Significance
Photosystem II (PSII) is a light-driven water:plastoquinone oxidoreductase in oxygenic photosynthetic organisms. Several inactive PSII intermediates are involved in the biogenesis of the multisubunit PSII complexes as well as in their repair after unavoidable oxidative damage targeted to PSII under light. Psb27 is one of the assembly factors associated with inactive PSII intermediate and plays important roles in the biogenesis/repair of PSII. Here, we report the structure of a dimeric Psb27-PSII from a thermophilic cyanobacterium Thermosynechococcus vulcanus by cryo-electron microscopy. Our results reveal the location and binding properties of Psb27 in the intermediate PSII and show the structural differences between the intermediate and native PSII. These results provide important clues for the roles of Psb27 in the biogenesis/repair of PSII.
Keywords: photosystem II, cryo-EM, assembly, repair, Psb27
Abstract
Photosystem II (PSII) is a multisubunit pigment-protein complex and catalyzes light-driven water oxidation, leading to the conversion of light energy into chemical energy and the release of molecular oxygen. Psb27 is a small thylakoid lumen-localized protein known to serve as an assembly factor for the biogenesis and repair of the PSII complex. The exact location and binding fashion of Psb27 in the intermediate PSII remain elusive. Here, we report the structure of a dimeric Psb27-PSII complex purified from a psbV deletion mutant (ΔPsbV) of the cyanobacterium Thermosynechococcus vulcanus, solved by cryo-electron microscopy. Our structure showed that Psb27 is associated with CP43 at the luminal side, with specific interactions formed between Helix 2 and Helix 3 of Psb27 and a loop region between Helix 3 and Helix 4 of CP43 (loop C) as well as the large, lumen-exposed and hydrophilic E-loop of CP43. The binding of Psb27 imposes some conflicts with the N-terminal region of PsbO and also induces some conformational changes in CP43, CP47, and D2. This makes PsbO unable to bind in the Psb27-PSII. Conformational changes also occurred in D1, PsbE, PsbF, and PsbZ; this, together with the conformational changes occurred in CP43, CP47, and D2, may prevent the binding of PsbU and induce dissociation of PsbJ. This structural information provides important insights into the regulation mechanism of Psb27 in the biogenesis and repair of PSII.
Photosystem II (PSII) is a multisubunit membrane protein complex located in the thylakoid membranes of cyanobacteria, various algae, and higher plants. It catalyzes the light-driven water oxidation and plastoquinone reduction (1–3), leading to the conversion of light energy into chemical energy and the evolution of oxygen, which are essential to sustain almost all life activities on the earth. A functional PSII core complex from cyanobacteria contains 20 protein subunits, including 17 intrinsic, transmembrane protein subunits and 3 membrane extrinsic subunits (3–5). The intrinsic subunits of the core, such as D1, D2, CP43, CP47, PsbE, PsbF, and PsbI, are highly conserved in all oxygenic photosynthetic organisms, whereas extrinsic subunits exhibit a large variety in different photosynthetic organisms (6–9). The three extrinsic subunits in cyanobacteria are PsbO, PsbU, and PsbV (10, 11), whereas red algae contain PsbQ′ (12, 13) and diatom PSII contains PsbQ′ plus Psb31 (14), in addition to the three cyanobacterial extrinsic proteins. These extrinsic proteins are changed to PsbO, PsbP, and PsbQ in the green lineage organisms (6–9, 15). In addition, CyanoP and CyanoQ are present in cyanobacteria; they have low sequence similarities with those of the green lineage organisms and may have regulatory functions in cyanobacterial PSII (16). The X-ray crystal structure or cryo-electron microscopy (cryo-EM) structure of PSII from different organisms has been reported (4, 5, 17–22); they revealed the locations of most of these subunits and a number of cofactors, including the Mn4CaO5 cluster, which catalyzes the water-splitting reaction.
In addition to the stable components of PSII mentioned above, there are several subunits that associate with the intermediate PSII complex during its assembly and are detached from PSII once it is matured (23–27). One such component is Psb27, which has been found to be associated with inactive cyanobacterial PSII that lacks the extrinsic proteins PsbO, PsbU, and PsbV (28–30). Psb27 is an ∼11-kDa, thylakoid lumen–localized lipoprotein and was found to facilitate the assembly of the Mn4CaO5 cluster by preventing binding of the extrinsic proteins before assembly of the Mn4CaO5 cluster (29–31). Psb27 was also found to play a significant role in the repair cycle of PSII after the damage of D1, which occurs under strong light illumination and is termed photodamage. In the cyanobacterium Synechocystis sp. PCC 6803, a psb27 deletion mutant showed impaired PSII repair activity after high-light photoinhibition, and the repair requires the presence of Psb27 (30, 32). The Psb27 protein was initially found to associate with the PSII monomer during assembly, but it was also found to associate with the PSII dimer, and its association provides a selective advantage for cyanobacterial cells under stress conditions, such as cold stress (33). In Arabidopsis thaliana, two different copies of Psb27 are present. One of them is required for D1 processing and involved in PSII biogenesis and stabilization, whereas the other copy played important roles in the efficient repair of photodamaged PSII and in enabling plants to adapt to fluctuating light intensity (34–36).
In the past few years, many different types of Psb27-PSII intermediates have been purified from cyanobacteria in a number of different biochemical preparations unable to evolve oxygen (30, 33, 37–40). These inactive Psb27-PSII complexes are deficient in the Mn4CaO5 cluster and three extrinsic subunits: PsbO, PsbV, and PsbU. The structure of Psb27 has been solved by both solution NMR (41, 42) and X-ray crystallography (43, 44), and its association with PSII assembly intermediate has been studied by chemical cross-linking analysis and mass spectrometry (45–48). These studies suggested that Psb27 binds to the large extrinsic E-loop of CP43 at the luminal side; however, there are discrepancies regarding the detailed binding sites of Psb27 in PSII (45–48). In addition, CP47 as well as the C terminuses of D1 and D2 were also reported to be the binding partners for Psb27 (45). Furthermore, it was reported that Psb27 partially occupies the binding sites of PsbO and PsbV, thereby preventing the binding of these extrinsic subunits (47). However, the exact binding site and functions of Psb27 is unknown since the structure of the Psb27-PSII complex has not been solved. We purified a Psb27-PSII complex from a psbV-deletion mutant (ΔPsbV) of the thermophilic cyanobacterium Thermosynechococcus vulcanus and analyzed its structure using single particle cryo-EM at an overall resolution of 3.78 Å. The structural location of Psb27 within the PSII intermediate, together with a number of structural differences between the intermediate and mature PSII, are revealed, which provide important insights into the roles of Psb27 in the biogenesis and repair cycle of PSII.
Results
Purification and Properties of the Dimeric Psb27-PSII Complex.
We made a mutant lacking PsbV in the cyanobacterium T. vulcanus, and purified PSII from the mutant by column chromatography with a Q Sepharose High Performance column eluted with a linear gradient of NaCl from 170 to 300 mM (Materials and Methods and SI Appendix, Fig. S1A) (49). Our previous studies have shown that PSII is eluted from the column in three fractions. The first two fractions eluted at around 420 mL and 650 mL elution volume shown in SI Appendix, Fig. S1A are PSII monomers without any extrinsic proteins but with the Psb27 and Psb28 bound, and the third one labeled with a vertical bar is the Psb27-PSII dimer that has no extrinsic proteins but binds Psb27 (49). This Psb27-PSII has no oxygen-evolving activity and may be the same Psb27-PSII dimer isolated by Grasse et al. (33), as both Psb27-PSII did not bind the three extrinsic proteins. No Psb28 was found in the purified Psb27-PSII dimers. It was also shown that the ΔPsbV contains PsbO in their cells, but most of PsbO was released from PSII during purification, and the oxygen-evolving activity of the mutant cells is around 50% of the wild-type strain (49). The Psb27-PSII obtained was characterized by size-exclusion chromatography, blue native polyacrylamide gel electrophoresis (PAGE), sodium dodecyl sulfate-PAGE (SDS-PAGE), and Western blotting (SI Appendix, Fig. S1 B–E). These analyses indicated that this complex exists as a homogenous dimer and is composed of the major transmembrane subunits of PSII core plus Psb27 but lacked the three extrinsic subunits PsbO, PsbV, and PsbU that were observed in the crystal structure of native PSII (4, 5).
