Significance
Cancer cells are frequently coated with chains of sugar molecules (glycans) that bind to inhibitory immune receptors, leading to suppression of anticancer immunity. While blocking these interactions may be therapeutically beneficial, the specific glycoprotein ligands that engage such receptors are often complex and difficult to characterize. To solve this problem, we used unbiased genome-wide screening to identify genes required for cell-surface presentation of ligands that bind members of the Siglec immune receptor family. This approach led to identification of the glycoprotein CD43 as a highly specific ligand for Siglec-7. Blocking the interaction between CD43 and Siglec-7 sensitizes leukemia cells to immune cell lysis, implying that targeting the CD43/Siglec-7 checkpoint could be therapeutically beneficial.
Keywords: Glycobiology, Tumor Immunology , CRISPR Screening
Abstract
Glyco-immune checkpoint receptors, molecules that inhibit immune cell activity following binding to glycosylated cell-surface antigens, are emerging as attractive targets for cancer immunotherapy. Defining biologically relevant ligands that bind and activate such receptors, however, has historically been a significant challenge. Here, we present a CRISPRi genomic screening strategy that allowed unbiased identification of the key genes required for cell-surface presentation of glycan ligands on leukemia cells that bind the glyco-immune checkpoint receptors Siglec-7 and Siglec-9. This approach revealed a selective interaction between Siglec-7 and the mucin-type glycoprotein CD43. Further work identified a specific N-terminal glycopeptide region of CD43 containing clusters of disialylated O-glycan tetrasaccharides that form specific Siglec-7 binding motifs. Knockout or blockade of CD43 in leukemia cells relieves Siglec-7-mediated inhibition of immune killing activity. This work identifies a potential target for immune checkpoint blockade therapy and represents a generalizable approach to dissection of glycan–receptor interactions in living cells.
Discovery of immune checkpoint ligands like PD-L1 and CD47 has revolutionized cancer treatment (1, 2). Such cell-surface proteins are broadly expressed in tumors and engage inhibitory receptors on immune cells, shielding cancer cells from detection and elimination by the immune system (1, 2). Blockade of these ligand–receptor interactions can generate profound, systemic antitumor immune responses (2, 3). Nevertheless, many patients fail to respond to checkpoint therapy (4). This is likely due to the fact that immune activity is finely regulated through integration of cellular signals deriving from numerous activating and inhibitory cell membrane receptors, which may be expressed heterogeneously in different patients and cancer subtypes (5, 6). Characterization of additional cancer-associated inhibitory immune ligands is therefore crucial for achieving better therapeutic manipulation of the anticancer immune response.
While research into immune checkpoints has historically focused on protein factors, recent work has also identified glycosylated antigens as pivotal players in cancer immune suppression. In particular, members of the Siglec (sialic acid-binding immunoglobulin-like lectin) family of glycan-binding immune receptors have emerged as potent targets for enhancing antitumor immunity in preclinical disease models (7–9). The human Siglecs are a large family of receptors that bind cells through complex interactions with a variety of glycans containing sialic acid residues (Fig. 1A) (10, 11). Upon binding to their glycan ligands, most Siglecs induce signaling through intracellular immunoreceptor tyrosine-based inhibitory motif (ITIM) and immunoreceptor tyrosine-based switch motif (ITSM) domains (motifs also found on classical immune checkpoint receptors like PD-1), delivering an inhibitory signal that quenches immune cell activity (Fig. 1A) (9, 11, 12). The Siglec family in humans has 14 members that are believed to bind a diverse range of glycoprotein and glycolipid substrates on the surface of cancer cells (11). Specific cancer-associated glycoproteins, for example, were recently shown to broadly suppress macrophage activity by engaging Siglec-9 (13). Other studies found that targeted blockade of macrophage Siglec-10 robustly enhances phagocytosis of breast cancer cells and slows tumor progression in mouse tumor models (7). Finally, broad desialylation of tumors through chemical and enzymatic approaches has also been recently validated as an effective strategy for neutralizing inhibitory Siglec signaling in vivo (14–16).
