Significance
Here we describe efficient, developmentally regulated stop codon readthrough of the Drosophila kelch gene. Stop codon readthrough is a form of translational recoding in which a UGA, UAG, or UAA codon is redefined as a sense codon, allowing translation to continue. We show that readthrough of the kelch stop codon occurs at highest efficiency specifically in neurons of the central nervous system (CNS), and we define a short sequence near the kelch ORF1 UGA codon that stimulates efficient readthrough. We also show that stop codon readthrough for three additional genes occurs with high efficiency in the CNS, suggesting that stop codon readthrough may contribute significantly to the neuronal proteome.
Keywords: stop codon readthrough, recoding, Drosophila, Kelch, central nervous system (CNS)
Abstract
Translational stop codon readthrough occurs in organisms ranging from viruses to mammals and is especially prevalent in decoding Drosophila and viral mRNAs. Recoding of UGA, UAG, or UAA to specify an amino acid allows a proportion of the protein encoded by a single gene to be C-terminally extended. The extended product from Drosophila kelch mRNA is 160 kDa, whereas unextended Kelch protein, a subunit of a Cullin3-RING ubiquitin ligase, is 76 kDa. Previously we reported tissue-specific regulation of readthrough of the first kelch stop codon. Here, we characterize major efficiency differences in a variety of cell types. Immunoblotting revealed low levels of readthrough in malpighian tubules, ovary, and testis but abundant readthrough product in lysates of larval and adult central nervous system (CNS) tissue. Reporters of readthrough demonstrated greater than 30% readthrough in adult brains, and imaging in larval and adult brains showed that readthrough occurred in neurons but not glia. The extent of readthrough stimulatory sequences flanking the readthrough stop codon was assessed in transgenic Drosophila and in human tissue culture cells where inefficient readthrough occurs. A 99-nucleotide sequence with potential to form an mRNA stem-loop 3′ of the readthrough stop codon stimulated readthrough efficiency. However, even with just six nucleotides of kelch mRNA sequence 3′ of the stop codon, readthrough efficiency only dropped to 6% in adult neurons. Finally, we show that high-efficiency readthrough in the Drosophila CNS is common; for many neuronal proteins, C-terminal extended forms of individual proteins are likely relatively abundant.
Context-dependent word meaning has a genetic counterpart in the dynamic redefinition of stop codons in specific locations. Dynamic codon reassignment occurs when competition between two alternative outcomes during translational decoding produces two distinct products in proportions that reflect the balance of competition. Gene expression is enriched by dynamic stop codon redefinition since, in addition to synthesis of protein encoded by the first open reading frame (ORF1), some of the product has a C-terminal extension. The extent of UGA, UAG, or UAA readthrough depends on the identity of the stop codon and on its flanking sequence, especially the 3′ mRNA sequence. UGA and to a lesser extent UAG are most frequently used for productive readthrough. Instead of involving tRNA, standard decoding of UGA, UAG, or UAA involves protein release factors and accessory proteins (1). A further distinctive feature is that in mammalian decoding, mammalian release factor 1 causes the nucleotide 3′ of the stop to be included in the ribosomal A site (2–4). Even where this feature does not occur, the identity of the base 3′ of a stop codon is relevant to its effectiveness for termination (5, 6).
Additional sequence, generally but not exclusively 3′, is relevant for the higher level of readthrough involved in evolutionarily selected occurrences. Many functionally utilized readthrough cassettes in plant viruses have particular 6- to 9-nt sequences 3′ of the stop codon (7–11), and counterpart 3′ adjacent short sequences have been studied in detail in yeast (12) and mammals (7, 13–15). The proximity of these types of stimulators to the stop codon implies they mediate their effects while within the ribosomal mRNA entrance channel. Nearby 3′ intra-mRNA structures have long been known to be important for readthrough of the Drosophila gene, headcase (16), as has a pseudoknot for the readthrough of the murine leukemia virus UAG gag terminator to yield the GagPol precursor that is the source of reverse transcriptase (17, 18). The effectiveness of the pseudoknot is aided by part of the readthrough product binding eRF1 and indirectly diminishing termination (19, 20).
Interestingly, regulation of readthrough of several plant viruses involves significant long-distance pairing of nucleotides just 3′ of the readthrough site with distant complementary nucleotides, in several cases with considerable complexity (21–24). The distant location of the 3′ component can have regulatory significance including avoidance of clashes between translating ribosomes and replicase (25, 26). Another study also showed the potential for relevant long-distance pairing for several vertebrate alphaviruses (27).
On completion of genome sequencing of 12 divergent Drosophila species, bioinformatics analyses provided the first evidence for abundant readthrough in insects and one crustacean (28, 29). In a thorough study, Jungreis et al. (29) estimated that the expression of more than 600 Anopheles gambiae mosquito genes and 900 Drosophila genes involves functional readthrough (30). Insects and probably crustaceans may be the greatest users of functional readthrough and so be the counterpart of what certain ciliates, e.g., Euplotes, are for frameshifting (31) and what squids and octopuses are for mRNA editing (32). Ribosome profiling revealed additional instances of Drosophila readthrough and experimental data on readthrough efficiency (33). The reason for the expression of an unusually large number of insect genes utilizing readthrough is unknown. One issue posed by this is whether there are tissue-specific differences in the level of readthrough.
Despite the variety, and in several cases the elaborate nature, of stimulatory signals 3′ of readthrough stop codons, in general the efficiency of utilized readthrough is substantially less than that of programmed frameshifting counterparts. In 1993, a striking exception with very high levels of readthrough was reported in expression of the Drosophila kelch gene (34, 35). The Kelch protein produced by termination at the first stop codon is 689 amino acids (aa). Like the first discovered occurrence of functional readthrough used by phage Qβ (36, 37), the kelch readthrough-derived C-terminal extension of 782 aa is long. The 689-aa Kelch protein is part of a ubiquitin ligase complex required for oocyte growth during oogenesis (38–40) and kelch mutants are female sterile (35). In addition to phenotypes during oogenesis, kelch mutations also result in defects in the larval peripheral nervous system, with loss of kelch causing a reduction in dendritic branching (41). Analysis of kelch expression revealed an unusually high level of kelch stop codon readthrough in larval imaginal discs and associated central nervous system (CNS) (34), despite no known role for Kelch in those tissues.
Here we examine in detail where kelch stop codon readthrough occurs and investigate how mRNA structure contributes to readthrough efficiency. We report exquisite tissue specificity with readthrough occurring at elevated levels in the CNS and imaginal discs. The full-length Kelch protein is abundant in larval and adult neurons, but not glial cells. We examined the contribution of cis-acting RNA features in stop codon readthrough efficiency and identified a predicted stem loop as a readthrough enhancer. Finally, we provide evidence that efficient stop codon readthrough in the CNS is widespread in Drosophila.
Results
Tissue-Specific Stop Codon Readthrough of kelch in CNS and Imaginal Discs.