From the SDS-PAGE analysis, the D1 subunit of the purified PSII exists as a mature form of the native PSII. To confirm either the presence or absence of a damaged D1, we analyzed the length of the D1 protein with mass spectrometry. Two types of the D1 protein with different C-terminal sequences were detected. One major type of the C-terminal sequence is the same as that of the mature D1 protein of PsbA1, ending in the sequence of NAHNFPLDLA (SI Appendix, Fig. S2A), whereas the other one is minor in amount and ends in ANLGMEVMHE (SI Appendix, Fig. S2B). This second D1 protein lacks the C-terminal amino acid sequence from Arg334 to Ala344. Although the second D1 protein form occupies around 1% of the total D1 protein, it may represent the sign of the D1 protein degradation and/or damage.
Overall Structure of the Dimeric Psb27-PSII Complex.
We collected 4,899 cryo-EM micrographs of the purified Psb27-PSII dimer and picked 1,783,091 particles for subsequent data processing (SI Appendix, Figs. S3 and S4). Although some of these particles may contain PSII without Psb27, after two-dimensional (2D) and three-dimensional (3D) classifications (SI Appendix, Figs. S3 and S4 and Table S1), particles with a major population were selected and processed, which yielded a cryo-EM map with an overall resolution of 3.78 Å for the whole Psb27-PSII complex (SI Appendix, Fig. S3).
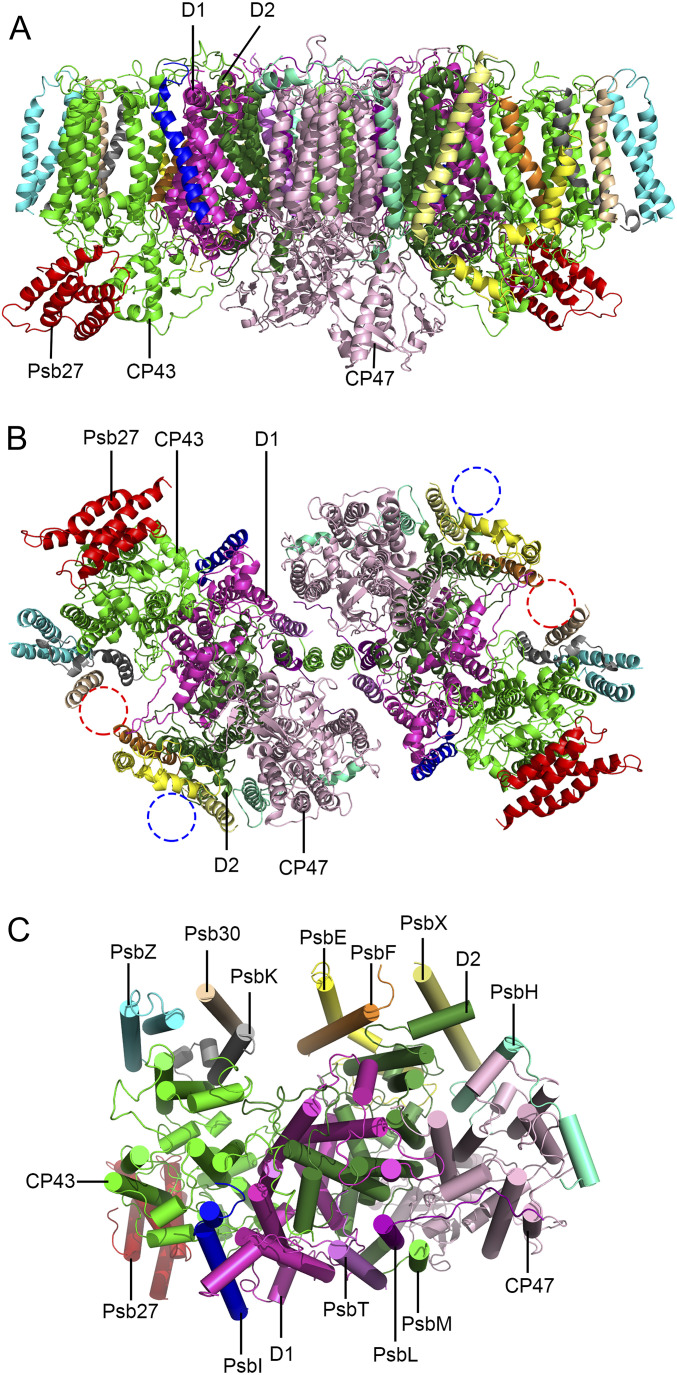
The overall structure of this Psb27-PSII complex contains two almost identical PSII monomers, forming a homodimer with a two-fold symmetry (Fig. 1). In the structure, each PSII monomer contains 16 protein subunits. These 16 subunits include 4 large intrinsic transmembrane subunits (D1, D2, CP43, and CP47), 11 low-molecular-mass intrinsic transmembrane subunits (PsbE, PsbF, PsbH, PsbI, PsbK, PsbL, PsbM, PsbT, PsbX, Psb30, and PsbZ), and 1 extrinsic subunit (Psb27) attached at the luminal surface (Fig. 1 and SI Appendix, Fig. S5). The rmsd values between the structures of the dimeric Psb27-PSII core and the native PSII (not including the extrinsic proteins) from the same cyanobacterium (Protein Data Bank [PDB] code: 3WU2) is 0.984 Å, suggesting a rather highly conserved core structure.
Fig. 1.
Overall structure of the Psb27-PSII complex from T. vulcanus. (A) Side view of the Psb27-PSII dimer along the membrane plane. (B) Top view of the Psb27-PSII dimer from the luminal side. The missing subunits PsbJ and PsbY are highlighted with a red and blue cycles, respectively. (C) Top view of a Psb27-PSII monomer from the stromal side. Subunit composition of the Psb27-PSII monomer is shown. Transmembrane α-helices are represented as ribbons (A and B) and cylinders (C), respectively.
The extrinsic subunits PsbO, PsbV, and PsbU present in the native cyanobacterial PSII (4, 5) are absent in the cryo-EM map of the current Psb27-PSII complex, consistent with the above SDS-PAGE analysis. PsbJ and PsbY are also not found in the cryo-EM structure, suggesting that they are loosely associated with the PSII core and lost during the sample purification (SI Appendix, Fig. S1F). The loose association of PsbY with PSII has been shown in native PSII (4, 5), whereas the loss of PsbJ may be caused by the association of Psb27 and/or the deletion of PsbV. In addition to the protein subunits, we identified 35 chlorophylls, 11 carotenoids, 2 pheophytins, 1 haem, and a number of lipids and other cofactors in each Psb27-PSII monomer (SI Appendix, Fig. S6 and Table S2). No electron density corresponding to the Mn4CaO5 cluster is observed, indicating the premature nature of the intermediate PSII and also explaining the loss of the oxygen-evolving activity.
The Interactions between Psb27 and Photosystem II Core.
The cryo-EM structure of Psb27 in the Psb27-PSII complex is a right-handed four-helix bundle with an up-down-up-down topology, which is nearly identical to the structures of recombinant Psb27 reported for cyanobacteria (41–43) or higher plant (44) and solved by NMR or X-ray crystallography (SI Appendix, Fig. S7). The Psb27 also contains a flexible N-terminal tail of seven residues; however, the so-called lipid modification that was found on the N-terminal cysteine of cyanobacterial Psb27 (30) was not observed in the current structure.
