Significance
Single molecule real-time (SMRT) sequencing theoretically offers the opportunity to directly assess certain base modifications of native DNA molecules without any prior chemical/enzymatic conversions and PCR amplification, using kinetic signals of a DNA polymerase. However, the kinetic signal changes caused by 5mC modification are extremely subtle. Hence, the robust genome-wide measurement of 5mC modification has not been achieved. We enhanced 5mC detection using SMRT sequencing by holistically analyzing kinetic signals of a DNA polymerase and sequence context for every base within a measurement window. We employed a convolutional neural network to train a methylation classification model, leading to genome-wide 5mC detection. The sensitivity and specificity reached 90% and 94%, with a 99% correlation of overall methylation level with bisulfite sequencing.
Keywords: third-generation sequencing, epigenetics, epigenomics, base modifications
Abstract
5-Methylcytosine (5mC) is an important type of epigenetic modification. Bisulfite sequencing (BS-seq) has limitations, such as severe DNA degradation. Using single molecule real-time sequencing, we developed a methodology to directly examine 5mC. This approach holistically examined kinetic signals of a DNA polymerase (including interpulse duration and pulse width) and sequence context for every nucleotide within a measurement window, termed the holistic kinetic (HK) model. The measurement window of each analyzed double-stranded DNA molecule comprised 21 nucleotides with a cytosine in a CpG site in the center. We used amplified DNA (unmethylated) and M.SssI-treated DNA (methylated) (M.SssI being a CpG methyltransferase) to train a convolutional neural network. The area under the curve for differentiating methylation states using such samples was up to 0.97. The sensitivity and specificity for genome-wide 5mC detection at single-base resolution reached 90% and 94%, respectively. The HK model was then tested on human–mouse hybrid fragments in which each member of the hybrid had a different methylation status. The model was also tested on human genomic DNA molecules extracted from various biological samples, such as buffy coat, placental, and tumoral tissues. The overall methylation levels deduced by the HK model were well correlated with those by BS-seq (r = 0.99; P < 0.0001) and allowed the measurement of allele-specific methylation patterns in imprinted genes. Taken together, this methodology has provided a system for simultaneous genome-wide genetic and epigenetic analyses.
DNA methylation is a biological process by which methyl groups are covalently added to DNA molecules. The most common form of this process occurs at the fifth position of the pyrimidine ring of cytosine: i.e., 5-methylcytosine (5mC). DNA methylation plays a number of essential roles in epigenetic regulation in cells, including genomic imprinting, X-chromosome inactivation, and carcinogenesis (1, 2). The most widely used method for detecting 5mC involves bisulfite treatment, followed by methods such as the PCR, or massively parallel DNA sequencing (i.e., bisulfite sequencing [BS-seq]) (3, 4). However, there are significant drawbacks to such bisulfite-based technologies. For instance, the harsh reaction conditions of bisulfite treatment could degrade the majority of the input DNA (5). Such DNA degradation renders long DNA molecule sequencing challenging. Another disadvantage is that bisulfite-induced DNA degradation preferentially acts on genomic regions enriched for unmethylated cytosines, resulting in an overestimation of global methylation and substantial variations at specific genomic regions among different bisulfite treatment protocols (4). Recently, a bisulfite-free method (called ten-eleven translocation (TET)-assisted pyridine borane sequencing, TAPS) for detecting 5mC has been published (6). This approach used milder conditions for converting 5mC to thymine, attempting to overcome the limitations present in BS-seq. However, TAPS involves multiple steps of enzymatic and chemical reactions, including TET oxidation, pyridine borane reduction, and PCR amplification. An undesired conversion efficacy occurring in any DNA conversion step would adversely affect the accuracy in 5mC analysis.
We envisioned that an ideal approach for measuring base modifications would be a method that could be directly applied to native DNA, without any chemical/enzymatic conversions of DNA and PCR amplification prior to sequencing. Third-generation sequencing technologies, such as nanopore sequencing (e.g., by Oxford Nanopore Technologies) and single molecule real-time (SMRT) sequencing (e.g., by Pacific BioSciences, PacBio), enable single molecule sequencing in real time, offering opportunities to explore such approaches for detecting base modifications. Liu et al. reported the feasibility of using nanopore sequencing to detect base modifications (7). However, the sequencing results were often accompanied by high sequencing errors, such as insertions and deletions (8). Such errors would cause the introduction of many loci missing the necessary signals for methylation analysis. Such a limitation of the current generation of nanopore sequencing may hamper the resolution of decoding methylation patterns at a single molecule level, especially for a large genome such as the human genome.
In contrast to nanopore sequencing that reads the DNA template at most twice (i.e., including both the Watson and Crick strands, also called two-dimensional [2D] sequencing), SMRT sequencing relies on the creation of circularized DNA templates that allow the molecules to be sequenced multiple times, thus greatly improving base calling accuracy (9). Base modifications would in theory influence the kinetics of a DNA polymerase during DNA synthesis. For example, the processivity of a DNA polymerase would be retarded at a thymine (T) incorporation opposite N6-methyladenine (6mA) on the template, leading to an increased time interval between the incorporations of the current and the next base (10). The pulse signals emitted from dye-labeled nucleotides could be used to monitor these changes in polymerization speed, thus enabling detection of base modifications (10). For example, the interpulse duration (IPD) (i.e., time interval between two successive fluorescence pulses) could be used to identify the N6-methylation of adenine (6mA). Unlike 6mA detection, to our knowledge, there is still no reported approach using SMRT sequencing to achieve practically meaningful accuracy for genome-wide detection of 5mC of native DNA molecules. The challenge for 5mC detection is caused by the subtle changes in the kinetics of a DNA polymerase by which a guanine is incorporated opposite 5mC. For example, Clark et al. reported that the detection rate of the 5mC using IPD at cytosines within CpG sites was low, ranging from 1.9 to 4.3% (11).
In this study, we attempted to develop an approach to achieve accurate detection of 5mC using SMRT sequencing, by holistically making use of sequence context and pulse signals associated with DNA polymerase kinetics, referred to as the holistic kinetic (HK) model. Based on the HK model, we utilized methylated and unmethylated datasets to train a convolutional neural network (CNN) to detect 5mC modifications.
Result
The Principle of the HK Model for 5mC Detection.
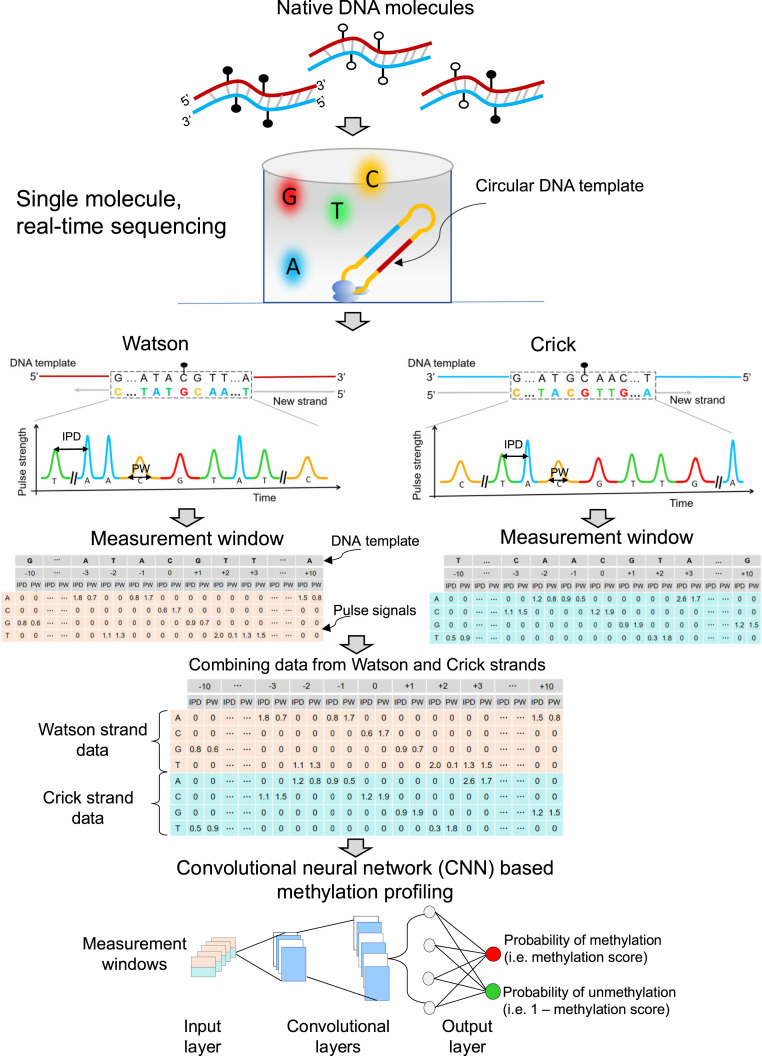
As shown in Fig. 1, double-stranded native DNA molecules were ligated with hairpin adapters, forming a topologically circular DNA template. Sequencing primers were annealed to circularized DNA templates via the complementary sites on hairpin adapters. Circularized DNA templates were bound to DNA polymerases, forming complexes each subsequently immobilized at the bottom of the zero-mode waveguides (ZMWs). A DNA polymerase molecule in a ZMW catalyzed the incorporation of nucleotides labeled with different fluorophores into the complementary strand of a DNA template. The kinetic changes of the DNA polymerase during polymerization can be monitored on a single-molecule basis.