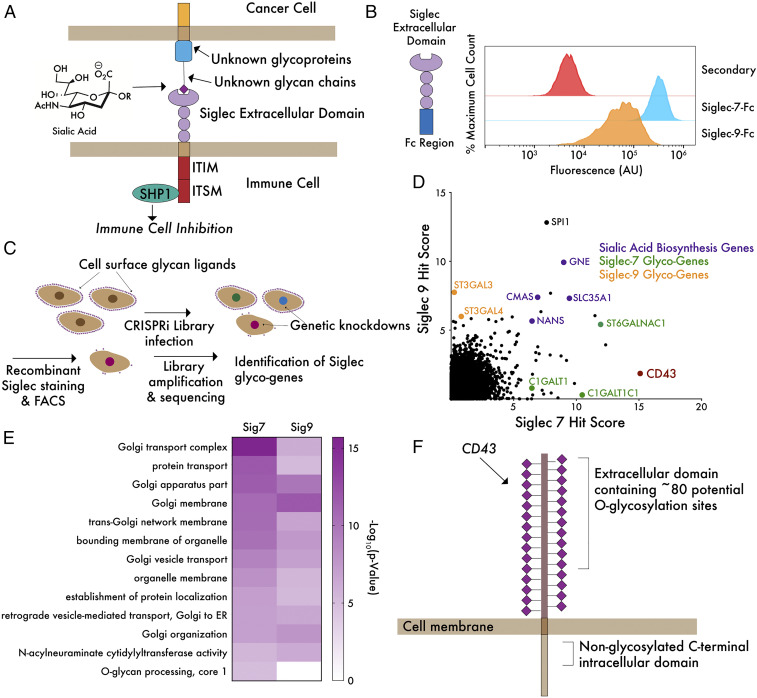
Fig. 1.
A genome-wide CRISPRi screen reveals core genes driving ligand expression for Siglec-7 and Siglec-9. (A) Siglecs possess an extracellular domain that binds to glycan ligands containing sialic acid. Ligand–receptor engagement initiates signaling through intracellular ITIM/ITSM domains, inhibiting immune activation. Sialic acid is incorporated into hundreds of glycan structures on thousands of individual proteins and lipids, making it challenging to define molecular details of ligand–receptor interactions. (B) K562 cells were incubated with 1 μg/mL Siglec-7–Fc and Siglec-9–Fc precomplexed to an Alexa Fluor 488 anti-huIgG secondary antibody and subjected to live cell flow cytometry. “Secondary” indicates noncomplexed Alexa Fluor 488 anti-huIgG. (C) A genome-wide library of ∼104,000 sgRNAs (five sgRNAs/gene) was transduced into K562-dCas9–KRAB cells and incubated with Siglec-7–Fc or Siglec-9–Fc as in B. FACS was performed to isolate a population of cells exhibiting at least a 10-fold reduction in ligand staining. Library amplification and sequencing was then performed to identify sgRNAs enriched in the low-staining population. (D) Hits for Siglec-7 and Siglec-9 were plotted and ranked by hit score (−log10[positive selection score]), where a higher value indicates a stronger enrichment of sgRNAs in the low-staining population. The screen identified a number of sialic acid biosynthesis genes (purple), glycotransferase genes specific for Siglec-7 and Siglec-9 (green and orange), as well as a single-cell surface glycoprotein, CD43, specific for Siglec-7 (red). (E) Enriched GO terms for hits in both screens show importance of Golgi and sialic acid biosynthesis genes. O-glycan biosynthesis is an enriched term in the Siglec-7 but not Siglec-9 screen. (F) The top hit of the Siglec-7 screen, CD43, is a mucin-type O-glycoprotein bearing many sialoglycan modification sites.
Therapeutic targeting of immune checkpoints has proceeded from a firm mechanistic understanding of how specific ligands engage their receptors (PD-1 binds PD-L1 and SIRP⍺ binds CD47) (2, 17). Precise definition of Siglec-binding ligand structures is therefore essential for both new biomarker discovery as well as development of Siglec-blocking therapeutic agents. Interactions between glycans and putative glycan-binding proteins (GBPs) are typically assessed by studying binding of recombinant GBPs to synthetic arrays containing a broad collection of defined glycan structures chemically linked to an inert surface (18, 19). Glycan arrays are useful tools that can point toward a class of isolated glycan structures that may form components of a biological binding motif. As such studies abstract glycans away from the key context of their native membrane and protein environment, however, they rarely capture the full range of structurally complex interactions that can mediate binding of a GBP to its physiological ligand (20, 21). As a case in point, recent work has found that inhibitory signaling through Siglec-10 is mediated almost entirely through specific binding to a single-cell surface ligand, the GPI-anchored protein CD24 (7). These examples suggest that interactions between Siglecs and their preferred ligands in physiological settings may be more specific than is often suggested by glycan array data.
