Patients differ from each other with regard to the disease subtype, number and location of the inflamed joints, time of developing new symptoms and complications, and treatment response. One of the major objectives of research in pediatric rheumatology is to seek for and determine effective markers (mainly serological and genetic) which may facilitate prognosing the disease course and therefore optimize JIA treatment. However, insufficient specificity is a frequent drawback of biomarkers utilized to measure the activity of inflammation. The most studied serological biomarkers in JIA include S100 proteins, particularly S100A8/A9 [56] and S100A12 [57], which are very sensitive in detecting high disease activity. Nevertheless, chronic neutrophil activation may be observed as well in other entities with a distinct origin. For instance, Hu et al. obtained similar S100A8/A9 values in both JIA and cystic fibrosis patients [42]. miRNAs as a class of molecules are not free of such limitations. Despite the potential usefulness of miR-23b in diagnosing rheumatic diseases, this molecule is also related to fibrogenesis within chronic liver injury and diabetic nephropathy [51]. Nevertheless, there are several miRNAs which have already been tested whether they are specific for systemic autoimmune diseases. Jin et al. reported three sequences (namely, miR-124, miR-448, and miR-551b) which had altered values in patients with RA, systemic lupus erythematosus, Sjögren's syndrome, and ulcerative colitis; the authors did not find significant differences of the miRNAs' levels between healthy controls and patients with pneumonia, sepsis, and HBV hepatitis [58].
There are several studies underlining the importance of a distinct route of miRNA expression in fibroblast-like synoviocytes (FLS) [15, 59]. Nziza et al. reported a set of miRNAs extracted from the synovial fluid (miR-6764-5p, miR-155, and miR-146a) that perfectly distinguish (AUC 1.0) children with JIA from patients with Kingella kingae septic arthritis [60]. However, such proceedings are more invasive than the examination of blood samples and exclude the possibility of comparison with healthy controls due to ethical issues. Therefore, further investigation of miRNAs in JIA should focus on the sequences that may ease the differentiation of patients basing on plasma and/or serum levels.
9. Conclusions
miRNAs seem to open a new chapter of diagnosing and monitoring activity of childhood arthropathies on the epigenetic level. They might provide the probable answer which patients are more likely to develop a more aggressive course of disease or respond properly to the administered therapy. The authors recommend to continue validation of the already well-known sequences (miR-16, miR-125a-5p, miR-146a, miR-155, and miR-223) and to further investigate the alternative promising molecules (miR-145, miR-23b, miR-27a, and miR-204). Studies including patients that are supposed to be recognized as early onset ANA-positive JIA are of utmost importance.
Acknowledgments
The study is a part of the National Science Centre research task MINIATURA 4 (decision No. 2020/04/X/NZ5/00117) and was supported by the Medical University of Lodz, Poland (grant No. 503/8-000-01/503-81-001-19-00).
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare no competing interests.
Authors' Contributions
KO collected literature and drafted the manuscript. ES supervised the overall study design and contributed to the manuscript preparation. All authors revised and approved the final version of the manuscript.
Supplementary Materials
Supplementary Table 1: list of microRNAs studied in rheumatology. ACPA: anti-citrullinated protein antibodies; ADA: adalimumab; ANA: antinuclear antibodies; AOSD: adult-onset Still's disease; AS: ankylosing spondylitis; BTRC: beta-transducin repeat containing gene; CD: cluster of differentiation; CF: cystic fibrosis; CIA: collagen-induced arthritis; CRP: C-reactive protein; DAS28: Disease Activity Score 28-Joint Count; DMARD: disease-modifying antirheumatic drug; ERA: enthesitis-related arthritis; ESR: erythrocytes sedimentation rate; FLS: fibroblast-like synoviocytes; foxp3: forkhead box P3; FMF: familial Mediterranean fever; GCS: glucocorticosteroids; IL: interleukin; IFNβ: interferon beta; INHBA: inhibin beta A; IRAK: interleukin-1 receptor-associated kinase; JADAS27: Juvenile Arthritis Disease Activity Score 27-Joint Count; JAK/STAT: Janus kinase/signal transducers and activators of transcription; JDM: juvenile dermatomyositis; JIA: juvenile idiopathic arthritis; JSA: juvenile spondyloarthropathy; miR: microRNA; MMP: matrix metalloproteinase; MRI: magnetic resonance imaging; MTX: methotrexate; NLRP3: NLR family pyrin domain containing protein 3; NFkB: nuclear factor kappa of activated B cells; OA: osteoarthritis; PBMC: peripheral blood mononuclear cell; PLT: platelet count; PsA: psoriatic arthritis; PTGS2: prostaglandin-endoperoxide synthase 2; RA: rheumatoid arthritis; RF: rheumatoid factor; RORyt: retinoic-acid-receptor-related orphan nuclear receptor gamma; RTX: rituximab; SLE: systemic lupus erythematosus; SNP: single-nucleotide polymorphism; SOCS3: suppressor of cytokine signaling 3; SoJIA: systemic onset of juvenile idiopathic arthritis; SS: Sjögren syndrome; SSZ: sulfasalazine; TAK1: transforming growth factor-beta-activated kinase 1; Th: helper T cells; TLR: Toll-like receptor; TNF: tumor necrosis factor; TRAF6: tumor necrosis factor receptor-associated factor 6; Treg: regulatory T cells; UC: ulcerative colitis; WBC: white blood count.