Fig. 1.
Schematic 5mC detection using single molecule sequencing and the HK model. Double-stranded DNA molecules were ligated with hairpin adapters, forming circular DNA templates. DNA polymerase in a ZMW would incorporate nucleotides labeled with different fluorophores into the complementary strand of a DNA template, thus emitting different fluorescent colors indicating nucleotide information: for example, red, yellow, green, and blue colors represented G, C, T, and A, respectively. The light pulse signals were reflective of DNA polymerase kinetics, depending on the base modifications. Pulse signals included IPD and PW. For a cytosine subjected to methylation analysis, IPDs, PWs, and sequence context surrounding that cytosine were organized into a data matrix, referred to as a measurement window. For illustration purposes, the 10 nt upstream and downstream of the cytosine within a CpG site in question were presented as 5′-G[CCATGC]ATACGTT[GATGCA]A-3′ for the Watson strand. The bases in the brackets were left out (denoted by “…”) for the sake of simplicity. In this case, the measurement window size, including the interrogated cytosine in the middle, was 21 nt. For a position of -3 corresponding to the base of adenine (“A”), the IPD (1.8) and PW (0.7) associated with “A” were filled in the corresponding cells between a column of “-3” and a row of “A.” The other cells in the same columns were filled by “0.” The remaining IPDs and PWs related to the 21-nt sequence context were filled in that measurement window based on the same rule. The kinetic signals and sequence context originating from the Crick strand (‘5-T[TTGCAT]CAACGTA[TGCATG]G-3′) were also processed similarly. The measurement windows for two CpG sites complementary to each other (i.e., the Watson strand and the Crick strand) were combined for downstream analysis. A number of combined measurement windows originating from methylated and unmethylated cytosines were used for training a CNN, so as to differentiate methylated and unmethylated cytosines in test samples. CNN involved input layer, convolutional layers, and output layer. The measurement windows were fed into the input layer, followed by the process of convolutional layers; then, the probability of methylation (range: 0 to 1) for a CpG was generated through the output layer based on a sigmoid function. This approach was referred to as the “holistic kinetic (HK) model” (HK model).
Different fluorescent dyes were used to determine the base content. For example, red, yellow, green, and blue colors represented G, C, T, and A, respectively (Fig. 1). The light pulse signals emitted from fluorescently labeled nucleotides were reflective of DNA polymerase kinetics, depending on the base modifications. Thus, the appropriate use of pulse signals would make it possible to determine whether a cytosine was methylated or not. Pulse signals included the IPD, that represented the time duration between two consecutive base incorporations, and the pulse width (PW), that represented the time duration of the emission of fluorescent signal associated with a base incorporation. The pulse signals were associated with the sequence context in which the polymerization reaction was occurring. Herein, we developed an approach for determining DNA methylation by using pulse signals, including IPDs, PWs and the sequence context. Sequence context referred to the base compositions (A, C, G, or T) and the base orders in a stretch of DNA. For a cytosine within a CpG site, IPDs, PWs, and sequence context surrounding that cytosine were organized into a data matrix, referred to as a measurement window (Fig. 1). As a molecule of a circular form could be sequenced multiple times, the mean IPD and PW value of each nucleotide within the measurement window were used for downstream analysis.
We would hereby use the data processing of kinetic signals and sequence context from the Watson strand as an example. The position of an interrogated cytosine within a CpG site in a template DNA was denoted as position 0. For illustration purposes, the Watson and Crick strand templates comprising 10 nucleotides (nt) upstream and downstream of the cytosine in question were presented as 5′-G[CCATGC]ATACGTT[GATGCA]A-3′ and 5′-T[TTGCAT]CAACGTA[TGCATG]G-3′, respectively. The bases in the brackets were left out in Fig. 1 for the sake of simplicity. In this case, the measurement window size including the interrogated cytosine itself (in the center) was 21 nt. For the position of -3 corresponding to the base of adenine (“A”), the IPD (1.8) and PW (0.7) associated with “A” were filled in the intersection places (called cells) between a column of “-3” and a row of “A.” The other cells between a column of -3 and rows of cytosine (“C”), guanine (“G”), and thymine (“T”) were filled by “0.” Other IPDs and PWs related to the 21-nt sequence context were filled in corresponding cells in that measurement window. The kinetic signals and sequence context originating from the Crick strand were similarly processed.
As nearly all methylated CpG sites would occur on both strands symmetrically (12), we combined the measurement window flanking a CpG site from the Watson strand with that flanking the paired CpG site from the Crick strand, forming a combined measurement window for downstream analysis. We utilized a number of combined measurement windows originating from methylated and unmethylated cytosines, to train a CNN. The trained CNN model would then be used for differentiating methylated and unmethylated cytosines in test samples. This analytic framework for 5mC detection was holistically taking advantage of kinetic signals of DNA polymerase across individual nucleotides within a measurement window, as well as sequence context (i.e., nucleotide information and orders), and was thus referred to as the “holistic kinetic (HK) model” (HK model). The HK model involved an input layer, convolutional layers, and an output layer. Data needed for the HK model (i.e., sequence context, IPD, and PW) from each measurement window were entered into the input layer and then processed by the convolutional layers (Fig. 1). The output, based on a sigmoid function, represented the probability of methylation, referred to as a methylation score for the cytosine in a CpG site, ranging from 0 to 1. As it was a binary classification, the probability of the cytosine within a CpG site being unmethylated would be 1 – methylation score. The larger the methylation score, the more likely a CpG site would be methylated. Based on the receiver operator characteristic (ROC) curve, a methylation score threshold was defined for classifying the methylation status for each CpG site residing within the analyzed DNA molecule. The details regarding the training and testing procedures are described in Materials and Methods.
Training the HK Model for 5mC Detection Using Amplified and M.SssI-Treated DNA.
To demonstrate the feasibility and performance of using the HK model to determine the methylation states in a genome-wide fashion, the model was trained and validated using SMRT sequencing datasets, including an unmethylated dataset (i.e., the negative dataset) and a methylated dataset (i.e., the positive dataset). The unmethylated dataset contained the sequencing results from amplified DNA that was prepared via whole genome amplification (WGA) (denoted as the WGA dataset). The use of unmodified nucleotides in the WGA resulted in the amplified DNA containing nearly no base modifications (with the exception of the small amount of input genomic DNA). The methylated dataset contained the sequencing results from DNA treated by the M.SssI (a CpG methyltransferase, isolated from a strain of Escherichia coli which contains the methyltransferase gene from Sprioplasma sp.strain MQ1, would methyalte all CpG sites in a double-stranded DNA) prior to sequencing (denoted as the M.SssI-treated dataset). M.SssI methyltransferase rendered CpG sites methylated (13). Among the sequenced CpG sites within the dataset of the M.SssI-treated sample, half was used for training the HK model. Within the WGA dataset, an equal number of CpG sites were randomly sampled for training the HK model. The remaining half of the CpG sites within the dataset of the M.SssI-treated sample and the same number from the WGA dataset were used for validation of the model. In this study, we used 1) the Sequel I sequencing kit 3.0 (Sequel sequencing kit 3.0 as its official name) on the PacBio Sequel I sequencer, and 2) the Sequel II sequencing kit 1.0 and Sequel II sequencing kit 2.0 on the PacBio Sequel II sequencer, obtaining WGA and M.SssI-treated DNA datasets for evaluating the HK model across different reagents and sequencers in this study.