An ideal platform for identification of novel GBP–ligand interactions would take into account the genetic complexity of glycoprotein synthesis, in which ligands are sometimes created through the combined activity of dozens of enzymes, and assess GBP–ligand interactions in a native environment, where factors such as clustering of adjacent glycan epitopes or secondary interactions with proteins or lipids are preserved (20, 22, 23). Indeed, recent years have seen the development of new strategies for assessment of GBP–ligand interactions in living cells. One study, for example, profiled GBP binding to panels of glyco-engineered cell lines featuring targeted knockouts and knock-ins of key glycosyltransferase genes (20). Such platforms can be powerful tools for targeted dissection of glycan-binding specificities. It can be difficult, however, to capture the enormous range of possible interactions that drive binding of a GBP to a specific glycoprotein or glycolipid ligand using targeted, array-based approaches. A truly unbiased, more high-throughput strategy for profiling such interactions in living cells would therefore be highly useful.
We therefore developed a complementary approach to identifying biological GBP ligands using CRISPR genomic screening technology. Briefly, we generated genome-wide CRISPR interference (CRISPRi) cell knockdown libraries and isolated cells that exhibited reduced binding to recombinant Siglec proteins. Using this strategy, we produced unbiased genetic maps of all the factors driving cell-surface ligand expression for these glyco-immune checkpoint receptors. This mode of Siglec ligand profiling captures both the cell-surface proteins and the intracellular biosynthetic machinery whose expression is required to create Siglec-binding antigens. We chose to apply this platform to two receptors of the family, Siglec-7 and Siglec-9. These molecules represent promising therapeutic targets that are known to inhibit activity of primary immune cells and to be induced on tumor-infiltrating lymphocytes in human tumor samples (9, 24, 25). Through this approach, we found that a specific glycoform of the mucin-type glycoprotein CD43 is the predominant ligand for Siglec-7, but not Siglec-9, on the surface of leukemia cells. Further biochemical and immunological studies identified a specific glycopeptide domain at the N terminus of CD43 that forms a Siglec-7–binding motif. These studies reveal a therapeutic target for glyco-immune checkpoint blockade and validate CRISPR genomic screening as a strategy for molecular definition of biological GBP–ligand interactions.
Results
Genome-Wide CRISPRi Screens Reveal Genetic Determinants of Siglec Ligand Expression in Cancer Cells.
We chose to conduct our CRISPR screening studies in the K562 chronic myeloid leukemia (CML) cell line, as staining of K562 cells with recombinant Siglec-7–Fc and Siglec-9–Fc fusion proteins revealed high expression of glycan ligands for each of these receptors by flow cytometry (Fig. 1B). We infected K562 cells expressing dCas9 fused to the Kruppel associated box (KRAB) transcriptional repression domain with a genome-wide library containing ∼104,000 CRISPRi single guide RNAs (sgRNAs; five sgRNAs/gene) (26). Cells were subsequently incubated with recombinant, fluorescently labelled Siglec-7–Fc and Siglec-9–Fc chimera proteins, and Siglec binding was determined by flow cytometry. Siglec-Fc fusion proteins are dimeric staining reagents that have been widely used to quantitate cell-surface expression of Siglec ligands, allowing straightforward comparison of our results with the existing literature (27). A population of cells exhibiting at least a 10-fold reduction in Siglec ligand expression was then isolated by fluorescence-activated cell sorting (FACS) (Fig. 1C). Sequencing of enriched sgRNAs in the low Siglec-staining population versus an unsorted control allowed identification of target genes that enable Siglec ligand expression. Downstream analysis yielded a list of 115 hit genes that significantly reduced Siglec-7 ligand expression and 55 hit genes that reduced Siglec-9 ligand expression, using a 10% false discovery rate cutoff (full results are given in Datasets S1 and S2). In both screens, top hits contained the core enzymes of the sialic acid biosynthesis machinery (CMAS, GNE, and NANS) and the transporter that imports sialic acid into the Golgi apparatus (SLC35A1) (Fig. 1D). Gene Ontology (GO) term analysis similarly revealed an enrichment of functional processes related to sialic acid biosynthesis (Fig. 1E and Datasets S3 and S4). The most common enriched GO terms in both screens related to Golgi vesicle transport. The key role of the Golgi apparatus in elaborating complex glycans and regulating glycosyltransferase activity provides a straightforward explanation for these results (28, 29). Interestingly, the top shared hit gene from both screens was the transcription factor SPI1, which is reported to be a regulator of lymphoid differentiation (Fig. 1D) (30). While we chose to focus on other aspects of this screening dataset for downstream validation, this result implies an interesting possible connection between the regulation of immune cell development and cellular glycosylation.
