Significance
Ocean warming is causing repeated mass coral bleaching, leading to catastrophic losses of coral reefs worldwide. Our ability to slow or revert this decline is hampered by an incomplete understanding of the processes underlying the breakdown of the coral–algal symbiosis. Here, we show that heat stress destabilizes the nutrient cycling between corals and their endosymbiotic algae long before bleaching becomes apparent. Notably, increased metabolic energy demands shift the coral–algal symbiosis from a nitrogen- to a carbon-limited state, reducing translocation and recycling of photosynthetic carbon. This effectively undermines the ecological advantage of harboring algal symbionts and directly contributes to the breakdown of the coral–algal symbiosis during heat stress.
Keywords: coral bleaching, endosymbiosis, metabolic interaction, resource competition, selfish symbiont
Abstract
Recurrent mass bleaching events are pushing coral reefs worldwide to the brink of ecological collapse. While the symptoms and consequences of this breakdown of the coral–algal symbiosis have been extensively characterized, our understanding of the underlying causes remains incomplete. Here, we investigated the nutrient fluxes and the physiological as well as molecular responses of the widespread coral Stylophora pistillata to heat stress prior to the onset of bleaching to identify processes involved in the breakdown of the coral–algal symbiosis. We show that altered nutrient cycling during heat stress is a primary driver of the functional breakdown of the symbiosis. Heat stress increased the metabolic energy demand of the coral host, which was compensated by the catabolic degradation of amino acids. The resulting shift from net uptake to release of ammonium by the coral holobiont subsequently promoted the growth of algal symbionts and retention of photosynthates. Together, these processes form a feedback loop that will gradually lead to the decoupling of carbon translocation from the symbiont to the host. Energy limitation and altered symbiotic nutrient cycling are thus key factors in the early heat stress response, directly contributing to the breakdown of the coral–algal symbiosis. Interpreting the stability of the coral holobiont in light of its metabolic interactions provides a missing link in our understanding of the environmental drivers of bleaching and may ultimately help uncover fundamental processes underpinning the functioning of endosymbioses in general.
Coral reef ecosystems are suffering a dramatic decline worldwide due to the effects of anthropogenic environmental change (1, 2). The steadily increasing rate of ocean warming is now the main driver of reef degradation due to recurrent mass bleaching of corals (3), that is, the expulsion or digestion of endosymbiotic algae by their coral host during extended periods of heat stress. Given that the rate of global warming appears to exceed the adaptive capacity of many corals, novel approaches are required to mitigate the effects of climate change on coral reefs (4). Importantly, the bleaching susceptibility of the coral holobiont, that is, the functional ecological unit composed of the coral host and its associated microbes (5), does not depend on temperature stress alone (6, 7). Rather, the response of the coral holobiont during heat stress is determined by the interactions between all holobiont members and must be understood in the context of their given environment (8). Consequently, differences in host identity, microbiome community composition, as well as abiotic and biotic environmental conditions result in differential bleaching susceptibility from organismal to ecosystem scales (9–11). This heterogeneity in bleaching susceptibility could be harnessed for active conservation measures to preserve the functioning of reefs in the future (12). Our ability to predict or mitigate the consequences of climate change on coral reefs thus requires an in-depth understanding of the processes underlying the breakdown of the coral–algal symbiosis during heat stress.
Molecular and ultrastructural evidence suggests that the loss of algal symbiont cells during bleaching in its essence is the result of an innate immune response of the coral host (13). Prevailing theories suggest that this immune response is initially triggered by the excess production and release of reactive oxygen species (ROS) by the algal symbionts due to photoinhibition (14). Indeed, heat stress does promote an increased ROS release by algal symbionts and the quenching of ROS via the addition of antioxidants has been shown to attenuate the severity of bleaching in corals (15, 16). Yet, recent observations have challenged the notion of photosynthetic ROS production as the primary driver of coral bleaching: 1) bleaching also occurs in the dark without photosynthetic ROS production (17); 2) oxidative stress in the coral host tissue during heat stress may precede the photoinhibition of algal symbionts (18); and 3) extracellular levels of superoxide do not correlate with symbiont abundance or bleaching status of corals during heat stress (19). Taken together, these studies suggest that the role of symbiotic ROS production in triggering coral bleaching is more complex than previously thought, and that additional cellular processes may be involved.
Efficient assimilation and recycling of organic and inorganic nutrients provide the functional basis of the coral–algal symbiosis and are the key to the ecological success of corals in highly oligotrophic waters (20). Photosynthates derived from the algal metabolism constitute the main energy source for the coral host besides heterotrophic feeding (21). Therefore, the ability of corals to cope with environmental stress depends (in part) on carbon translocation by algal symbionts (22). In a stable state, nitrogen limitation of algal symbionts causes the accumulation and subsequent release of excess photosynthetic carbon to the coral host, thereby ensuring the functioning of the symbiosis (23–26). Therefore, the stability of the symbiosis during stress may ultimately depend on the availability of environmental nitrogen and the ability of the host to control algal nitrogen uptake under these conditions (27, 28). Indeed, it is now apparent that the nutritional status and environmental nutrient availability can affect the bleaching susceptibility of corals during heat stress (29–33). Likewise, elevated temperatures at subbleaching levels have been linked to alterations in nitrogen uptake and carbon translocation in the coral–algal symbiosis (34–36). Taken together, these studies suggest that symbiotic nutrient exchange may play a critical role in the onset of coral bleaching (28). However, the underlying causes of altered nutrient cycling between coral and algae, the nature of these metabolic interactions, and their consequences for the functioning of the symbiosis remain poorly understood.
Here, we investigated the effect of heat stress on metabolic interactions in the coral–algal symbiosis and their consequences for holobiont functioning in a 3-wk-long heat stress experiment. To identify the primary processes initiating the destabilization of the symbiosis, we focused on early stress responses prior to any visual signs of bleaching, combining physiological and molecular approaches (coral host and algal symbiont gene expression) with quantification of nutrient fluxes (carbon and nitrogen) at the holobiont and cellular level.
Results and Discussion
Coral Bleaching Takes Time.
The Red Sea is home to some of the most thermotolerant corals on the planet and has been identified as a potential refuge for corals during climate change (37–41). We exposed five colonies of Stylophora pistillata from the central Red Sea to conditions reflecting the annual mean (29.1 °C; control) or summer maximum (32.9 °C; heat stress with 3 d of gradual temperature acclimation) temperature for the collection site in 2017 (SI Appendix, Fig. S1). After 21 d of heat stress, S. pistillata colonies showed clear signs of bleaching as reflected in a visual loss of pigmentation, corresponding to a 78% and 67% decline in algal symbiont densities and chlorophyll a content, respectively (see SI Appendix, Fig. S2 and Table S1 for statistical results). This observation confirms that corals from the central Rea Sea live close to their upper temperature tolerance limit and is consistent with the notion that the cumulative heat stress over time determines the bleaching threshold of a coral colony (42).
Oxidative Damage in the Coral Host prior to Coral Bleaching.
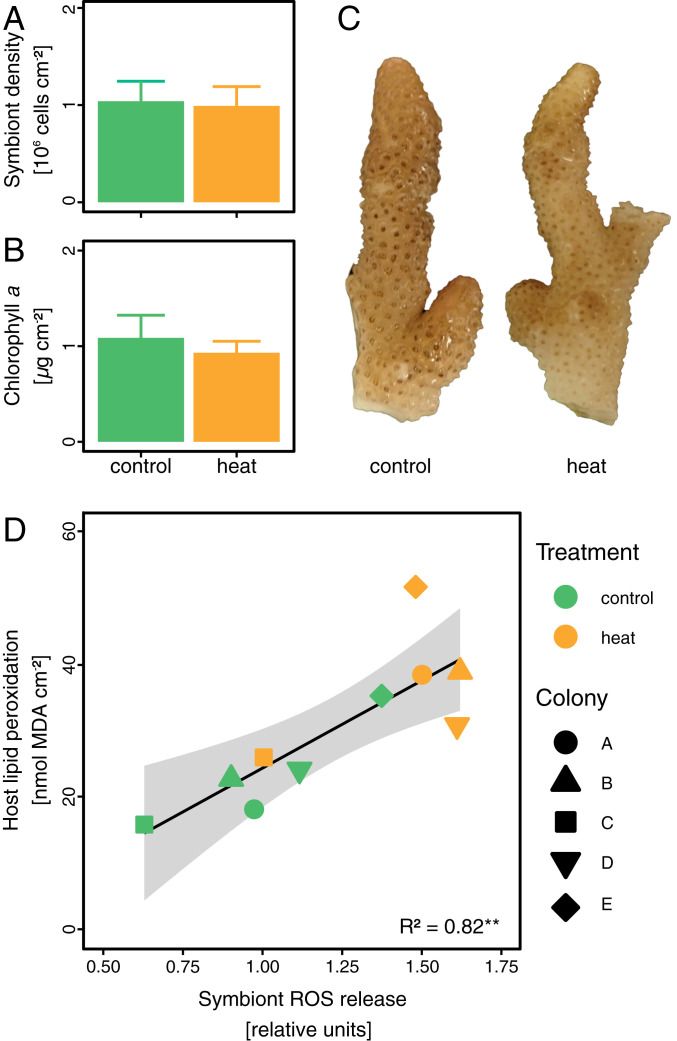
To understand the underlying processes that cause the breakdown of the coral–algal symbiosis during prolonged heat stress, we investigated the physiological performance and metabolic interactions of symbiotic partners on day 10 of the experiment, that is, after 7 d at maximum temperature but before visible bleaching commenced (SI Appendix, Fig. S1E). At this time point, S. pistillata colonies did not exhibit any visual change in pigmentation or decreases in symbiont density and chlorophyll a content between heat stress and control temperatures (Fig. 1 A–C). Despite the apparent integrity of the symbiosis at this time point, heat stress had pronounced effects on coral–algal interactions. Although maximum photosynthetic efficiency only exhibited a small reduction (7%) (SI Appendix, Fig. S3A), the relative ROS leakage from freshly isolated algal symbionts increased by 45% under heat stress conditions. This correlated with a significant increase in the level of oxidative damage in the coral host, as reflected in increased levels of lipid peroxidation (Fig. 1D). Such oxidative damage in response to algal symbiont ROS production is often considered to be the direct cellular trigger for symbiont loss during coral bleaching. Yet, in the present study, increased oxidative damage preceded the onset of visual bleaching [defined as the visual loss of pigmentation compared to control corals (43)] by 10 d. Consequently, coral bleaching does not appear to be directly and causally connected to the initial increased algal symbiont ROS production per se. If oxidative stress was indeed the main trigger of the subsequent bleaching, the resulting oxidative damage must have either gradually accumulated and/or gradually reduced—and eventually overwhelmed—the ability of the coral holobiont to cope with the oxidative stress over the course of the experiment.
