Abstract
Behavioral experiences activate the Fos transcription factor (TF) in sparse populations of neurons that are critical for encoding and recalling specific events1–3. However, there is limited understanding of the mechanisms by which experience drives circuit reorganization to establish a network of Fos-activated cells. It is also unknown if Fos is required in this process beyond serving as a marker of recent neural activity and, if so, which of its many gene targets underlie circuit reorganization. Here we demonstrate that when mice engage in spatial exploration of novel environments, perisomatic inhibition of Fos-expressing hippocampal CA1 pyramidal neurons by parvalbumin (PV)-interneurons (INs) is enhanced, while perisomatic inhibition by cholecystokinin (CCK)-INs is weakened. This bidirectional modulation of inhibition is abolished when the function of the Fos TF complex is disrupted. Single-cell RNA-sequencing, ribosome-associated mRNA profiling, and chromatin analyses, combined with electrophysiology, reveal that Fos activates the transcription of Scg2 (secretogranin II), a gene that encodes multiple distinct neuropeptides, to coordinate these changes in inhibition. As PV- and CCK-INs mediate distinct features of pyramidal cell activity4–6, the Scg2-dependent reorganization of inhibitory synaptic input might be predicted to affect network function in vivo. Consistent with this prediction, hippocampal gamma rhythms and pyramidal cell coupling to CA1 theta are significantly altered with loss of Scg2. These findings reveal an instructive role for Fos and Scg2 in establishing a network of Fos-activated neurons via the rewiring of local inhibition to form a selectively modulated state. The opposing plasticity mechanisms on distinct inhibitory pathways may support the consolidation of memories over time.
Neurons convert new experiences into stable representations in the brain to inform future actions. Mounting evidence suggests that sparse populations of neurons distributed across multiple regions of the brain form the neural substrates for a variety of behaviors1,2. A hallmark of these active neuronal ensembles is the transient expression of a set of genes, termed the immediate early genes (IEGs), one of which encodes the Fos TF3,7. A longstanding hypothesis has been that once activated by salient stimuli, Fos-expressing neurons undergo modifications that facilitate the encoding of specific features of an experience, such that subsequent reactivation of even a subset of these neurons is sufficient to elicit recall of the initial experience1,2. Yet whether these neuronal ensembles in fact become persistently modified, and if so, the nature of these changes and their underlying molecular mechanisms, remain unclear. Moreover, whether Fos induction, beyond serving as a proxy for recent neural activity, plays a causal role in coordinating circuit modifications required to encode an experience remains unresolved. Complicating progress in this regard is the fact that the Fos family of TFs is composed of seven at least partially functionally redundant members (Fos, Fosb, Fosl1, Fosl2, Jun, Junb, and Jund)3,7.
Fos-activated neurons in the hippocampal area CA1 have been shown to stably encode contextual information as compared to their non-Fos-activated counterparts2. As recurrent excitatory connectivity is weak within CA1, pyramidal cells (PCs) are known to be regulated in concert either via their common excitatory inputs8 or a local network of inhibitory GABAergic INs. Perisomatic-targeting INs, by virtue of their extensive axonal arborizations, are uniquely positioned to control spike frequency and duration in populations of PCs4,6. In this regard, two functionally distinct forms of perisomatic inhibition have been described, mediated by PV or CCK-INs. Whereas PV-INs display fast, non-adapting firing patterns and are predominantly activated in a feedforward fashion, CCK-INs fire regular, adapting trains of action potentials and provide predominantly feedback inhibition4–6,9,10. Perisomatic inhibition has also been shown to coordinate behavioral state-dependent network oscillations11,12. For example, PV-INs regulate gamma rhythms12, which are critical for transient synchrony of PCs, and both PV- and CCK-INs fire preferentially at different phases of theta5, which have been associated with memory encoding or retrieval13. By considering how inputs of each IN subtype are selectively modified onto Fos-activated neurons, we reasoned that it should be possible to gain mechanistic insights into how experience alters the temporal dynamics of network function to support long-term memories.
Bidirectional modulation of IN inputs
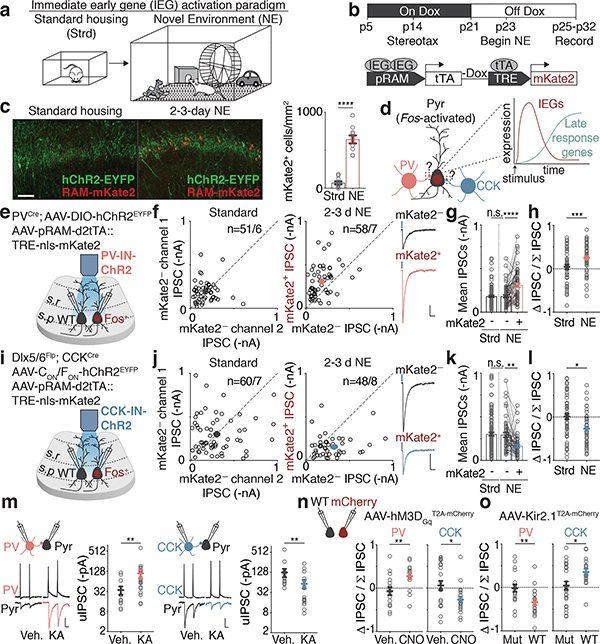
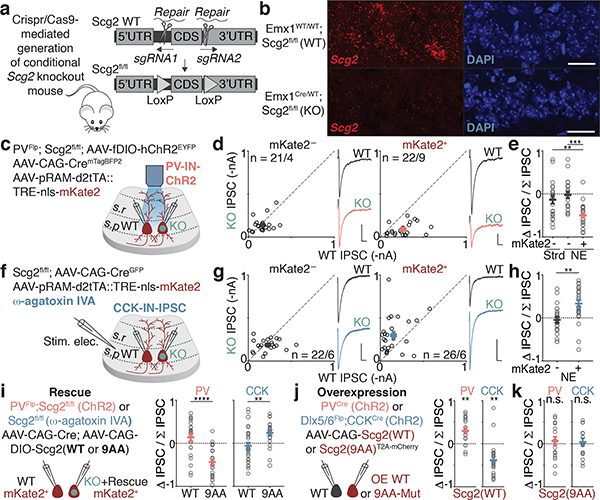
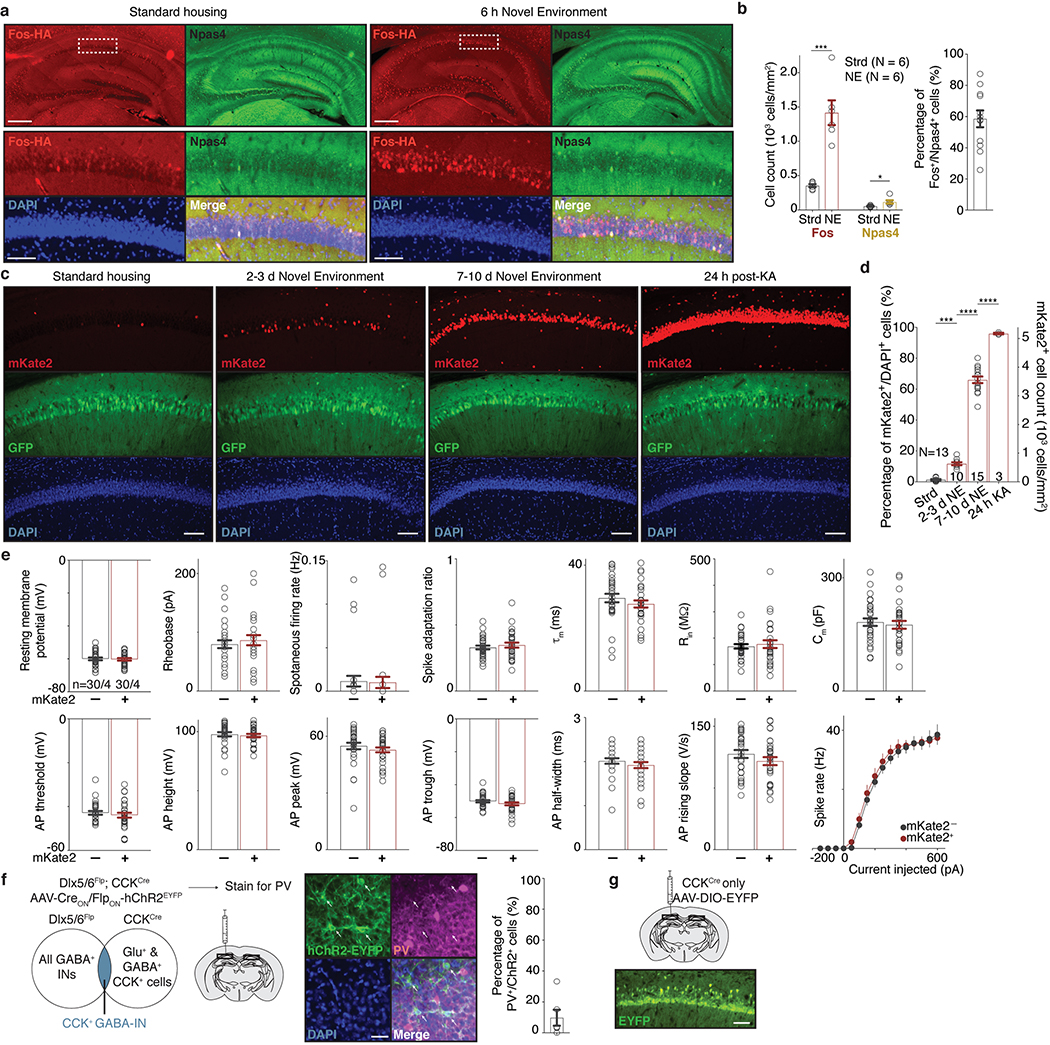
We first asked if either of these forms of perisomatic inhibition are differentially regulated onto Fos-expressing compared to neighboring non-Fos-expressing neurons. We exposed mice to a series of novel environments, which we found robustly activates Fos in a sparse subset of CA1 PCs (Fig. 1a, Extended Data Fig. 1a–d). To label these Fos-expressing neurons, we utilized a previously developed adeno-associated virus (AAV)-based reporter that expresses the fluorescent protein mKate2 selectively in recently activated neurons (Fig. 1b)14. Using this reporter, we detected a significant increase in the number of recently activated neurons (mKate2+) in mice exposed to 2–3 days (d) of novel environments (NE) compared to control mice housed under standard (Strd) conditions (Fig. 1c). We reasoned that this 2–3d timepoint would therefore be appropriate for assessing the long-lasting effects of Fos and its late-response target gene(s), which are usually activated within 1–12 hours (h) of stimulus onset (Fig. 1d).
Figure 1. Bidirectional modulation of IN inputs.
a. Schematic of standard housing (Strd) or novel environment (NE).
b. Experimental timeline and configuration of AAV-based activity reporter; mKate2 labeling is temporally controlled via doxycycline (Dox)14.
c. (Left) Representative images depicting Fos-activated neurons (red) and PV-IN-specific channelrhodopsin-2 (ChR2, green) in CA1 in Strd and 2–3d NE. (Right) Number of mKate2+ cells/mm2 in Strd (N=13 mice) and NE (N=10 mice). Scale:100 μm. ****p=2.6×10−10.
d. Schematic of Fos-activated CA1 PCs and its perisomatic-targeting inputs from PV- or CCK-INs. Schematic of activity-induced gene expression kinetics. In the early wave, immediate early genes such as Fos are expressed. Fos subsequently activates late-response genes.
e,i. Schematic of genetic strategy to introduce ChR2 into PV- or CCK-INs and measure light-evoked IPSCs.
f. Scatter plots of recorded pairs of (Left) mKate2— neurons in Strd (n=51/6) or (Right) mKate2+ and mKate2— pairs after 2–3d NE (n=58/7). Representative traces from a pair of neurons shown; blue marks depict light onset. Scale:100 pA;40 ms.
g. Mean PV-IPSC amplitudes from f. ****p=3.2×10−6.
h. Normalized differences in PV-IPSC amplitudes between pairs of neurons in f (Methods),***p=3.4×10−4.
j-l. As in f-h for CCK-IPSCs. Strd,n=60/7; NE,n=48/8. Scale:100 pA;40 ms. (k)**p=5.5×10−3. (l)*p=0.014.
m. IN-to-CA1 PC paired recording configuration, representative traces and uIPSC amplitudes for (Left) PV-to-CA1 (Vehicle(Veh.),n=13/6; KA,n=19/7; **p=0.003) or (Right) CCK-to-CA1 pairs (Veh.,n=16/9; KA,n=16/4; **p=0.010). Scale: 30 mV;20 pA;20 ms. Mann-Whitney test (two-sided).
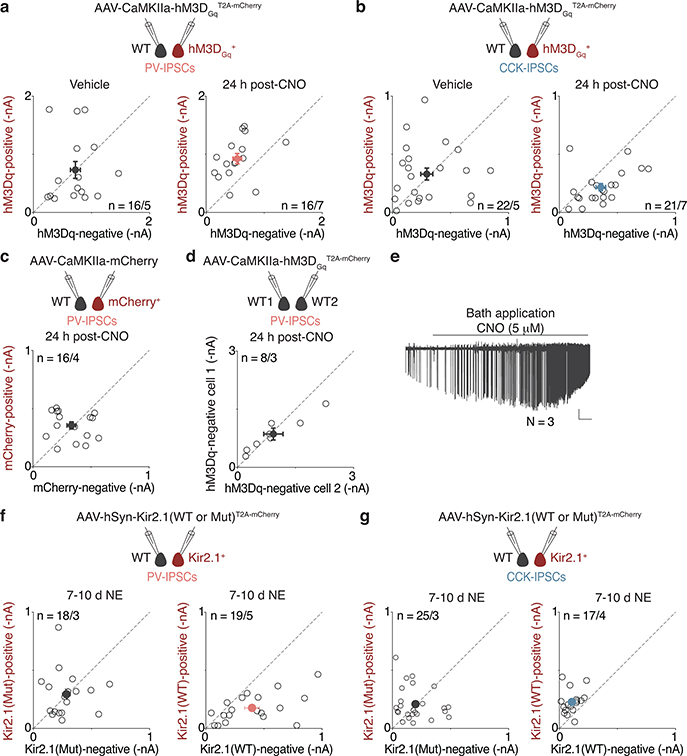
n. (Left) PV- and (Right) CCK-IPSC amplitudes of pairs of non-transduced (WT) and hM3DGq (mCherry+) neurons after 24h vehicle or CNO treatment. PV (Veh.,n=16/5; CNO,n=16/7; **p=0.006); CCK (Veh.,n=22/5; CNO,n=21/7; *p=0.014).
o. As in n but with Kir2.1. Control is a non-conducting mutant (KirMut). Mice were exposed to 7–10d NE, a period over which many CA1 PCs would have turned on Fos (Extended Data Fig. 1c,d). PV (KirMut,n=18/3; Kir2.1,n=19/5; **p=0.007); CCK (KirMut,n=25/3; Kir2.1,n=17/4; *p=0.023).
(f,h,j,l,m-o) Each open circle represents a pair of simultaneously recorded neurons.
(c,f-h,j-o) Mean±SEM.
(f,j,m-o) n=number of pairs/mice.
(c,k,l,n,o) Two-sided t-test. (g,k) Ordinary one-way ANOVA, multiple comparisons corrected.
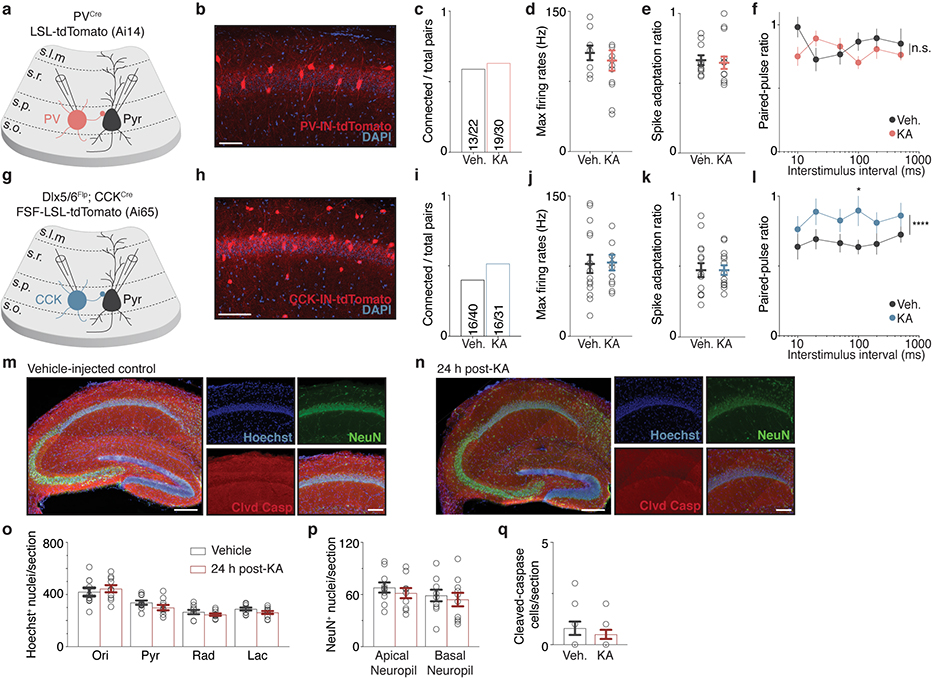
To assess PV-mediated inhibition, we expressed channelrhodopsin-2 (ChR2) via a Cre-dependent AAV in PVCre mice, which allowed for PV-mediated inhibitory postsynaptic currents (IPSCs) to be selectively evoked by focal photoactivation of ChR2-expressing PV-specific presynaptic boutons. We measured PV-IPSCs onto PCs by performing dual whole-cell voltage-clamp recordings on pairs of recently activated (Fos+/mKate2+) and neighboring non-activated (Fos—/mKate2—) CA1 PCs in acute hippocampal slices prepared 2–3d after initial NE exposure (Fig. 1e). We found that the mean PV-IPSC amplitude onto Fos+/mKate2+ (~310 pA) neurons is significantly larger by 1.7-fold relative to that of Fos—/mKate2— neurons in either Strd or NE conditions (~180 pA) (Fig. 1f–h), indicating that PV-mediated inhibition is strengthened onto Fos-expressing neurons. By contrast, other electrophysiological parameters are not significantly different between the two groups (Extended Data Fig. 1e).