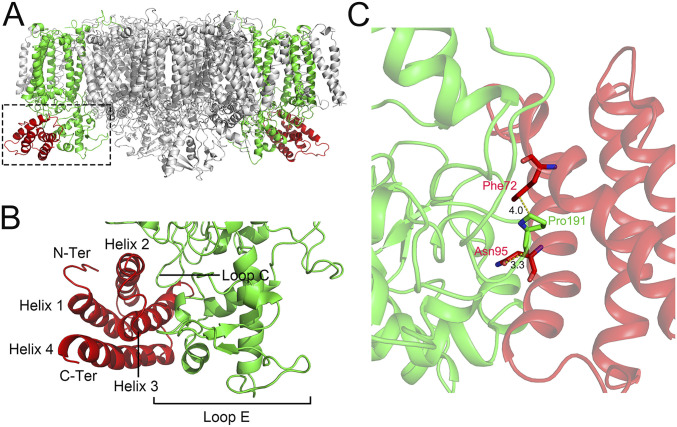
According to the structure of dimeric Psb27-PSII, Psb27 binds to PSII by directly associating with CP43 at the luminal side of PSII (Fig. 2 A and B), which is largely consistent with the previous results obtained by cross-linking and mass spectrometry studies (45–47) and biochemical (2D blue native/SDS-PAGE) analysis (48). However, the precise positions of binding and interaction between CP43 and Psb27 are not completely in agreement with any predictions of the previous studies (45–48). The main interactions between Psb27 and CP43 are found between Helix 2 and Helix 3 of Psb27, with a loop region between Helix 3 and Helix 4 (loop C) and the large luminal E-loop of CP43. Specifically, Pro191 of CP43 is protruded into a space between Helix 2 and Helix 3 of Psb27 and forms a hydrogen bond with Asn95 and interacts with Phe72 hydrophobically, both located in Helix 2 of Psb27 (Fig. 2C) (the numbering of amino acid residues of Psb27 and CP43 starts at the N-terminal residue of the mature proteins, respectively). However, the residues involved in the interactions between Psb27 and CP43 are rather limited, which may suggest a weak binding of Psb27 to PSII. This illustrates the loose, temporary association of Psb27 to the assembly intermediate of PSII and is an advantage for the biogenesis/repair of PSII, as Psb27 needs to be removed during the maturation/repair processes of PSII.
Fig. 2.
Interactions between Psb27 and the PSII core. (A) Side view of the whole Psb27-PSII complex, showing the binding site of Psb27 (highlighted with dotted box). (B) Enlarged view of the boxed area in A, showing the binding sites of Psb27 on CP43. (C) Interactions between Psb27 and CP43. The amino acid residues involved in the interactions are exhibited as sticks and labeled with red for Psb27 and green for CP43, respectively.
Differences in the Structures of Psb27-PSII and Native PSII.
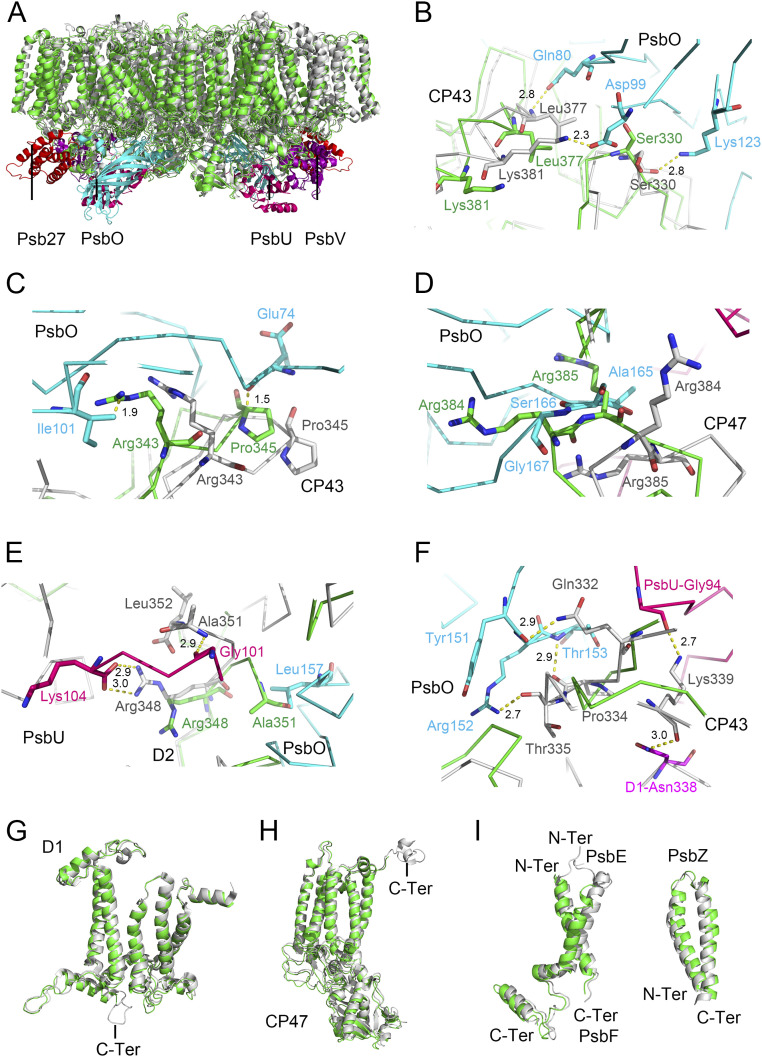
Although the overall structures of the intrinsic core subunits are similar with that of the native PSII (Figs. 1A and 3A), the dimeric Psb27-PSII shows some distinct differences in the structures of some part of the D1, D2, CP43, CP47, PsbE, PsbF, and PsbZ subunits compared with those of the native PSII (Fig. 3 B–I and SI Appendix, Tables S3 and S4). First, the large E-loop of CP43 is in contact with PsbO in native PSII; in particular, residues Ser330 and Leu377 in the E-loop of CP43 are hydrogen bonded to Lys123, Asp99, and Gln80 of PsbO in the native PSII (Fig. 3B). However, these residues change their side chain orientations in the Psb27-PSII, resulting in the breakage of these hydrogen bonds. Also, residues CP43-Arg362 and CP43-Glu348 interact with the N-terminal residues Asp8 and Gly12 of the PsbO subunit in the native PSII (4, 5, 50, 51), and these interactions are broken due to the structural changes of the residues in CP43. Furthermore, the region of Arg343 to Gly347 of CP43 are shifted toward the PsbO side, forming conflicts with the residues from PsbO between the pair of Arg343 and Pro345 of CP43 and the pair of Ile101 and Glu74 of PsbO (Fig. 3C). These changes result in the dissociation of PsbO in Psb27-PSII.
Fig. 3.
Structural comparisons between Psb27-PSII and native PSII (PDB code: 3WU2) from T. vulcanus. (A) Superposition of the overall structures of Psb27-PSII and native PSII. The core transmembrane subunits and Psb27 of Psb27-PSII are shown in green and red, respectively, whereas the core transmembrane subunits and the extrinsic proteins PsbO, PsbU, and PsbV of the native PSII are in gray, cyan, hot pink, and purple, respectively. (B) Comparison of binding sites of PsbO to the native CP43 with that to CP43 of Psb27-PSII. PsbO and native CP43 are depicted in cyan and gray, respectively, whereas CP43 in Psb27-PSII is depicted in green. The amino acid residues that are hydrogen-bonded between PsbO and native CP43 are depicted in sticks, with their distances depicted in Å. (C) Structural conflicts between CP43 and PsbO in Psb27-PSII. PsbO is in cyan, native CP43 is in gray, and CP43 in Psb27-PSII is in green, respectively. The amino acid residues that are in conflicts between PsbO and CP43 of Psb27-PSII are shown in sticks, with their distances depicted in Å. (D) Structural superposition of the binding site of PsbO on CP47, showing the conformational changes at the luminal loop region between Arg384 to Arg385 of CP47. CP47 in native PSII and Psb27-PSII is shown in gray and green, respectively, and PsbO is shown in cyan. The amino acid residues involved in the steric hindrance between PsbO and CP47 of Psb27-PSII are presented as sticks. (E) Structural changes of the D2 subunit that are in interactions with the PsbU subunit in the C-terminal region. PsbU is in hot pink, native D2 is in gray, and D2 of Psb27-PSII is in green, respectively. The amino acid residues involved in the interactions between the C-terminal regions of PsbU and D2 are shown in sticks and the distances are indicated in Å. (F) The region of Gln332–Lys339 in native CP43 that becomes more flexible in CP43 of Psb27-PSII. PsbO, PsbU, D1, and CP43 of native PSII are depicted in cyan, hot pink, magenta, and gray, respectively, and CP43 in Psb27-PSII is depicted in green. The amino acid residues that are hydrogen-bonded between PsbO and native CP43 are depicted in sticks, and their distances are depicted in Å. (G) Structural superposition of the D1 subunit between native PSII and Psb27-PSII, showing the missing of the C-terminal loop of D1 in Psb27-PSII. (H) Structural superposition of CP47 between native PSII and Psb27-PSII, showing the missing of the C-terminal helix of CP47 in Psb27-PSII. (I) Structural superposition of PsbE, PsbF, and PsbZ between native PSII and Psb27-PSII. In G, H, and I, the subunits in Psb27-PSII are depicted in green, and the subunits in native PSII are depicted in gray.