Fig. 1.
State of the coral-algal symbiosis on day 10 of heat stress. (A–C) Corals showed no significant differences in algal symbiont densities or chlorophyll a content and no visual signs of bleaching. (D) Relative release of ROS from freshly isolated symbionts increased during heat stress and correlated with levels of oxidative stress (measured as lipid peroxidation) in the coral host tissue. Bars indicate the mean ± SE. The line displays best-fitting linear regression. The shaded area indicates the 95% confidence intervals. The R2 value indicates the amount of variation explained by the linear regression.
RNA Sequencing Identifies Primary Heat Stress Responses in the Coral–Algal Symbiosis.
To further elucidate the early stress response, we investigated the transcriptomic response of the coral host and algal symbionts to heat stress on day 10 of the experiment, that is, prior to the onset of bleaching (SI Appendix, Fig. S1E). At this time point, overall gene expression profiles of the coral host and algal symbionts were largely determined by differences between mother colonies, accounting for 79% and 66% of the total variation, respectively (SI Appendix, Fig. S4 A and B and Tables S2 and S3). In contrast, heat stress only explained 12% and 17% of the variation in coral host and algal symbiont gene expression, reflecting the apparent integrity of the symbiosis at this time point. However, subsets of genes related to individual EuKaryotic Orthologous Groups (KOGs) showed a pronounced stress response. In particular, genes related to the KOG “energy production and conversion” showed strong differential expression due to heat stress in coral hosts and algal symbionts alike (SI Appendix, Fig. S4 C and D). This observation aligns with the predictions of the metabolic theory of ecology, which holds that increased metabolic turnover is a direct consequence of elevated temperatures in the coral holobiont (44). Consistently, the observed changes in gene expression indicated that altered energy metabolism is part of the initial response to heat stress in the coral–algal symbiosis.
Heat Stress Induces Coral Host Starvation and Amino Acid Degradation.
On day 10, a total of 3,426 of the 25,769 genes in S. pistillata (45) were significantly differentially expressed (1,699 up- and 1,727 down-regulated) (Dataset S1). Gene set enrichment analysis revealed 273 significantly enriched biological processes (Dataset S1). In addition to genes involved in well-characterized heat stress responses (stress sensing, protein folding, DNA replication and repair, and immune response), genes linked to energy and amino acid metabolism were among the most differentially expressed (SI Appendix, Fig. S4E). Notably, heat stress prompted the down-regulation of genes related to transport and metabolism of cellular energy substrates, such as lipids, fatty acids, and sugars. Genes associated with key processes in the cellular energy supply, such as the tricarboxylic acid (TCA) cycle and adenosine triphosphate (ATP) synthesis and metabolism, were also consistently down-regulated (Fig. 2D and SI Appendix, Fig. S4E). Taken together, the observed expression patterns point to a state of energy starvation in the coral host as a result of carbon limitation during heat stress.
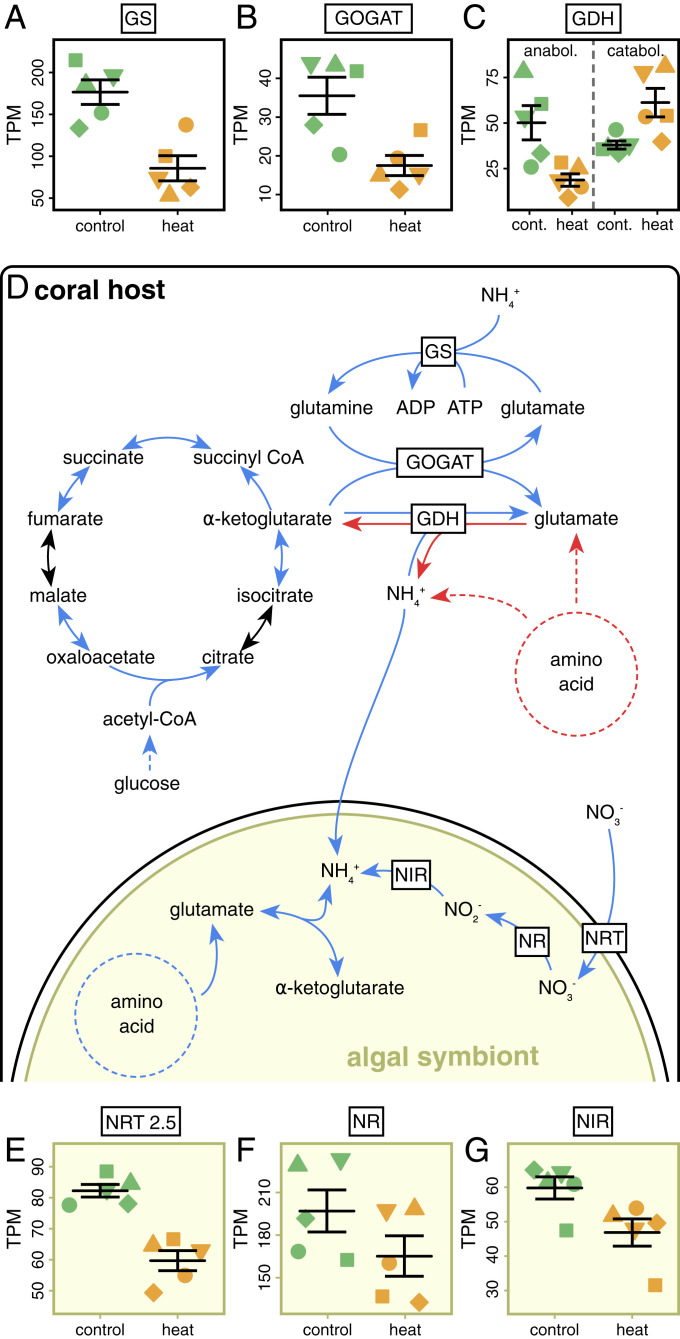
Fig. 2.
Regulation of coral host and algal symbiont gene expression on day 10 of heat stress. (A–C) Mean gene expression of significantly differentially expressed key genes in the amino acid metabolism of the coral host. (D) Overview of how highlighted metabolic pathways of the coral host and algal symbiont may interact to alter nutrient cycling in the symbiosis. (E–G) Mean gene expression of significant differentially expressed nitrate (NO3−) assimilation key genes of algal symbionts. Blue arrows indicate a significant down-regulation and red arrows indicate a significant up-regulation of gene expression during heat stress. Lines and error bars indicate mean ± SE. TPM = transcripts per million. For a complete list of differentially expressed genes as well as significant Gene Ontology terms see Dataset S1.
As a likely consequence of this energy limitation, patterns of altered amino acid cycling reflected an increased use of amino acids as alternative energy substrates during heat stress. Catabolic degradation pathways for several amino acids showed a strong up-regulation while key amino acid biosynthesis pathways were down-regulated (SI Appendix, Fig. S4E). In particular, glutamate metabolism, which plays a key role in the connection between anabolic and catabolic processes (46), exhibited strong differential regulation. Glutamate can be synthesized from α-ketoglutarate generated in the TCA cycle via the fixation of ammonium (NH4+). In a stable symbiosis, glutamate synthesis enables the coral host to utilize photosynthetically fixed and translocated carbon for amino acid and protein synthesis (24). Yet, we observed that, as a result of heat stress, the expression of two key coral host genes in the biosynthesis of glutamate, glutamine synthetase (GS) and glutamate synthase (GOGAT), were down-regulated (Fig. 2 A–D). At the same time, genes encoding glutamate dehydrogenase (GDH) showed either an up-regulation (two genes) or a down-regulation (one gene). GDHs facilitate the anabolic synthesis of glutamate as well as the catabolic degradation of glutamate to fuel the TCA cycle. Interestingly, in contrast to most animals, corals appear to possess distinct isoenzymes of GDH for the anabolic and catabolic reactions (47). The observed changes in expression of GDH genes may thus reflect opposing regulations of anabolic and catabolic reactions, that is, the down-regulation of anabolic and up-regulation of catabolic enzymes. Importantly, the pattern of GDH gene regulation observed in S. pistillata during heat stress was opposite to that observed in the comparison of symbiotic (nitrogen-limited) to symbiont-free (carbon-limited) Aiptasia sea anemones (24), suggesting down-regulation of anabolic glutamate synthesis and up-regulation of catabolic glutamate degradation. These alterations of the coral host glutamate metabolism and amino acid cycling observed on day 10 can thus be a direct consequence of energy limitation of the coral host’s metabolism due to heat stress. Such a starvation-driven shift from synthesis to degradation of amino acids is expected to result in greater production of NH4+ and reduced assimilation of environmental nitrogen by the coral host during heat stress (Fig. 2D). Consequently, starvation of the coral host may directly lead to increased nitrogen availability within the holobiont during heat stress.
Increased Ammonium Availability Alleviates Nitrogen Limitation of Algal Symbionts.