Additionally, both screens identified glycosyltransferases (GTs) that are known to generate specific glycan structures in the cell. ST3GAL3 and ST3GAL4, two enzymes that add sialic acid in an α-2,3–linked orientation to a variety of glycan structures bearing a terminal galactose, were two of the most significantly enriched GT hits in our Siglec-9 screen (31). We did not identify other components of more specific glycan biosynthesis machineries (for N-linked, O-linked, or lipid glycosylation, for example), likely indicating that Siglec-9 ligands are expressed redundantly on a range of different underlying scaffolds. Our Siglec-7 screen yielded strikingly different results. GO term analysis revealed O-glycosylation–related processes as significantly enriched in the Siglec-7 screen but not the Siglec-9 screen, indicating that Siglec-7 predominantly binds sialylated O-glycans (Fig. 1E).
Prior work has found that glycolipids (gangliosides) containing 2,8-linked sialic acids form ligands for Siglec-7 in certain contexts (32, 33). Our screen, however, did not recover any genes related to ganglioside biosynthesis. We also did not identify any members of the ST8SIA family of enzymes (which catalyze formation of 2,8-linked sialic acid structures) (34). Our finding that the predominant ligands for Siglec-7 are O-glycoproteins is supported by a number of recent studies. One work, for example, demonstrated that Siglec-7 ligands are almost completely depleted from a wide range of epithelial cancer cell lines by treatment with a protease (StcE) that specifically degrades mucin O-glycoproteins (35). Indeed, we found that treatment of K562s with this protease caused a loss in Siglec-7 ligand expression almost as complete as treatment with a sialidase enzyme (SI Appendix, Fig. S1). This result confirms that some form of O-glycoconjugate represents the dominant ligand for Siglec-7 in K562 cells. Independent cell-based glycan array studies have similarly shown that Siglec-7 preferentially binds O-linked glycans (20). However, our results do not preclude the possibility that gangliosides may be significant ligands for Siglec-7 in other select cell types, in particular neuronal cells that express such antigens at an unusually high density (36). Additionally, while we have mainly focused here on identification of trans ligands for Siglec-7, interaction between Siglec-7 and gangliosides in cis on certain immune cell types also remains a possibility.
The most significantly enriched hit in the Siglec-7 screen (but not the Siglec-9 screen) was unique among our top hit genes in not being a component of the general cellular glycosylation machinery or the Golgi trafficking network. Instead, we found a gene encoding a single-cell surface O-glycoprotein (the SPN gene). This result was surprising: as glycans are distributed on over 80% of secreted proteins, knockdown of a single-cell surface protein would not typically be expected to affect overall cell-surface binding of a receptor that primarily binds glycan ligands (37). The SPN gene encodes a cell-surface protein called CD43. CD43 is an extensively O-glycosylated mucin-domain glycoprotein, bearing over 80 putative sites for O-linked sialoglycans in its ∼270 amino acid extracellular domain (Fig. 1F) (38, 39). Most glycoforms of the CD43 protein are known to be extensively sialylated (39). CD43 has been reported to have multiple functions that include regulation of T cell activation, inhibition of cell–cell adhesion events, and lymphocyte tissue homing (40–42). This diverse and sometimes contradictory literature likely stems from the function of CD43 being primarily driven by presentation of glycan ligands whose chemical structure can vary among different cell types. For instance, CD43 has been shown to serve as a ligand for several different immune adhesion receptors, including sialoadhesin and E-selectin (which bind to glycoforms of CD43 containing α-2,3–linked sialic acid and Sialyl-Lewis X, respectively) (43, 44). However, no previous indication of a specific interaction between CD43 and Siglec-7, or indeed any immune checkpoint receptor, has ever been reported. Thus, we sought to explore whether there may be a selective biochemical and functional interface between these two proteins.
Siglec-7 Binds CD43 with High Selectivity.
