Abstract
Helicobacter pylori is a spiral-shaped bacterium, which plays a role in the aetiology of gastric diseases in humans. Non-H. pylori Helicobacter (NHPH) species naturally colonise the stomach of animals and also induce gastric lesions in humans, highlighting their zoonotic importance. We evaluated the gastric bacterial colonisation density and gastric lesions and sought to identify the main phylogenetic groups of the Helicobacter spp. obtained from dogs in the central region of Rio Grande do Sul, Brazil, with this study aiming to investigate the occurrence of Helicobacter spp. in saliva and gastric samples from these dogs. This study included 35 dogs and used analysis such as cytology, histopathology, PCR, rapid urease testing, and phylogenetic analysis. Of the dogs, 94.3% were positive for Helicobacter spp., and these bacteria were present in the stomach of 32 dogs and saliva of eight. Respectively, eight, 15, and nine dogs had mild, moderate, and severe colonisation. Lymphocytic-plasmacytic infiltrate was the main gastric lesion. However, the presence of Helicobacter and the density appeared to be unrelated to the gastric lesions. The samples possessed a high nucleotide identity with remarkably similar sequences among some of the species of NHPH such as H. heilmannii s.s., H. salomonis, H. felis, and H. bizzozeronii. The saliva of domestic dogs, even of those who appear clinically healthy, can cause Helicobacter infection in humans and other animals, with, in these dogs, increased density, occurrence rate, and predominance of NHPH of zoonotic importance being found in the stomach with a lower occurrence of Helicobacter spp. in the saliva.
1. Introduction
Helicobacter spp. are spiral-shaped mobile Gram-negative bacteria with tropism for the gastric mucosa of humans and animals [1]. Helicobacter pylori (H. pylori) was the first species isolated from the Helicobacter genus by Australian researchers Marshall and Warren in 1983 [2]. Further research has shown that humans are the natural host of H. pylori and have established this species as the primary aetiology of peptic ulcers and gastric neoplasms [3]. Years later, in samples of gastric mucosa from humans, another spiral-shaped bacteria with similar morphological features to H. pylori was documented, and this microbe was found to be remarkably similar to the spiral-shaped bacteria found in the gastric mucosa of domestic animals [4]. This new spiral bacteria was later classified by Solnick et al. [5] as Helicobacter heilmannii. However, phylogenetic studies were later employed utilising the 16 rRNA target gene, and these studies found instead that this bacteria belonged to a range of several Helicobacter species isolated from domestic and wild animals, such as H. felis, H. bizzozeronii, H. salomonis, Helicobacter heilmannii sensu stricto (H. heilmannii s.s.), H. bílis, H. cynogastricus, and H. baculiformis [6]. Therefore, in order to organise this large group of bacteria into a single term, they were consensually denominated as non-Helicobacter pylori Helicobacter (NHPH) [7].
NHPH are the target of various studies due to their relationship with upper digestive tract illnesses in humans and their zoonotic importance [8]. Dogs are the natural hosts of NHPH and harbour this bacteria in their gastric mucosa, gut, and oral cavity; thus, gastric juice, saliva, and faeces are possible sources of transmission for this bacteria to infect humans [9–11]. In dogs, the main species of NHPH found are H. heilmannii s.s, H. bizzozeronii, H. salomonis, H. felis, and H. canis [9, 11].
Chronic inflammation of the gastric mucosal tissue, peptic ulcers, and gastric mucosa associated lymphoid tissue lymphoma are the clinical alterations described in humans with NHPH infection [12, 13]. In human populations, NHPH has a prevalence of 0.5% in developed countries [14] and 6.2%–15% in underdeveloped countries [15, 16]. Thus, countries with lower socioeconomic development tend to have a higher prevalence of infected people with NHPH [17]. However, information regarding the importance of domestics dogs as reservoirs for this bacteria and the data related to the number of NHPH occurrences in the canine population of these countries has not yet been explicated [18].
Due to the zoonotic implications of NHPH, compounded by the high density of domestic animals, and sanitary problems affecting Brazil, it is important to elucidate this information to assist with future studies of public health. Moreover, the geographic variation could affect the prevalence of the Helicobacter species [11, 18]. Thus, the aim of this study was to investigate the presence of Helicobacter spp. from saliva and the gastric mucosa of domestic dogs. Moreover, this study documented the gastric bacterial colonisation density, the number of gastric lesions present and sought to identify the main phylogenetic groups of Helicobacter spp. found in dogs from the central region of Rio Grande do Sul.
2. Materials and Methods
2.1. Ethics Statement
All the procedures were performed with the consent of owners of the dogs and the study protocol was approved by the Animal Ethics Committee of Universidade Federal de Santa Maria (Approval number 2827081018) and conducted in accordance with national guidelines and regulations for the care and use of laboratory animals established by the National Council for Animal Experimentation Control (CONCEA), Brazil.
2.2. Animals
This study collected saliva and gastric mucosal samples from 35 client-owned domestic dogs. They included seven males and 28 females, ranging from seven months to 14 years of age. Twenty-three of these dogs were clinically normal (all females) and in these animals selected, any alteration or underlying disease were discarded through the history, physical examination, complete blood count, serum biochemistry profile (creatinine, blood urea nitrogen, alkaline phosphatase, albumin, and alanine aminotransaminase). In addition, these healthy animals were submitted to sterilization procedure in the counterpart of the upper digestive endoscopy and gastric biopsy sample collection. Twelve had a history of chronic vomiting (seven males and five females; seven dogs with chronic vomiting only, three with chronic vomiting accompanied with weight loss and alteration in appetite, one with chronic vomiting accompanied with diarrhea, and one with chronic vomiting accompanied with hematemesis). These symptomatic animals were only submitted to upper digestive endoscopy and gastric biopsy sample collection due to gastric symptoms. For selection of the dogs classified with chronic vomiting (related to chronic gastritis) in this study, the inclusion criteria employed was the confirmation of a history of clinical signs such as vomiting episodes accompanied or not with diarrhea, weight loss, and alteration in appetite, for a period superior to 3 weeks [19, 20]. Common differential diagnostics of chronic vomiting, such as drugs toxicity, foreign bodies, hepatic failure, or renal disease, were discarded by history, physical examination, complete blood count, serum biochemistry profile (creatinine, blood urea nitrogen, alkaline phosphatase, albumin and alanine aminotransaminase) and ultrasonography and/or radiography previous to procedure and by upper digestive endoscopy on the moment of study. These animals were arising from the municipalities of Santa Maria and Itaara, located in the central region of the State of the Rio Grande do Sul, Brazil. Preceding sample collection, the dogs did not receive effective eradication protocols for gastric Helicobacter spp. employing the use of the combination of amoxicillin and clarithromycin or metronidazole and proton pump inhibitor (omeprazole or lansoprazole) or famotidine for 14 days [21, 22]. However, antibiotics such as ampicillin, amikacin, metronidazole, and sulfamethoxazole-trimethoprim were used per three days in four dogs with clinical signs (dogs number: n.2 has received ampicillin and metronidazole; n.4 has received sulfamethoxazole and trimethoprim; n.21 has received metronidazole; and n.27 has received ampicillin, amikacin, and metronidazole) prior to sample collections.
