Significance
Beclin 1, an essential autophagy protein, is important for tumor suppression in mice, as well as in human breast and ovarian cancers. However, it is not well understood how Beclin 1 acts as a tumor suppressor. By performing a genetic screen to identify genes whose loss blocks the ability of Beclin 1 to inhibit the growth of breast cancer cells and follow-up biological analyses, we have identified a mechanism by which Beclin 1 prevents breast cancer growth. We found that Beclin 1 promotes the plasma membrane localization of E-cadherin, a breast tumor-suppressor molecule that restricts tumor growth and metastases only when present at the cell surface. These findings have important implications for understanding the cell biology of human breast cancer.
Keywords: Beclin 1, E-cadherin, breast cancer
Abstract
Beclin 1, an autophagy and haploinsufficient tumor-suppressor protein, is frequently monoallelically deleted in breast and ovarian cancers. However, the precise mechanisms by which Beclin 1 inhibits tumor growth remain largely unknown. To address this question, we performed a genome-wide CRISPR/Cas9 screen in MCF7 breast cancer cells to identify genes whose loss of function reverse Beclin 1-dependent inhibition of cellular proliferation. Small guide RNAs targeting CDH1 and CTNNA1, tumor-suppressor genes that encode cadherin/catenin complex members E-cadherin and alpha-catenin, respectively, were highly enriched in the screen. CRISPR/Cas9-mediated knockout of CDH1 or CTNNA1 reversed Beclin 1-dependent suppression of breast cancer cell proliferation and anchorage-independent growth. Moreover, deletion of CDH1 or CTNNA1 inhibited the tumor-suppressor effects of Beclin 1 in breast cancer xenografts. Enforced Beclin 1 expression in MCF7 cells and tumor xenografts increased cell surface localization of E-cadherin and decreased expression of mesenchymal markers and beta-catenin/Wnt target genes. Furthermore, CRISPR/Cas9-mediated knockout of BECN1 and the autophagy class III phosphatidylinositol kinase complex 2 (PI3KC3-C2) gene, UVRAG, but not PI3KC3-C1–specific ATG14 or other autophagy genes ATG13, ATG5, or ATG7, resulted in decreased E-cadherin plasma membrane and increased cytoplasmic E-cadherin localization. Taken together, these data reveal previously unrecognized cooperation between Beclin 1 and E-cadherin–mediated tumor suppression in breast cancer cells.
In 1999, Beclin 1 was identified as both an essential autophagy protein and a haploinsufficient tumor suppressor (1). It functions in autophagy as part of a class III phosphatidylinositol 3-kinase (PI3KC3) complex, including either PI3KC3-C1, which contains VPS34, VPS15, and ATG14 and mediates autophagic vesicle nucleation, or PI3KC3-C2, which contains VPS34, VPS15, and UVRAG and mediates autophagosomal maturation and endocytic trafficking (2, 3). Although the mechanisms underlying the tumor-suppressor activity of Beclin 1 are not well understood, there are multiple lines of evidence from human genetic, mouse genetic, and cellular studies indicating that it is an important tumor-suppressor protein.
BECN1 maps to a tumor susceptibility locus on chromosome 17q21 that is monoallelically deleted in ∼30% of human breast cancers (1, 4) and ∼77% of human ovarian cancers (5). In sporadic human breast cancer, decreases in BECN1 mRNA, rather than the nearby tumor suppressor BRCA1, increase the risk of aggressive subtypes and are associated with worse patient prognosis (4). Monoallelic deletion of Becn1 in C57/BL6J mice results in increased spontaneous age-related carcinomas and mammary premalignant lesions (6, 7), whereas monoallelic deletion of Becn1 in FVB mice results in frank mammary carcinoma with features of basal-like breast cancer (8). Moreover, allelic loss of Becn1 promotes early ovarian tumor formation in a mouse model of ovarian carcinoma driven by SV40 large T antigen (5). Thus, despite the frequent codeletion of BECN1 and BRCA1 in human breast and ovarian cancers, mouse genetic models indicate that allelic loss of Becn1 (but not of BRCA1) is sufficient to drive tumorigenesis (5–8). Moreover, mice with increased autophagy by virtue of a gain-of-function mutation in Beclin 1 (that decreases binding to its negative regulator Bcl-2) have decreased age-related spontaneous tumorigenesis (9) and decreased HER2-mediated mammary tumorigenesis (10).
In addition to allelic loss in hormonally-driven cancers, there are multiple other mechanisms for Beclin 1 inactivation in human cancer. These include indirect oncogenic signaling events that activate autophagy-suppressive mechanistic target of rapamycin (e.g., enhanced receptor tyrosine kinase, PI3K, AKT signaling), inactivation by overexpression of Beclin 1-binding Bcl-2 family members, and direct inhibitory phosphorylation of Beclin 1 by Akt, EGFR, and HER2 (11). We previously showed that Akt-mediated transformation requires the Beclin 1 target serine phosphorylation sites (S234 and S295) (12), and that EGFR-driven tumorigenesis of non-small cell lung carcinoma requires the Beclin 1 target tyrosine phosphorylation sites (Y229, Y233, and Y352) (13). Moreover, an autophagy-inducing peptide, Tat-Beclin 1 (14), which disrupts the HER2/Beclin 1 interaction, was found to be as effective as a clinically used tyrosine kinase inhibitor in the treatment of HER2-positive mammary xenografts in mice (10).
Allelic loss of Becn1 in immortalized mammary epithelial cells increases chromosomal instability (15). Similarly, BECN1 knockdown in ovarian cancer cells drives chromosome instability and enhances migration in ovarian tumorigenesis (5). Similar effects are also observed with knockdown/knockout of other essential autophagy genes, such as ATG3, ATG5, and ATG7 (16, 17). Furthermore, enforced expression of wild-type Beclin 1, but not an autophagy-defective Beclin 1 serine 90 mutant protein, suppresses the growth of MCF7 breast cancer cells to form xenografts in nude mice (18). Together, these results suggest a role for Beclin 1 and other autophagy proteins in preserving genomic stability and preventing growth of breast and ovarian epithelial tumor cells. However, the precise cellular mechanisms underlying the tumor-suppressor function of Beclin 1 remain unknown.
To investigate the mechanism of Beclin 1-dependent tumor suppression, we performed a genome-wide CRISPR/Cas9 screen to identify genes whose loss reverses Beclin 1-dependent inhibition of MCF7 breast carcinoma cell proliferation. Our results indicate that loss of two genes, CDH1 and CTNNA1, encoding members of the E-cadherin complex (E-cadherin and alpha-catenin, respectively) involved in cell adhesion and suppression of the epithelial-mesenchymal transition (EMT), reversed Beclin 1-dependent inhibition of MCF7 proliferation. Moreover, loss of CDH1 and CTNNA1, reversed Beclin 1-dependent inhibition of anchorage-independent growth and xenograft formation in NOD/SCID mice. Enforced Beclin 1 expression in MCF7 cells increased cell surface localization of E-cadherin, whereas knockout of UVRAG or BECN1, but not of ATG14 or other autophagy genes, increased E-cadherin cytoplasmic localization. As the lack of cell surface E-cadherin is strongly associated with invasion, increased tumor grade, metastasis, and poor patient prognosis in breast cancer (19–21), these findings identify a previously undescribed mechanism of Beclin 1 and UVRAG in mammary tumor suppression—namely, the suppression of tumor cell growth by promoting E-cadherin complex plasma membrane localization.