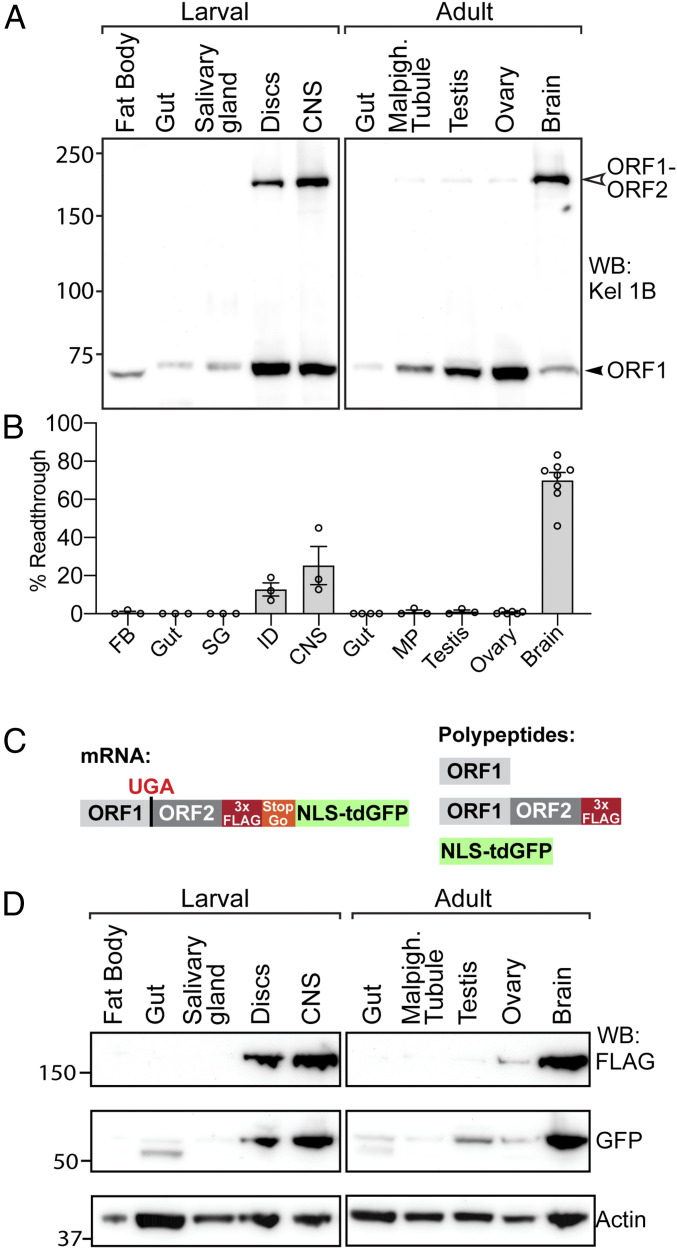
Our previous analysis of kelch revealed developmental and tissue-specific regulation of stop codon readthrough, with a high level of readthrough observed in a sample containing both imaginal discs and associated larval CNS (34). We reexamined Kelch expression in samples prepared from separated larval tissues and found the highest levels of readthrough product in both the CNS and imaginal discs. There was no detectable readthrough product in lysates of fat body, gut, and salivary gland tissue, though Kelch expression was low in these tissues (Fig. 1A). In adult tissues, brain samples had high levels of readthrough product, and malpighian tubules, testis, and ovary had very low levels (Fig. 1A). Strikingly, in adult brain samples up to 70% of Kelch proteins were detected as the readthrough product (Fig. 1B).
Fig. 1.
High-level readthrough of kelch mRNA in the Drosophila central nervous system. (A) Western blot of tissues dissected from third-instar larvae and 3- to 5-d-old adults probed with anti-Kelch. The predicted molecular masses for ORF1 and ORF1-ORF2 are 77 kDa and 160 kDa, respectively. (B) Quantification of the percent of ORF1-ORF2 in total Kelch protein present in immunoblots (n ≥ 3, error bars represent SEM). (C) Schematic of reporter system for detecting readthrough of endogenous kelch. Translation through the ORF1 UGA stop codon results in the Kelch ORF1-ORF2 readthrough product tagged at its C terminus with 3× FLAG and a distinct nuclear-localized tandem dimer GFP (NLS::tdGFP) polypeptide released during translation of a viral T2A/StopGo sequence. (D) Western blot of tissues dissected from third-instar larvae and adult animals homozygous for the readthrough reporter insertion in the kelch locus, probed with α-FLAG to detect the readthrough product, α-GFP to detect the released NLS::tdGFP, and α-actin as a loading control.
Our method to assess readthrough relied on an antibody (Kel 1B) against a Kelch antigen derived from ORF1 of kelch that detected both the ORF1 termination product and the readthrough product. Despite several attempts, we were unable to generate an antibody against antigens from the second open reading frame (ORF2) that would allow specific detection of the readthrough product. To investigate kelch readthrough more directly, we used CRISPR-Cas9-mediated homology-directed repair (HDR) to develop a reporter of endogenous kelch readthrough (Fig. 1C). This system allows for specific detection of the readthrough product using a 3×FLAG tag at the C terminus of ORF2, and also results in the production of a discrete nuclear-localized tandem dimer GFP (NLS::tdGFP), released via a viral T2A/StopGo sequence (42, 43) following readthrough translation of ORF2.
Importantly, the detection of the NLS::tdGFP protein served as a reliable assay of readthrough that is independent of the stability of the ORF1 or ORF1-ORF2 products. Western analysis of this reporter in adult and larval tissues showed elevated readthrough in the larval CNS and adult brain, with lower levels in the imaginal discs (Fig. 1D), similar to the results in immunoblot experiments (Fig. 1A). A very low level of readthrough was observed in the ovary with both the α-GFP and α-FLAG antibodies, again consistent with immunoblots probed for Kelch (Fig. 1A). Interestingly, whereas GFP was present in testis lysates, the FLAG antibody detected very little ORF1-ORF2 protein, suggesting that the ORF1-ORF2 product was less stable in the testis than in other tissues.
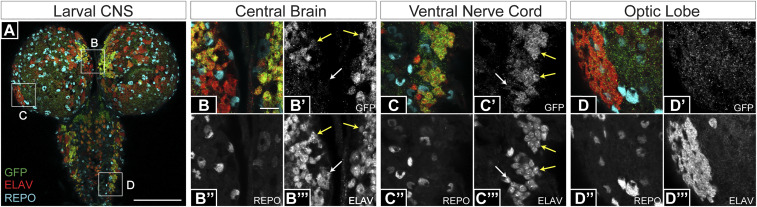
To further characterize tissue-specific kelch readthrough expression, we imaged NLS::tdGFP produced by the endogenous readthrough reporter. Imaging of larval CNS revealed abundant GFP-positive nuclei (Fig. 2). Within the CNS, the highest levels of NLS::tdGFP were observed in cells of the central brain (CB) and the ventral nerve cord (VNC), but not in the optic lobe (OL) (Fig. 2A). Colabeling with antibodies specific to neurons (ELAV) and glia (REPO) showed that the Kelch readthrough product accumulated specifically in neurons (yellow arrows) (Fig. 2 B and C). Within the CB and VNC, we found clusters of neurons (Fig. 2B′ and B′′′, white arrow) or single neurons (Fig. 2C′ and C′′′, white arrow) with no GFP above background, suggesting neuronal cell-type-specific regulation of kelch readthrough in the CNS. Neurons in the OL also had background levels of GFP labeling (Fig. 2D). We were unable to reliably image the tdGFP reporter in adult brains due to competing autofluorescent signals. Taken together, these results indicate a striking degree of tissue- and cell-specific regulation of kelch stop codon readthrough.
Fig. 2.
Visualization of endogenous kelch readthrough. (A) Analysis of larval CNS revealed the widespread presence of NLS::tdGFP produced following kelch readthrough translation. Three regions of the larval CNS were imaged at a higher magnification: central brain (B), ventral nerve cord (C), and optic lobe (D). NLS::tdGFP accumulated in neurons (ELAV, red) and not glial cells (REPO, blue) in the central brain (B–B′′′, yellow arrows) and in the ventral nerve cord (C–C′′′, yellow arrows). Some neurons (white arrows) in the central brain (B) and ventral nerve cord (C) did not show evidence of readthrough. (D–D′′′) NLS::tdGFP did not accumulate in optic lobe cells. (Scale bar for A, 100 µm; for B–D, 10 µm.)