2.3. Sample Collection
Following a fasting period of 12 hours, all dogs were anesthetically induced with propofol (4 mg/kg) and held in an anaesthetised state with isoflurane and 100% oxygen. Saliva collection was performed with a sterile swab prior to the endoscopic procedure (to prevent cross-contamination of saliva samples with gastric secretions), and the samples were stored in sterile normal saline and frozen at −80°C until further processing. The collection of gastric biopsies was performed with a flexible endoscope (Karl Storz GmbH & Co.KG., Tuttlingen, Germany) and 2.3 mm diameter biopsy forceps (Changzhou Jiuhong Medical Instrument Co. Ltd., Changzhou, China). Following the mucosal evaluation, four biopsy samples were collected from the body and gastric antrum. Impression smears of the two gastric biopsy samples from the body and antrum were prepared on an air-dried slide. Then impression smears from the same samples were placed into two tubes (one tube for each gastric zone) containing 10% formalin until further processing. One sample of each gastric zone was submitted for the rapid urease test. For molecular analysis, one sample from each gastric zone was stored in a tube containing sterile normal saline and frozen at −80°C until further processing.
In procedures using additional analgesic drugs and loco-regional anesthesia, sterilization in those asymptomatic female dogs was performed postgastroscopy employing the conventional open surgical spaying or laparoscopic-assisted method.
2.4. Rapid Urease Test (RUT)
RUT was employed with a Urease test kit (RenyLab, Barbacena, Brazil). Following collection, the biopsy samples for each gastric zone were placed together into a test tube and evaluated for 60 minutes. Colour transformation from yellow to pink within 60 minutes was considered positive, while, no colour transformation within 60 minutes was considered negative.
2.5. Cytology
Following the impression smears of the biopsy samples, the slides corresponding to both gastric zones were stained with a quick panoptic stain. The gastric bacterial colonisation densities (the mean of 10 microscopic fields at ×400) were recorded as follows (−0) no bacteria; (+1 = mild) less than 10 bacteria per field; (+2 = moderate) 10–50 bacteria per field, and (+3 = severe) more than 50 bacteria per field [23, 24].
2.6. Histopathology
The formalin-fixed gastric samples for the body and the antrum were sectioned and stained with haematoxylin and eosin and then processed routinely. The gastric samples were evaluated by using the World Small Animal Veterinary Association (WSAVA) gastrointestinal standardisation visual analogue scale [25], where the attributed scores are based on the gastric lesion severity and recorded as follows: (0) lesion not observed; (1) mild lesion; (2) moderate lesion; and (3) severe lesion. The values obtained in the evaluation of the three random fields for each gastric zone were calculated and expressed with the mean of the gastritis severity score.
2.7. DNA Extraction and PCR Amplification
Some animal samples, such as faeces and saliva, had PCR inhibitors. To minimize this issue, we used DNA extraction and PCR protocol used worldwide in the DNA microorganism detection. Thus, DNA was extracted from the saliva and gastric biopsies with the DNeasy Blood and Tissue Kit (Qiagen N.V., Hilden, Germany), following the manufacturer's instructions. Total DNA was extracted for each sample and submitted to PCR testing, which used the following oligonucleotide primers (F-5′- AAC GAT GAA GCT TCT AGC TTG CTA-3′; R-5′- GTG CTT ATT CST NAG ATA CCG TCA T-3′), which amplified a fragment of 399 base pairs (bp) for the 16S rRNA gene of the Helicobacter spp. [26]. PCR testing involved 1x buffer of PCR containing MgCl2, 10 mM of dNTPs (0,2 mM of each), 10 pmol of each primer, 1 U of Taq polymerase enzyme (Promega, Madison, Wisconsin, USA), and qsp water; following these conditions, the initial denaturation was 95°C per 5 minutes, followed by 32 cycles at 94°C–30 s; 62°C–30 s to annealing of the initiators and 72°C–30 s for chain extension; and final extension for 3 minutes at 72°C. The amplified product was analysed by electrophoresis in an agarose gel 1% (60V, 1 hour and 30 minutes), using GelRed® (Biotium, California, USA) and visualised by a transilluminator with ultraviolet light. In all of the amplifications, the positive control was obtained from the gastric mucosal samples from a dog known positive for Helicobacter spp. and the negative controls were ultrapure water samples.
2.8. Sequencing and Phylogenetic Analysis
The amplification products for PCR were submitted in duplicate for nucleotide sequencing via the Sanger method using Prism 3500 Genetic Analyzer equipment (Life Technologies, California, USA). The consensus sequence from the Staden Package program was used to start the nucleotide sequences [27]. Phylogenetic analysis utilised the consensus sequence for each of the amplified samples and the nucleotide sequences of the Helicobacter spp. obtained from the GenBank database (http://www.ncbi.nlm.nih.gov). The sequences were edited and aligned using the BioEdit Alignment Editor software suite, version 7.0.5.3 [28]. The phylogenetic tree was constructed using the MEGA X software [29].
2.9. Criteria for Positive Dogs with Helicobacter spp
A dog was considered positive for Helicobacter spp. when there was a positive result at least one test (cytology; RUT; or 16S rRNA PCR assay).
2.10. Statistical Analyses
Statistical analyses were then performed using the statistical software Action Stat Pro (Estatcamp, São Carlos, Brazil). Kruskal–Wallis test was employed to compare the bacterial colonisation density scores between the regions of the body and antrum. The Spearman test was used to evaluate the correlation between the scores of the histologic gastric lesions and the gastric bacterial colonisation density scores in both of the gastric zones. Fisher's exact test was used to determine if the Helicobacter infection was associated with gastric lesions in the histopathology findings. The significance level used was P < 0.05 for all statistic evaluations.
