Significance
We discovered that the histone/protein deacetylase SIRT7, which is the only member of the mammalian sirtuin family residing primarily in the nucleolus, stabilizes p53 in response to UV-induced DNA damage by enabling binding of deacetylated NPM to MDM2/HDM2. The study unveils a crucial mechanism by which the nucleolus controls cellular stress responses. Furthermore, it characterizes SIRT7 as a potent regulator of cellular stress and offers opportunities to manipulate cellular p53 levels, which is important for the development of antitumor therapies.
Keywords: nucleolus, sirtuins, nucleophosmin, p53, acetylation
Abstract
Adaptation to different forms of environmental stress is crucial for maintaining essential cellular functions and survival. The nucleolus plays a decisive role as a signaling hub for coordinating cellular responses to various extrinsic and intrinsic cues. p53 levels are normally kept low in unstressed cells, mainly due to E3 ubiquitin ligase MDM2-mediated degradation. Under stress, nucleophosmin (NPM) relocates from the nucleolus to the nucleoplasm and binds MDM2, thereby preventing degradation of p53 and allowing cell-cycle arrest and DNA repair. Here, we demonstrate that the mammalian sirtuin SIRT7 is an essential component for the regulation of p53 stability during stress responses induced by ultraviolet (UV) irradiation. The catalytic activity of SIRT7 is substantially increased upon UV irradiation through ataxia telangiectasia mutated and Rad3 related (ATR)-mediated phosphorylation, which promotes efficient deacetylation of the SIRT7 target NPM. Deacetylation is required for stress-dependent relocation of NPM into the nucleoplasm and MDM2 binding, thereby preventing ubiquitination and degradation of p53. In the absence of SIRT7, stress-dependent stabilization of p53 is abrogated, both in vitro and in vivo, impairing cellular stress responses. The study uncovers an essential SIRT7-dependent mechanism for stabilization of the tumor suppressor p53 in response to genotoxic stress.
The tumor suppressor p53 is a key player for cellular responses coping with DNA damage, metabolic stress, or other adverse incidents. p53 promotes cell-cycle arrest in response to stress, facilitates DNA repair, or triggers activation of apoptosis to eliminate cells that have accumulated high levels of DNA damage (1). Approximately 50% of human tumors contain mutations in the p53 gene, underscoring the importance of p53 in various signaling pathways implicated in tumorigenesis (1). In mice, loss of p53 leads to tumor development at a high frequency (2). Consistently, inhibition of p53 upon exposure to a variety of DNA-damaging agents, such as ultraviolet (UV) irradiation and environmental carcinogens, accelerates tumorigenesis (3–5).
Despite its undisputed role as a tumor suppressor, increased p53 concentrations appear harmful to cells, making tight regulation of p53 protein levels mandatory. p53 is maintained at low levels under basal conditions but cellular stressors lead to its stabilization, allowing p53 to elicit antitumor functions. The main regulator of p53 levels is the E3 ubiquitin ligase MDM2/HDM2 (in mice and humans, respectively). MDM2/HDM2 associates with and ubiquitinates p53, promoting its subsequent degradation through the proteasome (6). Disruption of the MDM2–p53 interaction provides a rapid means to stabilize p53 in response to stress (6). One of the important regulators of p53 activation is the abundant nucleolar protein nucleophosmin (NPM). NPM belongs to a large group of nucleolar proteins, which coordinate cellular responses to various extrinsic and intrinsic signals and participate in cell proliferation, apoptosis, DNA repair, and stress responses (7, 8). Under normal conditions, these molecules are sequestered in the nucleolus. In response to stressors, the nucleolus undergoes a profound reorganization leading to the release of nucleolar proteins to the nucleoplasm and the cytoplasm. As a consequence, several molecular pathways are activated that enable cellular adaptation to adverse conditions (7–9). Release of NPM from the nucleolus under stress plays a pivotal role for p53 activation. NPM associates with MDM2/HDM2, disrupts the MDM2/HDM2–p53 interaction, and inhibits MDM2/HDM2-dependent degradation of p53 (7, 10–12). The molecular basis underlying the relocalization of NPM and its regulation of the MDM2–p53 interaction is not well-understood.
Mammalian sirtuins comprise a family of seven proteins, SIRT1 to SIRT7, which share a conserved catalytic domain responsible for the sirtuin-specific enzymatic activity, NAD+-dependent protein deacetylation (13). Collectively, sirtuins are assumed to prevent age-dependent diseases and promote organismal homeostasis by enhancing cellular stress resistance (13). Hence, inactivation of different members of the sirtuin family often results in genomic instability and higher predisposition to cancer development (13, 14). SIRT7 is the only mammalian sirtuin that is primarily located in the nucleolus. SIRT7 is excluded from the nucleolus in response to various cellular stressors and localizes to sites of DNA damage where it promotes DNA repair mainly through deacetylation of H3K18 and desuccinylation of H3K122 (15–18).
Previous evidence demonstrates that SIRT7 plays an important role in the regulation of nucleolar functions. SIRT7 activates ribosomal DNA (rDNA) transcription and pre-rRNA processing (15, 19–21). Moreover, our group and others have recently shown that SIRT7 supports genomic stability by maintaining heterochromatin at rDNA genes (22, 23). Here, we demonstrate that SIRT7 is a phosphorylation substrate of ataxia telangiectasia mutated and Rad3 related (ATR) kinase, a major activator of the DNA damage response. ATR-mediated phosphorylation of SIRT7 increases its catalytic activity in response to genotoxic stress caused by UV irradiation. ATR-activated SIRT7 deacetylates NPM, thereby promoting its exclusion from nucleoli. Deacetylated NPM associates with MDM2/HDM2 and inhibits MDM2/HDM2-dependent ubiquitination and degradation of p53. Our results reveal an essential role of SIRT7 and NPM for stabilization of the tumor suppressor p53 in response to UV-induced genotoxic stress.
Results
SIRT7 Is Instrumental for NPM Exclusion from the Nucleolus following Genotoxic Stress.