Sequences 3′ of the UGA Codon Stimulate Readthrough.
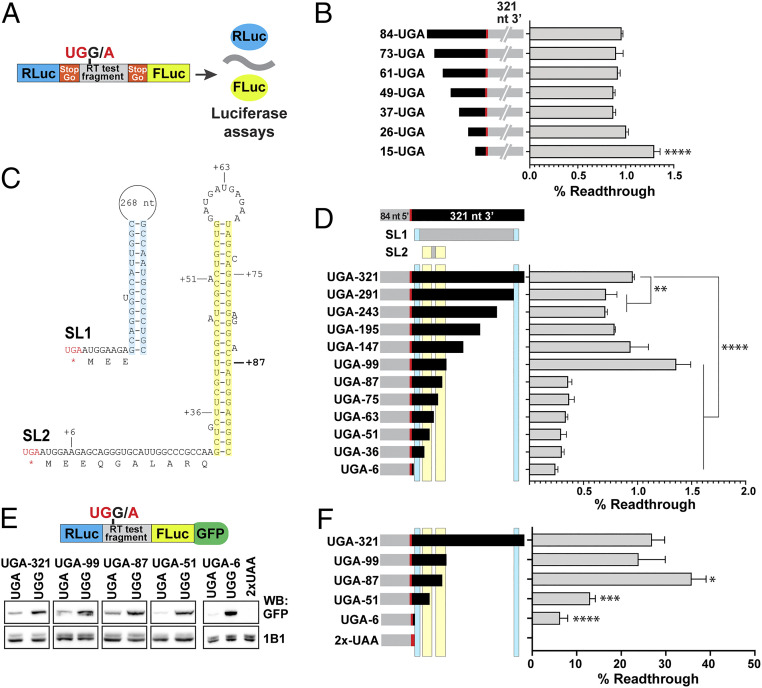
There are several examples of stop codon readthrough stimulation by 3' RNA secondary structures (18, 21, 27, 44). In kelch mRNA, a predicted stem loop (SL1) starting 8 nt 3′ of the ORF1 stop codon is supported by nucleotide conservation [Fig. 3C and SI Appendix, Fig. S1 (27)]. In addition, we identified the potential for a second stem loop (SL2) beginning 31 nt 3′ of the ORF1 stop (Fig. 3C) that is also widely conserved in Drosophila (SI Appendix, Fig. S1).
Fig. 3.
Analysis of the contribution to readthrough of cis-acting mRNA structural elements near the kelch stop codon. (A) Diagram of pSGDluc reporter construct. StopGo sequences from foot and mouth disease virus (FMDV) flanking the readthrough test fragment resulted in the translation of luciferase polypeptides free of residues encoded by kelch sequence; readthrough efficiency was calculated based on measured luciferase activities (46). (B) Sequences 5′ of the kelch stop codon were analyzed by transfecting the indicated 5′ truncation constructs into HEK-293T cells and measuring readthrough efficiency. (C) Diagram of predicted stem loop structures 3′ of the kelch ORF1 UGA codon. SL1 was previously described (27). For SL2, residues corresponding to the limits of 3′ deletions in D are indicated; location of 87 nt 3′ UTR is marked in bold. (D) Sequence requirements 3′ of the kelch UGA were analyzed as in B using the indicated 3′ deletion constructs. Predicted stem loops SL1 and SL2 are indicated above, with cyan or yellow shading corresponding to the shaded sequences in C. In B and D, n = 3 biological replicates. (E) Diagram of dual-luciferase construct for analysis of readthrough in transgenic Drosophila. GFP coding sequence was fused to the Renilla luciferase gene and readthrough efficiencies in F were calculated from the ratio of GFP produced by UGA constructs compared to matched in-frame UGG controls. Constructs were expressed in adult brains using nSybGal4, and lysates were prepared from heads. Representative Western blots of constructs analyzed are shown Below the diagram. The Top row shows immunoblotting with a GFP antibody and the Bottom row shows immunoblotting with an Adducin antibody (1B1) as a loading control. (F) Quantification of readthrough efficiencies in flies; n ≥ 3. In B, D, and F, mean and SD are plotted. Readthrough efficiencies of truncation constructs were compared to the corresponding in-frame control using ANOVA with multiple comparisons correction. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To test the possible stimulatory role of kelch sequences both upstream and downstream of the ORF1 stop codon, we generated an extensive series of dual luciferase reporters with 5′ and 3′ deletions and then assessed readthrough in HEK-293T cells (Fig. 3 A–D). This approach allowed us to survey an extensive series of deletion constructs so that we could perform more targeted experiments using transgenic Drosophila. We observed a readthrough efficiency of 1% from kelch reporters with 84 nt 5′ and 321 nt 3′ of the UGA stop codon (84-UGA-321), which encompasses the predicted stem loop structures. We did not detect a decrease in readthrough efficiency with any of the 5′ truncations tested but did observe a small increase for the reporter with 15 nt 5′ of the UGA codon (Fig. 3B). Furthermore, readthrough efficiency was unaffected by 3′ deletions that were expected to remove the 3′ component of the predicted SL1 stem loop (Fig. 3D). However, reporters with 87 nt or less of kelch sequence 3′ of the stop codon had reduced readthrough levels (Fig. 3D). These results suggest that in mammalian cells, SL2 is important for promoting readthrough.
Having identified a sequence supporting kelch readthrough in HEK-293T cells, we determined whether this sequence promotes readthrough in Drosophila by testing a subset of the truncation constructs. We modified a dual-luciferase readthrough reporter system so that GFP was fused to the C terminus of firefly luciferase, allowing readthrough to be monitored using GFP production. As kelch readthrough product was highest in the CNS, we assayed readthrough expression from lysates prepared from adult heads in which expression was driven by nSybGal4, a strong and specific pan-neuronal Gal4 driver (45). The readthrough efficiency in this reporter system for the 408 bp control fragment (84-UGA-321) was 30% (Fig. 3 E and F), significantly higher than we observed in HEK-293T cells, but consistent with the high level of readthrough product we observed when immunoblotting for Kelch in the CNS (Fig. 1A).
The dual-luciferase/GFP reporter constructs used in flies lacked the StopGo sequences that were present in our human cell culture assay system (Fig. 3A), raising the possibility that the lack of the StopGo sequences could affect the measured readthrough efficiencies in flies (46). To address this, we made additional constructs in which 84-UGA-321 and a corresponding UGG in-frame control (IFC) were flanked by StopGo (T2A) sequences and inserted between mCherry and GFP (SI Appendix, Fig. S2A). Expression of these constructs in larval neurons using nSybGal4 produced readily detectable GFP readthrough product (SI Appendix, Fig. S2 B and C). Fluorescence intensity measurements revealed a readthrough efficiency of 30% (SI Appendix, Fig. S2F), equal to that observed when analyzing the same 408-bp fragment in the dual-luciferase/GFP system. These results from two independent experimental approaches indicate that the baseline readthrough efficiency for the 408-bp fragment in Drosophila neurons is 30%.
We next examined the 3′ sequence requirements for readthrough in Drosophila. As was the case in HEK-293T cells, removal of the 3′ half stem of SL1 did not affect the readthrough efficiency (Fig. 3 E and F). In contrast to results in HEK-293T cells, 87 nt 3′ of the stop codon was sufficient to promote high-level readthrough in adult brains. When the sequence was truncated to 51 nt 3′ of the stop codon, completely destroying predicted SL2, we observed a significant decrease in readthrough efficiency (Fig. 3 E and F). The basis for the discrepancy between our cultured cell and Drosophila assays for the construct with 87 nt 3′ of the stop codon is not clear. One possibility could be that a Drosophila-specific SL2 binding protein may interact with the shortened 15-bp stem that could still form (Fig. 3C), or perhaps this shortened stem is sufficient to stimulate readthrough in fly but not in human cells. Despite the differences, both sets of results suggest that SL2 contributes to kelch readthrough efficiency.
