Significance
Multichannel single-molecule tracking is a powerful tool to probe molecular interactions with high spatial-temporal resolution. Current methods have at least two major limitations: the lack of high-performance intracellular single-molecule fluorescence probes and the difficulty of multispecies single-molecule labelling. Here, we employ a nanopore-electroporation technique to deliver organic dye-labelled proteins into living cells to achieve multi-color single molecule tracking. These organic dyes have better optical and chemical properties than the probes currently used. Therefore, we significantly expand the selection of intracellular single-molecule probes to widely available dyes. We demonstrate the application of this technique by revealing detailed spatial resolution and dynamics of membrane EGFR signaling transductions, which will shed light on the understanding of its fundamental mechanism.
Keywords: EGFR signaling, nanopore, electroporation, single molecule, ex vivo protein delivery
Abstract
Multicolor single-molecule tracking (SMT) provides a powerful tool to mechanistically probe molecular interactions in living cells. However, because of the limitations in the optical and chemical properties of currently available fluorophores and the multiprotein labeling strategies, intracellular multicolor SMT remains challenging for general research studies. Here, we introduce a practical method employing a nanopore-electroporation (NanoEP) technique to deliver multiple organic dye-labeled proteins into living cells for imaging. It can be easily expanded to three channels in commercial microscopes or be combined with other in situ labeling methods. Utilizing NanoEP, we demonstrate three-color SMT for both cytosolic and membrane proteins. Specifically, we simultaneously monitored single-molecule events downstream of EGFR signaling pathways in living cells. The results provide detailed resolution of the spatial localization and dynamics of Grb2 and SOS recruitment to activated EGFR along with the resultant Ras activation.
Single-molecule tracking (SMT) provides an unprecedented ability to directly observe molecular interactions with high spatial and temporal resolution, which has significantly advanced the understanding of physical molecular mechanisms underlying function in living cells (1–7). SMT becomes especially powerful when multiple molecular species are studied, as it allows direct observation of two or more types of molecules simultaneously, providing an informative insight into the study of the protein–protein interactions. Recently, much success has been made in the study of the receptor-G protein interactions and messenger RNA translation heterogeneity with the use of two- or three-color SMT (8, 9). However, because of the limitations in the optical and chemical properties of currently available fluorophores and the protein-labeling techniques, monitoring multiple proteins’ interactions in living cells remains challenging for general research applications.
Multicolor SMT has been possible to some extent with three different experimental strategies. First, photoactivatable or photoconvertible fluorescent fusion proteins, such as PAmcherry or mEOS3.2 (10, 11), are the most common probes for SMT as they are easily fused to the target proteins in cells genetically. However, the molecular brightness and photostability of fluorescent proteins are generally much lower than organic dyes (Table 1), thus limiting their applications for long-term molecule tracking. Second, inorganic fluorescence probes, such as quantum dots, have excellent optical properties, yet they require complex functionalization with biomolecules, which inherently introduces heterogeneity, defects, and undesirable multivalence. Besides, such large particles generally only work well on the extracellular side of cell membranes (2). Third, recent developments in membrane-permeable dyes have provided a promising way to label intracellular proteins in situ through specific fusion tags (12–16); however, the nonspecific association of dyes and the lipid bilayer is a general concern (17, 18) that needs to be validated in more context, especially in sensitive single-molecule experiments. In fact, many bright and lipid interaction-free organic dyes are membrane impermeable and thus far cannot be used for intracellular imaging (Table 1). Taken together, given current limitations in fluorescent probes and multispecies tagging methods, multicolor SMT in living cells still faces great challenges.
Table 1.
Comparison of commonly used dyes or fluorescent proteins for SMT*
| Channel | Dye or fluorescent protein | Extinction coefficient (M−1cm−1) | Quantum yield | Molecular brightness† | Membrane permeability | Dye-lipid bilayer interaction | Ref. |
| 488 | Alexa 488 | 73,000 | 0.92 | 67.2 | No | No | 17 |
| Atto 488 | 90,000 | 0.8 | 72.0 | No | No | 17 | |
| Oregon Green 488 | 85,000 | 0.92 | 78.2 | Yes | No | 53 | |
| PA-GFP | 17,400 | 0.79 | 13.7 | — | — | 54 | |
| 561 | Alexa 568 | 86,000 | 0.69 | 59.3 | No | No | 17 |
| PA-JF 549 | 98,900 | 0.85 | 84.1 | Yes | Yes‡ | 15 | |
| TMR | 78,000 | 0.41 | 32.0 | Yes | Yes | 16 | |
| PAmcherry1 | 18,000 | 0.46 | 8.3 | — | — | 10 | |
| mEOS3.2 | 32,200 | 0.55 | 17.7 | — | — | 11 | |
| 647 | Alexa 647 | 270,000 | 0.33 | 89.1 | No | No | 17 |
| PA-JF 646 | 120,000 | 0.53 | 63.6 | Yes | Yes‡ | 15 | |
| Cy5 | 250,000 | 0.28 | 70.0 | No | Yes | 17 | |
| iRFP 670 | 114,000 | 0.11 | 12.5 | — | — | 55 |
This table is not to summarize all single-molecule probes but only highlights some commonly used dyes or fluorescent proteins.
Molecular brightness is calculated by extinction coefficient × quantum yield/1,000.
Dye-lipid bilayer nonspecific interaction of PA-JF 549 and PA-JF 646 are not reported from the literature but were tested on our own. We observed nonspecific association of both dyes on plasma membranes in HeLa and COS7 cells. Those nonspecific associations can be removed almost completely by extensive washing steps.
An alternative solution to achieve multicolor SMT involves intracellular delivery of dye-labeled purified proteins. Microinjection and cuvette electroporation (CEP) are the most practical intracellular delivery strategies. While microinjection allows for the introduction of various types of exogenous macromolecules into living cells, it is labor intensive and requires a sophisticated experimental apparatus (19-21). Efforts have already been made to demonstrate that CEP allows intracellular delivery of dye-labeled molecules, including DNA oligos and proteins, for in vivo SMT (22-24). However, to the best of our knowledge, CEP has yet to be demonstrated to deliver multiple types of proteins to the same cells to achieve in vivo multicolor SMT. This could be due to the challenges of CEP: that it is inefficient for multitype cargo loading and is too invasive to conduct sequential delivery for different cargos. To overcome these challenges, we have demonstrated a simple nanopore electroporation (NanoEP) method built on a nanopore-embedded water filter to achieve nontoxic and highly efficient localized electroporation (25). In comparison with CEP, NanoEP porates a small fraction of the cell membrane, which not only preserves cell viability but also allows sequential delivery of various types of cargos. In addition, NanoEP employs electrophoresis as the main mass-transport mechanism to inject the charged cargos into cells, thus allowing for increases in cargo-loading efficiency of 10- to 100-fold, compared with the diffusion mechanism in CEP. We apply this method to deliver multiple organic dye-labeled proteins intracellularly to realize multicolor SMT in living cells. This method poses great experimental versatility. Purified proteins can be labeled with a broad selection of high-performance dyes that meet the optical criteria for SMT. As the protein labeling happens in a homogenous solution in vitro, this method circumvents the technical hurdles of the intracellular protein labeling with only membrane-permeable dyes. It can be easily expanded to three channels in commercial microscopes or be combined with other in situ labeling methods for broad research applications.