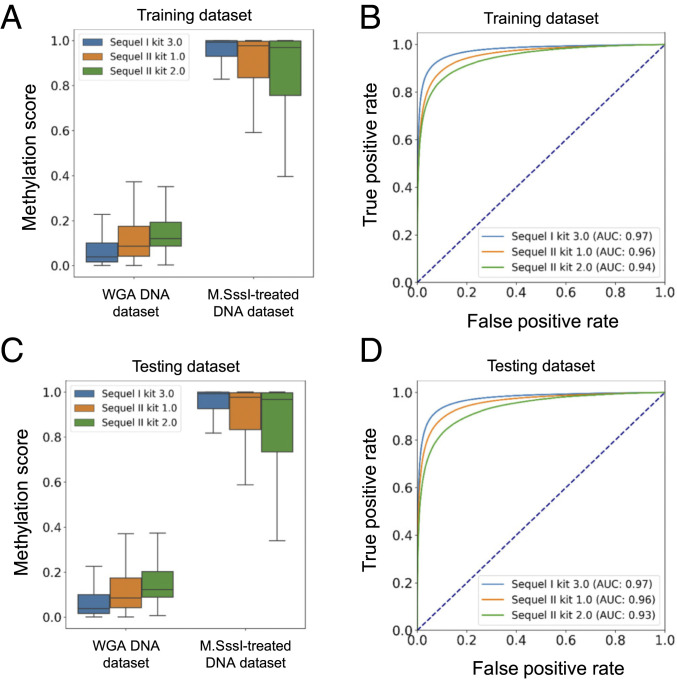
For the Sequel I sequencing kit 3.0, we used 328,233 CpG sites from an M.SssI-treated DNA sample (fully methylated) and 328,233 CpG sites from a WGA sample (fully unmethylated) to train the HK model. The methylation scores from the M.SssI-treated dataset (median: 0.99; interquartile range [IQR]: 0.93 to 1.0) were separated from the results from the WGA dataset (median: 0.04; IQR: 0.02 to 0.1) (P value: < 0.0001, Mann–Whitney U test) (Fig. 2A). The area under the ROC curve (AUC) was 0.97 (Fig. 2B).
Fig. 2.
The HK model training and validation using datasets generated from amplified DNA and M.SssI-treated DNA. (A) Box plots for methylation scores in training datasets derived from the whole genome amplified DNA (WGA DNA dataset) and M.SssI-treated DNA (M.SssI-treated DNA dataset) on the basis of different sequencing kits including Sequel I sequencing kit 3.0 and Sequel II sequencing kit 1.0 and 2.0. (B) ROC curves for training datasets on the basis of different sequencing kits. (C) Box plots for the methylation scores in testing datasets. (D) ROC curves for testing datasets.
We further analyzed the SMRT sequencing datasets prepared by different sequencing kits. The separation between WGA and M.SssI-treated datasets in terms of the methylation score was also clearly observable in both training datasets prepared by the Sequel II kit 1.0 and the Sequel II kit 2.0 (Fig. 2A), with the use of 11,272,552 and 325,780 CpG sites for the two datasets. The AUC values were 0.96 and 0.94 for datasets prepared by the Sequel II sequencing kit 1.0 and 2.0, respectively (Fig. 2B).
Performance of the HK Model for 5mC Detection Using Amplified and M.SssI-Treated DNA.
Fig. 2 C and D shows the performance of the HK model in the testing datasets. The AUC values were found to be 0.97, 0.96, and 0.93 for the Sequel I sequencing kit 3.0 and the Sequel II kit 1.0 and 2.0, respectively. These results suggested that the HK model could accurately determine the methylation states. The HK model was applicable to data produced by different sequencing kits and sequencers as long as the training and testing processes were based on the same experimental conditions.
The AUC values based on the HK model (range: 0.93 to 0.97) were much greater than the AUC values (0.53 to 0.67) based on IPD or PW values at CpG sites for all three testing datasets (SI Appendix, Fig. S1 A–C), suggesting that the HK model greatly outperformed the conventional methods using kinetic signals at a queried base.
We defined a methylation score cutoff for classifying the methylation status of CpG sites. We selected 0.5 as the methylation score cutoff, which was the point close to the top-left corner of each ROC curve in the training datasets. A CpG site with a methylation score above 0.5 was classified as methylated; otherwise, it would be classified as unmethylated. We could achieve 94% specificity and 90% sensitivity for datasets generated by Sequel I sequencing kit 3.0. For datasets generated by Sequel II sequencing kit 1.0, the specificity and sensitivity were 92% and 87%, respectively. For datasets generated by Sequel II sequencing kit 3.0, the specificity and sensitivity were 89% and 83%, respectively.
In addition to the CNN model, we attempted to evaluate the performance of 5mC detection using a hidden Markov model (HMM) for the sample BC01 with high-depth sequencing coverages by SMRT-seq (SI Appendix, Table S2). As a result, we found that the performance of HMM (83% sensitivity and 84% specificity) appeared to be worse than that of the HK model (87% sensitivity and 92% specificity). The details about the implementation of HMM are described in SI Appendix, Methods and Materials (SI Appendix, Figs. S2 and S3).
Effect of Window Size and Subread Depth on the Performance of 5mC Detection.
To study how the window size of the measurement window and subread depth affected the performance of the HK model, we varied the measurement window sizes, covering 1, 3, 5, 7, 9, 11, 21, 31, 41, 51, and 61 nt. For a particular measurement window size, we further varied the subread depths, covering 1, 2, 3, 4, 5, 10, 15, 20, 25, and 30×. The HK model was first trained using a training dataset comparing 100,000 measurement windows each from the WGA and M.SssI-treated datasets. For each combination of window size and subread depth, we randomly sampled 2,000 CpG sites from a full dataset that did not overlap with the training dataset, thus forming a testing dataset.
We analyzed datasets generated by the Sequel II sequencing kit 1.0 for which the subread depth was at 10×. The AUC was found to be 0.70 using a measurement window size of 1 (SI Appendix, Fig. S4A). As we increased the measurement window size to 3, 7, 21, and 31 nt, the AUC value increased to 0.84, 0.90, 0.93, and 0.93, respectively (SI Appendix, Fig. S4A). Besides, when applying a measurement window size of 21 nt, the AUC was found to be 0.75 using a subread depth of 1 (SI Appendix, Fig. S4B). As we increased the subread depth to 5 and 10, the AUC value was observed to increase to 0.85 and 0.93 (SI Appendix, Fig. S4B), respectively. These data suggested that the performance of the HK model could be improved by adjusting the measurement window size and subread depth requirement. SI Appendix, Fig. S4C shows that the performance of differentiating methylated cytosines from unmethylated cytosines reached a plateau with an AUC of 0.96, using a window size of 21 nt and a subread depth of 30×. The use of a measurement window size of 21 nt at a subread depth of 10× also allowed us to achieve an AUC of 0.93. To balance the number of molecules suitable for downstream analysis and accuracy, we adopted the window size of 21 nt and a subread depth of at least 10× as a default setting in this study. There are 28.2 million CpG sites in a human haploid genome, resulting in 28.2 million measurement windows. Of those, 69.4% of the 21-nt measurement windows harbored one CpG site. There were 21.2% and 6.5% of measurement windows that contained two and three CpG sites, respectively. Only 3% of the measurement windows contained more than three CpG sites. Thus, we believed that the majority of the measurements would not be affected by the potential interactions of kinetic signals caused by two nearby CpG sites residing within the same measurement window.