Second, conformational changes occurred in the loop region between Arg384 and Arg385 of CP47 (Fig. 3D) in Psb27-PSII, which leads to a structural conflict with the loop region between Ala165 and Gly167 of PsbO in the adjacent monomer of native PSII. This constitutes another reason for the release of PsbO in the Psb27-PSII. Additionally, D2-Ala351 is in conflict with PsbO-Leu157 (Fig. 3E) in Psb27-PSII, indicating the shift of D2.
Third, the C-terminus of D2 turns toward the direction of the luminal side, breaking the interactions between D2 and PsbU in Psb27-PSII. These interactions include hydrogen bonds between D2-Ala351 and PsbU-Gly101 and between D2-Arg348 and the C-terminal residue Lys104 of PsbU in native PSII (Fig. 3E). These may break the binding of PsbU to the PSII core. Moreover, the region of CP43-Gln332 to Lys339 has a lower electron density and their structure is shifted, probably due to increased flexibility in Psb27-PSII. This region is important for the binding of PsbO and PsbU in native PSII, and most of the interactions between this region and PsbO and PsbU are broken in Psb27-PSII (Fig. 3F). Finally, the C-terminal region of D1, starting from Arg334, is not visible in Psb27-PSII (Fig. 3G). As mentioned above, a very low amount of D1 lacks the C-terminal region starting from Arg334. However, the invisibility of this region in the cryo-EM densities is not due to its absence in the sequence but probably because of the low resolution and its higher flexibility in the absence of the Mn4CaO5 cluster in the Psb27-PSII.
In addition, for the low molecular weight subunits, slight shifts of helices are observed in the PsbE, PsbF, and PsbZ subunits (Fig. 3I and SI Appendix, Table S3). Eventually, the whole structure of this dimeric Psb27-PSII has become loose. This may constitute another reason for the loss of the PsbJ subunit in the complex (SI Appendix, Fig. S1F), as it is located in a pocket surrounded by PsbE/PsbF/PsbK in the native PSII (4, 5). The densities corresponding to the residues of the C-terminal tail of CP47 and N-terminal tails of PsbE and PsbF are also absent in the cryo-EM density maps (Fig. 3 H and I), which may reflect the increased flexibility of these terminal regions.
The Relative Location of Psb27 Compared to Other Extrinsic Proteins in the Luminal Side of PSII.
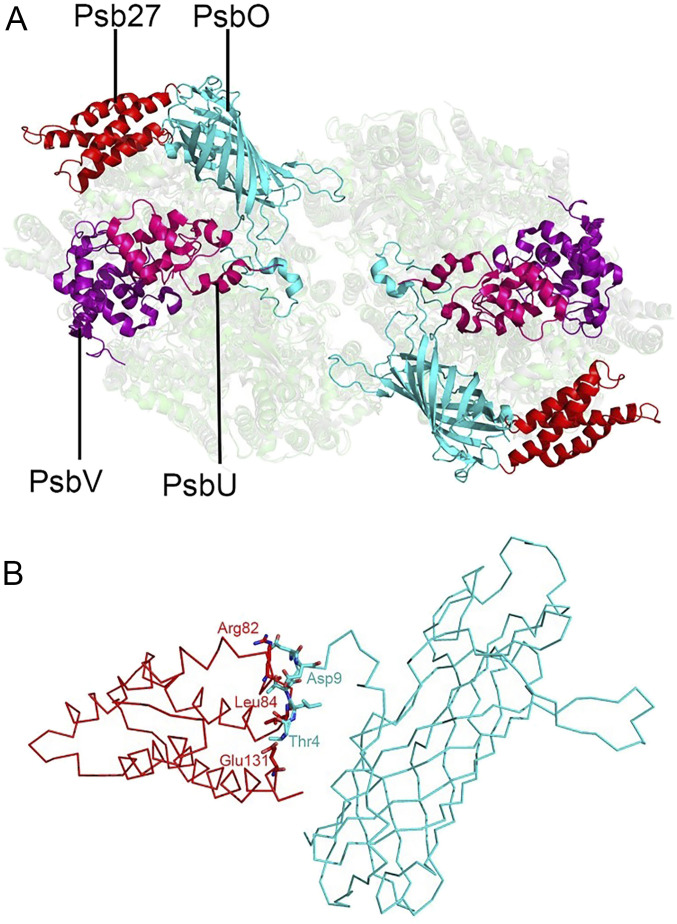
Structural comparison between Psb27-PSII and the native PSII from T. vulcanus shows that Psb27 does not occupy any position of the three extrinsic proteins PsbO, PsbV, and PsbU present in the crystal structure of native PSII (Fig. 4A). However, spatial conflicts were found between the N-terminal region from Thr4 to Asp9 of PsbO and the Arg82-Leu84 region of the loop between Helix 2 and Helix 3 of Psb27 as well as the C-terminal Glu131 residue of Psb27 (Fig. 4B). This suggests that the presence of Psb27 prevents the binding of the PsbO to the luminal side of PSII.
Fig. 4.
View of the luminal region after superposition of Psb27-PSII with the native PSII from T. vulcanus (PDB code: 3WU2). (A) Superposition of Psb27-PSII with the native PSII, showing the relative positions of Psb27, PsbO, PsbU, and PsbV in the luminal side. (B) Structural conflicts between PsbO and Psb27. Residues in conflicts between the N-terminal region of PsbO and the loop region (between Helix 2 and Helix 3) of Psb27, as well as a C-terminal residue Glu131 of Psb27, are depicted in sticks. The extrinsic Psb27 protein in Psb27-PSII, and PsbO, PsbU, and PsbV in native PSII, are shown in red, cyan, hot pink, and purple, respectively.