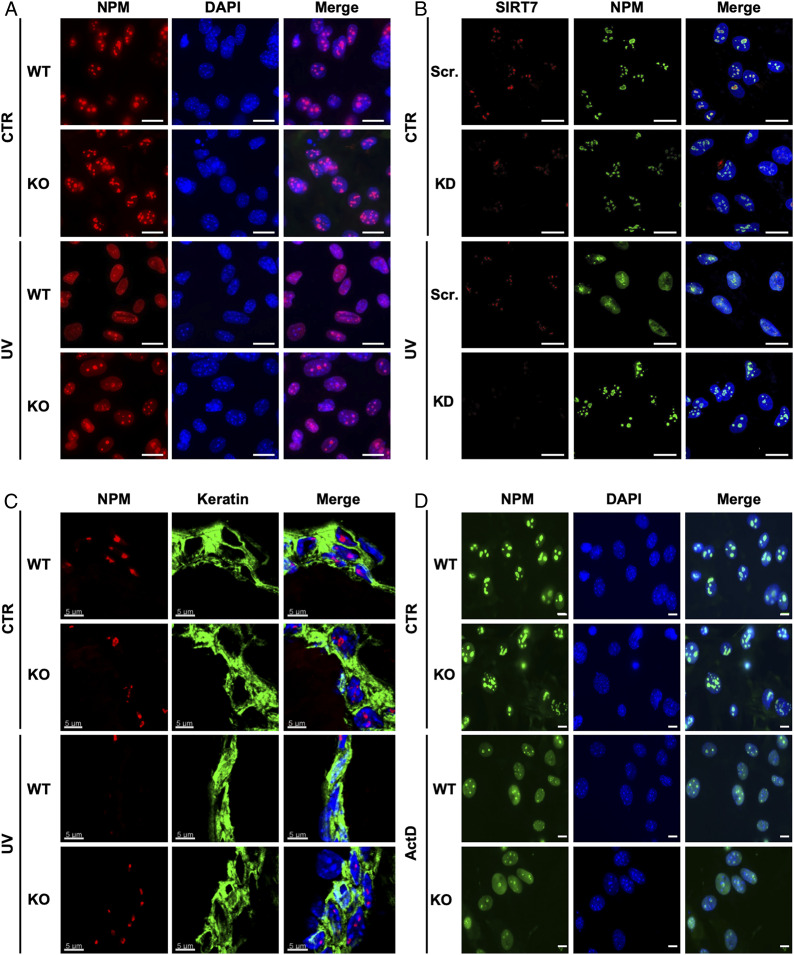
Since SIRT7 is the only member of the sirtuin family that is mainly localized in the nucleolus and interacts with NPM (19, 24), we speculated that SIRT7 is involved in controlling exclusion of NPM from the nucleolus following genotoxic stress. Thus, we monitored the subcellular localization of NPM in primary mouse embryonic fibroblasts (MEFs) derived from SIRT7 wild-type (WT) and knockout (KO) mice upon exposure to ultraviolet C (UVC) irradiation (Fig. 1A). Interestingly, NPM was retained in the nucleoli of UV-treated SIRT7 KO cells but relocalized to the nucleoplasm in WT cells, suggesting that SIRT7 is critical for this particular aspect of the cellular stress response (Fig. 1A and SI Appendix, Fig. S1A). Similar results were obtained in human U2OS cell lines stably expressing a short hairpin RNA (shRNA) targeting SIRT7 (knockdown; KD), indicating that the role of SIRT7 in controlling subcellular distribution of NPM is conserved between species (Fig. 1B). To investigate whether SIRT7 does also promote relocalization of NPM upon UV irradiation in vivo, we analyzed the localization of NPM in skin cells of SIRT7 WT and KO animals exposed to low-energy UV (UVA) irradiation. Importantly, SIRT7 KO mice showed impaired relocalization of NPM following UVA exposure as compared with WT littermates (Fig. 1C). Next, we asked whether SIRT7-mediated redistribution of NPM is a specific result of UV irradiation or represents a general response to DNA damage-inducing agents. Thus, we exposed WT and SIRT7 KO MEFs to different concentrations of doxorubicin, a well-established antitumor drug triggering release of NPM from nucleoli (25). We found that SIRT7 exerted dose-dependent effects in response to doxorubicin: at low concentrations (0.2 μg/mL), SIRT7 KO cells failed to release NPM from nucleoli, similar to the response to UV irradiation. However, at high doxorubicin concentrations (2 μg/mL), NPM was equally excluded from the nucleoli of WT and SIRT7 KO cells, suggesting that other, SIRT7-independent mechanisms are activated at high doxorubicin concentrations, which trigger exclusion of NPM (SI Appendix, Fig. S1B). Finally, we applied another inducer of genotoxic stress, actinomycin D (ActD), which inhibits ribosome biogenesis and strongly promotes release of NPM from the nucleoli (7, 26). ActD triggered NPM relocalization to a similar extent in WT and SIRT7 KO cells, indicating that the role of SIRT7 is context-dependent and only relevant for a distinct subset of stress-induced processes (Fig. 1D).
Fig. 1.
SIRT7 mediates release of NPM from nucleoli in response to genotoxic stress in vitro and in vivo. (A) Immunofluorescence (IF) staining for NPM of SIRT7-deficient primary MEFs 12 h after UVC irradiation (80 J/m2; UV) or without treatment (CTR) (n = 4). (Scale bars, 20 μm.) (B) IF staining of stable SIRT7 KD U2OS cells for NPM (green) and SIRT7 (red) 12 h after UVC irradiation (40 J/m2) or without treatment (n = 4). (Scale bars, 25 μm.) (C) IF staining of skin sections from age-matched WT and SIRT7 KO mice for NPM (red) and keratin (green), 12 h after exposure to 5,000 J/m2 UVA irradiation (n = 4). (Scale bars, 5 μm.) (D) IF staining of NPM in SIRT7 KO primary MEFs after exposure to 8 nM actinomycin D for 6 h (n = 3). (Scale bars, 10 μm.) Nuclei in A, B, and D were counterstained with DAPI. Scr., scrambled.
SIRT7 Promotes p53 Stabilization in Response to UV Irradiation.
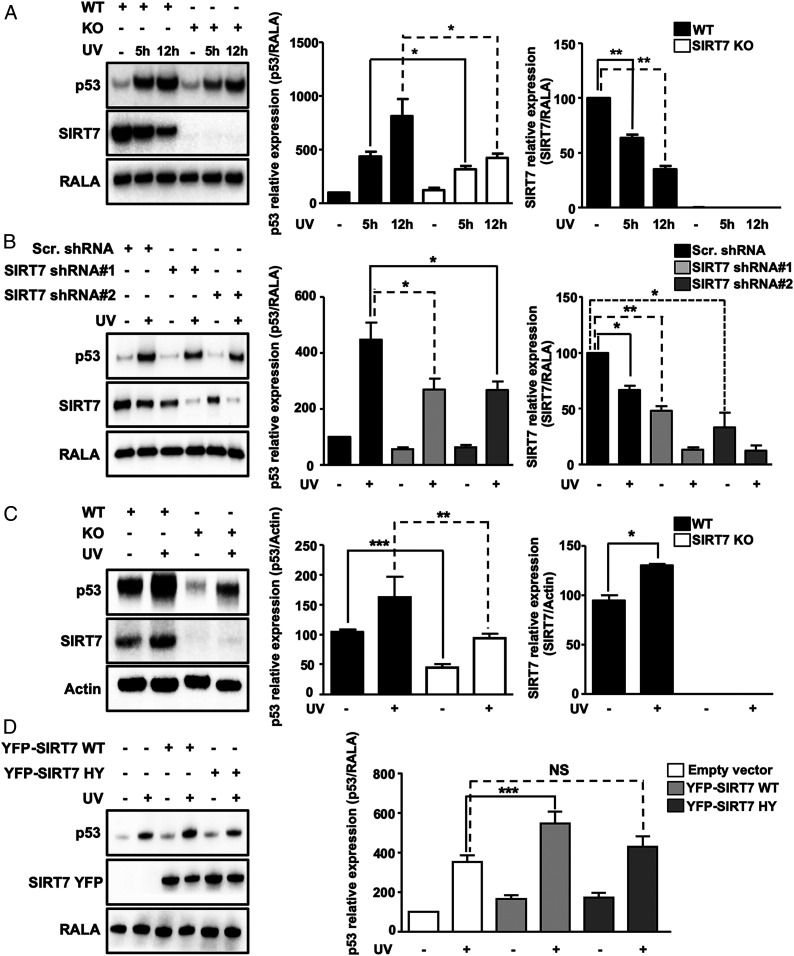
We hypothesized that impairment of UV-induced release of NPM in SIRT7-deficient cells might have an impact on stabilization of the tumor suppressor p53, which is a prominent target of NPM. Western blot analysis revealed a strong reduction of p53 accumulation under UVC irradiation in SIRT7 KO compared with WT MEFs (Fig. 2A). Likewise, U2OS cells stably expressing two different SIRT7-targeting shRNAs failed to stabilize p53 when exposed to UVC, while control cells transfected with scrambled shRNA accumulated p53 (Fig. 2B). Interestingly, SIRT7 protein levels were diminished in control cells following UVC irradiation compared with nonstressed cells (Fig. 2 A and B).
Fig. 2.
SIRT7 stabilizes p53 following UV irradiation. (A) Western blot analysis of p53 destabilization in SIRT7 knockout MEFs in response to UVC irradiation. Quantification of p53 and SIRT7 relative expression ± SD is shown (Right) (n = 7). (B) Western blot analysis of p53 destabilization in U2OS SIRT7 KD cell lines 12 h after UVC irradiation. Quantification of p53 and SIRT7 relative expression ± SD is shown (Right) (n = 9). (C) Western blot analysis of attenuated p53 stabilization in the skin of SIRT7 KO mice 12 h after UVA irradiation. Quantification of p53 and SIRT7 expression is shown (Right) (n = 4). (D) Western blot analysis of p53 levels 5 h after UVC irradiation in U2OS cells transfected with YFP-SIRT7 WT and YFP-SIRT7 HY (catalytically inactive SIRT7). Quantification of p53 relative levels ± SD is shown (Right) (n = 6). Ras-related protein Ral-A (RALA) or actin was used as loading control. *P < 0.05, **P < 0.01, ***P < 0.001; NS, not significant.