To assess CCK-mediated inhibition, we used an intersectional Flp- and Cre-dependent AAV15 in Dlx5/6Flp;CCKCre mice to drive the expression of ChR2 specifically in CCK-INs, as the CCKCre driver alone labels both glutamatergic and GABAergic neurons16 (Fig. 1i, Extended Data Fig. 1f,g). Using an analogous experimental paradigm to that described above, we found that in contrast to the selective increase in PV-mediated inhibition onto Fos+ CA1 PCs, the mean CCK-IPSC amplitude is significantly smaller onto Fos+ CA1 PCs (~170 pA) by 1.8-fold compared to that of Fos— CA1 PCs (~300 pA) (Fig. 1j–l).
These findings were corroborated by paired recordings of IN-to-CA1 PC to measure unitary IPSC (uIPSC) amplitudes. Recordings were performed using slices prepared from PVCre or Dlx5/6Flp; CCKCre tdTomato reporter mice 24h after exposure to kainic acid (KA) to synchronously and reliably activate nearly all CA1 PCs (Extended Data Fig. 1c,d). Consistent with our findings using light-evoked IPSC measurements, we found that PV-uIPSC amplitudes onto CA1 PCs are significantly larger by 3.2-fold, whereas CCK-uIPSC amplitudes are significantly smaller by 2.2-fold (Fig. 1m, Extended Data Fig. 2a–q).
These data indicate that NE exposure leads to selective, persistent bidirectional changes in perisomatic inhibition onto Fos-expressing neuronal ensembles, with PV-mediated inhibition strengthening and CCK-mediated inhibition weakening. We hereon refer to these modifications as “bidirectional perisomatic inhibitory plasticity.”
The bidirectional changes in perisomatic inhibition are a consequence of experience-driven neuronal activity, rather than a reflection of pre-existing differences between Fos+/mKate2+ and Fos—/mKate2— CA1 PCs insofar as they could be recapitulated by chemogenetic activation of neurons expressing the Gq-coupled DREADD17 receptor hM3Dq (Fig. 1n, Extended Data Fig. 3a–e). Conversely, silencing CA1 PCs via expression of an inwardly-rectifying potassium channel Kir2.118, but not a non-conducting mutant (KirMut), led to the inverse effects (Fig. 1o, Extended Data Fig. 3f,g).
Causal role for Fos family TFs
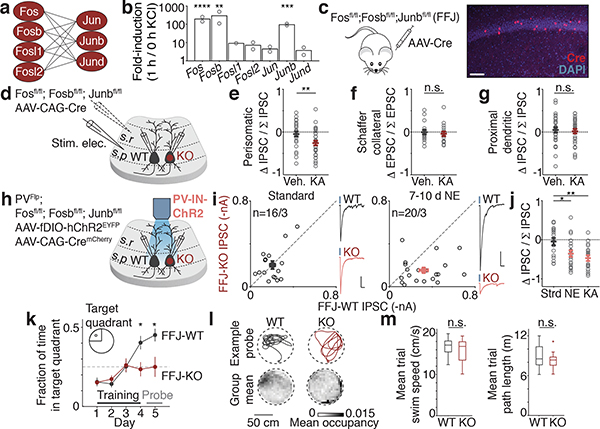
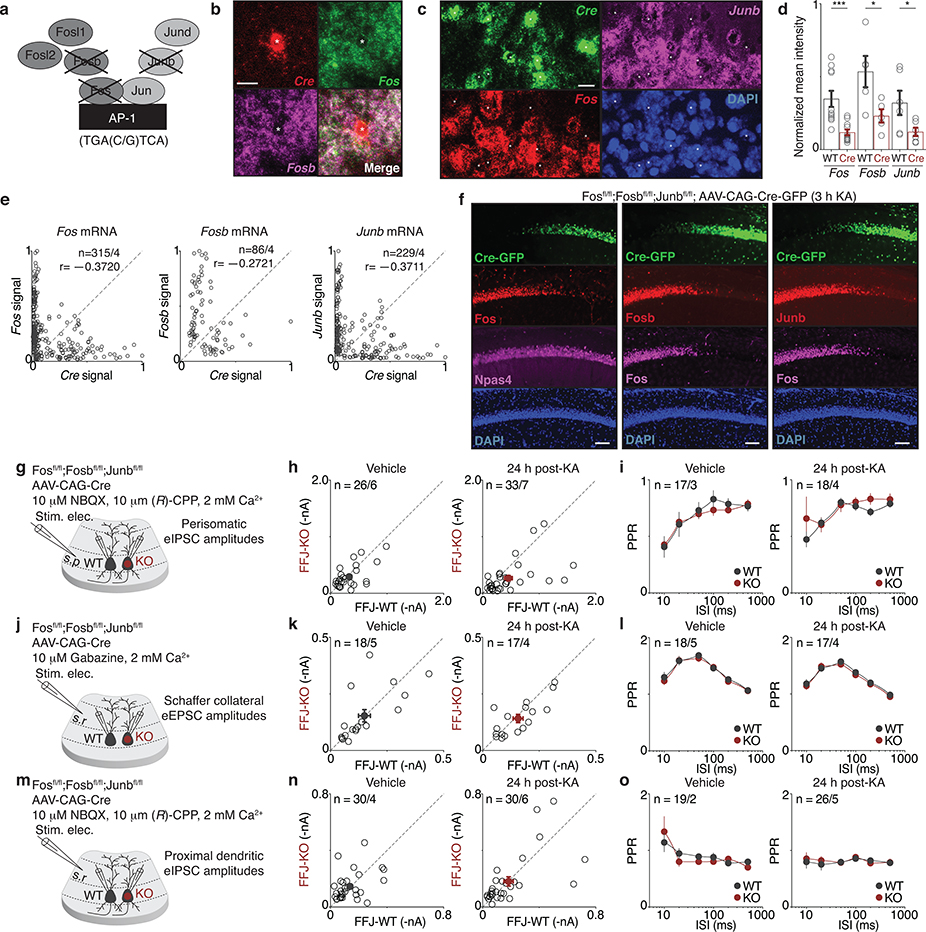
Since the induction of bidirectional perisomatic inhibitory plasticity occurs selectively onto Fos-expressing CA1 PCs, we considered the possibility that the Fos family of TFs, termed AP-1 factors, might mediate these changes. We first determined which of the seven members are induced in the hippocampus by neuronal activity (Fig. 2a). We found that Fos, Fosb, and Junb are induced by ~100-fold in membrane-depolarized hippocampal cultured neurons, while the four other AP-1 family members are significantly less responsive (Fig. 2b). We therefore developed a triple conditional knockout mouse line to enable the deletion of these strongly inducible AP-1 factors in a spatiotemporally-controlled manner (Fosfl/fl; Fosbfl/fl; Junbfl/fl, hereon FFJ)19 and verified the effective excision of these genes upon Cre expression in vivo by single-molecule RNA fluorescence in situ hybridization (smRNA-FISH) and immunostaining for each of these three proteins (Extended Data Fig. 4a–f).
Figure 2. Causal role for Fos family TFs.
a. Schematic depicting possible AP-1 homo- and heterodimers.
b. Mean fold-induction of each AP-1 member upon KCl-mediated depolarization in hippocampal neurons (bulk RNA-sequencing; Methods) showing significantly more induction of Fos (****p=9.1×10−5), Fosb(***p=0.008), and Junb(****p=2.2×10−7) compared to other four factors. n=2 biological replicates.
c. Schematic of Fosfl/fl;Fosbfl/fl;Junbfl/fl (FFJ) mouse transduced with AAV to sparsely express Cre (red). Representative CA1 image shown. Scale:100 μm.
d. Recording configuration with stimulus electrode placement in stratum pyramidale, to measure perisomatic eIPSCs, or stratum radiatum, for Schaffer-collateral eEPSCs or proximal dendritic eIPSCs.
e-g. Normalized differences in indicated pharmacologically-isolated current amplitudes between pairs of FFJ-WT and KO PCs, where (e) Veh.,n=26/6; KA,n=33/7; **p=0.005, (f) Veh.,n=18/5; KA,n=17/4, (g) Veh.,n=30/4; KA,n=30/6.
h. Schematic of strategy to introduce ChR2 into PV-INs and sparse Cre into the CA1 of PVFlp;FFJ.
i. Scatter plots of recorded pairs of FFJ-WT and -KO CA1 PCs in (Left) Strd (n=16/3) or (Right) 7–10d NE (n=20/3). Representative traces from pairs of neurons shown; blue marks depict light onset. Scale:50 pA(i) or 100 pA(j); 40 ms.
j. As in e-g for pairs depicted in i and 24h post-KA condition (n=19/3). *p=0.014(NE); **p=0.002(KA). Ordinary one-way ANOVA, multiple comparisons corrected.
k. Fraction of time spent swimming in target quadrant for FFJ-WTs (N=11 mice) and FFJ-KOs (N=12 mice). *p=0.014(Day 4); 0.016(Day 5).
l. (Top) Example probe trial swim traces. (Bottom) Mean probe trial occupancy maps, 5 cm bins.
m. Box plots of mean trial (Left) speed and (Right) path length; animals as in k. Center line, median; box limits, upper and lower quartiles; whiskers, min/max; “+” indicates outlier.
(e-g,i,j) Each open circle represents a pair of simultaneously recorded neurons; n=number of pairs/mice.
(e-g,i-k) Mean±SEM.
(b,e-g,k,m) Two-sided t-test.
After sparse deletion of Fos, Fosb, and Junb mediated by an AAV-Cre (Fig. 2c,d), we performed dual whole-cell recordings from FFJ-WT and neighboring FFJ-KO CA1 PCs while electrically stimulating perisomatic inhibitory axons. We found a significant 1.7-fold decrease in pharmacologically-isolated eIPSC amplitudes onto FFJ-KO compared to WT activated neurons (Fig. 2e, Extended Data Fig. 4g–i). By contrast, we observed no significant differences in CA3 Schaffer collateral-evoked excitatory postsynaptic current (eEPSC) or proximal dendritic eIPSC amplitudes between FFJ-WT and KO neurons under 24h post-vehicle or KA conditions (Fig. 2f,g, Extended Data Fig. 4j–o). Therefore, while in principle AP-1 could regulate Fos-expressing CA1 PCs by modulating their CA3 excitatory inputs or inhibition from distinct compartments, we show that AP-1 TFs are specifically required for the regulation of perisomatic inhibition.
To directly measure PV-mediated inhibition, we generated PVFlp/Flp;FFJ mice, which allowed for the expression of ChR2 specifically in PV-INs (Fig. 2h). Simultaneous slice recordings of ChR2-evoked PV-IPSCs in FFJ-WT and neighboring FFJ-KO neurons revealed no differences in Strd housed mice (Fig. 2i). By contrast, a significant decrease in PV-IPSC amplitudes onto FFJ-KO cells was observed in mice after 7–10d NE, with 90% of FFJ-KO cells showing smaller IPSC amplitudes compared to the average for WT cells (Fig. 2i,j). These data indicate that AP-1 is required for the experience-dependent recruitment of PV-mediated inhibition and identify a previously elusive function for AP-1 TFs in long-term plasticity.
Given that loss of AP-1 leads to significant defects in inhibition, we next asked if spatial learning and memory are affected under these conditions. FFJ mice were bilaterally injected with AAV expressing Cre (FFJ-KO) or a catalytically inactive ΔCre (FFJ-WT) in the CA1 region and assessed in the Morris water maze paradigm. In contrast to FFJ-WTs, the FFJ-KOs performed significantly worse on this spatial task and were unable to learn the location of the platform in the maze (Fig. 2k,l). We observed no statistically significant differences in mean swim speeds or path lengths between the two groups, arguing against a contribution of motor deficits in the FFJ-KOs (Fig. 2m). These results suggest that changes in perisomatic inhibitory plasticity of Fos-activated neuronal networks may contribute to hippocampal-dependent spatial learning.
Fos targets in CA1 pyramidal neurons
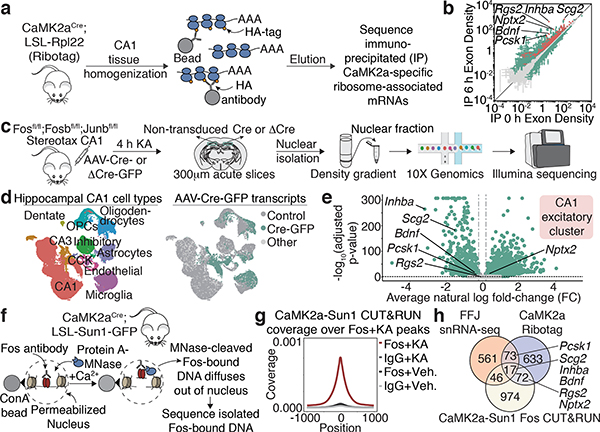
Although many activity-regulated genes (ARGs) have now been defined, difficulties in effectively disrupting AP-1 function in vivo have complicated the identification of genes that are specifically regulated by AP-1 TFs and thus might mediate the bidirectional modulation of perisomatic inhibition. The identification of AP-1 target genes has been further hampered by the pronounced neuronal cell-type-divergence of activity-dependent gene programs20, and it is unclear how AP-1 TFs, which are induced in nearly all cell types in the brain, contribute to this diversity. To address these challenges, we used a suite of genome-wide approaches to identify high-confidence AP-1 targets, focusing on CA1 PCs. We identified 1) ARGs in CA1 PCs, 2) genes that show reduced expression when AP-1 function is disrupted, and 3) genes that display activity-dependent Fos binding at nearby regulatory DNA elements. For these analyses, we subjected mice to KA treatment to strongly activate nearly all cells in CA1 and thus maximize signal-to-noise ratio for identification of genes. AP-1 target(s) of interest identified by this method were subsequently validated under more physiological conditions of NE exposure.
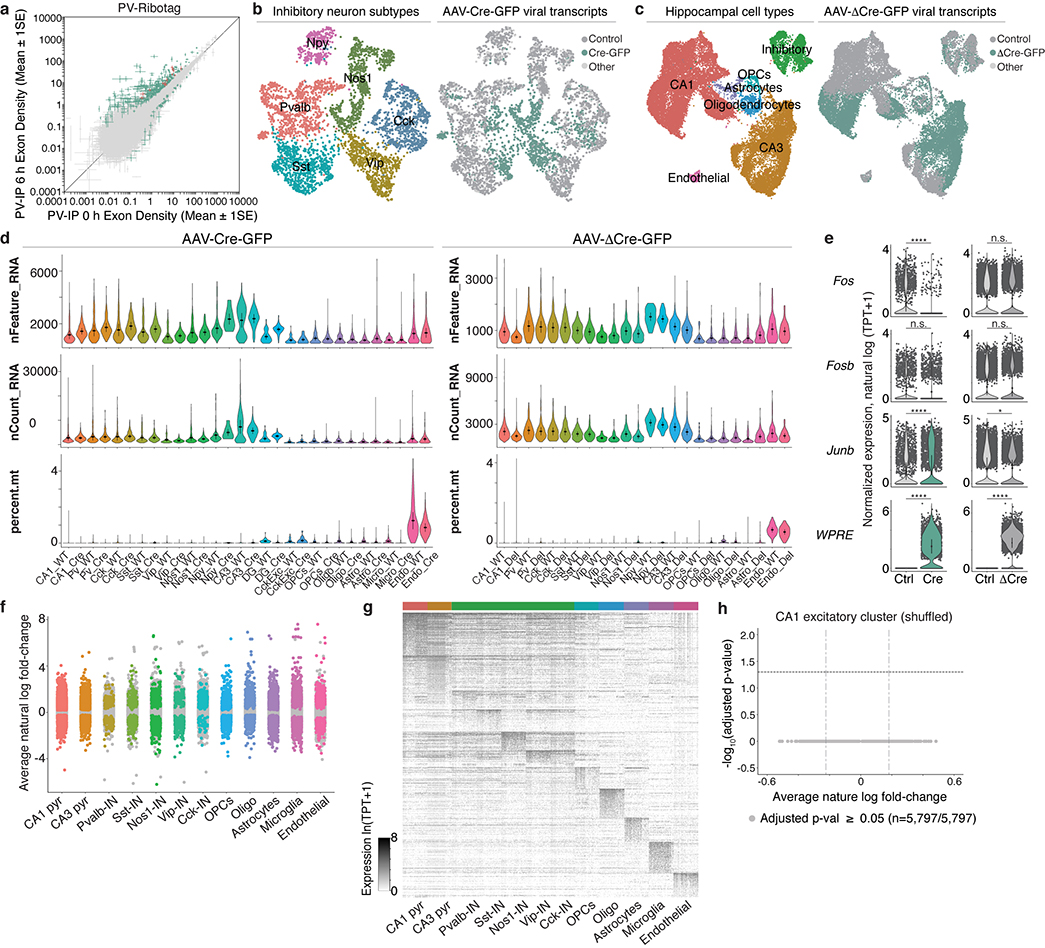
We first defined ARGs specific to CA1 PCs by ribosome-associated mRNA profiling (Fig. 3a). Using CA1 tissue from CaMK2aCre;Rpl22-HA(RiboTag)21 mice subjected to 6h vehicle or KA treatment, we immunoprecipitated and sequenced CaMK2a-specific ribosome-associated mRNAs. Differential gene expression (DGE) analysis identified 795 ARGs induced by at least 2-fold (FDR ≤ 0.005), of which 111 are significantly enriched in CaMK2a-positive neurons relative to other cell types, including PV-INs (Fig. 3b, Extended Data Fig. 5a).
