Significance
Plant DNA methylation, which occurs in three sequence contexts (CG, CHG, and CHH, where H refers to A, T, or C), is established and maintained by different mechanisms. In this study, we present genome-wide chromatin accessibility profiles of Arabidopsis mutants that are deficient in CG, CHG, and/or CHH methylation. Through a combination of DNA methylation, chromatin accessibility, and higher-order chromosome conformation profiling of these mutants, we uncover links between DNA methylation, chromatin accessibility, and 3D genome architecture. These results reveal the interplay between CG and non-CG methylation in heterochromatin maintenance and suggest that DNA methylation can directly impact chromatin structure.
Keywords: DNA methylation, epigenetics, chromatin accessibility
Abstract
DNA methylation is a major epigenetic modification found across species and has a profound impact on many biological processes. However, its influence on chromatin accessibility and higher-order genome organization remains unclear, particularly in plants. Here, we present genome-wide chromatin accessibility profiles of 18 Arabidopsis mutants that are deficient in CG, CHG, or CHH DNA methylation. We find that DNA methylation in all three sequence contexts impacts chromatin accessibility in heterochromatin. Many chromatin regions maintain inaccessibility when DNA methylation is lost in only one or two sequence contexts, and signatures of accessibility are particularly affected when DNA methylation is reduced in all contexts, suggesting an interplay between different types of DNA methylation. In addition, we found that increased chromatin accessibility was not always accompanied by increased transcription, suggesting that DNA methylation can directly impact chromatin structure by other mechanisms. We also observed that an increase in chromatin accessibility was accompanied by enhanced long-range chromatin interactions. Together, these results provide a valuable resource for chromatin architecture and DNA methylation analyses and uncover a pivotal role for methylation in the maintenance of heterochromatin inaccessibility.
DNA methylation is a conserved epigenetic mark that plays important roles in diverse biological processes, including gene regulation, transposable element (TE) silencing, imprinting, and X chromosome inactivation in eukaryotes. In plants, cytosine methylation occurs in three sequence contexts; CG, CHG, and CHH (where H refers to A, T, or C) (1–3). Methylation is mediated by METHYLTRANSFERASE 1 (MET1) (4) and DOMAINS REARRANGED METHYLASE 2 (DRM2) (5), orthologs of mammalian DNMT1 and DNMT3, respectively, as well as by two plant-specific DNA methyltransferases, CHROMOMETHYLASE2 (CMT2) (6) and CHROMOMETHYLASE3 (CMT3) (7). MET1 cooperates with a conserved cofactor VIM to maintain preexisting CG methylation during DNA replication (8), whereas DRM2, CMT3, and CMT2 control the maintenance of non-CG methylation (1). De novo DNA methylation is mediated by the plant-specific RNA-directed DNA methylation (RdDM) pathway that depends on DNA-dependent RNA polymerases, Pol IV and Pol V (1, 9, 10).
Pol IV produces transcripts (P4-RNAs) that are converted into double-stranded RNAs by RNA-DEPENDENT RNA POLYMERASE 2 (RDR2). These transcripts are diced into 24-nucleotides (nt) small interfering RNAs (siRNAs) by DICER-LIKE 3 (DCL3) and subsequently loaded into ARGONAUTE 4 (AGO4) or its homologs, AGO6 and AGO9 (11–13). Pol V produces long noncoding RNAs at target sites that pair with AGO4/siRNA complexes (12). Pol V chromatin occupancy requires the DNA-methylation reader proteins SUVH2 and SUVH9 [homologs of SU(VAR)3-9], as well as the DDR complex (13, 14). This latter complex consists of RNA-DIRECTED DNA METHYLATION 1 (RDM1), DEFECTIVE IN MERISTEM SILENCING 3 (DMS3), and DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1) (15, 16). The RdDM pathway ultimately recruits DRM2 to specific genomic sequences for de novo DNA methylation, as well as maintenance of non-CG methylation (17). Several other factors are involved in the RdDM pathway, though their functions remain less well characterized (18–21).
In eukaryotic organisms, genomic DNA forms chromatin, condensed arrangements of nucleoprotein complexes. The basic unit of chromatin is the nucleosome, which consists of ∼147 base pair (bp) of DNA wrapped around a histone octamer composed of one H3/H4 tetramer and two H2A/H2B dimers (22). DNA methylation is preferentially distributed over nucleosome regions and is less enriched over flanking nucleosome-depleted DNA, suggesting a link between nucleosome positioning and DNA methylation (23). Furthermore, DNA methylation has an important role in heterochromatin condensation and silencing (24). Indeed, mutations in both MET1 and the nucleosome remodeler DDM1 (DECREASE IN DNA METHYLATION 1) result in genome-wide loss of CG methylation, accompanied by visible chromatin decondensation at chromocenters, suggesting a connection between DNA methylation and chromatin compaction (25). However, whether CG, CHG, and CHH methylations have distinct relationships with chromatin accessibility, how they further affect the three-dimensional (3D) architecture of the genome, and the relationship between DNA methylation, chromatin accessibility, and transcription are unclear.
In Arabidopsis, various well-characterized DNA methylation mutants exist that show different levels or types of DNA methylation deficiencies (25). These mutants provide an excellent opportunity to explore the effects of different DNA methylation contexts on chromatin accessibility. Here, we profiled chromatin accessibility in 18 DNA methylation mutants, including met1, ddm1, fwa, nrpd1, nrpe1, nrpe1 nrpd1, dms3, drm1 drm2, cmt2, cmt3, cmt2 cmt3, drm1 drm2 cmt2 cmt3 (ddcc), drm3, idn2, idn2 idl1 idl2, suvr2, ago4, and frg1 frg2. These mutants decrease methylation in different sequence contexts, with met1 leading to loss of almost all CG methylation, cmt3 reducing CHG methylation, cmt2 impacting CHH methylation, and drm1 drm2 cmt2 cmt3 losing virtually all CHG and CHH methylation. The fwa line was derived from crossing a met1 homozygous mutant with wild type and selecting for a plant homozygous for wild-type MET1 alleles in the F2 population. This line has a chimeric genomic methylation pattern showing methylation losses at FWA and many other loci but has a wild-type methylation machinery (15), allowing the analysis of differentially methylated regions of the genome without the complication of possible indirect effects of mutations in DNA methylation machinery components. The other mutants impact the RdDM pathway and show decreases in DNA methylation to different degrees. Using a combination of DNA methylation profiling, chromatin accessibility profiling, and higher-order chromosome conformation profiles of these mutants, we find links between these parameters in Arabidopsis chromatin. This comprehensive characterization reveals the interplay of CG and non-CG methylation in chromatin accessibility.