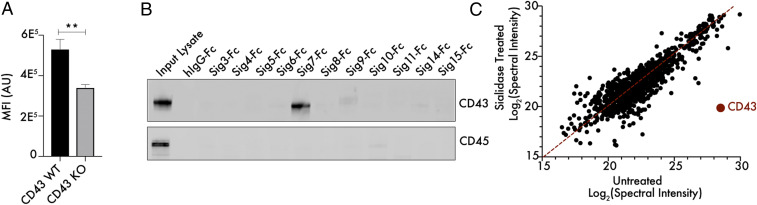
We first sought to validate our screening result by generating a K562 cell line with a targeted knockout in the SPN gene. CD43 knockout (KO) cells exhibited a 40 to 50% reduction in Siglec-7 binding by flow cytometry when compared to wild type (WT) cells, mirroring our screening dataset (Fig. 2A). Lentiviral transduction of a CD43 WT construct into CD43 KO cells restored WT levels of Siglec-7 ligand expression (SI Appendix, Figs. S2 and S3). Staining of cells with a mix of the SNA and MAL-II plant lectins, which bind to a broad range of sialylated glycans, did not reveal any distinction between CD43 WT and KO cells in overall sialic acid levels (SI Appendix, Fig. S4) (45, 46). We next assessed direct binding between CD43 and Siglec-7 using a pulldown assay. K562 cells were lysed under nondenaturing conditions, and cell lysates were passed over magnetic beads loaded with different recombinant Siglecs. Elution of Siglec-binding proteins followed by Western blotting showed that only Siglec-7, but not other Siglecs, strongly isolated CD43 from cell lysates (Fig. 2B). Enrichment of CD45 was not observed, which is surprising since it is also a densely O-glycosylated protein expressed in K562 cells (Fig. 2B). We also analyzed Siglec-7–binding proteins from cell lysates using mass spectrometry. Cell lysates were either left untreated or treated with a sialidase enzyme to remove sialic acid from cellular glycoproteins. Pulldown of Siglec-7–interacting proteins was then performed from both lysates, followed by tryptic digestion and identification of captured glycoproteins by tandem mass spectrometry (MS/MS). This approach showed CD43 as the dominant Siglec-7 binding protein that was enriched in a sialic acid–dependent manner (Fig. 2C).
Fig. 2.
CD43 binds Siglec-7 with unusual selectivity and specificity. (A) K562 CD43 KO cells were generated by CRISPR/Cas9 editing, incubated with 1 μg/mL Siglec-7–Fc/Alexa Fluor 488 anti-huIgG as previously described and subjected to live cell flow cytometry. n = 3, **P < 0.01. MFI indicates mean fluorescence intensity and AU indicates arbitrary units. (B) K562 cells were lysed under nondenaturing conditions, and lysate was passed over magnetic beads functionalized with recombinant Siglec-Fc proteins. The bead-binding protein fraction was eluted and subjected to Western blot to assess the interaction of these Siglecs with the mucin-type glycoproteins CD43 and CD45. (C) K562 cell lysates were either left untreated or treated with 100 nM VC-Sialidase for 1 h and then incubated with magnetic beads functionalized with recombinant Siglec7-Fc. Tryptic digestion and MS/MS-based identification of Siglec-7–binding proteins was then performed, and the total spectral intensity for each interacting protein was calculated.
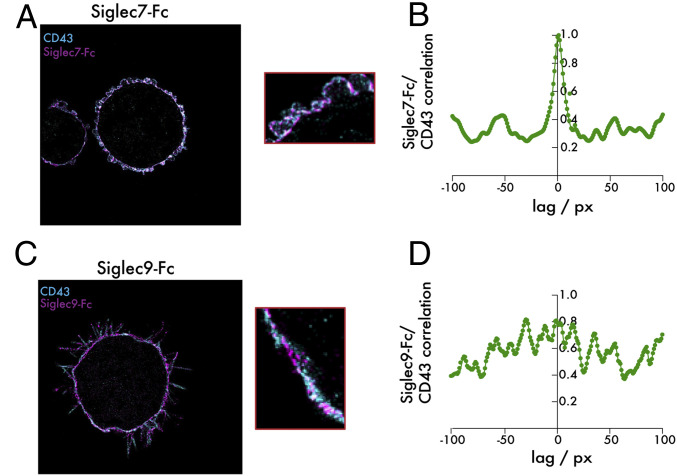
While these data strongly indicated a specific interaction between Siglec-7 and CD43, we wished to confirm this finding using a higher-resolution method that captured binding of the two proteins on an intact cell surface. We therefore costained fixed K562 cells with fluorescently labeled recombinant Siglec-7–Fc and an antibody targeting the extracellular domain of CD43. Imaging of these two stains by two-color super-resolution microscopy revealed extensive spatial correlation of the two proteins, with clusters of high CD43 density corresponding closely to clusters of high Siglec-7–Fc density on the membrane surface (Fig. 3A). A quantitative correlation analysis was performed that confirmed this qualitative observation. Using the super-resolution reconstructions, an intensity profile along the membrane was recorded of both the Siglec-7 and the CD43 signal. Then, a standard cross-correlation analysis of the two intensity traces was performed. The cross-correlation peak at low pixel lags is evidence of strong spatial correlation of Siglec-7 and CD43 (Fig. 3B). Notably, the same analysis using Siglec-9–Fc revealed no such spatial correlation of these glycan ligands with CD43 (Fig. 3 C and D). Taken together, these orthogonal methods all validate a previously undescribed specific binding relationship between Siglec-7 and the CD43 glycoprotein.