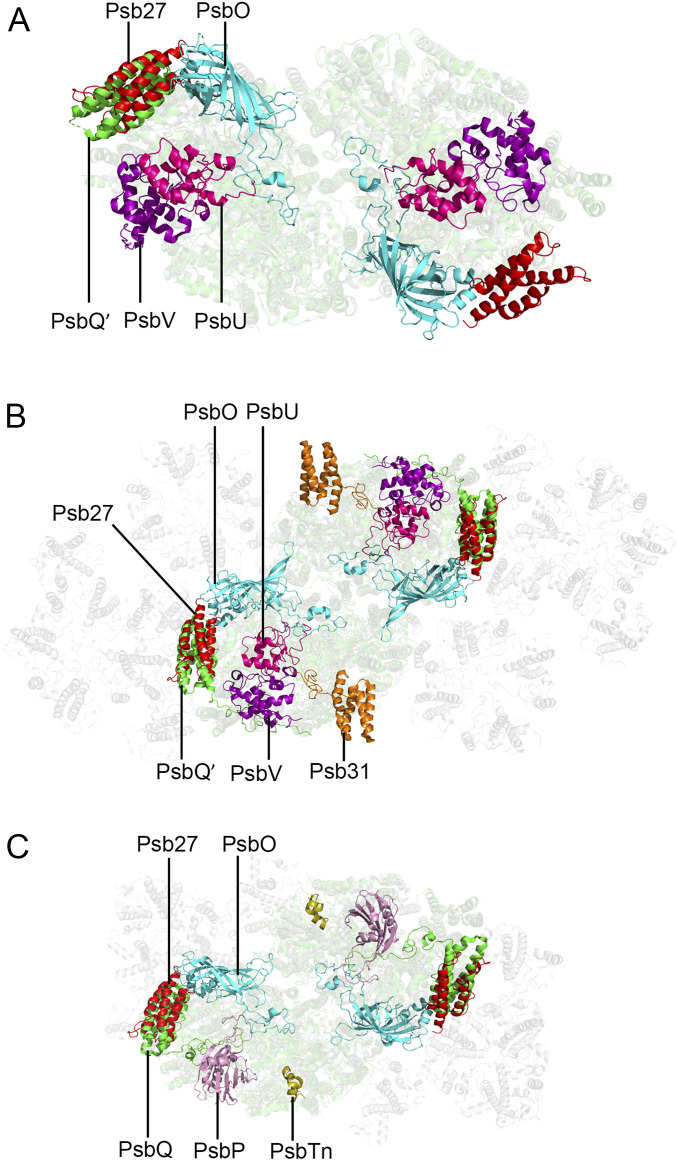
To further examine the location of Psb27 relative to other extrinsic proteins in the luminal side of PSII, we compared the structure of the Psb27-PSII complex with that of native PSII from red algae (17), diatom (18, 19), and spinach (20). We found that the binding site of Psb27 in the luminal surface of PSII largely overlaps with that of PsbQ′ and PsbQ in the PSII of red alga, diatom, and higher plant, respectively (Fig. 5 A–C and SI Appendix, Fig. S8). However, the N terminus and C terminus of Psb27 are oriented completely in the opposite direction compared to that of PsbQ′ or PsbQ, and the helices of Psb27 are slightly shifted relative to those of PsbQ′ and PsbQ in the PSII structures of red alga, diatom, and spinach (SI Appendix, Fig. S9).
Fig. 5.
View of the luminal region after superposition of Psb27 in Psb27-PSII with native PSII from other species. (A) Structural superposition of Psb27 with native PSII from the red alga Cyanidium caldarium (PDB code: 4YUU). (B) Structure superposition of Psb27 with native PSII from a diatom Chaetoceros gracilis (PDB code: 6JLU). (C) Structure superposition of Psb27 with native PSII from spinach (PDB code: 3JCU). The extrinsic proteins Psb27, PsbO, PsbU, and PsbV are shown in red, cyan, hot pink, and purple, respectively. The PsbQ and PsbQ′ are in green. The PsbP and PsbTn are in pink and wheat, respectively.
Discussion
Psb27-PSII is an assembly intermediate of PSII, which does not bind the three extrinsic proteins and the Mn4CaO5 cluster and has no oxygen-evolving activity. In this study, we analyzed the cryo-EM structure of the Psb27-PSII dimer complex from the cyanobacterium T. vulcanus, which was obtained from a ΔPsbV strain. The structure showed that the transiently associated Psb27 binds to CP43, with specific interactions formed between Helix 2 and Helix 3 of Psb27 and a loop region between Helix 3 and Helix 4 (loop C) and the large E-loop of CP43. These interaction sites were different from those reported previously by cross-linking and mass spectrometry analyses (45–48). The N-terminal region of Psb27, reported to be important for the association of Psb27 to CP43 previously (30), was not found in contact with CP43 in the present structure, which suggests that lipid modification, if present, is not essential for the tight interaction of Psb27 with PSII. There was also no direct interaction between Psb27 and CP47, D1, or D2, which is in contrast to the results reported previously by surface plasmon resonance spectroscopy (45).
Although Psb27 does not occupy any binding sites of PsbV and PsbU, its binding imposes some conflicts with the N-terminal region from Thr4 to Asp9 of PsbO. Together with the structural changes occurred in CP43, D2, and CP47 in Psb27-PSII, these result in the dissociation of PsbO from PSII. This is consistent with earlier studies showing that mutations around Arg384 and Arg385 of CP47 influence PSII assembly and activity (52–54). Because of the absence of PsbO, PsbU cannot bind to PSII stably (11). Furthermore, the C terminus of D2 has undergone some structural changes in Psb27-PSII (SI Appendix, Table S3), which also results in the inability of PsbU binding to PSII. In addition, PsbE, PsbF, and PsbZ have shifted their helices slightly compared with those of native PSII. Because PsbE and PsbF surround the binding site of PsbJ, these structural shifts may break down their interactions with PsbJ, resulting in the weak binding and loss of PsbJ in the purified Psb27-PSII. These results are mostly consistent with a very recent report showing the structure of an assembly intermediate of PSII possessing Psb27, Psb28, and an additional helix termed Psb34 isolated from a psbJ-deletion mutant (55). However, in that report, no conflict was found between Psb27 and PsbO, which is in contrast to the results reported in the present structure. This may be due to the limited resolutions of the present structure or lack of a more detailed examination of the structure of Zabret et al. (55). We should point out that the conflict of Psb27 with PsbO is consistent with the inability of binding PsbO to PSII in the presence of Psb27.
The position of Psb27 is far from the binding site of the Mn4CaO5 cluster in the luminal side of PSII, so it seems impossible that Psb27 assists assembly of the Mn4CaO5 cluster directly. It is more likely that Psb27 maintains the environment of the luminal side of PSII for the assembly of the Mn4CaO5 cluster. Since the C-terminal region of D1 is very important in the binding of the Mn4CaO5 cluster (4, 5), the absence of the Mn4CaO5 cluster may make this region flexible. The reason that we could not observe a part of the C-terminal region of D1 in our structure may be due to the low resolution of our structure, which makes the flexible C terminus of D1 difficult to be visible. At a higher resolution, the C-terminal region is fully visible, and they were indeed shifted away from the Mn4CaO5 cluster, leaving the binding site for the Mn4CaO5 cluster vacant (55). In the structure of Psb27-PSII-Psb28, an additional density close to the missing Mn4CaO5 cluster was observed, and this was suggested to represent a Mn or Ca ion or any other ions (55). We did not observe such a density in the present structure, which may, again, be due to the low resolution achieved or the loss of the ion in the present structure. It was reported that the Psb27 protein associated with the PSII precomplex facilitates the Mn4CaO5 cluster assembly by excluding the other extrinsic proteins in the repair cycle of PSII (30, 31, 40). Our results provide structural evidence to support the previous prediction and indicate that the binding and assembly of extrinsic proteins PsbO, PsbU, and PsbV are regulated by Psb27 in the luminal side of PSII.