Interestingly, a UGA construct with only 6 nt (AUGGAA) of 3′ sequence retained a readthrough efficiency of 6% when expressed in adult neurons (Fig. 3 E and F). In contrast, a reporter with tandem UAA stop codons (UAAUAA) showed no detectable readthrough in fly brains (Fig. 3 E and F). These data indicate that readthrough of the UGA-6 reporter likely represents a remarkably high basal level of kelch neuronal readthrough that is independent of stimulatory 3′ mRNA secondary structure.
kelch cDNA Readthrough Reporter Highlights Cell-Specific Regulation of Readthrough.
Our survey of endogenous kelch readthrough suggested that the CNS exhibited the highest levels of readthrough, while readthrough product was low or undetectable in other tissues. Furthermore, imaging of larval CNS with antibodies to neurons or glia revealed neuron-specific readthrough. To better understand which neurons support readthrough, we expressed the UGA-321 luciferase/GFP cDNA reporter constructs in distinct cell types. By comparing the pattern of Gal4-driven expression observed with the UGG IFC with the pattern of readthrough of the UGA reporter, we identified cell types that support readthrough to varying extents.
When expressed using the ubiquitous α-Tub84BGal4 driver, GFP from the UGG IFC construct revealed high-level expression in cells of the prothoracic gland (Fig. 4 A and B; outlined in yellow in Fig. 4A). Within the CNS, α-Tub84BGal4 drove expression in the OL, in clusters of cells in CB, and the VNC (Fig. 4A). Expression of the UGA readthrough reporter using α-Tub84BGal4 resulted in an approximate 10-fold reduction in overall fluorescence intensity. To compare the pattern of readthrough (Fig. 4C) to the pattern of expression driven by α-Tub84BGal4 (Fig. 4A), the white level for images of the UGA readthrough reporter was increased 10-fold. The readthrough reporter produced a markedly different pattern of GFP accumulation compared to the IFC. No GFP was observed in cells of the prothoracic gland (Fig. 4 D and D′), despite the high level of expression driven in these cells by α-Tub84BGal4 (Fig. 4 A and B′). This demonstrates that some cell types do not support readthrough, even for stop codon contexts that undergo high-level readthrough in other tissues. Within the CNS, GFP resulting from readthrough accumulated predominately in the VNC neuropil as well as in a subset of cell bodies in the CB region and VNC (Fig. 4C). GFP readthrough product did not accumulate in peripheral regions of the OL, where GFP produced from the IFC was present at high levels. Readthrough product was also absent from clusters of midline cells in the VNC, suggesting that readthrough may not occur in glial cells.
Fig. 4.
kelch cDNA reporter reveals cell types supporting kelch readthrough. (A–D) α-Tub84BGal4-driven expression in larval tissues of the UGG (in-frame control) reporter construct (A and B) and UGA readthrough reporter (C and D) in the dual-luciferase/GFP construct. Kelch cDNAs correspond to the 84-UGA/G-321 fragment described in Fig. 3.α-Tub84BGal4drove expression throughout the CNS (A) and at high levels in the endocrine prothoracic gland (PG), outlined in yellow and shown at higher magnification in B. (C and D) GFP intensity levels of the UGA readthrough reporter were increased 10-fold to allow the pattern of expression to be compared between the UGA and UGG constructs. (C) GFP produced by readthrough accumulated primarily in the central brain (CB) and the ventral nerve cord (VNC) neuropil. Green dotted line indicates the area occupied by the optic lobes (OL). GFP was not detected in the PG (C) and is shown at higher magnification in D and D′. (E–H) Images directly comparing GFP reporter expression in neurons or glia. Expression specifically in neurons using nSybGal4 (E) revealed readily detectable readthrough expression from the UGA readthrough reporter (F). Expression specifically in glial cells using repoGal4 (G) failed to produce detectable readthrough (region occupied by larval CNS outlined in white in H). (I) Western analysis of reporter expression in lysates prepared from heads of adult flies expressing reporter constructs specifically in neurons or glia. (J–S) UGG and UGA dual-fluorescent reporters expressed in adult brains reveal readthrough in five neuron subtypes; (J′–S′) GFP displayed with a color look-up table. (J and K) Pan-neural expression with nSybGal4 showed widespread readthrough in adult brains with highest levels in the mushroom bodies (J′ and K′, arrowheads). (L and M) Expression in GABA-producing neurons with Gad1Gal4. (N and O) Expression glutamate-producing neurons with VGlutGal4. (P and Q) Expression in dopamine-producing neurons with DdcGal4. A subset of dopaminergic neurons did not support readthrough (P′ and Q′, arrowheads). (R and S) Expression in serotonin receptor optic lobe neurons with 5-HT1AGal4. Brain structure in P–S is evident due to Bruchpilot antibody staining. (Scale bar for whole larval CNS images A, C, and E–H, 100 μm.) (Scale bar for prothoracic gland images B and D, 50 μm.) (Scale bar for adult brain images J–S, 200 µm.)
α-Tub84BGal4 drives expression in both neurons and glia, making it difficult to determine the identity of cells undergoing readthrough in the CNS. To assess the level of kelch readthrough supported in neurons, we used nSybGal4, a pan-neuronal driver (Fig. 4 E and F). When expressed exclusively in neurons, a readthrough product from the UGA reporter was readily observed (Fig. 4F); fluorescence intensity measurements indicated a readthrough efficiency of ∼30%. In contrast, when the reporters were expressed using a glial-specific repoGal4 driver (Fig. 4 G and H), readthrough was essentially undetectable (Fig. 4H). We confirmed these results by immunoblotting extracts from larval brains and observed high levels of readthrough product in neurons but not in glial cells (Fig. 4I). Taken together, this analysis shows that sequences flanking the kelch stop codon promote stop codon readthrough in a highly regulated cell-specific manner.
Since the transgenic readthrough reporters accurately recapitulated the results for endogenous NLS::tdGFP reporter in larval CNS (Fig. 2), we used the same fragment from the kelch gene in the Gal4-driven dual-fluorescent reporter (SI Appendix, Fig. S2) to analyze readthrough in adult brains. Strong neuronal drivers resulted in readily observable GFP readthrough product and allowed us to explore readthrough in distinct classes of neurons. Expression with a pan-neuronal Gal4 (nSybGal4) showed widespread expression throughout the brain from the UGG IFC (Fig. 4 J and K); GFP expressed from both the UGA readthrough reporter and the IFC accumulated prominently in the mushroom body neuropil (arrowheads in Fig. 4 J′ and K′).
We next determined whether readthrough could occur in neuronal types associated with use of specific neurotransmitters. Readthrough was clearly detected in GABAergic (Fig. 4 L and M), glutamatergic (Fig. 4 N and O), and dopaminergic (Fig. 4 P and Q) neurons, as well as neurons expressing Gal4 under the control of the serotonin receptor HT1A (Fig. 4 R and S). For all neuron classes examined, GFP readthrough product accumulated in a similar pattern as the IFC, with the exception of dopaminergic neurons, which appeared to have a subpopulation of neurons associated with the subesophageal ganglion with little readthrough apparent (arrowheads in Fig. 4 P′ and Q′). These results indicate that kelch readthrough can occur in many neuronal subtypes in the brain.
Highly Efficient Stop Codon Readthrough Is Common in the CNS.