The transcriptomic response of algal symbionts was closely linked to the response of the coral host. A total of 1,979 of the 49,109 genes of Symbiodinium microadriaticum (48) showed significant differential expression (861 up- and 1,118 down-regulated) on day 10 (Dataset S1). Gene set enrichment analysis revealed 82 significantly enriched biological processes (Dataset S1). In particular, nitrogen assimilation pathways were among the most affected processes in the algal symbionts (SI Appendix, Fig. S4F). In contrast to the coral host, algal symbionts possess the cellular machinery for assimilatory nitrate (NO3−) reduction (49). Yet, during heat stress, expression of genes encoding for high-affinity nitrate transporters (NRT), nitrate reductases (NR), and nitrite reductases (NIR) showed consistent down-regulation (Fig. 2 E–G). In combination with the down-regulation of several NH4+transporters, this indicated that algal symbionts invested fewer resources into the uptake of nitrogen from their environment. However, glutamate synthesis via the GS–GOGAT pathway showed no significant down-regulation, suggesting that active NH4+ fixation was maintained. Consequently, the observed alterations in algal nitrogen assimilation pathways point to increased NH4+ availability in the holobiont consistent with the heat stress-induced perturbation of the coral host nitrogen cycle discussed above (Fig. 2D).
Nitrogen limitation is critical to control the algal symbiont population in the coral holobiont (20, 27, 28). Without nitrogen limitation, the algal symbionts proliferate and consume the photosynthetically fixed carbon until no excess carbon is available for translocation to the coral host (23–25). Down-regulation of carbohydrate catabolism and ATP metabolism may reflect the reduced availability of excess carbon released by algal symbiont cells (SI Appendix, Fig. S4F). In contrast to the coral host, however, heat stress did not cause a consistent down-regulation of the TCA cycle in algal symbionts (SI Appendix, Fig. S4F). This suggests that photosynthetic carbon availability was still sufficient to fulfill algal symbiont energy demands and growth. In summary, algal symbiont gene expression during heat stress consistently pointed to increased nitrogen availability that stimulated the growth of algal symbionts and reduced availability of photosynthetic carbon for translocation to the coral host.
Retention of Photosynthates by Algal Symbionts Further Reduces Carbon Availability for the Coral Host.
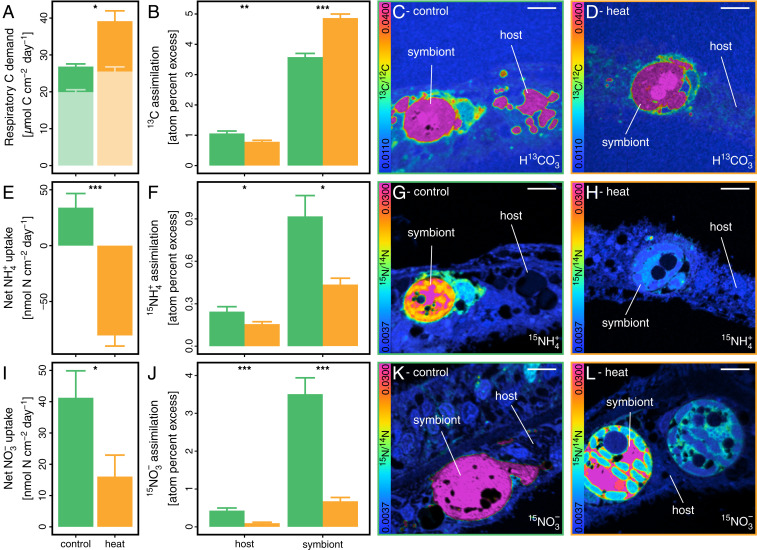
The altered expression of genes associated with key symbiotic nutrient cycling pathways on day 10 of heat stress was corroborated by drastic changes in the uptake and cycling of nutrients in the symbiosis. The respiration rate increased by 46% at the holobiont level, reflecting an increased metabolic turnover and higher energy demand during heat stress (Fig. 3A). The increased organic carbon demand as a consequence of increased respiratory energy metabolism was only partially compensated by a 27% stimulation of gross photosynthesis rates, likely due to increased CO2 availability (50). In light of the changes in coral host gene expression detailed above, this net increase in respiratory carbon consumption could not be sufficiently compensated by heterotrophic feeding, resulting in the consumption of energy reserves. This overall reduction in organic carbon availability further caused a 66% drop in the release of dissolved organic carbon (DOC) from the holobiont (SI Appendix, Fig. S3B).
Fig. 3.
Symbiotic assimilation and fate of carbon and nitrogen on day 10 of heat stress. (A) Respiratory carbon consumption of the holobiont derived from oxygen fluxes. Pale bars indicate carbon demand fulfilled by gross photosynthetic production. (B) Assimilation of 13C-bicarbonate (H13CO3−) into coral host and algal symbiont cells, respectively, based on NanoSIMS imaging (C and D). (E) Net ammonium (NH4+) uptake from seawater by coral fragments. Negative values indicate a net release of NH4+ from the holobiont. (F) Assimilation of 15NH4+ into coral host and algal symbiont cells, respectively, derived from NanoSIMS imaging (G and H). (I) Net nitrate (NO3−) uptake from seawater by coral fragments. (J) Assimilation of 15NO3− into coral host and algal symbiont cells derived from NanoSIMS imaging (K and L). Bars indicate mean ± SE. Asterisks indicated significant differences between treatments. (Scale bars in NanoSIMS images: 5 µm.)
Nanoscale secondary ion mass spectrometry (NanoSIMS) quantitative imaging of H13CO3− assimilation and photosynthate translocation revealed that these alterations in carbon availability were not evenly distributed between symbiotic partners. A 24-h incubation with H13CO3− on day 10 (see Materials and Methods for details) showed that while the 13C enrichment of algal symbiont cells increased by 36% under heat stress, the 13C enrichment of the surrounding coral host tissue decreased by 26% (Fig. 3 B–D). Due to dehydration and resin infiltration, most soluble cellular components (e.g., the cytosol) are lost during conventional sample processing for NanoSIMS analyses. Consequently, the measured enrichment levels reflect the net incorporation of isotopically enriched compounds into structural components of the cell. The increased 13C enrichment of algal symbiont cells during heat stress thus reflects the increased fixation of isotopically labeled carbon via photosynthesis and/or the increased retention of photosynthates for algal biomass synthesis. Likewise, the lower 13C enrichment in the coral host tissue during heat stress indicates reduced incorporation of photosynthates into host biomass. This is likely driven by the combined effects of reduced translocation of photosynthates by the algal symbionts as well as the increased respiratory consumption of organic carbon of the coral host metabolism (notably, a down-regulation of coral genes related to pyruvate carboxylation further supports reduced direct incorporation of bicarbonate by the host during heat stress, potentially having a minor contribution to overall enrichment levels). Consequently, the reduced availability of phototrophic carbon at the holobiont level was only detrimental to the coral host but not to the algal symbionts.
Increased Nitrogen Availability Promotes Growth of Algal Symbionts.
Changes in algal gene expression on day 10 suggest that the observed retention of photosynthates by algal symbionts is the direct consequence of increased nitrogen availability during heat stress. Indeed, heat stress resulted in a shift from a net uptake to a strong net release of NH4+ at the holobiont level at this time point, likely reflecting the increased production of NH4+ by the amino acid catabolism of the coral host (Fig. 3E). This excess availability of NH4+ was directly reflected in the assimilation of NH4+ from the surrounding seawater by both symbiotic partners. NanoSIMS imaging revealed a 53% and 36% reduction in 15NH4+ assimilation in algal symbionts and the surrounding coral host tissue, respectively, during a 24-h labeling incubation on day 10 (see Materials and Methods for details) (Fig. 3 F–H). This is consistent with an increase in the recycling of intrinsically generated unlabeled NH4+. In line with this, the reduced dependency on environmental nitrogen sources was further confirmed by a 61% decline in net nitrate (NO3−) uptake rates at the holobiont level during heat stress (Fig. 3I). NanoSIMS quantification of 15NO3− assimilation during a 24-h incubation on day 10 revealed an 81% and 88% decline in 15N enrichment levels of algal symbionts and coral host tissue, respectively.
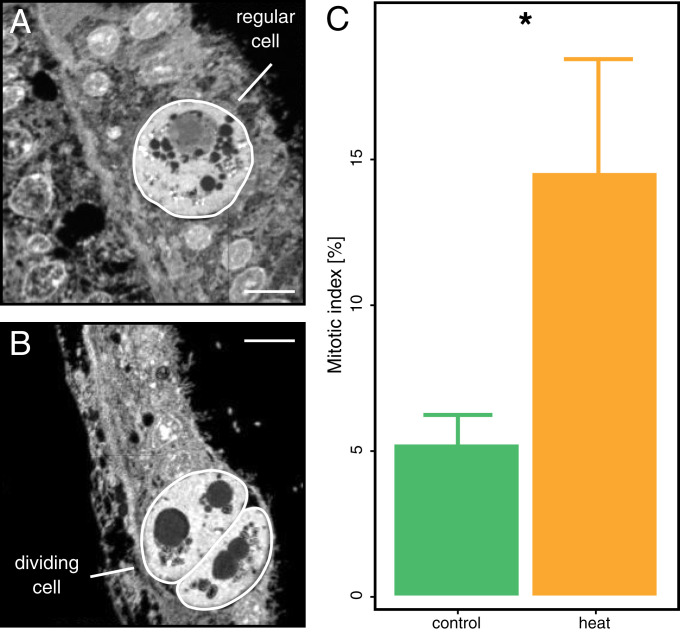
To assess the net effect of altered nitrogen cycling on algal symbionts, we estimated the proportion of dividing cells in hospite from NanoSIMS images (Fig. 4 A and B). The mitotic index of algal symbiont populations showed an approximately threefold increase during heat stress (Fig. 4C). This increase in algal proliferation during heat stress is consistent with an enhanced nitrogen availability for the algal symbionts and explains the increased retention of photosynthates for their growth. While the observation of increased algal cell division during heat stress may appear counterintuitive at first, a similar positive relation has been documented previously (51). In light of the stable algal symbiont density at day 10, this observation, therefore, suggests that the symbiont population must have temporarily increased between day 10 and the onset of visual bleaching, or that symbiont growth was balanced by continuous expulsion of excess symbionts even before the onset of visual bleaching.
Fig. 4.
