Abstract
Rationale:
Altered lipid metabolism has been implicated in heart failure (HF) development, but no prospective studies have examined comprehensive lipidomics data and subsequent risk of HF.
Objective:
We aimed to link single lipid metabolites and lipidomics networks to the risk of developing heart failure.
Methods and Results:
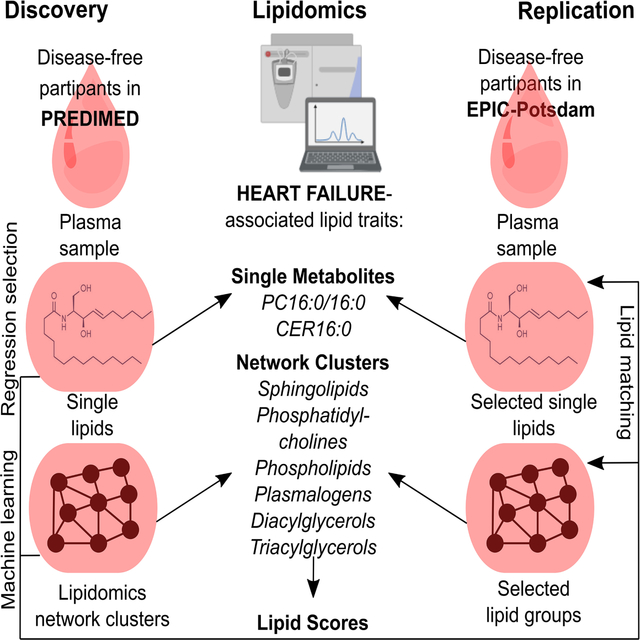
Discovery analyses were based on 216 targeted lipids in a case-control study (331 incident HF cases and 507 controls, matched by age, sex, and study center), nested within the PREDIMED study. Associations of single lipids were examined in conditional logistic regression models. Furthermore, lipidomics networks were linked to HF risk in a multi-step workflow, including machine learning-based identification of the HF-related network-clusters, and regression-based discovery of the HF-related lipid patterns within these clusters. If available, significant findings were externally validated in a subsample of the EPIC-Potsdam cohort (2414 at-risk-participants, including 87 incident HF-cases).
After confounder-adjustments, two lipids were significantly associated with HF risk in both cohorts: ceramide 16:0 (HR per SD in PREDIMED 1.28, 95%CI 1.13, 1.47) and phosphatidylcholine 32_0 (HR per SD in PREDIMED 1.23, 95%CI 1.08, 1.41). Additionally, lipid patterns in several network clusters were associated with HF risk in PREDIMED. Adjusted for standard risk factors, an internally cross-validated score based on the significant HF-related lipids that were identified in the network analysis in PREDIMED was associated with a higher HF risk (20 lipids, HR per SD 2.33, 95%CI 1.93, 2.81%). Moreover, a lipid score restricted to the externally available lipids was significantly associated with HF incidence in both cohorts (6 lipids, HRs per SD 1.30, 95%CI 1.14, 1.47 in PREDIMED, and 1.46, 95%CI, 1.17, 1.82 in EPIC-Potsdam).
Conclusions:
Our study identified and validated two lipid metabolites and several lipidomics patterns as potential novel biomarkers of HF risk. Lipid profiling may capture preclinical molecular alterations that predispose for incident HF.
Keywords: Heart failure, metabolism, lipidomics, biomarker, prospective human population study, lipids, chronic heart failure, biomarker discovery
Subject Terms: Biomarkers, Epidemiology, Heart Failure, Metabolism, Primary Prevention
Graphical Abstract
INTRODUCTION
Heart failure (HF) is a life-threatening disease, which in 2017 affected over 37 million individuals worldwide (1), and incidence rates are rising (2). The complex clinical syndrome is characterized by the inability of the heart to generate the output to meet physiological demands (reduced ejection fraction) or adequate cardiac output but only through compensatory neuro-humoral stimulation (preserved ejection fraction). Etiologically, idiopathic HF cases in older people are generally attributable to either one of the three major causes: coronary artery disease (CAD), pressure overload, and type 2 diabetes (T2D) (2). However, all types of HF confer a substantial burden to health-care systems and drastically impair the quality of life (3). Therefore, effective prevention strategies are urgently needed.
Epidemiological studies have established several risk factors for incident HF (4). Besides age and sex, the risk factors mostly comprise prevalent comorbidities, i.e., CAD, atrial fibrillation, T2D, hypertension, and chronic diseases of the lung and the kidney (4). At the molecular level, NTpro-BNP is an established HF-predictor but rather reflects activated compensation of decreasing cardiac performance (5). Other biomarkers are closely linked to the HF-related co-morbidities (4). Recently, the research interest shifted towards identifying biomarkers of pathophysiological processes in early HF-development (6).
The human heart has a continuously high energy demand. However, the failing heart is incapable of consistently generating the required energy for its adequate function. Among the potential molecular mechanisms, lipids are of central importance for short-term metabolic flexibility of the heart, and lipotoxic compounds could be a key factor that link metabolic stress to persistent damage in the myocardial tissue (7). In animal models, plasma lipidomic signatures reflect alterations in cardiac lipid metabolism that predispose for HF incidence (8). Moreover, disturbed lipid metabolism is indirectly connected to HF through the common comorbidities, including CAD (9) and T2D (10).
Evidence on the link between comprehensive lipid profiling data in humans and HF incidence is lacking. Here we present findings from a prospective case-control study nested within the PREDIMED (PREvención con DIeta MEDiterránea) trial (11, 12), in which we evaluated the association of baseline targeted lipidomics profiles with the risk of developing HF. Our lipidomics approach primarily targeted glycerophospholipids (PLs), sphingolipids (SLs), plasmalogens (PLGs), cholesterol esters (CEs), and tri-, di- and monoacylglycerols (TAGs, DAGs, and MAGs), as well as some specific signaling lipids. We explored single lipids as risk factors for HF incidence. We also generated a data-driven lipidomics network and investigated whether simultaneous consideration of network-based lipid clusters revealed additional associations between the baseline lipidomics profiles and future HF incidence. Finally, we evaluated the external validity of our findings in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam cohort.
METHODS
Expanded Methods are available online in the Supplemental Material.
Data availability.
The authors will be happy to provide access to the PREDIMED dataset (including data dictionaries), making possible the replication of the main analyses used for the present article. Due to the restrictions imposed by the Informed Consent and the Institutional Review Board, bona fide investigators interested in analyzing the PREDIMED dataset used for the present article may submit a brief proposal and statistical analysis plan to the corresponding authors. Upon approval from the PREDIMED Steering Committee and Institutional Review Boards, the data will be made available to them using an onsite secure access data enclave
Study populations.
The PREDIMED study was a multi-center dietary intervention trial that included 7,447 participants in three intervention arms (11, 12). The discovery analyses in PREDIMED included 331 participants with available blood samples and incident HF during an extended follow-up period (recruitment between June 2003 and June 2009, censoring date December 31st, 2017). For each case, between 1 and 3 controls were randomly selected from all participants at risk at the date of HF-diagnosis (incidence density sampling), matched by recruitment center, year of birth (± 5 years), and sex (n = 507controls, Online Figure I). The median follow-up time was 12y (IQR 9.9y-12.9y) among controls, and 7.2y (IQR 4.8y-9.7y) among cases. The study protocols were approved by the institutional review boards at all study locations (PREDIMED), and the Harvard TH Chan School of Public Health (PREDIMED case-control subproject). All participants gave written, informed consent.
The EPIC-Potsdam cohort included 27,548 participants (16,644 women and 10,904 men), recruited between 1994 and 1998 from the general population within the age range of 35–65 years (13). The replication analyses in EPIC-Potsdam were based on nested case-cohorts for cardiovascular disease (CVD) and T2D with available lipidomics data, including a random subsample (subcohort, n=1,262), all participants with incident T2D (n = 820) and all participants with incident primary CVD (n = 583). From the 2414 at-risk-participants, 87 developed incident HF during follow-up. The median follow-up time was 8.4y (IQR 7.6y-9.3y) among participants without incident HF, and 5.9y (IQR 3.2y-7.4y) among participants with an incident event. The study was approved by the Ethics Committee of the State of Brandenburg, Germany (EPIC-Potsdam). All participants gave written, informed consent.
The baseline examination in both studies included assisted assessment of prevalent diseases (including T2D and hypertension), education, smoking, and medication (14) in interviews and with validated questionnaires. Anthropometric variables and blood pressure were assessed by qualified medical personnel in the study centers (Online Note I).