3. Results
3.1. Detection of Helicobacter spp. and Gastric Bacterial Colonisation Density (Table 1)
Table 1.
Detection of Helicobacter spp., in different tests evaluated and gastric bacterial colonisation density scores.
| Animal no. | Chronic vomiting | Saliva | Gastric mucosa | |||
|---|---|---|---|---|---|---|
| 16S rRNA PCR | Score cytology | RUT | ||||
| assay | Body | Antrum | ||||
| 1 | P | + | − | − | − | N/P |
| 2 | P | − | − | − | − | N/P |
| 3 | P | + | + | ++ | ++ | + |
| 4 | P | − | − | − | − | − |
| 5 | P | + | + | + | ++ | + |
| 6 | P | − | − | ++ | + | + |
| 7 | P | − | + | ++ | ++ | + |
| 8 | P | − | + | ++ | + | + |
| 9 | A | − | + | + | +++ | + |
| 10 | A | − | + | ++ | + | + |
| 11 | A | − | + | +++ | ++ | + |
| 12 | A | − | + | ++ | N/P | + |
| 13 | A | − | − | + | ++ | + |
| 14 | A | − | + | ++ | +++ | + |
| 15 | A | − | + | + | N/P | + |
| 16 | P | − | + | ++ | + | + |
| 17 | A | − | + | ++ | + | + |
| 18 | A | − | − | + | N/P | + |
| 19 | A | − | − | ++ | ++ | + |
| 20 | P | − | + | + | + | + |
| 21 | P | − | + | + | + | + |
| 22 | A | + | − | ++ | +++ | + |
| 23 | A | − | + | + | + | + |
| 24 | A | + | + | + | + | + |
| 25 | A | + | + | +++ | +++ | + |
| 26 | A | − | + | +++ | N/P | + |
| 27 | P | + | + | + | + | + |
| 28 | A | − | + | ++ | +++ | + |
| 29 | A | + | + | ++ | + | + |
| 30 | A | − | + | ++ | ++ | + |
| 31 | A | − | + | ++ | ++ | + |
| 32 | A | − | + | ++ | +++ | + |
| 33 | A | − | + | + | ++ | + |
| 34 | A | + | − | + | − | − |
| 35 | A | − | + | +++ | ++ | + |
| Total | 9/35 | 26/35 | 32/35 | 27/35 | 31/35 | |
P: present; A: absent; RUT: Rapid Urease Test; N/P: not performed; −: negative; +: positive and mild score; ++: moderate score; +++: severe score.
In accordance with RUT and cytology, 88.5% (31/35) and 91.4% (32/35) of the dogs were positive, respectively. The spiral-shaped bacteria observed by cytology on all samples resembled NHPH morphology. Detection of the 16S rRNA, specific to the Helicobacter genus, was positive 25.7% in saliva (9/35) and 74.2% (26/35) in the gastric mucosa.
Through the evaluation of gastric bacterial colonisation density scores (Figure 1), 22.8% (8/35) of the dogs showed a mild score, 42.8% (15/35) a moderate score, and 25.7% (9/35) a severe score. In dogs with chronic vomiting, the gastric bacterial colonisation density was 25% mild score (3/12) and 50% moderate score (6/12), but in 25% of the dogs, the bacteria was not present (3/12). All asymptomatic dogs were positive for the presence of Helicobacter spp.; 21.8% of the dogs showed a mild score (5/23), 39.1% moderate score (9/23), and 39.1% a severe score (9/23). There were no statistically significant differences in comparison with scores between the body and the antrum (P=0.8).
Figure 1.
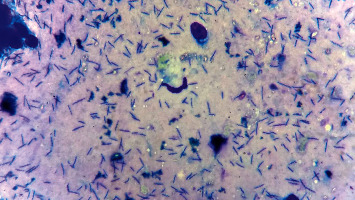
Impression smear of the gastric biopsy sample showing the presence of numerous Helicobacter organisms (Quick panoptic stain, ×1000).
3.2. Findings of the Upper Digestive Endoscopy and Histopathology
Endoscopic evaluation of three dogs without Helicobacter gastric presence showed macroscopic alterations, such as patchy erythema (2/3), spotty erythema (1/3), oedema (1/3), superficial irregularities (1/3), and vascular pattern visibility (1/3). Moreover, 32 dogs with Helicobacter gastric presence showed the following macroscopic alterations observed patchy erythema (3/32), spotty erythema (10/32), linear erythema (1/32), oedema (3/32), and vascular pattern visibility (3/32). Based on the WSAVA standards, for the three dogs without Helicobacter gastric presence, only one showed gastric lesions, while the 32 dogs with Helicobacter gastric presence, 21 showed gastric lesions, 20 mild gastritis, and one moderate gastritis (Table 2). The gastric lesion patterns found in the 35 dogs were lymphocytic-plasmacytic infiltrates (18/35), neutrophilic infiltrates (9/35), eosinophilic infiltrates (9/35), intraepithelial lymphocytes (4/35), fibrosis, and mucosal atrophy (3/35), and gastric lymphofollicular hyperplasia (3/35) (Tables 2 and 3) (Figure 2).
Table 2.
Findings of histologic gastric lesion on dogs colonised and noncolonised with Helicobacter in the stomach (mean of scores in both gastric zones).
| Histologic gastric lesion [25] | Total (n = 35) | Colonised (n = 32) | Noncolonised (n = 3) |
|---|---|---|---|
| Mild gastritis | 21 | 20 | 1 |
| Moderate gastritis | 1 | 1 | 0 |
| Fibrosis and mucosal atrophy | 3 | 2 | 1 |
| Lymphocytic-plasmacytic infiltrate | 18 | 17 | 1 |
| Neutrophilic infiltrate | 9 | 8 | 1 |
| Eosinophilic infiltrate | 9 | 8 | 1 |
| Gastric lymphofollicular hyperplasia | 3 | 3 | 0 |
| Intraepithelial lymphocytes | 4 | 4 | 0 |
Table 3.