Fig. 3.
Super-resolution imaging reveals selective cell-surface colocalization between CD43 and Siglec-7 ligands. (A) K562 cells were plated onto slides coated with fibronectin and incubated with 1 μg/mL Siglec-7–Fc/Alexa Fluor 647 anti-huIgG (purple) and a CD43-CF568 antibody (blue). An analysis of colocalization was then performed using super-resolution microscopy. (B) A correlation analysis assessing colocalization of Siglec-7–Fc and CD43 signal on the cell surface was performed. (C) The same experiment as in A was performed with Siglec-9–Fc. (D) The same analysis as in B was performed for Siglec-9–Fc. The total edge length of the images is 40.96 microns. One px corresponds to the bin width of the two-dimensional histogram used for the reconstruction, that is, 32 nm.
The Specificity of the Siglec-7/CD43 Interaction Is Mediated by a Complex Glycopeptide-Binding Motif.
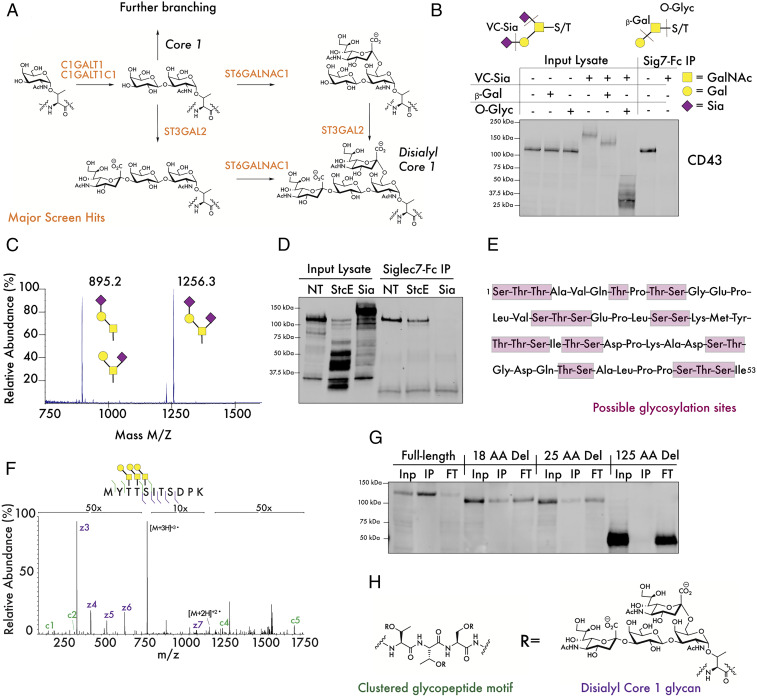
We next sought to interrogate the glycan structure on CD43 that might be bound by Siglec-7. Interestingly, our screening data identified a cluster of functional enzymes that immediately pointed toward a candidate epitope. Together the glycosyltransferases C1GALT1, C1GALT1C1, and ST6GALNAC1 were all among the top 10 hits in our Siglec-7 screen (Fig. 1D). C1GALT1 and C1GALTC1 (COSMC) form a complex that synthesizes the core 1 disaccharide structure from an initial serine/threonine (S/T)-linked GalNAc, while ST6GALNAC1 has been reported to add sialic acid to the GalNAc of this core 1 antigen (Fig. 4A) (47–49). These results imply that the disialyl core 1 tetrasaccharide (Fig. 4A), a motif commonly found on cell-surface O-glycoproteins, may be a component of the glycan epitope that binds Siglec-7 (50). The enzyme ST3GAL1 (which attaches sialic acid α-2,3–linked to galactose residues) has also been reported to be a key component of this biosynthetic pathway (51, 52). Our screen, however, did not identify ST3GAL1 as a driver of Siglec-7 ligand expression. Instead, the related enzyme ST3GAL2 was identified as a moderate strength hit (Dataset S1). ST3GAL2 has been reported to have similar biochemical activity toward O-linked glycans as ST3GAL1 in vitro, although this finding has never been confirmed in living cells (51, 53–55). Our results imply that ST3GAL2 could function redundantly with ST3GAL1 in cellular O-sialoglycan biosynthesis. Additional detailed studies are needed to reach this conclusion, however, given the complex genetic interactions between GT enzymes and the possibility of both false negative and false positive hits from large-scale screening studies.