Targeted plasma lipid profiles.
In PREDIMED, baseline blood samples were taken after an overnight fast by trained study personnel according to a standard protocol, fractioned, and the EDTA plasma was stored at −80°C in deep freezers. At the Broad Institute (Cambridge, MA, US), profiles of plasma polar and nonpolar lipids were assessed using a Nexera X2 U-HPLC system (Shimadzu Scientific Instruments; Marlborough, MA) coupled to an Exactive Plus orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA, US). In EPIC-Potsdam, blood was sampled by medical personnel (most participants did not fast before). The lipidomics data was generated with Metabolon (Morrisville, US) using the Metabolon® Complex Lipid Panel (Online Note II).
Ascertainment of heart failure.
In the PREDIMED study, HF was a prespecified secondary endpoint and the uniform diagnostic criteria were based on the 2005 guidelines on the diagnosis and treatment of acute and chronic HF of the European Society of Cardiology (15, 16). Both cohorts, PREDIMED and EPIC-Potsdam, used multiple sources for detection of incident HF, including medical records of all participants and consultation of the National Death Index (PREDIMED) and self-reports, death certificates, adhoc reviews of medical charts (PREDIMED), linkage to hospital records, and hints from other reported diseases or pharmacological treatments (EPIC-Potsdam). Potential cases of incident HF were validated by physicians. This study only included physician-verified incident HF (I50 of International Classification of Diseases, 10th revision) during the follow-up period (Online Note III).
Statistical analysis.
For lipids with a fraction of missingness below 0.25, we imputed missing values with half the minimal measured value. The inverse normal transformation, which generates a rank-based standard normal distribution (mean=0, SD=1), was applied to all lipid metabolites. We use the following annotation: XX_Y (XX = carbon atoms, Y = double bonds) for the number of carbon atoms and desaturations across several fatty acid- (FA)-residues; XX:Y for the number of carbon atoms and desaturations per single acyl chain.
Conditional logistic regression.
For conditional logistic regression analyses in PREDIMED, we used the clogit function in the survival R-package (https://CRAN.R-project.org/package=survival), stratifying by the matching identifier. We used the Efron-approximation (17) for partial likelihood estimators. According to the incidence density sampling (sampling with replacement), robust standard errors were generated clustering by the participant IDs. The overall model fit was evaluated with the robust score test (18).
Network and clustering.
To generate the lipidomics network, we used the conditional independence-based PC-algorithm (19). The algorithm was applied to the same lipidomics data in two nested case-control studies in the PREDIMED-trial. Then, we retained only edges that corresponded to partial correlations >0.1 in both case-control studies to generate a robust, biologically meaningful network (20, 21). In this network, we detected clusters with the walktrap-algorithm in the igraph R-package (http://igraph.org/).
Random forest-based evaluation of cluster importance.
We performed a machine-learning-based selection of the most important lipid clusters for HF prediction. To keep the matched case-control design, we prepared the lipidomics data by calculating deviations from matching-strata-specific means (22) and grew a random forest for HF-prediction (500 trees, sampling rate of 2/3) (23). Then, we evaluated the importance of lipid clusters for HF prediction in the out-of-bag sample. To this end, we assessed the predictive performance of the random forest model based on information on the full lipidomics data as compared to the lipidomics data with the joint permutation of lipid variables in each of the clusters (24). Clusters were ranked by the extent to which omitting the information in their variables hampered the predictive performance, with the largest increase in prediction error corresponding to the highest cluster importance.
Determining significant HF-predictors within important clusters.
To select the lipid patterns for high cluster importance, for each of the most important clusters, we simultaneously included all cluster-variables in a conditional logistic regression model with the model specification as described above. Then, we gradually removed the non-informative variables according to the highest p-value, until only significant (p-value <0.05) HF predictors remained (backwards selection) (25).
We also combined significant predictors across clusters into beta coefficient-weighted sum scores. Score1 was based on all selected within-cluster predictors of incident HF in PREDIMED. To derive the internally validated beta coefficients, we used 10-fold cross-validated elastic net regression [(26), (https://CRAN.R-project.org/package=clogitL1]. Score2 was based on externally validated within-cluster predictors, using the backwards selection as described above.
All the p-values in PREDIMED- analyses were derived from two-sided tests. An alpha-level of 0.05 was considered statistically significant. Where applicable we adjusted for multiple testing by controlling the false discovery rate (FDR) (27).
Validating the PREDIMED-discoveries in the EPIC-Potsdam cohort.
For lipids with two or more FA-residues, the lipidomics data in EPIC-Potsdam was more specific in terms of carbon atoms and desaturations contained in the single acyl-chains. Therefore, we first summed all lipids in EPIC-Potsdam corresponding to the sum formula in PREDIMED (Online Note IV). If the HF risk association was replicated on the sum level, we also evaluated which of the underlying lipids in EPIC-Potsdam drove the association. To this end, we simultaneously included all the EPIC-Potsdam lipids that matched the sum formula in PREDIMED and selected the most significant predictors using a backwards procedure.
HF risk associations in the EPIC-Potsdam cohort were assessed in Cox proportional hazards regression models, with age as the underlying time scale, applying the coxph function of the survival R-package (https://CRAN.R-project.org/package=survival). Ties were handled using the Efron method (17), and we used robust variance estimators clustering by participants’ ID. All models accounted for age and sex, as strata variables; further adjustments were similar to the models in PREDIMED. We considered directionally consistent (as compared to PREDIMED) estimates with a one-tailed p-value <0.05 as significant replications.
RESULTS
Descriptive statistics.
Overall, the PREDIMED-trial consisted of participants at increased cardiovascular risk due to a high prevalence of adiposity, T2D, and hypertension. Compared to the age- and sex-matched controls, participants with incident HF tended to have a higher BMI, and prevalent T2D and hypertension were more frequent among them (Table 1). Slight difference in the proportion of women between cases and controls arose from the varying number of controls per case. Compared to the PREDIMED sample, the included EPIC-Potsdam participants were younger, had a lower BMI (although overweight and obesity were also common), tended to have higher educational attainment and were more likely to be former or current smokers, but less likely to have prevalent T2D and hypertension (Online Table I). Over 97% of the PREDIMED participants and over 99.9% of the EPIC-Potsdam participants were white Europeans. Based on the covariance structure of the lipidomics data, we constructed a conditional independence lipid network with 26 densely connected clusters (Online Figure II).
Table 1:
Descriptive statistics of matched heart failure case-control samples in the PREDIMED
| Characteristics | Controls (n=507) | Cases (n=331) |
|---|---|---|
| Age [years] | 72 (66, 75)* | 71 (65, 75) |
| Women | 55% | 59% |
| BMI [kg/sqm] | 29.2 (26.9, 31.8) | 30.8 (28.5, 33.5) |
| Highest education | ||
| Primary school | 83% | 85% |
| Sec./high school | 11% | 11% |
| College/higher | 6% | 4% |
| Smoker | ||
| Never | 62% | 60% |
| Former | 11% | 14% |
| Current | 27% | 25% |
| Family history of CAD | 19% | 19% |
| Prevalent T2D† | 51% | 59% |
| Prevalent HT‡ | 84% | 88% |
| Diet intervention group§ | ||
| Low-fat (control) | 37% | 37% |
| MedDiet+EVOO | 37% | 31% |
| MedDiet+nuts | 26% | 32% |
Median (IQR), all such values
T2D: type 2 diabetes
HT: hypertension;
Participants were randomly assigned to one of three long-term intervention diets: low-fat (control), Mediterranean Diet (MedDiet) enriched with extra-virgin olive oil (+EVOO) or with nuts (+nuts).
Single lipid-HF associations.
After controlling the FDR (<0.05), only ethanolamide (EA) 18:1 was significantly associated with HF risk (RR 1.38, 95%CI 1.21, 1.57) in the minimally adjusted model. Further adjustment for other HF risk factors, namely prevalent T2D and hypertension at baseline, educational attainment, family history of early CAD, smoking, BMI, and intervention group attenuated the association (1.26, 95% CI 1.09, 1.46, FDR 0.066). This effect attenuation was largely observed after simultaneous adjustment for T2D and BMI, while the other factors played a minor role.