Results and Discussion
Genome-Wide Chromatin Accessibility Profiles of DNA Methylation–Deficient Mutants.
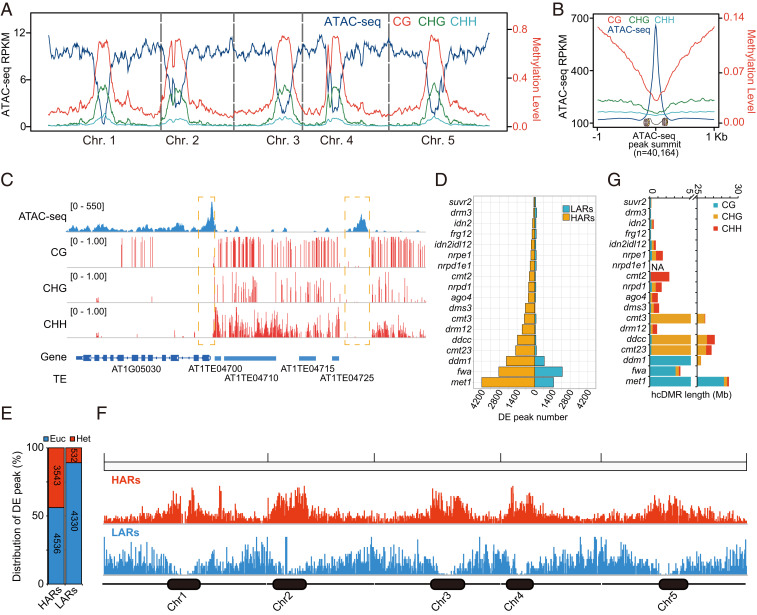
To investigate the relationship between DNA methylation and chromatin accessibility, we performed assay for transposase-accessible chromatin using sequencing (ATAC-seq) (26) using wild-type Col-0 floral buds of Arabidopsis. We first assessed the correlation of chromatin accessibility and DNA methylation by plotting CG, CHG, and CHH methylation and chromatin accessibility levels over chromosomes divided into 100 kilobase (kb) bins. As was previously reported, we observed that genome-wide accessible chromatin was enriched at euchromatin and depleted at heterochromatin, while CG, CHG, and CHH methylations are enriched at heterochromatin and depleted at euchromatin (Fig. 1A) (27). Using HMMRATAC (28), we defined 40,164 open chromatin peaks in wild type. We plotted CG, CHG, and CHH methylation levels over the summit of open chromatin peaks, including 1 kb of flanking sequence, and observed a gradual depletion of DNA methylation in all sequence contexts near open chromatin peaks (Fig. 1B). A higher-resolution (1 kb bins) correlation analysis also indicated that chromatin accessibility is anticorrelated with CG, CHG, and CHH methylation (SI Appendix, Fig. S1). This anticorrelation is also clearly evident at the level of individual genes and transposons (example genome browser view in Fig. 1C). In summary, these results show that chromatin accessibility anticorrelates with DNA methylation, as has been seen in previous studies (27, 29).
Fig. 1.
Whole-genome plot of DNA methylation and chromatin accessibility. (A) Genome-wide distribution of DNA methylation (CG, CHG, and CHH) and ATAC-seq signal. (B) DNA methylation (CG, CHG, and CHH) at ATAC-seq peak summits (n = 40,164). (C) Screenshot showing an example of chromatin accessibility, CG, CHG, and CHH methylation. (D) Number of differential peaks (DE; P value < 0.05, fold change > 2) in 18 mutants compared with wild type. (E) Distribution of HARs and LARs in euchromatin (Euc) and heterochromatin (Het) regions. (F) Chromosome view of HAR (red) and LAR (blue) peaks. Black dots represent heterochromatin regions. (G) Number of high-confidence CG, CHG, and CHH hypo-DMRs (hcDMR) in mutants.
To examine the relationship between different DNA methylation sequence contexts and chromatin accessibility, we conducted ATAC-seq in floral tissues of 18 representative backgrounds that are deficient in methylation in one or more sequence contexts (CG, CHG, and CHH), namely, met1, ddm1, fwa, nrpd1, nrpe1, nrpe1 nrpd1, dms3, drm1 drm2, cmt2, cmt3, cmt2 cmt3, drm1 drm2 cmt2 cmt3, drm3, idn2, idn2 idl1 idl2, suvr2, ago4, and frg1 frg2. To quantify chromatin accessibility changes, accessibility differences relative to wild type (fold change > 2 and P value < 0.05) were computed by comparing chromatin accessibility signals in each mutant with the wild-type control in a merged open chromatin peak dataset containing Col-0 and the 18 mutants (58,446 total peaks). We identified between 69 and 4,188 more highly accessible regions (hereafter referred to as HARs) and between 21 and 2,181 less accessible regions (hereafter referred to as LARs) in individual mutants compared with Col-0 (Fig. 1D). In total, we identified 8,079 peaks that are more accessible and 4,862 peaks that are less accessible in at least one mutant. The length of the majority of HARs and LARs were smaller than 500 bp, suggesting that DNA methylation affects chromatin accessibility of only a few nucleosomes in a region (SI Appendix, Fig. S2). The total number of open chromatin peaks (58,446 from all samples vs. 40,164 from Col-0) indicates that DNA methylation has a profound effect on genome-wide chromatin accessibility. We determined the chromosomal distributions of the differential peaks and found that 3,543 (43.85%) of HARs are located in heterochromatin regions, while the majority of LARs, 4,439 (89.24%), are located in euchromatic regions (Fig. 1 E and F).