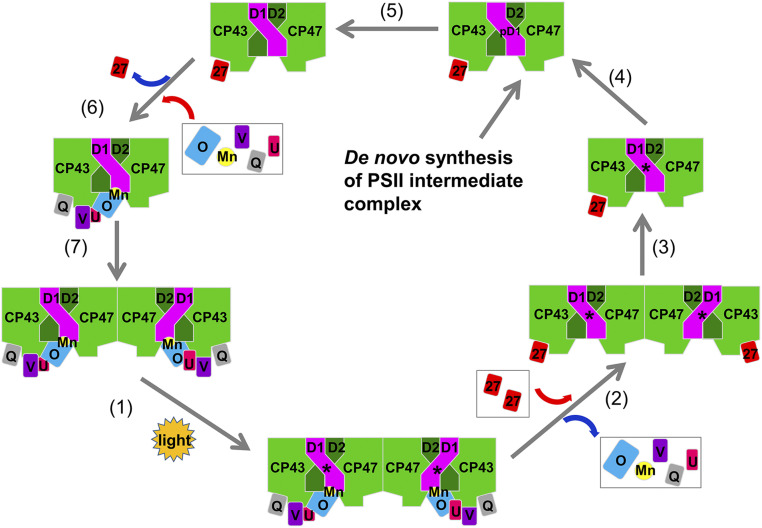
Based on the above results, we propose a schematic model for the assembly of the Mn4CaO5 cluster and extrinsic proteins in the luminal side of PSII (Fig. 6) by combining the current structural information with the results of previous studies (30–33, 40, 46). In the assembly/repair processes, upon light damage or when the PsbV subunit is deleted, the highly active dimeric PSII complex is transformed into a less active dimeric PSII and subsequently to an inactive one, with the three extrinsic proteins and the Mn4CaO5 cluster released. Subsequently, Psb27 is attached and the dimer is monomerized, followed by the replacement of the damaged D1 by a newly synthesized pre-D1. After the processing of the C-terminal region of D1, Psb27 is detached, followed by the assembly of the Mn4CaO5 and the attachment of the three extrinsic proteins. After completion of the assembly, the PSII monomer is linked to form an active dimer. However, it is not clear how Psb27 is bound and released in the process of the assembly. It was proposed that the conformational rearrangement of the luminal domain of CP43 plays an important role in the dissociation of Psb27 (47, 48), and the incorporation of the Mn4CaO5 cluster might trigger this dynamic rearrangement. Our results revealed the structural differences in a luminal region of CP43 between the native PSII and the nonfunctional Psb27-PSII, which could facilitate the release of Psb27 and the assembly of the Mn4CaO5 cluster. It should be noted that our data indicates the presence of steric conflicts between Psb27 and PsbO in the luminal side; these structural conflicts might be one of the factors to facilitate the detachment of Psb27 upon the binding of PsbO. It was proposed that PsbO is the first extrinsic protein recruited to PSII during donor side assembly (48).
Fig. 6.
A schematic model for the PSII repair/assembly, emphasizing the repair/assembly of the extrinsic part of PSII based on the structural information of the dimeric Psb27-PSII. CyanoQ is tentatively placed in the position of Psb27 after incorporation of manganese. D1 is shown in magentas, and the other core transmembrane subunits are in green. The “D1*” and “pD1” represent damaged D1 protein and premature D1 protein, respectively. The extrinsic proteins are PsbO (blue), Psb27 (red), PsbU (magenta), PsbV (purple), and CyanoQ (gray). The manganese cluster is shown in yellow.
Although the binding site of Psb27 does not overlap with any of the three extrinsic proteins in cyanobacterial PSII, our structure revealed that it is largely overlapped with the binding site of PsbQ′ in red algal and diatom PSII (17–19), or PsbQ in higher plant PSII (20). Although Psb27 and PsbQ (PsbQ′) do not share sequence similarity (SI Appendix, Fig. S10), they share a similar four helices bundle structure, and therefore it may be suggested that during evolution, Psb27 is replaced by PsbQ′ and PsbQ, which functions as a stable component of PSII. In this respect, it is worth noting that cyanobacteria have CyanoQ, which has a similar structure to Psb27 (56) and plays an important role in the optimal function of PSII (16, 57–60). Since CyanoQ is not a stable component of PSII and was not found in the crystal structure of native PSII, it may be related with the binding/removal of Psb27 during maturation and/or repair of PSII. However, CyanoQ has been shown to be associated with CP47 and PsbO in cyanobacterial PSII (61). If this is the case, the function of CyanoQ may not be related with Psb27, and its binding site may also be different with that in red algal, diatom, and higher plant PSII.
In conclusion, we have shown the exact binding site of Psb27 in the Psb27-PSII intermediate complex and its distinct structural features. Our results reveal the structural and functional relationships between Psb27 and PSII and provide important insights into the biogenesis/repair processes of native PSII.
Materials and Methods
Construction of the psbV-Deletion Mutant.
To generate the ΔPsbV strain of T. vulcanus, a plasmid vector, which includes a disrupted psbV gene from the 53rd nucleotide in the promoter region to the stop codon, a streptomycin-resistant gene cassette, as well as the upstream and downstream of psbV for homologous recombination, was constructed (49). The plasmid was introduced into the wild type cells of T. vulcanus by either electroporation or natural transformation as previously described (62, 63). After the transformants appeared in 2 to 5 wk, they were segregated by culturing on agar plates containing 10 µg/mL streptomycin. The mutant cells were confirmed by PCR and sequencing, and grown in a CO2 (3 to 5%)-enriched atmosphere at 45 °C under continuous light with an intensity of 40 μmol photons m−2 ⋅ s−1. Mutant strains were maintained in the presence of 10 µg/mL streptomycin but propagated in the absence of the antibiotics for cells to be harvested for the cryo-EM experiments.
Purification of the Psb27-PSII Complex.
Thylakoids, crude PSII complexes, and Psb27-PSII complexes were prepared as described previously (11, 64). The dimeric Psb27-PSII complexes were isolated from the crude PSII by ion exchange chromatography with a Q Sepharose High Performance column (Cytiva) eluted with a linear gradient from 170 to 300 mM NaCl (49) (SI Appendix, Fig. S1A). For cryo-EM studies, the sample in the eluted peak (labeled with a vertical bar in SI Appendix, Fig. S1A) was collected and concentrated to a chlorophyll concentration of 2.8 mg/mL using a 100 kDa cutoff membrane concentrator. The concentration of chlorophylls was determined according to Porra et al. (65).
Biochemical Analysis.
The protein compositions of purified Psb27-PSII complexes were analyzed by SDS-PAGE using a gel containing 16% polyacrylamide plus 7.5 M urea (66). For the size-exclusion chromatography analysis, the sample was subjected to gel filtration chromatography (Cytiva, Superose 6 Increase 10/300 GL) in a buffer containing 30 mM Mes-NaOH (pH 6.0), 3 mM CaCl2, and 0.015% (wt/vol) n-dodecyl-β-D-maltoside. For analysis of the PSII dimer and monomer, blue native PAGE was performed according to ref. 67. For immunoblotting analysis, proteins on the gel were transferred to a polyvinylidene fluoride membrane and incubated with antibodies against Psb27 or PsbJ. The subunit bands were visualized by enhanced chemiluminescence (Biostep, Germany) after incubation with a horseradish peroxidase-conjugated secondary antibody (Agrisera AS09602).
For mass spectrometry analysis, Coomassie Brilliant Blue-stained bands containing the targeted proteins were excised from SDS-PAGE gel and digested using sequencing grade-modified trypsin in 50 mM ammonium bicarbonate at 37 °C. The peptides were extracted twice with 1% trifluoroacetic acid in 50% acetonitrile aqueous solution for 30 min. For liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis, peptides were separated by a 120-min gradient elution at a flow rate of 0.300 μL/min with a Thermo-Dionex Ultimate 3,000 HPLC system, which was directly interfaced with the Thermo Orbitrap Fusion mass spectrometer. The analytical column was a fused silica capillary column (75 μm ID, 150-mm length; Upchurch, Oak Harbor, WA) packed with C-18 resin (300 A, 5 μm; Varian, Lexington, MA). The MS/MS spectra from each LC-MS/MS run were searched against the selected database using Proteome Discovery searching algorithm (version 1.4).
Cryo-EM Sample Preparation and Data Collection.
An aliquot of 4 μL the Psb27-PSII sample at a concentration of 2.8 mg Chl/mL was applied to glow-discharged Quantifoil holey carbon Cu grids (R 1.2/1.3, 400 mesh), and plunged into liquid ethane at around 100 K using an FEI Vitrobot Mark IV. Parameters were set as follows: blotting time 3 s, blotting force level 0, 100% humidity, and 8 °C. Sample screening was performed using a Tecnai Arctica 200 kV electron microscope equipped with an FEI Falcon II camera. The micrographs used for high resolution structure determination were collected with a 300 kV FEI Titan Krios electron microscope equipped with a K2 Summit direct electron detector (Gatan) in the super-resolution mode. A total of 4,899 micrographs were recorded at a nominal magnification of 22,500 times, yielding a pixel size of 1.30654 Å, with defocus values varied from −1.5 to −2.5 μm. Each exposure of 8 s was dose-fractionated to 32 movie frames, leading to a total dose rate of ∼50 e−/ Å2.