Figure 3. Fos targets in CA1 pyramidal neurons.
a,c,f. Workflow for Ribotag, FFJ snRNA-seq and Fos CUT&RUN (Methods).
b. Scatter plot showing CaMK2a-specific ARGs in 6h post-KA compared to vehicle conditions. Significantly different genes (green); FDR≤0.005. CaMK2a-enriched (IP over input) genes (red). Points represent mean±SE. n=4 mice/bioreplicate; 3 bioreplicates/condition.
d. UMAP visualization of nuclei from Cre+ and control FFJ snRNA-seq with (Left) cell type information or (Right) genotype assignments overlaid. “Control”: Cre— in control hemispheres; “Cre-GFP”: Cre+ in injected hemispheres; “Other”: Cre— or Cre+ in injected or control hemispheres, respectively. n=58,536 cells/6 mice.
e. Volcano plot for genes in CA1 excitatory cluster. Average natural-log fold-change (FC) comparing Cre+ and Cre— (x-axis); —log10 Bonferroni-corrected p-values (y-axis; Wilcoxon rank-sum, two-sided). Each point represents a gene detected in ≥5% of non-transduced cells, where light grey: p≥0.05 (n=3,429); darker grey: FC≤20% in either direction (n=42), green: p<0.05 and FC>20% (n=3,514).
g. Aggregate plot showing spike-in normalized Fos coverage per bin averaged across all Fos peaks (Methods). IgG serves as a specificity control. n=1 mouse/bioreplicate, 3 bioreplicates/condition.
h. Venn diagram showing intersection of significant CA1 PC-specific genes from CaMK2a-Ribotag (FC≥2), snRNA-seq (FC<−20%) and CUT&RUN (Fos peaks within 10 kb from TSS).
(c) Schematic images adapted with permission from Paxinos & Franklin (Elsevier), 10x Genomics and Illumina.
To determine which of these genes show reduced expression when AP-1 function is disrupted, we performed high-throughput single-nucleus RNA-sequencing (snRNA-seq) using the FFJ mice. Animals received either AAV Cre-GFP (Cre+) or ΔCre-GFP (ΔCre+) injected into one CA1 hemisphere, leaving cells in the contralateral hemisphere non-transduced as control. Mice were subjected to 4h KA, and CA1 nuclei were isolated and subsequently sorted using the 10X Genomics platform (Fig. 3c). We sequenced 83,750 single-cell transcriptomes isolated from 6 Cre+ and 4 ΔCre+ mice (Fig. 3d, Extended Data Fig. 5b–e). Nuclei were clustered into 12–15 cell types using the Seurat single-cell analysis pipeline (Fig. 3d). The presence of viral-derived transcripts was used to identify 17,027 Cre+ and 14,557 ΔCre+ nuclei. For each cell type, DGE analysis comparing Cre+ (or ΔCre+)-transduced nuclei to their respective non-transduced controls was used to identify a wide array of AP-1-regulated genes, many of which are cell-type-specific (Extended Data Fig. 5f,g). These data support the longstanding but previously untested hypothesis that AP-1 contributes to the cell-type-divergence of ARG expression. Specifically, within the CA1 excitatory neuron cluster, we identified 697 genes that are significantly downregulated by at least 20% with loss of AP-1 (Fig. 3e, Extended Data Fig. 5e–h).
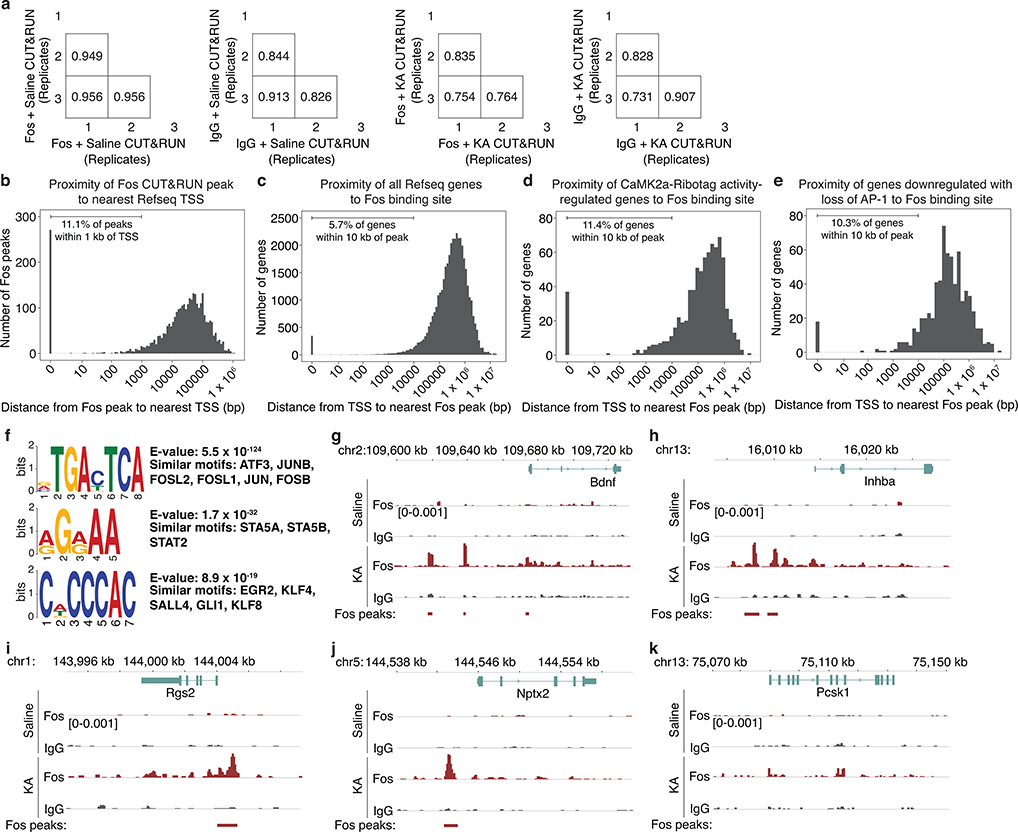
Finally, we identified genes that are likely direct targets of Fos in CA1 PCs using CUT&RUN, a chromatin profiling strategy in which in situ antibody-targeted controlled cleavage by micrococcal nuclease releases specific Fos protein-DNA complexes for sequencing22 (Fig. 3f). CaMK2a-expressing CA1 nuclei from CaMK2aCre;LSL-Sun1-sfGFP-Myc23 mice were isolated via sorting based on Cre-dependent expression of the GFP-tagged inner nuclear membrane protein, Sun1. We identified 3,295 Fos-bound activity-responsive loci from mice exposed to 2–3h KA as compared to vehicle treatment, with 1,109 genes containing at least one Fos-bound regulatory element within 10 kb of the transcription start site (TSS) (Fig. 3g, Extended Data Fig. 6a–f, Supplementary Fig. 1).
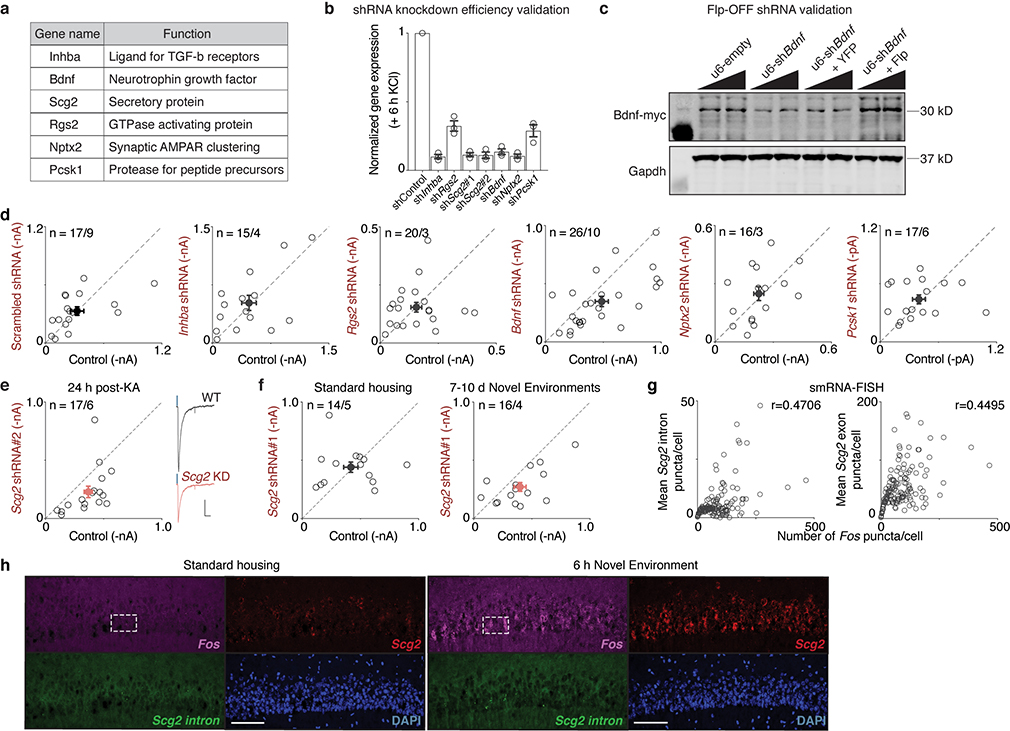
Intersection of the three datasets identified 17 genes that are inducible by activity in CA1 PCs (CaMK2a-Ribotag), show reduced expression with loss of AP-1 (FFJ snRNA-seq), and bind Fos at nearby regulatory elements (CaMK2a-Sun1 Fos CUT&RUN). An additional 191 genes are present in two of the three datasets (Fig. 3h, Supplementary Table 1). We focused our attention on three high-confidence AP-1-regulated candidate genes that display high fold-induction and whose expression is enriched in CA1 PCs (Inhba, Bdnf, and Scg2) and three other genes previously shown to contribute to inhibitory plasticity24 that are present in two of the three genomic datasets (Rgs2, Nptx2, and Pcsk1) (Extended Data Figs. 6g–k, 7a).
Fos-dependent effector of inhibition
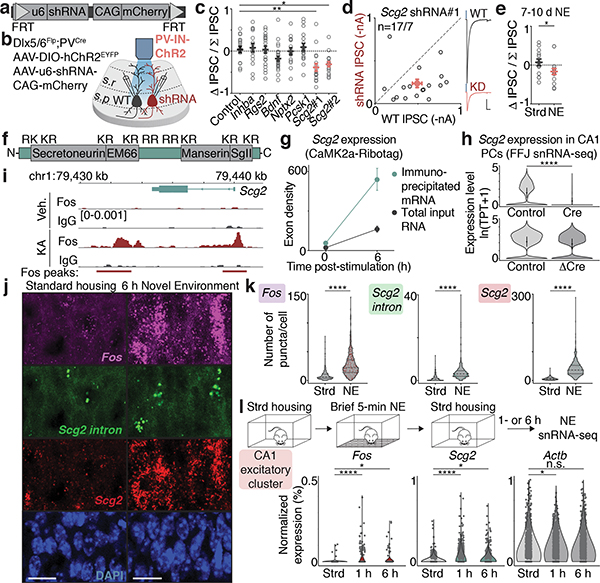
To identify molecular effector(s) of bidirectional perisomatic inhibitory plasticity downstream of Fos activation, short hairpin RNA (shRNA)-mediated gene knockdown was initially used to determine if any of the six candidate genes might mediate the activity-dependent strengthening of PV-mediated inhibition. After verifying the efficiency of knockdown in neurons (Extended Data Fig. 7b) and the absence of adverse effects on overall neuronal viability, individual shRNAs were cloned into a Flp-OFF AAV, allowing for Flp-dependent shRNA inactivation in GABAergic INs when using Dlx5/6Flp mice (Fig. 4a,b, Extended Data Fig. 7c, Supplementary Fig. 2a).
Figure 4. Fos-dependent effector of inhibition.
a,b. Schematic of FlpOFF u6-shRNA AAV construct used for recordings as depicted in b.
c. Normalized differences in PV-IPSC amplitudes between pairs of shRNA— or shRNA+ PCs post-24h KA treatment. Control,n=17/9; Inhba,n=15/4; Rgs2,n=20/3; Bdnf,n=26/10; Nptx2,n=16/3; Pcsk1,n=17/6; Scg2#1,n=17/7 (**p=0.002); shScg2#2,n=17/6 (*p=0.016). Ordinary one-way ANOVA, multiple comparisons corrected.
d. Scatter plot of recorded PV-IPSC amplitudes for Scg2#1 shRNA shown in c. Representative traces from a pair of neurons shown; blue marks depict light onset. Scale:100 pA;40 ms.
e. As in c for Scg2#1 shRNA in Strd (n=14/5) or 7–10d NE (n=16/4). *p=0.048.
f. Schematic of Scg2 protein depicting the four Scg2-derived neuropeptides and nine dibasic (KR or RR) cleavage residues.
g. Scg2 expression from CaMK2a-Ribotag in Fig. 3b showing significant induction and enrichment (relative to input) after 6h KA.
h. Violin plots depicting Scg2 expression in CA1 PCs in Cre or ΔCre compared to respective controls from FFJ snRNA-seq in Fig. 3e. TPT: tags per ten thousand. **** represents p=9.4×10−302 and >20% decrease. Mean ± 2 SD shown.
i. Tracks displaying Fos-binding sites surrounding the Scg2 locus from CUT&RUN in Fig. 3g. Y-axis shows spike-in normalized coverage scaled to maximum value (in brackets) observed at displayed locus.
j. Representative smRNA-FISH images of CA1 in Strd and 6h NE mice, probing for Fos (magenta), mature Scg2 (red), and intron-targeting Scg2 (green) transcripts (lower magnification shown in Extended Data Fig. 7h). Strd,N=4; NE,N=6 mice. Scale:20μm.
k. Violin plots of number of puncta per cell for smRNA-FISH in j. Dashed lines: medians and quartiles. Each point represents a cell. Strd,n=909; NE,n=1,548 cells. ****p=1×10−15.
l. (Top) Workflow of NE snRNA-seq. Mice were exposed to NE briefly (5 min), returned to Strd for 1h or 6h prior to CA1 dissection. (Bottom) Violin plots of normalized gene expression in CA1 PCs (n=1,659 cells after downsampling). Strd, N=2 mice; NE(1h, 6h), N=4 mice each. Fos (****p=4.2×10−9; *p=0.025), Scg2 (****p=2.2×10−16; *p=0.032), Actb (*p=0.014).
(c-e) Each open circle represents a pair of simultaneously recorded neurons, n=number of pairs/mice.
(c-e,g) Mean±SEM.
(e,k) Two-sided t-test. (h,l) Wilcoxon rank-sum (two-sided).
Upon sparse transduction of neurons, we simultaneously measured PV-IPSCs in neighboring pairs of shRNA-positive (mCherry+) and -negative (mCherry—) PCs by photostimulating PV-specific ChR2-expressing boutons in Dlx5/6Flp;PVCre mice that had been exposed to 24h KA (Fig. 4b). We observed no effects on PV-IPSC amplitudes upon expression of a control scrambled shRNA or shRNAs against Inhba, Rgs2, Nptx2, or Pcsk1, and only a slight decrease with knockdown of Bdnf (Fig. 4c, Extended Data Fig. 7d)25. By contrast, PV-mediated inhibition was significantly decreased by either of two independent shRNAs against Scg2 (Fig. 4c,d, Extended Data Fig. 7e). Similar results were also observed following more physiological conditions of NE exposure (Fig. 4e, Extended Data Fig. 7f), suggesting a prominent role for CA1 PC-derived Scg2 in the long-term regulation of PV-mediated inhibition.
Scg2 has previously been shown to be activity-regulated26 and to encode a neuropeptide precursor that undergoes endoproteolytic processing by Pcsk1/2 proteases27 to produce four distinct, non-overlapping neuropeptides: Secretoneurin, EM66, Manserin, and SgII (Fig. 4f); however, the functions of these peptides in the brain are largely unknown. We found that Scg2 is highly enriched in CA1 PCs (Fig. 4g), significantly downregulated upon AP-1 loss (Fig. 4h), and associated with several Fos-bound regulatory elements (Fig. 4i).
To confirm that Scg2 is expressed in the CA1 in an experience-dependent manner, we performed smRNA-FISH using mice exposed to 6h NE compared to Strd, probing for mature Fos and Scg2 RNA, as well as nascent intron-containing Scg2 transcripts (Fig. 4j). Fos and Scg2 showed correlated expression (Extended Data Fig. 7g,h), with both significantly induced in NE (Fig. 4k). We also found that a brief (5-min) NE exposure was sufficient to induce Fos and Scg2 in CA1 PCs when assessed by snRNA-seq 1- or 6-h after the exposure (Fig. 4l).