Since UVC is a very potent stress inducer that might provoke additional responses leading to degradation of SIRT7, we exposed SIRT7 WT and KO MEFs to UVA irradiation. In contrast to UVC, UVA exposure, which more closely mimics stress levels commonly encountered by the skin in vivo, induced an increase in SIRT7 protein levels (SI Appendix, Fig. S2A). Accumulation of p53 was reduced in SIRT7-deficient cells after UVA irradiation, although the effects were less pronounced compared with UVC (SI Appendix, Fig. S2A). To explore p53 accumulation and SIRT7 expression in vivo, WT and SIRT7 KO mice were exposed to low-energy, UVA irradiation. Notably, we also observed impaired stabilization of p53 in the skin of SIRT7-deficient mice (Fig. 2C), consistent with impaired exclusion of NPM from nucleoli (Fig. 1C). Furthermore, SIRT7 protein levels increased in the skin upon UVA irradiation in agreement with the in vitro findings, further confirming the critical role of SIRT7 in UV-induced cellular responses (Fig. 2C).
To explore the role of SIRT7 in p53 accumulation in response to doxorubicin treatment, we used two different doxorubicin concentrations as in the previous experiments. Administration of low doxorubicin to SIRT7 KO MEFs and to SIRT7-deficient U2OS cells prevented p53 accumulation, providing additional support for the critical role of SIRT7 in regulating p53 concentrations under genotoxic stress (SI Appendix, Fig. S2 B and C). In contrast, high doxorubicin resulted in enhanced accumulation of p53 when SIRT7 was depleted (Fig. 2B). The latter observation is consistent with results from a previous study, where only high doxorubicin concentrations were used (16). Interestingly, treatment with ActD led to accumulation of p53 in both SIRT7 WT and SIRT7 KO cells (SI Appendix, Fig. S2D), corroborating our assumption that SIRT7 regulates p53 stabilization only under distinct stress conditions.
To analyze whether the catalytic activity of SIRT7 is required for p53 stabilization, we transfected U2OS cells either with empty vector, mouse yellow fluorescent protein (YFP)-tagged wild-type SIRT7 (YFP-SIRT7 WT), or the YFP-tagged catalytically inactive mutant H188Y SIRT7 (YFP-SIRT7 HY). Cells were either left untreated after transfection or exposed to UVC and then subjected to Western blot analysis to determine p53 concentrations. Overexpression of SIRT7 WT but not the enzymatically inactive SIRT7 mutant significantly increased p53 levels upon UV irradiation, demonstrating that the catalytic activity of SIRT7 is required to achieve p53 stabilization under UVC stress (Fig. 2D).
SIRT7 Augments Association of NPM with HDM2 and Prevents p53 Ubiquitination under UV Irradiation.
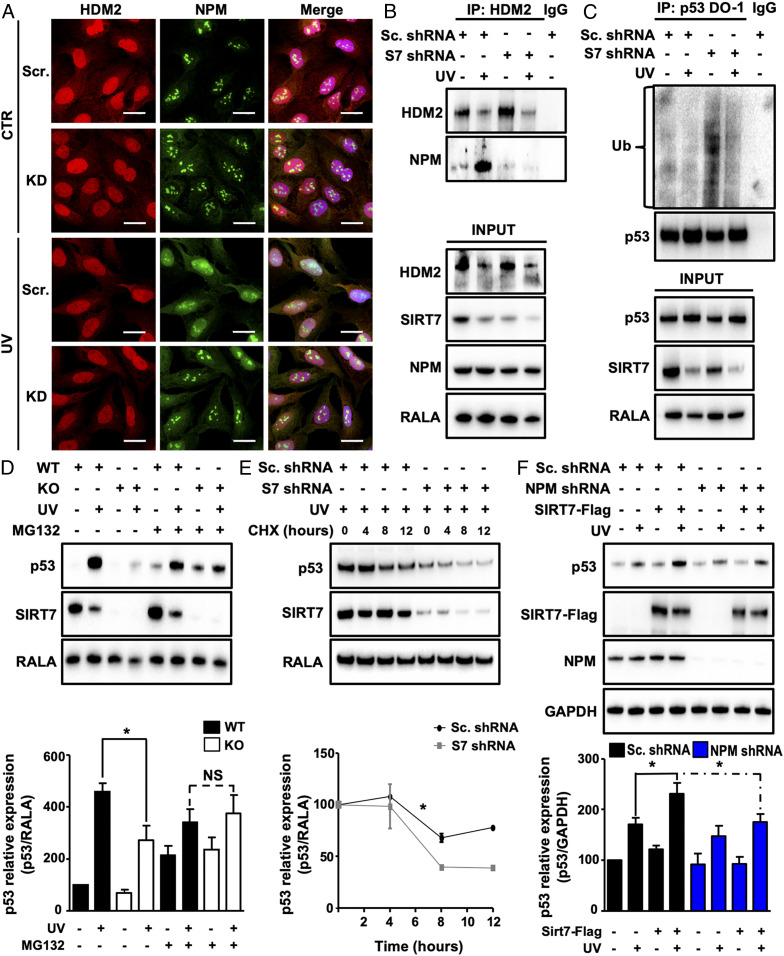
Stress-induced release of NPM from nucleoli promotes p53 stabilization. After translocation into the nucleoplasm, NPM associates with MDM2/HDM2 and prevents MDM2/HDM2-mediated ubiquitination and degradation of p53. Since we found that SIRT7 leads to p53 stabilization after UV irradiation, we wanted to know whether SIRT7 enhances the association of NPM with HDM2. Comparison of the subcellular localization of NPM and HDM2 in U2OS cells exposed to UVC irradiation revealed that NPM relocates into the nucleoplasm where HDM2 is present. Suppression of SIRT7 prevented the relocation of NPM into the nucleoplasm after UV exposure (Fig. 3A). Subsequent coimmunoprecipitation analysis confirmed that NPM/HDM2 binding is strongly increased in control cells upon UVC exposure but virtually abolished in SIRT7 KD cells (Fig. 3B). Since NPM inhibits HDM2-mediated ubiquitination of p53, we determined the level of p53 ubiquitination in SIRT7-deficient cells exposed to UVC irradiation. Endogenous p53 was immunoprecipitated and the degree of ubiquitination was measured by Western blot analysis using an anti–pan-ubiquitin antibody. SIRT7-deficient cells showed increased p53 ubiquitination under unstressed conditions and maintained higher ubiquitination levels after UV exposition, while p53 ubiquitination remained low in control cells transfected with scrambled shRNA (Fig. 3C). Importantly, inhibition of proteasomal activity in SIRT7 KO and WT cells by MG132 after UVC irradiation restored p53 levels in SIRT7-deficient cells to levels comparable to WT cells (Fig. 3D), further corroborating the critical role of SIRT7 in regulating ubiquitin-dependent proteasomal degradation of p53.
Fig. 3.