The results related to datasets generated by the Sequel I sequencing kit 3.0 and Sequel II sequencing kit 2.0 (SI Appendix, Figs. S5–S7) led to a consistent conclusion that the performance of the HK model would depend on the window sizes and subread depths. The increase of subread depth would generally increase AUC values in differentiating methylated and unmethylated cytosines. The measurement window size of 21 nt was a robust parameter for methylation analysis as such a window size appeared to reach a plateau value at a subread depth of 30× (SI Appendix, Fig. S7 A and B). Interestingly, the Sequel I sequencing kit 3.0, a relatively early reagent kit, paradoxically appeared to be superior to the other two updated kits for methylation analysis across a range of window sizes and subread depths. For example, with a measurement window size of 21 nt and a subread depth of 30, the AUC values were 0.98, 0.96, and 0.94, respectively, for the Sequel I sequencing kit 3.0 (SI Appendix, Fig. S7A) and Sequel II sequencing kit 1.0 (SI Appendix, Fig. S4C) and 2.0 (SI Appendix, Fig. S7B).
Effect of the Number of Sequence Contexts on the Performance of 5mC Detection.
There were a total of 28.2 million CpG sites in the human reference genome (University of California Santa Cruz hg19). Among them, a total of 20.7 million 21-nt sequence contexts centered on a CpG site were found. As shown in SI Appendix, Table S1, among the 20.7 million contexts, the percentages of sequence contexts used in the training of the HK model were 2.7%, 32.7%, and 1.3% for the datasets prepared by Sequel I kit v3, Sequel II kit v1, and Sequel II kit v2, respectively. Because we had obtained a much higher sequencing throughput for the training sample prepared by the Sequel II kit v1, there were many more sequence contexts empirically covered in that sample. Each testing dataset contained a similar amount of sequence contexts to its corresponding training dataset. Notably, even though a variable number of contexts were covered across different datasets during the training and testing processes, the performance of the resultant HK models appeared not to be varied much, with the area under the receiver operating characteristic curve (AUC) values ranging from 0.93 to 0.97.
To further investigate how the number of sequence contexts would affect the performance of the HK model, we carried out downsampling analysis of sequence contexts by randomly sampling 1,000, 5,000, 50,000, 100,000, 200,000, 300,000, 400,000, 500,000, 1,000,000, 5,000,000, and 10,000,000 sequence contexts. SI Appendix, Fig. S4D shows that the performance of the HK model progressively improved as the number of sequence contexts increased. For example, with the use of 1,000 sequence contexts, the AUC was 0.73 whereas the AUC increased to 0.95 with 300,000 sequence contexts. The plateau of the performance was reached at 300,000 sequence contexts. In other words, 1.45% of the 21-nt sequence contexts in the genome (i.e., 300,000/20.7 million × 100%) were sufficient to train the HK model well for distinguishing the methylated and unmethylated cytosines at CpG sites. We conjectured that many sequence contexts might have a similar impact on the kinetic features of DNA polymerase during SMRT sequencing. Therefore, there may be a certain degree of redundancy in genomic sequence contexts in such training.
The Analysis of Divergent Methylation States Using Human–Mouse Hybrid Fragments.
As the aforementioned validation process relied on WGA and M.SssI-treated DNA samples that were in theory homogenously methylated or unmethylated for a fragment, we further tested whether the HK model could be generalizable to fragments carrying heterogeneous methylation states (i.e., a fragment concurrently harboring methylated and unmethylated CpG sites). To this end, we generated two datasets comprising human–mouse hybrid fragments on the basis of restriction digestion (HindIII and NcoI, both being 6-base cutters) and DNA ligation (Materials and Methods), as illustrated in SI Appendix, Fig. S8. The first dataset contained the hybrid DNA molecules for which the human part was methylated by M.SssI and the mouse part was rendered unmethylated by WGA, named the human (meth)–mouse (unmeth) dataset. The second dataset contained the hybrid DNA molecules with opposite methylation patterns: i.e., the human part was unmethylated and the mouse part was methylated, named the human (unmeth)–mouse (meth) dataset. We used the Sequel II sequencer together with the Sequel II sequencing kit 1.0 to sequence sample H01 and H02, obtaining 5.7 million (median size: 1.3 kb; median subread depth: 10×) and 3.3 million (median size: 1.2 kb; median subread depth: 10.5×) molecules for the human (meth)–mouse (unmeth) and human (unmeth)–mouse (meth) datasets, respectively.
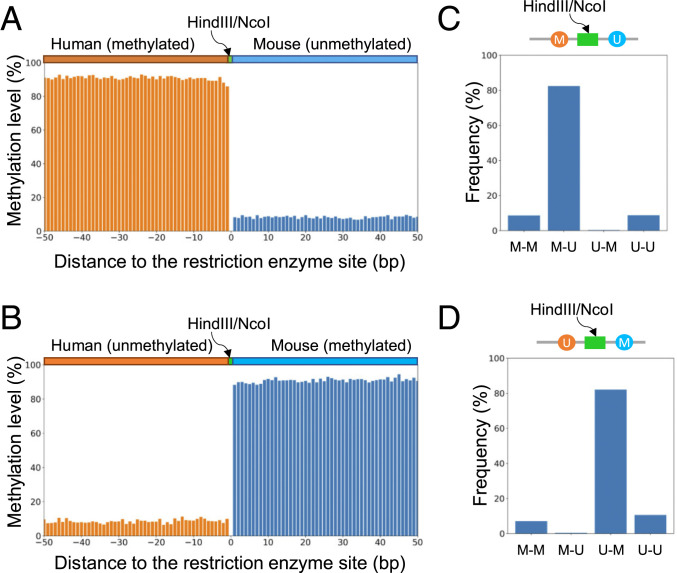
We applied the HK model trained from datasets with homogenous methylation states to determine the methylation states across CpG sites for each human–mouse hybrid DNA molecule in the human (meth)–mouse (unmeth) dataset. We pooled a total of 104,896 CpG sites within 50 base pairs (bp) upstream and downstream to restriction sites, according to relative positions (i.e., distances) to the nearest base of a restriction enzyme recognition site (HindIII or NcoI). Positions originating from the human part of a molecule were assigned as upstream (negative values) while those from the mouse part were assigned as downstream (positive values). The percentage of CpG sites determined to be methylated was deemed as the methylation level. Fig. 3A shows that the human part in this human (meth)–mouse (unmeth) dataset was shown to be methylated with a methylation level range of 85.9 to 93.0% whereas the mouse part was shown to be unmethylated with a methylation level range of 6.7 to 9.6%. Such patterns were found to be opposite in the human (unmeth)–mouse (meth) dataset (Fig. 3B).
Fig. 3.
Methylation pattern analysis for human–mouse hybrid fragments. (A) Methylation levels across CpG sites from human–mouse hybrid fragments present in the human (meth)–mouse (unmeth) dataset. CpG sites were pooled together according to the relative distance to the nearest base of a restriction cutting site (HindIII or NcoI). (B) Methylation levels across CpG sites from human–mouse hybrid fragments present in the human (unmeth)–mouse (meth) dataset. (C) Methylation patterns for the two nearest CpG sites immediately flanking a restriction cutting site (HindIII or NcoI) for human–mouse hybrid fragments present in the human (meth)–mouse (unmeth) dataset. (D) Methylation patterns for two CpG sites immediately flanking a restriction cutting site (HindIII/NcoI) for human–mouse hybrid fragments present in the human (unmeth)–mouse (meth) dataset. “M-M” represents that the first and second CpG sites in the human and mouse parts are both methylated. “M-U” represents that the first CpG site in the human part is methylated while the second CpG site in the mouse part is unmethylated. “U-M” represents that the first CpG site in the human part is unmethylated while the second CpG site in the mouse part is methylated. “U-U” represents that the first and second CpG sites in the human and mouse parts are both unmethylated.
We furthermore analyzed the two nearest CpG sites flanking restriction enzyme sites, evaluating the effect of the potential interactions of kinetic signals of neighboring CpG sites on the performance of the HK model. As the restriction enzyme recognition sites were 6 bp in length and did not contain CpG sites, the least number of nucleotides between two nearest CpG sites surrounding cutting sites was restricted to 6 bases (not including the 4 bases within the CpG sites) (SI Appendix, Fig. S9 A and B). The greatest number of nucleotides between the two nearest CpG sites was 17 (i.e., 21 − 4) because the window size of 21 nt was taken into account for this evaluation. For the human (meth)–mouse (unmeth) dataset, 82.4% of these two nearest CpG sites harbored the “M-U” pattern (Fig. 3C), indicating that the first cytosine within a CpG site from the human part was methylated (M) while the second cytosine within a CpG site was unmethylated (U). These results suggested that the HK model could robustly decode methylation for each CpG site in a DNA molecule even with divergent methylation states. Such a conclusion was further evidenced by the fact that 82.0% of these two nearest CpG sites harbored the “U-M” pattern in the human (unmeth)–mouse (meth) dataset (Fig. 3D).