To quantify the relationship between DNA methylation changes and chromatin accessibility changes, we compared the number of differentially methylated regions (DMRs) and differentially accessible peaks. In general, we observed that backgrounds with reduced CG methylation, such as met1, fwa, and ddm1, exhibited the most dramatic impact on chromatin accessibility (Pearson correlation R = 0.84, P value = 1.6E-5, Fig. 1 D and E). Mutants impacting CHG methylation, which include cmt3, drm1 drm2 cmt2 cmt3, and cmt2 cmt3, had a moderate impact on chromatin accessibility profiles, while cmt2 and RdDM mutants, including nrpe1 nrpd1, nrpd1, nrpe1, drm3, idn2, idn2 idl1 idl2, suvr2, ago4, and frg1 frg2, had the least impact on chromatin accessibility (Pearson correlation R = 0.24, P value = 0.33, Fig. 1 D and E). To assess the landscape of chromatin accessibility in these mutants, we plotted chromatin accessibility variation in bins of 100 kb along chromosomes, finding that most chromatin accessibility signal changes in these mutants occurred in heterochromatin regions (SI Appendix, Fig. S3).
Relationships between DNA Methylation and Chromatin Accessibility Changes.
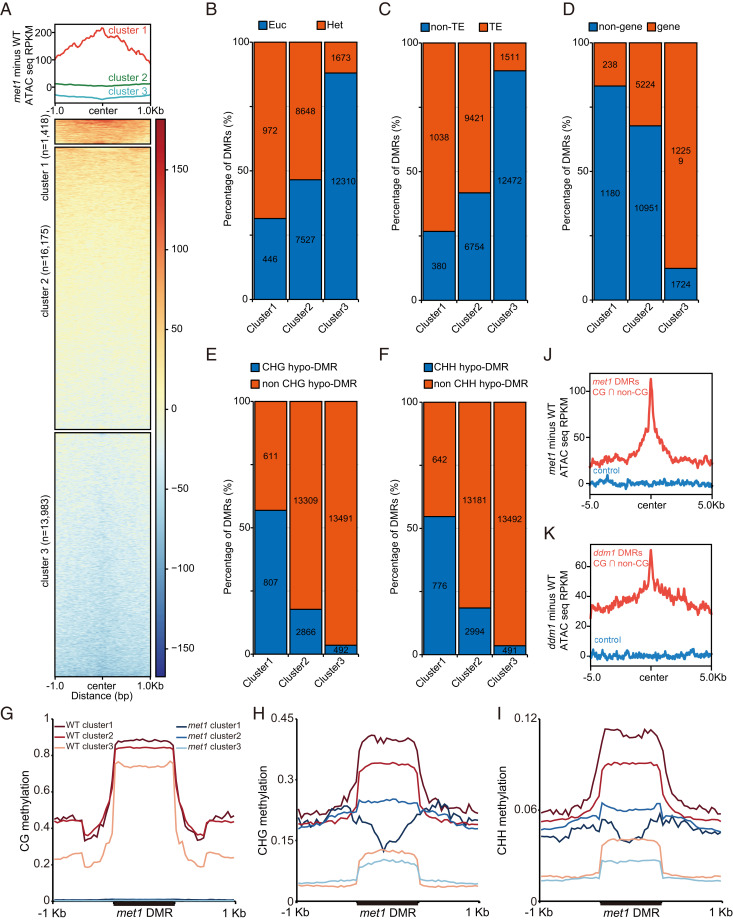
In plants, CG methylation tends to be found within the transcribed gene bodies of particular genes, while CG, CHG, and CHH methylations are found together at RdDM sites, TEs, and heterochromatin regions (30). Previous studies have shown that MET1 deficiency leads to genome-wide CG methylation loss and partial non-CG methylation loss (25). Interestingly, most met1 CG DMRs, especially those localized within gene bodies, showed no change or even showed decreased chromatin accessibility profiles, indicating that other factors may contribute to the maintenance of chromatin accessibility at these regions (SI Appendix, Fig. S4A). In agreement with previous studies, we detected an increase in CHG methylation over gene bodies in met1 (31) and also found that regions that gained CHG methylation became less accessible in met1 mutants (SI Appendix, Fig. S4 B–E). To distinguish CG DMRs that affect chromatin accessibility from those that do not, we analyzed met1 ATAC-seq data using K-means clustering. This method classified met1 CG hypo-DMRs into three clusters (cluster 1: more accessible cluster, cluster 2: unchanged cluster, cluster 3: less accessible cluster) (Fig. 2A). Cluster 1 contained 4.5% (1,418 out of the 31,576) of CG DMRs that showed increased chromatin accessibility in met1 mutants (Fig. 2A). The majority of cluster 1 CG DMRs were found in heterochromatin regions, while cluster 3 DMRs tended to be at genic regions, and cluster 2 DMRs were seen in both regions (Fig. 2 B–D). Interestingly, in addition to the loss of CG methylation at cluster 1 accessible regions, we found that cluster 1 CG DMRs showed higher overlap with met1 CHG and CHH hypo-DMRs, indicating that cluster 1 CG hypo-DMRs are also non-CG hypo-DMRs (Fig. 2 E and F). Consistent with this result, metaplots of DNA methylation over the three DMR clusters revealed that in addition to CG methylation loss, cluster 1 regions also completely or partially lost CHG and CHH methylation (Fig. 2 G–I). The simultaneous reduction of CG, CHG, and CHH methylations in cluster 1 CG DMRs, where chromatin accessibility is increased, suggested an interplay of CG and non-CG methylation in chromatin accessibility. Similarly, we observed that overlapped regions of CG and non-CG hypo-DMRs in met1 and ddm1 have increased chromatin accessibility (Fig. 2 J and K). Metaplots of DNA methylation in drm1 drm2 cmt2 cmt3, cmt2 cmt3, drm1 drm2, cmt3, dms3, ago4, nrpd1, nrpe1, cmt2, drm3, idn2, idn2 idl1 idl2, suvr2, and frg1 frg2 mutants also indicated that both CG and non-CG methylation were reduced over more accessible regions (SI Appendix, Fig. S5). The converse analysis showed a similar trend, where the ATAC-seq signal over CG, CHG, and CHH hypo-DMRs in these mutants showed that CG, CHG, and CHH hypo-DMRs became more accessible (SI Appendix, Fig. S6). Consistent with previous studies showing the overlap of different hypo-DMRs in different mutants, HARs and LARs also showed consistent overlaps (SI Appendix, Fig. S7) (25, 32). For example, strong RdDM mutants form a cluster, while weak RdDM mutants also cluster (SI Appendix, Fig. S7). Overall, these results suggest that DNA methylation plays an important role in chromatin accessibilty via an interplay or redundancy of CG and non-CG methylation.