A large number of genes are predicted to undergo stop codon readthrough in Drosophila (28–30, 47), including many that are expressed or known to function in the nervous system. Given the high-level readthrough we observed with kelch in neuronal tissues, we wondered whether neurons in Drosophila might generally support high-level readthrough. To explore this possibility, we obtained antibodies for genes known or predicted to undergo readthrough for which readthrough would result in a detectable shift in molecular weight. We probed immunoblots of lysates prepared from tissues that supported low-level kelch readthrough (ovary) or high-level neuronal kelch readthrough (adult brain or larval CNS) (Fig. 5 A–D). Western analysis of the RacGEF Sponge (48) revealed an additional product in brain lysate consistent with the predicted molecular weight of the annotated readthrough product (Fig. 5B). In adult brains, the apparent readthrough efficiency was 46%, while readthrough in ovary lysates was undetectable, similar to Kelch (Fig. 5A). We observed a similar result for the transmembrane phosphatase Ptp10D: 56% of Ptp10D protein was detected as readthrough product in brains, with only negligible readthrough detected in ovarian lysates (Fig. 5C). Readthrough of headcase has been characterized in detail (16), though its relative efficiency in different tissues has not been examined. We did not detect Headcase protein in lysates of adult brains, but we observed a similar level of readthrough (59%) as previously reported in lysates of larval CNS (16). Overall expression of Headcase in ovaries was significantly lower, but we were able to measure an apparent readthrough efficiency of 24% in ovarian lysates. Combined with our study of kelch readthrough, these results suggest that in Drosophila, neuronal tissues may be generally permissive for high-level stop codon readthrough.
Fig. 5.
Multiple Drosophila genes show high-level readthrough in neuronal tissue. (A–D) Western blots comparing readthrough translation detected in lysates prepared from ovaries and adult brains (A–C) or third instar larval (L3) CNS (D). Genes encoding each protein are diagrammed at Top; blue indicates ORF1 coding sequence, red indicates ORF2 coding sequence. Predicted molecular weights for ORF1 and ORF1-ORF2 readthrough products are listed. Ptp10D is a transmembrane receptor tyrosine phosphatase and is known to be glycosylated, which presumably accounts for the shift in apparent molecular weight (70). Blots were probed with antibodies against the ORF1 product of the indicated genes. Readthrough products are indicated with red arrowheads and ORF1 products are indicated with blue arrowheads. Apparent readthrough efficiencies are indicated beneath each lane, calculated from the ratio of readthrough product to the total quantity of protein detected.
Discussion
Highly Efficient Translational Readthrough of Drosophila Stop Codons.
Our results demonstrate that mRNAs from several Drosophila genes undergo high-level, tissue-specific stop codon readthrough in the CNS. For kelch, the highest levels of readthrough depend on sequences 3′ of the stop codon. When estimating kelch readthrough efficiency based on the steady-state levels of Kelch using an antibody to ORF1, we observed a remarkably high readthrough efficiency of ∼70% in the CNS. However, differential stability of the ORF1 and readthrough products could significantly bias this estimate. The Kelch ORF1 product is the substrate-binding component of a Cullin3-RING ubiquitin E3 ligase (CRL3Kelch) that can itself be degraded by the ubiquitin proteasome system (38), making it possible that the apparent high-efficiency readthrough was due in part to differential stability of the ORF1 and ORF1-ORF2 products.
To avoid the issue of ORF1 protein instability, we used two independent reporter-based approaches to assess kelch readthrough in the CNS. First, our endogenous readthrough reporter produced a discrete nuclear-localized tdGFP molecule each time readthrough occurred. The level of NLS::tdGFP served as a reliable indicator of relative readthrough when comparing cells and tissues. This reporter protein was most abundant in larval imaginal discs and CNS and in adult brains, indicating that the high apparent readthrough efficiency we observed using steady-state measurements with Kelch antibodies was not simply a result of tissue-specific differences in protein stability. Second, we analyzed kelch readthrough using cDNA reporter systems and measured a readthrough efficiency of 30% in the larval CNS and adult brains, demonstrating that the CNS supports high-level readthrough of the kelch ORF1 stop codon. The large difference in estimated readthrough efficiency observed with the ORF1 antibody in immunoblots compared to the cDNA reporter could be due to differential protein stability as discussed above, and/or a lack of sufficient flanking sequence in the reporter constructs necessary for maximal readthrough. Regardless, when taken together with our estimate of similar readthrough in the larval CNS or adult brain for sponge, Ptp10D, and headcase, these results suggest that in Drosophila, the CNS supports high-efficiency developmentally regulated stop codon readthrough.
Tissue- and Cell Type-Specific Regulation of Readthrough.
Analysis of both endogenous and cDNA-based readthrough reporter systems revealed a remarkable degree of tissue- and cell-specific readthrough. Readthrough efficiency was high in neurons, while glia and cells of the prothoracic gland could not support stop codon readthrough. Furthermore, using cell-type-specific expression, we found that diverse neuronal subtypes in the adult brain support readthrough. We note that headcase and Synapsin, two Drosophila genes for which readthrough has been characterized, exhibit high-efficiency readthrough in the CNS (this study and refs. 16, 49). In addition, genes identified with potential readthrough extensions based on codon conservation were found to be enriched for expression in the nervous system (29), and readthrough expression in the CNS was verified for at least one of these, Abd-B (28). Similarly, neuron-specific readthrough of a premature termination codon (PTC) in an apparent pseudogene, the Ir75a odorant receptor of Drosophila sechellia, results in production of a functional protein (50), and another analysis of PTC readthrough in Drosophila revealed highly efficient readthrough of PTCs in CNS neurons, but not glia (51).
Recent studies indicate that regulated translational readthrough in cellular gene decoding may be more widespread than previously appreciated. Cell-type-specific regulation of readthrough was observed in mice using ribosome profiling (52), suggesting broader translational mechanisms that permit cell-type-specific readthrough in flies and mammals. Human AQP4 encodes an aquaporin that undergoes efficient stop codon readthrough in cell culture (14) and for which readthrough efficiency may be regulated in a tissue-specific manner (53).
Possible Mechanisms Controlling Readthrough.
A number of factors are known to influence the efficiency of stop codon readthrough, including the local sequence context as well as the formation of RNA secondary structures 3′ of the stop codon. In addition, transacting factors have been shown to influence readthrough efficiency, including release factor abundance or modification, tRNA abundance, as well as other protein factors (54, 55).
Our analysis of kelch suggests that a predicted mRNA stem loop 3′ of the stop codon stimulates high-efficiency stop codon readthrough. Evidence for a stem loop structure promoting readthrough in Drosophila has also been described for headcase (16), and a conserved sequence 3′ of the Ptp10D stop codon is also predicted to form a stem loop (SI Appendix, Fig. S3 and ref. 28). Similar to kelch, readthrough of the headcase and Ptp10D stop codons appears to be developmentally regulated so that high-efficiency readthrough of these genes is promoted in the CNS, suggesting that stem loop structures may be important for promoting high-efficiency stop codon readthrough in the CNS.
One possible role of stem loops or other RNA secondary structures in enhancing readthrough could be precise pausing of ribosomes when a stop codon is within the A site, thus increasing dwell time and potentially nudging competition in favor of aminoacyl-tRNA decoding. Recently, cryo-EM studies revealed that during stop codon recognition by eukaryotic release factor 1 (eRF1), the nucleotide immediately 3′ of the stop codon is pulled into the A site to form a compact U turn (2–4). Perhaps a stable RNA secondary structure at the mRNA entrance tunnel could prevent eRF1 from pulling the fourth nucleotide into the A site and thus reduce termination efficiency. Another possibility is that a transacting factor could stimulate readthrough by binding to and stabilizing the RNA structure. Although there are no known examples of this type used for readthrough, recently two examples of viral ribosomal frameshifting signals were discovered where frameshifting is transactivated through the action of viral (cardiovirus 2A, arterivirus nsp1β) and cellular proteins [poly(C) binding protein] (56, 57).