Case by case of frequency of the gastric bacterial colonisation density, presence of 16rRNA Helicobacter genus on saliva and stomach, and histologic findings with degrees of WSAVA classification.
| Animal (no.) and history | 16S rRNA PCR assay | Gastric bacterial colonisation density score | Histologic gastric lesion and degree (WSAVA classification) |
|---|---|---|---|
| 1S | Saliva | Body: no bacteria Antrum: no bacteria |
Body: normal Antrum: normal |
| 2S | Negative | Body: no bacteria Antrum: no bacteria |
Body: mild, lymphocytic-plasmacytic, eosinophilic and neutrophilic infiltrate Antrum: mild, neutrophilic infiltrate and fibrosis and mucosal atrophy |
| 3S | Saliva, stomach | Body: moderate Antrum: moderate |
Body: normal Antrum: normal |
| 4S | Negative | Body: no bacteria Antrum: no bacteria |
Body: normal Antrum: normal |
| 5S | Saliva, stomach | Body: mild Antrum: moderate |
Body: normal Antrum: normal |
| 6S | Negative | Body: moderate Antrum: mild |
Body: severe, lymphofollicular hyperplasia; mild, lymphocytic-plasmacytic and neutrophilic infiltrate Antrum: mild, lymphocytic-plasmacytic infiltrate |
| 7S | Stomach | Body: moderate Antrum: moderate |
Body: moderate, lymphofollicular hyperplasia; mild, fibrosis and mucosal atrophy, lymphocytic-plasmacytic infiltrate and intraepithelial lymphocytes Antrum: normal |
| 8S | Stomach | Body: moderate Antrum: mild |
Body: normal Antrum: NP ∗ |
| 9A | Stomach | Body: mild Antrum: severe |
Body: normal Antrum: NP ∗ |
| 10A | Stomach | Body: moderate Antrum: mild |
Body: mild, lymphocytic-plasmacytic infiltrate Antrum: NP ∗ |
| 11A | Stomach | Body: severe Antrum: moderate |
Body: mild, lymphocytic-plasmacytic, neutrophilic and eosinophilic infiltrate and lymphofollicular hyperplasia Antrum: mild, lymphocytic-plasmacytic infiltrate |
| 12A | Stomach | Body: mild Antrum: N/P |
Body: mild, lymphocytic-plasmacytic infiltrate Antrum: NP |
| 13A | Negative | Body: mild Antrum: moderate |
Body: mild, lymphocytic-plasmacytic and neutrophilic infiltrate Antrum: mild, lymphocytic-plasmacytic and neutrophilic infiltrate |
| 14A | Stomach | Body: moderate Antrum: severe |
Body: NP ∗ Antrum: mild, lymphocytic-plasmacytic and neutrophilic infiltrate and intraepithelial lymphocytes |
| 15A | Stomach | Body: mild Antrum: N/P |
Body: mild, lymphocytic-plasmacytic, neutrophilic and eosinophilic infiltrate Antrum: NP |
| 16A | Stomach | Body: moderate Antrum: mild |
Body: normal Antrum: mild, lymphocytic-plasmacytic infiltrate |
| 17A | Stomach | Body: moderate Antrum: mild |
Body: mild, lymphocytic-plasmacytic and neutrophilic infiltrate Antrum: NP ∗ |
| 18A | Negative | Body: mild Antrum: N/P | Body: mild, lymphocytic-plasmacytic infiltrate Antrum: N/P |
| 19A | Negative | Body: moderate Antrum: moderate |
Body: normal Antrum: normal |
| 20S | Stomach | Body: mild Antrum: mild |
Body: normal Antrum: mild, lymphocytic-plasmacytic and eosinophilic infiltrate |
| 21S | Stomach | Body: mild Antrum: mild |
Body: normal Antrum: NP ∗ |
| 22A | Saliva | Body: moderate Antrum: severe |
Body: normal Antrum: normal |
| 23A | Stomach | Body: mild Antrum: mild |
Body: mild, eosinophilic infiltrate Antrum: normal |
| 24A | Saliva, stomach | Body: mild Antrum: mild |
Body: normal Antrum: normal |
| 25A | Saliva, stomach | Body: severe Antrum: severe |
Body: normal Antrum: NP ∗ |
| 26A | Stomach | Body: severe Antrum: N/P |
Body: mild, lymphocytic-plasmacytic infiltrate Antrum: NP |
| 27S | Saliva, stomach | Body: mild Antrum: mild |
Body: normal Antrum: NP ∗ |
| 28A | Stomach | Body: moderate Antrum: severe |
Body: normal Antrum: mild, lymphocytic-plasmacytic, neutrophilic and eosinophilic infiltrate |
| 29A | Saliva, stomach | Body: moderate Antrum: mild |
Body: normal Antrum: mild, intraepithelial lymphocytes |
| 30A | Stomach | Body: moderate Antrum: moderate |
Body: mild, lymphocytic-plasmacytic and eosinophilic infiltrate and intraepithelial lymphocytes normal Antrum: mild, lymphocytic-plasmacytic and eosinophilic infiltrate |
| 31A | Stomach | Body: moderate Antrum: moderate |
Body: normal Antrum: mild, lymphocytic-plasmacytic infiltrate |
| 32A | Stomach | Body: moderate Antrum: severe |
Body: mild, lymphocytic-plasmacytic infiltrate Antrum: mild, neutrophilic infiltrate |
| 33A | Stomach | Body: mild Antrum: moderate |
Body: normal Antrum: mild, eosinophilic infiltrate |
| 34A | Saliva | Body: mild Antrum: no bacteria |
Body: mild, eosinophilic infiltrate Antrum: mild, eosinophilic infiltrate |
| 35A | Stomach | Body: severe Antrum: moderate |
Body: normal Antrum: normal |
NP: not performed; ∗: unviable sample; A: asymptomatic (healthy); S: symptomatic (chronic vomiting).
Figure 2.