In addition, 26 lipids were marginally significantly associated with HF risk but with FDR > 0.05 (Online Table II), from which 15 were available in EPIC-Potsdam. Controlling for age and sex, 8 of these lipids were directionally consistently and significantly associated with HF risk in EPIC-Potsdam. Six of these 8 replicated associations were rendered non-significant after further adjustment for the other HF risk factors in PREDIMED, with similar effect attenuation in EPIC-Potsdam (Online Table III). However, the HF-associations of diacyl-phosphatidylcholine (PC) 32_0 and ceramide (CER) 16:0 were robust against these adjustments in both cohorts (Table 2). In both models, the FDR for these two lipid metabolites was significant (<0.05) after adjusting for the fifteen tested lipid metabolites in the replication study (Online Table III). Further adjustment for prevalent dyslipidemia (PREDIMED) or HDL- and total cholesterol and total TAGs (EPIC-Potsdam) did not attenuate the associations (Online Table IV). Leveraging the higher FA-resolution of the isomeric lipids in EPIC-Potsdam, we found that the association of PC 32_0 with HF risk was mainly attributable to PC 16:0/16:0 (Table 2).
Table 2:
Single lipid associations with heart failure risk
| PREDIMED | EPIC-Potsdam | ||||||
|---|---|---|---|---|---|---|---|
| Model | Lipid | RR (95% CI)* | p-value† | Matched lipid‡ | RR (95% CI)* | p-value|| | |
| M1 | PC32_0 | 1.21 (1.07, 1.36) | 0.00180 | PC 16:0/16:0 | 1.49 (1.19, 1.86) | 0.00024 | |
| M2 | 1.23 (1.08, 1.41) | 0.00224 | 1.43 (1.14, 1.8) | 0.00101 | |||
| M1 | CER16:0 | 1.20 (1.07, 1.34) | 0.00226 | CER 16:0 | 1.60 (1.29, 1.99) | 1.2E-05 | |
| M2 | 1.28 (1.13, 1.47) | 0.00017 | 1.48 (1.17, 1.87) | 0.00048 | |||
Two lipids were significantly (p-value<0.05) associated with HF risk in PREDIMED, and the association was replicated in EPIC-Potsdam in both, a minimally adjusted model (M1, adjusted for age, sex, and study center if applicable) and a confounder-adjusted model (M2, additionally adjusted for T2D- and hypertension prevalence, BMI, smoking status, educational attainment, family history of early CAD, and intervention group if applicable).
In PREDIMED, the relative risks (RR) correspond to OR from a conditional logistic regression model.
Two-tailed p-value in PREDIMED.
For isomeric lipids with multiple matches in EPIC-Potsdam, the best predictor among these lipids was selected according the lowest p-value (backwards selection).
In EPIC-Potsdam, RRs correspond to hazard ratios from a Cox model.
One-tailed p-value in EPIC-Potsdam (testing for directionally consistent estimates only).
Lipid network-HF associations.
We ranked the lipid network-clusters according to the joint importance of the cluster-variables for HF-prediction in a random forest model. Within each of the 8 top-ranking clusters, backwards selection identified a subset of lipids that were significantly associated with HF risk in PREDIMED: cluster 1, EA 16:1+MAG 16:1; cluster 2, seven SLs; cluster 3, 2 long-chain saturated fatty acid (SFA)-containing PCs; cluster 4, six PLs; cluster 5, one DAG with lc-SFA; cluster 6, five PLGs, cluster 7, two DAGs with unsaturated FA; and, cluster 8, four TAGs (Table 3 and Online Table V). Although not all the selected lipids were available, for 6 out of 8 clusters the best corresponding model was also significantly predictive for HF incidence in EPIC-Potsdam (Table 3).
Table 3:
Most important network clusters for heart failure prediction
| PREDIMED† | EPIC-Potsdam‡ | |||||
|---|---|---|---|---|---|---|
| Rank* | Cluster-characteristic Lipids | df | Model P | df | Model P | |
| 1 | MAGs & EA 16:1 | 2 | 0.00272 | 1 | 0.44730 | |
| 2 | Sphingolipids | 7 | 5.4E-08 | 6 | 0.00022 | |
| 3 | PCs with long-chain SFAs | 2 | 0.00051 | 2 | 0.00068 | |
| 4 | Phospholipids | 6 | 3.2E-09 | 0.04645 | ||
| 5 | DAGs with long-chain SFA | 1 | 0.00485 | - | - | |
| 6 | Plasmalogens | 5 | 0.00012 | 2 | 0.00248 | |
| 7 | DAGs with unsaturated FAs | 2 | 0.00014 | 2 | 0.02490 | |
| 8 | TAGs with long-chain FA | 4 | 0.00158 | 4 | 3.2E-06 | |
Cluster ranks correspond to the joint cluster-variable importance in a random forest model.
PREDIMED, df (degrees of freedom): number of retained significant HF-predictors; model P: robust score test-based p-value for the model fit.
EPIC-Potsdam, df: number of PREDIMED-selected HF predictors available for replication; model P:log-likelihood test-based p-value for the model fit.
By simultaneously considering the cluster-based lipid groups we detected the following lipid-HF risk associations in PREDIMED and replicated them in EPIC-Potsdam: CER 16:0, and sphingomyelin (SM) 18:0 (higher HF risk), and SM 18:1 (lower HF risk) in cluster 2; PC 32_0 (higher HF risk, attributed to PC 16:0/16:0 in EPIC-Potsdam) in cluster 3; PC 34_2 (higher HF risk, attributed to PC 18:1/16:1 in EPIC-Potsdam) in cluster 4; phosphatidylethanolamine- (PE-)PLG 36_1 (higher HF risk, attributed to PE-PLG 18:0/18:1 in EPIC-Potsdam) and PE-PLG (lower HF risk, attributed to PE-PLG 18:1/22:6 in EPIC-Potsdam) in cluster 6; DAG 34_1 (higher HF risk, attributed to DAG 16:1/18:0 in EPIC-Potsdam) in cluster 7; and, TAG 55_2 and TAG 56_5 1 (higher HF risk, attributed to TAG 55_2-fa18:1 and TAG 56_5-fa16:0 in EPIC-Potsdam), and TAG 58_7 (lower HF risk, attributed to TAG 58_7-fa16:0 in EPIC-Potsdam) (Table 4, Figure 1, and Figure 2). All replicated lipids were consistently associated with HF risk if we used the sum of all isomeric lipids in EPIC-Potsdam (Online Table VI) instead of selecting the strongest predictor among all possible matches.
Table 4:
Lipid clusters and heart failure incidence
| PREDIMED | EPIC-Potsdam | ||||||
|---|---|---|---|---|---|---|---|
| Model | Lipid | RR (95% CI)* | p-value† | Matched lipid‡ | RR (95% CI)§ | p-value|| | |
| Cluster 2: Sphingolipids | |||||||
| M1 | CER16:0 | 1.24 (1.06, 1.46) | 0.00777 | CER16:0 | 1.81 (1.24, 2.63) | 0.00102 | |
| M2 | 1.27 (1.06, 1.51) | 0.00848 | 1.72 (1.14, 2.6) | 0.00507 | |||
| M1 | SM18:0 | 1.61 (1.16, 2.24) | 0.00470 | SM18:0 | 1.71 (0.93, 3.16) | 0.04294 | |
| M2 | 1.43 (1.01, 2.02) | 0.04242 | 1.54 (0.82, 2.9) | 0.09111 | |||
| M1 | SM18:1 | 0.57 (0.39, 0.85) | 0.00604 | SM18:1 | 0.53 (0.3, 0.96) | 0.01734 | |
| M2 | 0.66 (0.43, 1.02) | 0.05918 | 0.63 (0.34, 1.15) | 0.06556 | |||
| Cluster 3: lc-SFA PCs | |||||||
| M1 | PC32_0 | 1.27 (1.12, 1.45) | 0.00021 | PC16:0/16:0 | 1.31 (0.99, 1.73) | 0.02996 | |
| M2 | 1.25 (1.08, 1.44) | 0.00216 | 1.34 (1.01, 1.79) | 0.02200 | |||
| Cluster 4: Phospholipids | |||||||
| M1 | PC34_2 | 1.66 (1.38, 2) | 1.1E-07 | PC18:1/16:1 | 1.82 (1.25, 2.63) | 0.00097 | |
| M2 | 1.54 (1.25, 1.89) | 4.3E-05 | 1.82 (1.24, 2.67) | 0.00141 | |||
| Cluster 6: Plasmalogens | |||||||
| M1 | PE-PLG36_1 | 1.16 (1.03, 1.32) | 0.01811 | PE-PLG18:0/18:1 | 1.48 (1.17, 1.86) | 0.00052 | |
| M2 | 1.12 (0.98, 1.28) | 0.08904 | 1.39 (1.1, 1.76) | 0.00336 | |||
| M1 | PE-PLG40_7 | 0.65 (0.51, 0.82) | 0.00026 | PE-PLG18:1/22:6 | 0.76 (0.6, 0.97) | 0.01178 | |
| M2 | 0.68 (0.53, 0.87) | 0.00216 | 0.79 (0.62, 1) | 0.02319 | |||
| Cluster 7: Unsaturated FA-DAGs | |||||||
| M1 | DAG34_1 | 1.63 (1.3, 2.03) | 1.7E-05 | DAG16:1/18:0 | 1.45 (1.02, 2.06) | 0.02004 | |
| M2 | 1.31 (1.03, 1.67) | 0.02780 | 1.35 (0.94, 1.93) | 0.05001 | |||
| Cluster 8: TAGs | |||||||
| M1 | TAG55_2 | 1.16 (1, 1.34) | 0.04918 | TAG55_2-fa18:1 | 1.42 (0.99, 2.03) | 0.02867 | |
| M2 | 1.13 (0.96, 1.33) | 0.13176 | 1.43 (0.95, 2.17) | 0.04394 | |||
| M1 | TAG56_5 | 1.26 (1.09, 1.45) | 0.00208 | TAG56_5-fa16:0 | 3.06 (1.69, 5.55) | 0.00011 | |
| M2 | 1.2 (1.03, 1.41) | 0.02159 | 2.6 (1.41, 4.78) | 0.00107 | |||
| M1 | TAG58_7 | 0.55 (0.4, 0.76) | 0.00024 | TAG58_7-fa16:0 | 0.38 (0.23, 0.63) | 8.4E-05 | |
| M2 | 0.49 (0.35, 0.7) | 0.00010 | 0.38 (0.23, 0.65) | 0.00017 | |||
PREDIMED: relative risks (RRs) based on OR from a conditional logistic regression model.