Scg2 mediates PV- and CCK-IN plasticity
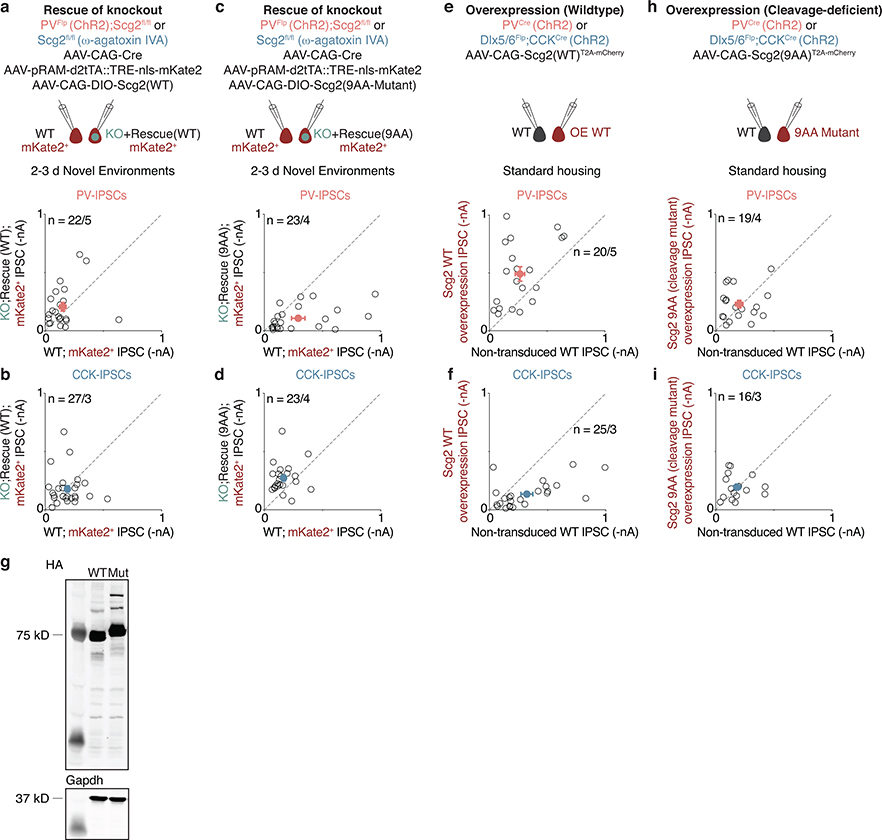
To investigate further the requirement of Scg2 for bidirectional perisomatic inhibitory plasticity, we generated and verified an Scg2 conditional knockout mouse line (Scg2fl/fl; Fig. 5a,b, Extended Data Fig. 8a). After crossing with PVFlp mice, the resulting PVFlp/Flp;Scg2fl/fl animals were sparsely transduced with AAV expressing Cre and co-injected with the AAV RAM-mKate2 reporter and a separate Flp-dependent AAV to localize ChR2 expression to PV-INs (Fig. 5c). We then exposed these mice to 2–3d NE and subsequently recorded light-evoked PV-IPSC amplitudes simultaneously in neighboring Fos-activated neurons that were Cre-positive (Scg2-KO Cre+/Fos+) or Cre-negative (Scg2-WT Cre—/Fos+) (Fig. 5c). Consistent with the data obtained by shRNA-mediated knockdown of Scg2, we found that PV-IPSC amplitudes onto activated Scg2-KO neurons are on average significantly smaller by 3-fold compared to Scg2-WT (Fig. 5d,e). This effect was not observed in non-activated (mKate2—) neurons in either Strd or NE (Fig. 5d,e). Thus, Fos-activated CA1 PCs require Scg2 to induce plasticity of PV-IN synapses.
Figure 5. Scg2 mediates PV- and CCK-IN plasticity.
a. Schematic depicting strategy for generation of Scg2fl/fl line using CRISPR/Cas9.
b. smRNA-FISH validation of Scg2fl/fl crossed to Emx1Cre to excise Scg2 in all excitatory cells. N=2 mice/line. Scale:20 μm.
c. Schematic of strategy to introduce ChR2 into PV-INs in PVFlp;Scg2fl/fl mice, mark recently active cells with RAM-mKate2, and sparsely transduce Cre into CA1 PCs.
d. Scatter plots of recorded (Left) mKate2— (n=21/4) or (Right) mKate2+ (n=22/9) pairs of Scg2-WT and -KO neurons after 2–3d NE. Representative traces from pairs of neurons shown; blue marks depict light onset. Scale:50 pA;40 ms.
e. Normalized differences in PV-IPSC amplitudes between pairs of neurons in d and mKate2— pairs from Strd (n=22/5). **p=0.004, ***p=1.4×10−4. Ordinary one-way ANOVA, multiple comparisons corrected.
f. Schematic of pharmacological strategy used to isolate CCK-INs in Scg2fl/fl mice. NBQX, (R)-CPP, and ω-agatoxin IVA (to block PV-IPSCs) used.
g,h. As in d,e for CCK-IPSCs. mKate2—,n=22/6; mKate2+,n=26/6. Scale:100 pA;40 ms. **p=0.001.
i-k. As in e,h for pairs of neurons depicted in i,j, where (i) PV (WT,n=22/5; 9AA,n=23/4; ****p=1.2×10−5); CCK (WT,n=27/3; 9AA,n=23/4, **p=0.005), (j) PV (n=20/5; **p=0.001); CCK (n=25/3; **p=0.004), (k) PV (n=19/4); CCK (n=16/3).
(d,e,g-k) Each open circle represents a pair of simultaneously recorded neurons; mean±SEM shown; n=number of pairs/mice.
(h,i) Two-sided t-test, (j,k) One-sample t-test (two-sided) with hypothetical mean of 0.
We next asked if Scg2 also regulates CCK-mediated inhibition. Given that a CCK-IN-only Flp-driver line is not available, we used two orthogonal approaches to measure CCK-IPSCs. First, we employed a pharmacological strategy in which CCK-IPSCs were specifically measured by blocking PV-IPSCs using ω-agatoxin IVA4,10. Simultaneously recording from pairs of Scg2-WT (Cre—/Fos+) and KO (Cre+/Fos+) neurons after 2–3d NE, we found that the mean CCK-IPSC amplitude onto Scg2-KO neurons is significantly larger by 2-fold relative to WT specifically in Fos-activated neurons (Fig. 5f–h). Similar results were also obtained using an independent approach involving our intersectional genetic strategy using Dlx5/6Flp; CCKCre mice in conjunction with Scg2 shRNA-mediated knockdown (Extended Data Fig. 8b–f). Thus, a single experience-regulated AP-1 target, Scg2, couples the bidirectional regulation of PV- and CCK-mediated inhibition onto Fos-activated neurons.
These findings were further corroborated through a series of rescue and overexpression experiments. Notably, we found that the defects in both PV- and CCK-mediated inhibition are restored to control levels when Scg2 is re-expressed under either shRNA or Scg2fl/fl conditions (Fig. 5i, Extended Data Figs. 8g, 9a–d). In addition, we compared light-evoked PV- or CCK-IPSC amplitudes onto Scg2-overexpressing (Scg2-OE) and neighboring control (Scg2-WT) neurons, and found that gain-of-function of Scg2 is sufficient to strengthen PV- and weaken CCK-mediated inhibition, respectively, in the absence of neural activity (Fig. 5j, Extended Data Fig. 9e,f).
Cleavage of the Scg2 precursor is known to give rise to multiple neuropeptides with potentially distinct functions (Fig. 4f). Given that Scg2 cleavage is directed by a series of internal dibasic residues, we generated a cleavage-resistant form of Scg2 in which the nine dibasic sequences were mutated to alanine (9AA-Mut). Having first verified that these sequence changes do not affect Scg2 expression levels (Extended Data Fig. 9g, Supplementary Fig. 2b), we found that expression of this cleavage-deficient Scg2 fails to recapitulate the effects of wildtype Scg2-OE (Fig. 5k, Extended Data Fig. 9h,i) or rescue the effects of loss of Scg2 (Fig. 5i, Extended Data Fig. 9c,d). Thus, while we cannot formally exclude other modes of Scg2 action, such as the contribution to dense core vesicle biogenesis or the packaging of neuropeptides into dense core vesicles27, our results suggest that the processing of Scg2 precursor protein to mature peptides may be required for experience-dependent bidirectional perisomatic inhibitory plasticity, raising the intriguing possibility that distinct Scg2-derived peptides might coordinate aspects of bidirectional plasticity.
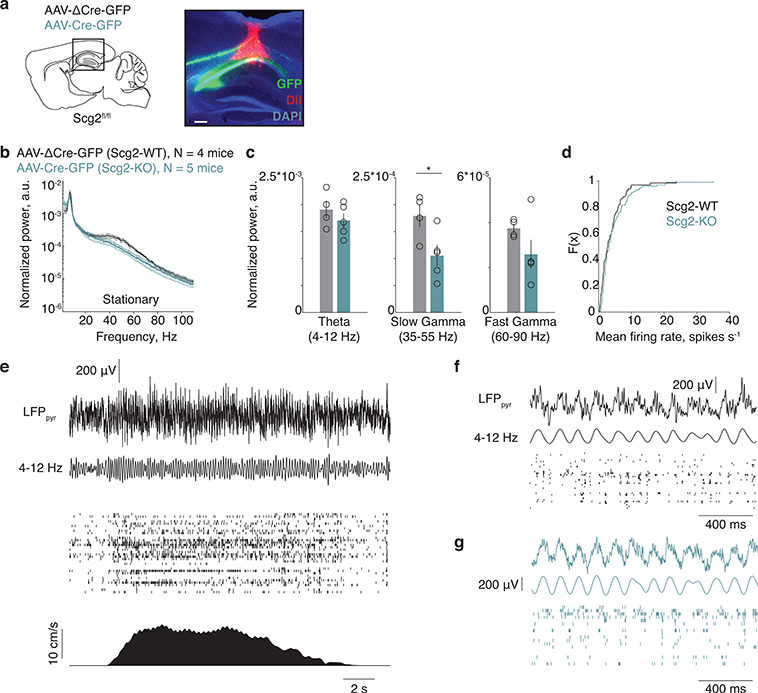
Scg2 crucial for network rhythms in vivo
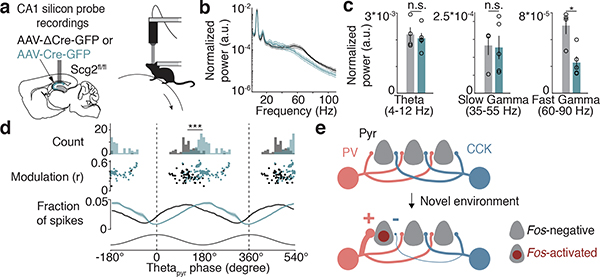
To determine whether the Fos-Scg2 pathway alters the function of hippocampal networks in vivo, we assessed the effects of disrupting Scg2 function on hippocampal network oscillations. We performed silicon probe recordings in awake head-fixed mice running on an air-supported ball (Fig. 6a). Scg2fl/fl mice were injected with AAV expressing ΔCre- (Scg2-WT) or Cre-GFP (Scg2-KO) bilaterally into the CA1 (Extended Data Fig. 10a). We found that the frequency spectra in the gamma range are altered, with Scg2-KOs displaying significantly lower fast gamma (60–90 Hz) power compared to Scg2-WTs when running (Fig. 6b,c, Extended Data Fig. 10b,c). By contrast, the power of theta rhythms (4–12 Hz) recorded in CA1 stratum pyramidale (thetapyr) and mean spike rates are not significantly different between Scg2-WTs and Scg2-KOs (Fig. 6b,c, Extended Data Fig. 10b–g).
Figure 6. Scg2 crucial for network rhythms in vivo.
a. (Left) Schematic of silicon probe placement in CA1 pyramidal layer and (Right) head-fixed awake-behaving setup. After AAV injections, mice were exposed to NE daily for 1–2 weeks prior to recordings.
b. Normalized power spectrum of network oscillations in running Scg2-WT (N=4 mice) or Scg2-KO (N=5 mice); one session per mouse.
c. Mean of the normalized power spectra within theta, slow gamma, and fast gamma bands during running as in b. *p=0.009 (Two-sided t-test).
d. Theta phase modulation of putative CA1 PCs. Two cycles of theta shown. (Top) Mean spike-triggered theta phase distributions for Scg2-WT (grey, n=67 units) and KO (green, n=103 units) units. ***p<0.001 bootstrap significance test of difference between circular means of the two distributions; 1000 shuffles. (Middle) Mean theta phase and mean resultant length for each unit. (Bottom) Fraction of spikes in each theta phase bin (10° bins).
e. Model depicting experience-dependent reorganization of perisomatic IN networks upon activation of Fos in CA1 PCs (Pyr), where weights of PV and CCK-IN synaptic inputs are bidirectionally modulated.
(b-d) Mean±SEM.
(a) Schematic image (left) adapted with permission from Paxinos & Franklin (Elsevier),
Additionally, we found that PCs in Scg2-KOs fired at a significantly different preferred thetapyr phase compared to Scg2-WT cells (Fig. 6d). Scg2-KO cells tend to fire later in the thetapyr cycle, corresponding to the ascending phase of thetapyr, whereas Scg2-WT cells on average fire at the descending phase of thetapyr (WT: 120.6° and KO: 187.3° relative to peak of thetapyr defined at 0°). These results are consistent with the observed change in the balance between PV- and CCK-IN inputs upon loss of Scg2, as PV- and CCK-INs themselves have been observed to fire during the descending and ascending phases of theta oscillations, respectively5,6.
Discussion
Despite the prevalence of Fos-activated neuronal networks across many regions of the brain, there is limited understanding of the circuit and molecular mechanisms by which these networks become persistently modified to support the consolidation of experiences over time. Moreover, whether Fos has a causal role in orchestrating circuit modifications, and which of its many targets underlie these processes, are not known. Here we discover a bidirectional perisomatic inhibitory plasticity mechanism by which Fos-activated circuits are selectively reorganized in response to experience (Fig. 6e). We show that a Fos-to-Scg2 pathway is critical for this reorganization, and further define a role for Scg2 neuropeptidergic modulation in the entrainment of PC activity relative to theta phase and the regulation of gamma rhythms. These results, together with our finding that Fos is necessary for spatial learning, raise the possibility that Fos-dependent circuit reorganization is required to establish a network of cells for encoding and recalling memories.
Despite the broad axonal arborizations of PV- and CCK-INs within the CA1 pyramidal layer, distinct mechanisms appear to specifically reorganize and establish Fos-activated microcircuits compared to non-Fos-activated networks. That PV- and CCK-IN synaptic strengths are oppositely regulated by novel experience suggests functional consequences for this reorganization beyond a strictly homeostatic role in which increased PC activity is balanced by increased perisomatic inhibition within the network. Future work will be needed to understand how this experience-dependent shift in inhibitory control alters the temporal dynamics of network function in behaviorally adaptive ways.
For example, the peak and trough phases of theta rhythm measured in the CA1 pyramidal layer have been associated with memory encoding and recall, respectively, as the dominant source of inputs to CA1 cycles between entorhinal cortex and CA311,13. Fos-mediated reorganization of inputs from PV- and CCK-INs, which themselves fire at different theta phases, could provide a potential mechanism for altering a cell’s eligibility to take part in these processes. We also find that Scg2-expressing PCs preferentially fire at the descending phase of theta, where PV-INs also tend to fire5,6, raising the possibility that the Fos-dependent recruitment of PV-mediated inhibition is critical for the formation of functional PV-pyramidal cell ensembles to support the consolidation of memories. In addition, Scg2-dependent regulation of gamma rhythms may be critical for transiently synchronizing the activity of populations of neurons within and across brain regions to facilitate information processing12.
Additional distinctions in the molecular and physiological properties of PV- and CCK-INs may also contribute to the functional consequences of this shift. For example, experience-dependent strengthening of PV-mediated inhibition onto PCs may increase their spike threshold and impose narrower time windows for synaptic integration, which may allow them to better synchronize their firing6,9. It will also be interesting to investigate whether Fos or Scg2 contributes to endocannabinoid signaling involving presynaptic CCK-INs28,29.
While the specific in vivo cellular and learning-related activity features that lead to the induction of Fos during natural behaviors is a subject of active investigation1,2, our findings indicate that Fos expression plays an instructive role in orchestrating persistent circuit modifications, beyond serving as a marker of recent neural activity. In particular, Fos coordinates neuropeptidergic networks to modulate connectivity through its regulation of Scg2. In the brain, Scg2 has been reported to be mostly processed into its distinct neuropeptides27,30, suggesting that individual Scg2-derived peptides likely mediate bidirectional perisomatic inhibitory plasticity. Although alternative mechanisms of Scg2-mediated inhibitory plasticity should also be examined27, future work characterizing the specific Scg2-derived peptides that are involved, their pre- or post-synaptic sites of action, and the identity of their cognate G-protein coupled receptors will be critical for further assessing the physiological functions of Fos-Scg2 signaling and pathological consequences when this pathway is disrupted.
Methods
Animals.
Animals were handled according to protocols approved by the Harvard University Standing Committee on Animal Care and were in accordance with federal guidelines. The following mouse lines were used: PV-Cre (JAX 017320), CCK-Cre (JAX 012706), PV-Flpo (JAX 022730), C57BL/6J (JAX 000664), Ai14 (JAX 007914), Ai65 (JAX 021875), CaMK2a-Cre (JAX 005359), Rpl22/Ribotag (JAX 029977), LSL-Sun1-sfGFP-Myc (JAX 021039), Emx1-Cre (JAX 005628), Dlx5/6-Flpe31, Fosfl/fl;Fosbfl/fl;Junbfl/fl19, Fos-FlagHA19, Npas4-FlagHA32, C57BL/6N (Charles River Laboratories; for embryonic cultured neurons), and Scg2fl/fl (this paper).