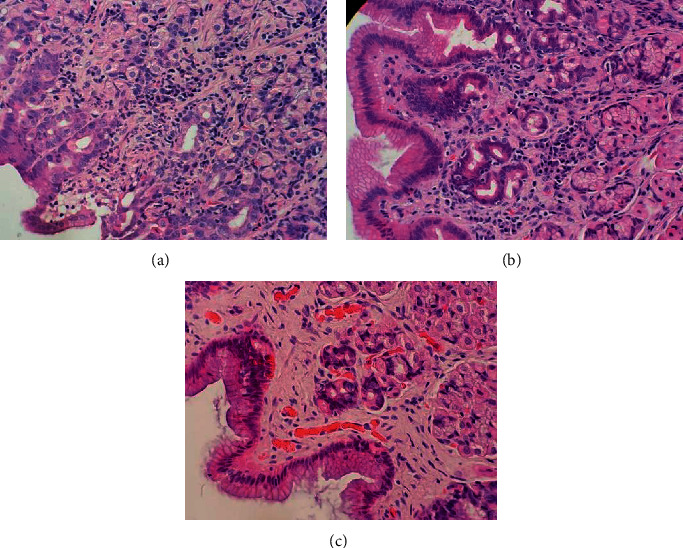
Gastric mucosa (body, A and B; antrum, C) biopsy sections. (a) Lamina propia with a high quantity of polymorphonuclear cells (asterisk), mainly eosinophilic infiltrate. (b) Lamina propia showed a high quantity of mononuclear cells (asterisk), with a predominance of plasmocytes. (c) Image of the normal gastric mucosa. (Haematoxylin and eosin stain, ×400).
Although the number of lymphocytic-plasmacytic infiltrates was high in the animals with Helicobacter gastric presence (Table 2); statistically, this was not associated with a known pattern of gastric lesions (P > 0.05). In addition, there was no correlation between the scores of histologic lesions with gastric bacterial colonisation density scores in the body (P=0.3) and antrum (P=0.9).
3.3. Nucleotide Identity and Phylogenetic Analysis
The identity of the nucleotides between the sequences obtained varied by 94.3 to 100%. The identity was high too when these sequences were compared with sequences of H. heilmannii (HM625818; 95.5 to 100%), H. salomonis (U89351; 94.3 to 100%), H. felis (AY686607; 94.3 to 100%), and H. bizzozeronii (NR026372; 94.9 to 99.3%). The identity of the nucleotide changed from 89.1% to 94.2% when compared with the sequences of H. pylori (U00679) samples (Figure 3).
Figure 3.
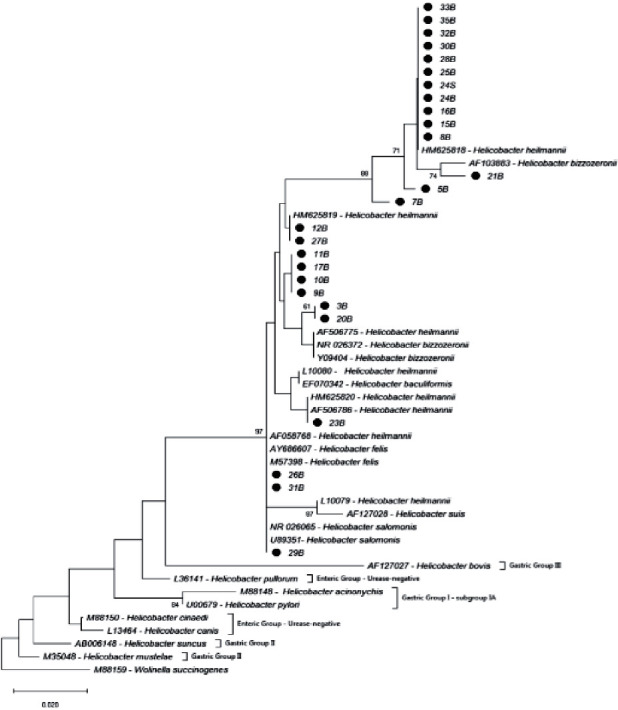
Phylogenetic tree based in the nucleotide sequence of 16S rRNA gene of Helicobacter spp. The tree was build employing the Neighbor-Joining method with a bootstrap of 1000 replicates. The evolutionary distances were computed using of Kimura-2 method. Values above 60% are showed. The sequences obtained of this study are highlighted with a black circle; n°B, animal number/gastric biopsy sample; n°S, animal number/saliva sample.
4. Discussion
The occurrence of Helicobacter spp. (94.3%) in the dogs belonging to the Brazilian geographic region of this study was higher than that of the dogs from Japan (34.7%) [22], South Korea (78.4%) [30], Denmark (76.7%) [31], Germany (82%) [32], and Portugal (87%) [33]. However, our results were similar with dogs of Poland (96.7%) [11], Iran (95%) [34], Venezuela (95%) [18], and Costa Rica (95%) [35]. Interestingly, these results were different from those observed in human populations, whose occurrence of Helicobacter spp. increased according to the socioeconomic underdevelopment of the region [17], with the aforementioned results demonstrating the high level of geographic variation for these bacteria in domestic dog populations, regardless of socioeconomic level. However, additional data from developed and underdeveloped countries are required in order to correlate the Helicobacter spp. prevalence in populations of domestic dogs from different geographic regions [18, 33].
Through the evaluation of gastric bacterial colonisation density scores, 22.8% (8/35) of the dogs showed a mild score, 42.8% (15/35) a moderate score, and 25.7% (9/35) a severe score. Thus, more than half of the subject dogs were reservoirs of a substantial gastric colonisation density for NHPH.
During the gastroscopy, the main alteration observed was erythema; however, none of the dogs showed the presence of erosions or ulcerations that were described by Kubota-Aizawa et al. [22] and Suárez-Esquivel et al. [35]. Furthermore, some of the gastroscopy findings failed to match the histological findings. For instance, when microscopic lesions were observed, the macroscopic evaluation did not always exhibit alterations; however, these findings are not abnormal [19]. Therefore, the diagnosis of gastritis should not be based solely upon gastroscopy visualisation, as biopsy collection is indispensable to confirm the diagnosis of gastritis [36].
The main histologic changes observed in the dogs of this study were the presence of lymphocytic-plasmacytic infiltrates, which were commonly observed when NHPH was present [33]. Nevertheless, NHPH factors regarding the gastric presence and gastric bacterial colonisation density were not found to be correlated with the gastric lesions and severity scores, and this result was similar to other authors' results [9, 18]. Moreover, the virulence and pathogenicity could vary between the different Helicobacter species and strains within an NHPH infection [37]. Kubota-Aizawa et al. [22] suggest in vitro cultivation of each NHPH species and experimental infection of canine gastric cell lines or dogs to investigate the pathogenicity of each NHPH species.