Two-tailed p-value in PREDIMED.
For isomeric lipids with multiple matches in EPIC-Potsdam, the best predictor among these lipids was selected according the lowest p-value.
In EPIC-Potsdam, RRs correspond to HRs from a Cox model.
One-tailed p-value.
d.i., directionally inconsistent
M1: controlled for age, sex, and study center (if applicable).
M2: additionally adjusted for T2D- and hypertension prevalence, BMI, smoking status, educational attainment, family history of early CAD, and intervention group (if applicable).
Figure 1:
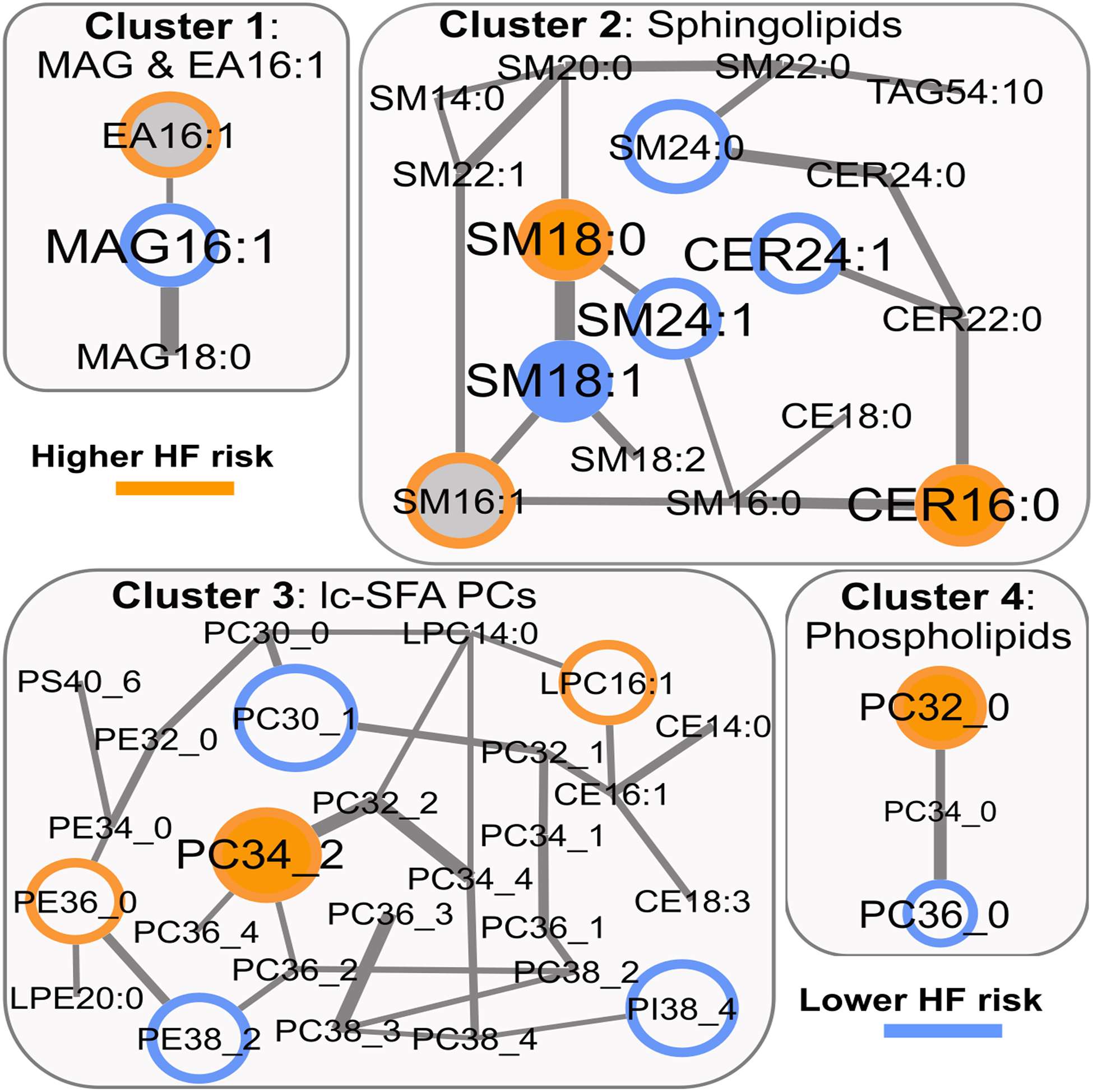
Lipidomics network – selected clusters 1 to 4 and HF risk. Colored border: HF risk association in PREDIMED; Colored filling: HF risk association in EPIC-Potsdam; Grey filling: not available in EPIC-Potsdam; Edge-width: partial correlation strength between lipids, adjusted for all other lipids.