Here, we take this strategy to look at epidermal growth factor receptor (EGFR) signaling transduction on plasma membranes. Despite decades of intense studies on structural and functional details of EGFR, there remains little consensus on the fundamental mechanism by which receptor triggering is coupled to its downstream Ras-Erk activation. The canonical view of EGFR activation considers the monomeric receptor to dimerize upon the addition of epidermal growth factor (EGF) ligands (26), which involves a strong cooperativity between the extracellular, transmembrane, and intracellular domains (27). Recently, questions were raised that dimerization was not sufficient to activate Ras (28). Weakened EGFR dimers induced by low-affinity ligands even result in a more sustained Erk activation (29). Other studies have observed that EGFR predominately existed in nanoclusters upon activation (30) and identified possible oligomerization interfaces through extracellular cis interactions (31, 32). Such models argue that the oligomerization is essential to organizing kinase domains in a way to fully boost receptor phosphorylation. While EGFR nanoclustering was mostly explained by its own molecular structures, the intracellular domain continuously interacts with adaptor proteins, kinases and phosphatases, and actin cytoskeleton, which could participate in remodeling the receptor organizations. In a comparative T cell receptor-signaling system, an interesting finding shows that a scaffold protein, LAT, an adaptor protein, Grb2, and a guanine exchange factor, SOS, together form a condensation phase separation through multivalent interactions (33–35). Notably, the phase-separated clusters are signaling hubs to activate SOS through kinetic proofreading, which eventually leads to Ras activation (36). It is not clear whether such a mechanism applies to EGFR. Only with better spatial and temporal resolution could it help unravel these outstanding questions. In this study, we simultaneously monitored two or three protein species, including Grb2, SOS, Ras, and Src, which are single molecules downstream of the EGFR signaling pathway in HeLa and COS-7 cell lines. The results reveal that Grb2 and SOS are recruited to the plasma membrane coordinately to form nanoclusters after EGF stimulation, while Ras and Src are more uniformly distributed. Finally, we extend the application of NanoEP to primary T cells.
Results and Discussion
Live Cell Imaging through Intracellular Protein Delivery.
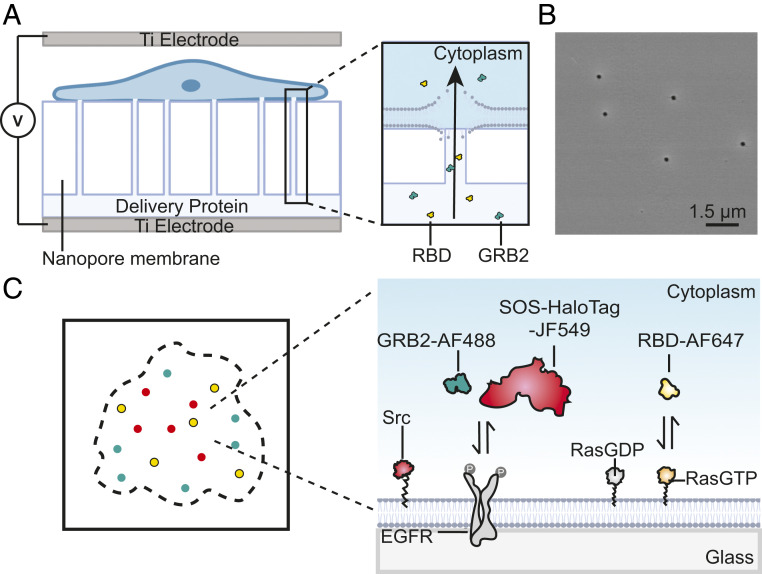
To perform multicolor SMT in living cells, the targeted proteins are purified and labeled with desirable organic dyes through N-terminal amine-NHS (N-HydroxySuccinimide) ester conjugation. Next, the dye-labeled proteins are delivered into the cytoplasm by NanoEP as described previously (25) (Fig. 1A). The NanoEP device consists of two flat titanium electrodes and a polydimethylsiloxane (PDMS) holder with a track-etched polycarbonate (PC) water-filter membrane embedded with nanopores of about 100 nm in diameter (Fig. 1B). Cells are cultured on the PC membrane and precoated with fibronectin so that the cell membrane makes close contact to the nanopores. When applied with direct current electric pulses through the nanopores, transient pores form on the local cell membrane that makes close contact to the nanopores, allowing for delivering of the dye-labeled proteins into cells with minimal cellular toxicity. The delivered cells are then replaced on the integrin ligand Arg–Gly–Asp (RGD)-coated glass coverslip for imaging with total internal reflection fluorescence (TIRF) microscopy. In this study, we focus on EGFR signaling transductions. After activation by EGF, Grb2 and SOS are recruited to EGFR, which further catalyzes the Ras-GDP exchange to Ras-GTP, and activates MAPK signaling cascade (37). The GTP-loaded Ras can be detected by a Ras-binding domain (RBD) truncated from Raf (36). EGFR also undergoes Src-dependent tyrosine phosphorylation in several residuals that are different from autophosphorylation sites (38), which adds another layer of signaling modulations (Fig. 1C). Simultaneously monitoring of these molecules by multicolor SMT would give an unpreceded resolution of how they are dynamically coupled on membranes.
Fig. 1.
Schematic of multicolor single-molecule imaging enabled by NanoEP delivery. (A) A schematic of the NanoEP device. A nanopore-embedded PC water-filter membrane is sealed at the base of PDMS holder to connect delivery proteins and cell membranes. (B) An SEM image of randomly distributed 100-nm nanopores on the PC membrane. (C) A schematic of multichannel single-molecule imaging of the EGFR signaling pathway. Dye-labeled Grb2 and RBD are delivered into cells, while SOS and Src are expressed by plasmid transfection.