Results
CRISPR Screen for Regulators of Beclin 1-Dependent Tumor Suppression.
The genome-wide clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 screen is a powerful tool to systematically elucidate gene functions in tumor cells. To investigate genes and pathways that contribute to Beclin 1-dependent tumor suppression, we performed a genome-wide CRISPR/Cas9 screen using MCF7 breast cancer cells with or without enforced Beclin 1 expression. For this purpose, we used two previously well-characterized human breast cancer cell lines, MCF7.beclin 1 cells that stably express tetracycline-repressible Beclin 1 and MCF7.control cells stably transfected with an empty vector (22, 23). The MCF7.control and MCF7.beclin 1 cells were transduced with a pooled human genome-scale CRISPR/Cas9 knockout Brunello small guide (sg) RNA library (24) (SI Appendix, Fig. S1A). We confirmed that transduction with the CRISPR library did not interfere with Beclin 1-enforced expression in MCF7.beclin 1 cells and did not interfere with Beclin 1-dependent tumor suppression (SI Appendix, Fig. S1 B and C). For each cell line, we assessed the abundance of sgRNAs by next-generation sequencing at the start of the screen (T = 0) and after 4 wk of proliferation (T = 4). Two independent algorithms were used to calculate the enrichment or depletion of sgRNAs. The log2 fold change (LFC) in sgRNA abundance after 4 wk (T = 4) compared to baseline (T = 0) was calculated, and differentially enriched sgRNAs in MCF7.control and MCF7.beclin 1 cells are represented as volcano plots (Fig. 1). A complementary algorithm, Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) (25) showed a similar list of positively enriched sgRNAs (SI Appendix, Fig. S2).
Fig. 1.
Whole-genome CRISPR/Cas9 screen to identify drivers of breast cancer cell proliferation. (A and B) Volcano plots of the CRISPR screen analyses demonstrating enriched sgRNAs (targeting the listed genes) for MCF7.control cells (A) and MCF7.beclin 1 cells (B) after 4 wk of cell proliferation. Genes targeted by the most significantly enriched sgRNAs are highlighted in red. The x-axis represents the average LFC of all the sgRNAs that target a gene, and the y-axis represents the average –log10 P values for all the sgRNAs targeting a gene. P values were calculated using hypergeometric distribution. See also SI Appendix, Fig. S2.
CRISPR Screen for Regulators of Proliferation Identifies the Interferon Signaling Pathway.
After 4 wk of proliferation, MCF7.control cells were enriched for sgRNAs targeting well-known interferon (IFN) signaling pathway molecules (Fig. 1A and SI Appendix, Table S1). The seven most highly selected sgRNAs were directed at IFNAR2, STAT2, STAT1, IFNAR1, TYK2, IRF9, and JAK1, components of the type I IFN JAK-STAT pathway (26). MAGeCK analysis (false discovery rate [FDR] <0.05) confirmed enrichment of the same sgRNAs (SI Appendix, Fig. S2 A and B and Table S2). None of those sgRNAs was enriched in MCF7.beclin 1 cells, and these sgRNAs were also enriched in a direct comparison between MCF7.beclin 1 cells at T = 4 and MCF7.control cells at T = 4 (SI Appendix, Fig. S2E). Thus, knockout of IFN pathway genes confers a growth advantage for control MCF7 breast cancer cells, but not those with enforced Beclin 1 expression.
Analysis of endogenous mRNA expression of IFN pathway genes revealed significantly higher expression of STAT1, STAT2, and IRF9 (which encode transcription factors involved in IFN signaling) in MCF7.control cells compared to MCF7.beclin 1 cells, while JAK1 expression was higher in MCF7.beclin 1 cells (SI Appendix, Fig. S3A). Moreover, expression of two well-described type I IFN JAK-STAT target genes, ISG15 and OAS1, was markedly lower in MCF7.beclin 1 cells compared with MCF7.control cells (SI Appendix, Fig. S3B), as well as in xenografts derived from MCF7.beclin 1 cells compared with those derived from MCF7.control cells (SI Appendix, Fig. S3C). As it is established that Beclin 1, as well as other autophagy proteins, down-regulate IFN signaling (27, 28), one possible interpretation of our data is that there is suppression of the IFN pathway with enforced expression of Beclin 1, obviating the need for specific enrichment of sgRNAs that target IFN signaling during in vitro growth. Thus, taken together, our CRISPR analyses confirm that repression of IFN signaling, a pathway known to suppress tumorigenesis (29), promotes MCF7 breast cancer cell proliferation and indicate that down-regulation of IFN signaling may represent a mechanism by which enforced Beclin 1 expression suppresses breast cancer cell proliferation.
CRISPR Screen for Regulators of Beclin 1-Dependent Tumor Suppression Identifies the Cadherin/Catenin Complex.
As our major goal was to dissect the mechanisms by which Beclin 1 functions to inhibit tumor cell proliferation, we focused primarily on genes whose loss reversed Beclin 1-dependent growth inhibition—specifically, sgRNAs that were most enriched in the MCF7.beclin 1 CRISPR screen (T = 4 vs. T = 0) (Fig. 1B and SI Appendix, Table S3). We discovered highly enriched sgRNAs that target genes involved in crucial biological processes relevant to tumorigenesis, including adherens junction formation or stability and cell-cell adhesion (CDH1 and CTNNA1) (30), tight junctions (CLDN7, F11R, and CLDN3) (31, 32), G protein-coupled receptor signaling (RIC8A and GNA13) (33, 34), and EMT and cell growth (CDH1, CTNNA1, KDF1, and CSK) (19, 35–38). A parallel analysis using the MAGeCK algorithm (FDR <0.05) confirmed RIC8A, CDH1, GNA13, and CTNNA1 as the top hits from the screen (SI Appendix, Fig. S2C and Table S4), and overall there was substantial overlap among the top-scoring sgRNAs identified using the two methods (SI Appendix, Fig. S2D). Thus, our CRISPR screen revealed genes associated with important biological processes involving the formation of adherens junctions, cell adhesion, and EMT that on loss conferred a selective advantage for cell proliferation in MCF7.beclin 1 cells. We measured endogenous mRNA expression of the target genes of the top-ranked sgRNAs in both MCF7.control and MCF7.beclin1 cells and observed no major differences (SI Appendix, Fig. S4A). Therefore, the enrichment of sgRNAs encoding these genes is due not to increased transcription in MCF7.beclin 1 cells, but rather to roles for such genes in mediating the tumor-suppressor function of Beclin 1.