References
- 1.Cirillo F., Lazzeroni P., Sartori C., Street M. Inflammatory diseases and growth: effects on the GH-IGF axis and on growth plate. International Journal of Molecular Sciences. 2017;18(9):p. 1878. doi: 10.3390/ijms18091878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karami J., Aslani S., Tahmasebi M. N., et al. Epigenetics in rheumatoid arthritis; fibroblast-like synoviocytes as an emerging paradigm in the pathogenesis of the disease. Immunology and Cell Biology. 2020;98(3):171–186. doi: 10.1111/imcb.12311. [DOI] [PubMed] [Google Scholar]
- 3.Yan L., Liang M., Hou X., et al. The role of microRNA-16 in the pathogenesis of autoimmune diseases: a comprehensive review. Biomedicine & Pharmacotherapy. 2019;112, article 108583 doi: 10.1016/j.biopha.2019.01.044. [DOI] [PubMed] [Google Scholar]
- 4.Iwamoto N., Kawakami A. Recent findings regarding the effects of microRNAs on fibroblast-like synovial cells in rheumatoid arthritis. Immunological Medicine. 2019;42(4):156–161. doi: 10.1080/25785826.2019.1695490. [DOI] [PubMed] [Google Scholar]
- 5.Ormseth M. J., Solus J. F., Sheng Q., et al. Development and validation of a microRNA panel to differentiate between patients with rheumatoid arthritis or systemic lupus erythematosus and controls. The Journal of Rheumatology. 2020;47(2):188–196. doi: 10.3899/jrheum.181029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamiya Y., Kawada J., Kawano Y., et al. Serum microRNAs as potential biomarkers of juvenile idiopathic arthritis. Clinical Rheumatology. 2015;34(10):1705–1712. doi: 10.1007/s10067-015-2922-1. [DOI] [PubMed] [Google Scholar]
- 7.Romo-García M. F., Bastian Y., Zapata-Zuñiga M., et al. Identification of putative miRNA biomarkers in early rheumatoid arthritis by genome-wide microarray profiling: a pilot study. Gene. 2019;720, article 144081 doi: 10.1016/j.gene.2019.144081. [DOI] [PubMed] [Google Scholar]
- 8.Mi S., Zhang J., Zhang W., Huang R. S. Circulating microRNAs as biomarkers for inflammatory diseases. MicroRNA. 2013;2(1):64–72. doi: 10.2174/2211536611302010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhanji R. A., Eystathioy T., Chan E. K., Bloch D. B., Fritzler M. J. Clinical and serological features of patients with autoantibodies to GW/P bodies. Clinical Immunology. 2007;125(3):247–256. doi: 10.1016/j.clim.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lashine Y. A., Salah S., Aboelenein H. R., Abdelaziz A. I. Correcting the expression of miRNA-155 represses PP2Ac and enhances the release of IL-2 in PBMCs of juvenile SLE patients. Lupus. 2014;24(3):240–247. doi: 10.1177/0961203314552117. [DOI] [PubMed] [Google Scholar]
- 11.Hammaker D., Firestein G. S. Epigenetics of inflammatory arthritis. Current Opinion in Rheumatology. 2018;30(2):188–196. doi: 10.1097/BOR.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Świdrowska-Jaros J., Orczyk K., Smolewska E. Macrophages - silent enemies in juvenile idiopathic arthritis. Postępy Higieny i Medycyny Doświadczalnej. 2016;70:743–750. doi: 10.5604/17322693.1208887. [DOI] [PubMed] [Google Scholar]
- 13.Zhou R., Li X., Hu G., Gong A. Y., Drescher K. M., Chen X. M. miR-16 targets transcriptional corepressor SMRT and modulates NF-kappaB-regulated transactivation of interleukin-8 gene. PLoS One. 2012;7(1, article e30772) doi: 10.1371/journal.pone.0030772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R., Hu G., Gong A. Y., Chen X. M. Binding of NF-kappaB p65 subunit to the promoter elements is involved in LPS-induced transactivation of miRNA genes in human biliary epithelial cells. Nucleic Acids Research. 2010;38(10):3222–3232. doi: 10.1093/nar/gkq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murata K., Yoshitomi H., Tanida S., et al. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Research & Therapy. 2010;12(3) doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demir F., Çebi A. H., Kalyoncu M. Evaluation of plasma microRNA expressions in patients with juvenile idiopathic arthritis. Clinical Rheumatology. 2018;37(12):3255–3262. doi: 10.1007/s10067-018-4277-x. [DOI] [PubMed] [Google Scholar]