Fig. 4.
Siglec-7 binds a specific glycopeptide motif at the N terminus of CD43. (A) The synthesis of the disialyl core 1 tetrasaccharide is catalyzed by enzymes that were significantly enriched hits in the Siglec-7 screen. (B) K562 lysates were digested with the indicated combinations of sialidase from Vibrio Cholerae (VC-Sia), bovine β1-3,4–Galactosidase (β-Gal), and O-Glycosidase from S. pneumoniae (O-Glyc). The digestion specificities of each of these enzymes are indicated, with the dotted lines indicating the preferred enzymatic cleavage point. GalNAc = N-Acetylgalactosamine, Gal = galactose, and Sia = sialic acid. (C) CD43 was isolated from CD43-myc–FLAG-expressing cells and subjected to glycomic analysis through β-elimination and liquid chromatography/MS. (D) K562 lysate was treated with 100 ng/mL StcE or 100 nM VC-Sia for 30 min at 37 °C, passed over magnetic beads functionalized with recombinant Siglec-7–Fc, and subjected to Western blot with an antibody binding the nonglycosylated CD43 intracellular domain. (E) The N terminus of CD43 contains repeated clusters of serine/threonine residues that can serve as points for glycan attachment. (F) CD43-myc–FLAG was isolated and digested with VC-Sia, GluC, and trypsin. Intact glycopeptides were subsequently analyzed by MS/MS. A representative electron transfer dissociation spectrum of a glycopeptide from the N terminus of CD43 with indicated glycans attached to adjacent residues is shown. (G) K562 cells expressing CD43-myc–FLAG constructs exhibiting truncations of the indicated number of amino acids (AA) from the N terminus of the CD43 extracellular domain were lysed, passed over Siglec-7–Fc-functionalized beads, and subjected to analysis by Western blot with an anti-myc antibody. Inp = input lysate, IP = bead-binding fraction, FT = flow-through. (H) The putative Siglec-7 binding structure on CD43.
Taken together, these data led us to investigate whether CD43 might be glycosylated with disialyl core 1 antigens. Indeed, treatment of cell lysates with a sialidase enzyme as well as enzymes specific for cleaving terminal galactose (β1-3,4–galactosidase) and the core 1 disaccharide (Streptococcus pneumoniae O-glycosidase) caused significant shifts in the molecular weight (MW) of CD43 by Western blot (Fig. 4B). Sialidase treatment of the protein also abolished pulldown of CD43 by Siglec-7, further confirming that Siglec-7 binding requires CD43 sialylation (Fig. 4B). Sialidase treatment also was found to increase binding to the plant lectin PNA, which selectively interacts with unsubstituted core 1 structures (SI Appendix, Fig. S5) (56). To more precisely quantify glycosylation on CD43, we overexpressed a tagged CD43 construct in K562 cells and isolated CD43 protein from 5 × 108 cells by affinity purification. Mass spectrometry-based glycomic analysis of CD43-linked glycans indicated that the predominant O-linked structures present on the protein were indeed disialyl core 1 structures (Fig. 4C). A significant fraction of monosialylated core 1 was also seen; further analysis of these species by MS/MS indicated that that these glycoforms of CD43 represent a mix of the α-2,3–linked and α-2,6–linked isomers (SI Appendix, Fig. S6).