Proportion of dividing algal symbiont cells on day 10 of heat stress. NanoSIMS images for 12C14N− were used to quantify the abundance of (A) regular and (B) dividing algal symbiont cells in the coral tissue sections. (C) These data were used to calculate the proportion of dividing cells in the algal symbiont population. Note that this mitotic index likely reflects an underestimation of the true proportion of dividing cells due to the two-dimensional nature of NanoSIMS images. Bars indicate mean ± SE. Asterisks indicated significant differences between treatments. (Scale bars in NanoSIMS images, 5 µm.)
Dynamic Nutritional States in the Coral–Algal Symbiosis.
The perturbed nutrient cycling observed in the present study paints a clear picture of the metabolic state of the S. pistillata–Symbiodiniaceae symbiosis on day 10 of heat stress, that is, prior to the onset of (visual) bleaching. At the same time, our results allow for the identification of putative feedback loops directly affecting the functioning of this symbiosis over time as detailed in the following.
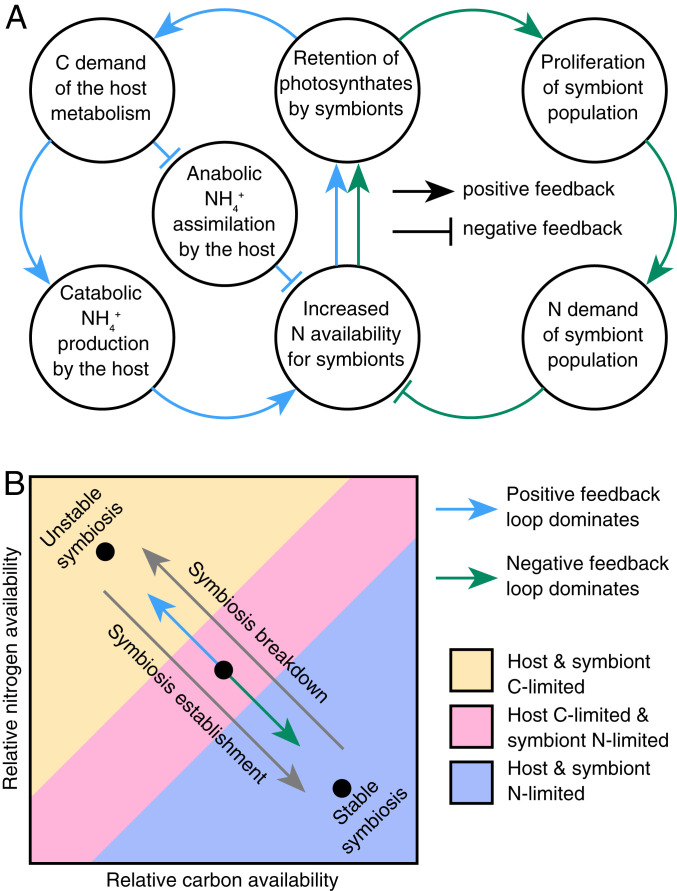
A positive (self-amplifying) feedback loop destabilizes nutrient cycling in the symbiosis during heat stress. First, heat stress reduces energy availability for the host. Second, the resulting shift in the host metabolism from amino acid anabolism to catabolism increases nitrogen availability for the algal symbionts, which begin to grow and divide faster. Third, this, in turn, reduces carbon translocation to the coral host, exacerbating its state of starvation (Fig. 5A). While elevated temperatures may initially only mildly affect energy availability in the coral host, the positive feedback of these processes will gradually lead to more pronounced alterations in the nutrient cycling status between the coral host and algal symbiont during prolonged heat stress. Over time, these processes eventually lead to a collapse of carbon translocation by the symbiont, which undermines the ecological advantage of this symbiosis for the host.
Fig. 5.
Model outlining the dynamic transition between a stable and unstable state of the coral–algal symbiosis based on metabolic interactions between the coral host and its algal symbionts. (A) This model posits that the nutritional status of host and symbionts is passively controlled by a positive and a negative feedback loop arising from the nutrient exchange between the host and its algal symbionts. (B) In a stable state, carbon (C) translocation by algal symbionts fulfils or exceeds the metabolic C demands of the coral host, thereby arresting the symbiosis in a nitrogen (N)-limited state (purple shaded area). Environmental stressors, such as rapid warming, may cause a proportional increase of host C requirements, resulting in a feedback loop imbalance in favor of the positive feedback loop that destabilizes C cycling in the symbiosis. This imbalance will first affect the coral host metabolism resulting in C limitation, while algal symbionts remain N-limited (pink shaded area). If conditions persist, eventually the entire symbiosis shifts towards a C-limited stage (yellow shaded area). Likewise, the rapid proliferation of algal symbionts during the (re)establishment of the symbiosis will accordingly result in a feedback loop imbalance in favor of the negative feedback loop that effectively increases competition for available N between algal symbionts. Thereby, the symbiosis gradually shifts towards an N-limited state in which C translocation and recycling are maximized.
These processes can be potentially compensated by a negative (self-attenuating) feedback loop that may stabilize symbiotic nutrient cycling during elevated temperatures: 1) The alteration of the coral host metabolism increases nitrogen availability for algal symbionts during heat stress; 2) the increased nutrient availability stimulates algal cell division rates and population growth; and 3) as more algal cells compete for the available nitrogen, population growth effectively limits nitrogen availability for individual algal cells and eventually inhibits their growth (23, 24, 52–54). Translocation of algal photosynthates to the coral host could thus be maintained at a level that prevents severe starvation of the coral host and an increase in nitrogen availability during heat stress (Fig. 5A). By maintaining the nitrogen-limited state of algal symbionts, this feedback loop may stabilize photosynthate translocation from the algae to the host.
Our findings suggest that the state of nutrient cycling is dependent on the dynamic equilibrium between the outlined positive and negative feedback loops (Fig. 5B). In a stable state, the coral host and algal symbionts compete for available NH4+, thereby arresting the symbiosis in a state in which carbon recycling is maximized. Moderate rates of warming increase the metabolic energy demand of the coral host and NH4+ availability, but this may be balanced by stimulated algal symbiont growth. In contrast, rapid or severe warming results in pronounced coral host starvation and high levels of available NH4+ that cannot be compensated for by algal symbiont proliferation (as outlined above). Under such conditions, carbon translocation from the algal symbionts will gradually decrease, ultimately shifting both symbiotic partners towards a carbon-limited state. Interestingly, the same processes that destabilize nutrient exchange during heat stress may, in reverse order, lead to the onset of nutrient exchange during the (re)infection of the coral host with algal symbionts. Symbiont-free cnidarians are in a carbon-limited state (24, 53, 55). As such, initial coral-colonizing symbionts can rapidly proliferate due to high nitrogen availability. As algal symbiont densities rise, increasing competition for available nitrogen within the symbiont population will effectively limit nitrogen uptake by individual algal cells (23, 52). Being nitrogen-limited in their growth, algal symbionts will start releasing excess organic carbon to the coral host, thereby gradually shifting the system to a nitrogen-limited state that is characteristic of a stable symbiosis (Fig. 5B).
Taken together, the onset, maintenance, and breakdown of the cnidarian–Symbiodiniaceae symbiosis directly relates to the underlying symbiotic nutrient cycling and can be explained as a consequence of metabolic interactions arising from resource competition between the cnidarian host and its algal symbiont population. As the ability to assimilate NH4+ appears to be a ubiquitous feature of symbiotic cnidarians and Symbiodiniaceae, the findings of this study are putatively relevant to most, if not all, cnidarian–Symbiodiniaceae symbioses, as well as other symbioses with a similar potential for symbiotic resource competition (24, 53).
Symbiotic Nutrient Cycling Explains Patterns of Bleaching on Coral Reefs.
While the instability of symbiotic nutrient cycling appears to be an inherent feature of cnidarian–Symbiodiniaceae symbioses, heat stress responses can be different between coral species and even individual colonies (8, 36). The conceptual framework developed here can directly advance our understanding of the complex processes underlying this heterogeneity of bleaching susceptibility in light of the underlying symbiotic nutrient cycling.
The bleaching susceptibility of a coral colony depends on the identity of the symbiotic partners (10, 56). Our findings suggest that algal symbiont population growth could compensate for increased nitrogen availability and stabilize symbiotic nutrient cycling during heat stress. Indeed, previous studies reported an increase of symbiont densities during moderate seasonal warming (57). Further, corals hosting fast-growing algal symbionts of the genus Durusdinium maintained higher rates of photosynthate translocation during heat stress and showed a reduced bleaching susceptibility (57, 58). Hence, fast symbiont population growth appears to support host nutrition and reduce bleaching susceptibility of corals. In addition, heterotrophic feeding provides another significant energy source for the host’s metabolism (59). As the majority of heterotrophically acquired nitrogen appears to be incorporated into host biomass (60, 61), heterotrophic feeding could allow coral hosts to avoid starvation and maintain a state of nitrogen limitation for their algal symbionts during heat stress. Indeed, previous studies have shown that corals that are able to increase their feeding rates can stabilize symbiotic nutrient cycling during heat stress (62–64). Furthermore, corals with high heterotrophic capacity appear to be less susceptible to bleaching and have higher resilience to heat stress (29, 65, 66). Hence, the conceptual model developed here can help explain how functional traits of algal symbionts and their coral hosts directly affect the functioning of the symbiosis during heat stress.
Besides coral host and algal symbiont traits, the response of coral colonies to heat stress is shaped by local environmental conditions (8, 11). In this context, previous studies reported that environmental nitrogen availability can affect the bleaching susceptibility of corals (31, 67). While moderate increases in seawater NH4+ concentrations do not impair the symbiosis during heat stress (30, 68), increased NO3− concentrations reduce translocation of photosynthates by algal symbionts and enhance bleaching susceptibility of corals (25, 69). In contrast to NH4+, in hospite NO3− availability cannot be regulated via symbiotic resource competition as the host lacks the cellular machinery for nitrate reduction (70). The enhanced NO3− uptake by algal symbionts in the present study may thus be a consequence of host regulation. While this efficient NO3− assimilation by algal symbionts may support holobiont nutrition under oligotrophic conditions, elevated NO3− levels may promote the selfish retention of photosynthates by algal symbionts as outlined above. Coupled with the effects of heat stress, NO3− may thus further destabilize symbiotic nutrient cycling in corals. Likewise, previous studies showed that symbiotic cnidarians from areas with strong diurnal temperature variations are more thermotolerant than their counterparts in areas of low temperature variability (41, 71, 72). Here we found that heat stress stimulates both respiration as well as photosynthesis rates. Consequently, the observed depletion of host energy reserves during heat stress is likely more pronounced at night in the absence of photosynthesis. As diurnal temperature variability commonly involves a cooling at night and warming during the day (72), this implies that strong diurnal temperature variations will reduce the effects of heat stress on the energy budget of the host compared to areas with similar mean temperature but lower diurnal variability. Hence, high diurnal temperature variations may stabilize symbiotic nutrient cycling during heat stress.