The conditional Scg2fl/fl knockout mouse was generated with the help of the Harvard Genome Modification Facility (Lin Wu). Briefly, LoxP sites were introduced flanking the entire coding exon of Scg2. Cas9 mRNA, two sgRNAs each targeting a site for LoxP insertion, and two 150–200 bp single-stranded oligonucleotides for repair were injected into C57BL/6J mouse zygotes. Correct cis insertion of both LoxP sites were verified by standard PCR and Sanger sequencing. A founder male was bred to C57BL/6J mice for at least three generations before experimental use.
Mice were housed in ventilated micro-isolator cages in a temperature- and humidity-controlled environment under a standard 12 h light/dark cycle, with food and water provided ad libitum. Both male and female littermate mice were used in similar proportions and divided between control and experimental groups for all experiments conducted. For in vivo silicon probe recordings and Morris water maze experiments, only male littermate mice, housed in a reverse 12 h light/dark cycle, were used.
Novel environment paradigm.
Animals at weaning age and above (>P21) were placed in a large opaque cage (0.66 m x 0.46 m x 0.38 m) in a group with other mice, equipped with an assortment of enrichment including a running wheel, mazes, tunnels, ladders, huts, swings, and different kinds of animal bedding. Rodent pellets were hidden in mazes to encourage spatial exploration. Mice were placed in a specific environment for 12–24 h. The environments were subsequently significantly changed daily to provide novel multisensory experiences and the transcriptional activation of a larger proportion of neurons.
Intraperitoneal (i.p.) injections.
For experiments in which seizures were induced, kainic acid (5–10 mg/kg for electrophysiology or 15–20 mg/kg for genomic or histological analyses) (Sigma Aldrich, K0250) reconstituted in 0.001 N NaOH in PBS was injected (i.p.). We used 1–1.5 h or 2–3 h KA as the timepoint for capturing the peak of immediate early gene (e.g., Fos) RNA or protein induction, respectively. We used 4 h KA as the timepoint for capturing the peak of nascent RNA induction for late-response genes, as nascent RNA molecules are first present in the nuclei (FFJ snRNA-seq). Subsequently, for ribosome-associated mature RNA from late-response genes, we used a 6 h KA timepoint as we reasoned that more mature RNA tends to associate with ribosomes at this later timepoint (Ribotag). For electrophysiology, mice were sacrificed 24 h after KA injection to allow sufficient time for the expression and action of activity-dependent genes but far in advance of any measurable seizure-related cellular toxicity (see Extended Data Fig. 2m–q).
For chemogenetic activation experiments, clozapine N-oxide (CNO; Sigma C0832) reconstituted in 0.4% DMSO in PBS was injected (i.p.) at 5 mg/kg in mice 24 h prior to electrophysiology.
Stereotaxic surgery.
For acute hippocampal slice recordings, mice aged P13–15 of equal proportion male and female were anesthetized by isoflurane inhalation (2% induction, 1% maintenance) and positioned within a stereotaxic frame (Kopf Model 963). Animal temperature was maintained at 37°C by a heat pad. All surgeries were performed according to protocols approved by the Harvard University Standing Committee on Animal Care and were in accordance with federal guidelines. Fur around the scalp area was removed using a shaver and sterilized with three alternating washes with betadine and 70% ethanol. A burr hole was drilled through the skull above the CA1 region of hippocampus (medial/lateral, ML: ± 2.9 mm; anterior/posterior, AP: −2.4 mm; dorsal/ventral, DV: −2.8 mm) to allow for specific targeting of this region with a glass pipette pulled to a tip diameter of roughly 50 μm. AAV virus (1000 nL) was injected at 150 nL/min and the pipette was left in place for 5 min upon completion of viral infusion to allow for viral spreading. All animals were given postoperative analgesic (flunixin, 2.5 mg/kg) as well as additional injections at 12 h-intervals for the 72 h following surgery.
Viral vectors and titers.
All AAVs used were prepared in the Boston Children’s Hospital Viral Core and were of serotype AAV2/1. For sparse transductions, viruses were injected at 1E+8 genome copies per hippocampal hemisphere. For dense transductions, viruses were injected at 2E+9 genome copies per hippocampal hemisphere. The viral vectors and original titers are as follows: pAAV-EF1a-DIO-hChR2(H134R)-EYFP (Addgene 20298, 1.75E+13 gc/mL), pAAV-EF1a-fDIO-hChR2(H134R)-EYFP (Addgene 55639, 1.39E+13 gc/mL), pAAV-hSyn-Con/Fon-hChR2-EYFP (Addgene 55645, 2.25E+14 gc/mL), pAAV-pRAM-tTA::TRE-NLS-mKate2-WPREpA (Addgene 84474, 2.25E+13 gc/mL), pAAV-CAG-Cre-GFP (M. During, Ohio State University, 1.75E+13 gc/mL), pAAV-CAG-Cre-mCherry (this paper, 9.10E+12 gc/mL), pAAV-CAG-Cre-mTagBFP2 (this paper, 2.97E+12 gc/mL), pAAV-CAG-deltaCre-GFP (this paper, 2.79E+12 gc/mL), pAAV-FlpOFF-u6-shRNA-CAG-mCherry (this paper): Scrambled control shRNA (ACTTACGCTGAGTACTTCG) (5.08E+13 gc/mL), Inhba (CCTTCCACTCAACAGTCATT) (4.62E+13 gc/mL), Bdnf (GAATTGGCTGGCGATTCATA) (6.97E+13 gc/mL), Pcsk1 (GATAATGATCATGATCCATT) (6.02 E+12 gc/mL), Nptx2 (GAAGACATTGCCTGAGCTGT) (1.30E+12 gc/mL), Scg2#1 (GCAGACAAGCACCTTATGAA) (8.11E+11 gc/mL), Scg2#2 (CCCTTGATTCTCAGTCTATT) (2.75E+13 gc/mL), Rgs2 (GCTCCCAAAGAGATAAACAT) (6.14E+12 gc/mL), pAAV-CaMKIIa-mCherry (this paper, 3.80E+12 gc/mL), pAAV-CaMKIIa-hM3DGq-T2A-mCherry (this paper, 1.20E+12 gc/mL), pAAV-hSyn-FlpOFF-Kir2.1-T2A-mCherry (this paper, 2.26E+12 gc/mL), pAAV-hSyn-FlpOFF-Kir.2.1(Mutant)-T2A-mCherry18 (this paper, 1.28E+12 gc/mL), pAAV-u6(Frt)-shRNA#31-CAG-Scg2-rescue (shRNA-resistant)-1xHA-T2A-mCherry-Frt-SV40 (this paper, 1.88E+12 gc/mL), pAAV-CAG-DIO-Scg2(WT)-3xHA-bGH polyA (this paper, 8.22E+13 gc/mL), pAAV-CAG-DIO-Scg2(9AA Mutant)-3xHA-bGH polyA (this paper, 6.13E+13 gc/mL), pAAV-CAG-Frt-Scg2(WT)-1xHA-T2A-mCherry-Frt-bGH polyA (this paper, 1.08E+13 gc/mL), and pAAV-CAG-Scg2(9AA Mutant)-1xHA-T2A-mCherry-Frt-bGH polyA (this paper, 3.71E+12 gc/mL).
For lentiviral production of shRNAs, lentiviral backbone pSicoR (Addgene 11579) was used for cloning all shRNAs. A total of 10 mg of lentiviral plasmid was transfected into 293T cells in a 10-cm dish along with third generation packaging vectors pMD2.G (Addgene 12259), pRSV-rev (Addgene 12253) and pMDLg/pRRE (Addgene 12251). At 12–16 h following transfection, 293T cells were switched to Neurobasal media containing B27 supplement, and supernatant containing virus was collected at 36 h post-transfection and spun down to remove cellular debris at 1,000xg for 5 min.
Acute slice preparation.
Transverse hippocampal slices were prepared from mice aged P23-P32. Animals were anaesthetized with ketamine/xylazine and transcardially perfused with ice-cold choline-based artificial cerebrospinal fluid (choline-ACSF) equilibrated with 95% O2/5% CO2 consisting of (in mM): 110 choline chloride, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 7 MgCl2, 25 glucose, 0.5 CaCl2, 11.6 sodium L-ascorbate, and 3.1 sodium pyruvate. Cerebral hemispheres were quickly removed and placed into ice-cold choline-ACSF. Tissue was rapidly blocked and transferred to a vibratome (Leica VT1000). Dorsal hippocampal slices of 300 μm thickness were collected in a holding chamber containing ACSF consisting of (in mM): 127 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 2.5 KCl, 1 MgCl2, 10 glucose, and 2 CaCl2. For all solutions, pH was set to 7.2 and osmolarity to 300 mOsm. Slices were incubated at 32°C for 20 min and maintained at room temperature (RT, 22°C) for 30 min before recordings began. All recordings were performed at RT within 4–5 h of slice preparation. AAV transduction was assessed by epifluorescence. For experiments where sparse transduction of CA1 was intended, slices with 10–30% of CA1 neurons infected were used, and slices showing > 30% of CA1 neurons infected were discarded from further analysis. For optogenetic stimulation experiments, slices showing channelrhodopsin-2 (ChR2) spread across the entire CA1 were used, and slices showing partial expression of ChR2 across CA1 were discarded from further analysis. For all experiments, slices were discarded if AAV transduction spread to CA3 and/or dentate gyrus regions.
Ex vivo electrophysiology.
For whole-cell voltage-clamp recordings, a CsCl-based internal solution consisting of (in mM): 135 CsCl, 3.3 QX314-Cl, 10 HEPES, 4 MgATP, 0.5 NaGTP, 8 Na2-phosphocreatinine, 1.1 EGTA, and 0.1 CaCl2 (pH 7.2, 290 mOsm) was used for all IPSC measurements. A Cs+-methanesulfonate internal solution consisting of (in mM): 127 CsMeSO3, 10 CsCl, 10 HEPES, 0.5 EGTA, 2 MgCl2, 0.16 CaCl2, 2 MgATP, 0.4 NaGTP, 14 Na2-phosphocreatinine, and 2 QX314-Cl (pH 7.2, 295 mOsm) was used for all EPSC measurements. A K+-based internal solution consisting of (in mM): 142 K+-gluconate, 4 KCl, 10 HEPES, 4 MgATP, 0.3 NaGTP, 10 Na2-phosphocreatinine, and 1.1 EGTA (pH 7.2, 280 mOsm) was used for all current-clamp recordings. Membrane potentials were not corrected for liquid junction potential (which were experimentally measured as −5 mV for CsCl-based internal solution, and 60 mV for K-gluconate-based internal solution). In all recordings, neurons were held at −70 mV with patch pipettes made with borosilicate glass with filament (Sutter BF150–86-7.5) with 2–4 MΩ open pipette resistance. For all dual whole-cell recordings of pairs of CA1 pyramidal neurons, recording from neighboring neurons increased the probability that both neurons received synaptic inputs from the same population of inhibitory axons, and ensured that both neurons were exposed to an identical stimulus magnitude and intensity.
Recordings were made on an upright Olympus BX51 WI microscope with an infrared CCD camera (Dage-MTI IR-1000) and 60X water immersion objective (Olympus Lumplan Fl/IR 60X/0.90 numerical aperture). Neurons were visualized using video-assisted infrared differential interference contrast, and fluorescence was identified by epifluorescence driven by a light-emitting diode (Excelitas XCite LED120). For photostimulation of ChR2-expressing boutons, 470 nm blue light was delivered from the LED through the reflected light fluorescence illumination port and the 60X objective. Pulses were delivered at 0.4 Hz. Pulse duration (0.1–0.2 ms) and intensity (1.3–5.9 mW/mm2) were adjusted for each recording to evoke small but reliable monosynaptic IPSCs. No pharmacology was used for optogenetic stimulation experiments.
For electrical stimulation experiments, electrical current was delivered via theta glass stimulation electrode placed in the center of stratum pyramidale or stratum radiatum within 150–200 μm of the recorded neuron pair. The stimulus strength was the minimum required to generate small but reliable currents in both neurons. IPSCs were pharmacologically isolated via the addition of 10 μM NBQX (Tocris 1044) and 10 μM (R)-CPP (Tocris 0247) to the ACSF perfusion. For pharmacological isolation of CCK-IPSCs specifically, in addition to blocking excitatory currents, PV-IPSCs were blocked using 0.4 μM of ω-agatoxin IVA, a selective antagonist for P/Q-type calcium channels (Peptides International, PAG-4256-s). EPSCs were pharmacologically isolated by adding 10 μM gabazine (Tocris 1262).
For simultaneous dual whole-cell recordings, we determined that the IPSCs measured were monosynaptic as the addition of NBQX and (R)-CPP in the bath did not alter the onset latency of the IPSCs. For the paired interneuron-to-CA1 pyramidal neuron recordings, the monosynaptic nature of the IPSCs was confirmed based on the expected onset latency of 1–3 ms in slice.
Data acquisition and analysis.
Data were low-pass filtered at 4 kHz and sampled at 10 kHz with an Axon Multiclamp 700B amplifier, and digitized with an Axon Digidata 1440A data acquisition system controlled using Clampex 10.6 (Molecular Devices). Experiments were discarded if holding current exceeded −500 pA, or if series resistance was greater than 30 MΩ. For the dual whole-cell recordings of CA1 pyramidal neurons, recordings were discarded if series resistance differed by more than 30% between the two neurons. The recorded traces were analyzed using Clampfit 10.6 software (Molecular Devices) or Axograph (1.7.6). All current amplitude measurements are expressed as mean ± SEM, or as differences in amplitudes between a pair of neurons normalized to the total amplitudes of both neurons (Δ IPSC / Σ IPSC). The differences (Δ IPSC) are calculated between a fluorescently labeled (i.e., manipulated) cell minus a control (i.e., non-manipulated) cell, such that a positive number indicates a larger IPSC amplitude in the manipulated cell compared to the control cell, and vice versa.
Sample sizes were not predetermined and are similar to those reported in the literature18,24,32. Previous work suggests that in general, approximately 15–20 pairs of neurons (n) collected from 3–5 animals (N) are sufficient for each experiment. Most data, except where specified (Fig. 4c–e), was not collected blind to genotype or conditions, but all offline analyses were conducted blind. All statistical analyses were performed using Prism 8 (Graphpad). Data were tested for normality using the D’Agostino-Pearson, Shapiro-Wilk, and Kolmogorov-Smirnov normality tests. For simultaneous dual whole-cell recordings of pyramidal neurons, parametric t-tests or non-parametric Wilcoxon rank-sum tests (two-sided) were used. For recordings of unitary connections, non-parametric Mann-Whitney tests (two-sided) were used. A mixed model was used to confirm that findings were not driven by a single mouse. The numbers of cells recorded per animal were capped to ensure even sampling across mice comprising a dataset (e.g., if n = 20 pairs were obtained using N = 4 mice, 4–6 pairs were used per animal).
Histology.
Mice were anaesthetized with 10 mg/mL ketamine and 1 mg/mL xylazine in PBS via i.p. injection. When fully anaesthetized, the animals were transcardially perfused with 5 mL ice-cold PBS followed by 20 mL of cold 4% PFA in PBS. Brains were dissected and post-fixed for 1 h at 4°C in 4% PFA, followed by three washes (each for 30 min) in cold PBS. Coronal sections (40 μm thick) were subsequently cut using a Leica VT1000 vibratome and stored in PBS in 4°C. For immunostaining, slices were permeabilized for 30 min at RT in PBS containing 0.3% Triton X-100. Slices were blocked for 1 h at RT with PBS containing 0.3% Triton X-100, 2% normal donkey serum and 0.1% fish gelatin. Slices were incubated in primary antibodies diluted in blocking solution at 4°C for 48 h: rabbit anti-Fos antibody (Synaptic Systems 226003, 1:3000), mouse anti-Fos (Abcam ab208942, 1:1000), rabbit anti-Npas433 (1:1000), rabbit anti-Fosb (Cell Signaling Technology 2251S, 1:1000), rabbit anti-Junb (Cell Signaling Technology 3753S, 1:1000), rat anti-HA (Sigma ROAHAHA, 1:500), rabbit anti-parvalbumin (Swant PV27, 1:10,000), rabbit anti-cleaved Caspase-3 (Cell Signaling Technology 9661S, 1:1000), and mouse monoclonal anti-NeuN (Millipore Sigma, MAB377, 1:1000). Slices were then washed three times with PBS each for 10 min at RT, incubated for 2 h at RT with secondary antibodies conjugated to Alexa dye (Life Technologies; rat Alexa 555 (A21434), rabbit Alexa 488 (A21206), rabbit Alexa 555 (A31572), rabbit Alexa 647 (A31573), mouse Alexa 555 (A31570), mouse Alexa 647 (A31571), 1:250), and washed three times with PBS. Slices were then mounted in DAPI Fluoromount-G (Southern Biotech) and imaged on a virtual slide microscope (Olympus VS120).
Single-molecule RNA fluorescence in situ hybridization (smRNA-FISH).
For sample preparation, hippocampal hemispheres from mice were fresh- or fixed-frozen in Tissue-Tek Cryo-OCT compound (Fisher Scientific) on dry ice and stored in −80°C until further use. Hippocampi were sectioned at a thickness of 15–20 μm and RNAs were detected by RNAscope (Advanced Cell Diagnostics) using the manufacturer’s protocol. Probes for Fos, Fosb, and Junb were custom designed with Advanced Cell Diagnostics specifically to detect exons excised upon Cre recombinase expression. The following probes were used: Mm-Cre (Cat. #546951), Mm-Fos (Cat. #584741), Mm-Fosb (Cat. #584751), Mm-Junb (Cat. #584761), Mm-Scg2 (Cat. #477691), and Mm-Scg2 intron (Cat. #859141). All in situ hybridizations were imaged using a confocal microscope (Zeiss Imager Z2) and analyzed in ImageJ (Fiji v1.0).