Fig. 2.
ATAC-seq profile of met1 CG DMRs. (A) K-means clustering of met1 CG hypo-DMRs into three clusters. (B) Overlap of the three met1 CG hypo-DMR clusters with Euc and Het regions. (C) Overlap of the three met1 CG hypo-DMR clusters with TE and non-TE regions. (D) Overlap of the three clusters with genic and nongenic regions. (E) Overlap of the three met1 CG hypo-DMR clusters with met1 CHG hypo-DMRs and non-CHG hypo-DMRs. (F) Overlap of the three met1 CG hypo-DMR clusters with met1 CHH hypo-DMRs and non-CHH hypo-DMRs. Metaplot of CG (G), CHG (H), and CHH (I) methylation in the three met1 CG hypo-DMR clusters. Increase in chromatin accessibility in regions of overlapping CG and non-CG hypo-DMRs in met1 (J) and ddm1 (K). n = 3,510 and 6,053, respectively. WT represents wild-type Col-0.
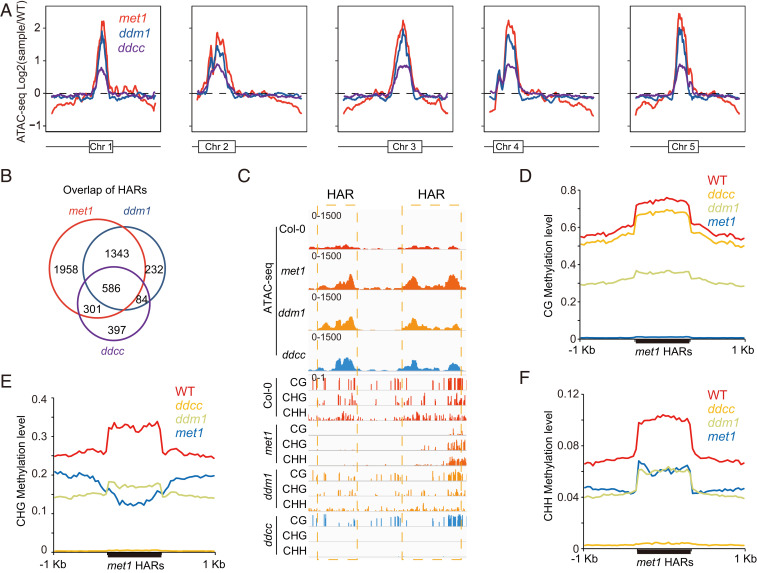
To further explore the interplay of CG and non-CG methylations on chromatin accessibility, we next compared met1, ddm1, and drm1 drm2 cmt2 cmt3 (hereafter termed ddcc) in detail since these mutants showed the most dramatic changes in chromatin accessibility. Genome-wide chromatin accessibility analysis indicated that these three mutants exhibited similar patterns of increased chromatin accessibility within heterochromatin regions, though ddcc showed a smaller overall effect (Fig. 3A). Venn diagram analysis of the accessible peaks showed a high degree of overlap between the HARs in met1, ddm1, and ddcc mutants (Fig. 3B). Consistent with this trend, the HARs in the met1 mutant exhibited a decrease in CG and non-CG methylation at these regions in all three mutant backgrounds (Fig. 3 C–F).
Fig. 3.
Connection between DNA methylation and chromatin accessibility. (A) Genome-wide pattern of chromatin accessibility variation in met1, ddm1, and ddcc. Variation in ATAC-seq signal (log2 mutant vs. Col-0) is depicted on the y axis. The box in each chromosome represents the pericentromeric heterochromatin region. (B) Overlap of peaks with more accessibility identified by ATAC-seq in met1, ddm1, and ddcc. (C) Screenshot showing simultaneous reduction of CG, CHG, and CHH methylations in peaks with more accessibility in met1, ddm1, and ddcc. Average distribution of CG (D), CHG (E), and CHH (F) methylation over peaks with more accessibility in met1.