Figure 2:
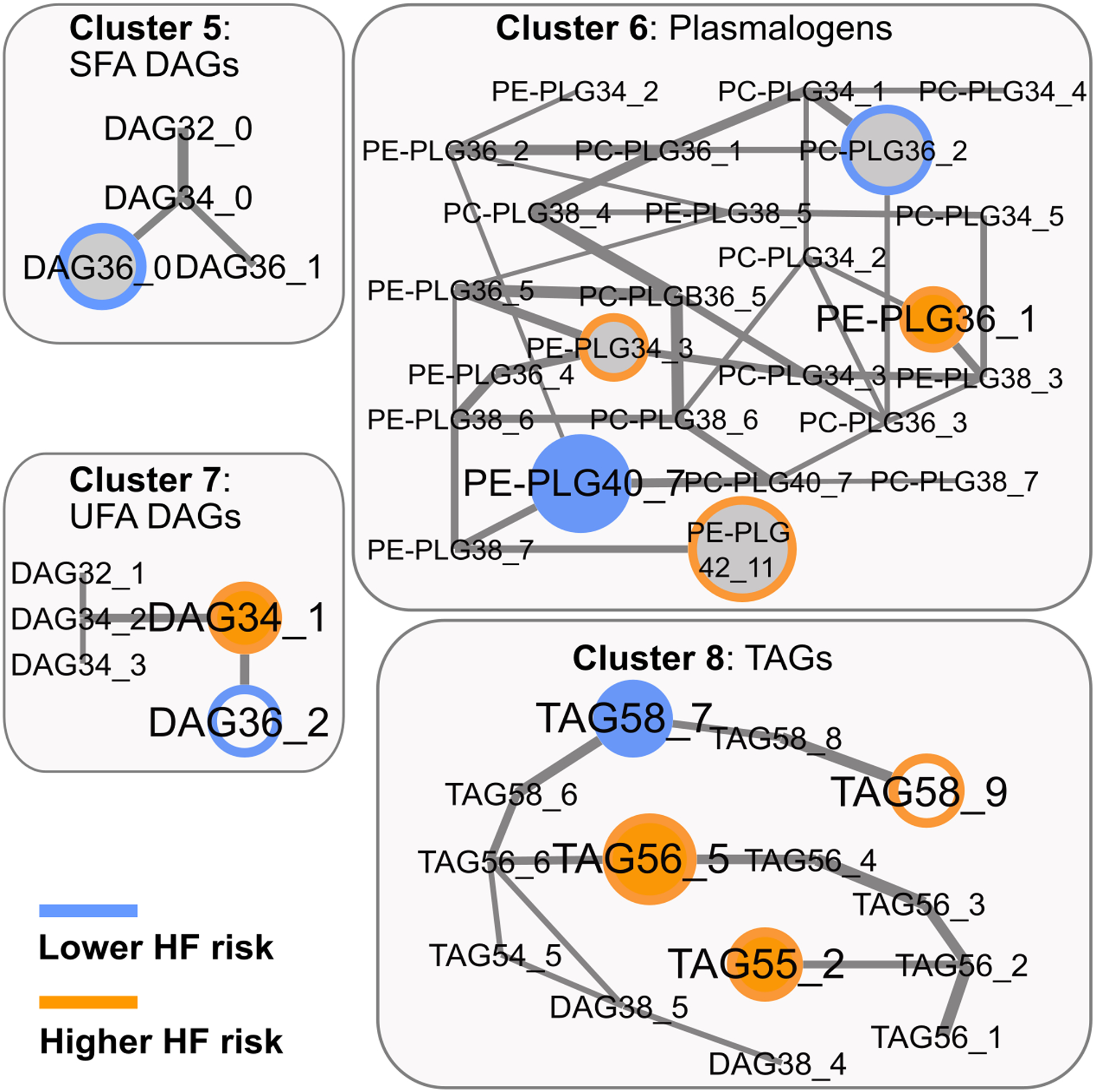
Lipidomics network – selected clusters 5 to 8 and HF risk. Colored border: HF risk association in PREDIMED; Colored filling: HF risk association in EPIC-Potsdam; Grey filling: not available in EPIC-Potsdam; Edge-width: partial correlation strength between lipids, adjusted for all other lipids.
Several lipid-HF risk associations in PREDIMED were not replicated in EPIC-Potsdam. Cluster 1 contained two oppositely directed predictors in PREDIMED, MAG 16:1 (lower HF risk) and EA 16:1 (higher HF risk); the latter was not available in EPIC-Potsdam and MAG 16:1 was not significantly associated with HF risk without adjustment for EA 16:1, in neither of the cohorts. Cluster 5 contained a single significant predictor, DAG 36_0, which was not available in EPIC-Potsdam (Online Table V). The external validity of the risk associations of six lipids, namely, EA 16:1, SM 16:1, PE-PLG 34_3 and PE-PLG 42_11 (higher HF risk), and DAG 36_0 and PC-PLG 36_2 (lower HF risk), could not be tested because these lipids were not available in the lipidomics dataset in EPIC-Potsdam (Online Table V). Moreover, the network cluster-based analysis suggested 12 HF predictors in PREDIMED that were not statistically significantly associated with HF risk in EPIC-Potsdam (Online Table V).
Network-based lipid scores and HF risk.
Out of the 29 significant within-cluster predictors, 20 were selected as independent HF predictors based on cross-validated elastic net regression: EA 16:1, MAG 16:0, CER 16:0, SM 16:1, CER 24:1, SM 24:0, SM 24:1, PC 32_0, LPC 16:1, PC 30_1, PC 34_2, PE 38_2, PI 38_4, DAG 36_0, PEPLG 34_3, PC-PLG 36_2, PE-PLG 40_7, PE-PLG 42_11, TAG 56_5, TAG 58_7. We summarized the HF association of these lipids in a sum score, weighted with the cross-validated betas (Score1). Higher Score1 points were associated with markedly increased HF risk (RR per SD 2.38, 95%CI 1.99, 2.85). Adjustment for the prevalence of T2D, hypertension, BMI, family history of CVD, intervention group, and smoking status did not alter the association of Score1 with HF incidence (RR per SD 2.33, 95%CI 1.93, 2.81) (Table 5), and no test for interaction by one of these covariables was statistically significant (data not shown).
Table 5:
Lipid scores and heart failure incidence
| PREDIMED* | EPIC-Potsdam† | |||||
|---|---|---|---|---|---|---|
| RR (95%CI) | PPREDIMED | RR (95%CI) | PEPIC | |||
| Score1 (cross-validated) | M1 | 2.38 (1.99, 2.85) | <2.2E-16 | |||
| M2 | 2.33 (1.93, 2.81) | <2.2E-16 | ||||
| Score2 (replicated) | M1 | 1.39 (1.24, 1.57) | 6.0E-08 | 1.55 (1.25, 1.92) | 6.96E-05 | |
| M2 | 1.30 (1.14, 1.47) | 8.2E-05 | 1.46 (1.17, 1.82) | 8.01E-04 | ||
Score1: 20 lipid-markers, selected from all within-cluster predictors in PREDIMED based on cross-validated elastic net regression (Formula: 0.18*EA 16:1 – 0.04*MAG 16:0 + 0.14*CER 16:0 + 0.24*SM 16:1 – 0.05*CER 24:1 – 0.18*SM 24:0 – 0.11*SM 24:1 + 0.33* PC 32_0 + 0.17*LPC 16:1 −0.19* PC 30_1 + 0.04* PC 34_2 – 0.01*PE 38_2 – 0.35*PI 38_4 – 0.17*DAG 36_0 + 0.16* PEPLG 34_3 – 0.25* PCPLG 36_2 – 0.22* PEPLG 40_7 + 0.20* PEPLG 42_11 + 0.19* TAG 56_5 – 0.08* TAG 58_7).
Score2: 6 mutually independent lipid-markers selected from the 11 externally validated HF-predictors with conditional logistic regression-based backwards selection: (Formula: 0.14* PEPLG 36_1 + 0.14* TAG 56_5 – 0.19*PEPLG 40_7- 0.21*TAG 55_2 + 0.25*DAG 34_1 + 0.21*PC 32_0).
M1: controlled for age, sex, and study center (if applicable).
M2: additionally adjusted for T2D- and hypertension prevalence, BMI, smoking status, educational attainment, family history of early CAD, and intervention group.
We also selected 6 mutually independent HF-predictors from the 11 within-cluster predictors that were replicated in EPIC-Potsdam (PEPLG 36_1, TAG 56_5, TAG 40_7, TAG 55_2, DAG 34_1, and PC 32_0) and constructed a score, weighted with the beta coefficients in PREDIMED (Score2). Higher Score2 was consistently associated with increased HF risk in PREDIMED (RR per SD 1.39, 95%CI 1.24–1.57) and in EPIC-Potsdam (RR per SD 1.55, 95%CI 1.25–1.93) (Table 5). The association of the replicated Score2 in PREDIMED was only slightly attenuated after adjustment for all these risk factors (RR per SD 1.30, 95%CI 1.14, 1.47). This modest effect attenuation was mostly attributable to adjustment for baseline BMI, and none of the tests for multiplicative interaction was significant. In EPIC-Potsdam, adjustment of the Score2-HF risk association for the analogous covariables had comparably small impact on the effect estimate (RR per SD 1.46, 95% CI 1.14, 1.86) (Table 5).
Sensitivity analyses.
We adjusted the scores for standard lipid markers (prevalence of dyslipidemia in PREDIMED, total and HDL-cholesterol and total TAGs in EPIC-Potsdam), which had no appreciable effect on the risk estimates (Online Table VII). We also evaluated whether the marked difference in effect sizes between Score1 (based on all within-cluster hits in PREDIMED) and Score2 (based only on the lipids that were available and replicated in EPIC-Potsdam) was rather attributable to the lack of information (lipids not available in EPIC-Potsdam) or to the failed replications. To this end, we used the same workflow as for Score1 (10-fold cross-validated elastic net regression) to construct another score in PREDIMED, but removing EA 16:1, SM 16:1, DAG 36_0, PC-PLG 36_2, PE-PLG 34_3, PE-PLG 42_11 from the lipid set because we had no corresponding measurements in EPIC-Potsdam. In PREDIMED, the cross-validated lipid score based on lipids available in EPIC (regardless of the replication results) was associated with a RR for HF of 1.46 (1.30, 1.63) per SD, a rather minor difference compared to the risk estimate of the replicated Score2.