SIRT7 enhances association of NPM with HDM2 and prevents p53 ubiquitination under UV. (A) IF staining for NPM and HDM2 of SIRT7 KD and control U2OS cells 12 h after UVC irradiation. Nuclei were counterstained with DAPI (n = 3). (Scale bars, 25 µm.) (B) Coupled immunoprecipitation (IP) (HDM2 antibody) and Western blot analysis (NPM antibody) of SIRT7 KD and control (scrambled) U2OS cell lysates 3 h after UVC irradiation (n = 3). (C) Coupled IP (p53-DO-1 antibody) and Western blot analysis (ubiquitin antibody) of SIRT7 KD and control (scrambled) U2OS cell lysates 2 h after UVC irradiation followed by a 5-h incubation with 10 µM MG-132 (n = 3). The membrane was reprobed with anti-p53 antibody to ensure equal IP of p53 (Top). (D) Western blot analysis of p53 levels in WT and SIRT7 KO MEFs 12 h after UVC irradiation, followed by a 5-h treatment with 10 µM MG-132. RALA was used as loading control. Quantification of p53 relative levels ± SD is given (Bottom) (n = 3). (E) Western blot analysis of p53 levels in control (scrambled) and SIRT7 KD U2OS cell lysates 12 h after UVC irradiation followed by treatment with cycloheximide (CHX; 50 µg/mL). Quantification of p53 relative levels ± SEM is given (Bottom) (n = 3; two-way ANOVA). (F) Western blot analysis of p53, SIRT7, and NPM levels in NPM KD and control (scrambled) U2OS cells transfected with Flag-SIRT7 5 h after UVC irradiation. GAPDH was used as a loading control. Quantification of p53 relative levels ± SD is given (Bottom) (n = 3). *P < 0.05.
To exclude the possibility that SIRT7 regulates transcription of the p53 gene and thereby alters p53 protein levels, we determined p53 mRNA expression in SIRT7 KO and WT MEFs. No significant changes were observed by qRT-PCR analysis, indicating that SIRT7 regulates p53 posttranslationally (SI Appendix, Fig. S3A). Similar results were obtained in stable U2OS cells expressing scrambled or SIRT7-targeting shRNA (SI Appendix, Fig. S3B). In addition, we investigated the levels of p53 protein in UVC-irradiated scrambled and SIRT7 KD cells after treatment with the translation inhibitor cycloheximide. Cells lacking SIRT7 showed lower stability of p53 protein in response to UV irradiation compared with control cells (Fig. 3E). Taken together, the data indicate that the absence of p53 accumulation in SIRT7-deficient cells is primarily caused by increased ubiquitination of p53 and subsequent proteasomal degradation.
To unequivocally demonstrate that SIRT7-dependent association of NPM with HDM2 is responsible for efficient stabilization of p53 under UV stress, we suppressed expression of NPM. Western blot analysis of p53 revealed that overexpression of SIRT7 in NPM KD but not control U2OS cells failed to increase p53 levels, indicating that NPM is required for SIRT7-mediated stabilization of p53 following UV irradiation (Fig. 3F).
ATR-Mediated Phosphorylation of SIRT7 Increases Its Catalytic Activity under UV Stress and Is Responsible for Enhanced NPM Deacetylation.
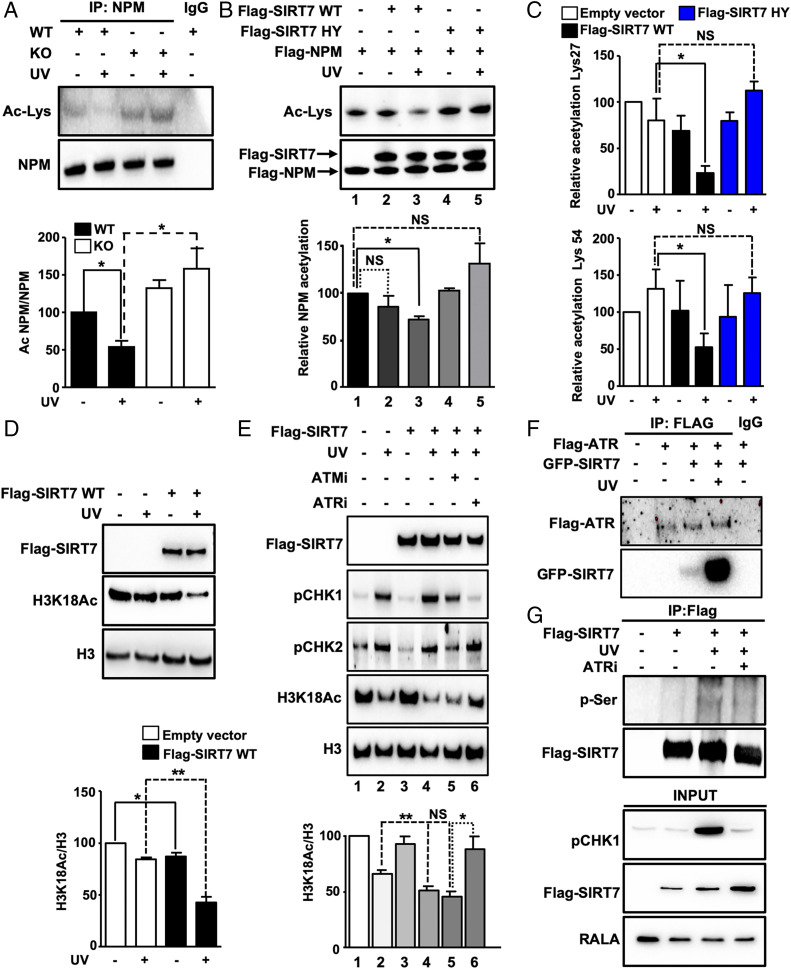
To gain more insights into the molecular mechanism governing SIRT7-mediated exclusion of NPM from nucleoli leading to p53 stabilization upon UV irradiation, we searched for SIRT7 interaction partners. Coupled immunoprecipitation and mass spectrometry analysis indicated that SIRT7 interacts with NPM (SI Appendix, Fig. S4A), which was further confirmed by coimmunoprecipitation and colocalization experiments demonstrating that SIRT7 and NPM form a molecular complex in nucleoli (SI Appendix, Fig. S4 B and C). Interestingly, UVC irradiation resulted in more efficient association of SIRT7 with NPM compared with unstressed conditions (SI Appendix, Fig. S4D). Moreover, we found that NPM serves as a deacetylation substrate for SIRT7 and that UVC exposure of cells increased the deacetylation activity of SIRT7. UV irradiation led to strong reduction of NPM acetylation whereas SIRT7 KO cells maintained a high level of acetylated NPM both under normal conditions and after UV exposure (Fig. 4A).
Fig. 4.
UV irradiation stimulates the enzymatic activity of SIRT7 required for efficient deacetylation of NPM. (A) Coupled IP (anti-NPM antibody) and Western blot analysis (anti-acetyllysine antibody) of SIRT7 WT and KO MEFs 5 h after UVC irradiation in the presence of trichostatin A (n = 3). Equal IP of NPM was verified by Western blotting. Quantification of acetylated NPM normalized to immunoprecipitated NPM protein ± SD (Bottom) (n = 4). (B) In vitro deacetylation assay using precipitated Flag-SIRT7 wild type (Flag-SIRT7 WT), SIRT7 catalytically inactive mutant (Flag-SIRT7 HY), and Flag-NPM after overexpression in 293T HEK cells followed by UVC exposure. NPM deacetylation was determined using an anti-acetyllysine antibody (Ac-Lys). Quantification of relative NPM acetylation is shown (Bottom) (n = 3). (C) Relative acetylation levels ± SD of NPM at lysine 27 (Top) and lysine 54 (Bottom) as determined by mass spectrometry of immunoprecipitated NPM using 293T HEK cells transfected with an empty vector, Flag-SIRT7 WT, and Flag-SIRT7 HY 5 h after UVC irradiation (n = 4). (D) Western blot analysis of H3K18Ac in U2OS cells transfected with an empty vector or with Flag-SIRT7 WT 5 h after exposure to UVC. Quantification (Bottom) (n = 3). (E) Western blot analysis of H3K18Ac in 293T HEK cells transfected with an empty vector or with Flag-tagged wild-type SIRT7 (Flag-SIRT7) after exposure to UVC. Cells were pretreated with vehicle, ATR inhibitor (ATRi; 4 µM), and ATM inhibitor (ATMi; 10 µM) prior to exposure to 40 J/m2 UVC, cultivated for 5 h with inhibitors, and analyzed by Western blot. Inhibition of ATR and ATM was assessed by determining phosphorylation levels (p) of CHK1 and CHK2. Quantification of H3K18Ac level normalized to total histone 3 (H3) is shown (Bottom) (n = 3). (F) Coupled IP (anti-Flag antibody) and Western blot analysis (anti-Flag and anti-GFP antibodies) of 293T HEK cells transfected with GFP-SIRT7 and Flag-ATR 5 h after UVC irradiation (n = 3). (G) Coupled IP (anti-Flag M2 affinity beads) and Western blot analysis (anti-phosphorylated serine and anti-Flag antibodies) of 293T HEK cells treated with vehicle or ATRi 5 h after UVC irradiation (n = 3). *P < 0.05, **P < 0.01.