Taken together, our model helps to explain how differences in algal symbiont traits, coral host heterotrophic capacity, and local environmental conditions affect the stability of symbiotic nutrient cycling during heat stress. Furthermore, these observations suggest that the stability of symbiotic nutrient cycling is directly linked to the stability of the coral–Symbiodiniaceae symbiosis itself.
What Triggers Coral Bleaching?
Importantly, at this point the cellular triggers of algal symbiont expulsion remain unknown. Previous studies proposed that ROS levels and oxidative damage in the coral host tissue may be an important cue for coral bleaching (15, 16, 73). The level of oxidative damage depends on the interplay of ROS production, antioxidant capacity, and cellular repair processes (13). Therefore, alterations in cellular energy availability, as observed here, will directly affect the ability of the host to cope with oxidative stress and thereby indirectly determine the thermal threshold of bleaching in corals. In addition, Hill and Hill (74) proposed that the arrest of maturation of the phagosome surrounding the endosymbiont depends on its release of photosynthates to mimic the digestion of prey. In this light, altered symbiotic nutrient cycling, that is, the cessation of photosynthate transfer, may be the direct cause for symbiont expulsion. Our findings suggest that heat stress gradually reduces carbon translocation by the algal symbionts, ultimately shifting the symbiotic system to a carbon-limited state. Eventually, carbon release by the algal symbiont may be insufficient to maintain phagosome arrest. As the phagosome matures, the symbiont would either be digested or expelled from the coral host cell (75). While this sanctioning of less beneficial symbionts by the host may be critical to maintaining the functioning of the symbiosis in a stable state (76), the same processes underlying this control could explain the collapse of the symbiont population during prolonged heat stress.
Importantly, the loss of algal symbionts is not the direct cause of coral mortality during bleaching (77). Rather, mortality is caused by the starvation of the coral host that follows a decrease in phototrophic carbon input by the algae (77). Here, we have shown that heat stress destabilizes symbiotic carbon translocation before the actual loss of algal symbionts, that is, prior to any visible signs of bleaching. Consequently, corals can experience severe carbon limitation even before algal symbiont abundance is reduced. Such energy starvation of the host has previously been described in corals following bleaching unless compensated by enhanced heterotrophy (65, 78, 79). As such, the observed mass mortality of corals during global heat anomalies may not be the direct consequence of the loss of algal symbionts per se but more fundamentally reflect the starvation of the coral host due to the collapse of symbiotic nutrient cycling itself.
Conclusion
Our efforts to preserve coral reefs in the Anthropocene are hampered by an incomplete understanding of the processes underpinning susceptibility and resilience of corals to heat stress and bleaching. Our study demonstrates that heat stress destabilizes symbiotic nutrient cycling well before the actual breakdown of the coral–algal symbiosis. As the functioning of symbiotic nutrient exchange appears to either directly or indirectly contribute to the breakdown of the coral-Symbiodiniaceae symbiosis, the thermotolerance of corals may be a reflection of the stability of the underlying nutrient cycling. The nutritional status of the host may thus be a better proxy of the resilience of corals to heat stress than symbiont-centered metrics or the thermal bleaching threshold itself (although any of these measures represent interconnected holobiont responses to varying degrees). Taken together, our findings suggest that the functional onset, maintenance, and breakdown of the coral-Symbiodiniaceae symbiosis can be understood in light of the underlying symbiotic nutrient cycling. Further, the passive and self-sustaining regulation of the symbiosis arising from resource competition between the coral host and its algal symbionts implies that the findings of this study may be directly transferable to other (photo)symbiotic systems. As such, the negative consequences of global warming on symbiotic interactions are unlikely to be restricted to coral holobionts but may pose a threat to marine and terrestrial symbioses at large.
Materials and Methods
Sampling Site, Coral Collection, and Experimental Design.
Corals were collected from Abu Shosha reef (22°18ʹ16.3ʺN; 39°02ʹ57.7ʺE) close to the Saudi Arabian central Red Sea coast (SI Appendix, Fig. S1). Temperatures were monitored at a water depth of ∼5 m throughout the year 2017 with HOBO pendant temperature loggers (Onset) to identify annual mean (29.1 °C) and absolute maximum temperatures (32.9 °C) representative for a nonbleaching year in the region (80). In September 2018, five colonies of S. pistillata (⌀ >30 cm) were collected at the same location and depth with a minimal distance of 10 m between colonies and immediately transported to the indoor aquaria facility at the Coastal and Marine Resources core laboratory at King Abdullah University of Science and Technology (KAUST, 22°18′20.5″N; 39°06′14.3″E). Colonies were fragmented into 20 similar-sized nubbins (mean surface area = 21.1 cm2) each, which were distributed over two 150-L aquaria per colony (one tank assigned for each treatment in the experiment; a total of 10 aquaria). Each tank was equipped with a temperature controller (D-D The Aquarium Solution Ltd), a 600-W heater (Schego), current pump (Tunze), and a Radion light system (Ecotech Marine Inc.). All tanks were filled with freshly collected seawater from Abu Shosha reef (salinity = 40.1 ± 0.2; NH4+ = 0.48 µM ± 0.03; NO3− = 0.19 ± 0.05 µM; PO43- = 0.03 ± 0.00 µM) with a daily water renewal rate of 25% and maintained at a 12-h:12-h light/dark regime resembling in situ conditions (mean daytime irradiation = 380 µmol quanta⋅m−2⋅s−1, peak daytime irradiation = 750 µmol quanta⋅m−2⋅s−1) and constant temperature of 29.1 °C. Daily replenishment with reef water was chosen to mimic in situ reef conditions and support heterotrophic feeding from naturally occurring plankton sources. Notably, no supplemental feeding was provided (59, 64). However, given the low density of coral fragments in the aquaria, the level of water exchange was sufficient to ensure stable water parameters (salinity = 40.2 ± 0.1; NH4+ = 0.41 ± 0.09 µM; NO3− = 0.21 ± 0.09 µM; PO43- = 0.05 ± 0.01 µM) and to maintain coral pigmentation levels that resembled their natural reef site counterparts.
After 7 d of acclimation, the five aquaria assigned to the heat stress treatment were gradually ramped to a final constant temperature of 32.9 °C (absolute maximum temperature of 2017 and 0.8 °C above the maximum monthly mean temperature of 2017) over the course of 3 d, matching observed maximum daily warming rates from 2017 (80). Concomitantly, the remaining five aquaria were maintained at a temperature of 29.1 °C (mean temperature of 2017) as control conditions (SI Appendix, Fig. S1). Corals were sampled after 10 d (i.e., 7 d at maximum temperature) and after 21 d (when visual bleaching was observed). In this context, we defined bleaching as the visible loss of pigmentation resulting in the pale appearance of coral holobionts according to Douglas (43). On day 10, a range of response parameters was assessed to identify primary physiological, molecular, and metabolic responses of the coral–algal symbiosis to heat stress (see below for details). In contrast, on day 21 only symbiont density and chlorophyll a content was recorded to quantify the extent of bleaching. For all response parameters, one nubbin per colony and treatment was used (i.e., n = 5 for each treatment; SI Appendix, Fig. S1). All incubation measurements were performed on the day of sampling. For all other measurements, fragments were snap-frozen in liquid N2 and stored at −80 °C for later processing.
Algal Symbiont Density and Chlorophyll a Content.
Frozen coral fragments were placed into individual Ziploc bags and doused in 7.5 mL of ice-cold phosphate-buffered saline (PBS) buffer (1×). All tissue was removed from the coral skeleton using air pressure and the resulting tissue slurry was transferred into a Falcon tube and homogenized for 30 s with a T-18 UltraTurrax (IKA) on ice. Symbiont cells were washed in three cycles of centrifugation (3,000 × g) and resuspension in PBS. For symbiont density, three technical replicates of 200 µL were transferred into a 96-well plate through a cell strainer (40-µm mesh size). Symbiont concentrations were quantified by flow cytometry using the BD LSRFortessa (BD Biosciences) with an excitation wavelength of 488 nm and fluorescence emission detection at 695/40 nm. Subsequent gating of recorded events was done in FlowJo v.10.5.3 based on forward scatter and chlorophyll autofluorescence. For chlorophyll content analysis, 1 mL of washed symbiont cell suspension was transferred into an Eppendorf tube, pelleted, and resuspended in acetone (100%). Samples were incubated in the dark at 4 °C for 24 h before three technical replicates of 200 µL were transferred into a 96-well plate. The absorption of samples was immediately recorded at 630, 664, and 750 nm using a SpectraMax Paradigm microplate reader (Molecular Devices). Chlorophyll a content was calculated following Jeffrey and Humphrey (81):
Algal symbiont cell and chlorophyll a concentrations were corrected for sample volume and normalized to the surface area of the coral fragment.
Oxidative Stress.
Symbiotic ROS production was quantified from freshly isolated symbiont cells (82). For this, the tissue was removed from freshly sampled corals and homogenized as described above using sterile seawater. Washed symbiont cells were split into three technical replicates of 1 mL each in Eppendorf tubes and incubated for 30 min in the light (daily mean irradiance; 380 µmol photons⋅m−2⋅s−1) according to treatment temperature conditions. CellROX orange (Life Technologies) was added to the tubes at a final concentration of 5 µM, and cells were removed by centrifugation. Two-hundred-microliter aliquots of the supernatant were immediately transferred into 96-well plates and incubated in the dark at 37 °C for 30 min. Relative ROS release (CellROX fluorescence) was quantified using a SpectraMax Paradigm microplate reader (Molecular Devices) at 545-nm excitation and 565-nm emission wavelength and normalized to the coral surface area (discussed below).