This study was the first conducted in Brazil and investigated the presence of Helicobacter in the saliva of dogs. The saliva may exert an important role transmission source of NHPH to humans and other domestic animals [11]. In addition, some authors have suggested that the oral cavity could be a potential niche of Helicobacter spp. colonisation [10]. In this study, the results revealed divergent findings from those of other studies. Commonly, a high prevalence of 71.1% to 100% of Helicobacter spp. was detected in the saliva of canine populations and documented [9, 10]; however, the present study observed an occurrence significantly lower, at 25.7% (9/35). Another novel finding was that one dog was positive for NHPH in the saliva and negative in the gastric mucosa, which has not been reported by another study in a canine population. This observation has been previously described in cats [24]. However, we cannot affirm this based only on a case, but two hypotheses arose with this finding: It is possible that dogs with a gastric absence of Helicobacter spp. can harbour this bacteria in the oral cavity; or, this finding could be resultant of the very low number of Helicobacters distributed in the gastric mucosa of this dog, resulting in the absence of positivity by different methods of detection in the samples evaluated (false negative). The cross-contamination was discharged because of the saliva samples being collected prior to the endoscopic procedure and collection of gastric biopsies.
The phylogenetic analysis failed to determine the species of the Helicobacter detected in the individual samples. However, based on the closeness of the samples of this study with strains of Helicobacter species such as H. heilmannii s.s, H. salomonis, H. felis, and H. bizzozeronii, we can infer similarity between our samples with these NHPH species. These species of Helicobacter are important as they are considered to share close evolutionary relationships and genomic similarities with H. pylori [38–40]. This might explain the capacity of these NHPH to readily adapt to the gastric environment of humans and to induce gastric diseases, highlighting the zoonotic nature of these bacterial species [39]. The phylogenetic analysis of saliva samples was only possible to dog number 24, and the other samples were not performed due to a weak fluorescence signal.
Due to Helicobacter infection as an important risk factor, currently in Brazil the high mortality rate associated with gastric cancer, it was the fourth-largest cause of death due to neoplasms for men, and the sixth among women [41] and the prevalence and mortality rates due to peptic ulcers have increased as the population age increases [42]. However, the real percentage of these cases related to NHPH infection is unkown. Notably, as the human population increases their contact with domestic animals, a directly proportional increase in NHPH infected people could result relative to the socioeconomic underdevelopment of the country they inhabit [14]. Furthermore, saliva and faeces of dogs with NHPH may be potential sources of infection for human populations [9, 11]. Thus, this is what motivated the authors to initiate and develop this study, as we were faced with a growing canine population while poor sanitary conditions are still a reality in our country. Due to the zoonotic importance of NHPH bacteria, future cohort studies evaluating the prevalence of Helicobacter spp. in the Brazilian human population that could be directly associated with their pets would be extremely relevant in order to elucidate whether NHPH is a concern to public health.
5. Conclusions
The results of the present study demonstrated a low occurrence of Helicobacter spp. in the saliva, and a high occurrence, predominance, and density of zoonotic importance in the stomach of domestic dogs of the central region of Rio Grande do Sul, Brazil. The saliva of the domestic dogs may be a transmission reservoir of Helicobacter, even in animals without bacterial colonisation. The Helicobacter gastric presence and gastric colonisation density failed to show an association with the gastric lesions pattern observed during histopathology.
Acknowledgments
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brazil (CAPES) (Grant number: 001).
Abbreviations
- NHPH:
Non-H. pylori Helicobacter species.
Data Availability
The occurrence data of Helicobacters used to support the findings of this study are included within the article. The sequence data used to support findings of this article were deposited in GenBank; the accession numbers are Sample 3B: Genbank accession MN966439/Sample 5B: Genbank accession MN966440/Sample 7B: Genbank accession MN966441/Sample 8B: Genbank accession MN966442/Sample 9B: Genbank accession MN966443/Sample 10B: Genbank accession MN966444/Sample 11B: Genbank accession MN966445/Sample 12B: Genbank accession MN966446/Sample 15B: Genbank accession MN966447/Sample 16B: Genbank accession MN966448/Sample 17B: Genbank accession MN966449/Sample 20B: Genbank accession MN966450/Sample 21B: Genbank accession MN966451/Sample 23B: Genbank accession MN966452/Sample 24B: Genbank accession MN966453/Sample 24S: Genbank accession MN966454/Sample 25B: Genbank accession MN966455/Sample 26B: Genbank accession MN966456/Sample 27B: Genbank accession MN966457/Sample 28B: Genbank accession MN966458/Sample 29B: Genbank accession MN966459/Sample 30B: Genbank accession MN966460/Sample 31B: Genbank accession MN966461/Sample 32B: Genbank accession MN966462/Sample 33B: Genbank accession MN966463/Sample 35B: Genbank accession MN966464.
Additional Points
Limitations of This Study. Unfortunately, the limitation in this study was the number of animals employed, which resulted in the limited duration of the study (14 months) and weak routine over this time. Samples from the antrum region were not possible to be collected due to difficult endoscopic access of the pylorus in four animals, and for histopathological analysis, some samples were not viable due to the insufficient amount of tissue or absence of the sample.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
DDGS contributed to design, sample collection, clinical evaluation, and selection of the dogs, tabulation, statistical analysis, data interpretation, and writing of the article; CBEM performed preparation of slides of the impression smear of cytology and interpretation of gastric bacterial colonisation density; JFC performed extraction of DNA, execution of the polymerase chain reaction assay, phylogenetic analysis, and construction of phylogenetic tree; MMF performed histopathological processing and interpretation of gastric lesion severity; LFP performed the endoscopic procedure and the gastric biopsy samples collection; BNA, VM, and OGV performed clinical evaluation and selection of the dogs and sterilization procedure of the female dogs; LRM performed anesthesia of the dogs; STLPF was an advisor and contributed to the design, review, and assistance on the elaboration of the article.