To validate these findings, we performed in vitro deacetylation assays using purified human Flag-tagged SIRT7 WT, the SIRT7-inactivating mutation SIRT7-H187Y, and NPM. SIRT7-Flag proteins were isolated from 293T HEK cells kept under normal conditions and after UVC irradiation, while NPM-Flag was isolated from the same cells without stress exposure. Only SIRT7 WT isolated from UV-irradiated cells efficiently deacetylated NPM in vitro (Fig. 4B). Neither SIRT7 WT from unstressed cells nor SIRT7 mutant proteins showed any noteworthy deacetylation activity on NPM (Fig. 4B). To identify lysine residues of NPM that are specifically deacetylated by SIRT7 in response to UV exposure, we transfected 293T HEK cells with empty vector and with WT and HY mutant SIRT7, which were either exposed to UV irradiation or left untreated. Measurement of immunoprecipitated NPM by mass spectrometry revealed that SIRT7 WT specifically deacetylated NPM at two lysine residues, lysines 27 (Lys-27) and 54 (Lys-54), after UV irradiation, while the catalytically inactive mutant did not alter acetylation at these residues (Fig. 4C). We also identified several other acetylated lysines, which, however, did not depend on SIRT7 (SI Appendix, Fig. S4E).
To analyze whether the increased enzymatic activity of SIRT7 after UV irradiation is restricted to Lys-27 and Lys-54 of NPM, we employed H3K18Ac as a different substrate (27). We found that without UV irradiation, overexpression of SIRT7 in U2OS cells only slightly affected acetylation of H3K18. In contrast, SIRT7 overexpression in UV-irradiated cells dramatically reduced H3K18Ac levels compared with cells expressing an empty vector, indicating that UV-induced cellular stress globally increases the enzymatic activity of SIRT7 (Fig. 4D). Increased SIRT7-dependent H3K18 deacetylation under UV was observed also in a similar experiment using 293T HEK cells (SI Appendix, Fig. S5A). In marked contrast, ActD failed to enhance SIRT7 catalytic activity, indicating that SIRT7 activation is not caused by inhibition of ribosome biogenesis (SI Appendix, Fig. S5A).
Ataxia telangiectasia mutated (ATM) and ATM and ATR kinases are key mediators of the DNA damage response and control cellular reactions following exposure to genotoxic stress (28, 29). We wondered whether these kinases might be required for SIRT7 activation and analyzed the capacity of SIRT7 to deacetylate H3K18 after UVC irradiation in the presence of ATM- and ATR-specific inhibitors. Notably, inhibition of ATR but not of ATM significantly impaired SIRT7 activation (Fig. 4E). Conversely, ATR overexpression enhanced SIRT7 activity under UV (SI Appendix, Fig. S5B). Further experiments demonstrated that ATR and SIRT7 efficiently coimmunoprecipitate after UVC irradiation (Fig. 4F) and that the ATR inhibitor abolishes UV-induced SIRT7 phosphorylation (Fig. 4G), supporting the importance of ATR-dependent SIRT7 phosphorylation in response to UV irradiation. Mass spectrometry analysis revealed that ATR phosphorylates human SIRT7 at serines 134 and 136 (SI Appendix, Fig. S5 C–E). In the mouse, serine 136 corresponds to Ser-137, while mouse Ser-134 is replaced by a proline at position 135 (SI Appendix, Fig. S5F). To validate the functional relevance of ATR-mediated phosphorylation, we replaced Ser-137 in mouse SIRT7 by aspartic acid, which mimics phosphorylation at this residue. Strikingly, the SIRT7 S137D mutant increased deacetylation of the prototypical SIRT7 target H3K36Ac (30), providing compelling evidence that ATR-mediated phosphorylation activates SIRT7 (SI Appendix, Fig. S5G).
SIRT7-Dependent Deacetylation of NPM Promotes p53 Stabilization.
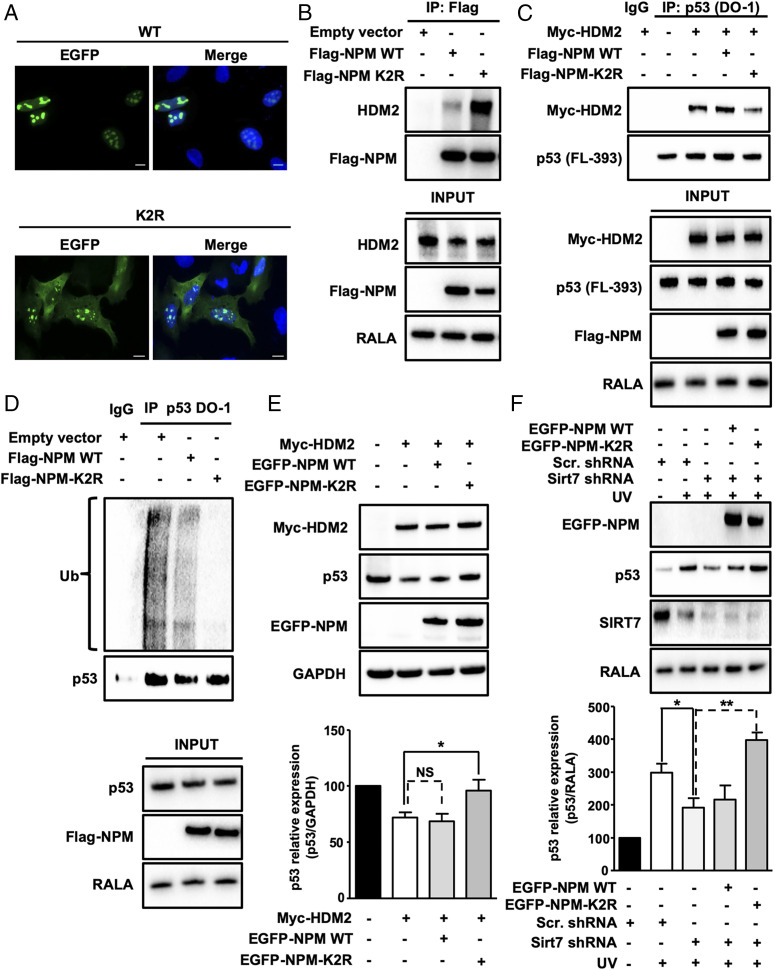
In order to test whether deacetylation of NPM at lysines 27 and 54 is crucial for NPM exclusion from nucleoli, we replaced Lys-27 and Lys-54 in mouse NPM-EGFP (enhanced green fluorescent protein) by arginines to mimic deacetylation at these residues (K2R) and transfected the corresponding construct into human U2OS cells, in which endogenous NPM expression was suppressed by shRNA-mediated KD. Immunofluorescence revealed reduced nucleolar localization of the K2R NPM deacetylation mimetic mutant and a shift toward the nucleoplasm compared with WT NPM-EGFP (Fig. 5A). Importantly, inhibition of ATR impaired NPM translocation under UVC irradiation, providing additional proof for the role of ATR in SIRT7 activation and NPM deacetylation (SI Appendix, Fig. S6A).
Fig. 5.