Lipid peroxidation (quantified as total malondialdehyde content) was used as a proxy of oxidative stress in the host tissue. Coral tissue was removed from frozen coral fragments and homogenized as outlined above using ice-cold PBS buffer (1×). Symbiont cells were removed by centrifugation and three 100-µL aliquots of supernatant for each biological replicate were transferred into new Eppendorf tubes. The concentration of malondialdehyde in the samples was measured colorimetrically using the Lipid Peroxidation (MDA) Assay Kit (Abcam) according to the manufacturer’s instructions. Malondialdehyde content was corrected for sample volume and normalized to the coral surface area.
Photosynthetic Performance and Respiration.
The maximum photosynthetic efficiency of coral holobionts was assessed via pulse-amplitude modulation (PAM) fluorometry. For this, coral fragments were dark-adapted for 30 min and the ratio between variable and maximum chlorophyll fluorescence (Fv/Fm) was recorded using the red version of the MINI-PAM-II (Walz) with a 5.5-mm fiber optic.
Oxygen (O2) production and consumption measurements were used to quantify net photosynthesis and respiration rates during light and dark incubations, respectively. For this purpose, individual coral fragments were transferred into 320-mL Nalgene incubation chambers filled with sterile seawater. Chambers were submerged in a water bath to maintain a constant temperature according to treatment conditions and stirred using magnetic stirrers. Specimens were left to acclimate for 30 min in the dark. Subsequently, O2 concentrations were recorded every second for ∼1 h at a constant light level of 380 µmol photons⋅m−2⋅s−1 (mean irradiation during 12-h light phase) followed by ∼1 h in the dark using FireSting O2 optical oxygen meters (PyroScience).
Local linear regressions were performed using the LoLinR R package v0.0.9 to objectively identify the best-fitting linear regression to calculate O2 fluxes during light and dark incubations, respectively (83). O2 fluxes were corrected for seawater controls, normalized to the coral surface area (discussed below), and converted into their carbon equivalents using photosynthetic and respiration quotients of 1.1. and 0.9, respectively (84). Gross photosynthesis rates were calculated as the sum of net photosynthesis (derived from light incubations) and respiration (derived from dark incubations) rates. The daily net photosynthetic carbon budget was calculated as the difference between daily fixation of carbon via gross photosynthesis (12 h) and the loss of fixed carbon via respiration (24 h).
Nutrient Uptake/Release.
To assess net nutrient uptake and release rates, corals were incubated in 1-L incubation jars filled with 750 mL of artificial seawater for 24 h. Artificial seawater composition was adapted from Harrison et al. (85) to mimic Red Sea conditions (salinity = 39, pH = 8.1, 492.5 mM NaCl, 46.23 mM MgCl2, 10.8 mM Na2SO4, 9.0 mM CaCl2, 7.9 mM KCl, 2.5 mM NaHCO3). Inorganic nutrients were added at elevated concentrations (5 µM NH4+, 5 µM NO3−, 2 µM PO43−) compared to oligotrophic Red Sea water to allow for accurate measurements of nutrient depletion. Incubation water in jars was stirred using magnetic stirrers and maintained in a water bath mimicking aquaria conditions (light levels and temperature) according to treatment. Changes in nutrient concentrations during incubations were assessed by comparing start and end-point measurement, corrected for seawater controls and normalized to the surface area of the nubbins (discussed below). For analysis of inorganic nutrient concentrations, 50 mL of seawater were collected with a syringe, filtered (precombusted GF/F, 0.45 µm), transferred into a Falcon tube, and immediately flash-frozen for later analysis. For analysis of DOC concentrations, 10 mL of seawater was collected with a syringe, filtered (PSF, 0.22 µm), transferred into precombusted glass vials, acidified with concentrated phosphoric acid (pH <2), and immediately sealed gas-tight for later analysis. Seawater samples were analyzed in triplicate using a SA3000/5000 nutrient auto-analyzer (Skalar) and a TOC-L analyzer (Shimadzu) for inorganic and organic nutrient concentrations, respectively.
Isotope Labeling and Sample Preparation.
Corals were labeled with 13C and 15N to allow visualization and quantification of inorganic nutrient uptake and translocation in the coral–algae symbiosis using NanoSIMS imaging. Isotope labeling incubations were performed identically to nutrient uptake/release incubations explained above except that either HCO3− and NH4+ or NO3− were replaced with 13C- or 15N-enriched (∼99.8%) counterparts during artificial seawater preparation. After 24 h of incubation, small pieces (∼5 mm length) were clipped off the tip of coral fragments and immediately transferred into a fixative solution (1.25% glutaraldehyde and 0.5% paraformaldehyde in 0.1 M phosphate buffer). After 24 h of fixation at 4 °C, the samples were washed in PBS (1×) and decalcified using 0.1 M ethylenediaminetetraacetic acid (4 °C, exchanged daily for 14 d). The samples were dissected into small pieces containing a row of individual polyps. The tissues were dehydrated in a series of increasing ethanol concentrations (50, 70, 90, and 100%) followed by 100% acetone. The tissues were then gradually infiltrated with SPURR resin of increasing concentrations (25, 50, 75, and 100%). Subsequently, tissues were embedded in SPURR resin and cut into 170-nm sections using an Ultracut E microtome (Leica Microsystems) and mounted on silicon wafers for NanoSIMS imaging.
NanoSIMS Imaging and Analysis.
Silicon wafers with attached sample sections were gold-coated and imaged with the NanoSIMS 50 ion probe (58) at the Center for Microscopy, Characterisation and Analysis at the University of Western Australia. Surfaces of samples were bombarded with a 16-keV primary Cs+ beam focused to a spot size of about 100 nm with a current of ca. 2 pA. Secondary molecular ions 12C12C−, 12C13C−, 12C14N−, and 12C15N− were simultaneously collected in electron multipliers at a mass resolution (M/∆M) of about 8,000. Charge compensation was not necessary. At least 15 images of different areas within the gastrodermis of the polyp tissue (30-µm raster with 256 × 256 pixels) were recorded for each coral nubbin for all targeted secondary molecular ions by rastering the primary beam across the sample with a dwell time of 9 ms per pixel; six planes were recorded for each area. Image processing was performed using the ImageJ plugin OpenMIMS (National Resource for Imaging Mass Spectrometry, https://github.com/BWHCNI/OpenMIMS/wiki). After drift correction, the individual planes were summed and the 13C/12C or 15N/14N maps were expressed as a hue-saturation-intensity image, where the color scale represents the isotope ratio. Assimilation of the isotope labels (atom % enrichment compared to unlabeled controls) was quantified for 25 (for 15NH4+-labeled samples) and 15 (for 15NO3−-labeled samples; 5 in the case of one colony) symbiont cells per coral fragment by circling individual regions of interest (ROIs) based on the 12C14N− silhouette of the symbiont cells. To minimize the potential bias of varying symbiont densities (23), enrichment of the host tissue was measured only in the direct vicinity of each symbiont cell. For this, round ROIs were drawn with a radius of ca. 7 µm around the centroid of the symbiont cell and symbiont and symbiosome content was removed, resulting in a doughnut-shaped ROI for the host tissue. To estimate the proportion of dividing or freshly divided symbiont cells, the mitotic index of the symbiont population was derived from 12C14N− images. Specifically, the occurrence of symbiont cells in a dividing stage was counted in relation to the total occurrence of symbiont cells in the section of each coral colony (Fig. 3). Notably, the two-dimensional nature of NanoSIMS images may lead to an underestimation of the true mitotic index as not all dividing cells can be detected. However, this underestimation should be identical for all samples and thus not affect relative differences in the mitotic index between treatments.
RNA Sequencing.
Frozen coral fragments were placed into a Ziploc bag on ice and covered in 1.5 mL of ice-cold RLT tissue lysis buffer. Tissue was removed from the coral skeleton using air pressure for 1 min and the resulting tissue slurry in RLT buffer was immediately transferred into an Eppendorf tube and snap-frozen in liquid nitrogen. Total RNA was extracted from 500 µL defrosted coral slurry with the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. The concentration of total RNA was determined using Qubit BR RNA Assay Kit (Thermo Scientific), and its integrity was evaluated using an Agilent 2100 Bioanalyzer and Nano Agilent RNA 6000 kit (Agilent Technologies), according to the manufacturer’s instructions. PolyA+ selection and subsequent messenger RNA (mRNA) library preparation were done using the TruSeq Stranded mRNA Library Prep kit (Illumina), according to the manufacturer’s instructions. Resulting libraries (average fragments of 390 bp) were sequenced using the Illumina HiSeq4000 platform at the Bioscience Core laboratory facilities at KAUST to obtain paired-end reads of 150 bp.
Transcriptome Analysis.
Sequencing yielded an average of ∼70 million sequence reads per sample. Sequence reads were quality-trimmed and Illumina adapters were removed using Trimmomatic v.0.39 (86). The successful removal of adapters from paired reads was confirmed using FastQC v.0.11.5 (87). To separate host and symbiont reads sequence reads, processed sequence reads were split based on their best match to the gene models of S. pistillata (45) and S. microadriaticum [the dominant algal symbiont of S. pistillata on shallow Red Sea reefs (48, 88, 89)] using BBsplit and subsequently mapped to the respective gene models using BBmap (BBtools v.37.10) (90). Gene expression was quantified with Salmon v.1.0.0 (91) using the alignment-based mode. This produced on average ∼30 million and ∼8 million mapped read pairs per sample for the coral host and algal symbiont, respectively. Effective counts were used for identifying significantly differentially expressed genes (P < 0.05) between control and heat stress treatments using DeSeq2 v.1.26.0 (92). Differentially expressed genes were used to perform gene set enrichment analysis with topGO v.2.38.1 using the weight01 algorithm and no multiple test correction (93). Pathways of interest were further investigated by mapping differentially expressed genes to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways using KEGG mapper v.3.2 (94). Genes were assigned to KOGs categories using EggNOG 4.5.1 (95). Variance stabilizing transformation was applied to count data for principal component analysis and visualization of similarity between transcriptome-wide expression profiles as implemented in DeSeq2.