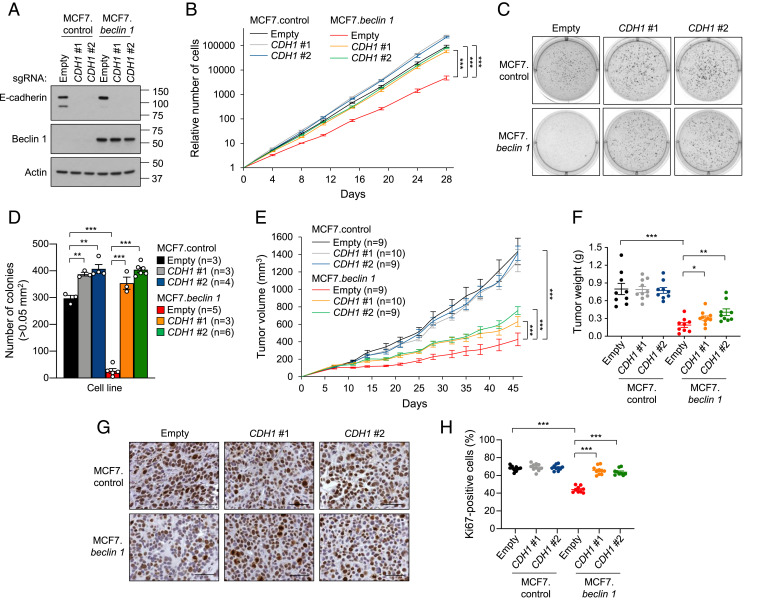
We next directed our attention to two genes targeted by the highly enriched sgRNAs in MCF7.beclin 1 cells, CDH1 and CTNNA1. These genes play pivotal roles in the function of adherens junctions, cell adhesion, and EMT, and their loss or inactivation has been associated with the progression of breast cancer (20, 21, 30, 35, 36). To corroborate the CRISPR screen results for CDH1, we selected two individual sgRNAs per gene and generated CDH1 CRISPR/Cas9 knockout cells (Fig. 2A). CDH1 knockout in MCF7.beclin 1 cells reversed the inhibitory effects of Beclin 1 on cellular proliferation, resulting in growth curves similar to those of MCF7.control cells (Fig. 2B). As reported previously, enforced Beclin 1 expression inhibited anchorage-independent growth in soft agar colony formation assays (1, 22), but colony growth was rescued in MCF7.beclin 1 cells on knockout of CDH1 (Fig. 2 C and D). CDH1 knockout also led to an increase in colony formation in MCF7.control cells; however, the absolute number of colonies was similar in MCF7.control cells and MCF7.beclin 1 cells with CDH1 depletion (Fig. 2D), indicating that CDH1 depletion per se was sufficient to completely reverse Beclin 1-dependent suppression of anchorage-independent growth. Importantly, depletion of CDH1 enhanced the tumor growth of MCF7.beclin 1 xenografts in NOD/SCID mice but had no effect on the growth of MCF7.control xenografts (Fig. 2E). At the end of the 46-d observation period, tumors derived from MCF7.beclin 1 cells depleted of CDH1 were larger than those derived from MCF7.beclin 1 cells transduced with empty vector (Fig. 2F). Furthermore, the percentage of Ki-67–positive nuclei (a marker of proliferation) was significantly lower in MCF7.beclin 1 compared to MCF7.control xenografts, but with CDH1 knockout completely reverted to the percentage observed in MCF7.control xenografts (Fig. 2 G and H). The percentage of Ki-67–positively stained cells was not increased in MCF7.control xenografts with CDH1 knockout. Depletion of CDH1 in MCF7.beclin 1 cells did not completely recapitulate the tumor growth of MCF7.control cells. A possible explanation for this phenotype may be the selection of different cell populations. Therefore, we analyzed the mRNA expression of cancer stem cell genes and found that expression of both CD44 and CD24 was significantly decreased in MCF7.beclin 1 cells depleted of CDH1 compared to MCF7.control cells (SI Appendix, Fig. S5). This suggests that the reduction of cancer stem cell markers in MCF7.beclin 1 cells with CDH1 knockout could explain in part the failure of completely recapitulating tumor growth of BECN1 haploinsufficiency. Taken together, depletion of CDH1 in MCF7.beclin 1 cells increases tumor cell proliferation, anchorage-independent growth, and tumor growth, demonstrating that E-cadherin is required for Beclin 1 tumor-suppressor activity in breast cancer cells.
Fig. 2.
E-cadherin deletion impairs Beclin 1-dependent inhibition of cell proliferation, anchorage-independent growth, and xenograft tumor growth. (A) Western blot analysis of E-cadherin in indicated cells after transduction with a CRISPR/Cas9 lentivirus containing either no (empty) or two sgRNAs that target CDH1. (B) Cellular proliferation assays of cells indicated in A. Data points are mean ± SEM for triplicate samples. Similar results were observed in three independent experiments. ***P < 0.001 for indicated comparisons, linear mixed-effect model. (C and D) Representative images (C) and quantitation (D) of soft agar colonies formed by MCF7.control and MCF7.beclin 1 cells transduced with indicated vectors. Bars represent mean ± SEM of three to six replicate samples. Similar results were observed in three independent experiments. **P < 0.01; ***P < 0.001, unpaired two-tailed Student’s t test. (E) Xenograft tumor growth in NOD/SCID mice of MCF7.control and MCF7.beclin 1 cells transduced with CRISPR/Cas9 lentivirus containing either empty vector or two sgRNAs targeting CDH1. Data points represent mean ± SEM tumor volume for indicated number of mice per genotype. Similar results were observed in three independent experiments. ***P < 0.001 for indicated comparison, linear mixed-effect model. (F) Tumor weights at end of xenograft study in E. Data are mean ± SEM. Each data point represents a different mouse. *P < 0.05; **P < 0.01; ***P < 0.001, unpaired two-tailed Student’s t test. (G and H) Representative images (G) and quantitation (H) of percentage Ki-67–positive nuclei per high-power field of indicated MCF7 xenograft tumor genotype. Data are mean ± SEM for 9 to 10 xenografts per genotype (at least 10 randomly selected fields analyzed per xenograft by an observer blinded to genotype). Each data point represents a different mouse. ***P < 0.001, unpaired two-tailed Student’s t test. (Scale bars: 50 μm.)
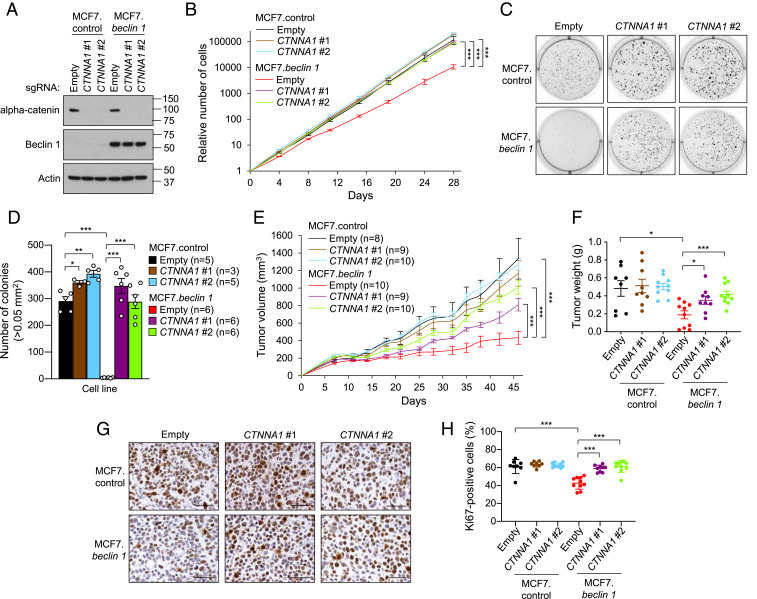
Similar to E-cadherin, mutations or down-regulated expression of alpha-catenin has been observed in multiple human cancers (36). Loss of CTNNA1 (which encodes alpha-catenin) in MCF7.beclin 1 cells (Fig. 3A) increased proliferation rates (Fig. 3B) and anchorage-independent growth (Fig. 3 C and D), and both cellular proliferation and the numbers of soft agar colonies were similar in MCF7.control cells and MCF7.beclin 1 cells depleted of CTNNA1. Compared to MCF7.beclin 1 cells transduced with empty vector, xenografts derived from MCF7.beclin 1 cells with CTNNA1 knockout had enhanced tumor growth (Fig. 3E), increased tumor weight (Fig. 3F) and a higher percentage of Ki-67–positive nuclei (Fig. 3 G and H). Thus, in addition to E-cadherin, the cadherin/catenin complex protein alpha-catenin, is essential for the tumor-suppressor function of Beclin 1 in breast cancer cells.