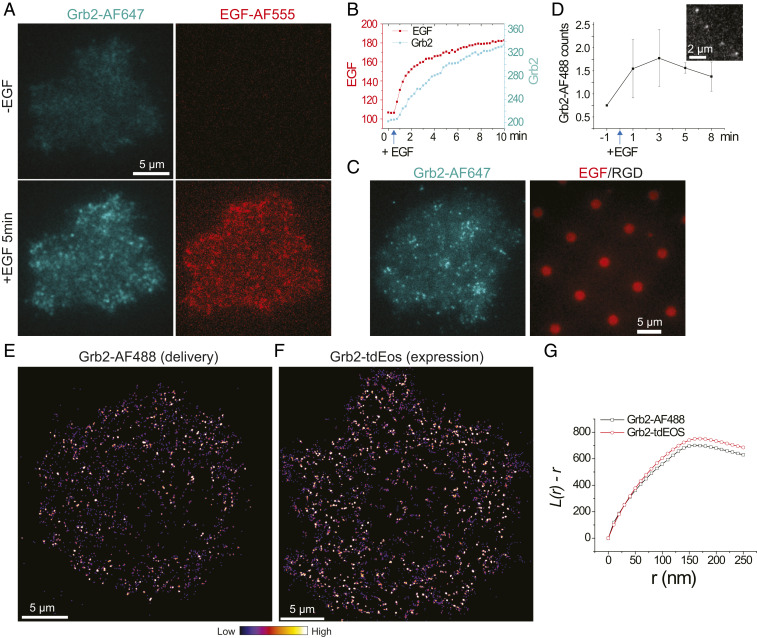
To confirm that NanoEP-delivered purified proteins are fully functional, we first studied Alexa Fluor (AF) 647–labeled Grb2 activities in response to EGF stimulation in living HeLa cells at bulk concentration. Immediately after addition of EGF ligands, the delivered Grb2-AF647 were recruited to plasma membranes, suggesting its association onto activated EGFR (Fig. 2A and Movie S1). Grb2-AF647 intensity gradually increased along with bound EGF in the next 10 min (Fig. 2B), which is consistent with the observation using plasmid expression as reported previously (39). The functionality of Grb2-AF647 was further confirmed by their enrichment on the EGF region when cells were spread on EGF/RGD-micropatterned substrates, while the RGD region also recruited Grb2 through integrin receptors to a lesser degree (40)(Fig. 2C). These results suggest that delivered Grb2-AF647 behave similarly to native proteins. This method can be easily extended to other dyes and cell lines. We replaced the protein-labeling dye from AF647 to AF488. The delivered Grb2-AF488 were also functional in COS7 cells, as observed by their colocalization with ephrinA1-displayed lipid bilayers through EphA2 receptor-signaling pathways (SI Appendix, Fig. S1), which is also consistent with previous results using Grb2-tdEos expression (41).
Fig. 2.
Delivery of Grb2 into HeLa cells for bulk- and single-molecule imaging. (A) Fluorescent images of AF647, labeled Grb2, recruitment to plasma membrane after EGF (50 ng/mL) stimulation. (B) Quantification of time-course Grb2 and EGF fluorescence intensity. (C) Fluorescent images of a Grb2-AF647-delivered HeLa cell spreading on EGF patterned substrate. (D) Counts of Grb2-AF488 molecule appearances in each 1.5-s recording period as response to EGF stimulation, with an insertion of a representative single-molecule image. n = 6 cells; error bar shown as SD. (E) A reconstructed frequency image of Grb2-AF488 single-molecule recruitment after EGF stimulation. It is generated by assembly of the first spots of every single-molecule trajectory. Each spot is plotted as Gaussian point spread function with 30-nm pixel size and 90-nm FWHM. (F) A reconstructed frequency image of Grb2-tdEos single-molecule recruitment after EGF stimulation. (G) A cluster analysis of Grb2-AF488 and Grb2-tdEos using Ripley’s K-function (L(r)-r).
NanoEP Enabled Multicolor SMT.
The major advantage of the NanoEP strategy is to probe single molecules inside living cells with high-performance, membrane-impermeable fluorophores. By diluting Grb2-AF488 protein concentration, we are able to deliver a trace amount of proteins into HeLa cells so that single-molecule resolution can be achieved. As shown in Movie S2, shortly after EGF stimulation, delivered Grb2-AF488 single molecules clearly emerged in the periphery of the cell. Its recruitment rate (Kon) increased and peaked at about 3 min after EGF (Fig. 2D), suggesting maximal EGFR phosphorylation at this time point. The coordinates of every Grb2-AF488 molecule at its first trajectory position were assembled to reconstruct a binding frequency image [also termed sptPALM (42)], revealing its clustering characteristic (Fig. 2E). This is consistent with the result using a photoconvertible tdEos fusion protein (39) (Fig. 2F). Both images showed that Grb2 has an average cluster radius at around 160 nm after EGFR activation, analyzed by Ripley’s K-function (43) (Fig. 2G). Importantly, the amount of delivered proteins are almost negligible compared with endogenous expression levels, thus posing little effect on changing native signaling transductions, which is generally a concern with plasmid overexpression. Therefore, NanoEP-enabled protein delivery is a potent method to study single-molecule dynamics in living cells.