In our experiments, human cells appeared more sensitive to perturbation of SL2 than fly cells; however, the much lower levels of kelch readthrough in mammalian cells is likely a confounding factor. A deletion that affects a part of the stem (UGA-87) caused a drop in readthrough efficiency in human cells while the same deletion produced slightly better readthrough in flies. Full removal of the 3′ portion of the stem in flies (UGA-51) resulted in a nearly threefold drop in readthrough. It is noteworthy that the predicted loop of SL2 is maintained in the UGA-87 reporters, perhaps allowing any fly-specific transacting factor to still interact. In any case, we view the activity of kelch SL2 in fly neurons as the more relevant with regard to readthrough stimulation.
Evasion of Nonsense-Mediated Decay Does Not Explain Tissue-Specific Readthrough.
A consideration with stop codons that are read through, especially when the stop codon is followed by a second long ORF, is nonsense-mediated decay (NMD). Indeed, avoidance of NMD has been documented in a few instances of readthrough in RNA viruses (20, 58). Proximity of the stop codon to a 3′ splice junction, and so to exon junction complex (EJC) proteins on the mRNA is relevant in mammals where any stop greater than 50 nt upstream of an EJC is marked for NMD (59). However, it is unlikely that differential tissue-specific resistance to NMD due to proximity to an EJC could explain our observations since the Ptp10D stop codon is 1,045 nt from a 3′ splice site. In addition, splice junctions and EJC proteins do not appear to be involved with PTC recognition for NMD in Drosophila (60, 61) and an instance of PTC suppression found to occur in the CNS is not due to tissue-specific differences in the NMD response (51). Furthermore, publicly available Drosophila RNAseq data available at FlyBase and FlyAtlas2 reveal comparable transcript levels in low-readthrough (ovary, salivary gland) versus high-readthrough (brain) tissues, for both kelch and the other readthrough genes (62, 63). Thus, it is unlikely that our results can be explained by a lower level of NMD in neurons.
Recently, it has become clear that the position of a stop codon within the mRNA can affect termination efficiency (64) with stop codons distant from the polyA tail having lower termination efficiency, typically resulting in NMD (59). Intriguingly, a novel genetic code was recently discovered where all three standard stop codons (TAA, TAG, and TGA) specify amino acids in Condylostoma magnum. These codons are decoded as amino acids at internal positions but specify translation termination when in close proximity to an mRNA 3′ end (65, 66). However, the length of the 3′ untranslated region (UTR) after the first stop codon in Drosophila readthrough genes ranges widely from a few nucleotides to thousands (28, 33). For example, kelch mRNA is abundant in ovarian cells where readthrough is nearly undetectable even though the 3′ UTR following the first stop codon is over 3,000 nt. This constellation of readthrough genes in the Drosophila genome argues against a correlation between 3′ UTR length and evasion of NMD in favor of readthrough.
ORF2 Function.
Evidence for extensive stop codon readthrough in Drosophila comes from ribosome footprinting experiments (33) and analysis of codon conservation downstream of the first stop codon in genes (28–30, 47). Based on these data, there are now over 400 genes annotated in FlyBase as readthrough genes with highly conserved extensions among Drosophila species. In contrast, there are only a few dozen readthrough genes documented in the mouse/human genome (14, 33, 52, 67, 68), suggesting the Drosophila genome finds readthrough particularly useful. Curiously, the peptide extensions produced by readthrough genes in Drosophila vary considerably in length from fewer than 10 amino acids to over 800. Remarkably, many of these readthrough extensions have been conserved over hundreds of millions of years of insect evolution (30), indicating that they are under positive selection and function to increase the fitness of these insects. That readthrough is exceptionally and dramatically efficient in the central nervous system is intriguing.
Materials and Methods
Drosophila Stocks.
Flies were maintained at 25 °C on cornmeal/molasses medium. Gal4 drivers were obtained from the Bloomington Drosophila Stock Center (BDSC); abbreviated descriptions used in the text refer to the following stocks: tubGal4: P[tubP-GAL4]LL7, RRID:BDSC_5138; HT1AGal4: P[GMR54B06-GAL4]attP2, RRID:BDSC_49589; Gad1Gal4: P[Gad1-GAL4.3.098]2, RRID:BDSC_51630; nSybGal4: P[nSyb-GAL4.S]3, RRID:BDSC_51635; DdcGal4: P[Ddc-GAL4.L]4.3D, RRID:BDSC_7010; VGlutGal4: P[VGlut-GAL4.D]1, RRID:BDSC_24635.
Cloning and Molecular Biology.
For the endogenous kelch readthrough reporter, we designed a homology donor (HD) construct with a readthrough reporter cassette located immediately 5′ of the kelch ORF2 stop codon. The reporter cassette consisted of a 3×FLAG epitope tag, a T2A StopGo sequence, and a nuclear-localized tandem green fluorescent protein (NLS::tdGFP) reporter gene. The 5′ and 3′ homology arms were 1,501 bp and 1,100 bp, respectively. A 3×P3-DsRed transformation marker flanked by PiggyBAC transposon termini (derived from pScarlessHD-DsRed; gift of Kate O’Connor-Giles (Brown University, Providence, RI), Addgene plasmid #64703) was inserted between duplicated TTAA PiggyBAC target sequences in the 3′ homology arm, 42 nt 3′ of the ORF2 stop codon. A 1.5-kb fragment containing the readthrough reporter cassette and adjacent sequences was synthesized (Genscript) and assembled with remaining 5′ and 3′ homology sequences and the DsRed transformation marker using conventional cloning techniques. The protospacer adjacent motif (PAM) of the guide RNA (gRNA) site used for targeting was mutated from AGG to AGA in this vector. Oligos encoding a gRNA targeting a site 40 nucleotides 5′ of the ORF2 TAA stop codon (gRNA: gGCTGTTGGTAGTGCTT—GGA; — indicates cleavage site. Initial “g” residue is not present in genomic sequence) were cloned into pBFvU6.2 (69). The homology donor construct and U6-gRNA plasmids were injected into fly embryos expressing transgenic Cas9 at BestGene. DsRed+ transformants were isolated, and the DsRed marker gene was subsequently excised by crossing to a PiggyBac transposase transgenic line (RRID:BDSC 8285). Initial HDR events and DsRed-excision lines were verified by PCR for correct targeting and integration. For the generation of dual-luciferase expression constructs for mammalian cell expression, inserts were amplified by PCR using a kelch synthetic DNA (gBlock, Integrated DNA Technologies) as template and cloned into pSGDluc (46) using standard techniques. For Drosophila constructs, readthrough test fragments flanked by dual-luciferase coding sequences were cloned into a modified pJFRC28, a PhiC31 UAS-GFP expression vector (Addgene plasmid #36431). Plasmids were integrated at the attP2 site on chromosome 3L by injection at Rainbow Transgenics or Genetivision. All constructs were verified by DNA sequencing.
Immunoblotting.