Methylation Determination Using the HK Model for Biological Samples.
To further validate whether the trained HK model could be used for analyzing real biological samples, we sequenced 11 tissue DNA samples using the Sequel II sequencer together with the Sequel II sequencing kit 1.0 (PacBio) (SI Appendix, Table S2). We obtained a median of 6 million sequenced molecules, with a median of 5.9 kilobases (kb) in size. The median subread depth was 4.3× (IQR: 3.6 to 6.7×). Each sample was also sequenced by BS-seq to a median of 50 million paired reads. The methylation states across CpG sites were determined by the Methy–Pipe software (14).
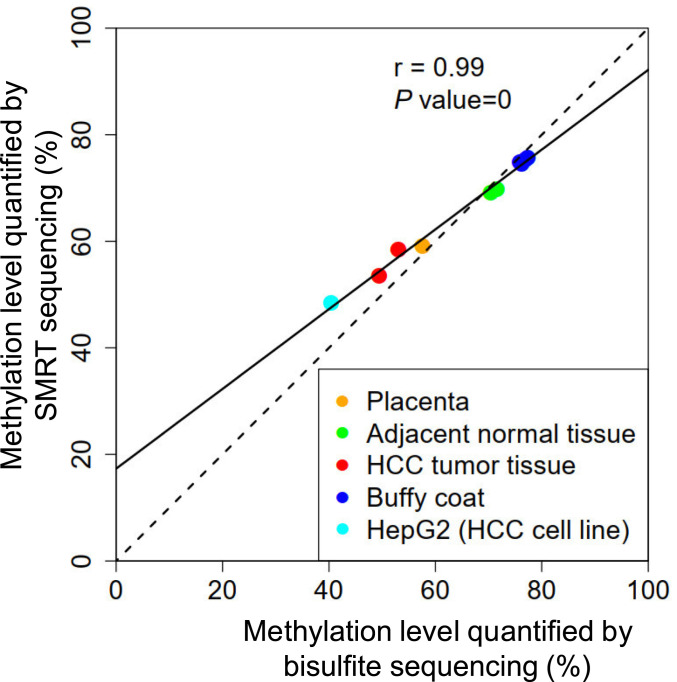
We compared the overall methylation levels between two measurements by the HK model and BS-seq. The overall methylation levels were defined as the percentage of CpG sites determined to be methylated among all sequenced CpG sites. Fig. 4 shows that the overall methylation levels across samples analyzed by the HK model correlated well with those quantified by BS-seq (r = 0.99; P value < 0.0001). The methylation levels concerning placental DNA (sample PL01), hepatocellular carcinoma (HCC) tumor tissue DNA (HCC01 and HCC02), and HepG2 cell line DNA were lower (range: 48.4 to 58.4%) than the counterparts of adjacent nontumoral DNA (NT01 and NT02) and buffy coat DNA (BC01 to BC05) (range: 69.0 to 75.7%). The hypomethylation observed in placental DNA, HCC tumor tissue DNA, and HepG2 cell line DNA was in agreement with previous studies (15–18), further suggesting the robustness of the HK model for differentiating methylated and unmethylated cytosines in native DNA molecules from various biological samples.
Fig. 4.
Correlation of overall methylation levels quantified by BS-seq and the HK model. Each dot represents one sample.
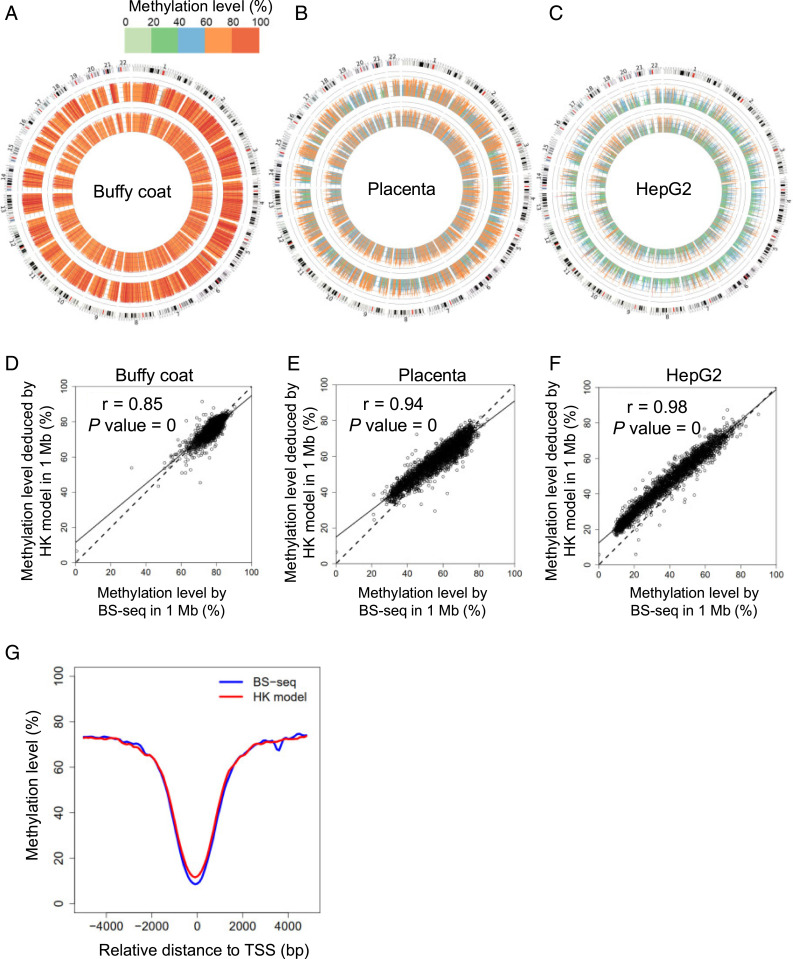
In addition to the methylation levels in a whole genome, we further analyzed the methylation levels at 1-megabase (Mb) resolution. From Circos plots (19) showing the analysis for buffy coat DNA, placental DNA, and HepG2 cell line DNA samples (Fig. 5 A–C), the methylation level profile across 1-Mb genomic bins deduced by the HK model (Fig. 5 A–C, inner ring) was highly concordant with that determined by BS-seq (Fig. 5 A–C, outer ring). The concordance between the HK model and BS-seq was further evidenced in the scatter plots (Fig. 5 D–F), showing a correlation coefficient of 0.85, 0.94, and 0.98 for buffy coat DNA, placental DNA, and HepG2 cell line DNA samples, respectively. The results for HCC tumor samples and their paired adjacent nontumoral tissue samples are shown in SI Appendix, Figs. S10 and S11.
Fig. 5.
Methylation levels quantified by BS-seq and the HK model at 1-Mb resolution. Circos plots show methylation levels determined by the HK model (inner ring) and BS-seq (outer ring) across different 1-Mb regions of human genome for buffy coat (A), placenta (B), and the HepG2 HCC cell line (C). Scatter plots show correlations of methylation level in each 1-Mb genomic region determined by the HK model and BS-seq for buffy coat (D), placenta (E), and the HepG2 HCC cell line (F). (G) Methylation patterns surrounding TSSs.
It was well known that lower methylation densities would be observed in regions near transcription start sites (TSSs) (12). Notably, a “valley pattern” concerning methylation levels surrounding TSS regions was indeed seen in results determined by the HK model, which were confirmed in the BS-seq results (Fig. 5G).
Methylation Correlation at Single-Base Resolution between the HK Model and BS-Seq.
To compare the correlation at single-base resolution, we calculated the methylation level for each CpG site covered by at least 20 sequenced molecules in both the SMRT-seq and BS-seq results for the sample BC01. As there were a large number of CpG sites, a smoothed scatter plot was used for visualizing the correlation of methylation levels deduced by the HK model and BS-seq (SI Appendix, Fig. S12). A Pearson’s correlation coefficient of 0.8 (P value < 0.0001) was observed between the HK model and BS-seq.