Moreover, we assessed the HF risk association of the main lipid markers (CER 16:0, PC 32_0, Score1, and Score2) across strata according to diet intervention group. For all selected lipid markers, the HF risk estimates were very similar across the intervention groups (Online Table VIII). Excluding the 35 participants who developed HF during the first three years of follow-up in PREDIMED (lag-time analysis) had no appreciable effects on the HF risk estimates of the main lipid markers. The confounder-adjusted HRs per SD (95%CI) were: CER 16:0, 1.27 (1.12, 1.45); PC 32_0, 1.24 (1.07, 1.44); Score1, 2.21 (1.80, 2.72) and Score2 1.24 (1.07, 1.42). Excluding the 21 PREDIMED-participants who developed HF after an acute myocardial infarction had negligible effects, rendering the following confounder-adjusted HR per SD (95%CI): CER 16:0, 1.24 (1.08, 1.43); PC 32_0, 1.28 (1.10, 1.48); Score1, 2.20 (1.82, 2.67) and Score2 1.30 (1.13, 1.49).
DISCUSSION
In this prospective HF case-control study nested within the PREDIMED trial, we assessed baseline concentrations of 216 targeted lipids. The associations of CER 16:0 and PC 32_0 with higher HF risk were robust against confounder adjustment and externally replicated in the EPIC-Potsdam cohort, where auxiliary analyses attributed the latter association to PC 16:0/16:0. We also discovered eight HF-related lipidomics network clusters in PREDIMED: EA 16:1+MAG 16:1; seven SLs; two long-chain SFA-containing PCs; six PLs; five PLGs; three DAGs from two distinct clusters; and four TAGs. Albeit not all the lipids were available, for six out of eight patterns, the best-matching models also significantly predicted HF incidence in EPIC-Potsdam. After adjustment for other HF risk factors, a cross-validated lipid score based on all the significantly HF incidence-related lipids in PREDIMED was associated with a 133% (95%CI 93%−185%) higher HF risk per SD. A lipid score restricted to the externally available and replicated lipids was also associated with HF incidence, 30% (95%CI 14%−47%) higher risk per SD in PREDIMED and 46% (95%CI 17%−82%) higher risk per SD in EPIC-Potsdam.
Our results linked enrichment of palmitate-containing lipids (PC16:0/16:0 and CER16:0) to higher HF risk, robust against adjustment for other risk factors for HF and classical blood lipid markers. The simultaneous analysis of interconnected lipid groups corroborated these findings and further identified metabolites with palmitoleic acid (C16:1) and stearate (C18:0) in several lipid classes (SMs, PCs, PLGs, and DAGs) as markers for higher HF risk. To our knowledge, this is the first study that links a lipidomics screen in disease-free participants to future HF incidence. Previous targeted assessments of a limited number of SLs consistently detected elevated HF risk with high CER 16:0 plasma concentrations (28, 29). However, these studies did not measure the SMs 16:1 and 18:0, which we identified as complementary high-risk-markers. A FA-profiling study in men linked high relative palmitate abundance and a high C16:1/C16:0-ratio in plasma phospholipids to higher HF risk (30). We assessed higher HF-risk with high concentrations of C16:0- and C16:1-containing lipids, namely PCs 16:0/16:0 and 18:1/16:1 and DAG 16:1/18:0, which is overall coherent but more specific than the previous reports.
We observed associations of very-long-chain-FA-containing SLs (CER 24:1, SM 24:0, and SM 24:1) with lower HF risk in PREDIMED when models were mutually adjusted for high-risk SLs. Consistently, higher plasma concentration of CER 24:0 (28) and SM 24:0 (29) were associated with lower HF risk in other studies, also dependent on simultaneously accounting for the high-risk SLs, and high relative lignoceric acid (C24:0) abundance in total circulating plasma lipids was related to lower HF risk (31). However, the SL-studies did not measure SM C18:1, for which we replicated the inverse HF risk association in the EPIC-Potsdam cohort. On the pattern-level, we further discovered and replicated lower HF risk associated with the polyunsaturated FA (PUFA)-containing lipids PE-PLG 18:1/22:6 and TAG 58_7-fa16:0. Several lines of research link PUFAs to HF incidence (32–34), but the evidence was hitherto not specific in terms of their localization in lipid classes. Metabolites from the same lipid classes with shorter acyl chains and fewer double-bonds (PE-PLG 18:0/18:1; TAG 55_2-fa18:1, TAG 56_5-fa16:0) were associated with higher HF risk in PREDIMED and EPIC-Potsdam. To our knowledge, this was the first analysis that linked PLG- and TAG-profiles to HF incidence.
Distinct reasons may explain why some PREDIMED-findings were not replicated in EPIC-Potsdam. The probability of false discoveries among the replicated associations is very low, but a considerable fraction of the non-replicated marginally significant HF risk-associations in PREDIMED were probably due to chance. However, some of the divergent associations may also reflect differences in the source populations between both cohorts, for example, in terms of genetic background, diet, lifestyle, and socio-economic background (as reflected in the marked differences in the distribution of highest educational attainment); or differences between the applied lipidomics platforms.
The different platforms certainly precluded the replication of lipids that were not assessed in EPIC-Potsdam. For example, EA 16:1, SM 16:1, DAG 36_0, and several PLGs were integral components of the HF-related lipid clusters in PREDIMED with no available measurements in EPIC-Potsdam. In the replication-analyses, this may have also compromised the sensitivity for the associations of other lipids within the same pattern. Overall, it is remarkable that, despite the different lipidomics platform used in EPIC-Potsdam, most model-associations were replicated on the lipid pattern-level.
We summarized the joint effect of the selected lipid-patterns on HF-risk in weighted scores. Score1 was based on all the significant within-cluster predictors in PREDIMED, deriving beta weights with cross-validated elastic net regression to avoid overfitting. While this should have produced internally robust risk estimates, we could not test the external generalizability of the very strong association (138% higher HF risk per SD). Score2 relied on the replicated lipids only, a conservative approach. In both cohorts, higher Score2 was associated with a markedly increased HF risk, 39% per SD in PREDIMED, and 55% per SD in EPIC-Potsdam. The slightly stronger estimate for Score2 in EPIC-Potsdam suggests advantages of the more specific measurements for isomeric lipids for risk assessment in the replication cohort.
Our sensitivity analyses revealed that the marked difference in risk estimates between Score1 and Score2 was almost entirely attributable to removing the six lipids that were not assessed in EPIC-Potsdam. These results underpin that our multi-marker approach relied on simultaneous information on all components. In lipid metabolism, metabolically closely related compounds can have opposite systemic effects, for example initiating versus resolving inflammatory responses, likely producing divergent disease associations of closely correlated metabolites. Such interdependent risk relations are only detectable with statistical workflows that consider interrelated lipids simultaneously.
Independent of the underlying cause, decreased FA-oxidation is a major metabolic characteristic of the failing heart (35). The selection of palmitate-containing lipids as markers for high HF risk is coherent with cardiac lipotoxic effects of palmitate in mechanistic studies (36). The plasma lipidome might integrate information on the heart’s metabolic flexibility and the circulating substrate availability, possibly reflecting the myocardial susceptibility to lipotoxic damage. Furthermore, palmitate-containing lipid metabolites, particularly CER 16:0, were implicated in systemic inflammatory and metabolic signaling (37), which regulate cardiac remodeling and contractile mechanics and are, therefore, critical in heart failure progression (38). Plasma lipid profiles were also linked to T2D and CAD (9, 10, 39, 40), which are among the major underlying conditions of HF. For example, we have previously shown an association of CER 16:0 with higher CVD-risk in PREDIMED (41).Therefore, part of the HF-related lipid profiles may reflect general cardiometabolic health, which is linked to HF risk through mediating clinical conditions.
From a translational point of view, lipid-based multi-biomarker panels may capture information on common etiological mechanism of several important cardiometabolic endpoints. In addition to the above-discussed link to cardiometabolic risk, lipid profiles reflect individual traits including genome and microbiome and environmental disease determinants including the diet. The PREDIMED trial and other studies have demonstrated that lipid profiles are modifiable through dietary interventions (41, 42). However, the Mediterranean diet intervention in the PREDIMED study did not substantially affect HF risk and, concordantly, the risk associations of the selected lipid profiles did not differ between the dietary intervention arms. Studies are warranted that investigate if other diet and lifestyle interventions affect the HF-associated lipid metabolites, and in turn possibly prevent HF incidence (16).