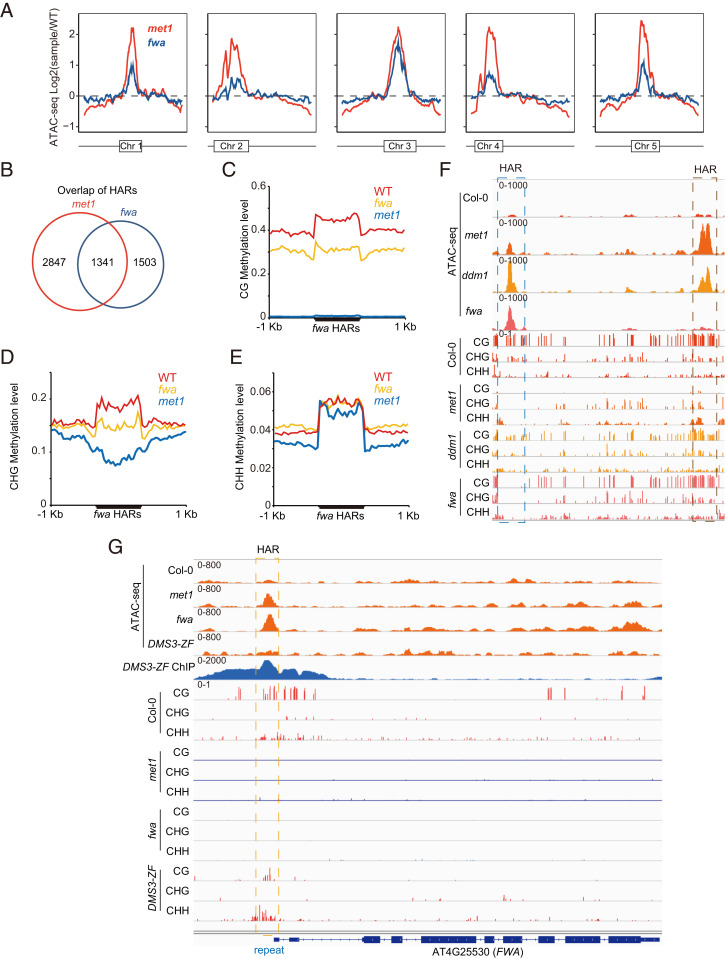
fwa is a background with wild-type methylation machinery but heritably reduced DNA methylation at many genomic regions and was created by crossing met1 with Col-0 and then selecting for the wild-type MET1 alleles (33). As expected, fwa DNA methylation levels were less affected than in met1 (SI Appendix, Fig. S8); however, fwa did display dramatic changes in chromatin accessibility when compared with Col-0 (Fig. 1 D and G). We found that chromatin accessibility and DNA methylation of most euchromatin regions in the fwa epiallele were comparable to Col-0, while heterochromatin regions showed lower DNA methylation and chromatin accessibility states than wild type (Fig. 4A and SI Appendix, Fig. S8 D–F). HARs showed high overlap between met1 and fwa (Fig. 4B). Metaplots of DNA methylation over fwa HARs showed that these regions have lower CG and CHG than Col-0 (Fig. 4 C–E). Chromatin accessibility was restored to wild-type levels only when CG and non-CG methylations were both comparable to Col-0 (Fig. 4F). Loss of CG and CHH methylations at the short repeats in the FWA promoter creates heritable fwa epialleles which show ectopic FWA expression and a late flowering phenotype (33). Inspection of these short repeats revealed that both met1 and fwa showed a small, accessible peak which is inaccessible in wild type (Fig. 4G). We hypothesized that this small, accessible peak should become less accessible if DNA methylation at this region is reestablished. To test this hypothesis, we conducted ATAC-seq in an artificial zinc finger DMS3 (ZF-DMS3) transgenic plant line that targets DNA methylation to the short repeat region at the FWA locus (9). Indeed, we found that when the short repeat region regained CG and CHH methylations, it became inaccessible (Fig. 4G). In conclusion, these data further support the hypothesis that CG and non-CG methylations promote chromatin inaccessibility.
Fig. 4.
Connection between DNA methylation and chromatin accessibility. (A) Genome-wide pattern of chromatin accessibility variation in met1 and fwa. Variation in ATAC-seq (log2 mutant vs. Col-0) is depicted on the y axis. The box in each chromosome represents the pericentromeric heterochromatin region. (B) Overlap of peaks with more accessibility identified by ATAC-seq in met1 and fwa. Average distribution of CG (C), CHG (D), and CHH (E) methylation over peaks with higher accessibility in fwa. (F) Screenshot showing CG, CHG, and CHH methylations and ATAC-seq data in different mutants. (G) Screenshot of the FWA locus showing that targeted methylation by DMS3-ZF results in the gain of CG and CHH methylations and the loss of the open chromatin region upstream of FWA.
Increased Chromatin Accessibility Is Frequently Not Associated with Transcription Changes.
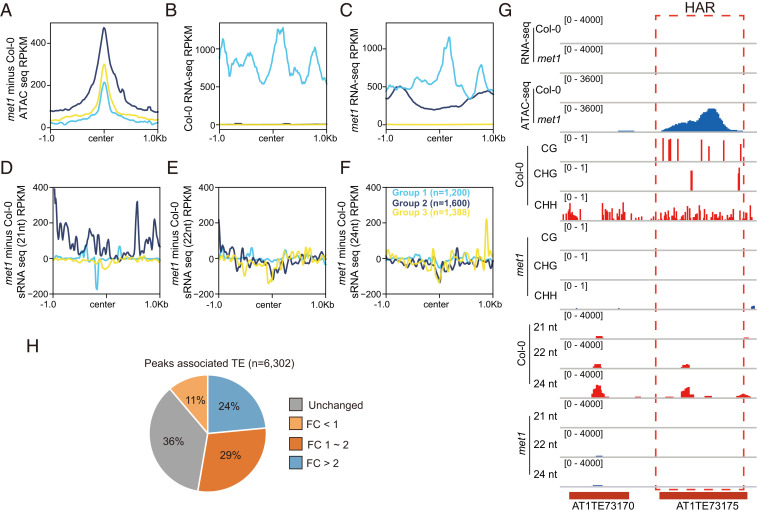
Given that increased chromatin accessibility is associated with loss of DNA methylation in many transcriptionally silent heterochromatin regions, a possibility is that increased accessibility is solely due to increases in transcription. To test this, we assessed whether the increased chromatin accessibility was correlated with an increase in transcription by analyzing RNA sequencing (RNA-seq) and small RNA-seq profiles at HARs, including 1 kb of flanking sequence, in met1 and Col-0. We observed that the 4,188 HARs in met1 could be classified into three groups based on the expression level of the locus affected (Fig. 5 A–C). For group 1 (n = 1,200), increased chromatin accessibility did not impact expression or siRNA levels, and this group already had high expression levels in wild-type plants (Fig. 5 A–F). For group 2 (n = 1,600), increased chromatin accessibility was accompanied by derepression of mRNA expression in met1 mutants. Small RNA-seq metaplots indicated that the derepression of group 2 was also accompanied by an increase in 21 nt small RNAs, suggesting that RDR6-mediated RNA interference was active in these regions (Fig. 5 A–F) (34). Most interestingly, group 3 (n = 1,388) loci showed no expression in either wild type or met1, and there was no increase in siRNA levels at these loci (Fig. 5 A–F). Instead, we observed that 21 nt, 22 nt, and 24 nt siRNAs were actually decreased in group 3 HARs, likely because RdDM activity is reduced at these sites (Fig. 5 D–F and SI Appendix, Fig. S9).
Fig. 5.