- 17.Ma X., Wu F., Xin L., et al. Differential plasma microRNAs expression in juvenile idiopathic arthritis. Modern Rheumatology. 2016;26(2):224–232. doi: 10.3109/14397595.2015.1060663. [DOI] [PubMed] [Google Scholar]
- 18.Filková M., Aradi B., Šenolt L., et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Annals of the Rheumatic Diseases. 2014;73(10):1898–1904. doi: 10.1136/annrheumdis-2012-202815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schönauen K., Le N., von Arnim U., Schulz C., Malfertheiner P., Link A. Circulating and fecal microRNAs as biomarkers for inflammatory bowel diseases. Inflammatory Bowel Diseases. 2018;24(7):1547–1557. doi: 10.1093/ibd/izy046. [DOI] [PubMed] [Google Scholar]
- 20.Wang M., Wang L., Zhang X., et al. Overexpression of miR-31 in peripheral blood mononuclear cells (PBMC) from patients with ankylosing spondylitis. Medical Science Monitor. 2017;23:5488–5494. doi: 10.12659/msm.905238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulert G. S., Fall N., Harley J. B., et al. Monocyte microRNA expression in active systemic juvenile idiopathic arthritis implicates microRNA-125a-5p in polarized monocyte phenotypes. Arthritis & Rhematology. 2016;68(9):2300–2313. doi: 10.1002/art.39694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Do T., Tan R., Bennett M., et al. MicroRNA networks associated with active systemic juvenile idiopathic arthritis regulate CD163 expression and anti-inflammatory functions in macrophages through two distinct mechanisms. Journal of Leukocyte Biology. 2018;103(1):71–85. doi: 10.1002/JLB.2A0317-107R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leong J. Y., Guan Y. J., Albani S., Arkachaisri T. Recent advances in our understanding of the pathogenesis of juvenile idiopathic arthritis and their potential clinical implications. Expert Review of Clinical Immunology. 2018;14(11):933–944. doi: 10.1080/1744666X.2018.1529757. [DOI] [PubMed] [Google Scholar]
- 24.Evangelatos G., Fragoulis G. E., Koulouri V., Lambrou G. I. MicroRNAs in rheumatoid arthritis: from pathogenesis to clinical impact. Autoimmunity Reviews. 2019;18(11, article 102391) doi: 10.1016/j.autrev.2019.102391. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y. M., Lin K. Y., Chen Y. Q. Diverse functions of miR-125 family in different cell contexts. Journal of Hematology & Oncology. 2013;6(1):p. 6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duroux-Richard I., Pers Y. M., Fabre S., et al. Circulating miRNA-125b is a potential biomarker predicting response to rituximab in rheumatoid arthritis. Mediators of Inflammation. 2014;2014:9. doi: 10.1155/2014/342524.342524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lai N. S., Koo M., Yu C. L., Lu M. C. Immunopathogenesis of systemic lupus erythematosus and rheumatoid arthritis: the role of aberrant expression of non-coding RNAs in T cells. Clinical & Experimental Immunology. 2017;187(3):327–336. doi: 10.1111/cei.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D., Duan M., Feng Y., Geng L., Li X., Zhang W. MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA. Molecular Immunology. 2016;77:205–212. doi: 10.1016/j.molimm.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Huang C., Liu X. J., QunZhou, et al. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. International Immunopharmacology. 2016;32:46–54. doi: 10.1016/j.intimp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Hou J., Wang P., Lin L., et al. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. Journal of Immunology. 2009;183(3):2150–2158. doi: 10.4049/jimmunol.0900707. [DOI] [PubMed] [Google Scholar]