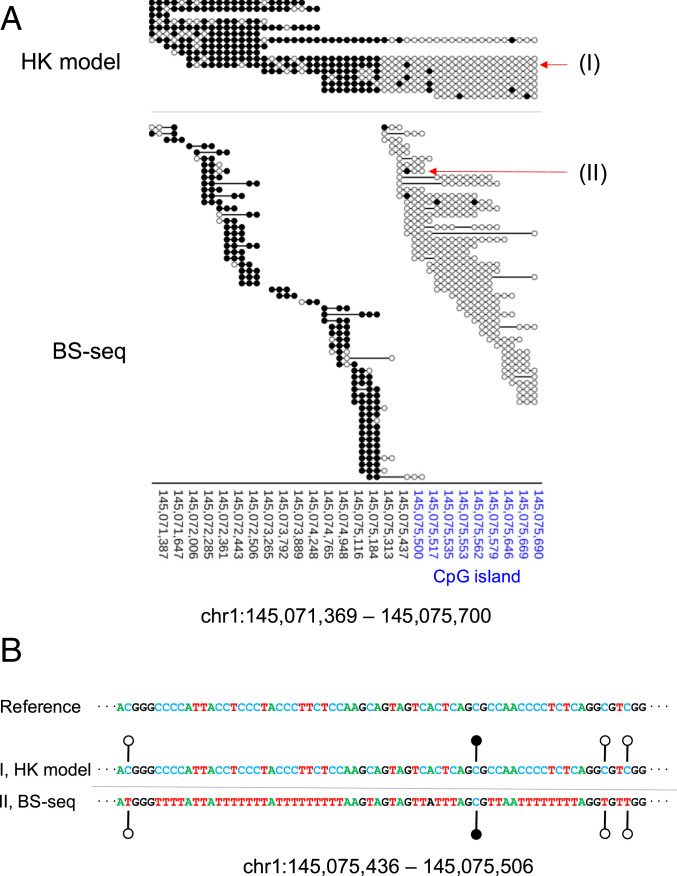
A representative region (chr1: 145,071,369 to 145,075,700) with a relatively high sequencing depth was used for illustrating the comparison between the HK model and BS-seq at single-base resolution. As shown in Fig. 6A, 16 sequenced molecules were from this region, which were subjected to analysis by the HK model, with a median read length of 3,103 nt (range: 1,484 to 8,490 nt). The portion of a molecule overlapping with the CpG island (CGI) region was mainly determined to be unmethylated whereas the portion of a molecule outside the CGI region (i.e., CGI shore) tended to be methylated (Fig. 6A). Such distinct patterns were confirmed in the result by BS-seq, with 102 resulting sequences (median size: 163 nt; range: 30 to 599 nt). Fig. 6B illustrates that the HK model could provide full genotype information, including A, C, G, and T (i.e., four-letter information) and methylation states at CpG dinucleotides. However, for BS-seq, the genotype information was mainly restricted to three-letter information (i.e., A, G, and T).
Fig. 6.
Methylation patterns at single-base resolution. (A) Methylation patterns for the region chr1: 145,071,369 to 145,075,700 overlapping the CGI. The genomic coordinates of the CGI are highlighted in blue. “(I)” and “(II)” represent two sequence reads that are used to highlight the difference in the readout between the HK model and BS-seq. (B) Genetic and epigenetic information generated using the HK model (denoted “I”) and BS-seq (denoted “II”). For the ease of visualization, A, C, T, and G are denoted in different colors. For the HK model, the original genomic sequence and methylation information are directly and simultaneously read out from the results. For BS-seq, the interpretation of a “TG” readout (i.e., whether the T means an unmethylated cytosine, or whether a T is present at that position in the genome) can only be made after comparison with the reference genomic sequence. Filled lollipops, methylated C; unfilled lollipops, unmethylated C.
Methylation Determination for Representative Imprinted Genes.
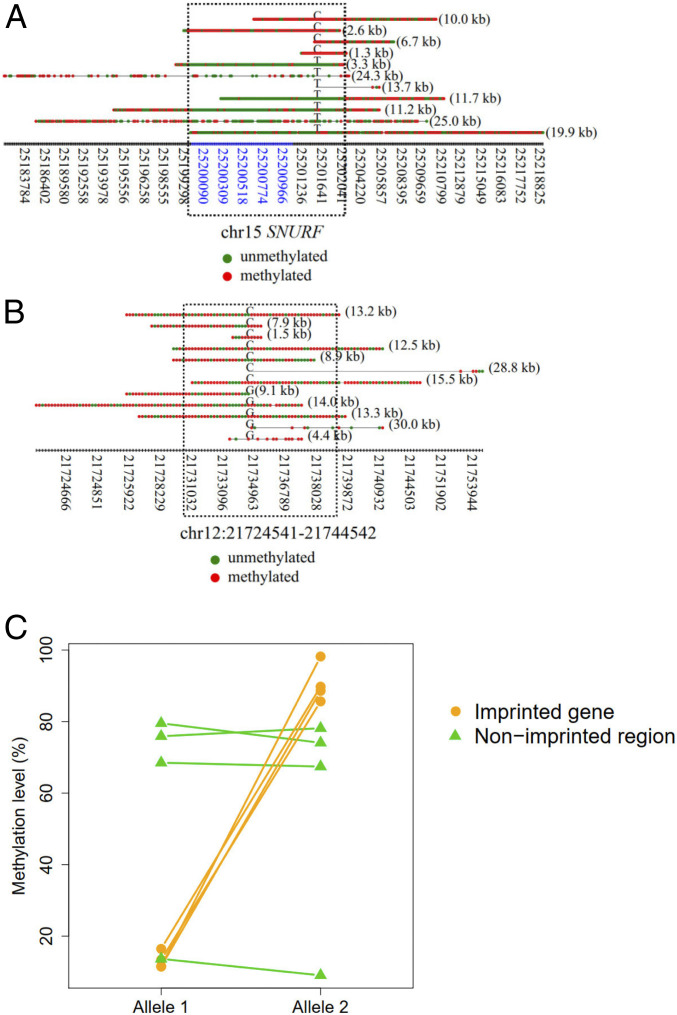
DNA methylation is important for establishing imprinting marks on either paternal or maternal alleles (20), often displaying allele-specific methylation patterns. Therefore, we expected that the SMRT sequencing would enable analysis of allele-specific methylation patterns in a single molecule resolution using the HK model. We selected four representative imprinted genes, SNURF, PLAGL1, NAP1L5, and ZIM2, which were commonly imprinted across various tissues reported in a study (21). We applied the HK model to determine the methylation states of those molecules overlapping with these four imprinted genes in the sample BC01, as this sample had a relatively high sequencing depth (SI Appendix, Table S2). As an example, the imprinted gene, SNURF, displayed allele-specific methylation patterns spanning a known imprinted control region (22) ranging from 25,200,004 to 25,201,976 on chromosome 15 (Fig. 7A). The fragments linked to the “C” allele were methylated on that imprinted control region whereas the fragments linked to the “T” allele were unmethylated. The differential methylation patterns between alleles were generally not observable in nonimprinted regions, such as a region (chr12: 21,729,541 to 21,739,542) randomly selected from the genome (Fig. 7B). In contrast to nonimprinted regions, all four imprinted genes had differentially methylated regions between two alleles (Fig. 7C). SI Appendix, Fig. S13 shows the methylation patterns for each DNA molecule covering the other three imprinted genes (NAP1L5, ZIM2, and PLAGL1), all exhibiting allele-specific methylation patterns.
Fig. 7.
Methylation patterns for each single molecule derived from imprinted regions. (A) An example showing the methylation patterns for each DNA molecule in association with imprinted regions of gene SNURF. The x axis indicates the coordinates of CpG sites. The coordinates highlighted in blue indicate CGIs. Red dots indicate methylated CpG sites. Green dots indicate unmethylated CpG sites. The alphabet embedded among each horizontal series red and green dots (i.e., CpG sites) indicates the allele at the SNP site. The numbers in parentheses on the right of each horizontal series of dots indicate the size of a fragment. The dashed rectangle indicates the regions overlapped with the known imprinting control region. (B) An example showing the methylation patterns for each DNA molecule originating from nonimprinted regions. The dashed rectangle indicates a region surrounding the SNP site highlighted for comparison. (C) Methylation levels between imprinted and nonimprinted regions.