Validation of loss of Fos, Fosb, and Junb in the Fosfl/fl;Fosbfl/fl;Junbfl/fl (FFJ) conditional knockout mouse line.
Efficient excision of Fos, Fosb, and Junb upon Cre expression was confirmed at the RNA level using smRNA-FISH and at the protein level using immunostaining for each of the three genes. The Fos conditional knockout allele allows for deletion of three exons, including the last exon encoding the 3’ UTR, upon Cre expression, whereas the Fosb and Junb conditional knockout alleles are single-exon deletions (Exon 2 of 4 for Fosb; coding region only for Junb). As such, for smRNA-FISH, the probes were custom designed to specifically target the excised exons. Note that snRNA-seq detects the 3’ ends of transcripts, resulting in comparatively sparse coverage of full transcripts particularly at the 5’ end of genes. This approach can therefore be used to confirm the deletion of Fos but not Fosb and Junb due to the design of the conditional knockout alleles, which leaves intact the 3’ transcripts of Fosb and Junb upon Cre excision, resulting in non-trivial tags during library preparation.
In vitro hippocampal neuronal culture and RNA isolation for qRT–PCR or bulk RNA-sequencing.
Embryonic hippocampi from C57BL/6N (Charles River Laboratories) or Scg2fl/fl were dissected at age E16.5 or P0, respectively, then dissociated with papain (Sigma Aldrich 10108014001). Cultures were generated by combining multiple embryos of both males and females (mixed sex cultures). Papain digestion was terminated with the addition of ovomucoid (trypsin inhibitor; Worthington). Cells were gently triturated through a P1000 pipette and passed through a 40 μm filter. Neurons were plated onto cell culture dishes pre-coated overnight with poly-D-lysine (20 mg/mL) and laminin (4 mg/mL). Neurons were grown in Neurobasal medium (GIBCO) containing B27 supplement (2%), penicillin-streptomycin (50 U/mL penicillin and 50 U/mL streptomycin) and gluta-MAX (1 mM). Neurons were grown in incubators maintained at 37°C and a CO2 concentration of 5%. In all experiments, independent replicates were generated from dissections of mice on different days. Neurons were cultured in 6-well dishes at 1 million neurons per well. Neurons were transduced with lentiviral supernatant on days in vitro 2 (DIV2) by replacing one third of Neurobasal media with lentiviral supernatant. Fresh media was added at DIV4 (one fourth total volume). At DIV7, neurons were depolarized with 55 mM potassium chloride (KCl) for 1- or 6 h to assess immediate early or late-response activity-dependent genes, respectively, and RNA was subsequently harvested by gentle agitation with Trizol (Life Technologies 15596026) at RT for 2 min. The RNeasy Micro kit (Qiagen 74004) was used according to the manufacturer’s instructions to purify DNA-free RNA. For qRT-PCR, RNA was converted to cDNA using 200 ng of RNA with the high-capacity cDNA reverse transcription kit (Life Technologies 4374966). qRT-PCR was performed with technical triplicates and mapped back to relative RNA concentrations using a standard curve built from a serial dilution of cDNA. Data were collected using a QuantStudio 3 qPCR machine (Applied Biosystems). For bulk RNA-sequencing, 100 ng of RNA was used to generate libraries following rRNA depletion (NEBNext, E6310X) according to the manufacturer’s instructions (NEBNext, E7420). 75 bp reads were generated on the Illumina Nextseq 500 and subsequently analyzed using a standardized RNA-seq data analysis pipeline34.
Morris water maze behavioral paradigm.
8–14-week-old littermate Fosfl/fl;Fosbfl/fl;Junbfl/fl mice were injected with AAV-Cre-GFP or AAV-ΔCre-GFP bilaterally into the CA1 (stereotaxic coordinates of AP −2 mm, ML ±1.5 mm, DV −1.3 mm from bregma). Mice were given injections of dexamethasone and buprenorphine SR, and allowed to recover for 1–2 weeks before behavioral training. The maze (97 cm in diameter) was filled with RT water made opaque by the addition of tempera to a height of 40 cm. A hidden platform of 7 cm-diameter was placed 14 cm from the edge of the maze and submerged 1 cm below the water level. Distal cues were placed on all four walls of the testing room. Mice were trained in two blocks per day for four consecutive days (days 1–4). Each block consisted of four trials. In each trial, mice were placed at one of eight (randomized) start positions spaced evenly along half of the circumference of the pool opposite the half of the pool that contained the hidden platform. Mice were given 60 s to find the platform. If mice did not find the platform within this time, they were guided to the platform by the experimenter and allowed to sit for 10 s. Mice were subsequently removed from the platform and placed in a warmed cage to dry. Two 40 s probe trials were conducted one day after training (day 5) during which the platform was removed. The swim paths of the mice were recorded by a video camera suspended several feet above the center of the maze. The experimenter was blinded to the genotype of the mice. Mice that did not swim (“floaters”) were excluded from further analysis.
Analysis.
All video tracking and analysis was carried out using custom MATLAB code. Swim trajectories for each trial were tracked semi-automatically and manually corrected. For one mouse in the study, due to tracking issues the trials in the second block on the first day (trials 5–8) were excluded from the analysis – therefore for that mouse only four trials were considered in the performance metric on day 1. For analyses of swim speeds and path lengths, the mean was computed for each mouse across all trials on the first two days in order to control for similar levels of exploration.
Ribosome-associated mRNA profiling.
Hippocampal tissue was rapidly dissected from mice and subsequently used for isolation of ribosome-bound mRNAs. Immunopurification of ribosome-bound mRNAs was performed as previously described21 with 10 mM Ribonucleoside Vanadyl Complex (NEB S1402S) present in the lysis buffer and using the mouse monoclonal anti-HA antibody (Sigma HA-7, H3663, 12 μg/IP). A small fraction of lysate before the immunoprecipitation was used as input for each sample. All RNA samples (20 ng for CaMK2a; 2.5 ng for PV) with sufficient integrity as analyzed by 2100 Bioanalyzer were SPIA-amplified with the Ovation RNA-seq System V2 (NuGEN). Subsequently, SPIA-amplified cDNA (1 μg) was fragmented to a length of ~400 bp using a Covaris S2 sonicator (Acoustic Wave Instruments). Fragmented cDNA (100 ng) was used to generate Illumina-compatible sequencing libraries using the Ovation Ultralow System V2 (NuGEN). Libraries were sequenced on the Illumina NextSeq 500 (Basespace) for 75 bp single-end reads to a depth of 20–40 million reads per sample.
Analysis.
Analyses of Ribotag sequencing were performed for each sample at each stimulation time point as previously described35. Briefly, raw sequencing reads ≤ 75 bp in length were 3′-trimmed to a uniform 70 bp (ignoring the ~0.1% of reads that were shorter than this) and filtered for quality control. These were then mapped strand-nonspecifically to the mm10 genome (GRCm38) using the Burrows-Wheeler Aligner (bwa), allowing up to 2 mismatches and no gaps. In addition to the usual assembled chromosomes, alignment targets included mitochondrial DNA and a library of ~7 million short splice-junction sequences. Typically, 75–80% of reads were mappable; nonuniquely mapped reads were discarded, as were any that mapped to loci of rRNA genes (from RepeatMasker).
Genic features were based on exonic loci from the NCBI RefSeq annotation for mm10. Mean expression density across a gene’s exons was taken as a proxy for its expression level. (However, noncoding genes, some of which expressed quite highly and variably from one sample to the next, were excluded from these analyses.) The splice-junction target sequences for each gene comprised subsequences of minimal length of all possible concatenations of two or more ordered exons such that their boundaries would be crossed by 70 bp reads. This provided an exhaustive, nonredundant set of predictable exon-junction-spanning loci which typically accounted for ~ 20% of all exonic reads from mature messages.
Differential expression analyses employed edgeR in R to compare transcript levels in all biological-replicate samples at 6 h of KA stimulation to the unstimulated samples. A gene’s expression level was flagged as significantly changed if (a) the Benjamini-Hochberg-corrected p value (q value) for the change, as calculated by edgeR, was consistent with a false discovery rate (FDR) of ≤ 0.005, and (b) it passed a modest background filter (total number of reads ≥ 4 across all compared samples).
Nuclei isolation.
Hippocampal tissue from mice was rapidly dissected and dounce homogenized. Dounce homogenization was performed in Buffer HB (0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM Tricine-KOH, pH 7.8 supplemented with protease inhibitors, 1 mM DTT, 0.15 mM spermine and 0.5 mM spermidine) with a tight pestle for 20 strokes in a 1.5 mL total volume. Tissue was then supplemented with 96 uL 5% IGEPAL CA-630 and dounced an additional 5 strokes with a tight pestle. Homogenate was then filtered through a 40 μm strainer to remove large debris and collected in a 15 mL conical tube prior to the addition of 3.5 mL of Buffer HB and 5 mL of working solution (50% iodixanol with 25 mM KCl, 5 mM MgCl2, 20 mM Tricine-KOH pH 7.8 supplemented with protease inhibitors, DTT, spermine and spermidine). Homogenized tissue was underlaid with 1 mL of 30% iodixanol and 1 mL of 40% iodixanol (diluted from working solution) solutions. Samples were centrifuged at 10,000xg for 18 min. Nuclei were collected in a 70 uL or 250 uL volume at the 30/40% iodixanol interface for 10X Genomics and CUT&RUN protocols, respectively.
FFJ single-nucleus RNA-sequencing (snRNA-seq).
FFJ snRNA-seq was performed with the 10X Genomics Chromium Single Cell Kit (v3). Approximately 7,000–10,000 nuclei were added to the RT mix prior to loading on the microfluidic chip. Each snRNA-seq sample consists of pooled nuclei from 2 mice. All downstream steps for the cDNA synthesis, cDNA amplification and library preparation were performed according to the manufacturer’s instructions (10X Genomics). All samples were sequenced on Illumina NextSeq 500 (Basespace) with 58 bp (read 1), 26 bp (read 2) and 8 bp (index).
Analysis.
Initial FASTQ files were generated using the standard bcl2fastq Illumina pipeline, and gene expression tables for each barcode were generated using the CellRanger (v3.0.0) pipeline according to instructions provided by 10X Genomics. AAV transduced cells were detected by the presence of mRNA species mapping to the WPRE-bGH polyA sequence present in all AAVs used in this study. WPRE transcripts were PCR amplified using custom primers. Gene expression tables were then imported into R and analyzed using custom written functions as well as the Seurat (v3) package. Exclusion criteria: Nuclei were removed from the dataset if they contained fewer than 500 discovered genes or had greater than 5% of reads mapping to mitochondrial genes. General analysis parameters: Raw UMI counts were normalized to 104 UMIs per cell (i.e., tags per ten thousand, TPT). Nuclei from all Cre (or all ΔCre) mice were merged for the purposes of dimensionality reduction and clustering. Highly variable genes were identified using the FindVariableFeatures function (selection.method = ‘vst’, nFeatures = 2000), which identifies the 2,000 most variable genes amongst the analyzed nuclei. Principal component analysis based on the 2,000 most variable genes was performed using the RunPCA function to reduce the dimensionality of the dataset. The top 20 principal components were retained and projected into a 2-dimensional space using the uniform manifold approximation and projection (UMAP) algorithm implemented using the RunUMAP function (n.neighbors = 50, min.dist = 0.5). The following genes were used as a guide to assign cell type identities to the graph-based clusters: pan-excitatory (Slc17a7); CA1 excitatory neurons (Fibcd1, Mpped1); CA3 excitatory neurons (Spock1, Cpne4); excitatory dentate gyrus (Prox1, C1ql2); pan-inhibitory (Gad2, Slc32a1); Sst+ interneurons (Sst); Pvalb+ interneurons (Pvalb); Vip+ interneurons (Vip); Cck+ interneurons (Cck); Nos1+ interneurons (Nos1), Npy+ interneurons (Npy), Oligodendrocytes (Aspa, Opalin, Gjb1); Oligodendrocyte precursor cells (Gpr17, C1ql1); Microglia (Cx3cr1, C1qc); Endothelial cells (Ly6c1, Cldn5); Astrocytes (Cldn10, Gjb6, Gfap)20,36,37. Differential gene expression (DGE) analysis: Statistical significance of gene expression changes for all genes detected in greater than 5% of respective non-transduced control cells for Cre or ΔCre samples was calculated using the Wilcoxon rank-sum test implemented through the FindMarkers function (logfc.threshold = 0.01, pseudocount.use = 0.001). Violin plots were generated using the VlnPlot function with default parameters and heatmaps were generated using a custom written function in R. Heatmaps display the normalized gene expression values from 100 randomly selected cells from each indicated cell identity, and genes displayed are AP-1 targets showing reduced expression by at least 20% in the FFJ KO (Cre+) and whose expression is detected in at least 25% of analyzed nuclei.
CUT&RUN.
Hippocampal nuclei from CaMK2aCre/+; LSL-Sun1-sfGFP-Myc/+ mice injected with saline or 2–3 h KA were isolated as described above. Isolated nuclei were diluted two-fold with CUT&RUN wash buffer supplemented with 4 mM EDTA and stained with DRAQ5 (Abcam ab108410) at a 1:500 dilution. CaMK2a+ (GFP+) nuclei, resulting from CaMK2a-Cre-mediated expression of Sun1-sfGFP-Myc, were isolated by flow cytometry using a Sony SH800Z Cell Sorter and subsequently analyzed using FlowJo (10.6). Sorted nuclei were resuspended in 1 mL cold CUT&RUN wash buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 0.2% Tween-20, 1 mg/mL BSA, 10 mM sodium butyrate, and 0.5 mM spermidine supplemented with protease inhibitors), using 50,000 nuclei for each reaction. Nuclei were bound to magnetic Concanavalin-A (ConA) beads (Bangs Laboratories) that had been washed with CUT&RUN binding buffer (20 mM HEPES-KOH pH 7.9, 10 mM KCl, 1 mM CaCl2, 1 mM MnCl2). ConA-bead-bound nuclei were then incubated overnight in cold CUT&RUN antibody buffer (CUT&RUN wash buffer supplemented with 0.1% Triton X-100 and 2 mM EDTA) and an in-house rabbit polyclonal anti-Fos antibody (affinity eluted #1096, 1:100) or rabbit IgG antibody (Cell Signaling Technology #2729, 1:100).
After antibody incubation, ConA-bead-bound nuclei were washed with CUT&RUN antibody buffer, resuspended in CUT&RUN Triton-wash buffer (CUT&RUN wash buffer supplemented with 0.1% Triton X-100), and Protein-A-MNase was added at a final concentration of 700 ng/mL. Samples were incubated at 4°C for 1 h. The ConA-bead-bound nuclei were then washed twice with CUT&RUN Triton-wash buffer and ultimately resuspended in 100 uL of CUT&RUN Triton-wash buffer. 3 uL of 100 mM CaCl2 was added to each sample and samples were incubated on ice for 30 min. The reaction was stopped by the addition of 100 uL of 2x STOP buffer (340 mM NaCl, 20 mM EDTA, 4 mM EGTA, 0.04% Triton X-100, 20 pg/mL yeast spike-in DNA, and 0.1 μg/mL RNase A) and incubation at 37°C for 20 min. After incubation, ConA beads were captured using a magnet and supernatants containing DNA fragments released by Protein-A-MNase were collected. Supernatants were then treated with 2 uL of 10% SDS and 2 uL of 20 mg/mL Proteinase-K and incubated at 65°C with gentle shaking for 1 h. DNA was then purified using standard phenol/chloroform extraction with ethanol precipitation. DNA was resuspended in 30 uL of 0.1x TE buffer. CUT&RUN sequencing libraries were generated essentially as previously described38, with the following modifications: Adapter ligation to end-repaired, and A-tailed DNA was performed using Rapid T4 DNA ligase (Enzymatics). PCR-amplified libraries were purified from adapter dimers using a 1.1x ratio of AMPure XP beads, eluting in 20 uL of 10 mM Tris pH 8.0. All CUT&RUN libraries were sequenced on a NextSeq 500 (Basespace) using paired-end 40 bp reads.
Analysis.
After demultiplexing, sequencing reads were trimmed for quality and remaining adapter sequence using Trimmomatic v0.36 and kseq. Trimmed reads were aligned to the mm10 genome using Bowtie2 v2.2.9 with the following parameters: --local --very-sensitive-local --no-unal --dovetail --no-mixed --no-discordant --phred33 -I 10 -X 700. Trimmed reads were also aligned to the sacCer3 genome with the same parameters to recover reads corresponding to yeast spike-in DNA used in CUT&RUN. Genome-wide coverage of CUT&RUN fragments was generated using Bedtools v2.27.1 genomecov, normalizing to the number of yeast spike-in reads obtained for each sample. Normalized coverage tracks were visualized using IGV v2.4.10 and represent the average signal across all three biological replicates. CUT&RUN coverage over 100 bp bins genome-wide was determined using Deeptools v.3.0.2 multiBigwigSummary and was used to calculate Pearson correlation between pairs of replicate samples for each antibody and stimulus condition. Peaks were identified for Fos CUT&RUN using SEACR v.1.1 using the following parameters: norm, relaxed. CUT&RUN performed using rabbit IgG was used as the negative control sample for peak calling. Peaks were subsequently filtered to identify peaks found in all three biological replicates for each condition, creating a conservative set of Fos-bound sites. Peaks within 150 bp of each other were then merged using Bedtools v2.27.1 merge. Plots of spike-in normalized CUT&RUN coverage over peaks were generated by first centering peaks on the maximum of CUT&RUN signal within the peak. CUT&RUN coverage over 50 bp bins spanning 1,000 bp upstream and downstream of the peak center was calculated using Deeptools v.3.0.2 computeMatrix. Coverage in each bin was averaged across all peaks, and average per-bin coverage was plotted in R using ggplot2.