For blots analyzing readthrough in different tissues (Fig. 1), samples were dissected in physiological buffers (phosphate buffered saline [PBS] or Schneider’s S2 medium [Thermo Fisher]), flash frozen on dry ice, and stored at −80 °C until use. Ovarian tissue was lysed in sodium dodecyl sulfate (SDS) sample buffer using plastic pestles and a motorized homogenizer (Kontes). All other tissues were lysed by freeze-thaw treatment, pipetting and incubation at 95C in SDS sample buffer. Quantities of tissue used for equal loading were determined empirically; equal loading was verified by amido black staining. Proteins were separated on either a NuPAGE 4 to 12% gradient gel (Fig. 1D) or 7.5% polyacrylamide gels (all other figures). Separated proteins were transferred to nitrocellulose and membranes were blocked and probed with the following antibodies: mouse α-Kelch (Kel 1B, Developmental Studies Hybridoma Bank [DSHB], 1:25), mouse α-Adducin (1B1, DSHB, 1:50), mouse α-FLAG (M2, Sigma F1804, 1:1,000), rabbit α-GFP (Invitrogen A-11122, 1:1,000), mouse α-actin (JLA20, DSHB, 1:50), mouse α-Headcase (HDC U33, 1:40), and guinea pig α-Sponge (gift from Erika Geisbrecht, Kansas State University, Manhattan, KS; 1:1,000). Blots were washed, probed with corresponding secondary horseradish peroxidase (HRP)-conjugated secondary antibodies (Pierce), washed again, incubated with enhanced chemiluminescent (ECL) reagents (Clarity, Bio-Rad), and imaged using a charged coupled device (CCD) (ProteinSimple).
For semiquantitative immunoblotting of GFP readthrough products, lysates were prepared from heads expressing UAS reporter constructs. Ten heads per biological replicate were flash frozen on dry ice, homogenized in 50 μL of SDS sample buffer, and incubated at 95 °C for 5 min. A total of 10 μL was loaded per lane, separated on 7.5% polyacrylamide gels, and blotted as described above. Intensities of GFP and Adducin bands were measured in ImageJ/FIJI, and GFP was normalized for loading based on Adducin levels. Readthrough efficiency was calculated as the fraction of GFP detected in the UGA readthrough sample relative to its matched UGG in-frame control.
Immunofluorescence.
Tissues were dissected in PBS or Schneider’s S2 medium, fixed in paraformaldehyde and washed four times in PBTx (PBS + 0.1% Triton X-100, 0.5% bovine serum albumin [BSA]). Larval CNS expressing fluorescent proteins from cDNA reporter constructs were incubated with 0.5 µg/mL 4,6-diamino-2-phenylindole (DAPI), washed, and mounted in 90% glycerol/20 mM Tris pH 8.0. The following primary antibodies were used for immunofluorescence as indicated in the figure legends: rabbit anti-GFP (Thermo Fisher A11122, 1:500), rat anti-ELAV (DSHB, 7E8A10, 1:20), mouse anti-REPO (DSHB 8D12, 1:25), and mouse anti-Bruchpilot (DSHB nc82, 1:40). Samples were washed in PBTx and then incubated with α-rabbit 488 (Thermo Fisher A11034, 1:500), α-rat 568 (Thermo Fisher A11077, 1:500), and α-mouse 647 (Thermo Fisher A21236, 1:500), washed and mounted in ProLong (Thermo Fisher).
Imaging and Analysis.
Samples were imaged using either a Leica SP8 scanning confocal system with a 40× Plan Apo 1.30 NA objective or a Zeiss Axio observer 7 inverted microscope with a CrEST X-light v2 spinning disk system, Photometrics BSi sCMOS camera and either a 20× Plan Apo 0.8 NA objective or a 40× C-Apo 1.2 NA water-immersion objective. Images were processed and analyzed using ImageJ/FIJI. For image sets in which fluorescence intensities were compared, images were collected under identical imaging conditions using the Zeiss/sCMOS system. Images were processed in FIJI and levels were set identically except where indicated in the figure legend. For quantification of readthrough of the mCherry/GFP readthrough reporter, summed-intensity projection images of larval brains expressing mCherry and GFP were analyzed. Masks outlining each CNS were generated by autothresholding the mCherry channel of each image in FIJI (n = 4 for each construct), and GFP intensities within the masked region were measured. Readthrough was calculated based on the ratio of the summed intensity of GFP in the TGA readthrough construct to the TGG in-frame control.
Cell Culture and Transfections.
HEK293T cells (ATCC) were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1 mM l-glutamine and antibiotics. Cells were transfected with Lipofectamine 2000 reagent (Invitrogen), using the 1-d protocol in which suspended cells are added directly to the DNA complexes in half-area 96-well plates. The following were added to each well: 25 ng of each plasmid plus 0.2 μL Lipofectamine 2000 in 25 μL Opti-Mem (Gibco). The transfecting DNA complexes in each well were incubated with 3 × 104 cells suspended in 50 μL DMEM + 10% FBS at 37 °C in 5% CO2 for 24 h.
Dual-Luciferase Assay.
Firefly and Renilla luciferase activities were determined using the Dual-Luciferase Stop & Glo Reporter Assay System (Promega). Relative light units were measured on a Veritas Microplate Luminometer with two injectors (Turner Biosystems). Transfected cells were lysed in 12.6 μL of 1× passive lysis buffer (PLB) and light emission was measured following injection of 25 μL of either Renilla or firefly luciferase substrate. Readthrough efficiencies (% readthrough) were determined by calculating relative luciferase activities (firefly/Renilla) of TGA constructs and dividing by relative luciferase activities from replicate wells of control TGG constructs. The number of biological replicates for each experiment is indicated in each figure legend.
Supplementary Material
Acknowledgments
We thank Jenna O’Neil for preliminary work on this project at Yale University. N.M.W. and J.F.A. thank Ray Gesteland for facilitating the Utah component of this work. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. Monoclonal antibodies were obtained from the Developmental Studies Hybridoma Bank (University of Iowa) or were the kind gift of Erika Geisbrecht (Kansas State University). This research was supported by Irish Research Council Advanced Laureate (IRCLA/2019/74) to J.F.A. and NIH grant R01 GM0043301 to L.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012793118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Ivanov A., et al. , Polyadenylate-binding protein-interacting proteins PAIP1 and PAIP2 affect translation termination. J. Biol. Chem. 294, 8630–8639 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown A., Shao S., Murray J., Hegde R. S., Ramakrishnan V., Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matheisl S., Berninghausen O., Becker T., Beckmann R., Structure of a human translation termination complex. Nucleic Acids Res. 43, 8615–8626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shao S., et al. , Decoding mammalian ribosome-mRNA states by translational GTPase complexes. Cell 167, 1229–1240.e15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown C. M., Stockwell P. A., Trotman C. N., Tate W. P., Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 18, 6339–6345 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCaughan K. K., Brown C. M., Dalphin M. E., Berry M. J., Tate W. P., Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. U.S.A. 92, 5431–5435 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzalone A. V., Zairis S., Lin A. J., Rabadan R., Cornish V. W., Interrogation of eukaryotic stop codon readthrough signals by in vitro RNA selection. Biochemistry 58, 1167–1178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beier H., Grimm M., Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 29, 4767–4782 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth A. E., Brierley I., Non-canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrell L., Melcher U., Atkins J. F., Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res. 30, 2011–2017 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skuzeski J. M., Nichols L. M., Gesteland R. F., Atkins J. F., The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J. Mol. Biol. 218, 365–373 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Namy O., Hatin I., Rousset J. P., Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2, 787–793 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cridge A. G., Crowe-McAuliffe C., Mathew S. F., Tate W. P., Eukaryotic translational termination efficiency is influenced by the 3′ nucleotides within the ribosomal mRNA channel. Nucleic Acids Res. 46, 1927–1944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loughran G., et al. , Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res. 42, 8928–8938 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajput B., Pruitt K. D., Murphy T. D., RefSeq curation and annotation of stop codon recoding in vertebrates. Nucleic Acids Res. 47, 594–606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steneberg P., Samakovlis C., A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep. 2, 593–597 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Houck-Loomis B., et al. , An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480, 561–564 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wills N. M., Gesteland R. F., Atkins J. F., Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl. Acad. Sci. U.S.A. 88, 6991–6995 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orlova M., Yueh A., Leung J., Goff S. P., Reverse transcriptase of Moloney murine leukemia virus binds to eukaryotic release factor 1 to modulate suppression of translational termination. Cell 115, 319–331 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Tang X., et al. , Structural basis of suppression of host translation termination by Moloney Murine Leukemia Virus. Nat. Commun. 