Discussion
We have developed an approach for holistically making use of kinetic signals and sequence context to realize the genome-wide detection of cytosine methylation by SMRT sequencing. The robustness of the approach has allowed us to decipher 5mC patterns in the human genome. Several previous studies attempted to use SMRT sequencing to detect base modifications. However, practically meaningful accuracy for genome-wide detection of 5mC using SMRT sequencing has not previously been achieved. For example, Clark et al. reported that the detection rate of the 5mCs in a native DNA using the IPD metric was very low, ranging from 1.9 to 4.3% (11). It was concluded that the prior conversion of 5mC to 5-carboxylcytosine (5caC) using Tet proteins would be required to improve the sensitivity of 5mC (11) as the change of IPD induced by 5caC was much greater than 5mC. In a more recent report by Blow et al., the IPD ratio-based method was used to detect the base modifications in 217 bacterial and 13 archaeal species with 130-fold read coverage per organism (23). Among all the base modifications they identified, 5mCs only accounted for 5% and was much lower than expected (11), suggesting the low sensitivity of single-molecule real-time sequencing for detecting 5mC. Hence, we believe that our methodology has addressed an unmet need in the field.
In this study, we developed a methodology of utilizing kinetic features of DNA polymerase for every base within the measurement window (e.g., 10 nt upstream and downstream flanking a CpG site in question). The measurement window allowed the representation of various kinetic signals in combination with sequence context in a way analogous to an image with different pixel patterns: i.e., in the form of a 2D matrix. Thus, CNN, a class of deep learning algorithms, could be used for differentiating methylated and unmethylated cytosines after training, leading to a robust performance (AUC: 0.97). To the best of our knowledge (10, 24–26), there was no prior study reporting the simultaneous consideration of IPD, PW, and sequence context for base modification analysis. The lack of an effective way of using kinetic signals and sequence context might be one reason why the previously reported performance of 5mC detection had not achieved the practically useful accuracy, hampering translation of SMRT sequencing to real-word applications of 5mC detection of native DNA.
Using the HK model, we have dramatically improved the detection rate for 5mC up to 90% at a specificity of 94% in the validation datasets generated from amplified DNA and M.SssI-treated DNA. Several factors were considered to be informative for further improvement of 5mC detection. First, the actual efficiency of CpG methyltransferase (M.SssI) would determine the likelihood of being methylated for CpG sites in the M.SssI-treated dataset. If the methylation efficacy of M.SssI was 90%, 10% of CpG sites that were unmethylated would be falsely considered as methylated CpG sites in the training of the HK model, perhaps leading to the detection rate below 100%. Second, for the WGA dataset, the methylation status of original input DNA prior to the WGA process would add noise during the training of the HK model. In the future, it would be interesting to explore the use of other training datasets (e.g., using synthetic oligonucleotides with known methylation states) for enhancing the overall performance of the HK model. Third, the sequencing kits would be another factor affecting the performance. Notably, we found that the newer sequencing kits (the Sequel II sequencing kit) were inferior to the old-generation sequencing kit (the Sequel I sequencing kit 3.0). It might imply that the base modification detection using SMRT sequencing could be further optimized through engineering DNA polymerases and reagents.
From results regarding human–mouse hybrid fragments, the aggregate methylated levels of DNA from the unmethylated part were observed to systematically above a methylation level of 0% whereas the aggregate methylated levels of DNA from the methylated part were observed to below a methylation level of 100%. Such a difference from the expected values might be likely attributed to the suboptimal conversation rate of M.SssI treatment, affecting the accuracy of the HK model. Such deviations in methylation estimation were also present in native DNA molecules from biological DNA samples, when compared to that measured by BS-seq. However, the methylation levels deduced by the HK model were highly correlated with those values determined by BS-seq. Such a deviation between the measurement by the HK model and BS-seq could in the future be harmonized by recalibration between studies.
Theoretically, in a measurement window with more than one CpG site, the kinetic signals from these CpG sites might interact with one another. To investigate this possibility, we used the HK model to classify the methylation status of the human–mouse hybrid fragments in which the human and murine portions of the hybrid fragment possessed opposing methylation status. The data demonstrated that the HK model was able to decode the divergent methylation status of CpG sites separated by at least 6 nt. As the length of the restriction site (i.e., 6 bp) involved in the human–mouse hybrid fragment assay limited the least distance between CpG sites that we could assess (SI Appendix, Fig. S9), the performance for CpG sites separated by nucleotides less than 6 nt would warrant future research. In the further study, the synthetic oligonucleotides carrying multiple CpG sites characteristic of different methylation status within a measurement window would be informative to enhance the HK model in the training process, in an attempt to address the methylation status of CpG sites near one another in a testing sample. On the other hand, as the methylation status among CpG sites within a close genomic distance (<50 bp) tended to be comethylated or counmethylated (27), we believe that the current version of the HK model would be broadly applicable to analyze DNA from various biological samples. Such a hypothesis was in part evidenced by the fact that the methylation patterns surrounding TSS regions (commonly overlapping with CGIs) appeared to be very consistent between the HK model and BS-seq.
In addition to the methylcytosines, other oxidized derivatives of cytosine, such as 5-hydroxymethylcyotsine (5hmC), 5-formylcytosine (5fC), and 5caC, had been reported to be present in mammalian genomes (28–30). However, we believe that the actual impact on the performance of the HK might be small, because of the low abundance of these other modified cytosines in tissues other than the brain (28). A future follow-up effort would be to enhance the HK model by incorporating these other modified cytosines during the training process.
It was previously reported that the genomic variations, including single nucleotide variants, insertions, and deletions, would introduce “quantification bias” of methylation levels in the step of alignment for BS-seq reads (31). For example, if a sample genome has a “CG-to-TG” variant relative to the reference genome sequence, standard alignment approaches would consider a “TG” dinucleotide in a read to be derived from an unmethylated CpG dinucleotide, resulting in an underestimation of methylation level (31). Such a quantification bias would lead to inaccurate data interpretation when comparing methylation patterns between species and human normal-cancer datasets with divergent genotypes (31). In addition to a dedicated bioinformatics approach for mitigating the quantification bias (31), the HK model presented in this study would provide an opportunity to address the quantification bias due to the issue of mappability in the standard alignment approaches for BS-seq. In this regard, we wish to highlight two attributes of the approach described in our study. 1) The HK model used the kinetic values of a DNA polymerase derived from subreads generated by SMRT-seq for base modification analysis. Subreads could be used for constructing the circular consensus sequences (CCSs), providing highly accurate sequence information (accuracy: 99.8%) on the DNA template (32); 2) The nature of multikilobase long-read sequencing would facilitate elucidation of haplotype information. Thus, the HK model allows one to simultaneously analyze epigenetics and genetics for each DNA molecule (Fig. 6B). For the traditional BS-seq, when one sees a sequence “TG,” one would not know whether the “T” is the result of bisulfite conversion on an unmethylated “C” until one looks at the reference genomic sequence. In contrast, using the HK model, the methylation status is determined based on the PW, IPD, and sequence context without alignment to a reference genome (Fig. 6B).
Another advantage of the HK model is its ability to elucidate the methylation states across a long DNA molecule (tens of kilobases). For example, the short fragments (50 to 600 bp)-based BS-seq were not suitable for decoding the methylation states of imprinted regions as the short DNA fragment lacked the ability to efficiently link the methylation states to individual haplotypes. In contrast, the HK model-based analysis for SMRT sequencing has made it possible to effectively link methylation states across CpG sites to parental haplotypes using long DNA molecules. We believe that the HK model-based methylation analysis would open up many new possibilities for studying the genetics and epigenetics in different organisms and may be useful in many molecular diagnostic applications (e.g., in oncology).
Materials and Methods
Sample Recruitment and Processing.
HCC patients and pregnancy samples were recruited from the Department of Surgery and the Department of Obstetrics and Gynecology, respectively, of the Prince of Wales Hospital, Hong Kong. The study was approved by the Joint Chinese University of Hong Kong–Hospital Authority New Territories East Cluster Clinical Research Ethics Committee. Written informed consent forms were obtained from the patients. The details are described in SI Appendix, Methods and Materials.