Cryo-EM Image Processing.
For the data collected on the Titan Krios electron microscopy, the beam-induced motion of the whole micrograph with 32 movie frames was corrected by MotionCor2 (68), and the defocus values and parameters of astigmatism were estimated by CTFFIND4 (69) using the corrected micrographs without dose weighting. A small dataset of 2,789 particles were manually picked and processed by reference-free 2D classification using RELION (70). Six good 2D averages were selected as references for the auto-picking of all 4,899 micrographs, and a total of 1,783,091 particles were automatically picked using RELION. In order to speed up the data processing, the whole 1,783,091 particles were evenly split into four subsets and then subjected to the reference-free 2D classification. After two rounds of 2D classification, 1,625,956 particles were selected from all of the four subsets and subjected to 3D classification without imposing any symmetry. A density map generated from an atomic model of PSII core (PDB code: 3WU2) without the three extrinsic proteins was low-pass filtered to 60 Å as the initial model for the 3D classification. After two rounds of 3D classification, 341,426 particles were selected for the further 3D auto-refinement. Further improvements of the density map were achieved by a set of soft-masks for dose-weighted micrographs and by applying a two-fold symmetry. The final resolution of the density map is 3.37 Å based on the gold-standard Fourier Shell Correlation (FSC) = 0.143 criteria (71), while the density map of the Psb27 subunit is much worse than the PSII core. To improve the resolution of Psb27, the 341,426 particles were used again for one more 3D classification without image alignment. A total of 87,473 particles of one class with good map quality of the Psb27 subunit was selected for 3D auto-refinement and postprocessing. The final resolution of Psb27-PSII with a high-quality density map of the Psb27 subunit was 3.78 Å based on the gold-standard FSC = 0.143 criteria.
Model Building and Refinement.
The 3.37 Å resolution cryo-EM map was used for the model building of the PSII core. The crystal structure of PSII core from T. vulcanus PSII (PDB code: 3WU2) was first manually fitted into the 3.37 Å resolution cryo-EM map using UCSF Chimera (72), then the model of the PSII core was first manually adjusted against the cryo-EM map with Crystallographic Object-Oriented Toolkit (COOT) (73), and then refined in real space by Python-based Hierarchical Environment for Integrated Xtallography (PHENIX) (74). The refined model was corrected again in COOT until there were no more improvements in both MolProbity score and geometry parameters.
The 3.78 Å resolution cryo-EM map was used for model building of the Psb27-PSII. The crystal structure of Psb27 from T. elongatus (PDB code: 2Y6X) and the crystal structure of the PSII core from T. vulcanus (PDB code: 3WU2) were fitted into the 3.78 Å resolution cryo-EM map in Chimera, and then COOT was used for manual adjustment and real space refinement for both proteins and cofactors on the basis of the cryo-EM density map, followed by real-space automatic refinement against the cryo-EM map by PHENIX (74). The final statistics for data processing and structure refinement are summarized in the SI Appendix, Table S1.
Supplementary Material
Acknowledgments
We thank J. Lei and the staff at the Tsinghua University Branch of the National Center for Protein Sciences Beijing for providing facility support, the “Explorer 100” cluster system of the Tsinghua National Laboratory for Information Science and Technology for providing computation resources, and Prof. Haiteng Deng and Xianbin Meng in Proteinomics Facility at Technology Center for Protein Sciences, Tsinghua University, for protein MS analysis. This work was supported by the National Key R&D Program of China (2017YFA0503700, 2016YFA0501101, 2017YFA0504600, and 2019YFA0906300), the National Natural Science Foundation of China (31470339), the Strategic Priority Research Program of Chinese Academy of Science (CAS) (XDB17000000), a CAS Key Research program for Frontier Science (QYZDY-SSW-SMC003), and the Youth Innovation Promotion Association of CAS (2020081).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2018053118/-/DCSupplemental.
Data Availability.
The cryo-EM density map and atomic model of the Psb27-PSII complex at 3.78 Å have been deposited in the Electron Microscopy Data Bank (EMD) and PDB (EMD code EMD-30511 and PDB code 7CZL). All other data are included in the article and/or supporting information.
References
- 1.Nelson N., Yocum C. F., Structure and function of photosystems I and II. Annu. Rev. Plant Biol. 57, 521–565 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Barber J., Photosystem II: The engine of life. Q. Rev. Biophys. 36, 71–89 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Shen J.-R., The structure of photosystem II and the mechanism of water oxidation in photosynthesis. Annu. Rev. Plant Biol. 66, 23–48 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Umena Y., Kawakami K., Shen J.-R., Kamiya N., Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Suga M., et al. , Native structure of photosystem II at 1.95 Å resolution viewed by femtosecond X-ray pulses. Nature 517, 99–103 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Enami I., et al. , Structures and functions of the extrinsic proteins of photosystem II from different species. Photosynth. Res. 98, 349–363 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Bricker T. M., Roose J. L., Fagerlund R. D., Frankel L. K., Eaton-Rye J. J., The extrinsic proteins of photosystem II. Biochim. Biophys. Acta 1817, 121–142 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Ifuku K., Localization and functional characterization of the extrinsic subunits of photosystem II: An update. Biosci. Biotechnol. Biochem. 79, 1223–1231 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Roose J. L., Frankel L. K., Mummadisetti M. P., Bricker T. M., The extrinsic proteins of photosystem II: Update. Planta 243, 889–908 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Shen J.-R., Ikeuchi M., Inoue Y., Stoichiometric association of extrinsic cytochrome c550 and 12 kDa protein with a highly purified oxygen-evolving photosystem II core complex from Synechococcus vulcanus. FEBS Lett. 301, 145–149 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Shen J.-R., Inoue Y., Binding and functional properties of two new extrinsic components, cytochrome c-550 and a 12-kDa protein, in cyanobacterial photosystem II. Biochemistry 32, 1825–1832 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Enami I., et al. , Isolation and characterization of a photosystem II complex from the red alga Cyanidium caldarium: Association of cytochrome c-550 and a 12 kDa protein with the complex. Biochim. Biophys. Acta 1232, 208–216 (1995). [DOI] [PubMed] [Google Scholar]
- 13.Enami I., Kikuchi S., Fukuda T., Ohta H., Shen J.-R., Binding and functional properties of four extrinsic proteins of photosystem II from a red alga, Cyanidium caldarium, as studied by release-reconstitution experiments. Biochemistry 37, 2787–2793 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Nagao R., et al. , Binding and functional properties of five extrinsic proteins in oxygen-evolving photosystem II from a marine centric diatom, Chaetoceros gracilis. J. Biol. Chem. 285, 29191–29199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ifuku K., The PsbP and PsbQ family proteins in the photosynthetic machinery of chloroplasts. Plant Physiol. Biochem. 81, 108–114 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Thornton L. E., et al. , Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem ii activity in the cyanobacterium Synechocystis 6803. Plant Cell 16, 2164–2175 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ago H., et al. , Novel features of eukaryotic photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 291, 5676–5687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pi X., et al. , The pigment-protein network of a diatom photosystem II-light-harvesting antenna supercomplex. Science 365, aax4406 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Nagao R., et al. , Structural basis for energy harvesting and dissipation in a diatom PSII-FCPII supercomplex. Nat. Plants 5, 890–901 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Wei X., et al. , Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534, 69–74 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Shen L., et al. , Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 116, 21246–21255 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheng X., et al. , Structural insight into light harvesting for photosystem II in green algae. Nat. Plants 5, 1320–1330 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J., Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komenda J., Sobotka R., Nixon P. J., Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Nickelsen J., Rengstl B., Photosystem II assembly: From cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Järvi S., Suorsa M., Aro E.