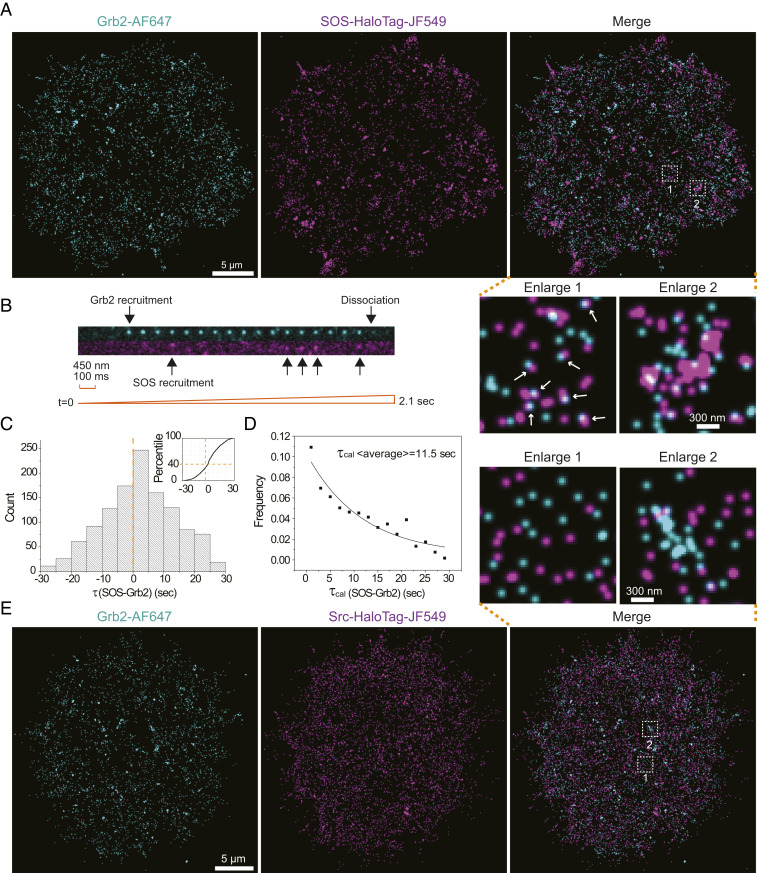
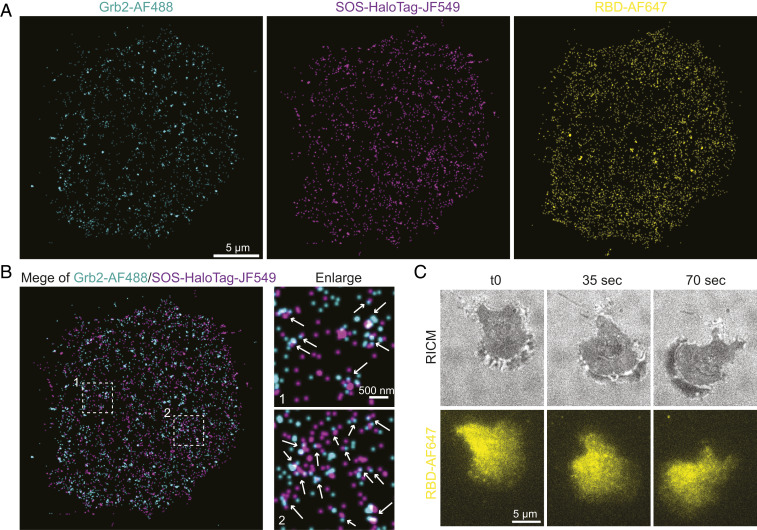
The NanoEP method can be combined with available in situ labeling strategy for multicolor SMT. Many large proteins, such as SOS (∼160 kDa), and membrane proteins, such as Src, are difficult to purify and still maintain normal activity in water-soluble conditions and thus cannot be delivered by NanoEP. In some cases, purifying proteins with correct posttranslational modifications can be challenging. Genetic expression of these proteins with a Halo-tag allows in situ labeling with membrane-permeable dyes. As studied here, Halo-tagged SOS were expressed in HeLa cells, labeled with photoactivatable dye JF549, and the cells were subsequently delivered with Grb2-AF647 to achieve two-color SMT. Single-channel imaging first confirmed dynamic recruitment of SOS-HaloTag-JF549 by EGFR activation, with Kon peaks at 3 min (SI Appendix, Fig. S2A and Movie S3). Next, simultaneous two-color tracking indicated correlative recruitment of Grb2-AF647 and SOS-HaloTag-JF549 onto plasma membranes (Fig. 3A and Movie S4). Formation of Grb2-AF647 and SOS-HaloTag-JF549 doublets were found throughout the cell (Fig. 3A, enlarge 1), as well as their coalescences (Fig. 3A, enlarge 2), suggesting that EGFR, Grb2, and SOS form complexes, which further assemble into clusters. SOS-HaloTag-JF549 appeared to have a slightly larger cluster size than Grb2-AF647 (SI Appendix, Fig. S2B), possibly reflecting the features from integrin adhesions and signaling.
Fig. 3.
Two-color single-molecule imaging by combining NanoEP delivery and in situ labeling. (A) Reconstructed frequency images of Grb2-AF647 and SOS-HaloTag-JF549 single molecules after EGF stimulation, with indicated enlarged regions. The arrows indicate Grb2 and SOS doublets. (B) An image montage constructed from the two-color Grb2-AF647 and SOS- HaloTag-JF549 single-molecule movie, with arrows indicating molecule recruitment and dissociation. (C) Quantification of recruitment delay τ(SOS-Grb2) in each of the SOS and Grb2 doublets, with bin size set as 5 s. (D) τcal(SOS-Grb2) is calculated by subtracting the negative portion from its correspondent positive values, with bin size set as 2 s. The data are fitted by a single exponential decay function in a solid line. (E) A reconstructed image of Grb2-AF647 and Src- HaloTag-JF549 single molecules after EGF stimulation, with indicated enlarged regions.
Simultaneous two-color SMT is a powerful method to detect molecular timing, which could provide insightful information to understand signaling propagations, such as that shown in the discovery of kinetic proofreading mechanism of SOS activation in an in vitro reconstitution system (36). However, such a method remains difficult to implement into living cells. Here, we seek to directly measure the time delay between Grb2 and SOS recruitment downstream of EGFR signaling pathway in HeLa cells. As shown in the snapshots (Movie S4), the two-color single-molecule movie clearly indicated recruitment of a Grb2-AF647 molecule onto membranes, followed by dynamic recruitment of SOS-HaloTag-JF549 in the same place, before they both dissociated or were photobleached (Fig. 3B). The first trajectory coordinates of such Grb2-AF647:SOS-HaloTag-JF549 doublets were mapped out throughout the cell to calculate recruitment time difference τ(SOS-Grb2), with the criteria that a paired Grb2-AF647 and SOS-HaloTag-JF549 are less than 30 nm apart (SI Appendix, Fig. S2C). To avoid complexity from clustering, a Grb2 or SOS with multiple neighboring molecules was excluded in this analysis. Statistically, SOS-HaloTag-JF549 were found to be recruited later than Grb2-AF647, consistent with the model that SOS bind EGFR through Grb2 adaptor (Fig. 3C). Notably, the observed single-molecule events were surrounded by numerous nonfluorescent molecules. As EGFR requires a minimum of dimerization, or multimerization, to become activated, the recruited Grb2-AF647 and SOS-HaloTag-JF549 in each doublet possibly bind to different receptors in the same complex, resulting the apparent negative values in τ(SOS-Grb2) distributions. Suppose the background molecular-binding sequence follows a Gaussian random distribution, the time delay between Grb2-AF647 and SOS-HaloTag-JF549 could then be inferred by subtracting the negative portion from its correspondent positive values. As a result, calculated τcal(SOS-Grb2) fits well with a single exponential kinetic model, given an average time delay of about 11.5 s (Fig. 2D). Such a long time could not be the waiting period of a SOS molecule binding onto an individual Grb2, because the average dwell time of Grb2 onto a comparative LAT molecule (35), or the dwell time of SH2 domain of Grb2 onto EGFR (39), were both measured to be subsecond level. It is more likely to represent an ensembled value of a larger complex, suggesting that even the well resolved doublets were predominately located in condensed nanoclusters.