Coral Surface Area.
All rate measurements (O2, ROS, and nutrient fluxes) were normalized to coral fragment surface area calculated by three-dimensional (3D) computer modeling (96). For this, ∼40 photos were taken from coral fragments from different angles in front of a white background. The photos were uploaded to the Autodesk Photo-to-3D cloud service (Autodesk) to generate 3D models of each fragment. The surface of 3D models was analyzed using ReCap Photo version 4.2.0.2 (Autodesk).
Statistical Analysis.
All statistical analyses were performed in R v.3.6.2 (97). Differences in physiological responses across treatments were analyzed in a paired design based on colony replicates. Normally distributed data (Shapiro–Wilk test P < 0.05) were tested with a paired two-sided t test. Otherwise, data were analyzed using a Wilcoxon signed-rank test. The relationship between symbiotic ROS production and host lipid peroxidation was assessed via a correlation analysis using Pearson's product moment correlation coefficient. NanoSIMS measurements of relative 13C and 15N abundance in the host and symbiont tissue/cells were analyzed using linear mixed models with treatment as a fixed and colony as a random effect using the lme4 R package v.1.1-21 (98). NanoSIMS data were log-transformed prior to analysis to account for skewing of data. Differences in expression profiles were analyzed with permutational multivariate analysis of variance with treatment and colony as explanatory variables as implemented in the vegan R package v.2.5-6 (99).
Supplementary Material
Acknowledgments
We thank Dr. Zenon B. Batang and Dr. Nabeel M. Alikunhi for their continuous support and assistance with aquaria maintenance. Further, Ioannis Georgakakis, Mustafa Altunkaya, Gabriela Perna, and Prof. Matt Kilburn are acknowledged for their help and support with sample processing and data analysis. N.R., C.P., A.C., M.P., J.-B.R., and C.R.V. were supported by the KAUST competitive research grant URF/1/3400-01-01. C.R.V. also acknowledges funding from KAUST and the German Research Foundation (DFG), grant 433042944. A.M. is supported by Swiss National Science Foundation, grant 200021_179092.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022653118/-/DCSupplemental.
Data Availability.
Detailed results of gene expression analyses are provided as Dataset S1. All physiological and NanoSIMS raw data are deposited in the Zenodo data repository (https://zenodo.org/record/4429583#.X_xfoS1Q30o) (100). Additionally, raw sequencing data are deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA638625.
References
- 1.Knowlton N., Jackson J. B. C., Shifting baselines, local impacts, and global change on coral reefs. PLoS Biol. 6, e54 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoegh-Guldberg O., et al. , Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Hughes T. P., et al. , Global warming and recurrent mass bleaching of corals. Nature 543, 373–377 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hughes T. P., et al. , Coral reefs in the Anthropocene. Nature 546, 82–90 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Rohwer F., Seguritan V., Azam F., Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10 (2002). [Google Scholar]
- 6.Bellworthy J., Fine M., Beyond peak summer temperatures, branching corals in the Gulf of Aqaba are resilient to thermal stress but sensitive to high light. Coral Reefs 36, 1071–1082 (2017). [Google Scholar]
- 7.Pogoreutz C., et al. , Sugar enrichment provides evidence for a role of nitrogen fixation in coral bleaching. Glob. Change Biol. 23, 3838–3848 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Suggett D. J., Smith D. J., Coral bleaching patterns are the outcome of complex biological and environmental networking. Glob. Change Biol. 26, 68–79 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg E., Kushmaro A., Kramarsky-Winter E., Banin E., Yossi L., The role of microorganisms in coral bleaching. ISME J. 3, 139–146 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Baird A. H., Bhagooli R., Ralph P. J., Takahashi S., Coral bleaching: The role of the host. Trends Ecol. Evol. 24, 16–20 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Ziegler M., Seneca F. O., Yum L. K., Palumbi S. R., Voolstra C. R., Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 8, 14213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Oppen M. J. H., Oliver J. K., Putnam H. M., Gates R. D., Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weis V. M., Cellular mechanisms of Cnidarian bleaching: Stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Smith D. J., Suggett D. J., Baker N. R., Is photoinhibition of zooxanthellae photosynthesis the primary cause of thermal bleaching in corals? Glob. Change Biol. 11, 1–11 (2005). [Google Scholar]
- 15.Lesser M. P., Oxidative stress causes coral bleaching during exposure to elevated temperatures. Coral Reefs 16, 187–192 (1997). [Google Scholar]
- 16.Lesser M. P., Elevated temperatures and ultraviolet radiation cause oxidative stress and inhibit photosynthesis in ymbiotic dinoflagellates. Limnol. Oceanogr. 41, 271–283 (1996). [Google Scholar]
- 17.Tolleter D., et al. , Coral bleaching independent of photosynthetic activity. Curr. Biol. 23, 1782–1786 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Krueger T., et al. , Differential coral bleaching—Contrasting the activity and response of enzymatic antioxidants in symbiotic partners under thermal stress. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 190, 15–25 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Diaz J. M., et al. , Species-specific control of external superoxide levels by the coral holobiont during a natural bleaching event. Nat. Commun. 7, 13801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muscatine L., Porter J. W., Reef corals: Mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460 (1977). [Google Scholar]
- 21.Falkowski P. G., Dubinsky Z., Muscatine L., Porter J. W., Light and the bioenergetics of a symbiotic coral. Bioscience 34, 705–709 (1984). [Google Scholar]
- 22.Lesser M. P., Using energetic budgets to assess the effects of environmental stress on corals: Are we measuring the right things? Coral Reefs 32, 25–33 (2013). [Google Scholar]
- 23.Krueger T., et al. , Intracellular competition for nitrogen controls dinoflagellate population density in corals. Proc. Biol. Sci. 287, 20200049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui G., et al. , Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PLoS Genet. 15, e1008189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ezzat L., Maguer J.-F., Grover R., Ferrier-Pagès C., New insights into carbon acquisition and exchanges within the coral-dinoflagellate symbiosis under NH4+ and NO3- supply. Proc. Biol. Sci. 282, 20150610 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kopp C., et al. , Subcellular investigation of photosynthesis-driven carbon assimilation in the symbiotic reef coral Pocillopora damicornis. mBio 6, e02299-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rädecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J., Wild C., Nitrogen cycling in corals: The key to understanding holobiont functioning? Trends Microbiol. 23, 490–497 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Morris L. A., Voolstra C. R., Quigley K. M., Bourne D. G., Bay L. K., Nutrient availability and metabolism affect the stability of coral–Symbiodiniaceae symbioses. Trends Microbiol. 27, 678–689 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Conti-Jerpe I. E., et al. , Trophic strategy and bleaching resistance in reef-building corals. Sci. Adv. 6, eaaz5443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Béraud E., Gevaert F., Rottier C., Ferrier-Pagès C., The response of the scleractinian coral Turbinaria reniformis to thermal stress depends on the nitrogen status of the coral holobiont. J. Exp. Biol. 216, 2665–2674 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Wiedenmann J., et al. , Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 3, 160–164 (2012). [Google Scholar]
- 32.Ezzat L., Maguer J.-F., Grover R., Ferrier-Pagès C., Limited phosphorus availability is the Achilles heel of tropical reef corals in a warming ocean. Sci. Rep. 6, 31768 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeCarlo T. M., et al. , Nutrient-supplying ocean currents modulate coral bleaching susceptibility. Sci. Adv. 6, eabc5493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker D. M., Freeman C. J., Wong J. C. Y., Fogel M. L., Knowlton N., Climate change promotes parasitism in a coral symbiosis. ISME J. 12, 921–930 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbin E. M., et al. , Short-term thermal acclimation modifies the metabolic condition of the coral holobiont. Front. Mar. Sci. 5, 10 (2018). [Google Scholar]
- 36.Hoadley K. D., et al. , Physiological response to elevated temperature and pCO2 varies across four Pacific coral species: Understanding the unique host+symbiont response. Sci. Rep. 5, 18371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fine M., Gildor H., Genin A., A coral reef refuge in the Red Sea. Glob. Change Biol. 19, 3640–3647 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Osman E. O., et al. , Thermal refugia against coral bleaching throughout the northern Red Sea. Glob. Change Biol. 24, e474–e484 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Krueger T., et al. , Common reef-building coral in the Northern Red Sea resistant to elevated temperature and acidification. R. Soc. Open Sci. 4, 170038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grottoli A. G., Tchernov D., Winters G., Physiological and biogeochemical responses of super-corals to thermal stress from the Northern Gulf of Aqaba, Red Sea. Front. Mar. Sci. 4, 215 (2017). [Google Scholar]
- 41.Voolstra C. R.et al., Standardized short-term acute heat stress assays resolve historical differences in coral thermotolerance across microhabitat reef sites. Glob. Change Biol. 26, 4328–4343 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Berkelmans R., Time-integrated thermal bleaching thresholds of reefs and their variation on the Great Barrier Reef. Mar. Ecol. Prog. Ser. 229, 73–82 (2002). [Google Scholar]
- 43.Douglas A. E., Coral bleaching–How and why? Mar. Pollut. Bull. 46, 385–392 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Brown J. H., Gillooly J. F., Allen A. P., Savage V. M., West G. B., Toward a metabolic theory of ecology. Ecology 85, 1771–1789 (2004). [Google Scholar]