-M., Photosystem II repair in plant chloroplasts–Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847, 900–909 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Heinz S., Liauw P., Nickelsen J., Nowaczyk M., Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857, 274–287 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kashino Y., et al. , Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Roose J. L., Pakrasi H. B., Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J. Biol. Chem. 279, 45417–45422 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Nowaczyk M. M., et al. , Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18, 3121–3131 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roose J. L., Pakrasi H. B., The Psb27 protein facilitates manganese cluster assembly in photosystem II. J. Biol. Chem. 283, 4044–4050 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Mabbitt P. D., Wilbanks S. M., Eaton-Rye J. J., Structure and function of the hydrophilic photosystem II assembly proteins: Psb27, Psb28 and Ycf48. Plant Physiol. Biochem. 81, 96–107 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Grasse N., et al. , Role of novel dimeric photosystem II (PSII)-Psb27 protein complex in PSII repair. J. Biol. Chem. 286, 29548–29555 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei L., et al. , LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J. Biol. Chem. 285, 21391–21398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., et al. , A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol. Biol. 61, 567–575 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Hou X., Fu A., Garcia V. J., Buchanan B. B., Luan S., PSB27: A thylakoid protein enabling Arabidopsis to adapt to changing light intensity. Proc. Natl. Acad. Sci. U.S.A. 112, 1613–1618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mamedov F., Nowaczyk M. M., Thapper A., Rögner M., Styring S., Functional characterization of monomeric photosystem II core preparations from Thermosynechococcus elongatus with or without the Psb27 protein. Biochemistry 46, 5542–5551 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Liu H., Roose J. L., Cameron J. C., Pakrasi H. B., A genetically tagged Psb27 protein allows purification of two consecutive photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J. Biol. Chem. 286, 24865–24871 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowaczyk M. M., et al. , Deletion of psbJ leads to accumulation of Psb27-Psb28 photosystem II complexes in Thermosynechococcus elongatus. Biochim. Biophys. Acta 1817, 1339–1345 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Weisz D. A., et al. , A novel chlorophyll protein complex in the repair cycle of photosystem II. Proc. Natl. Acad. Sci. U.S.A. 116, 21907–21913 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cormann K. U., et al. , Structure of Psb27 in solution: Implications for transient binding to photosystem II during biogenesis and repair. Biochemistry 48, 8768–8770 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Mabbitt P. D., et al. , Solution structure of Psb27 from cyanobacterial photosystem II. Biochemistry 48, 8771–8773 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Michoux F., et al. , Crystal structure of the Psb27 assembly factor at 1.6 Å: implications for binding to photosystem II. Photosynth. Res. 110, 169–175 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Xingxing C., et al. , Crystal structure of Psb27 from Arabidopsis thaliana determined at a resolution of 1.85 Å. Photosynth. Res. 136, 139–146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cormann K. U., Möller M., Nowaczyk M. M., Critical assessment of protein cross-linking and molecular docking: An updated model for the interaction between photosystem II and Psb27. Front. Plant Sci. 7, 157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H., Huang R. Y.-C., Chen J., Gross M. L., Pakrasi H. B., Psb27, a transiently associated protein, binds to the chlorophyll binding protein CP43 in photosystem II assembly intermediates. Proc. Natl. Acad. Sci. U.S.A. 108, 18536–18541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu H., et al. , Mass spectrometry-based footprinting reveals structural dynamics of loop E of the chlorophyll-binding protein CP43 during photosystem II assembly in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 288, 14212–14220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Komenda J., et al. , The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 158, 476–486 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiao Y., et al. , Role of PsbV-Tyr137 in photosystem II studied by site-directed mutagenesis in the thermophilic cyanobacterium Thermosynechococcus vulcanus. Photosynth. Res. 146, 41–54 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Eaton-Rye J. J., Murata N., Evidence that the amino-terminus of the 33 kDa extrinsic protein is required for binding to the photosystem II complex. Biochim. Biophys. Acta 977, 219–226 (1989). [DOI] [PubMed] [Google Scholar]
- 51.Popelka H., Yocum C., Probing the N-terminal sequence of spinach PsbO: Evidence that essential threonine residues bind to different functional sites in eukaryotic photosystem II. Photosynth. Res. 112, 117–128 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Eaton-Rye J. J., Vermaas W. F., Oligonucleotide-directed mutagenesis of psbB, the gene encoding CP47, employing a deletion mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 17, 1165–1177 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Putnam-Evans C., Bricker T. M., Site-directed mutagenesis of the CPa-1 protein of photosystem II: Alteration of the basic residue pair 384,385R to 384,385G leads to a defect associated with the oxygen-evolving complex. Biochemistry 31, 11482–11488 (1992). [DOI] [PubMed] [Google Scholar]
- 54.Gleiter H. M., et al. , Involvement of the CP47 protein in stabilization and photoactivation of a functional water-oxidizing complex in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry 34, 6847–6856 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Zabret J., et al. , How to build a water-splitting machine: Structural insights into photosystem II assembly. bioRxiv [Preprint]. 10.1101/2020.09.14.294884. Accessed 15 September 2020. [DOI]
- 56.Jackson S. A., Fagerlund R. D., Wilbanks S. M., Eaton-Rye J. J., Crystal structure of PsbQ from Synechocystis sp. PCC 6803 at 1.8 A: Implications for binding and function in cyanobacterial photosystem II. Biochemistry 49, 2765–2767 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Summerfield T. C., Shand J. A., Bentley F. K., Eaton-Rye J. J., PsbQ (Sll1638) in Synechocystis sp. PCC 6803 is required for photosystem II activity in specific mutants and in nutrient-limiting conditions. Biochemistry 44, 805–815 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Kashino Y., Inoue-Kashino N., Roose J. L., Pakrasi H. B., Absence of the PsbQ protein results in destabilization of the PsbV protein and decreased oxygen evolution activity in cyanobacterial photosystem II. J. Biol. Chem. 281, 20834–20841 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Roose J. L., Kashino Y., Pakrasi H. B., The PsbQ protein defines cyanobacterial photosystem II complexes with highest activity and stability. Proc. Natl. Acad. Sci. U.S.A. 104, 2548–2553 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michoux F., et al. , Crystal structure of CyanoQ from the thermophilic cyanobacterium Thermosynechococcus elongatus and detection in isolated photosystem II complexes. Photosynth. Res. 122, 57–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu H., et al. , MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc. Natl. Acad. Sci. U.S.A. 111, 4638–4643 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mühlenhoff U., Chauvat F., Gene transfer and manipulation in the thermophilic cyanobacterium Synechococcus elongatus. Mol. Gen. Genet. 252, 93–100 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Iwai M., Katoh H., Katayama M., Ikeuchi M., Improved genetic transformation of the thermophilic cyanobacterium, Thermosynechococcus elongatus BP-1. Plant Cell Physiol. 45, 171–175 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Shen J. R., Kamiya N., Crystallization and the crystal properties of the oxygen-evolving photosystem II from Synechococcus vulcanus. Biochemistry 39, 14739–14744 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Porra R. J., Thompson W. A., Kriedemann P. E., Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 (1989). [Google Scholar]
- 66.Ikeuchi M., Inoue Y., A new 4.8-kDa polypeptide intrinsic to the PS II reaction center, as revealed by modified SDS-PAGE with improved resolution of low-molecular-weight proteins. Plant Cell Physiol. 29, 1233–1239 (1988). [Google Scholar]
- 67.Kawakami K., Iwai M., Ikeuchi M., Kamiya N., Shen J.-R., Location of PsbY in oxygen-evolving photosystem II revealed by mutagenesis and X-ray crystallography. FEBS Lett. 581, 4983–4987 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Zheng S. Q., et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scheres S. H. W., Chen S., Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map and atomic model of the Psb27-PSII complex at 3.78 Å have been deposited in the Electron Microscopy Data Bank (EMD) and PDB (EMD code EMD-30511 and PDB code 7CZL). All other data are included in the article and/or supporting information.