Chromatin accessibility and transcription. (A) Metaplot of ATAC-seq data over three groups of met1 HARs (n = 4,188). (B) Expression level of three groups of met1 HARs in Col-0. (C) Expression level of three groups of met1 HARs in met1. (D) The 21 nt siRNA variation over three groups of met1 HARs. (E) The 22 nt siRNA variation over three groups of met1 HARs. (F) The 24 nt siRNA variation over three groups of met1 HARs. (G) Screenshot showing a TE with increased chromatin accessibility but no expression in met1. (H) Expression fold change of TEs with HARs in met1 (n = 6,302).
We also examined the relationship between chromatin accessibility increases and transcriptional changes at annotated TEs, which are frequently up-regulated in met1 (35). A heat map of RNA-seq data indicated that some of the derepressed TEs in met1 gained open chromatin peaks (SI Appendix, Fig. S10). However, only 53% of HARs associated with TEs exhibited transcriptional up-regulation, while 36% remained unexpressed (Fig. 5 G and H). We also examined which families of TEs were enriched in silenced (no expression change, SI Appendix, Table S1) or derepressed (fold expression change > 2, SI Appendix, Table S2) TEs (Fig. 5H) for TEs overlapping with HARs. This showed a strong enrichment for a few families of TEs, including AtCOPIA38A and AtGP2N, showing transcriptional derepression, while many different families, with little enrichment for any particular TEs families, showed no expression change. Thus, many types of TEs gain chromatin accessibility in met1 but are not transcriptionally up-regulated.
Increased Chromatin Accessibility Is Associated with Changes in 3D Genome Architecture.
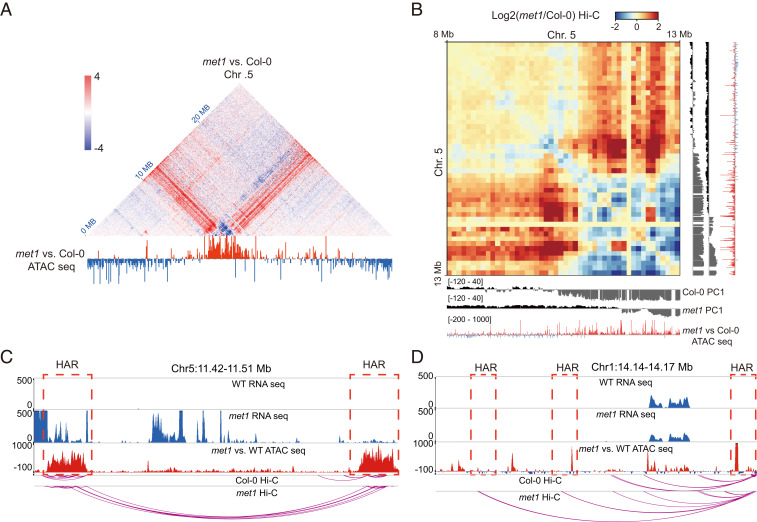
To test whether increased chromatin accessibility is associated with chromosome conformation variation, we performed high-throughput chromosome conformation capture (Hi-C) sequencing in representative mutants that display different levels of DNA methylation loss, including cmt2 cmt3 and ddcc, and combined this analysis with previously published Hi-C data for met1, cmt3, and ddm1 (36). Consistent with previous analyses, met1 and ddm1 showed similar patterns of chromosome conformation changes in heterochromatin regions from our reanalysis (SI Appendix, Fig. S11). Additionally, we observed that cmt3, cmt2 cmt3, and ddcc mutants showed similar patterns of chromosome conformation changes in heterochromatin regions, though not as dramatically as those in met1 and ddm1 (SI Appendix, Fig. S11). This result is consistent with the ATAC-seq changes in these mutants and indicates that CG methylation loss has a stronger impact on heterochromatin accessibility than non-CG methylation loss (Fig. 1 D and G). When comparing Hi-C and ATAC-seq data at higher resolution, we observed that the conformation variations detected by Hi-C were highly correlated with the chromatin accessibility variations detected by ATAC-seq, suggesting that the chromatin accessibility changes are accompanied by redistribution of chromatin interactions (SI Appendix, Fig. S12). For example, we observed that chromosome 5 exhibited more accessibility in heterochromatin regions in met1, and these regions showed more long-range interactions in Hi-C data (Fig. 6A). Furthermore, a comparison of the first principal component (PC1) values between Col-0 and met1 suggested that chromatin accessibility increases were associated with the conversion of regions from the inactive compartment to the active compartment in met1 (Fig. 6B). By closely inspecting heterochromatin regions, it was apparent that HARs in met1 showed increased interactions (Fig. 6C). This was also true for HARs that showed no up-regulation of transcription in met1 (Fig. 6D), suggesting that loss of methylation has a direct impact on chromatin accessibility and 3D genome organization.
Fig. 6.
Chromosome conformation signal redistribution in met1. (A) Redistribution of chromosome conformation in chromosome 5 in met1 related to chromatin accessibility variation. Red indicates level of Hi-C or ATAC-seq is higher in met1. Blue indicates level of Hi-C or ATAC-seq is lower in met1. (B) Heat map showing Hi-C variation of met1 in 100 kb resolution from 8 Mb to 13 Mb on chromosome 5. Tracks represent PC1 value in Col-0, PC1 value in met1, and ATAC-seq signal changes in met1 vs. Col-0. (C) Screenshot showing derepression of TE, increase of chromatin accessibility, and gain of long-range interactions in met1. (D) Screenshot showing increase of chromatin accessibility and gain of long-range interactions in met1.
Conclusion
Our profiling of 18 different DNA methylation mutants showed that reduction of DNA methylation caused increases in chromatin accessibility. We also found that chromatin accessibility increases were accompanied by local changes in chromosome conformation profiles with an increase of long-range chromatin interactions. CG methylation loss led to the most significant effect on chromatin accessibility, as observed in met1, fwa, and ddm1 backgrounds. In some cases, however, we also observed decreases in accessibility; for instance, we found that although gene body CG methylation is lost in met1 mutants, these regions became less accessible, likely due to increased CHG methylation.