7, 12070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown C. M., Dinesh-Kumar S. P., Miller W. A., Local and distant sequences are required for efficient readthrough of the barley yellow dwarf virus PAV coat protein gene stop codon. J. Virol. 70, 5884–5892 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller W. A., Jackson J., Feng Y., Cis- and trans-regulation of luteovirus gene expression by the 3′ end of the viral genome. Virus Res. 206, 37–45 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newburn L. R., White K. A., Atypical RNA elements modulate translational readthrough in tobacco necrosis virus D. J. Virol., 91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y., et al. , A stem-loop structure in potato leafroll virus open reading frame 5 (ORF5) is essential for readthrough translation of the coat protein ORF stop codon 700 bases upstream. J. Virol., 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cimino P. A., Nicholson B. L., Wu B., Xu W., White K. A., Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog. 7, e1002423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller W. A., White K. A., Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 44, 447–467 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Firth A. E., Wills N. M., Gesteland R. F., Atkins J. F., Stimulation of stop codon readthrough: Frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 39, 6679–6691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jungreis I., et al. , Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res. 21, 2096–2113 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin M. F., et al. , Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res. 17, 1823–1836 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jungreis I., et al. , Evolutionary dynamics of abundant stop codon readthrough. Mol. Biol. Evol. 33, 3108–3132 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobanov A. V., et al. , Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nat. Struct. Mol. Biol. 24, 61–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liscovitch-Brauer N., et al. , Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell 169, 191–202.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunn J. G., Foo C. K., Belletier N. G., Gavis E. R., Weissman J. S., Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2, e01179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson D. N., Cooley L., Examination of the function of two kelch proteins generated by stop codon suppression. Development 124, 1405–1417 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Xue F., Cooley L., kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72, 681–693 (1993). [DOI] [PubMed] [Google Scholar]
- 36.Hofstetter H., Monstein H. J., Weissmann C., The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim. Biophys. Acta 374, 238–251 (1974). [DOI] [PubMed] [Google Scholar]
- 37.Weiner A. M., Weber K., Natural read-through at the UGA termination signal of Q-beta coat protein cistron. Nat. New Biol. 234, 206–209 (1971). [DOI] [PubMed] [Google Scholar]
- 38.Hudson A. M., Cooley L., Drosophila Kelch functions with Cullin-3 to organize the ring canal actin cytoskeleton. J. Cell Biol. 188, 29–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson A. M., Mannix K. M., Cooley L., Actin cytoskeletal organization in Drosophila germline ring canals depends on kelch function in a cullin-RING E3 ligase. Genetics 201, 1117–1131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hudson A. M., Mannix K. M., Gerdes J. A., Kottemann M. C., Cooley L., Targeted substrate degradation by Kelch controls the actin cytoskeleton during ring canal expansion. Development, 146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Djagaeva I., Doronkin S., COP9 limits dendritic branching via Cullin3-dependent degradation of the actin-crosslinking BTB-domain protein Kelch. PLoS One 4, e7598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atkins J. F., et al. , A case for “StopGo”: Reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (stop) after addition of glycine and then allowing continued translation (Go). RNA 13, 803–810 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diao F., White B. H., A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics 190, 1139–1144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brault V., et al. , Aphid transmission of beet western yellows luteovirus requires the minor capsid read-through protein P74. EMBO J. 14, 650–659 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan C. C., et al. , Systematic discovery of Rab GTPases with synaptic functions in Drosophila. Curr. Biol. 21, 1704–1715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loughran G., Howard M. T., Firth A. E., Atkins J. F., Avoidance of reporter assay distortions from fused dual reporters. RNA 23, 1285–1289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mudge J. M., et al. , Discovery of high-confidence human protein-coding genes and exons by whole-genome PhyloCSF helps elucidate 118 GWAS loci. Genome Res. 29, 2073–2087 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biersmith B., Wang Z. H., Geisbrecht E. R., Fine-tuning of the actin cytoskeleton and cell adhesion during Drosophila development by the unconventional Guanine nucleotide exchange factors myoblast city and sponge. Genetics 200, 551–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klagges B. R., et al. , Invertebrate synapsins: A single gene codes for several isoforms in Drosophila. J. Neurosci. 16, 3154–3165 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prieto-Godino L. L., et al. , Olfactory receptor pseudo-pseudogenes. Nature 539, 93–97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y., et al. , Premature termination codon readthrough in Drosophila varies in a developmental and tissue-specific manner. Sci. Rep. 10, 8485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sapkota D., et al. , Cell-type-specific profiling of alternative translation identifies regulated protein isoform variation in the mouse brain. Cell Rep. 26, 594–607.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palazzo C., et al. , Tissue distribution of the readthrough isoform of AQP4 reveals a dual role of AQP4ex limited to CNS. Int. J. Mol. Sci., 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baranov P. V., Atkins J. F., Yordanova M. M., Augmented genetic decoding: Global, local and temporal alterations of decoding processes and codon meaning. Nat. Rev. Genet. 16, 517–529 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Hellen C. U. T., Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb. Perspect. Biol., 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu C., et al. , A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Napthine S., et al. , A novel role for poly(C) binding proteins in programmed ribosomal frameshifting. Nucleic Acids Res. 44, 5491–5503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.May J. P., Yuan X., Sawicki E., Simon A. E., RNA virus evasion of nonsense-mediated decay. PLoS Pathog. 14, e1007459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurosaki T., Popp M. W., Maquat L. E., Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gatfield D., Unterholzner L., Ciccarelli F. D., Bork P., Izaurralde E., Nonsense-mediated mRNA decay in Drosophila: At the intersection of the yeast and mammalian pathways. EMBO J. 22, 3960–3970 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Metzstein M. M., Krasnow M. A., Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2, e180 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gerstein M. B., et al. , Comparative analysis of the transcriptome across distant species. Nature 512, 445–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leader D. P., Krause S. A., Pandit A., Davies S. A., Dow J. A. T., FlyAtlas 2: A new version of the Drosophila melanogaster expression atlas with RNA-seq, miRNA-seq and sex-specific data. Nucleic Acids Res. 46, D809–D815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ivanov A., et al. , PABP enhances release factor recruitment and stop codon recognition during translation termination. Nucleic Acids Res. 44, 7766–7776 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heaphy S. M., Mariotti M., Gladyshev V. N., Atkins J. F., Baranov P. V., Novel ciliate genetic code variants including the reassignment of all three stop codons to sense codons in condylostoma magnum. Mol. Biol. Evol. 33, 2885–2889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Swart E. C., Serra V., Petroni G., Nowacki M., Genetic codes with No dedicated stop codon: Context-dependent translation termination. Cell 166, 691–702 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loughran G., et al. , Stop codon readthrough generates a C-terminally extended variant of the human vitamin D receptor with reduced calcitriol response. J. Biol. Chem. 293, 4434–4444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schueren F., et al. , Peroxisomal lactate dehydrogenase is generated by translational readthrough in mammals. eLife 3, e03640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kondo S., Ueda R., Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tian S. S., Tsoulfas P., Zinn K., Three receptor-linked protein-tyrosine phosphatases are selectively expressed on central nervous system axons in the Drosophila embryo. Cell 67, 675–685 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.