References
- 1.Haesebrouck F., Pasmans F., Flahou B., et al. Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clinical Microbiology Reviews. 2009;22(2):202–223. doi: 10.1128/cmr.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall B., Warren J. R. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. The Lancet. 1984;323(8390):1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 3.Dunn B. E., Cohen H., Blaser M. J. Helicobacter pylori. Clinical Microbiology Reviews. 1997;10(4):720–741. doi: 10.1128/cmr.10.4.720-741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dent J., McNulty C., Uff J., Wilkinson S. P., Gear M. W. L. Spiral organisms in the gastric antrum. The Lancet. 1987;330(8550):p. 96. doi: 10.1016/s0140-6736(87)92754-1. [DOI] [PubMed] [Google Scholar]
- 5.Solnick J. V., O’Rourke J., Lee A., Paster B. J., Dewhirst F. E., Tompkins L. S. An uncultured gastric spiral organism is a newly identified Helicobacter in humans. Journal of Infectious Diseases. 1993;168(2):379–385. doi: 10.1093/infdis/168.2.379. [DOI] [PubMed] [Google Scholar]
- 6.O’Rourke J. L., Solnick J. V., Neilan B. A., et al. Description of ‘Candidatus Helicobacter heilmannii’ based on DNA sequence analysis of 16S rRNA and urease genes. International Journal of Systematic Evolution Microbiology. 2004;54(6):2203–2211. doi: 10.1099/ijs.0.63117-0. [DOI] [PubMed] [Google Scholar]
- 7.Haesebrouck F., Pasmans F., Flahou B., Smet A., Vandamme P., Ducatelle R. Non-Helicobacter pylori Helicobacter species in the human gastric mucosa: a proposal to introduce the terms H. heilmannii sensu lato and sensu stricto. Helicobacter. 2011;16(4):339–340. doi: 10.1111/j.1523-5378.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 8.Mladenova-Hristova I., Grekova O., Patel A. Zoonotic potential of Helicobacter spp. Journal of Microbiology, Immunology and Infection. 2017;50(3):265–269. doi: 10.1016/j.jmii.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Ekman E., Fredriksson M., Trowald-Wigh G. Helicobacter spp. in the saliva, stomach, duodenum and faeces of colony dogs. The Veterinary Journal. 2013;195(1):127–129. doi: 10.1016/j.tvjl.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Recordati C., Gualdi V., Tosi S., et al. Detection of Helicobacter spp. DNA in the oral cavity of dogs. Veterinary Microbiology. 2007;119(2–4):346–351. doi: 10.1016/j.vetmic.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Jankowski M., Spużak J., Kubiak K., Glińska-Suchocka K., Biernat M. Detection of Helicobacter spp. in the saliva of dogs with gastritis. Polish Journal of Veterinary Sciences. 2016;19(1):133–140. doi: 10.1515/pjvs-2016-0017. [DOI] [PubMed] [Google Scholar]
- 12.Stolte M., Kroher G., Meining A., Morgner A., Bayerdörffer E., Bethke B. A comparison ofHelicobacter pyloriandH. heilmanniiGastritis: a matched control study involving 404 patients. Scandinavian Journal of Gastroenterology. 1997;32(1):28–33. doi: 10.3109/00365529709025059. [DOI] [PubMed] [Google Scholar]
- 13.Morgner A., Lehn N., Andersen L. P., et al. Helicobacter heilmannii-associated primary gastric low-grade MALT lymphoma: complete remission after curing the infection. Gastroenterology. 2000;118(5):821–828. doi: 10.1016/s0016-5085(00)70167-3. [DOI] [PubMed] [Google Scholar]
- 14.O’Rourke J., Grehan M., Lee A. Non-pylori helicobacter species in humans. Gut. 2001;49(5):601–606. doi: 10.1136/gut.49.5.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y. L., Yamada N., Wen M., Matsuhisa T., Miki M. Gastrospirillum hominis and Helicobacter pylori infection in Thai individuals—comparison of histopathological changes of gastric mucosa. Pathology International. 1998;48(7):507–511. doi: 10.1111/j.1440-1827.1998.tb03941.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu J., He L., Haesebrouck F., et al. Prevalence of coinfection with gastric non-Helicobacter pylori Helicobacter(NHPH) species inHelicobacter pylori-infected patients suffering from gastric disease in beijing, China. Helicobacter. 2015;20(4):284–290. doi: 10.1111/hel.12201. [DOI] [PubMed] [Google Scholar]
- 17.Chow J., Tang H., Mazmanian S. K. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Current Opinion In Immunology. 2011;23(4):473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polanco R., Salazar V., Reyes N., García-Amado M. A., Michelangeli F., Contreras M. High prevalence of DNA from Non-H. pylori Helicobacters in the gastric mucosa of venezuelan pet dogs and its histological alterations. Revista do Instituto de Medicina Tropical de São Paulo. 2011;53(4):207–212. doi: 10.1590/s0036-46652011000400006. [DOI] [PubMed] [Google Scholar]
- 19.Washabau R. J., Day M. J., Willard M. D., et al. Endoscopic, biopsy, and histophatologic guidelines for the evaluation of gastrointestinal inflammation in companion animals. Journal of Veterinary Internal Medicine. 2010;24(1):10–16. doi: 10.1111/j.1939-1676.2010.0520.x. [DOI] [PubMed] [Google Scholar]
- 20.Dossin O. Clinical history. In: Steiner J. M., editor. Small Animal Gastroenterology. 1. Hannover, Germany: Schlütersche; 2008. [Google Scholar]
- 21.Kubota S., Ohno K., Tsukamoto A., et al. Value of the 13C-urea breath test for detection of gastric Helicobacter spp. infection in dogs undergoing endoscopic examination. Journal of Veterinary Medical Science. 2013;75(8):1049–1054. doi: 10.1292/jvms.12-0528. [DOI] [PubMed] [Google Scholar]
- 22.Kubota-Aizawa S., Ohno K., Fukushima K., et al. Epidemiological study of gastric Helicobacter spp. in dogs with gastrointestinal disease in Japan and diversity of Helicobacter heilmannii sensu stricto. The Veterinary Journal. 2017;225:56–62. doi: 10.1016/j.tvjl.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Akhtardanesh B., Jamshidi S., Sasani F., Mohammadi M., Bokaee S., Salehi T. Z. Helicobacter infection and gastric lesions in domestic and stray cats. Veterinarski Arhiv. 2006;76(6):479–488. [Google Scholar]