SIRT7-mediated deacetylation of NPM at lysines 27 and 54 promotes p53 stabilization by inhibiting HDM2. (A) IF staining of NPM KD U2OS cells transfected with WT EGFP-NPM and K2R EGFP-NPM using an anti-EGFP antibody. Cell nuclei were counterstained with DAPI (n = 3). (Scale bars, 10 µm.) (B) Coupled IP (anti-Flag M2 affinity beads) and Western blot analysis (antibody against endogenous HDM2) of 293T HEK cells transfected with empty vector, Flag-NPM WT, and Flag-NPM K2R mutant. Inputs are shown (Bottom) (n = 3). (C) Coupled IP (p53-DO-1 antibody) and Western blot analysis (antibody against Myc-HDM2) of 293T HEK cells transfected with Myc-HDM2, Flag-tagged WT, and K2R NPM followed by a 5-h treatment with MG-132. p53 concentrations were assessed using an anti-p53 antibody. Inputs are shown (Bottom) (n = 3). (D) Coupled IP (p53-DO-1 antibody) and Western blot analysis (anti-ubiquitin antibody) of U2OS cells transfected with empty vector, Flag-tagged WT and K2R NPM followed by a 5-h treatment with 10 µM MG-132. Inputs are shown (Bottom) (n = 3). (E) Western blot analysis of p53 levels in U2OS cells transfected with empty vector, Myc-tagged HDM2, Flag-tagged WT, and K2R NPM. GAPDH was used as a loading control. The graph represents the average of relative p53 levels ± SD (n = 4). (F) Western blot analysis of p53 in scrambled and SIRT7 KD cells transfected with EGFP-tagged WT and K2R NPM following UVC irradiation 48 h posttransfection and harvested 5 h after irradiation (n = 4). *P < 0.05, **P < 0.01.
Coimmunoprecipitation of WT or K2R NPM with HDM2 in 293T HEK cells revealed that K2R NPM but not WT NPM efficiently coprecipitated with endogenous HDM2 (Fig. 5B). The strong association of the NPM K2R mutant with HDM2 suggested that NPM deacetylation is required for preventing binding of HDM2 to p53. To prove this assumption, we transfected Flag-tagged WT and K2R NPM mutant constructs together with Myc-tagged HDM2 in 293T HEK cells and immunoprecipitated endogenous p53 protein. Importantly, K2R NPM but not WT NPM efficiently reduced the interaction between HDM2 and p53 (Fig. 5C). Furthermore, we observed that K2R NPM but not WT NPM strongly inhibited p53 ubiquitination (Fig. 5D). In addition, we monitored the levels of p53 in U2OS cells by Western blotting after transfection of Myc-tagged HDM2 alone or in combination with WT and K2R NPM. As expected, the K2R NPM deacetylation mimetic mutant inhibited HDM2-induced degradation of p53 while WT NPM exerted almost no effects under these conditions (Fig. 5E). To further confirm the role of SIRT7-mediated deacetylation of NPM in p53 stabilization, we analyzed the levels of p53 protein expression in SIRT7 KD cells exposed to UVC irradiation in the presence of WT or K2R mutant NPM overexpression. We found that K2R but not WT NPM prevented the decline of p53 in SIRT7 KD cells (Fig. 5F). Finally, inhibition of ATR impaired p53 stabilization following UVC irradiation in primary WT MEFs while only minor effects were observed in SIRT7-deficient cells (SI Appendix, Fig. S6B).
SIRT7-Mediated Deacetylation of NPM Enhances p53-Dependent Transcriptional Responses.
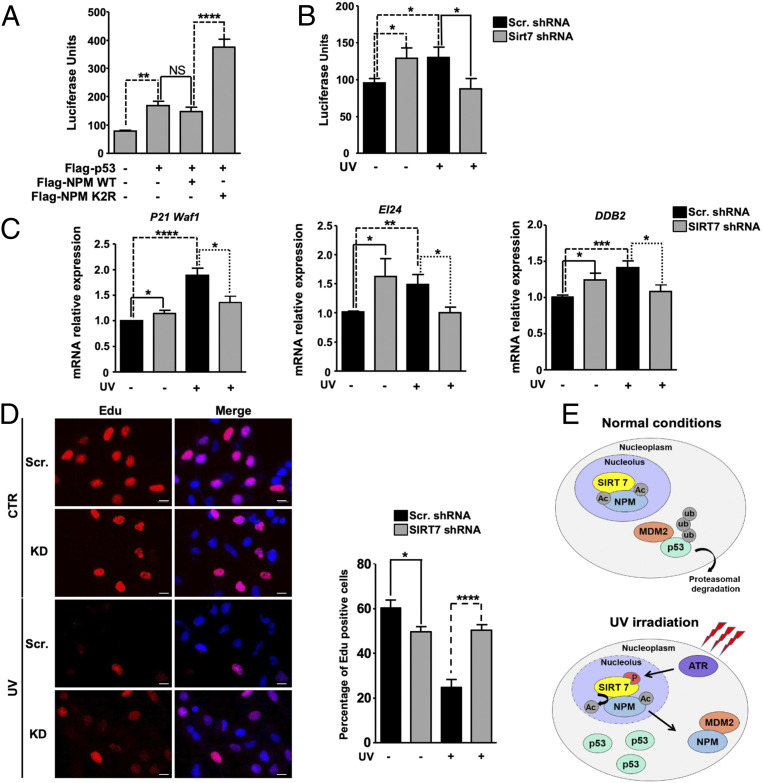
Since SIRT7-mediated deacetylation of NPM promotes p53 stabilization, we next asked whether the deacetylated NPM has an impact on p53-dependent transcriptional responses. Transfection of a luciferase reporter construct containing p53 binding sites in U2OS cells together with p53 alone or in combination with WT and K2R mutant NPM revealed that K2R NPM dramatically stimulated p53 transcriptional responses while WT NPM failed to do so (Fig. 6A).
Fig. 6.
SIRT7-mediated deacetylation of NPM stimulates p53 transcriptional activity. (A) p53-dependent luciferase activity in U2OS cells transfected with p53, WT, and K2R NPM (n = 6). (B) Luciferase assay for p53 activity in scrambled and SIRT7 KD stable U2OS cells 5 h after exposure to 10 J/m2 UVC (n = 6). (C) qRT-PCR analysis of mRNA expression of p53 target genes (P21Waf1, EI24, and DDB2) in control (scrambled) and SIRT7 KD U2OS cells 5 h after UVC irradiation (10 J/m2). β-Actin was used as a loading control. Quantification of average mRNA levels relative to β-actin ± SD is shown in the histograms (n = 6). (D) EdU (5-ethynyl-2′-deoxyuridine) staining of scrambled and SIRT7 KD cells either left untreated or following exposure to UVC irradiation (10 J/m2, 5 h). Cell nuclei were counterstained with DAPI. (Scale bars, 20 μm.) The percentage of EdU-positive cells is shown in the histograms. A minimum of 100 cells was analyzed for each experiment (n = 3). (E) Scheme depicting the putative mechanisms of NPM regulation by SIRT7 following UV-induced genotoxic stress. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Consistent with attenuated stabilization of p53 in the absence of SIRT7, we observed a reduced stimulation of the p53 reporter in UVC-irradiated SIRT7-deficient cells (Fig. 6B). However, the transcriptional activity of p53 was higher in SIRT7-deficient than in control cells when no UV stress was applied (Fig. 6B). Moreover, unstressed SIRT7-deficient cells showed a reduced proliferation rate, which was reverted by inhibition of p53 (SI Appendix, Fig. S7), indicating that increased p53 activity in SIRT7-deficient cells is responsible for the slower proliferation rate. The different levels of p53 activity in nonstressed and stressed SIRT7-deficient cells suggest that SIRT7 controls p53 activity by different means under baseline versus stress conditions. The differential expression of the p53 targets P21, EI24, and DDB2 supports this assumption: Expression of p53 target genes was higher in SIRT7 KD cells without stress but lower under UV irradiation (Fig. 6C). Accordingly, SIRT7-deficient cells were unable to inhibit cell-cycle progression under UV irradiation (Fig. 6D).