Fig. 3.
Alpha-catenin deletion impairs Beclin 1-dependent inhibition of cell proliferation, anchorage-independent growth, and xenograft tumor growth. (A) Western blot analysis of alpha-catenin in indicated cells after transduction with a CRISPR/Cas9 lentivirus containing either no (empty) or two sgRNAs targeting CTTNA1. (B) Cellular proliferation of indicated cells in A. Data points are mean ± SEM for triplicate samples. Similar results were observed in three independent experiments. ***P < 0.001 for indicated comparison, linear mixed-effect model. (C and D) Representative images (C) and quantitation (D) of soft agar colonies formed by MCF7.control and MCF7.beclin 1 cells transduced with indicated vectors. Bars are mean ± SEM of three to six replicate samples. Similar results were observed in three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001, unpaired two-tailed Student’s t test. (E) Xenograft tumor growth in NOD/SCID mice of MCF7.control and MCF7.beclin 1 cells transduced with CRISPR/Cas9 lentivirus containing either empty vector or two sgRNAs targeting CTTNA1. Data points represent mean ± SEM tumor volume for the indicated number of mice per genotype. Similar results were observed in three independent experiments. ***P < 0.001 for indicated comparison, linear mixed-effect model. (F) Tumor weights at the end of the xenograft study in E. Data are mean ± SEM. Each data point represents a different mouse. *P < 0.05; ***P < 0.001, unpaired two-tailed Student’s t test. (G and H) Representative images (G) and quantitation (H) of percentage Ki-67–positive nuclei per high-power field of the indicated MCF7 xenograft tumor genotype. Data are mean ± SEM for 8 to 10 xenografts per genotype (at least 10 randomly selected fields analyzed per xenograft by an observer blinded to genotype). Each data point represents a different mouse. ***P < 0.001, unpaired two-tailed Student’s t test. (Scale bars: 50 μm.)
Enforced Beclin 1 Expression Increases Membrane Localization of the E-Cadherin Complex.
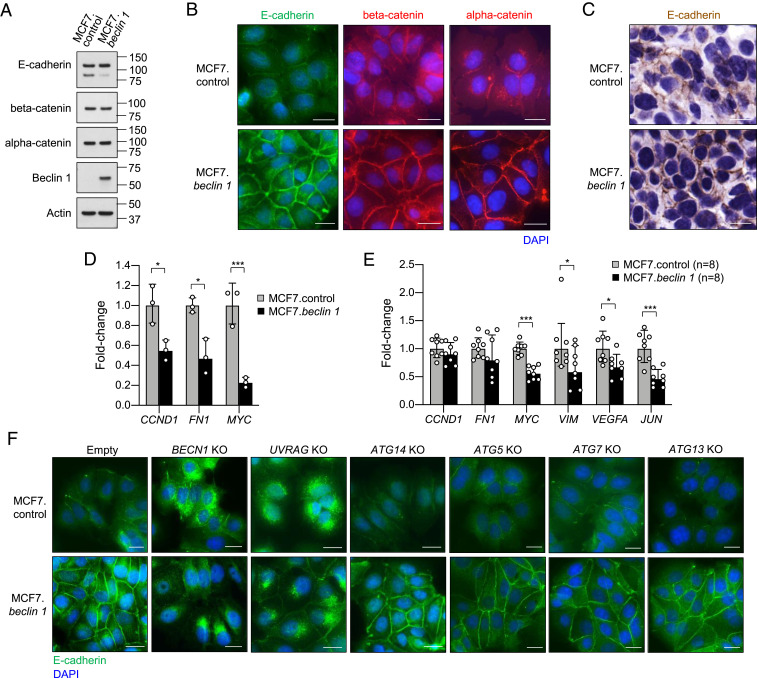
E-cadherin and alpha-catenin are connected by beta-catenin and function as key components of the adherens junctions (30). To gain further insight into how E-cadherin and alpha-catenin might cooperate with Beclin 1 to facilitate tumor suppression, we compared the protein levels and subcellular localization of E-cadherin, alpha-catenin, and beta-catenin in MCF7.control and MCF7.beclin 1 cells. Full-length E-cadherin, alpha-catenin, and beta-catenin protein expression levels were similar in MCF7.control and MCF7.beclin 1 cells (Fig. 4A), although increased amounts of a lower molecular weight band for E-cadherin were present in MCF7.control cells (Fig. 4A and SI Appendix, Fig. S6). No differences were observed in mRNA expression for genes encoding E-cadherin, alpha-catenin, or beta-catenin in MCF7.control cells compared to MCF7.beclin 1 cells (SI Appendix, Fig. S4).
Fig. 4.
Beclin 1 and UVRAG are required for the membrane localization of the E-cadherin complex in MCF7 breast cancer cells. (A) Western blot expression of indicated proteins in MCF7.control and MCF7.beclin 1 cells. (B) Representative immunofluorescence images of E-cadherin, beta-catenin, and alpha-catenin staining in MCF7.control and MCF7.beclin 1 cells. (Scale bars: 20 μm.) (C) Representative images of immunohistochemical staining of E-cadherin in xenograft tumors derived from indicated MCF7 cell line. Similar results were observed in 8 to 10 mice analyzed per group. (Scale bars: 20 μm.) (D) qRT-PCR analysis of beta-catenin/Wnt signaling target gene expression in MCF7.control and MCF7.beclin 1 cells. CCND1, Cyclin D1; FN1, Fibronectin. Bars represent mean ± SEM of triplicate samples. Similar results were observed in three independent experiments. *P < 0.05; ***P < 0.001, unpaired two-tailed Student’s t test. (E) qRT-PCR analysis of beta-catenin/Wnt signaling target gene expression gene expression in xenograft tumors derived from MCF7.control and MCF7.beclin 1 cells. VIM, Vimentin. Bars represent mean ± SEM of tumors from eight mice per genotype. *P < 0.05, unpaired two-tailed Student’s t test. (F) Representative immunofluorescence images of E-cadherin staining in MCF7.control and MCF7.beclin 1 cells transduced with either empty vector or sgRNAs targeting BECN1, UVRAG, ATG14, ATG5, ATG7, or ATG13. (Scale bars: 20 μm.)