To determine distances between transcription start sites (TSS) and Fos binding sites, positions of TSSes for Refseq, CaMK2a-Ribotag activity-regulated, and CA1 excitatory neuron-specific AP-1-regulated genes were obtained from the UCSC table browser. Distances between Fos binding sites and the nearest TSS39 were calculated using Bedtools v.2.27.1 closest. Histograms of distances between Fos-bound sites and TSSes were plotted in R using ggplot2. We determined the statistical significance of the differences between the distributions of distances for Refseq, CaMK2a-Ribotag activity-regulated, and CA1 excitatory neuron-specific AP-1-regulated genes using a Wilcoxon rank-sum test in R.
To identify enriched transcription factor motifs within Fos binding sites, genomic sequences 250 bp upstream and downstream of Fos peak centers were retrieved using Bedtools v.2.27.1 getfasta and used as input to MEME-ChIP. Motifs were searched against the HOCOMOCO Mouse v11 CORE database, allowing for multiple occurrences of motifs per sequence and using default settings for all other parameters. The three motifs with the lowest E-value were reported.
Novel environment single-nucleus RNA-sequencing (NE snRNA-seq).
C57BL/6J mice were exposed to a brief 5-min novel environment stimulus and subsequently returned to their home cages for 1 h or 6 h prior to hippocampal tissue collection. Nuclei were isolated from hippocampal tissue as described above and snRNA-seq was performed using the 10X Genomics or inDrops40 platform. A total of 23,610 nuclei, with a range of 700–15,000 RNA molecule counts per cell and 200–2,500 unique genes per cell, were clustered into ~13 cell types using the UMAP algorithm. The genes Slc17a7, Fibcd1, and Pex5l were used as a guide to assign cell type identity to the dorsal CA1 excitatory neuron cluster. Raw UMI counts for each gene were normalized to total UMI counts per cell. Differential gene expression and statistical significance were measured using the Wilcoxon rank-sum test. A down-sampled total of 1,659 CA1 excitatory nuclei were used per condition.
Immunoblotting.
Whole-cell extracts from 293T cells were generated by rapid lysis of cells in boiling Laemmli SDS lysis buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue, 0.125 M Tris HCl pH 6.8). Protein extracts were resolved on 4–12% Bis-Tris gradient (Extended Data Fig. 7c) or 8% Tris-Glycine gels (Extended Data Fig. 9g) and subsequently transferred onto nitrocellulose membranes. Membranes were incubated overnight in the following primary antibodies: mouse anti-Myc (Developmental Studies Hybridoma Bank 9E10 in Extended Data Fig. 7c, 1:1000) or mouse anti-HA (Sigma HA-7, H3663 in Extended Data Fig. 9g, 1:1000) and rabbit anti-Gapdh (Sigma G9545, 1:2000). Following washes, membranes were incubated with secondary antibodies conjugated to IRDye 800CW (LI-COR; mouse (926–32210), rabbit (926–32211), 1:5000) for imaging with the LI-COR Odyssey system.
In vivo silicon probe recordings.
For all in vivo electrophysiology recordings, 8–10-week-old Scg2fl/fl mice underwent two stereotaxic surgeries. In the first surgery, AAV-Cre-GFP or AAV-ΔCre-GFP was injected into the CA1, and future silicon probe sites over CA1 (stereotaxic coordinates approximately AP −2 mm and ML ±1.8 mm from bregma) were marked on the surface of the skull. Metabond (Parkell) was used to attach a titanium headplate and cover the remaining exposed skull. Mice were given injections of dexamethasone and buprenorphine SR, and allowed to recover for 1–2 weeks, during which they were exposed to novel environments daily and habituated to head-fixation on the air-supported Styrofoam ball. On the day of recording, a second surgery (craniotomy) was performed at one of the marked locations on either the left or right hemisphere. The craniotomy was covered with Kwik-Sil (World Precision Instruments) and the mouse was allowed to recover fully from anesthesia for at least 4 h. 64-channel silicon probes (Neuronexus) were inserted into the cortex and slowly lowered ~1.25–1.5 mm below the surface of the pia to the pyramidal layer of CA1. In some cases, melted agarose (2% w/v) was applied to the headplate well to stabilize the probe. Probe advancement was stopped in the pyramidal layer of CA1, as evidenced by the presence of theta oscillations and appearance of multiple units in high density across multiple channels. All data were digitized and acquired at 20 kHz (Intan Technologies RHD2000 Recording system).
Analysis.
All data analysis41 was carried out with custom MATLAB scripts. Channels that were outside of CA1 were excluded from analysis. Spike sorting was performed using Kilosort2 (https://github.com/MouseLand/Kilosort2 )42, followed by manual inspection and curation of clustering using Phy2 (https://github.com/cortex-lab/phy )43. Only well-isolated units were chosen for further analysis. Additionally, single units had to meet the following criteria: detected on fewer than 20 channels, half-maximum spike width of less than 1 ms, at least 1,000 spikes detected in the session, and overall firing rate of > 0.01 spikes per second. Units were divided into putative excitatory and inhibitory subclasses based on the spike trough to peak duration, as described previously44, using a cutoff of 0.7 ms, below which units were labeled as inhibitory interneurons. Due to the low number of inhibitory interneurons recorded, these were excluded from analyses. For local field potential (LFP) analysis, data from each channel was filtered and downsampled to 1000 Hz. For theta phase-locking analysis, only periods during running were used in the analysis. A single channel within the stratum pyramidale was chosen as the reference. LFPs were filtered and the Hilbert transform was used to determine the phase. The preferred phase of each neuron was computed as the circular mean of the phase at each spike using the CircStats toolbox in MATLAB45. For comparison of single cell properties in the WT and KO groups, cells were pooled across mice. Power spectra were computed between 1 and 120 Hz using the multitaper method (timebandwidth=5, tapers=3) in the Chronux toolbox46. Power at frequencies between 58–62 Hz were excluded from all subsequent analyses due to 59–61 Hz notch filtering applied (2nd order butterworth filter) to remove noise. Power spectra were computed for each channel individually and averaged across channels. To compare across mice and sessions, individual session power spectra were normalized by the sum over all frequencies in the power spectra (1–120 Hz range). Fraction of spikes as a function of theta phase was computed on an individual unit basis by summing spikes in each 10° bin during running and then dividing by the sum of spikes across all bins.
Extended Data
Extended Data Figure 1. Characterization of novel environment paradigm, AAV-based activity reporter RAM-mKate2, and intersectional genetic strategy for CCK-INs.
a. (Top) Representative immunostaining images of Fos and Npas4 in hippocampus obtained from (Left) mice housed under standard (Strd) conditions or (Right) exposed to novel environment (NE) for 6 h. Scale: 400 μm. (Bottom) Higher magnification of insets. Scale: 100 μm. To immunostain for both Fos and Npas4 proteins in the same sections, mice where Fos or Npas4 has been endogenously-tagged with a Flag-HA tag (Fos-FlagHA19 and Npas4-FlagHA32) were used with a rat anti-HA antibody, while the reciprocal protein was probed with a rabbit polyclonal antibody (Methods).
b. (Left) Number of Fos+ and Npas4+ nuclei in the CA1 of Strd or 6 h NE mice. Strd, N = 6 mice; NE, N = 6 mice. Note that within CA1, significantly fewer Npas4+ cells were detected, indicating that the AAV-based reporter RAM-mKate2 mainly labels Fos-activated neurons. Two-sided t-test; ***p=1.6×10−4, *p=0.033. (Right) Quantification of number of Npas4+ cells that are also Fos+.
c. Representative images of mKate2+ neurons across different timepoints and conditions as in d. An AAV encoding GFP was used as a control for the viral injections. Scale: 100 μm.
d. Percentages of mKate2+ neurons over total number of DAPI+ cells (left y-axis) or density of mKate2+ neurons (right y-axis). The average percentages of mKate2+ neurons are 1%, 12%, 66% and 96% under Strd (N = 13 mice), 2–3 d NE (N = 10 mice, ***p=2.7×10−4), 7–10 d NE (N = 15 mice, ****p<1×10−15), and 24 h post-KA injection (N = 3 mice, ****p=7.3×10−10), respectively. Ordinary one-way ANOVA, multiple comparisons corrected. Note that data for Strd and 2–3 d NE are replotted from Fig. 1c.
e. Bar plots of additional electrophysiological parameters for mKate2— and mKate2+ neurons. n = 30 pairs/4 mice per group. Two-sided t-test; not significant (n.s.) for all parameters.
f. (Left) Schematic of intersectional strategy involving Dlx5/6Flp;CCKCre mice transduced with a dual Cre/Flp recombinase-dependent ChR2EYFP fusion protein necessary to target specifically CCK-INs. (Middle) Representative immunostaining for PV in magenta; ChR2EYFP in green. (Right) Percentage of ChR2+ cells in the CA1 field showing overlap with PV expression is low, indicating that the Dlx5/6Flp;CCKCre line is suited for genetic targeting of CCK-INs. N = 4 mice. Scale: 40 μm.
g. Representative image of CA1 region of CCKCre mice transduced with AAV encoding Cre-dependent EYFP depicting widespread EYFP expression in the CA1 and underscoring the necessity of the intersectional strategy in f for targeting CCK-INs specifically. N = 2 mice. Scale: 100 μm.
(b,d-f) Mean ± SEM.
(f,g) Schematic image (left) adapted with permission from Paxinos & Franklin (Elsevier),
Extended Data Figure 2. IN-to-CA1 PC paired recordings and cell health parameters in 24 h post-KA condition.
a,g. Schematic of genetic strategy to label PV-INs (PVCre;Ai14) or CCK-INs (Dlx5/6Flp;CCKCre;Ai65).
b,h. Representative images of tdTomato fluorescence in the CA1 field. Scale: 100 μm. N = 2 mice per line.
c,i. Quantification of the fraction of (c) PV- or (i) CCK-to-CA1 PC synaptically-connected pairs from the overall number of pairs recorded in both vehicle (Veh.) and 24 h post-KA mice. (c) Veh., n = 13/22; KA, n = 19/30; (i) Veh., n = 16/40; KA, n = 16/3, where n = connections/total pairs.
d,j. Quantification of maximum firing rate of (d) PV- or (j) CCK-INs from connected pairs. (d) Veh., n = 10/6; KA, n = 14/7; (j) Veh., n = 15/9; KA, n = 14/4, where n = cells/mice.
e,k. Quantification of spike adaptation ratio of (e) PV- or (j) CCK-INs from connected pairs as in d,j.
f,l. Quantification of paired pulse ratios (PPRs) of uIPSCs at the indicated interstimulus intervals (ISI) for (f) PV- (Veh., n = 13/6; KA, n = 19/7) or (l) CCK- (Veh., n = 16/9; KA, n = 16/4) to-CA1 PC connected pairs, where n = pairs/mice. Two-sided t-tests performed at each ISI or for all ISIs comparing Veh. and 24 h KA conditions; *p=0.039, ****p=4.4×10−5.
m,n. Representative hippocampal images from (m) Veh. and (n) 24 h post-KA conditions. Sections were immunostained for NeuN (green) and cleaved-caspase 3 (red), and counterstained with Hoechst (blue). Scale: 200 μm (left); 100 μm (right, CA1 field). N = 2 mice per condition.
o-q. Quantification of (o) Hoechst+ nuclei, (p) NeuN+ nuclei, and (q) Cleaved-caspase+ cells per 40-μm section in all layers of CA1. Results suggest that KA injection does not induce cell death within 24 h. Veh. and KA, n = 10 sections/2 mice, respectively.
(d-f,j-l,o-q) Mean ± SEM.
Extended Data Figure 3. Chemogenetic activation of CA1 PCs recapitulated bidirectional changes in perisomatic inhibition while silencing of CA1 PCs led to inverse effects.
a-d,f,g. (Top) Schematic of recording configuration. (Bottom) Scatter plots of (a,c,d,f) PV- or (b,g) CCK-IPSCs recorded from non-transduced WT and indicated viral-transduced neighboring CA1 PCs. (a) Veh., n = 16/5; CNO, n = 16/7; (b) Veh., n = 22/5; CNO, n = 21/7; (c) CNO, n = 16/4; (d) Note: pairs of non-transduced cells, CNO, n = 8/3; (f) KirMut, n = 18/3; Kir2.1, n = 19/5; (g) KirMut, n = 25/3; Kir2.1, n = 17/4, where n = number of pairs/mice and each open circle represents a pair of simultaneously recorded neurons, with closed circles representing mean ± SEM.
e. Representative trace of spikes detected from a CA1 PC in cell-attached mode in slice after bath application of CNO. As expected, addition of CNO led to firing rate increases in hM3DGq-expressing neurons, providing further confidence that CNO injection (via i.p.) in mice in vivo chemogenetically activates hM3DGq-expressing neurons in the CA1. N = 3 cells/3 mice. Scale: 50 pA, 60 s.
Extended Data Figure 4. Validation of Fosfl/fl;Fosbfl/fl;Junbfl/fl (FFJ) mouse line and additional electrophysiological parameters in FFJ-WT and KO cells.
a. Schematic representation of the AP-1 members conditionally deleted in FFJ line.
b,c. Representative images of smRNA-FISH validating loss of Fos and Fosb (and Junb in c) upon Cre expression in the CA1 field of 1–1.5 h post-KA-injected FFJ mice. Scale: 20 μm. N = 4 mice.
d. (Right) Normalized pixel intensity for Cre-negative and Cre-positive cells. Each point represents average for individual sections across N = 4 mice. Fos, ***p=7.7×10−4; Fosb, *p=0.031; Junb, *p=0.047.
e. Scatter plots of normalized pixel intensities of Cre signal against Fos, Fosb or Junb signals for each cell. Pearson correlation coefficients (r) shown. Fos, n = 315; Fosb, n = 86; Junb, n = 229 cells from N = 4 mice.
f. Representative images of Cre-injected FFJ sections immunostained for Fos, Fosb, Junb, and Npas4 proteins in the CA1 field of 3 h post-KA. Scale: 100 μm. N = 3 mice.
g,j,m. Schematic of stimulus electrode placement in stratum pyramidale to stimulate perisomatic inhibitory axons (g), or stratum radiatum to stimulate Schaffer collaterals (j) or proximal dendritic inhibitory axons (m).
h,k,n. Scatter plots of recorded pairs of FFJ-WT and FFJ-KO CA1 PCs in 24 h post-vehicle (Left) or -KA injected (Right) mice, where (h) Veh., n = 26/6; KA, n = 33/7; (k) Veh., n = 18/5; KA, n = 17/4; (n) Veh., n = 30/4; KA, n = 30/6.
i,l,o. Quantification of PPRs for indicated currents, where (i) Veh., n = 17/3; KA, n = 18/4; (l) Veh., n = 18/5; KA, n = 17/4; (o) Veh., n = 19/2; KA, n = 26/5.
(h,i,k,l,n,o) n = number of pairs/mice.
(d,h,i,k,l,n,o) Mean ± SEM.
Extended Data Figure 5. RNA-sequencing to identify CA1 pyramidal neuron-specific Fos targets.
a. Scatter plot showing PV-specific ARGs identified by comparing 6 h post-KA to vehicle-injected conditions. Significantly different genes (green); FDR ≤ 0.005. PV-enriched (IP over input) genes (red). Points represent mean ± SE. n = 9–10 mice/biological replicate; 4 biological replicates per condition.
b. UMAP visualization of IN subtypes using only Gad2-expressing (“Inhibitory”) cells from Fig. 3d.
c. UMAP visualization of ΔCre+ and respective control nuclei with (Left) cell type information or (Right) genotype assignments overlaid. “Control”: ΔCre— in control hemispheres; “ΔCre-GFP”: ΔCre+ in injected hemispheres; “Other”: ΔCre— or ΔCre+ in injected or control hemispheres, respectively. n = 25,214 cells/4 mice.
d. Quality control metrics for each transcriptionally distinct cell type identified by snRNA-seq in both Cre+ and ΔCre+ (“Del”) samples as in c and Fig. 3d. (Top) Number of unique genes per cell; (Middle) Number of RNA molecules per cell; (Bottom) Percentage of reads that map to mitochondrial genome.
e. Violin plots depicting CA1 PC-specific expression of Fos (****p=9.7×10−127), Fosb, Junb (****p=7.2×10−26; *p=0.003), and viral-derived WPRE (****p=0). Note that the design of the FFJ line renders snRNA-seq validation of excision of Fosb and Junb suboptimal (see Extended Data Fig. 4b–f and Methods). TPT, tags per ten thousand.
f. Strip plot displaying DGE between Cre+ and control samples for each transcriptionally distinct cell type. Colored points represent significant genes (Bonferroni-corrected p-value < 0.05, with average natural log FC > 20%); grey points represent non-significant genes.
g. Heatmap depicting normalized gene expression values from 100 randomly selected cells from each indicated cell type identity. Genes are cell-type-enriched AP-1 targets downregulated by at least 20% with loss of AP-1, and whose expression is detected in at least 25% of non-transduced cells.
h. Volcano plot of shuffled data where Cre+ and control CA1 excitatory nuclei are randomly assigned between two groups, showing no significant gene expression differences (light grey; Bonferroni-corrected p-value > 0.05), thus further indicating that the expression differences observed between Cre+ and control were due to presence of Cre.
(d,e) Mean ± 2 SD.
(e-h) Wilcoxon rank-sum (two-sided).