SMRTbell Template Library Preparation, Sequencing, and Alignment.
SMRT sequencing was performed using the Sequel Systems (PacBio) according to the manufacturer’s instructions. This study involved both the Sequel I and Sequel II systems. Specifications about reagent kits used for SMRT-seq are detailed in SI Appendix, Methods and Materials. SI Appendix, Table S2 summarizes which kits were applied for each sample. Sequencing reads were aligned to the human reference genome (hg19) using BWA aligner (33).
SMRT Sequencing Datasets for Amplified DNA and M.SssI-Treated DNA.
We used the Sequel I sequencer together with the Sequel I sequencing kit 3.0 to sequence sample W01 and M01, obtaining 0.74 and 0.74 million sequenced molecules, with a median 319 and 296 bp in size, respectively. The circularized DNA template was sequenced multiple times, thus generating many readouts from the same DNA template. A readout that began at one adapter sequence and ended at the other adapter sequence was defined as a subread. One full cycle of a circularized molecule passing through the DNA polymerase would generate two subreads. The mean number of subreads per strand covering a site was defined as the subread depth. The median subread depth was 11× and 10.5× for WGA and M.SssI-treated datasets, respectively. We used the Sequel II sequencer together with the Sequel II sequencing kit 1.0 to sequence W02 and M02, obtaining 3.0 million (median size: 4.4 kb) and 2.1 million (median size: 3.7 kb) sequenced molecules for WGA and M.SssI-treated datasets, respectively. The median subread depth was 3.5× and 5× for the WGA and M.SssI-treated datasets, respectively. In addition, we used the Sequel II sequencer together with the Sequel II sequencing kit 2.0 to sequence W03 and M03, obtaining 0.26 million (median size: 728 bp; median subread depth: 30.5×) and 0.26 million (median size: 392 bp; median subread depth: 41.5×) sequenced molecules for WGA and M.SssI-treated datasets, respectively.
Human–Mouse Hybrid Fragment Generation.
Human and mouse DNA was whole-genome amplified with Phi29 polymerase (NEB) and random hexamers (ThermoFisher) to create unmethylated DNA (unmeth), or treated with M.SssI (NEB) to become methylated DNA (meth). The hybrid fragments were created in a way that the DNA species mentioned in the previous sentence were each subjected to double restriction enzyme digestion (HindIII and NcoI) (NEB), 1:1 mixing of unmethylated and methylated DNA, followed by DNA ligation via T4 DNA ligase (NEB). The cleavage sites of HindIII and NcoI were 5′-A^AGCTT-3′ and 5′-C^CATGG-3′ (“^” denotes the restriction enzyme cutting locus), respectively. Two sets of hybrid DNA were generated: human (unmeth)–mouse (meth) and human (meth)–mouse (unmeth).
CNN.
The CNN model made use of two one-dimensional (1D)-convolutional layers, each having 64 filters with a kernel size of 4. The activation function of the rectified linear unit (ReLU) was used for those convolutional layers. A batch normalization layer was applied subsequently, followed by a dropout layer with a dropout rate of 0.5. A maximum pooling layer with a pool size of 2 was used. A flattened layer was further added, followed by a fully connected layer comprising 10 neurons with the use of the ReLU activation function. The output layer with one neuron was finally applied, with a sigmoid activation function to yield the probabilistic score for a CpG site of being methylated (i.e., methylation score). The program for the CNN model was implemented on the basis of the Keras deep learning framework (https://keras.io/).
Procedures for Training and Testing the HK Model.
The measurement windows associated with methylated CpG sites (the M.SssI-treated DNA dataset) and those associated with unmethylated CpG sites (the WGA DNA dataset) were used for training the HK model through CNN. Data within each measurement window flanking a cytosine within a CpG context, including the sequence context, mean IPDs, and PWs originating from subreads across individual nucleotides, were entered into the HK model. Each target output (i.e., analogous to a dependent variable value) for a CpG site in M.SssI-treated DNA datasets was assigned as “1” while each target output for a CpG site in WGA DNA datasets was assigned as “0.” The patterns present in the measurement windows of methylated and unmethylated CpG sites were used for training CNN to determine the parameters (often called weights) of the HK model. The details are described in SI Appendix, Methods and Materials.
Supplementary Material
Acknowledgments
This work was supported by the Research Grants Council of the Hong Kong Special Administrative Region Government under the Theme-based research scheme (T12-403/15-N and T12-401/16-W). Y.M.D.L. is supported by an endowed chair from the Li Ka Shing Foundation.
Footnotes
Competing interest statement: A patent application on the described technology has been filed and licensed to Take2 Holdings Limited, founded by the research team.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019768118/-/DCSupplemental.
Data Availability.
Sequence data for the subjects studied in this work have been deposited at the European Genome-Phenome Archive (EGA), https://www.ebi.ac.uk/ega/, hosted by the European Bioinformatics Institute (EBI) (accession no. EGAS00001004642).
References
- 1.Feinberg A. P., The key role of epigenetics in human disease prevention and mitigation. N. Engl. J. Med. 378, 1323–1334 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Smith Z. D., Meissner A., DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Hofer A., Liu Z. J., Balasubramanian S., Detection, structure and function of modified DNA bases. J. Am. Chem. Soc. 141, 6420–6429 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Olova N., et al. , Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biol. 19, 33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grunau C., Clark S. J., Rosenthal A., Bisulfite genomic sequencing: Systematic investigation of critical experimental parameters. Nucleic Acids Res. 29, E65 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., et al. , Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 37, 424–429 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Liu Q., et al. , Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 10, 2449 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodwin S., McPherson J. D., McCombie W. R., Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eid J., et al. , Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Flusberg B. A., et al. , Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat. Methods 7, 461–465 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark T. A., et al. , Enhanced 5-methylcytosine detection in single-molecule, real-time sequencing via Tet1 oxidation. BMC Biol. 11, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister R., et al. , Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greer E. L., et al. , DNA methylation on N6-adenine in C. elegans. Cell 161, 868–878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang P., et al. , Methy-pipe: An integrated bioinformatics pipeline for whole genome bisulfite sequencing data analysis. PLoS One 9, e100360 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feinberg A. P., Vogelstein B., Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92 (1983). [DOI] [PubMed] [Google Scholar]
- 16.Lun F. M. F., et al. , Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin. Chem. 59, 1583–1594 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Chan K. C. A., et al. , Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. U.S.A. 110, 18761–18768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anwar S. L., et al. , LINE-1 hypomethylation in human hepatocellular carcinomas correlates with shorter overall survival and CIMP phenotype. PLoS One 14, e0216374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzywinski M., et al. , Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zink F., et al. , Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet. 50, 1542–1552 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Baran Y.et al.; GTEx Consortium , The landscape of genomic imprinting across diverse adult human tissues. Genome Res. 25, 927–936 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Court F., et al. , Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 24, 554–569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blow M. J., et al. , The epigenomic landscape of prokaryotes. PLoS Genet. 12, e1005854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schadt E. E., et al. , Modeling kinetic rate variation in third generation DNA sequencing data to detect putative modifications to DNA bases. Genome Res. 23, 129–141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng Z., et al. , Detecting DNA modifications from SMRT sequencing data by modeling sequence context dependence of polymerase kinetic. PLOS Comput. Biol. 9, e1002935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark T. A., et al. , Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40, e29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Affinito O., et al. , Nucleotide distance influences co-methylation between nearby CpG sites. Genomics 112, 144–150 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Nestor C. E., et al. , Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 22, 467–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachman M., et al. , 5-Formylcytosine can be a stable DNA modification in mammals. Nat. Chem. Biol. 11, 555–557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X., et al. , Base-resolution maps of 5-formylcytosine and 5-carboxylcytosine reveal genome-wide DNA demethylation dynamics. Cell Res. 25, 386–389 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wulfridge P., Langmead B., Feinberg A. P., Hansen K. D., Analyzing whole genome bisulfite sequencing data from highly divergent genotypes. Nucleic Acids Res. 47, e117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wenger A. M., et al. , Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data for the subjects studied in this work have been deposited at the European Genome-Phenome Archive (EGA), https://www.ebi.ac.uk/ega/, hosted by the European Bioinformatics Institute (EBI) (accession no. EGAS00001004642).