The data conclusively demonstrate that UV irradiation enhances SIRT7 activity through ATR-mediated SIRT7 phosphorylation, which results in deacetylation of NPM at Lys-27 and Lys-54, leading to release of NPM from nucleoli and association with HDM2. The enhanced binding of NPM to HDM2 prevents HDM2-mediated proteasomal degradation of p53, causing increased activation of the p53 transcriptional program (Fig. 6E).
Discussion
The discovery of the critical role of the nucleolus in cellular protection against stress has attracted much attention in the last decade. The release of NPM from the nucleolus represents a crucial event that leads to stabilization of the tumor suppressor p53 (11). So far, the molecular mechanisms governing this process have not been fully characterized (10–12). Here, we demonstrate that the enzymatic activity of the histone/protein deacetylase SIRT7 increases in response to UV irradiation due to phosphorylation at serines 134 and 136 by ATR. Activated SIRT7 deacetylates NPM, which leaves nucleoli and associates with HDM2, thereby disrupting the HDM2–p53 interaction and stabilizing p53. Loss of SIRT7 prevents p53 stabilization both in vitro and in vivo.
Notably, overexpression of K2R NPM in SIRT7-deficient cells was sufficient to restore stabilization of p53 to normal levels, confirming the decisive role of deacetylated NPM for p53 stabilization under UV stress. It is interesting to note that deacetylation of NPM was not as efficient as UV irradiation to expel NPM K2R from the nucleolus, suggesting that other factors such as dephosphorylation and S-glutathionylation of NPM contribute to NPM translocation (11, 31). Our data uncovered a critical role of ATR in phosphorylating and thus enhancing SIRT7 activity in response to stress. In addition, inhibition of ATR suppresses p53 stabilization in SIRT7-deficient cells, at least to some extent, indicating a SIRT7-independent action of ATR. It seems reasonable to assume that ATR regulates the UV response within the SIRT7–NPM–p53 axis at several levels. Besides the activation of SIRT7, ATR may promote NPM exclusion from nucleoli by direct phosphorylation (32) and, finally, phosphorylation of p53 by ATR contributes to p53 stabilization as well (33).
In contrast to SIRT7, SIRT1 inhibits exclusion of NPM from nucleoli through a mechanism requiring interaction with NPM but not NPM deacetylation (34). We previously demonstrated that SIRT7 and SIRT1 associate in the nucleolus to stabilize rDNA heterochromatin (22). The interplay between SIRT1 and SIRT7 might constitute an additional regulatory level to control subcellular localization of NPM following genotoxic stress, although further work is needed to assess the exact role of this interaction in this pathway. Finally, SIRT6 deacetylates NPM too, although the functional role of this deacetylation remains unexplored, especially in the context of cellular stress responses (24). Taken together, the available data suggest an interplay of nuclear sirtuins, constituting a highly complex molecular network that fine-tunes redistribution of NPM outside the nucleolus following specific stress stimuli, thereby controlling p53 stabilization.
Our work uncovered that the ATR/SIRT7/NPM/MDM2 pathway only plays a major role for some but not all stress responses. The ATR/SIRT7/NPM/MDM2 pathway clearly reinforces p53 stabilization under low and high UV intensities as well as under low doses of doxorubicin. In contrast, high doses of doxorubicin or ActD treatment, which inhibit ribosome biogenesis, do not seem to involve the ATR/SIRT7/NPM/MDM2 pathway. Instead, SIRT7 directly suppresses the function of p53 under high concentrations of doxorubicin as described previously (16). Other mechanisms might be employed as well, depending on the stressor and the cell type. For example, SIRT7 inhibits expression of p53 in neurons exposed to ischemia/reperfusion injury and cardiomyocytes subjected to hypoxia/reoxygenation injury but has no effect on p53 levels in cell lines derived from hepatocellular carcinomas (35–37). During the preparation of this manuscript, Lu and collaborators demonstrated that SIRT7 promotes accumulation of p53 by destabilizing MDM2 in response to glucose starvation (38). In contrast to these findings, we did not observe significant changes in MDM2/HDM2 expression in SIRT7-deficient cells exposed to UV. Notwithstanding, both studies agree that SIRT7 is a potent inhibitor of MDM2 either by promoting degradation of MDM2 or by preventing its association with p53 through deacetylation of NPM. SIRT7 might employ both mechanisms either separately or in a synergistic manner to boost activation of p53, according to cellular needs.
Our study highlights the indirect regulation of p53 by SIRT7, but we did not ignore that SIRT7 is able to directly deacetylate p53 for suppression of apoptosis in response to stress (36, 37, 39). However, at present, it is not clear how this interaction influences the function of p53 under UVC. UVC evokes a rather unique type of p53 response that is still not well-characterized. Instead of promoting apoptosis, p53 exerts prosurvival functions under UVC, probably by favoring DNA repair (40). The potential role of direct SIRT7-mediated deacetylation of p53 under UV warrants further research.
Our data clearly demonstrate that SIRT7-mediated deacetylation of NPM and subsequent stabilization of p53 is a pivotal response to UV-induced genotoxic stress both in vitro and in vivo in the skin. Exposure to solar UV irradiation is the major cause of skin cancer and p53 plays a crucial role in this tumor entity. Inactivation of p53 enhances the incidence of spontaneous and UV-induced skin tumors in murine models and inactivating mutations of the p53 gene are often found in human skin cancers (41). Hence, it is reasonable to assume that SIRT7 acts as a tumor suppressor in UV-induced skin cancer through stabilization of p53.
Stabilization of p53 and thereby activation of apoptosis is exploited by numerous antitumor drugs (8). On the other hand, stabilization of p53 in tumors harboring oncogenic mutations in the p53 gene might be detrimental (42). Thus, pharmacological approaches are under development to destabilize mutant p53 (42). Therapeutic manipulation of the SIRT7–NPM–HDM2–p53 axis in cancer cells might help to achieve this goal. For example, pharmacological inhibition of SIRT7 in tumors harboring mutant p53 will promote p53 destabilization while activation of SIRT7 in tumors expressing WT p53 will lead to its stabilization, in particular when combined with drugs inducing cellular stress responses.
Materials and Methods
Mice and UV Irradiation of the Skin.
Generation of C57BL/6 SIRT7−/− mice was described previously (39). UV irradiation of the skin was performed as described in SI Appendix.
Isolation of Primary Mouse Embryonic Fibroblasts, Cell Culture, and Treatments.
Primary MEFs were isolated from WT and SIRT7−/− embryos as described previously (22). Primary MEFs and U2OS and 293T HEK cells were cultivated and treated as described in SI Appendix.
Generation of Stable KD Cell Lines by Lentivirus-Driven shRNA.
Stable SIRT7 and NPM KD cells were generated as described previously (22). All shRNA sequences are listed in SI Appendix, Table S1.
Plasmids and Cloning.
All plasmids and cloning details are described in SI Appendix.
Western Blot, Immunoprecipitation, In Vitro Deacetylation Assay, and Mass Spectrometry Analysis.
Western blot analysis was performed as described (22) using antibodies listed in SI Appendix, Table S2. Immunoprecipitation experiments were performed using specific antibodies (SI Appendix, Table S2) or anti-Flag M2 affinity beads (Sigma-Aldrich) as described (22, 43). To minimize detection of immunoglobulin, TrueBlot horseradish peroxidase-conjugated secondary antibodies (Rockland) were used. For detection of ubiquitinated p53, cell lysates were immunoprecipitated with p53 Do-1 antibody, resolved by gel electrophoresis, and transferred onto polyvinylidene fluoride membranes (Thermo Fisher). Membranes were boiled in double-distilled water for 30 min and blocked for an additional 30 min in 0.5% (weight/volume [vol]) porcine skin gelatin (Sigma-Aldrich)/0.05% (vol/vol) Tween-20 prior to incubation with an anti-ubiquitin antibody. In vitro deacetylation assay and mass spectrometry analysis for the identification of NPM acetylated lysines were performed as described in SI Appendix.