- 31.Pauley K. M., Satoh M., Chan A. L., Bubb M. R., Reeves W. H., Chan E. K. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Research & Therapy. 2008;10(4) doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., Han Y., Qu H., Fang J., Ye M., Yin W. Correlation of microRNA expression profile with clinical response to tumor necrosis factor inhibitor in treating rheumatoid arthritis patients: a prospective cohort study. Journal of Clinical Laboratory Analysis. 2019;33(7, article e22953) doi: 10.1002/jcla.22953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monticelli S., Ansel K. M., Xiao C., et al. MicroRNA profiling of the murine hematopoietic system. Genome Biology. 2005;6(8) doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mookherjee N., El-Gabalawy H. S. High degree of correlation between whole blood and PBMC expression levels of miR-155 and miR-146a in healthy controls and rheumatoid arthritis patients. Journal of Immunological Methods. 2013;400-401:106–110. doi: 10.1016/j.jim.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Singh S., Rai G., Aggarwal A. Association of microRNA-146a and its target gene IRAK1 polymorphism with enthesitis related arthritis category of juvenile idiopathic arthritis. Rheumatology International. 2014;34(10):1395–1400. doi: 10.1007/s00296-014-3001-7. [DOI] [PubMed] [Google Scholar]
- 36.Xiao Y., Liu H., Chen L., Wang Y., Yao X., Jiang X. Association of microRNAs genes polymorphisms with arthritis: a systematic review and meta-analysis. Bioscience Reports. 2019;39(7) doi: 10.1042/BSR20190298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh A., Patro P. S., Aggarwal A. MicroRNA-132, miR-146a, and miR-155 as potential biomarkers of methotrexate response in patients with rheumatoid arthritis. Clinical Rheumatology. 2019;38(3):877–884. doi: 10.1007/s10067-018-4380-z. [DOI] [PubMed] [Google Scholar]
- 38.Niu X., Schulert G. S. Functional regulation of macrophage phenotypes by microRNAs in inflammatory arthritis. Frontiers in Immunology. 2019;10 doi: 10.3389/fimmu.2019.02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez-Nunez R. T., Louafi F., Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Rα1) The Journal of Biological Chemistry. 2011;286(3):1786–1794. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurowska-Stolarska M., Alivernini S., Ballantine L. E., et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proceedings of the National Academy of Sciences. 2011;108(27):11193–11198. doi: 10.1073/pnas.1019536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vigorito E., Perks K. L., Abreu-Goodger C., et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Z., Jiang K., Frank M. B., Chen Y., Jarvis J. N. Complexity and specificity of the neutrophil transcriptomes in juvenile idiopathic arthritis. Scientific Reports. 2016;6(1) doi: 10.1038/srep27453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qian B. P., Ji M. L., Qiu Y., et al. Identification of serum miR-146a and miR-155 as novel noninvasive complementary biomarkers for ankylosing spondylitis. Spine. 2016;41(9):735–742. doi: 10.1097/BRS.0000000000001339. [DOI] [PubMed] [Google Scholar]
- 44.Su L. C., Huang A. F., Jia H., Liu Y., Xu W. D. Role of microRNA-155 in rheumatoid arthritis. International Journal of Rheumatic Diseases. 2017;20(11):1631–1637. doi: 10.1111/1756-185X.13202. [DOI] [PubMed] [Google Scholar]
- 45.Fulci V., Scappucci G., Sebastiani G. D., et al. miR-223 is overexpressed in T-lymphocytes of patients affected by rheumatoid arthritis. Human Immunology. 2010;71(2):206–211. doi: 10.1016/j.humimm.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Chen Q., Wang H., Liu Y., et al. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS One. 2012;7(8, article e42971) doi: 10.1371/journal.pone.0042971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bauernfeind F., Rieger A., Schildberg F. A., Knolle P. A., Schmid-Burgk J. L., Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. Journal of Immunology. 2012;189(8):4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- 48.Lu M. C., Yu C. L., Chen H. C., Yu H. C., Huang H. B., Lai N. S. Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clinical and Experimental Immunology. 2014;177(3):641–651. doi: 10.1111/cei.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y. T., Chen S. Y., Wang C. R., et al. Brief report: amelioration of collagen-induced arthritis in mice by lentivirus-mediated silencing of microRNA-223. Arthritis & Rheumatism. 2012;64(10):3240–3245. doi: 10.1002/art.34550. [DOI] [PubMed] [Google Scholar]