This two-color SMT capability was also verified by simultaneously imaging delivered Grb2-AF647 and expressed nonreceptor kinase Src-HaloTag-JF549. Src phosphorylates several tyrosine residues of EGFR (38). In contrast to SOS, Src-HaloTag-JF549 were much more diffusive on plasma membrane and appeared uniformly distributed without clear correlation with Grb2 (Fig. 3E and Movie S5). This suggests that the Src interaction with EGFR is rather fast, which is consistent with the transient nature of kinase-substrate interactions: the negative charge of the added phosphate on tyrosine frequently repels the kinase (44). Together, a combination of NanoEP delivery and in situ labeling enables multicolor SMT for both cytosolic and membrane proteins.
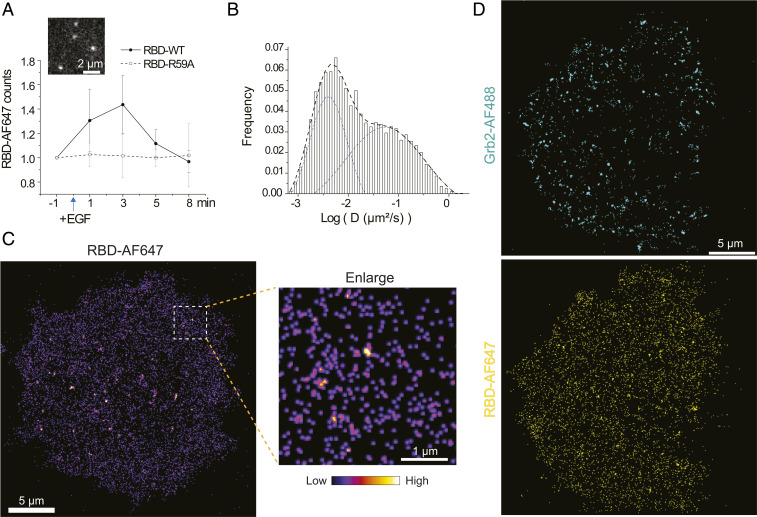
NanoEP also allows delivery of multiple proteins into the same cell. We further applied this system to track Ras activation, which is downstream of Grb2 and SOS. Ras undergoes GDP to GTP exchange after activation, which can be visualized by our recently developed RBD sensor that reversibly bind Ras-GTP (36). RBD-AF647 was verified to bind Ras-GTP in an in vitro assay (SI Appendix, Fig. S3). When delivered into cells, RBD-AF647 exhibited prominent membrane-recruitment dynamics (Movie S6). Its recruitment rate had a pulsed increase after EGF stimulation and then quickly reverted to basal level (Fig. 4A), suggesting fast turnover of Ras-GTP. Whereas in the negative control, a point-mutation RBD-R59A that has 29-fold lower Ras-GTP binding affinity (45), did not change its binding rate after EGF (Movie S7). RBD-AF647 had both immobile and mobile populations on the membranes (Fig. 4B). Their recruitment was largely uniform although embedded with tiny “hot spots” (Fig. 4C), reflecting membrane diffusion of Ras-GTP, as well as the possible formation of Ras nanoclusters as reported previously (46, 47). The immobile fraction of RBD-AF647 increased by about 5% after EGF stimulation, which could be the increase of Ras nanoclusters (SI Appendix, Fig. S4A). Noteworthily, a few big cluster-like structures mostly located in the central region of the cell probably correspond to endocytic/recycling vesicles, whose membranes are also rich in Ras-GTP (48). Similar with Grb2, RBD labeling can be easily switched to another dye, such as Janelia fluorophore (JF) 646, and was verified in COS7 cells (SI Appendix, Fig. S4B). Having validated single-molecule imaging of RBD, we combined Grb2-AF488 and RBD-AF647 delivery in the same HeLa cell for two-color SMT. While Grb2-AF488 showed robust clustering, RBD-AF647 were uniformly distributed (Fig. 4D), suggesting that Ras was quickly disseminated after activation from EGFR:Grb2:SOS complexes.
Fig. 4.
Delivery of RBD and Grb2 for two-color single-molecule imaging. (A) Counts of AF647-labeled RBD-WT or RBD-R59A mutant molecules in each 1.5-s acquisition period as response to EGF stimulation, with an insertion of a representative RBD-WT-AF647 single-molecule image. n = 6 cells for each group; error bar shown as SD. (B) Distribution of RBD-AF647 diffusion coefficient (D) from each molecular trajectory after EGF stimulation. The D value is normalized by log (D), shown in X-axis. The distribution is well fitted by a two-peak Gaussian distribution function in dashed lines. n = 10 cells. (C) A reconstructed frequency image of RBD-AF647 single molecules after EGF stimulation. (D) Two-color reconstructed frequency images of Grb2-AF488 and RBD-AF647 single molecules after EGF stimulation.
We then delivered both Grb2-AF488 and RBD-AF647 into SOS-HaloTag-JF549 expressing cells to demonstrate three-color SMT. Consistent with previous results, Grb2-AF488 and SOS-HaloTag-JF549 showed clear clustering after EGF treatment, whereas RBD-AF647 were more uniform, although embedded with tiny “hot spots” (Fig. 5A and Movie S8). Merging of Grb2-AF488 and SOS-HaloTag-JF549 images indicated coassembly of these molecules (Fig. 5B). Taken together, simultaneous three-color SMT has revealed the spatial heterogeneity of EGFR signaling: Grb2 and SOS tend to assemble with EGFR into higher order clusters, which then transduce signals into membrane diffusive Ras. Therefore, we anticipate that the further downstream Raf, which is activated by Ras-GTP, is very likely to spatially decouple from EGFR activation sites.
Fig. 5.