Interestingly, we found that chromatin accessibility increases were not always associated with transcriptional derepression. For instance, we found large increases in chromatin accessibility at some TE sequences that were not associated with any transcription. These regions also showed increases in 3D chromatin interactions from Hi-C data. This suggests that DNA methylation, in addition to its role in regulating transcription, has a separate effect on chromatin accessibility and 3D genome architecture (see model in SI Appendix, Fig. S13). While we do not understand the mechanisms at play, it seems possible that methylation might recruit chromatin modifiers and/or nucleosome remodelers that directly impact the association of DNA with nucleosomes and thus affect DNA accessibility. This is consistent with findings in both plants and animals that DNA methylation regions have a higher level of nucleosome occupancy (23, 37–40). DNA methylation likely also regulates the binding of transcription factors and other DNA binding proteins to DNA, which likely contributes to chromatin accessibility changes.
Materials and Methods
Plant Materials.
All Arabidopsis plants used in this study were of the Col-0 ecotype and were grown at 22 °C under long-day conditions (16 h light, 8 h dark). The following Arabidopsis mutant lines were used: met1-3 (CS16394) (41), ddm1-2 (seventh-generation inbred) (42), fwa-4 epiallele (15), nrpe1 nrpd1 (crossing nrpd1-4 [SALK_083051] and nrpe1-11 [SALK_029919]) (43), nrpd1-4 (SALK_083051) (44), nrpe1-12 (SALK_033852), cmt2 cmt3 (crossing WISCDSLOX7E02 and SALK_148381), dms3-4 (SALK_125019C), drm1 drm2 (crossing drm1-2 [SALK_031705] and drm2-2 [SALK_150863]), drm1 drm2 cmt2 cmt3 (6), cmt2-7 (WISCDSLOX7E02), cmt3-11 (SALK_148381), drm3-1 (SALK_136439), idn2-1 (SALK_012288), idn2 idl1 idl2 (crossing SALK_075378 and SALK_012288) (21), suvr2-1 (SAIL_832_E07) (18), ago4-5 (45), frg1 frg2 (crossing SALK_027637 and SALK_057016) (18), and DMS3-ZF (9).
Whole-Genome Bisulfite Sequencing and Analysis.
Bisulfite sequencing reads were obtained from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) as accession numbers GSE62801 (18), GSE39901 (25), and GSE51304 (6) and mapped to the TAIR10 reference genome using bsmap (version 2.90) and allowing two mismatches and one best hit (-v 2 -w 1) (SI Appendix, Table S3) (46). Reads with three or more consecutively methylated CHH sites were considered to be nonconverted reads and were removed from the analysis. DNA methylation levels were calculated by #methylated cytosines/(#cytosines + #thymines). DMRs were called by hcDMR caller with P < 0.01 and at least 33 libraries (out of 54) used as supported controls for each bin for where the difference in CG, CHG, and CHH methylations is at least 0.4, 0.2, and 0.1, respectively (32). DMRs within 200 bp of each other were merged.
ATAC-Seq and Analysis.
Inflorescence tissues of 1-mo-old Col-0 and mutant plants were collected for nuclei extraction as described previously (47). Approximately 5 g of inflorescence tissue were collected in ice-cold grinding buffer and ground with an Omni International General Laboratory Homogenizer. Samples were filtered twice through a two-layer Miracloth and a 40 µm nylon mesh Cell Strainer (Fisher) and collected into a 50 mL tube. Samples were spun for 10 min at 3,000 g. After centrifugation, the supernatant was discarded, and the pellet was washed and resuspended with 25 mL of grinding buffer using a Dounce homogenizer. The centrifugation, wash, and resuspension steps were repeated twice. Then nuclei were resuspended with 0.5 mL of freezing buffer. Collected nuclei were used for Tn5 transposition reaction (Illumina) with 25 µL of 2× dimethylformamide mixed with 2.5 µL Tn5 and 22.5 µL nuclei suspension at 37 °C for 0.5 h and purified with a ChIP DNA Clean & Concentrator Kit (Zymo). ATAC-seq libraries were generated with Phusion High-Fidelity DNA Polymerase (New England Biolabs). We generated four biological replicates for wild-type Col-0 and at least two biological replicates for mutants. ATAC-seq reads adaptors were trimmed with trim_galore before mapping to the Arabidopsis thaliana reference genome TAIR10 using Bowtie (version 1.2.3, -X 2000 -m 1). Duplicated reads were deduplicated with SAMtools rmdup (version 1.9). Reads that aligned to chloroplast and mitochondrial DNA were filtered out for the following analyses. ATAC-seq peaks were called by HMMRATAC (version 1.2.9) with minimum length of 50 bp for each replicate, and consensus set of peaks of each replicates were merged by bedtools (version 2.26.0) intersect while allowing 10 base pairs of distance (28, 48). To call differential accessible peaks, the R package edgeR (version 3.30.0) was used (49).
Hi-C and Analysis.
Hi-C libraries were prepared according to previous protocols (36, 50). Previously published Hi-C data prepared by the same protocol were downloaded from the NCBI Sequence Read Archive (SRA) as accession number SRP043612 (36). Paired-end Hi-C reads were aligned to TAIR10 with HiC-Pro (version 2.11.1) (51). The whole-genome Hi-C heat map was converted with juicer_tools (version 1.13.02) and visualized with Juicebox (version 1.11.08) (52). Hi-C loops were called with analyzeHiC in Homer2 with 200 bp resolution and P value < 1E-10 (53). The WashU EpiGenome Browser version 46.2 (https://epgg-test.wustl.edu/browser/) was used to visualize Hi-C loops and ATAC-seq data (54). PC1 values of Hi-C data were calculated with Homer2 (53). Regional Hi-C visualization was performed by hicexplorer (version 3.4.3) (55).
met1 RNA-Seq and Small RNA-Seq Analysis.
RNA-seq reads were downloaded from the NCBI GEO as accession number GSE93584 (35). Cleaned short reads were aligned to reference genome TAIR10 by Bowtie2 (56), and expression abundance was calculated by RSEM with default parameters (57). Heat maps were visualized with the R package pheatmap (58). TEs that were located within an accessible peak or in the 1,000 bp sequence flanking each side of a peak were defined as accessible peak associated TEs. For small RNA-seq analysis, small RNA-seq reads were downloaded from the same study (35). Adaptor sequence was trimmed with cutadapt (version 2.5), and trimmed reads were mapped to the reference genome TAIR10 using Bowtie (version 1.2.3) with only one unique hit (-m 1) and zero mismatches (-v 0) (59).