- 24.Tabrizi A. S., Jamshidi S., Oghalaei A., Salehi T. Z., Eshkaftaki A. B., Mohammadi M. Identification of Helicobacter spp. in oral secretions vs. gastric mucosa of stray cats. Veterinary Microbiology. 2010;140(1-2):142–146. doi: 10.1016/j.vetmic.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 25.Day M. J., Bilzer T., Mansell J., et al. Histopathological standards for the diagnosis of gastrointestinal inflammation in endoscopic biopsy samples from the dog and cat: a report from the World Small animal veterinary association gastrointestinal standardization group. Journal of Comparative Pathology. 2008;138:S1–S43. doi: 10.1016/j.jcpa.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Germani Y., Dauga C., Duval P., et al. Strategy for the detection of Helicobacter species by amplification of 16S rRNA genes and identification of H. felis in a human gastric biopsy. Research in Microbiology. 1997;148(4):315–326. doi: 10.1016/s0923-2508(97)81587-2. [DOI] [PubMed] [Google Scholar]
- 27.Staden R. The Staden sequence analysis package. Molecular Biotechnology. 1996;5(3):233–241. doi: 10.1007/bf02900361. [DOI] [PubMed] [Google Scholar]
- 28.Hall T. A. BioEdit: a user-friendly biological sequence alignment edi-tor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–99. [Google Scholar]
- 29.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang C. Y., Han H. R., Youn H. Y. Prevalence and clinical characterization of gastric Helicobacter species infection of dogs and cats in Korea. Journal of Veterinary Science. 2002;3(2):123–133. doi: 10.4142/jvs.2002.3.2.123. [DOI] [PubMed] [Google Scholar]
- 31.Wiinberg B., Spohr A., Dietz H. H., et al. Quantitative analysis of inflammatory and immune responses in dogs with gastritis and their relationship to Helicobacter spp. infection. Journal of Veterinary Internal Medicine. 2005;19(1):4–14. doi: 10.1892/0891-6640(2005)19<4:qaoiai>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Hermanns W., Kregel K., Breuer W., Lechner J. Helicobacter-like organisms: histopathological examination of gastric biopsies from dogs and cats. Journal of Comparative Pathology. 1995;112(3):307–318. doi: 10.1016/s0021-9975(05)80083-0. [DOI] [PubMed] [Google Scholar]
- 33.Amorim I., Smet A., Alves O., et al. Presence and significance of Helicobacter spp. in the gastric mucosa of Portuguese dogs. Gut Pathogens. 2015;7:p. 12. doi: 10.1186/s13099-015-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asl A. S., Jamshidi S., Mohammadi M., Soroush M. H., Bahadori A., Oghalaie A. Detection of atypical cultivable canine gastric Helicobacter strain and its biochemical and morphological characters in naturally infected dogs. Zoonoses and Public Health. 2010;57(4):244–248. doi: 10.1111/j.1863-2378.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- 35.Suárez-Esquivel M., Alfaro-Alarcón A., Guzmán-Verri C., Barquero-Calvo E. Analysis of the association between density of Helicobacterspp and gastric lesions in dogs. American Journal of Veterinary Research. 2017;78(12):1414–1420. doi: 10.2460/ajvr.78.12.1414. [DOI] [PubMed] [Google Scholar]
- 36.Redéen S., Petersson F., Jönsson K. A., Borch K. Relationship of gastroscopic features to histological findings in gastritis and Helicobacter pylori infection in a general population sample. Endoscopy. 2003;35(11):946–950. doi: 10.1055/s-2003-43479. [DOI] [PubMed] [Google Scholar]
- 37.Joosten M., Blaecher C., Flahou B., Ducatelle R., Haesebrouck F., Smet A. Diversity in bacterium-host interactions within the species Helicobacter heilmannii sensu stricto. Veterinary Research. 2013;44(1):p. 65. doi: 10.1186/1297-9716-44-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gueneau P., Loiseauxdegoer S. Helicobacter: molecular phylogeny and the origin of gastric colonization in the genus. Infection, Genetics and Evolution. 2002;1(3):215–223. doi: 10.1016/s1567-1348(02)00025-4. [DOI] [PubMed] [Google Scholar]
- 39.Schott T., Kondadi P. K., Hänninen M. L., Rossi M. Comparative Genomics of Helicobacter pylori and the human-derived Helicobacter bizzozeronii CIII-1 strain reveal the molecular basis of the zoonotic nature of non-pylori gastric Helicobacter infections in humans. BMC Genomics. 2011;12(534):p. 534. doi: 10.1186/1471-2164-12-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Øverby A., Yamagata Murayama S., Matsui H., Nakamura M. In the aftermath of. Digestion. 2016;93(4):260–265. doi: 10.1159/000445399. [DOI] [PubMed] [Google Scholar]
- 41. Instituto Nacional do Câncer. Estatísticas do Câncer. https://www.inca.gov.br/numeros-de-cancer.
- 42.Oliveira A. F., Carvalho J. R., Costa M. F. S., Lobato L. C. P., Silva R. S., Schramm J. M. A. Estimativa da prevalência e da mortalidade por complicações da úlcera péptica, Brasil, 2008: uma proposta metodológica. Epidemiologia e Serviço da Saúde. 2015;24(1):145–154. doi: 10.5123/s1679-49742015000100016. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The occurrence data of Helicobacters used to support the findings of this study are included within the article. The sequence data used to support findings of this article were deposited in GenBank; the accession numbers are Sample 3B: Genbank accession MN966439/Sample 5B: Genbank accession MN966440/Sample 7B: Genbank accession MN966441/Sample 8B: Genbank accession MN966442/Sample 9B: Genbank accession MN966443/Sample 10B: Genbank accession MN966444/Sample 11B: Genbank accession MN966445/Sample 12B: Genbank accession MN966446/Sample 15B: Genbank accession MN966447/Sample 16B: Genbank accession MN966448/Sample 17B: Genbank accession MN966449/Sample 20B: Genbank accession MN966450/Sample 21B: Genbank accession MN966451/Sample 23B: Genbank accession MN966452/Sample 24B: Genbank accession MN966453/Sample 24S: Genbank accession MN966454/Sample 25B: Genbank accession MN966455/Sample 26B: Genbank accession MN966456/Sample 27B: Genbank accession MN966457/Sample 28B: Genbank accession MN966458/Sample 29B: Genbank accession MN966459/Sample 30B: Genbank accession MN966460/Sample 31B: Genbank accession MN966461/Sample 32B: Genbank accession MN966462/Sample 33B: Genbank accession MN966463/Sample 35B: Genbank accession MN966464.