Despite similar levels of gene and protein expression, there were remarkable differences in the subcellular localization of E-cadherin, alpha-catenin, and beta-catenin between MCF7.control cells and MCF7.beclin 1 cells (Fig. 4B). In MCF7.beclin 1 cells, all three proteins were predominantly localized at the plasma membrane, whereas substantially weaker plasma membrane localization and greater cytoplasmic accumulation was present in MCF7.control cells. Moreover, there was also a marked increase in plasma membrane E-cadherin staining in xenograft tumors derived from MCF7.beclin 1 cells compared to those derived from MCF7.control cells (Fig. 4C). To further strengthen these findings, we transiently transfected Beclin 1 in another breast cancer cell line and found that enforced Beclin 1 expression also increased plasma membrane E-cadherin staining in MDA-MB-468 cells (SI Appendix, Fig. S7 A and B). Consistent with the known function of cell surface E-cadherin in cell adhesion (30), MCF7.beclin 1 cells were more tightly adherent and displayed more uniform cell-cell junctions (Fig. 4B). This was substantiated by an increase in the membrane localization of the junctional proteins claudin 3, claudin 7, and junctional adhesion protein-1 (JAM-1, also known as F11R) in MCF7.beclin 1 cells, while there was no clear difference in the distribution of claudin 4 (SI Appendix, Fig. S7C).
Increased membrane localization of beta-catenin prevents its function as a signal transducer in the activation of gene transcription in the Wnt signaling pathway, which is one mechanism by which the loss of cell surface E-cadherin complex promotes tumor progression (19, 39). Therefore, we examined selected target genes in the beta-catenin/Wnt signaling pathway in MCF7.control and MCF7.beclin 1 cells and xenografts. We observed reduced transcription of Wnt pathway target genes CCND1 and MYC and the mesenchymal marker FN1 in MCF7 cells with enforced Beclin 1 expression (Fig. 4D). Furthermore, in xenografts derived from MCF7.beclin 1 cells, the expression of MYC, mesenchymal marker VIM, and metastasis-associated genes VEGFA and JUN were all reduced compared to xenografts derived from MCF7.control cells (Fig. 4E). Thus, enforced Beclin 1 expression increases the cell surface localization of the E-cadherin complex, where it both functions in cell-cell adhesion and prevents the activation of signal transduction pathways involved in tumor growth and metastases.
Beclin 1 and UVRAG, but Not ATG14, Are Required for E-Cadherin Membrane Localization.
To confirm whether E-cadherin membrane accumulation in MCF7.beclin 1 cells is due to Beclin 1, we generated CRISPR/Cas9-mediated BECN1 knockout cell lines and performed immunofluorescence staining for E-cadherin. Depletion of BECN1 in MCF7.beclin 1 cells greatly diminished membrane staining. This was accompanied by cytoplasmic accumulation of E-cadherin in a subpopulation of cells that appeared as dense perinuclear regions (Fig. 4F and SI Appendix, Fig. S6A). Even in the MCF7.control cell line (which expresses low levels of endogenous Beclin 1), there was a transition from diffuse cytoplasmic staining to accumulation of E-cadherin close to the nucleus in some cells with BECN1 knockout. This altered pattern of E-cadherin staining is similar to that observed on dissociation of adherens junctions (40).
We next asked whether the promotion of E-cadherin cell surface localization is specific to Beclin 1 or is a more general function of autophagy proteins. To address this question, we created CRISPR/Cas9 knockouts for additional autophagy-related genes in MCF7.control and MCF7.beclin 1 cells. We generated knockouts for ATG14, a component of the PI3KC3-C1 complex; UVRAG, a component of the PI3KC3-C2 complex, and the autophagy genes ATG5, ATG7, and ATG13 (SI Appendix, Fig. S6 B–F) and performed immunofluorescence analyses for E-cadherin. No changes in subcellular location of E-cadherin were observed on knockout of autophagy genes ATG5 or ATG7, which encode proteins in the conjugation system involved in autophagosomal membrane expansion, or ATG13, which encodes a component of the preinitiation complex (Fig. 4F). Similarly, knockout of the PI3KC3 C1-associated ATG14 had no impact on E-cadherin staining. However, a marked effect was observed in UVRAG knockout cells either with or without enforced Beclin 1 expression with an almost complete absence of membrane E-cadherin and dense perinuclear accumulation of E-cadherin (Fig. 4F). Taken together, these data demonstrate that Beclin 1 and UVRAG, but not ATG14, ATG13, ATG5, or ATG7 are essential for E-cadherin membrane localization in MCF7 cells.
We also performed immunofluorescence analysis of beta-catenin in BECN1 and UVRAG knockouts. A marked loss of beta-catenin cell surface localization and increased cytoplasmic staining was observed in MCF7.beclin 1 cells on knockout of BECN1 or UVRAG. However, we did not detect a clear increase in the nuclear localization of beta-catenin in either MCF7.control or MCF7.beclin 1 cells with BECN1 or UVRAG knockout (SI Appendix, Fig. S8A).
To gain further insight into the mechanism by which Beclin 1 and UVRAG regulate the cell surface localization of E-cadherin, we evaluated the distribution of growth factor receptors HGFR and FGFR1, which play key roles in the endocytosis of E-cadherin (41). We found that localization of both HGFR and FGFR1 is similar in MCF7.control and MCF7.beclin 1 cells, with HGFR located mostly in the cytoplasm and FGFR1 displaying punctate staining in both the cytoplasm and nucleus (SI Appendix, Fig. S8 B and C). In addition, knockout of BECN1 or UVRAG did not alter the distribution of these growth factor receptors in either MCF7.control or MCF7.beclin 1 cells, suggesting that endocytosis of E-cadherin through these growth factor receptors probably does not explain how Beclin 1 and UVRAG promote the membrane localization of E-cadherin.
Discussion
Beclin 1 is a haploinsufficient tumor suppressor in breast cancer, but the precise mechanism by which it suppresses breast cancer cell growth is unknown. Here we conducted an unbiased genome-wide CRISPR/Cas9 screen for drivers of cellular proliferation in breast cancer cells and identified the cell-cell adhesion protein E-cadherin and the cadherin/catenin complex protein, alpha-catenin, as key mediators of Beclin 1-dependent tumor suppression. Our findings demonstrate that Beclin 1 and the PI3KC3-C2–specific member of the PI3KC3 complex, UVRAG, promote the membrane localization of E-cadherin and components of the E-cadherin/catenin complex, thereby increasing cell adhesion and preventing the transcription of beta-catenin–signaling target genes, including genes involved in tumor growth and tumor progression.
We found that CRISPR/Cas9-mediated knockout of either CDH1 and CTNNA1 reversed Beclin 1-mediated inhibition of proliferation, anchorage-independent growth, and tumor xenograft growth in MCF7 breast cancer cells. We speculate that CDH1 and CTNNA1 deletion were genetically selected for in our CRISPR/Cas9 screen, as enforced Beclin 1 expression promotes the cell surface localization of the E-cadherin complex, where it is known to function in tumor suppression. Endogenous Beclin 1 and UVRAG, but not other autophagy genes, including ATG14, ATG13, ATG5, and ATG7, promote E-cadherin cell surface localization, as demonstrated by CRISPR-Cas9–mediated deletion in both MCF7.control and MCF7.beclin 1 cells. Thus, this specific tumor-suppressor function of Beclin 1 is not related to autophagy, but rather is likely a specific trafficking function of the Beclin 1/UVRAG-containing PI3KC3-C2 complex. This is consistent with the observation that UVRAG exerts a similar tumor-suppressor function as Beclin 1 in cancer cells (42). Further studies are needed to determine the precise cellular mechanism by which Beclin 1 and UVRAG regulate the plasma membrane localization of E-cadherin. This is not likely to be other previously described autophagy-independent functions of Beclin 1, such as LC3-associated phagocytosis, LC3-associated endocytosis, or autophagy-gene dependent secretion, given the lack of a requirement for ATG5 and ATG7 (43–45).