Several limitations of this study must be acknowledged. The HF diagnosis was based on the 2005 (time of study design) guidelines of the European Society of Cardiology (15, 16). Ever since, the diagnostic criteria for HF have been refined, and future studies should investigate whether the updated case definition affects the risk association of the selected lipid metabolites. However, more specific HF detection is expected to strengthen true risk associations. It would also be interesting to evaluate if the prospective associations for the selected lipids differ between types of HF (for example, preserved vs. reduced ejection fraction), but this information was not available for many of HF cases in the PREDIMED trial. Moreover, we used different lipidomics platforms in the discovery and the replication cohort. The effect attenuation of the replicated score due to missing information on some lipids was discussed above. But for the remaining lipids, our results suggest the robustness of measurements despite different laboratories and analytical platforms, both important preconditions for clinical implementation. Still, replicability studies are warranted to systematically investigate the comparability of lipidomics measurements between two of the most frequently used platforms. Our study relied on moderate sample-sizes. We used a statistical workflow that included resampling and cross-validation to avoid overfitting in the discovery cohort, and we externally validated our main findings, indicating their generalizability. We also increased the sensitivity for true associations by considering data-driven lipid groups. However, similar studies with higher statistical power may find additional robust HF-risk-associations among the lipids that we analyzed. Moreover, investigation of the generalizability of our findings to other races and ethnicities is warranted as we only demonstrated external validity of our findings in middle-aged to older, white populations.
In summary, we identified specific lipid metabolites and lipidomics patterns that were significantly associated with future HF risk in two independent cohorts. The available evidence on the role of lipid metabolism and lipotoxicity in HF pathogenesis suggests that the identified lipidomics traits reflect early molecular mechanisms of HF development. Another possible underlying mechanism connects the identified lipids to HF incidence through mediating cardiometabolic conditions, including T2D and CAD. Our results suggest that lipid profiling may provide novel tools for HF risk prediction and risk stratification and thus facilitate personalized prevention efforts.
Supplementary Material
NOVELTY AND SIGNIFICANCE.
What Is Known?
Disturbed lipid metabolism was implicated in the pathogenesis of chronic heart failure through in vitro studies and animal models.
Plasma lipidomics profiles were associated with prevalent heart failure in cross-sectional studies.
What New Information Does This Article Contribute?
Baseline plasma concentrations of two palmitate-containing lipid metabolites (phosphatidylcholine C16:0/C16:0 and ceramide C16:0) were associated with a higher risk of subsequent heart failure.
Lipidomics network clusters that included sphingolipids, two diacyl phosphatidylcholine-containing clusters, plasmalogens, diacylglycerols, and triacylglycerols were associated with the risk of developing heart failure.
Lipid scores based on the clusters were associated with a markedly higher heart failure risk.
A link between plasma lipid profiles and heart failure risk is supported by cross-sectional investigations and experimental evidence. However, prospective human studies that relate comprehensive lipidomics screens to the risk of developing heart failure were lacking. Herein, we showed that baseline plasma concentrations of diacyl phosphatidylcholine C16:0/C16:0 and ceramide C16:0 were associated with higher heart failure risk in two independent cohorts. We also demonstrated that considering a data-driven lipidomics network revealed additional associations of interrelated lipid clusters with heart failure incidence. In clusters of sphingolipids, diacyl phosphatidylcholines, plasmalogens, diacylglycerols, and triacylglycerols, we detected patterns of lipid metabolites that were significantly associated with heart failure risk. The weighted combination of the selected lipid predictors resulted in scores that were strongly associated with heart failure risk. Our study establishes for the first time a link between circulating lipidomics profiles at baseline and subsequent occurrence of heart failure. Our results encourage mechanistic studies into the biological role of the selected lipid predictors in heart failure etiology and suggest that lipid metabolites may improve risk prediction and facilitate risk stratification for targeted heart failure prevention.
ACKNOWLEDGMENTS
We thank all participants of the PREDIMED trial and the EPIC-Potsdam study for their collaboration. We acknowledge the excellent assistance of the PREDIMED personnel, and the Human Study Center at the German Institute for Human Nutrition Potsdam-Rehbruecke, led by Dr. Manuela Bergmann.
SOURCES OF FUNDING
This study is supported by the NIH grants R01HL118264 and R01DK102896.The PREDIMED trial was supported by the official funding agency for biomedical research of the Spanish government, the Instituto de Salud Carlos III, through grants provided to research networks specifically developed for the trial (RTIC G03/140 to R.E. and RTIC RD 06/0045 to Dr. Miguel A. Martínez-González) and through the Centro de Investigación Biomédica en Red de Fisiopatología de la Obesidad y Nutrición and by grants from: the Centro Nacional de Investigaciones Cardiovasculares (CNIC 06/2007); the Fondo de Investigación Sanitaria-Fondo Europeo de Desarrollo Regional (PI04-2239, PI 05/2584, CP06/00100, PI07/0240, PI07/1138, PI07/0954, PI 07/0473, PI10/01407, PI10/02658, PI11/01647, P11/02505 and PI13/00462); the Ministerio de Ciencia e Investigación (AGL-2009-13906-C02 and AGL2010-22319-C03); the Fundación Mapfre 2010, Consejería de Salud de la Junta de Andalucía (PI0105/2007); the Public Health Division of the Department of Health of the Autonomous Government of Catalonia, Generalitat Valenciana (ACOMP06109, GVA-COMP2010-181, GVACOMP2011-151, CS2010-AP-111, and CS2011-AP-042); and the Regional Government of Navarra (P27/2011).
The recruitment phase of the EPIC-Potsdam Study was supported by the Federal Ministry of Science, Germany (01 EA 9401) and the European Union (SOC 95201408 05F02). The follow-up of the EPIC-Potsdam Study was supported by German Cancer Aid (70-2488-Ha I) and the European Community (SOC 98200769 05F02). The lipidomics measurements in EPIC-Potsdam were supported by a grant from the German Ministry of Education and Research (BMBF) and the European Union in the frame of the Joint Programming Initiative ‘A Healthy Diet for a Healthy Life’ to the Fatty Acid Metabolism - Interlinking Diet with Cardiometabolic Health (FAME)-consortium (01EA1704) and a grant from the German Ministry of Education and Research (BMBF) and the State of Brandenburg (DZD grant 82DZD00302).
J. Salas-Salvadó, gratefully acknowledges the financial support by ICREA under the ICREA Academia programme. Clemens Wittenbecher was supported by an individual fellowship from the German Research Foundation (DFG).
Nonstandard Abbreviations
- CAD
coronary artery disease
- CE
cholesterol ester
- CER
ceramide
- CVD
cardiovascular disease
- DAG
diacylglycerol
- EA
ethanolamide
- EPIC
European Prospective Investigation into Cancer and Nutrition
- FA
fatty acid
- FDR
false discovery rate
- HF
heart failure
- MAG
monoacylglycerol
- PC
diacyl-phosphatidylcholine
- PE
phosphatidylethanolamine
- PLG
plasmalogen
- PL
glycerophospholipid
- PREDIMED
PREvención con DIeta MEDiterránea
- PUFA
polyunsaturated fatty acid
- SFA
saturated fatty acid
- SL
sphingolipid
- SM
sphingomyelin
- T2D
type 2 diabetes
- TAG
triacylglycerol
Footnotes
Publisher's Disclaimer: This article is published in its accepted form. It has not been copyedited and has not appeared in an issue of the journal. Preparation for inclusion in an issue of Circulation Research involves copyediting, typesetting, proofreading, and author review, which may lead to differences between this accepted version of the manuscript and the final, published version.
ISRCTN Registry: https://doi.org/10.1186/ISRCTN35739639.
DISCLOSURES
None.