Three-color single-molecule imaging. (A) Three-color reconstructed frequency image of Grb2-AF647, SOS- HaloTag-JF549, and RBD-AF647 single molecules after EGF stimulation. (B) A merging of Grb2-AF647 and SOS- HaloTag-JF549, with arrows indicating formation of doublets or clusters. (C) Snapshots of a RBD-AF647 delivered primary T cell crawling on ICAM supported lipid bilayers. RICM stands for reflective interference contrast microscopy.
There is one caveat for such three-color imaging experiments. Unexpectedly, we detected a noticeable diffusion difference when tracking Src-HaloTag-JF549 molecules under single 561-nm laser illumination or alternating with 488-nm laser exposure for an extended period of time, possibly due to photo-induced oxidation of JF549 dyes by cross-illumination; but this did not happen in the case of 647 nm laser coexposure (SI Appendix, Fig. S5). A compromised solution is to image 488 nm and other channels simultaneously for a short time to reduce cross-effect, at the cost of recording fewer molecules. Prospective tests of more dyes and understanding of their photochemical properties are necessary to find the optimal combination in three-color SMT.
Application of NanoEP in Primary T Cells.
The NanoEP strategy not only promotes multicolor SMT in cell lines, but also enables single molecule imaging in hard-to-transfect primary T cells by intracellularly delivering dye-labeled proteins with minimum toxicity. As it is known, T cells are extremely sensitive to ligand stimulation. SMT provides an ideal way to study individual ligands: T cell receptor recognition and signaling propagation (4, 6). However, primary T cells generally rely on virus-based transfection to image, which is troublesome and has a broad variation in transfection efficiency. To tackle the transfection hurdle, we demonstrate that NanoEP is able to successfully deliver RBD-AF647 into mouse T cells for imaging at bulk concentration at a high efficiency (∼60%) (Movie S9), as well as at single-molecule levels (Movie S10). The cells were robustly crawling on intercellular adhesion molecule (ICAM) functionalized supported lipid bilayers (Fig. 5C and Movie S9), suggesting that the delivery did not affect cellular activities. We envision NanoEP-facilitated SMT will be a promising method to study single molecules in versatile primary cells.
Conclusion
In summary, we employed nanopore-mediated protein delivery to realize multicolor SMT in living cells. This strategy significantly expands intracellular single-molecule probes to optical high performance and lipid interaction–free organic dyes in all wavelength bands. With this system, we simultaneously tracked two or three molecules downstream of the EGFR signaling pathway and provided detailed resolution of the spatial localization and dynamics in the receptor signaling transductions. We found that Grb2 and SOS predominately form relatively stable clusters in response to EGFR activation, possibly suggesting their coassembly with the receptors into higher-order structures. Ras and Src only interacted with EGFR transiently, suggesting that even downstream Raf activation is spatially decoupled from EGFR clusters. Additionally, this method is promising for single-molecule imaging in primary cells in the future.
Materials and Methods
Assembly of the Nanopore Device.
The experimental protocol was described previously (25). The NanoEP device consists of two titanium-plate electrodes, a 20-μm-thick track-etched PC water-filter membrane with nanopore density of 2 × 108 nanopores per cm2 (Whatman) and a PDMS (Dow Corning) square (1.5 × 1.5 × 0.2 cm) with a hole 0.5 cm in diameter in the middle. We chose the 100-nm nanopore PC membrane for its strength and its adequate pore size for the passage of our target biomolecules. We used uncured PDMS with the ratio of 10 (base):1- (curing agent) to attach the PC membrane onto one end of the PDMS hole to make a well and cured the construct at 90 °C for 5 min.
Protein Labeling.
The NHS-ester labeling process of Grb2-AF647 (Sigma), Grb2-AF488 (Sigma), RBD-AF647 (Sigma), and RBD-JF646 (TOCRIS) followed the vendor’s manual, at pH 8.3 and at room temperature for 1 h. Specifically, the protein:dye molar ratio is controlled to be 1:3 during conjugation reaction to avoid overlabeling, which results in the final dye-to-protein ratio of Grb2-AF647 (0.8), Grb2-AF488 (0.9), RBD-AF647 (0.8), and RBD-JF646 (0.6). The proteins are prepared in the concentration of 3 to 5 μM PBS (phosphate-buffered saline) solution for delivery.
Protein Delivery by Nanopore Electroporation.
To perform protein delivery, the NanoEP device is first coated with 20 μL 100 ng/mL fibronectin for 2 h. After two washes with deionized water and one wash with cell culture medium, the device is ready to use. About 15,000 cells of interest are pipetted into the device well in 80 μL corresponding culture medium. To form a tight cell–nanopore interface, adherent cells are grown on the PC surface overnight under 5% CO2 at 37 °C. For suspension-primary T cells, cells are centrifuged at 150 × g for 5 min to establish tight cell contact with the nanopores before delivery. To perform delivery, square-wave DC pulses of 20 Hz, 200 μs, and a range of voltage intensities from 20 to 45 V are generated by a square-pulse stimulator (Grass Instruments) and applied between the two titanium electrodes for 20 to 60 sec.
Electrophoresis is considered to be the dominant mechanism to transport biomolecules across the nanopore membrane. Therefore, electric field polarity is primarily determined by the charges on delivery molecules. As different dyes have different charges, the charge on a protein can be greatly altered by dye labeling, thus the ultimate charges on a dye-labeled protein cannot be assessed by summing up the charges of each amino acid on a protein. A preliminary delivery test is preformed to determine which electric field polarity drives the charged dye-labeled protein into cells. Briefly, cells of interest are cultured on the NanoEP device as described above. A total of 5 µL dye-labeled protein solution is loaded at ∼10 nM concentration on the bottom titanium electrode, and the NanoEP device is immediately placed on top of the protein solution. Both electric field polarities (cathode as the top electrode and anode as the top electrode) are used to deliver the dye-labeled protein. The proper electric-field polarity for the protein of interest is determined by comparing the protein-loading efficiency from both electric-field polarities. Delivery duration is another parameter that can be tuned to control delivery dosage of proteins. Longer duration allows for more protein loading and vice versa. The detailed mathematical model of the cargo mass transport mechanism is described in our previous work (49). Since electroporation physically prorates the cell membrane, the variation of cell membrane composition between different cell types does not strongly affect the performance of NanoEP as long as the cell membrane has tight contact with nanopores during delivery. The delivery conditions for each dye-labeled protein are summarized in Table 2.
Table 2.