The observation that Beclin 1 and UVRAG promote E-cadherin cell surface localization is directly relevant to human breast cancer, as there is extensive evidence that cell surface E-cadherin functions in tumor suppression, and that its loss promotes breast cancer progression. Loss of E-cadherin by down-regulation of expression, loss of heterozygosity, and/or inactivating mutations and/or loss of cell surface E-cadherin localization have been associated with tumor progression and poor patient outcome in human breast cancer, including invasive lobular carcinoma and invasive ductal cancer (19–21). E-cadherin prevents tumor progression through various mechanisms, including regulation of contact-mediated inhibition of proliferation through Hippo signaling, suppression of beta-catenin/Wnt signaling, and promotion of EMT—the underlying process for metastasis that allows the transformation of epithelial cells to a more mesenchymal phenotype, including loss of cell-cell adhesion and increases in migratory capacity and invasiveness (19, 35, 46). It will be interesting to determine whether allelic loss of BECN1, which occurs in ∼30% of human breast cancers and ∼77% of human ovarian cancers, contributes to tumor progression through the loss of cell surface E-cadherin.
Our identification of an autophagy-independent role for E-cadherin in mediating the tumor-suppressor effects of Beclin 1 is not incompatible with additional effects of autophagy proteins, including the lysosomal degradation pathway of autophagy itself, in preventing breast cancer progression. Of note, a recent study found that several different autophagy genes, including ATG5 and ATG12, prevent the metastatic growth of breast carcinoma cells through autophagic degradation of the protein NBR1 (47). We speculate that the autophagy machinery, an ancient stress response protein network, evolved multiple mechanisms by which it can act to restrict the growth of established tumor cells.
Materials and Methods
Cell Culture.
MDA-MB-468 cells were obtained from the American Type Culture Collection (ATCC) and cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (vol/vol) FBS and 100 U/mL penicillin-streptomycin. MCF7.control and MCF7.beclin 1 tet-off cell lines were described previously (22, 23) and maintained in DMEM supplemented with 10% (vol/vol) Tet System Approved FBS (Takara), 2 mM l-glutamine, 1 mg/mL G418, 0.2 mg/mL hygromycin B, 10 μg/mL insulin, 50 μg/mL gentamicin, and 1 μg/mL doxycycline. Doxycycline was removed 5 d prior to experiments to induce expression of Beclin 1. Proliferation assays were performed in triplicate, and cells were counted with a Bio-Rad TC20 automated cell counter.
CRISPR/Cas9-Based Genetic Screen.
A CRISPR/Cas9 genetic screen in MCF7.control and MCF7.beclin 1 cells was performed with the Brunello sgRNA library (24). Lentivirus containing the Brunello sgRNA library was purchased from the Broad Institute. Optimal transduction conditions were determined for the lentivirus in MCF7.control and MCF7.beclin 1 cells to achieve 30 to 50% infection efficiency, corresponding to a multiplicity of infection of ∼0.5 to 0.65 (24). For CRISPR library screening, 1.35 × 108 MCF7.control and MCF7.beclin 1 cells were seeded in triplicate and transduced with the Brunello lentivirus in the presence of 8 μg/mL polybrene and 1 μg/mL doxycycline for 17 h before changing to regular growth medium. After 48 h, cells harboring CRISPR constructs were subjected to selection with 1 μg/mL puromycin and 1 μg/mL doxycycline. After 7 d of selection, both puromycin and doxycycline were removed from the medium, and cells were cultured for an additional 7 d to induce the expression of Beclin 1 in the MCF7.beclin 1 cells. At least 4 × 107 cells were then harvested for time point T = 0 wk, and cell pellets were frozen at −80 °C. Cells were cultured for an additional 4 wk and then harvested for time point T = 4 wk. A schematic overview of the CRISPR screen SI Appendix, Fig. S1A shows the experimental timeline. To maintain a representation of 500 cells per sgRNA in the library, at least 4 × 107 cells per biological triplicate were maintained throughout the CRISPR screen. Genomic DNA (gDNA) was extracted using the QIAamp DNA Blood Maxi Kit (Qiagen) according to the manufacturer’s instructions, and DNA concentrations were measured by UV spectrophotometry using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Illumina sequencing was performed by the Broad Institute as described previously (24). PCR of gDNA was performed to attach sequencing adaptors and barcode samples, which were divided into multiple 100-μL reactions (total volume) containing a maximum of 10 μg gDNA. A PCR master mix per 96-well plate consisted of 150 μL of ExTaq DNA Polymerase (Clontech), 1,000 μL of 10× Ex Taq buffer, 800 μL of dNTP provided with the enzyme, 50 μL of P5 stagger primer mix (stock at 100 μM concentration), and 2,000 μL of water. Each well consisted of 50 μL of gDNA plus water, 40 μL of PCR master mix, and 10 μL of a uniquely barcoded P7 primer (stock at 5 μM concentration). PCR cycling conditions were as follows: an initial 1 min at 95 °C; then 28 cycles of 30 s at 94 °C, 30 s at 52.5 °C, and 30 s at 72 °C; and a final 10-min extension at 72 °C. P5/P7 primers were synthesized at Integrated DNA Technologies. Samples were purified with Agencourt AMPure XP SPRI beads (Beckman Coulter; A63880) according to the manufacturer’s instructions. Samples were sequenced on an Illumina HiSeq 2500 at the Broad Institute.
Screen Analysis.
Read counts, assignment to experimental conditions, and normalization were performed as described previously (24). For the generation of volcano plots, we first calculated the LFC of the normalized reads for each sgRNA between two conditions (i.e., Beclin 1 at T = 4 wk – Beclin 1 at T = 0 wk) and then averaged these values for each triplicate sample. The sgRNAs were then ranked by LFC in either in ascending or descending order, and P values were calculated by using the hypergeometric distribution without replacement based on the rank order of the LFC of the perturbations.
MAGeCK was performed as described previously (25). In brief, read counts across all samples were first subjected to median normalization. Mean variance modeling was applied to describe the relationship of mean and variance in the triplicates. The mean-variance model was then used to calculate the statistical significance of each sgRNA (i.e., sgRNA ranking). Essential genes were then identified by looking for genes whose sgRNAs were ranked consistently higher (by significance) using robust rank aggregation (RRA). Finally, enriched pathways were identified by applying the RRA algorithm to the ranked list of genes. Enriched genes were selected using a false discovery rate (FDR) <0.05.
CRISPR/Cas9-Mediated Generation of Knockout Cells.