- 45.Voolstra C. R., et al. , Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Sci. Rep. 7, 17583 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahav O., Dubinsky Z., Achituv Y., Falkowski P. G., Smith D. C., Ammonium metabolism in the zooxanthellate coral, Stylophora pistillata. Proc. R. Soc. Lond. B Biol. Sci. 236, 325–337 (1989). [Google Scholar]
- 47.Dudler N., Miller D. J., Characterization of two glutamate dehydrogenases from the symbiotic microalga Symbiodinium microadriaticum isolated from the coral Acropora formosa. Mar. Biol. 97, 427–430 (1988). [Google Scholar]
- 48.Aranda M., et al. , Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 39734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ocaña-Pallarès E., Najle S. R., Scazzocchio C., Ruiz-Trillo I., Reticulate evolution in eukaryotes: Origin and evolution of the nitrate assimilation pathway. PLoS Genet. 15, e1007986 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rädecker N., Pogoreutz C., Wild C., Voolstra C. R., Stimulated respiration and net photosynthesis in Cassiopeia sp. during glucose enrichment suggests in hospite CO2 limitation of algal endosymbionts. Front. Mar. Sci. 4, 267 (2017). [Google Scholar]
- 51.Strychar K. B., Coates M., Sammarco P. W., Loss of Symbiodinium from bleached Australian scleractinian corals (Acropora hyacinthus, Favites complanata and Porites solida). Mar. Freshw. Res. 55, 135–144 (2004). [Google Scholar]
- 52.McIlroy S. E., Wong J. C. Y., Baker D. M., Competitive traits of coral symbionts may alter the structure and function of the microbiome. ISME J. 14, 2424–2432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang T., et al. , Symbiont population control by host-symbiont metabolic interaction in Symbiodiniaceae-cnidarian associations. Nat. Commun. 11, 108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falkowski P. G., Dubinsky Z., Muscatine L., McCloskey L., Population control in symbiotic Corals. Bioscience 43, 606–611 (1993). [Google Scholar]
- 55.Rädecker N., et al. , Using Aiptasia as a model to study metabolic interactions in cnidarian-Symbiodinium symbioses. Front. Physiol. 9, 214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sampayo E. M., Ridgway T., Bongaerts P., Hoegh-Guldberg O., Bleaching susceptibility and mortality of corals are determined by fine-scale differences in symbiont type. Proc. Natl. Acad. Sci. U.S.A. 105, 10444–10449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cunning R., Baker A. C., Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 3, 259–262 (2012). [Google Scholar]
- 58.Baker D. M., Andras J. P., Jordán-Garza A. G., Fogel M. L., Nitrate competition in a coral symbiosis varies with temperature among Symbiodinium clades. ISME J. 7, 1248–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Houlbrèque F., Ferrier-Pagès C., Heterotrophy in tropical scleractinian corals. Biol. Rev. Camb. Philos. Soc. 84, 1–17 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Piniak G. A., Lipschultz F., McClelland J., Assimilation and partitioning of prey nitrogen within two anthozoans and their endosymbiotic zooxanthellae. Mar. Ecol. Prog. Ser. 262, 125–136 (2003). [Google Scholar]
- 61.Krueger T., et al. , Temperature and feeding induce tissue level changes in autotrophic and heterotrophic nutrient allocation in the coral symbiosis - A NanoSIMS study. Sci. Rep. 8, 12710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tremblay P., Gori A., Maguer J. F., Hoogenboom M., Ferrier-Pagès C., Heterotrophy promotes the re-establishment of photosynthate translocation in a symbiotic coral after heat stress. Sci. Rep. 6, 38112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferrier-Pagès C., Rottier C., Beraud E., Levy O., Experimental assessment of the feeding effort of three scleractinian coral species during a thermal stress: Effect on the rates of photosynthesis. J. Exp. Mar. Biol. Ecol. 390, 118–124 (2010). [Google Scholar]
- 64.Palardy J. E., Rodrigues L. J., Grottoli A. G., The importance of zooplankton to the daily metabolic carbon requirements of healthy and bleached corals at two depths. J. Exp. Mar. Biol. Ecol. 367, 180–188 (2008). [Google Scholar]
- 65.Grottoli A. G., Rodrigues L. J., Palardy J. E., Heterotrophic plasticity and resilience in bleached corals. Nature 440, 1186–1189 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Tagliafico A., et al. , Lipid-enriched diets reduce the impacts of thermal stress in corals. Mar. Ecol. Prog. Ser. 573, 129–141 (2017). [Google Scholar]
- 67.Donovan M. K., et al. , Nitrogen pollution interacts with heat stress to increase coral bleaching across the seascape. Proc. Natl. Acad. Sci. U.S.A. 117, 5351–5357 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandes de Barros Marangoni L., Ferrier-Pagès C., Rottier C., Bianchini A., Grover R., Unravelling the different causes of nitrate and ammonium effects on coral bleaching. Sci. Rep. 10, 11975 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ezzat L., Towle E., Irisson J.-O., Langdon C., Ferrier-Pagès C., The relationship between heterotrophic feeding and inorganic nutrient availability in the scleractinian coral T. reniformis under a short-term temperature increase. Limnol. Oceanogr. 61, 89–102 (2016). [Google Scholar]
- 70.Grover R., Maguer J.-F., Allemand D., Ferrier-Pagés C., Nitrate uptake in the scleractinian coral Stylophora pistillata. Limnol. Oceanogr. 48, 2266–2274 (2003). [Google Scholar]
- 71.Safaie A., et al. , High frequency temperature variability reduces the risk of coral bleaching. Nat. Commun. 9, 1671 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klein S. G., et al. , Night-time temperature reprieves enhance the thermal tolerance of a symbiotic cnidarian. Front. Mar. Sci. 6, 453 (2019). [Google Scholar]
- 73.Downs C. A., et al. , Oxidative stress and seasonal coral bleaching. Free Radic. Biol. Med. 33, 533–543 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Hill M., Hill A., The magnesium inhibition and arrested phagosome hypotheses: New perspectives on the evolution and ecology of Symbiodinium symbioses. Biol. Rev. Camb. Philos. Soc. 87, 804–821 (2012). [DOI] [PubMed] [Google Scholar]
- 75.Bieri T., Onishi M., Xiang T., Grossman A. R., Pringle J. R., Relative contributions of various cellular mechanisms to loss of algae during cnidarian bleaching. PLoS One 11, e0152693 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frederickson M. E., Rethinking mutualism stability: Cheaters and the evolution of sanctions. Q. Rev. Biol. 88, 269–295 (2013). [DOI] [PubMed] [Google Scholar]
- 77.Anthony K. R. N., Connolly S. R., Hoegh-Guldberg O., Bleaching, energetics, and coral mortality risk: Effects of temperature, light, and sediment regime. Limnol. Oceanogr. 52, 716–726 (2007). [Google Scholar]
- 78.Hughes A. D., Grottoli A. G., Pease T. K., Matsui Y., Acquisition and assimilation of carbon in non-bleached and bleached corals. Mar. Ecol. Prog. Ser. 420, 91–101 (2010). [Google Scholar]
- 79.Cunning R., Muller E. B., Gates R. D., Nisbet R. M., A dynamic bioenergetic model for coral-Symbiodinium symbioses and coral bleaching as an alternate stable state. J. Theor. Biol. 431, 49–62 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Roth F., et al. , High rates of carbon and dinitrogen fixation suggest a critical role of benthic pioneer communities in the energy and nutrient dynamics of coral reefs. Funct. Ecol. 34, 1991–2004 (2020). [Google Scholar]
- 81.Jeffrey S. W., Humphrey G. F., New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 167, 191–194 (1975). [Google Scholar]
- 82.Cziesielski M. J., et al. , Multi-omics analysis of thermal stress response in a zooxanthellate cnidarian reveals the importance of associating with thermotolerant symbionts. Proc. Biol. Sci. 285, 20172654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olito C., White C. R., Marshall D. J., Barneche D. R., Estimating monotonic rates from biological data using local linear regression. J. Exp. Biol. 220, 759–764 (2017). [DOI] [PubMed] [Google Scholar]
- 84.Muscatine L., McCloskey L. R., Marian R. E., Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnol. Oceanogr. 26, 601–611 (1981). [Google Scholar]
- 85.Harrison P. J., Waters R. E., Taylor F. J. R., A broad spectrum artificial sea water medium for coastal and open ocean phytoplankton. J. Phycol. 16, 28–35 (1980). [Google Scholar]
- 86.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andrews S., FastQC: A quality control tool for high throughput sequence data (2010). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 88.Ziegler M., et al. , Biogeography and molecular diversity of coral symbionts in the genus Symbiodinium around the Arabian Peninsula. J. Biogeogr. 44, 674–686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ziegler M., Arif C., Voolstra C. R., “Symbiodiniaceae diversity in Red Sea coral reefs & coral bleaching” in Coral Reefs of the Red Sea, Voolstra C. R., Berumen M. L., Eds. (Springer International Publishing, 2019), pp. 69–89. [Google Scholar]
- 90.Bushnell B., BBTools software package (2014). sourceforge. net/projects/bbmap.
- 91.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Alexa A., Rahnenfuhrer J., topGO: Enrichment analysis for Gene Ontology (R package version 2) (2010). https://bioconductor.org/packages/release/bioc/html/topGO.html.
- 94.Kanehisa M., Sato Y., KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 29, 28–35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huerta-Cepas J., et al. , eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lavy A., et al. , A quick, easy and non‐intrusive method for underwater volume and surface area evaluation of benthic organisms by 3D computer modelling. Methods Ecol. Evol. 6, 521–531 (2015). [Google Scholar]
- 97.R. Core Team , R: A Language and Environment for Statistical Computing (R Foundation for statistical computing, Vienna, 2020). [Google Scholar]
- 98.Bates D., Fitting linear mixed models in R. R News 5, 27–30 (2005). [Google Scholar]
- 99.Oksanen J., et al. , The vegan package. Community Ecol. Package 10, 631–637 (2007). [Google Scholar]
- 100.Rädecker N., et al. , Data for "Heat stress destabilizes symbiotic nutrient cycling in corals." Zenodo. https://zenodo.org/record/4429583#.X_xgfS1Q30p. Deposited 11 January 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed results of gene expression analyses are provided as Dataset S1. All physiological and NanoSIMS raw data are deposited in the Zenodo data repository (https://zenodo.org/record/4429583#.X_xfoS1Q30o) (100). Additionally, raw sequencing data are deposited in the NCBI Sequence Read Archive under BioProject accession number PRJNA638625.