Summary of NanoEP delivery parameters of different proteins
| Dye-labeled protein name | Top electrode polarity | Voltage intensity (V) | Delivery duration (s) |
| Grb2-AF647 | Negative | 35 | 30 |
| Grb2-AF488 | Positive | 35 | 60 |
| RBD-AF647 | Positive | 30 | 15 |
| RBD-JF646 | Negative | 30 | 20 |
To deliver two or more dye-labeled proteins, each protein specie is delivered into the cells independently, one after another. To combine with in situ labeling strategy, cells are transfected with SOS-Halo/Src-Halo 1 d before usage. On the day of usage, SOS-HaloTag/Src-HaloTag are labeled with membrane-permeable dye PA-JF549 (a generous gift from Luke Lavis, Janelia Research Campus, HHMI, Ashburn, VA) at single molecule density (∼10 nM dye for 30 min). After that, single Grb2-AF647 or Grb2-AF488 plus RBD-AF647 are delivered into cells. After delivery, the delivered cells are detached by enzyme-free cell stripper (Thermo-Fisher), washed four times, and replated to a PLL-PEG-Biotin + Neutravidin + Biotin-RGD precoated glass-slip cell culture chamber (41). The delivered cells are then imaged between 1 to 3 h at 37 °C in imaging buffer (25 mM Tris, 140 mM NaCl, 3 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 5.5 mM D-glucose).
Plasmid Constructs.
The C terminus of SOS and Src sequences is attached to a linker (GGGGSGGGDP) followed by the sequence of HaloTag fusion protein. The combined gene is cloned into the mammalian expression vector (pCAGGS). Both sequences of SOS-Halo and Src-Halo are further confirmed by DNA sequencing.
Micropatterning.
Micropatterning of supported lipid-bilayer ephrinA1 and RGD, or immobilized EGF and RGD were done by photolithography, as reported in our previous paper (41).
Cell Transfection and EGF Treatment.
HeLa and COS7 cells were transfected by Lipofectamine 2000 (Thermo Fisher) 1 d before usage, following vendor’s manual. During the experiment, 50 ng/mL EGF was added into the imaging chamber to stimulate cells. The cells were then quickly imaged and recorded for single-molecule movies.
Mouse T Cell Preparation.
CD4+ T cells from the lymph nodes and spleens of 6- to 20-wk-old AND × B10.BR transgenic mice (Jackson Laboratory) were stimulated with MCC (moth cytochrome C) peptide in vitro, as described previously (4). All animal work was performed with prior approval by Lawrence Berkeley National Laboratory Animal Welfare and Research Committee under the approved protocol 17702.
Single-Molecule Imaging.
An Eclipse Ti inverted microscope (Nikon) with a TIRF system and iXon EMCCD (Electron Multiplying Charged-Coupled Device) camera (Andor Technology) was used for live-cell imaging. TIRF microscopy was performed with a 100× TIRF objective with a numerical aperture of 1.49 (Nikon) and a U-N4S 4-laser unit as a laser source (Nikon). To increase tracking accuracy, cells with the density of individual molecules less than ∼0.5/µm2 were chosen for imaging.
To obtain time-dependent recruitment rate of Grb2-AF488, SOS-JF549, and RBD-AF549, the properly delivered or transfected cells were imaged at 20 fps (frames per sec) video rate, for a total of 30 frames (1.5 s) at each time point before or after EGF treatment. To obtain reconstructed frequency images, cells were imaged 1 to 2 min after EGF treatment at 20 fps video rate, for a total of 1,000 frames (50 s) for future analysis.
Multichannel single-molecule images were acquired using a “Triggering TIRF” mode in Nikon microscope, in which multilasers were alternating to illuminate the sample, and the photons from each fluorescence signal were recorded by a single camera sequentially. In this mode, a single multiband filter was used for all colors so that the filter does not need to be switched during imaging. Digital switching of different lasers allows for the fast alternating illumination and essentially simultaneous multichannel single-molecule recording. For both two- and three-color imaging, the exposure time of each image is 50 ms; two-color imaging has a 10 fps video rate and three-color imaging has a 6.67 fps video rate.
Image Analysis.
Single-molecule movies were tracked by an ImageJ plugin “TrackMate” (50) to obtain molecular spots and trajectories. The total counted molecule numbers per time is calculated as binding frequency (Kon). To get diffusion coefficient (D), the first four spots from each trajectory were linearly fitted by the equation MSD = 4Dt, in which MSD is mean square displacement, and t is time delay between each image (50 ms). Individual D values were log normalized in the distribution plot to clearly show immobile and mobile populations (1). The distribution was fitted by two-peak normal distributions with Origin software to get immobile and mobile fractions.
The frequency image reconstruction was done by an ImageJ plugin “Octane” developed by Ji Yu’s Lab (51). The first spots of every molecular trajectory (with trajectory length more than two spots) were assembled to generate binding frequency image. All x-y coordinates were replaced as Gaussian point spread function with 30 nm pixel size and 90 nm FWHM (full width at half maximum).
The spatial dispersion of these x-y coordinates was tested using modified Ripley’s K-function, so called L(r) function, considering the edge affect correction (52). Counting the number of points within a circle with radius (r), we computed L values as function of r with 10 nm bin. Both Grb2-AF488 and Grb2-tdEos had peak L values at r = 160 nm, corresponding to the size of 320 nm cluster.
Grb2 and SOS doublets in two-color single-molecule imaging were isolated with the criteria that the distance between a paired Grb2 and SOS is less than 30 nm. For simplicity, a Grb2 or SOS molecule with multiple neighboring molecules that is within 30 nm were excluded in this analysis. All detected Grb2 and SOS doublets were isolated pairs. Further, the time delay τ(SOS-Grb2) quantification excluded doublets that were longer than 30 s because these only contributed to a very small fraction of the total data points, and they were most likely to be resulted from coincident colocalization during 100 s long imaging of the cell.