- 50.Sun J., Feng M., Wu F., et al. Plasma miR-26a as a diagnostic biomarker regulates cytokine expression in systemic juvenile idiopathic arthritis. The Journal of Rheumatology. 2016;43(8):1607–1614. doi: 10.3899/jrheum.150593. [DOI] [PubMed] [Google Scholar]
- 51.Liu X., Ni S., Li C., et al. Circulating microRNA-23b as a new biomarker for rheumatoid arthritis. Gene. 2019;712:p. 143911. doi: 10.1016/j.gene.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giancane G., Alongi A., Ravelli A. Update on the pathogenesis and treatment of juvenile idiopathic arthritis. Current Opinion in Rheumatology. 2017;29(5):523–529. doi: 10.1097/BOR.0000000000000417. [DOI] [PubMed] [Google Scholar]
- 53.Martini A., Ravelli A., Avcin T., et al. Toward new classification criteria for juvenile idiopathic arthritis: first steps, pediatric rheumatology international trials organization international consensus. The Journal of Rheumatology. 2019;46(2):190–197. doi: 10.3899/jrheum.180168. [DOI] [PubMed] [Google Scholar]
- 54.Ringold S., Angeles-Han S. T., Beukelman T., et al. 2019 American College of Rheumatology/Arthritis Foundation guideline for the treatment of juvenile idiopathic arthritis: therapeutic approaches for non-systemic polyarthritis, sacroiliitis, and enthesitis. Arthritis & Rheumatology. 2019;71(6):846–863. doi: 10.1002/art.40884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sode J., Krintel S. B., Carlsen A. L., et al. Plasma microRNA profiles in patients with early rheumatoid arthritis responding to adalimumab plus methotrexate vs methotrexate alone: a placebo-controlled clinical trial. The Journal of Rheumatology. 2018;45(1):53–61. doi: 10.3899/jrheum.170266. [DOI] [PubMed] [Google Scholar]
- 56.Bojko J. Measurement of blood calprotectin (MRP-8/MRP-14) levels in patients with juvenile idiopathic arthritis. Reumatologia. 2017;1(1):18–22. doi: 10.5114/reum.2017.66682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Orczyk K., Smolewska E. A granulocyte-specific protein S100A12 as a potential prognostic factor affecting aggressiveness of therapy in patients with juvenile idiopathic arthritis. Journal of Immunology Research. 2018;2018:7. doi: 10.1155/2018/5349837.5349837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin F., Hu H., Xu M., et al. Serum microRNA profiles serve as novel biomarkers for autoimmune diseases. Frontiers in Immunology. 2018;9, article 2381 doi: 10.3389/fimmu.2018.02381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zakeri Z., Salmaninejad A., Hosseini N., et al. MicroRNA and exosome: key players in rheumatoid arthritis. Journal of Cellular Biochemistry. 2019;120:10930–10944. doi: 10.1002/jcb.28499. [DOI] [PubMed] [Google Scholar]
- 60.Nziza N., Jeziorski E., Delpont M., et al. Synovial-fluid miRNA signature for diagnosis of juvenile idiopathic arthritis. Cells. 2019;8(12, article 1521) doi: 10.3390/cells8121521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duroux-Richard I., Jorgensen C., Apparailly F. miRNAs and rheumatoid arthritis - promising novel biomarkers. Swiss Medical Weekly. 2011;141, article w13175 doi: 10.4414/smw.2011.13175. [DOI] [PubMed] [Google Scholar]