A lentivirus vector, pXPR_023 (pLentiCRISPRv2; Addgene; 52961) (48), containing hSpCas9 was used (provided by J.G.D., Broad Institute). For each knockout cell line, five sgRNAs targeting individual genes were designed using the Genetic Perturbation Platform (GPP) sgRNA designer (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). Cloning of the sgRNAs into the pXPR_023 vector was performed according to the online instructions on the GPP web portal (https://portals.broadinstitute.org/gpp/public/resources/protocols). For virus production, 8 million Phoenix cells (ATCC) were seeded per 100-mm dish at 1 d prior to transfection. pXPR_023 lentivirus vector containing either no sgRNA (empty) or an sgRNA targeting an individual gene was cotransfected with the packaging plasmid pCMV-d8.91 and the envelope plasmid pMDG in Phoenix cells using the transfection reagent Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 4 to 6 h, the medium was changed to 1× DMEM supplemented with 30% Tet System Approved FBS. Medium containing virus particles was harvested at 48 h after transfection and filtered through a 0.45-μm membrane. MCF7.control and MCF7.beclin 1 cells were transduced with lentivirus in the presence of 8 μg/mL polybrene for 3 h. After 72 h, infected cells were subjected to puromycin selection (1 μg/mL) for at least 4 d, or until control cells without virus were all dead, and knockout efficiency of the cell pools was tested by Western blotting. For each gene knockout, two cell line pools with the highest knockdown efficiency were selected and used for subsequent experiments. All sgRNAs used in this study are listed in SI Appendix, Table S5.
Soft Agar Colony Formation Assay.
Assays were performed in six-well culture plates. Base agar consisted of 0.5% agar, 1× DMEM, 10% (vol/vol) Tet System Approved FBS, 2 mM l-glutamine, 1 mg/mL G418, 0.2 mg/mL hygromycin B, 10 μg/mL insulin, and 50 μg/mL gentamicin. Top agar consisted of 0.3% agar, 1× DMEM, 10% (vol/vol) Tet System Approved FBS, 2 mM l-glutamine, 1 mg/mL G418, 0.2 mg/mL hygromycin B, 10 μg/mL insulin, and 50 μg/mL gentamicin with 4 × 104 MCF7 cells per well. A feeder layer of 0.3% agar, 1× DMEM, 10% (vol/vol) Tet System Approved FBS, 2 mM l-glutamine, 1 mg/mL G418, 0.2 mg/mL hygromycin B, 10 μg/mL insulin, and 50 μg/mL gentamicin was added weekly. Cells were incubated for 21 d at 37 °C with 5% CO2. Colonies were stained overnight with 1 mg/mL Nitro blue tetrazolium chloride solution (Sigma-Aldrich) prior to imaging. Grayscale images of the colonies were captured with the ChemiDoc MP Imaging System (Bio-Rad). Colonies ≥0.05 mm2 were counted using the “Analyze Particles” algorithm in ImageJ.
Xenograft Tumor Growth Assay.
A 60-d slow-release 1.7-mg estrogen pellet (Innovative Research of America) was injected subcutaneously in the neck region of 6-wk-old female NOD/SCID mice. After 1 wk, 5 × 106 MCF7.control or MCF7.beclin 1 cells transduced with pLentiCRISPRv2 containing the indicated sgRNA sequences were resuspended in 200 μL of PBS:Matrigel matrix (Corning; 1:1; vol/vol) and injected into the right flank of each mouse. Tumor volumes were measured twice weekly until the end of the experiment. After 46 d, all mice were euthanized, and the tumors were dissected and weighed. Each group contained 8 to 10 mice. All animal experiments were performed in accordance with institutional guidelines and approved by the UT Southwestern Medical Center’s Institutional Animal Care and Use Committee.
Transient Transfection of Beclin 1.
A pBicep vector (Sigma-Aldrich) containing 3xFLAG-Beclin 1 was used. For transient transfection of Beclin 1, either 1.5 × 104 MDA-MB-468 cells were seeded per well in eight-well chamber slides (Labtek) or 2 × 105 MDA-MB-468 cells were seeded per well in a 12-well plate. After 24 h, cells were either mock transfected or transfected with a pBicep-3xFLAG-Beclin 1 vector using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 4 to 6 h, medium was changed to DMEM supplemented with 10% (vol/vol) FBS. At 24 h after transfection, cells were prepared for either immunofluorescence or Western blot analysis as described below.
Immunofluorescence Analysis.
For immunofluorescence staining, 1.75 × 104 MCF7 cells were seeded per well in eight-well chamber slides (Lab-Tek). After 2 d, cells were fixed in 2% paraformaldehyde for 10 min, followed by methanol permeabilization for 10 min at −20 °C. Cells were blocked in 2% BSA in PBS for 1 h and then probed with anti-E-cadherin (Santa Cruz Biotechnology; sc-8426, 1:100 dilution), anti–E-cadherin (Cell Signaling Technology; 3195; 1:250 dilution), anti–alpha-catenin (Thermo Fisher Scientific; 13-9700, 1:250 dilution), anti–beta-catenin (BD Biosciences; 610153; 1:250 dilution), anti-claudin 3 (Thermo Fisher Scientific; 34-1700, 1:100 dilution), anti-claudin 4 (Thermo Fisher Scientific; 32-9400, 1:100 dilution), anti-claudin 7 (Thermo Fisher Scientific; 34-9100, 1:100 dilution), anti–JAM-1 (Santa Cruz Biotechnology; sc-53624, 1:50 dilution), anti-HGFR (Santa Cruz Biotechnology; sc-10, 1:100 dilution) or anti-FGFR1 (Abcam; ab10646, 1:250 dilution) antibodies overnight at 4 °C. Alexa Fluor 488 (Invitrogen; A21202, 1:500 dilution) or Alexa Fluor 594 (Invitrogen; A21203, 1:500 dilution) anti-mouse secondary antibodies were then added for 45 min at room temperature in the dark, after which the slides were mounted with ProLong Diamond Antifade Mountant with DAPI (Invitrogen). Immunofluorescent images were captured using a Zeiss Axioplan 2 microscope.
Immunohistochemistry.
MCF7 tumor xenografts were fixed in formalin, embedded in paraffin, and sectioned at 5-μm thickness. For antigen retrieval, sections were incubated in citrate buffer (10 mM citric acid, pH 6.0) at 50 °C for 13 min. Immunohistochemical staining of sections was performed using anti–Ki-67 (Abcam; ab16667, 1:200 dilution) and anti–E-cadherin (Cell Signaling Technology; 3195, 1:200 dilution) antibodies overnight at 4 °C, followed by detection with the ABC Elite Immunoperoxidase Kit (Vector Laboratories) according to the manufacturer’s instructions. Images were captured using a Zeiss Axio Imager Z2 microscope. Both the number of Ki-67–positive cells and total number of cells were counted in sections of each mouse to calculate the percentage of Ki-67–positive cells per unit area.
Western Blot Analyses.
Cells were washed in ice-cold PBS and then scraped in ice-cold lysis buffer (50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100) containing cOmplete protease (Roche) and Halt phosphatase (Thermo Fisher Scientific) inhibitor mixtures and rotated for at least 30 min at 4 °C. Lysates were centrifuged at 14,000 × g for 10 min at 4 °C. Cleared lysates were diluted in 2× Laemmli buffer supplemented with beta-mercaptoethanol and then boiled for 10 min. Whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on precast 4 to 20% Mini-Protean TGX gels (Bio-Rad), transferred to PVDF membranes, and probed with the indicated antibodies.