Supplementary Material
Acknowledgments
This work was supported by NIH U01 (CA202241) and the W. M. Keck Foundation. Z.C. is also funded by Shanghai Municipal Science and Technology Major Project ZJLab (2018SHZDZX01).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012229118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Sako Y., Kusumi A., Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J. Cell Biol. 125, 1251–1264 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung I., et al. , Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 464, 783–787 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Sakon J. J., Weninger K. R., Detecting the conformation of individual proteins in live cells. Nat. Methods 7, 203–205 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Donoghue G. P., Pielak R. M., Smoligovets A. A., Lin J. J., Groves J. T., Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. eLife 2, e00778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chong S., et al. , Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science 361, eaar2555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin J. J. Y., et al. , Mapping the stochastic sequence of individual ligand-receptor binding events to cellular activation: T cells act on the rare events. Sci. Signal. 12, eaat8715 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusumi A., Tsunoyama T. A., Hirosawa K. M., Kasai R. S., Fujiwara T. K., Tracking single molecules at work in living cells. Nat. Chem. Biol. 10, 524–532 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Boersma S., et al. , Multi-color single-molecule imaging uncovers extensive heterogeneity in mRNA decoding. Cell 178, 458–472.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sungkaworn T., et al. , Single-molecule imaging reveals receptor-G protein interactions at cell surface hot spots. Nature 550, 543–547 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Subach F. V., et al. , Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang M., et al. , Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 9, 727–729 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Los G. V., et al. , HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Keppler A., Pick H., Arrivoli C., Vogel H., Johnsson K., Labeling of fusion proteins with synthetic fluorophores in live cells. Proc. Natl. Acad. Sci. U.S.A. 101, 9955–9959 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L., et al. , A general strategy to develop cell permeable and fluorogenic probes for multicolour nanoscopy. Nat. Chem. 12, 165–172 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Grimm J. B., et al. , Bright photoactivatable fluorophores for single-molecule imaging. Nat. Methods 13, 985–988 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Grimm J. B., et al. , A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250, 3, 250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes L. D., Rawle R. J., Boxer S. G., Choose your label wisely: Water-soluble fluorophores often interact with lipid bilayers. PLoS One 9, e87649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z., Yomo D., Gradinaru C., Choosing the right fluorophore for single-molecule fluorescence studies in a lipid environment. Biochim. Biophys. Acta Biomembr. 1859, 1242–1253 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Hallett M. B., Campbell J. S., Laffafian I., Dewitt S., Microinjection and micropipette-controlled phagocytosis methods for neutrophils. Methods Mol. Biol. 2087, 117–125 (2020). [DOI] [PubMed] [Google Scholar]
- 20.von Dassow G., Valley J., Robbins K., Microinjection of oocytes and embryos with synthetic mRNA encoding molecular probes. Methods Cell Biol. 150, 189–222 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Jones V. A. S., Bucher M., Hambleton E. A., Guse A., Microinjection to deliver protein, mRNA, and DNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Sci. Rep. 8, 16437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crawford R., et al. , Long-lived intracellular single-molecule fluorescence using electroporated molecules. Biophys. J. 105, 2439–2450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Paolo D., Afanzar O., Armitage J. P., Berry R. M., Single-molecule imaging of electroporated dye-labelled CheY in live Escherichia coli. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun C., Cao Z., Wu M., Lu C., Intracellular tracking of single native molecules with electroporation-delivered quantum dots. Anal. Chem. 86, 11403–11409 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Cao Y., et al. , Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. Proc. Natl. Acad. Sci. U.S.A. 116, 7899–7904 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlessinger J., Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110, 669–672 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Endres N. F., et al. , Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152, 543–556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang S. I., et al. , Phosphorylated EGFR dimers are not sufficient to activate Ras. Cell Rep. 22, 2593–2600 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freed D. M., et al. , EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell 171, 683–695.e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clayton A. H. A., Orchard S. G., Nice E. C., Posner R. G., Burgess A. W., Predominance of activated EGFR higher-order oligomers on the cell surface. Growth Factors 26, 316–324 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Needham S. R., et al. , EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat. Commun. 7, 13307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y., et al. , Molecular basis for multimerization in the activation of the epidermal growth factor receptor. eLife 5, e14107 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Houtman J. C. D., et al. , Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat. Struct. Mol. Biol. 13, 798–805 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Su X., et al. , Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W. Y. C., et al. , Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc. Natl. Acad. Sci. U.S.A. 113, 8218–8223 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W. Y. C., et al. , A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi T., et al. , Conservation of protein abundance patterns reveals the regulatory architecture of the EGFR-MAPK pathway. Sci. Signal. 9, rs6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Z., et al. , EGFR family and Src family kinase interactions: Mechanics matters? Curr. Opin. Cell Biol. 51, 97–102 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Oh D., et al. , Fast rebinding increases dwell time of Src homology 2 (SH2)-containing proteins near the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 109, 14024–14029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng S. Y. S., Sun G., Schlaepfer D. D., Pallen C. J., Grb2 promotes integrin-induced focal adhesion kinase (FAK) autophosphorylation and directs the phosphorylation of protein tyrosine phosphatase α by the Src-FAK kinase complex. Mol. Cell. Biol. 34, 348–361 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z., et al. , Spatially modulated ephrinA1:EphA2 signaling increases local contractility and global focal adhesion dynamics to promote cell motility. Proc. Natl. Acad. Sci. U.S.A. 115, E5696–E5705 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manley S., et al. , High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods 5, 155–157 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Prior I. A., Muncke C., Parton R. G., Hancock J. F., Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Oliveira P. S. L., et al. , Revisiting protein kinase-substrate interactions: Toward therapeutic development. Sci. Signal. 9, re3 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Jaitner B. K., et al. , Discrimination of amino acids mediating Ras binding from noninteracting residues affecting raf activation by double mutant analysis. J. Biol. Chem. 272, 29927–29933 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y., Hancock J. F., Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim. Biophys. Acta Mol. Cell Res. 1853, 841–849 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y., et al. , Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168, 239–251.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gomez G. A., Daniotti J. L., H-ras dynamically interacts with recycling endosomes in CHO-K1 cells: Involvement of Rab5 and Rab11 in the trafficking of H-Ras to this pericentriolar endocytic compartment. J. Biol. Chem. 280, 34997–35010 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Cao Y., et al. , Universal intracellular biomolecule delivery with precise dosage control. Sci. Adv. 4, eaat8131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tinevez J.-Y., et al. , TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Jadwin J. A., et al. , Time-resolved multimodal analysis of Src Homology 2 (SH2) domain binding in signaling by receptor tyrosine kinases. eLife 5, e11835 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goreaud F., Pélissier R., On explicit formulas of edge effect correction for Ripley’s K-function. J. Veg. Sci. 10, 433–438 (1999). [Google Scholar]
- 53.Sabinis R. W., “Oregon Green 488 carboxylic acid” in Handbook of Fluorescent Dyes and Probes (2015), pp. 302–303. [Google Scholar]
- 54.Patterson G. H., Lippincott-Schwartz J., A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 (2002). [DOI] [PubMed] [Google Scholar]
- 55.Shcherbakova D. M., Verkhusha V. V., Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 10, 751–754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.