SUPPLEMENTAL MATERIALS
Expanded Materials & Methods (Online Notes I-IV)
Online Tables I-VIII
Online Figures I and II
REFERENCES
- 1.Savarese G, Lund LH. Global Public Health Burden of Heart Failure. Card Fail Rev. 2017;3:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas S, Rich MW. Epidemiology, pathophysiology, and prognosis of heart failure in the elderly. Heart Fail Clin. 2007;3:381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziaeian B, Fonarow GC. Epidemiology and aetiology of heart failure. Nat Rev Cardiol. 2016;13:368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahle BW, Owen AJ, Chin KL, Reid CM. Risk Prediction Models for Incident Heart Failure: A Systematic Review of Methodology and Model Performance. J Card Fail. 2017;23:680–687. [DOI] [PubMed] [Google Scholar]
- 5.Butler J, Kalogeropoulos A, Georgiopoulou V, et al. Incident heart failure prediction in the elderly: the health ABC heart failure score. Circ Heart Fail. 2008;1:125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Savic-Radojevic A, Pljesa-Ercegovac M, Matic M, Simic D, Radovanovic S, Simic T. Novel Biomarkers of Heart Failure. Adv Clin Chem. 2017;79:93–152. [DOI] [PubMed] [Google Scholar]
- 7.Goldberg IJ, Trent CM, Schulze PC. Lipid metabolism and toxicity in the heart. Cell Metab. 2012;15:805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halade GV, Kain V, Tourki B, Jadapalli JK. Lipoxygenase drives lipidomic and metabolic reprogramming in ischemic heart failure. Metabolism. 2019;96:22–32. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Canela M, Hruby A, Clish CB, Liang L, Martinez-Gonzalez MA, Hu FB. Comprehensive Metabolomic Profiling and Incident Cardiovascular Disease: A Systematic Review. J Am Heart Assoc. 2017;6:e005705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guasch-Ferre M, Hruby A, Toledo E, Clish CB, Martinez-Gonzalez MA, Salas-Salvado J, Hu FB. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care. 2016;39:833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estruch R, Ros E, Salas-Salvado J, et al. Primary Prevention of Cardiovascular Disease with a Mediterranean Diet Supplemented with Extra-Virgin Olive Oil or Nuts. N Engl J Med. 2018;378:e34. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Gonzalez MA, Corella D, Salas-Salvado J, et al. Cohort profile: design and methods of the PREDIMED study. International journal of epidemiology. 2012;41:377–85. [DOI] [PubMed] [Google Scholar]
- 13.Boeing H, Korfmann A, Bergmann MM. Recruitment procedures of EPIC-Germany. European Investigation into Cancer and Nutrition. Annals of nutrition & metabolism. 1999;43:205–215. [DOI] [PubMed] [Google Scholar]
- 14.Schulze MB, Hoffmann K, Boeing H, et al. An Accurate Risk Score Based on Anthropometric, Dietary, and Lifestyle Factors to Predict the Development of Type 2 Diabetes. Diabetes Care. 2007;30:510–515. [DOI] [PubMed] [Google Scholar]
- 15.Nieminen MS, Bohm M, Cowie MR, et al. Executive summary of the guidelines on the diagnosis and treatment of acute heart failure: the Task Force on Acute Heart Failure of the European Society of Cardiology. Eur Heart J. 2005;26(4):384–416. [DOI] [PubMed] [Google Scholar]
- 16.Papadaki A, Martinez-Gonzalez MA, Alonso-Gomez A, et al. Mediterranean diet and risk of heart failure: results from the PREDIMED randomized controlled trial. Eur J Heart Fail. 2017;19:1179–1185. [DOI] [PubMed] [Google Scholar]
- 17.Hertz-Picciotto I, Rockhill B. Validity and efficiency of approximation methods for tied survival times in Cox regression. Biometrics. 1997;53:1151–1156. [PubMed] [Google Scholar]
- 18.Wooldridge JM. Selection corrections for panel data models under conditional mean independence assumptions. Journal of Econometrics. 1995;68:115–132. [Google Scholar]
- 19.Maathuis MH, Colombo D, Kalisch M, Bühlmann P. Predicting causal effects in large-scale systems from observational data. Nature Methods. 2010;7:247–248. [DOI] [PubMed] [Google Scholar]
- 20.Wittenbecher C Linking whole-grain bread, coffee, and red meat to the risk of type 2 diabetes: Using metabolomics networks to infer potential biological mechanisms [Monograph]. Potsdam: Potsdam University; 2017. [Google Scholar]
- 21.Iqbal K, Dietrich S, Wittenbecher C, et al. Comparison of metabolite networks from four German population-based studies. International journal of epidemiology. 2018;47:2070–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanfill B, Reehl S, Bramer L, Nakayasu ES, Rich SS, Metz TO, Rewers M, Webb-Robertson BJ, Group TS. Extending Classification Algorithms to Case-Control Studies. Biomed Eng Comput Biol. 2019;10:1179597219858954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breiman L Random forests. Machine learning. 2001;45:5–32. [Google Scholar]
- 24.Nicodemus KK, Malley JD, Strobl C, Ziegler A. The behaviour of random forest permutation-based variable importance measures under predictor correlation. BMC Bioinformatics. 2010;11:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrell F Regression Modeling Strategies: With Applications to Linear Models, Logistic Regression and Survival Analysis. 2001. New York Springer-Verlag New York. Inc. [Google Scholar]
- 26.Ogutu JO, Schulz-Streeck T, Piepho H-P, editors. Genomic selection using regularized linear regression models: ridge regression, lasso, elastic net and their extensions BMC proceedings; 2012: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal statistical society: series B (Methodological). 1995;57:289–300. [Google Scholar]
- 28.Peterson LR, Xanthakis V, Duncan MS, et al. Ceramide Remodeling and Risk of Cardiovascular Events and Mortality. J Am Heart Assoc. 2018;7:e007931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemaitre RN, Jensen PN, Hoofnagle A, et al. Plasma Ceramides and Sphingomyelins in Relation to Heart Failure Risk. Circ Heart Fail. 2019;12:e005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Djousse L, Weir NL, Hanson NQ, Tsai MY, Gaziano JM. Plasma phospholipid concentration of cis-palmitoleic acid and risk of heart failure. Circ Heart Fail. 2012;5:703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaitre RN, McKnight B, Sotoodehnia N, et al. Circulating Very Long-Chain Saturated Fatty Acids and Heart Failure: The Cardiovascular Health Study. J Am Heart Assoc. 2018;7:e010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Block RC, Liu L, Herrington DM, Huang S, Tsai MY, O’Connell TD, Shearer GC. Predicting Risk for Incident Heart Failure With Omega-3 Fatty Acids: From MESA. JACC Heart Fail. 2019;7:651–661. [DOI] [PubMed] [Google Scholar]
- 33.Handelsman Y, Shapiro MD. Triglycerides, Atherosclerosis, and Cardiovascular Outcome Studies: Focus on Omega-3 Fatty Acids. Endocr Pract. 2017;23:100–112. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Y, Yu B, Alexander D, Manolio TA, Aguilar D, Coresh J, Heiss G, Boerwinkle E, Nettleton JA. Associations between metabolomic compounds and incident heart failure among African Americans: the ARIC Study. American journal of epidemiology. 2013;178:534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato T, Niizuma S, Inuzuka Y, et al. Analysis of metabolic remodeling in compensated left ventricular hypertrophy and heart failure. Circ Heart Fail. 2010;3:420–430. [DOI] [PubMed] [Google Scholar]
- 36.Akoumi A, Haffar T, Mousterji M, Kiss RS, Bousette N. Palmitate mediated diacylglycerol accumulation causes endoplasmic reticulum stress, Plin2 degradation, and cell death in H9C2 cardiomyoblasts. Exp Cell Res. 2017;354:85–94. [DOI] [PubMed] [Google Scholar]
- 37.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schirone L, Forte M, Palmerio S, et al. A Review of the Molecular Mechanisms Underlying the Development and Progression of Cardiac Remodeling. Oxid Med Cell Longev. 2017;2017:3920195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fretts AM, Jensen PN, Hoofnagle A, et al. Plasma Ceramide Species Are Associated with Diabetes Risk in Participants of the Strong Heart Study. J Nutr. 2020;150:1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hilvo M, Meikle PJ, Pedersen ER, et al. Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur Heart J. 2020;41:371–380. [DOI] [PubMed] [Google Scholar]
- 41.Wang DD, Toledo E, Hruby A, et al. Plasma Ceramides, Mediterranean Diet, and Incident Cardiovascular Disease in the PREDIMED Trial (Prevencion con Dieta Mediterranea). Circulation. 2017;135:2028–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guasch-Ferre M, Bhupathiraju SN, Hu FB. Use of Metabolomics in Improving Assessment of Dietary Intake. Clin Chem. 2018;64:82–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors will be happy to provide access to the PREDIMED dataset (including data dictionaries), making possible the replication of the main analyses used for the present article. Due to the restrictions imposed by the Informed Consent and the Institutional Review Board, bona fide investigators interested in analyzing the PREDIMED dataset used for the present article may submit a brief proposal and statistical analysis plan to the corresponding authors. Upon approval from the PREDIMED Steering Committee and Institutional Review Boards, the data will be made available to them using an onsite secure access data enclave