Supplementary Material
Acknowledgments
We thank S.E.J. laboratory members for helpful discussions. High-throughput sequencing was performed at the Broad Stem Cell Research Center BioSequencing Core Facility at the University of California, Los Angeles, with the help of Mahnaz Akhavan. Work in the S.E.J. laboratory was supported by NIH Grant R35 GM130272. Work in the W.L. laboratory was supported by the Fundamental Research Funds for the Central Universities Grant K20200099. S.H.D. was supported by NIH Grant K99-GM135515.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023347118/-/DCSupplemental.
Data Availability.
The sequences reported in this paper have been deposited in the GEO database (accession no. GSE155503).
References
- 1.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson I. R., Jacobsen S. E., Epigenetic inheritance in plants. Nature 447, 418–424 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Chan S. W.-L., Henderson I. R., Jacobsen S. E., Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6, 351–360 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Finnegan E. J., Dennis E. S., Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 21, 2383–2388 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao X., Jacobsen S. E., Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12, 1138–1144 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Stroud H., et al. , Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E., Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Woo H. R., Dittmer T. A., Richards E. J., Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis. PLoS Genet. 4, e1000156 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallego-Bartolome J., et al. , Co-targeting RNA polymerases IV and V promotes efficient de novo DNA methylation in Arabidopsis. Cell 176, 1068–1082.e1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzke M. A., Kanno T., Matzke A. J., RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Wierzbicki A. T., Ream T. S., Haag J. R., Pikaard C. S., RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41, 630–634 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wierzbicki A. T., Haag J. R., Pikaard C. S., Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong X., et al. , DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat. Struct. Mol. Biol. 19, 870–875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wongpalee S. P., et al. , CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation. Nat. Commun. 10, 3916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson L. M., et al. , SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation. Nature 507, 124–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z.-W., et al. , The SET domain proteins SUVH2 and SUVH9 are required for Pol V occupancy at RNA-directed DNA methylation loci. PLoS Genet. 10, e1003948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong X., et al. , Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157, 1050–1060 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groth M., et al. , SNF2 chromatin remodeler-family proteins FRG1 and -2 are required for RNA-directed DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 111, 17666–17671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong X., et al. , Domains rearranged methyltransferase3 controls DNA methylation and regulates RNA polymerase V transcript abundance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, 911–916 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ausin I., Mockler T. C., Chory J., Jacobsen S. E., IDN1 and IDN2 are required for de novo DNA methylation in Arabidopsis thaliana. Nat. Struct. Mol. Biol. 16, 1325–1327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ausin I., et al. , INVOLVED IN DE NOVO 2-containing complex involved in RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 8374–8381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arents G., Burlingame R. W., Wang B.-C., Love W. E., Moudrianakis E. N., The nucleosomal core histone octamer at 3.1 A resolution: A tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. U.S.A. 88, 10148–10152 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chodavarapu R. K., et al. , Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rountree M. R., Selker E. U., DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity 105, 38–44 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Stroud H., Greenberg M. V., Feng S., Bernatavichute Y. V., Jacobsen S. E., Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152, 352–364 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ricci W. A., et al. , Widespread long-range cis-regulatory elements in the maize genome. Nat. Plants 5, 1237–1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tarbell E. D., Liu T., HMMRATAC: A hidden Markov ModeleR for ATAC-seq. Nucleic Acids Res. 47, e91 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potok M. E., et al. , Arabidopsis SWR1-associated protein methyl-CpG-binding domain 9 is required for histone H2A.Z deposition. Nat. Commun. 10, 3352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cokus S. J., et al. , Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deleris A., et al. , Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 8, e1003062 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., et al. , Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E1069–E1074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soppe W. J., et al. , The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 6, 791–802 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Nuthikattu S., et al. , The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oberlin S., Sarazin A., Chevalier C., Voinnet O., Marí-Ordóñez A., A genome-wide transcriptome and translatome analysis of Arabidopsis transposons identifies a unique and conserved genome expression strategy for Ty1/Copia retroelements. Genome Res. 27, 1549–1562 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng S., et al. , Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 55, 694–707 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Razin A., CpG methylation, chromatin structure and gene silencing—A three-way connection. EMBO J. 17, 4905–4908 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jimenez-Useche I., et al. , DNA methylation effects on tetra-nucleosome compaction and aggregation. Biophys. J. 107, 1629–1636 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wollmann H., et al. , The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 18, 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collings C. K., Anderson J. N., Links between DNA methylation and nucleosome occupancy in the human genome. Epigenetics Chromatin 10, 18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saze H., Mittelsten Scheid O., Paszkowski J., Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34, 65–69 (2003). [DOI] [PubMed] [Google Scholar]
- 42.Jeddeloh J. A., Stokes T. L., Richards E. J., Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Pontier D., et al. , Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19, 2030–2040 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herr A. J., Jensen M. B., Dalmay T., Baulcombe D. C., RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Greenberg M. V., et al. , Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics 6, 344–354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xi Y., Li W., BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10, 232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hetzel J., Duttke S. H., Benner C., Chory J., Nascent RNA sequencing reveals distinct features in plant transcription. Proc. Natl. Acad. Sci. U.S.A. 113, 12316–12321 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moissiard G., et al. , MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336, 1448–1451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Servant N., et al. , HiC-pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 16, 259 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durand N. C., et al. , Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S., et al. , Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536.e1522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X., et al. , Exploring long-range genome interactions using the WashU Epigenome Browser. Nat. Methods 10, 375–376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolff J., et al. , Galaxy HiCExplorer: A web server for reproducible Hi-C data analysis, quality control and visualization. Nucleic Acids Res. 46, W11–W16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kolde R., Pheatmap: Pretty Heatmaps. Available at http://CRAN.R-project.org/package=pheatmap (2012).
- 59.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences reported in this paper have been deposited in the GEO database (accession no. GSE155503).