- 62.Cirillo F., Lazzeroni P., Catellani C., Sartori C., Amarri S., Street M. E. MicroRNAs link chronic inflammation in childhood to growth impairment and insulin-resistance. Cytokine & Growth Factor Reviews. 2018;39:1–18. doi: 10.1016/j.cytogfr.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 63.Nziza N., Duroux-Richard I., Apparailly F. MicroRNAs in juvenile idiopathic arthritis: can we learn more about pathophysiological mechanisms? Autoimmunity Reviews. 2019;18(8):796–804. doi: 10.1016/j.autrev.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Mu N., Gu J., Huang T., et al. A novel NF-κB/YY1/microRNA-10a regulatory circuit in fibroblast-like synoviocytes regulates inflammation in rheumatoid arthritis. Scientific Reports. 2016;6, article 20059 doi: 10.1038/srep20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng Z. T., Li J., Ren J., Lv Z. Expression of miR-146a and miR-16 in peripheral blood mononuclear cells of patients with rheumatoid arthritis and their correlation to the disease activity. Nan Fang YiKe Da XueXueBao. 2011;31(2):320–323. [PubMed] [Google Scholar]
- 66.Talari M., Kapadia B., Kain V., et al. MicroRNA-16 modulates macrophage polarization leading to improved insulin sensitivity in myoblasts. Biochimie. 2015;119:16–26. doi: 10.1016/j.biochi.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 67.Paradowska-Gorycka A., Stypińska B. The role of cell-free circulating microRNA in diagnostics in patients with rheumatoid arthritis. Reumatologia. 2016;3(3):95–96. doi: 10.5114/reum.2016.61207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y. H., Liu W., Xue B., et al. Upregulated expression of microRNA-16 correlates with Th17/Treg cell imbalance in patients with rheumatoid arthritis. DNA and Cell Biology. 2016;35(12):853–860. doi: 10.1089/dna.2016.3349. [DOI] [PubMed] [Google Scholar]
- 69.Akhtar N., Singh A. K., Ahmed S. MicroRNA-17 suppresses TNF-α signaling by interfering with TRAF2 and cIAP2 association in rheumatoid arthritis synovial fibroblasts. Journal of Immunology. 2016;197(6):2219–2228. doi: 10.4049/jimmunol.1600360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H. W., Xie Y., Li F., Sun G. C., Chen Z., Zeng H. S. Effect of miR-19a and miR-21 on the JAK/STAT signaling pathway in the peripheral blood mononuclear cells of patients with systemic juvenile idiopathic arthritis. Experimental and Therapeutic Medicine. 2016;11(6):2531–2536. doi: 10.3892/etm.2016.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sheedy F. J., Palsson-McDermott E., Hennessy E. J., et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nature Immunology. 2010;11(2):141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 72.Wang Y. Y., Sun G., Luo H., et al. MiR-21 modulates hTERT through a STAT3-dependent manner on glioblastoma cell growth. CNS Neuroscience & Therapeutics. 2012;18(9):722–728. doi: 10.1111/j.1755-5949.2012.00349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ouboussad L., Hunt L., Hensor E. M. A., et al. Profiling microRNAs in individuals at risk of progression to rheumatoid arthritis. Arthritis Research & Therapy. 2017;19(1):p. 288. doi: 10.1186/s13075-017-1492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng J., Jiang H. Y., Li J., et al. MicroRNA-23b promotes tolerogenic properties of dendritic cells in vitro through inhibiting Notch1/NF-κB signalling pathways. Allergy. 2012;67(3):362–370. doi: 10.1111/j.1398-9995.2011.02776.x. [DOI] [PubMed] [Google Scholar]
- 75.Murata K., Furu M., Yoshitomi H., et al. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS One. 2013;8(7, article e69118) doi: 10.1371/journal.pone.0069118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niederer F., Trenkmann M., Ospelt C., et al. Down-regulation of microRNA-34a∗ in rheumatoid arthritis synovial fibroblasts promotes apoptosis resistance. Arthritis and Rheumatism. 2012;64(6):1771–1779. doi: 10.1002/art.34334. [DOI] [PubMed] [Google Scholar]