Extended Data Figure 6. CaMK2a-Sun1 Fos CUT&RUN revealed Fos binding sites across genome.
a. Pairwise Pearson correlation between CaMK2a-Sun1 Fos CUT&RUN biological replicates for each antibody and stimulus condition.
b. Histogram plotting distribution of distances between CaMK2a-Sun1 Fos CUT&RUN peaks and the nearest Refseq transcription start site (TSS). Peaks with a distance of 0 overlap the TSS. As expected39, ~90% of Fos-bound sites are distal to the TSS.
c-e. Histograms plotting distributions of distances between the TSS of (c) all Refseq genes, (d) CaMK2a-Ribotag ARGs, or (e) CA1 excitatory genes downregulated with AP-1 loss, and the nearest Fos binding site. A distance of 0 indicates overlap of a Fos peak with the TSS. Notably, both CaMK2a-specific ARGs (d) and putative AP-1 targets downregulated with AP-1 loss in FFJ snRNA-seq (e) are significantly enriched for Fos-bound sites, which are significantly closer to the TSS when compared to all genes (c) (p < 2.2×10−16, Wilcoxon rank-sum, two-sided), providing further support that these genes are direct targets of Fos.
f. Top three enriched motifs identified by MEME-ChIP from CaMK2a-Sun1 Fos CUT&RUN peaks. E-values and matching transcription factor motifs are displayed to the right of each enriched motif. Fos CUT&RUN peaks identified therefore shows significant enrichment for the AP-1 motif.
g-k. Tracks displaying Fos or IgG binding under 2–3 h post-vehicle or KA conditions for genomic regions surrounding the (g) Bdnf, (h) Inhba, (i) Rgs2, (j) Nptx2, or (k) Pcsk1 genes (see Fig. 4i for Scg2). Y-axis shows spike-in normalized CUT&RUN coverage. Tracks are scaled to the maximum value observed for all samples for the displayed genomic locus, shown in brackets.
Extended Data Figure 7. Analyses of AP-1-regulated candidate genes to identify molecular effector of bidirectional perisomatic inhibitory plasticity.
a. Table of AP-1-regulated candidate genes analyzed and their known functions.
b. qRT-PCR validation of shRNA efficacy using cultured hippocampal neurons transduced with lentivirus encoding the indicated shRNA. n = 3 biological replicates for each shRNA. Mean ± SEM.
c. Western blot confirmation of the efficacy of the FlpOFF u6-shRNA strategy, where Bdnf shRNA-containing plasmid was transfected in 293T cells along with Bdnf-myc, and excision of the shRNA expression cassette via introduction of Flp recombinase was confirmed. Loading controls (Gapdh) were run on a separate blot (see Supplementary Fig. 2a for full scans). 100- or 500-ng transfections of indicated u6-plasmid were loaded side-by-side on blot. n = 2 biological replicates.
d-f. Scatter plots of recorded PV-IPSC amplitudes from non-transduced shRNA— (“Control”) and neighboring shRNA+ CA1 PCs from mice 24 h post-KA injection. The shRNA target is shown on the y-axis: (d) Scrambled, n = 17/9; Inhba, n = 15/4; Rgs2, n = 20/3; Bdnf, n = 26/10; Nptx2, n = 16/3; Pcsk1, n = 17/6; (e) Scg2 shRNA#2, n = 17/6. Representative traces from a pair of neurons shown; blue marks depict light onset. Scale: 100 pA, 40 ms; (f) Scg2 shRNA#1, Strd, n = 14/5; 7–10 d NE, n = 16/4, where n = number of pairs/mice. Each open circle represents a pair of simultaneously recorded neurons; closed circles represent mean ± SEM.
g. smRNA-FISH scatter plots as in Fig. 4k depicting the correlation between Fos and (Left) Scg2 intron or (Right) Scg2 mRNA expression. Each point represents the mean number of Scg2 puncta/cell within a bin, with a bin width of 1 Fos punctum/cell. Pearson correlation coefficients (r) are shown.
h. Lower magnification images of smRNA-FISH as in Fig. 4j. Scale: 100 μm.
Extended Data Figure 8. Scg2 is a molecular effector of bidirectional perisomatic inhibitory plasticity.
a. qRT-PCR validation of conditional Scg2fl/fl line, where normalized (Left) Scg2 and (Right) Fos RNA levels in cultured hippocampal neurons derived from Scg2fl/fl mice are shown. Cultures were transduced with lentiviral Cre or ΔCre and membrane depolarized with KCl for 0 h or 6 h. n = 3 biological replicates. Mean ± SEM. Two-sided t-test, **p=0.002.
b. Schematic of intersectional genetic strategy to introduce ChR2 into CCK-INs and sparsely introduce shRNAs specifically into CA1 PCs of Dlx5/6Flp;CCKCre mice.
c. Normalized differences in CCK-IPSC amplitudes between pairs of Scg2 shRNA— and shRNA+ PCs depicted in d-f. Strd, n = 30/4; NE, n = 24/3; KA, n = 19/4. Ordinary one-way ANOVA, multiple comparisons corrected; NE, **p=0.005; KA, **p=0.002.
d-f. Scatter plots of CCK-IPSC amplitudes of pairs as in c. Representative traces from pairs of neurons shown; blue marks depict light onset. Scale: 100 pA, 40 ms.
g. (Top) Schematic of recording configuration. Scatter plots of (Bottom left) PV-IPSC or (Bottom right) CCK-IPSC amplitudes recorded from pairs of neurons of which one was non-transduced (WT) and the other expressed a Scg2 shRNA with an shRNA-resistant full-length Scg2 rescue construct. Normalized differences in IPSC amplitudes between pairs of neurons shown to the right of each scatter plot. PV, n = 19/6; CCK, n = 19/4. One-sample t-test (two-sided) with hypothetical mean of 0, *p=0.011.
(c-g) Each open circle represents a pair of simultaneously recorded neurons; closed circles represent mean ± SEM; n = number of pairs/mice.
Extended Data Figure 9. A series of rescue and overexpression analyses suggest a critical role for the processing of Scg2.
a,b. Scatter plots of PV-IPSC (a) and CCK-IPSC (b) amplitudes recorded from mKate2+ pairs that are either Cre— (WT) or Cre+ (KO). Scg2-KO neurons also expressed a Cre-dependent full-length Scg2 construct (Rescue WT) to rescue the loss of Scg2. PV, n = 22/5; CCK, n = 27/3.
c,d. As in a,b but using a Cre-dependent non-cleavable Scg2 mutant (Rescue 9AA) instead, which failed to rescue the loss of Scg2. PV, n = 23/4; CCK, n = 23/4.
e,f. Scatter plots of PV-IPSC (e) and CCK-IPSC (f) amplitudes recorded from non-transduced (WT) and neighboring full-length Scg2-overexpressing CA1 PCs (OE WT), showing that gain-of-function of Scg2 is sufficient to induce bidirectional perisomatic inhibitory plasticity in the absence of neural activity. PV, n = 20/5; CCK, n = 25/3.
g. Western blot confirmation of stable expression of Scg2 and the non-cleavable Scg2 mutant (9AA-Mutant) constructs containing an HA-tag in 293T cells. Expression levels were measured by immunoblot analysis with HA antibody. Loading controls (Gapdh) were run on a separate blot (see Supplementary Fig. 2b for full scans). n = 2 biological replicates.
h,i. As in e,f with overexpression of the non-cleavable Scg2 mutant (9AA Mutant) instead, which failed to induce changes in inhibition. PV, n = 19/4; CCK, n = 16/3.
(a-f,h,i) Each open circle represents a pair of simultaneously recorded neurons; closed circles represent mean ± SEM; n = number of pairs/mice.
Extended Data Figure 10. Silicon probe recordings in Scg2-WT and Scg2-KO mice to assess effects on network oscillations.
a. (Left) Schematic of stereotaxic injection and recording site in CA1 pyramidal layer. (Right) Representative image of silicon probe placement in CA1 pyramidal layer with Cre-GFP (green) and Dil (red). N = 4 mice. Scale: 200 μm.
b. Normalized power spectrum of network oscillations in Scg2-WT or KO mice during stationary periods. Average across Scg2-WT (grey, N = 4) or Scg2-KO (green, N = 5) mice, one session per mouse. Mean ± SEM.
c. Mean of the normalized power spectra within theta, slow gamma, and fast gamma bands during stationary periods as shown in b. Two-sided t-test, * p = 0.037. Mean ± SEM.
d. Cumulative histogram of mean firing rate for all Scg2-WT and Scg2-KO units. Mean firing rate is not significantly different (two-sided t-test, p = 0.2138). Scg2-WT (n = 67 units) and Scg2-KO (n = 103 units).
e. Example local field potential (LFP), single-unit activity, and running speed in a Scg2-WT mouse. From top to bottom: Denoised and downsampled LFP, 4–12 Hz bandpass filtered LFP, population spiking activity raster plot, and smoothed running speed.
f. Expanded snippet of data from the example in e. From top to bottom: Denoised and downsampled LFP, 4–12 Hz bandpass filtered LFP, and population spiking activity raster plot.
g. As in f with example data from a Scg2-KO mouse.
(a) Schematic image (left) adapted with permission from Paxinos & Franklin (Elsevier),
Supplementary Material
Acknowledgements.
We thank all members of the Greenberg laboratory for discussions and critical feedback during the course of this work. We also thank all members of the Harvey laboratory for discussions and critical feedback. We thank W. Regehr, D. Ginty, R. Wilson, C.Tzeng, J. Green, E. Pollina, and T.Vierbuchen for feedback and critical evaluation of the data and/or manuscript. We thank G. Fishell for the Dlx5/6Flp mice. We thank L. Wu and the Harvard Genome Modification Facility for aiding in the generation of the Scg2fl/fl mice. We thank C. Wang and the Boston Children’s Hospital Viral Core for generation of AAVs. We thank the Harvard Neurobiology Imaging Facility (NINDS P30 Core Center grant NS072030) for imaging support. We also thank O. Mazor and P. Gorelik at the Harvard Research Instrumentation Core for technical design and support. This work was supported by NIH grant R37 NS028829 (M.E.G.), NIH grant R01 NS089521 (C.D.H.), T32 NS007473 (C.P.D.), F32 NS112455 (C.P.D.), Stuart H.Q. and Victoria Quan fellowship (E-L.Y. and N.L.P.), and Harvard Department of Neurobiology graduate fellowship (E-L.Y.).
Footnotes
Competing interests. The authors declare no competing interests.
Code availability
Custom code will be provided upon reasonable request.
Data availability
Ribotag, FFJ snRNA-seq and CUT&RUN data are deposited into the public repository Gene Expression Ombinus (GEO) with accession number GSE158843. All other data will be shared upon reasonable request.
References
- 1.Josselyn SA & Tonegawa S Memory engrams: Recalling the past and imagining the future. Science 367, doi: 10.1126/science.aaw4325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka KZ et al. The hippocampal engram maps experience but not place. Science 361, 392–397, doi: 10.1126/science.aat5397 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Greenberg ME & Ziff EB Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311, 433–438 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Freund TF & Katona I Perisomatic inhibition. Neuron 56, 33–42, doi: 10.1016/j.neuron.2007.09.012 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Klausberger T et al. Complementary roles of cholecystokinin- and parvalbumin-expressing GABAergic neurons in hippocampal network oscillations. J Neurosci 25, 9782–9793, doi: 10.1523/JNEUROSCI.3269-05.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartos M & Elgueta C Functional characteristics of parvalbumin- and cholecystokinin-expressing basket cells. J Physiol 590, 669–681, doi: 10.1113/jphysiol.2011.226175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yap EL & Greenberg ME Activity-Regulated Transcription: Bridging the Gap between Neural Activity and Behavior. Neuron 100, 330–348, doi: 10.1016/j.neuron.2018.10.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryan TJ, Roy DS, Pignatelli M, Arons A & Tonegawa S Memory. Engram cells retain memory under retrograde amnesia. Science 348, 1007–1013, doi: 10.1126/science.aaa5542 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glickfeld LL & Scanziani M Distinct timing in the activity of cannabinoid-sensitive and cannabinoid-insensitive basket cells. Nat Neurosci 9, 807–815, doi: 10.1038/nn1688 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hefft S & Jonas P Asynchronous GABA release generates long-lasting inhibition at a hippocampal interneuron-principal neuron synapse. Nat Neurosci 8, 1319–1328, doi: 10.1038/nn1542 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Buzsaki G Theta oscillations in the hippocampus. Neuron 33, 325–340, doi: 10.1016/s0896-6273(02)00586-x (2002). [DOI] [PubMed] [Google Scholar]
- 12.Buzsaki G & Wang XJ Mechanisms of gamma oscillations. Annu Rev Neurosci 35, 203–225, doi: 10.1146/annurev-neuro-062111-150444 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasselmo ME & Stern CE Theta rhythm and the encoding and retrieval of space and time. Neuroimage 85 Pt 2, 656–666, doi: 10.1016/j.neuroimage.2013.06.022 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorensen AT et al. A robust activity marking system for exploring active neuronal ensembles. Elife 5, doi: 10.7554/eLife.13918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenno LE et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods 11, 763–772, doi: 10.1038/nmeth.2996 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taniguchi H et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013, doi: 10.1016/j.neuron.2011.07.026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth BL DREADDs for Neuroscientists. Neuron 89, 683–694, doi: 10.1016/j.neuron.2016.01.040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue M, Atallah BV & Scanziani M Equalizing excitation-inhibition ratios across visual cortical neurons. Nature 511, 596–600, doi: 10.1038/nature13321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vierbuchen T et al. AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol Cell 68, 1067–1082 e1012, doi: 10.1016/j.molcel.2017.11.026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hrvatin S et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21, 120–129, doi: 10.1038/s41593-017-0029-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanz E et al. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc Natl Acad Sci U S A 106, 13939–13944, doi: 10.1073/pnas.0907143106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skene PJ & Henikoff S An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6, doi: 10.7554/eLife.21856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo A et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384, doi: 10.1016/j.neuron.2015.05.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bloodgood BL, Sharma N, Browne HA, Trepman AZ & Greenberg ME The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature 503, 121–125, doi: 10.1038/nature12743 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hensch TK Bistable parvalbumin circuits pivotal for brain plasticity. Cell 156, 17–19, doi: 10.1016/j.cell.2013.12.034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nedivi E, Hevroni D, Naot D, Israeli D & Citri Y Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature 363, 718–722, doi: 10.1038/363718a0 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Fischer-Colbrie R, Laslop A & Kirchmair R Secretogranin II: molecular properties, regulation of biosynthesis and processing to the neuropeptide secretoneurin. Prog Neurobiol 46, 49–70, doi: 10.1016/0301-0082(94)00060-u (1995). [DOI] [PubMed] [Google Scholar]
- 28.Foldy C, Malenka RC & Sudhof TC Autism-associated neuroligin-3 mutations commonly disrupt tonic endocannabinoid signaling. Neuron 78, 498–509, doi: 10.1016/j.neuron.2013.02.036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartzell AL et al. NPAS4 recruits CCK basket cell synapses and enhances cannabinoid-sensitive inhibition in the mouse hippocampus. Elife 7, doi: 10.7554/eLife.35927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiler R et al. Chromogranins in rat brain: characterization, topographical distribution and regulation of synthesis. Brain Res 532, 87–94, doi: 10.1016/0006-8993(90)91746-4 (1990). [DOI] [PubMed] [Google Scholar]
References
- 31.Miyoshi G et al. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci 30, 1582–1594, doi: 10.1523/JNEUROSCI.4515-09.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma N et al. ARNT2 Tunes Activity-Dependent Gene Expression through NCoR2-Mediated Repression and NPAS4-Mediated Activation. Neuron 102, 390–406 e399, doi: 10.1016/j.neuron.2019.02.007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y et al. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455, 1198–1204, doi: 10.1038/nature07319 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ataman B et al. Evolution of Osteocrin as an activity-regulated factor in the primate brain. Nature 539, 242–247, doi: 10.1038/nature20111 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mardinly AR et al. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature 531, 371–375, doi: 10.1038/nature17187 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Habib N et al. Div-Seq: Single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science 353, 925–928, doi: 10.1126/science.aad7038 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cembrowski MS, Wang L, Sugino K, Shields BC & Spruston N Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. Elife 5, e14997, doi: 10.7554/eLife.14997 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hainer SJ & Fazzio TG High-Resolution Chromatin Profiling Using CUT&RUN. Curr Protoc Mol Biol 126, e85, doi: 10.1002/cpmb.85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malik AN et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci 17, 1330–1339, doi: 10.1038/nn.3808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klein AM et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201, doi: 10.1016/j.cell.2015.04.044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yatsenko D et al. DataJoint: managing big scientific data using MATLAB or Python. bioRxiv, doi: 10.1101/031658 (2015). [DOI] [Google Scholar]
- 42.Pachitariu M, Steinmetz N, Kadir S, Carandini M & Harris KD Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels. bioRxiv, doi: 10.1101/061481 (2016). [DOI] [Google Scholar]
- 43.Rossant C et al. Spike sorting for large, dense electrode arrays. Nat Neurosci 19, 634–641, doi: 10.1038/nn.4268 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartho P et al. Characterization of neocortical principal cells and interneurons by network interactions and extracellular features. J Neurophysiol 92, 600–608, doi: 10.1152/jn.01170.2003 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Berens P CircStat: A MATLAB Toolbox for Circular Statistics. Journal of Statistical Software 31, doi: 10.18637/jss.v031.i10 (2009). [DOI] [Google Scholar]
- 46.Bokil H, Andrews P, Kulkarni JE, Mehta S & Mitra PP Chronux: a platform for analyzing neural signals. J Neurosci Methods 192, 146–151, doi: 10.1016/j.jneumeth.2010.06.020 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Ribotag, FFJ snRNA-seq and CUT&RUN data are deposited into the public repository Gene Expression Ombinus (GEO) with accession number GSE158843. All other data will be shared upon reasonable request.