The following antibodies were used for Western blot analysis. Anti–beta-actin HRP (sc-47778, 1:10,000 dilution), anti-Beclin 1 (sc-11427, 1:2,000 dilution), and anti–E-cadherin (sc-7870, 1:2,000 dilution) antibodies were obtained from Santa Cruz Biotechnology. Anti-Atg5 (NB110-53818; 1:500 dilution) antibody was obtained from Novus Biologicals. Anti-Atg7 (A2856, 1:1,000 dilution) and anti-FLAG (F1804, 1:500 dilution) antibodies were purchased from Sigma-Aldrich. Anti-Atg13 (13468S, 1:500 dilution) and anti-UVRAG (13115S, 1:2,000 dilution) antibodies were obtained from Cell Signaling Technologies. Anti-Atg14 (M184-3; 1:500 dilution) antibody was purchased from MBL International. Anti–alpha-catenin (610194, 1:500 dilution) and anti–beta-catenin (610153, 1:2,000 dilution) antibodies were obtained from BD Biosciences.
qRT-PCR.
To assess mRNA expression of CRISPR screen hits, beta-catenin/Wnt target genes, and cancer stem cell genes, total RNA was extracted from MCF7 cells or MCF7 xenografts using the RNeasy Plus Mini Kit and QIAshredder columns (Qiagen) according to the manufacturer’s instructions. Reverse transcription of 2 μg of RNA was performed using the iScript cDNA Synthesis Kit (Bio-Rad). cDNAs were diluted 10-fold prior to use in qRT-PCR analyses, which was performed with the QuantiFast SYBR Green PCR Kit (Qiagen). Gapdh and beta-actin were used as housekeeping genes. Relative fold expression was calculated using the 2-ΔΔCt method and normalized to the average of the MCF7.control samples. Primer sequences are listed in SI Appendix, Table S6.
Statistical Analyses.
Experimental data were analyzed using GraphPad Prism 8 and R software. Statistically significant differences between groups were determined using the unpaired two-tailed Student’s t test to analyze data from qRT-PCR, soft agar colony formation assays, tumor weights, and Ki-67–positive staining of tumors. For analysis of proliferation assays and tumor growth, a mixed linear regression model on log-scale cell number with an interaction term of time (days) and group for each comparison was used. As there were three repeated measurements for cell number at each time point, a random intercept was added for measurement in each model. P values and 95% confidence intervals for the interaction term for the significance level of the difference in growth rate between each pair of groups are reported.
Supplementary Material
Acknowledgments
We thank L. Nguyen for expert technical assistance, all members of the B.L. laboratory for helpful discussions, and H. Smith for assistance with manuscript preparation. This work was supported by NIH Grants U19 AI109725 and U19 AI142784 (to B.L.) and by Cancer Prevention Research Institute of Texas Grant RP120718 (to B.L.).
Footnotes
Competing interest statement: B.L. is a scientific cofounder of Casma Therapeutics, Inc.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020478118/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Liang X. H., et al. , Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T., Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2, 330–335 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilanges B., Posor Y., Vanhaesebroeck B., PI3K isoforms in cell signalling and vesicle trafficking. Nat. Rev. Mol. Cell Biol. 20, 515–534 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Tang H., et al. , Decreased BECN1 mRNA expression in human breast cancer is associated with estrogen receptor-negative subtypes and poor prognosis. EBioMedicine 2, 255–263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delaney J. R., et al. , Autophagy gene haploinsufficiency drives chromosome instability, increases migration, and promotes early ovarian tumors. PLoS Genet. 16, e1008558 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu X., et al. , Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 112, 1809–1820 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yue Z., Jin S., Yang C., Levine A. J., Heintz N., Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 100, 15077–15082 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cicchini M., et al. , Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy 10, 2036–2052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernández A. F., et al. , Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature 558, 136–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vega-Rubín-de-Celis S., et al. , Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 115, 4176–4181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine B., Liu R., Dong X., Zhong Q., Beclin orthologs: Integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol. 25, 533–544 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang R. C., et al. , Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y., et al. , EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 154, 1269–1284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shoji-Kawata S., et al. , Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karantza-Wadsworth V., et al. , Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 21, 1621–1635 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathew R., et al. , Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 21, 1367–1381 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nassour J., et al. , Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565, 659–663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei Y., et al. , The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife 4, e05289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeanes A., Gottardi C. J., Yap A. S., Cadherins and cancer: How does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berx G., Van Roy F., The E-cadherin/catenin complex: An important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res. 3, 289–293 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Yin S., Zhang L., Liu W., Chen B., Prognostic value of reduced E-cadherin expression in breast cancer: A meta-analysis. Oncotarget 8, 16445–16455 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang X. H., Yu J., Brown K., Levine B., Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 61, 3443–3449 (2001). [PubMed] [Google Scholar]
- 23.Furuya N., Yu J., Byfield M., Pattingre S., Levine B., The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 1, 46–52 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W., et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivashkiv L. B., Donlin L. T., Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang Q., et al. , Crosstalk between the cGAS DNA sensor and Beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe 15, 228–238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B., Mizushima N., Virgin H. W., Autophagy in immunity and inflammation. Nature 469, 323–335 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zitvogel L., Galluzzi L., Kepp O., Smyth M. J., Kroemer G., Type I interferons in anticancer immunity. Nat. Rev. Immunol. 15, 405–414 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Hartsock A., Nelson W. J., Adherens and tight junctions: Structure, function and connections to the actin cytoskeleton. Biochim. Biophys. Acta 1778, 660–669 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krause G., et al. , Structure and function of claudins. Biochim. Biophys. Acta 1778, 631–645 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Mandell K. J., McCall I. C., Parkos C. A., Involvement of the junctional adhesion molecule-1 (JAM1) homodimer interface in regulation of epithelial barrier function. J. Biol. Chem. 279, 16254–16262 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Wilkie T. M., Kinch L., New roles for G alpha and RGS proteins: Communication continues despite pulling sisters apart. Curr. Biol. 15, R843–R854 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Strathmann M. P., Simon M. I., G alpha 12 and G alpha 13 subunits define a fourth class of G protein alpha subunits. Proc. Natl. Acad. Sci. U.S.A. 88, 5582–5586 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R., Weinberg R. A., The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benjamin J. M., Nelson W. J., Bench to bedside and back again: Molecular mechanisms of alpha-catenin function and roles in tumorigenesis. Semin. Cancer Biol. 18, 53–64 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee S., Kong Y., Weatherbee S. D., Forward genetics identifies Kdf1/1810019J16Rik as an essential regulator of the proliferation-differentiation decision in epidermal progenitor cells. Dev. Biol. 383, 201–213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang F., et al. , PRL3 promotes cell invasion and proliferation by down-regulation of Csk leading to Src activation. J. Biol. Chem. 282, 5413–5419 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Onder T. T., et al. , Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68, 3645–3654 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Kam Y., Quaranta V., Cadherin-bound beta-catenin feeds into the Wnt pathway upon adherens junctions dissociation: Evidence for an intersection between beta-catenin pools. PLoS One 4, e4580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delva E., Kowalczyk A. P., Regulation of cadherin trafficking. Traffic 10, 259–267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang C., et al. , Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat. Cell Biol. 8, 688–699 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Heckmann B. L., Green D. R., LC3-associated phagocytosis at a glance. J. Cell Sci. 132, jcs222984 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heckmann B. L., et al. , LC3-associated endocytosis facilitates beta-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell 178, 536–551.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cadwell K., Debnath J., Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J. Cell Biol. 217, 813–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim N. G., Koh E., Chen X., Gumbiner B. M., E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. U.S.A. 108, 11930–11935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marsh T., et al. , Autophagic degradation of NBR1 restricts metastatic outgrowth during mammary tumor progression. Dev. Cell 52, 591–604.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.