- 77.Hu Q., Gong W., Gu J., et al. Plasma microRNA profiles as a potential biomarker in differentiating adult-onset Still’s disease from sepsis. Frontiers in Immunology. 2019;9, article 3099 doi: 10.3389/fimmu.2018.03099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wei Q., Lv F., Zhang H., et al. MicroRNA-101-3p inhibits fibroblast-like synoviocyte proliferation and inflammation in rheumatoid arthritis by targeting PTGS2. Bioscience Reports. 2020;40(1, article BSR20191136) doi: 10.1042/BSR20191136. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Anaparti V., Smolik I., Meng X., Spicer V., Mookherjee N., El-Gabalawy H. Whole blood microRNA expression pattern differentiates patients with rheumatoid arthritis, their seropositive first-degree relatives, and healthy unrelated control subjects. Arthritis Research & Therapy. 2017;19(1):p. 249. doi: 10.1186/s13075-017-1459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan Z. D., Cao Q., Huang N., et al. MicroRNA-125b regulates Th17/Treg cell differentiation and is associated with juvenile idiopathic arthritis. World Journal of Pediatrics. 2020;16(1):99–110. doi: 10.1007/s12519-019-00265-z. [DOI] [PubMed] [Google Scholar]
- 81.Kim E., Cook-Mills J., Morgan G., Sredni S. T., Pachman L. M. Increased expression of vascular cell adhesion molecule 1 in muscle biopsy samples from juvenile dermatomyositis patients with short duration of untreated disease is regulated by miR-126. Arthritis & Rheumatism. 2012;64(11):3809–3817. doi: 10.1002/art.34606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lagos D., Pollara G., Henderson S., et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nature Cell Biology. 2010;12(5):513–519. doi: 10.1038/ncb2054. [DOI] [PubMed] [Google Scholar]
- 83.Bandyopadhyay S., Lane T., Venugopal R., et al. MicroRNA-133a-1 regulates inflammasome activation through uncoupling protein-2. Biochemical and Biophysical Research Communications. 2013;439(3):407–412. doi: 10.1016/j.bbrc.2013.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu C., Pan A., Chen X., Tu J., Xia X., Sun L. MiR-5571-3p and miR-135b-5p, derived from analyses of microRNA profile sequencing, correlate with increased disease risk and activity of rheumatoid arthritis. Clinical Rheumatology. 2019;38(6):1753–1765. doi: 10.1007/s10067-018-04417-w. [DOI] [PubMed] [Google Scholar]
- 85.Lai N. S., Yu H. C., Tung C. H., Huang K. Y., Huang H. B., Lu M. C. The role of aberrant expression of T cell miRNAs affected by TNF-α in the immunopathogenesis of rheumatoid arthritis. Arthritis Research & Therapy. 2017;19(1):p. 261. doi: 10.1186/s13075-017-1465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andonian B. J., Chou C.-H., Ilkayeva O. R., et al. Plasma microRNAs in established rheumatoid arthritis relate to adiposity and altered plasma and skeletal muscle cytokine and metabolic profiles. Frontiers in Immunology. 2019;10, article 1475 doi: 10.3389/fimmu.2019.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y., Wang X., Yang M., et al. miR-145-5p increases osteoclast numbers in vitro and aggravates bone erosion in collagen-induced arthritis by targeting osteoprotegerin. Medical Science Monitor. 2018;24:5292–5300. doi: 10.12659/MSM.908219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen X., Ba Y., Ma L., et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Research. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 90.Niimoto T., Nakasa T., Ishikawa M., et al. MicroRNA-146a expresses in interleukin-17 producing T cells in rheumatoid arthritis patients. BMC Musculoskeletal Disorders. 2010;11:p. 209. doi: 10.1186/1471-2474-11-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Q., Haupt S., Kreuzer J. T., et al. Decreased expression of miR-146a and miR-155 contributes to an abnormal Treg phenotype in patients with rheumatoid arthritis. Annals of the Rheumatic Diseases. 2015;74(6):1265–1274. doi: 10.1136/annrheumdis-2013-204377. [DOI] [PubMed] [Google Scholar]
- 92.Lin S. H., Ho J. C., Li S. C., Chen J. F., Hsiao C. C., Lee C. H. MiR-146a-5p expression in peripheral CD14+ monocytes from patients with psoriatic arthritis induces osteoclast activation, bone resorption, and correlates with clinical response. Journal of Clinical Medicine. 2019;8(1):p. 110. doi: 10.3390/jcm8010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stanczyk J., Leslie Pedrioli D. M., Brentano F., et al. Altered expression of microRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis and Rheumatism. 2008;58(4):1001–1009. doi: 10.1002/art.23386. [DOI] [PubMed] [Google Scholar]