RNA Extraction and Quantitative Reverse-Transcription PCR.
RNA extraction and qRT-PCR were performed as described (43). Primer sequences are given in SI Appendix, Table S3.
Immunofluorescence Analysis.
Immunofluorescence analysis was performed as described (43) using the antibodies listed in SI Appendix, Table S4.
Luciferase Assay.
p53 transcriptional activity was measured using a dual-luciferase assay as described in SI Appendix.
Statistical Analysis.
Data are expressed as the mean ± SD of at least three independent biological replicates. Statistical significance was assessed by Student’s t test unless otherwise specified using GraphPad Prism 5.0 software. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; NS, not significant.
Supplementary Material
Acknowledgments
We thank M. Euler for technical assistance. This work was supported by the Max Planck Society, the Excellence Initiative “Cardiopulmonary Institute,” Deutsche Forschungsgemeinschaft Collaborative Research Center SFB1213 (TP A02 and B02), Klinische Forschungsgruppe 309 TP 08, German Center for Cardiovascular Research and European Research Area Network on Cardiovascular Diseases Project CLARIFY, and the Spanish Ministry of Economy and Competitiveness Ministerio de Economía y Competitividad (SAF2014-55964R and SAF2017-88975R to A.V.) cofounded by Fondo Europeo de Desarrollo Regional funds and the European Regional Development Fund—A Way to Make Europe.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015339118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or SI Appendix.
References
- 1.Vousden K. H., Lu X., Live or let die: The cell’s response to p53. Nat. Rev. Cancer 2, 594–604 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Liu J., Zhang C., Hu W., Feng Z., Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 356, 197–203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ziegler A., et al. , Sunburn and p53 in the onset of skin cancer. Nature 372, 773–776 (1994). [DOI] [PubMed] [Google Scholar]
- 4.Salim E. I., et al. , Carcinogenicity of dimethylarsinic acid in p53 heterozygous knockout and wild-type C57BL/6J mice. Carcinogenesis 24, 335–342 (2003). [DOI] [PubMed] [Google Scholar]
- 5.De Flora S., et al. , Molecular alterations and lung tumors in p53 mutant mice exposed to cigarette smoke. Cancer Res. 63, 793–800 (2003). [PubMed] [Google Scholar]
- 6.Nag S., Qin J., Srivenugopal K. S., Wang M., Zhang R., The MDM2-p53 pathway revisited. J. Biomed. Res. 27, 254–271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I., The nucleolus under stress. Mol. Cell 40, 216–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quin J. E., et al. , Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta 1842, 802–816 (2014). [DOI] [PubMed] [Google Scholar]
- 9.James A., Wang Y., Raje H., Rosby R., DiMario P., Nucleolar stress with and without p53. Nucleus 5, 402–426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colombo E., Marine J. C., Danovi D., Falini B., Pelicci P. G., Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4, 529–533 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Yang K., et al. , A redox mechanism underlying nucleolar stress sensing by nucleophosmin. Nat. Commun. 7, 13599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurki S., et al. , Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5, 465–475 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Ianni A., Yuan X., Bober E., Braun T., Sirtuins in the cardiovascular system: Potential targets in pediatric cardiology. Pediatr. Cardiol. 39, 983–992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosch-Presegué L., Vaquero A., Sirtuins in stress response: Guardians of the genome. Oncogene 33, 3764–3775 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Chen S., et al. , Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell 52, 303–313 (2013). [DOI] [PubMed] [Google Scholar]
- 16.Kiran S., Oddi V., Ramakrishna G., Sirtuin 7 promotes cellular survival following genomic stress by attenuation of DNA damage, SAPK activation and p53 response. Exp. Cell Res. 331, 123–141 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Vazquez B. N., et al. , SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 35, 1488–1503 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., et al. , SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 7, 12235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford E., et al. , Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 20, 1075–1080 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iyer-Bierhoff A., et al. , SIRT7-dependent deacetylation of fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Rep. 25, 2946–2954.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Chen X., et al. , Inhibition of Wnt/β-catenin signaling suppresses bleomycin-induced pulmonary fibrosis by attenuating the expression of TGF-β1 and FGF-2. Exp. Mol. Pathol. 101, 22–30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ianni A., Hoelper S., Krueger M., Braun T., Bober E., Sirt7 stabilizes rDNA heterochromatin through recruitment of DNMT1 and Sirt1. Biochem. Biophys. Res. Commun. 492, 434–440 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Paredes S., et al. , The epigenetic regulator SIRT7 guards against mammalian cellular senescence induced by ribosomal DNA instability. J. Biol. Chem. 293, 11242–11250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee N., et al. , Comparative interactomes of SIRT6 and SIRT7: Implication of functional links to aging. Proteomics 14, 1610–1622 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Avitabile D., et al. , Nucleolar stress is an early response to myocardial damage involving nucleolar proteins nucleostemin and nucleophosmin. Proc. Natl. Acad. Sci. U.S.A. 108, 6145–6150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donati G., Peddigari S., Mercer C. A., Thomas G., 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 4, 87–98 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barber M. F., et al. , SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487, 114–118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cimprich K. A., Cortez D., ATR: An essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9, 616–627 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stokes M. P., et al. , Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 104, 19855–19860 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W. W., et al. , A click chemistry approach reveals the chromatin-dependent histone H3K36 deacylase nature of SIRT7. J. Am. Chem. Soc. 141, 2462–2473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanellis D. C., Bursac S., Tsichlis P. N., Volarevic S., Eliopoulos A. G., Physical and functional interaction of the TPL2 kinase with nucleophosmin. Oncogene 34, 2516–2526 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Maiguel D. A., Jones L., Chakravarty D., Yang C., Carrier F., Nucleophosmin sets a threshold for p53 response to UV radiation. Mol. Cell. Biol. 24, 3703–3711 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavin M. F., Gueven N., The complexity of p53 stabilization and activation. Cell Death Differ. 13, 941–950 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Bi X., et al. , Inhibition of nucleolar stress response by Sirt1: A potential mechanism of acetylation-independent regulation of p53 accumulation. Aging Cell 18, e12900 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv J., Tian J., Zheng G., Zhao J., Sirtuin7 is involved in protecting neurons against oxygen-glucose deprivation and reoxygenation-induced injury through regulation of the p53 signaling pathway. J. Biochem. Mol. Toxicol. 31, e21955 (2017). [DOI] [PubMed] [Google Scholar]
- 36.Sun M., et al. , MicroRNA-148b-3p is involved in regulating hypoxia/reoxygenation-induced injury of cardiomyocytes in vitro through modulating SIRT7/p53 signaling. Chem. Biol. Interact. 296, 211–219 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Zhao J., et al. , SIRT7 regulates hepatocellular carcinoma response to therapy by altering the p53-dependent cell death pathway. J. Exp. Clin. Cancer Res. 38, 252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y. F., et al. , SIRT7 activates p53 by enhancing PCAF-mediated MDM2 degradation to arrest the cell cycle. Oncogene 39, 4650–4665 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vakhrusheva O., et al. , Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 102, 703–710 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Lo P. K., Huang S. Z., Chen H. C., Wang F. F., The prosurvival activity of p53 protects cells from UV-induced apoptosis by inhibiting c-Jun NH2-terminal kinase activity and mitochondrial death signaling. Cancer Res. 64, 8736–8745 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Benjamin C. L., Ananthaswamy H. N., p53 and the pathogenesis of skin cancer. Toxicol. Appl. Pharmacol. 224, 241–248 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue X., et al. , Mutant p53 in cancer: Accumulation, gain-of-function, and therapy. J. Mol. Biol. 429, 1595–1606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang J., et al. , Sirt7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of Sirt1. Proc. Natl. Acad. Sci. U.S.A. 114, E8352–E8361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.