- 94.Chen X. M., Zhao Y., Wu X. D., et al. Novel findings from determination of common expressed plasma exosomal microRNAs in patients with psoriatic arthritis, psoriasis vulgaris, rheumatoid arthritis, and gouty arthritis. Discovery Medicine. 2019;28(151):47–68. [PubMed] [Google Scholar]
- 95.Alivernini S., Tolusso B., Petricca L., et al. Synovial predictors of differentiation to definite arthritis in patients with seronegative undifferentiated peripheral inflammatory arthritis: microRNA signature, histological, and ultrasound features. Frontiers in Medicine. 2018;5:p. 186. doi: 10.3389/fmed.2018.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fernández-Ruiz J. C., Ramos-Remus C., Sánchez-Corona J., et al. Analysis of miRNA expression in patients with rheumatoid arthritis during remission and relapse after a 5-year trial of tofacitinib treatment. International Immunopharmacology. 2018;63:35–42. doi: 10.1016/j.intimp.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 97.Ormseth M. J., Solus J. F., Vickers K. C., Oeser A. M., Raggi P., Stein C. M. Utility of select plasma microRNA for disease and cardiovascular risk assessment in patients with rheumatoid arthritis. The Journal of Rheumatology. 2015;42(10):1746–1751. doi: 10.3899/jrheum.150232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Legesse-Miller A., Elemento O., Pfau S. J., Forman J. J., Tavazoie S., Coller H. A. let-7 overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. The Journal of Biological Chemistry. 2009;284(11):6605–6609. doi: 10.1074/jbc.C900002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: list of microRNAs studied in rheumatology. ACPA: anti-citrullinated protein antibodies; ADA: adalimumab; ANA: antinuclear antibodies; AOSD: adult-onset Still's disease; AS: ankylosing spondylitis; BTRC: beta-transducin repeat containing gene; CD: cluster of differentiation; CF: cystic fibrosis; CIA: collagen-induced arthritis; CRP: C-reactive protein; DAS28: Disease Activity Score 28-Joint Count; DMARD: disease-modifying antirheumatic drug; ERA: enthesitis-related arthritis; ESR: erythrocytes sedimentation rate; FLS: fibroblast-like synoviocytes; foxp3: forkhead box P3; FMF: familial Mediterranean fever; GCS: glucocorticosteroids; IL: interleukin; IFNβ: interferon beta; INHBA: inhibin beta A; IRAK: interleukin-1 receptor-associated kinase; JADAS27: Juvenile Arthritis Disease Activity Score 27-Joint Count; JAK/STAT: Janus kinase/signal transducers and activators of transcription; JDM: juvenile dermatomyositis; JIA: juvenile idiopathic arthritis; JSA: juvenile spondyloarthropathy; miR: microRNA; MMP: matrix metalloproteinase; MRI: magnetic resonance imaging; MTX: methotrexate; NLRP3: NLR family pyrin domain containing protein 3; NFkB: nuclear factor kappa of activated B cells; OA: osteoarthritis; PBMC: peripheral blood mononuclear cell; PLT: platelet count; PsA: psoriatic arthritis; PTGS2: prostaglandin-endoperoxide synthase 2; RA: rheumatoid arthritis; RF: rheumatoid factor; RORyt: retinoic-acid-receptor-related orphan nuclear receptor gamma; RTX: rituximab; SLE: systemic lupus erythematosus; SNP: single-nucleotide polymorphism; SOCS3: suppressor of cytokine signaling 3; SoJIA: systemic onset of juvenile idiopathic arthritis; SS: Sjögren syndrome; SSZ: sulfasalazine; TAK1: transforming growth factor-beta-activated kinase 1; Th: helper T cells; TLR: Toll-like receptor; TNF: tumor necrosis factor; TRAF6: tumor necrosis factor receptor-associated factor 6; Treg: regulatory T cells; UC: ulcerative colitis; WBC: white blood count.
Data Availability Statement
The data used to support the findings of this study are included within the article.