While these data indicate that the disialyl core 1 motif is a component of the Siglec-7 binding structure, our findings did not fully explain the specificity of Siglec-7 binding to CD43. While Siglec-7 has previously been cocrystallized with glycans bearing a branched alpha 2,6-linkage, unbiased glycan array studies have not revealed this specific disialyl core 1 structure to be a particularly strong binder of Siglec-7 in vitro (19, 57). Additionally, disialyl core 1 is a fairly common glycan in a range of O-glycosylated proteins, not just CD43 (50). We therefore hypothesized that the way in which these glycan ligands are presented on the CD43 protein backbone confers specificity for Siglec-7. We first assessed whether Siglec-7 binding to CD43 required a specific region of the CD43 protein. We performed a limited proteolysis of CD43 in cell lysates using the mucin-specific protease StcE, generating truncated fragments of CD43 that could be detected with an antibody directed to the intracellular domain of the protein (Fig. 4D) (35). StcE cleaves mucin-type glycoproteins specifically at glycosylated residues, allowing for the generation of truncated CD43 fragments with an intact (nonglycosylated) intracellular domain (Fig. 4D) (35). Pulldown of CD43 from StcE-treated lysates showed that Siglec-7 only bound full-length CD43 fragments containing a fully intact N-terminal domain (Fig. 4D).
Specific GBP–ligand binding events have previously been demonstrated to occur upon formation of “clustered saccharide patches,” unique cell-surface structures in which repeated glycan ligands are presented in a specific orientation and topology to mediate high-affinity binding (58). Even cells that present similar overall glycan content can exhibit differential binding to GBPs if glycan epitopes are organized in these specific binding domains (58). Indeed, while CD43 is adorned with potential O-glycosylation sites throughout its extracellular domain, we noted a particularly high density of possible O-glycosylation sites in the first ∼50 amino acids of the protein, with four motifs that contained three S/T residues in a row (Fig. 4E). The remainder of the protein, while still heavily enriched in S/T residues, did not exhibit the same clustering of putative glycosylation sites. We therefore hypothesized that the closely spaced presentation of disialyl core 1 ligands at the N terminus of CD43 could form this type of clustered binding domain for Siglec-7.
Next, we localized particular sites of glycosylation in CD43 via glycoproteomic analysis by higher energy collision dissociation–triggered electron transfer dissociation MS/MS, allowing us to fully confirm over 50 individual sites of glycosylation in the protein (SI Appendix, Fig. S7). In these studies, we sequenced glycopeptides from the CD43 N terminus that showed full occupancy of clustered S/T residues with core 1 glycans (Fig. 4F). We next generated and expressed CD43 truncation constructs exhibiting sequential deletion of protein regions containing these densely glycosylated patches, starting from the N terminus of the protein. Deletion of even a small peptide region of CD43 was seen to significantly weaken pulldown of CD43 by Siglec-7 (Fig. 4G). Full deletion of approximately one-third of the extracellular domain, encompassing all of these putative clustered glycosylation motifs, totally abolished binding (Fig. 4G). Notably, binding was lost despite this truncated CD43 still being extensively glycosylated in its remaining extracellular region (SI Appendix, Fig. S7). These data indicate that strong interaction of Siglec-7 with disialyl core 1 tetrasaccharides requires display of these glycans on densely glycosylated peptide sequences that are specifically enriched at the N terminus of this mucin O-glycoprotein (Fig. 4H).
CD43 Interacts with Siglec-7 at the Immune Synapse and Drives Suppression of Immune Activity.
We next assessed whether CD43 acts as a specific ligand for Siglec-7 in live cells. We first studied the interaction of the two proteins at synapses (contact points) between primary immune cells and cancer cells. Prior studies have shown that inhibition of immune activity by Siglec-7 requires recruitment of the receptor to these synaptic structures, where its intracellular signaling domain can mediate inactivation of other activating receptors that may be engaged at the synapse (59). Natural killer (NK) cells, the majority of which constitutively express Siglec-7, were isolated and cocultured with K562 cells (9). Cells were subsequently fixed and stained with antibodies against CD43 and Siglec-7. Synapses between the cell types were visualized using super-resolution microscopy, revealing accumulation of Siglec-7 at the contact point between the two cells (Fig. 5A). CD43 accumulation was similarly observed at synaptic contact points and colocalized with Siglec-7 (SI Appendix, Fig. S8). Further detailed analysis of CD43 synaptic localization and colocalization with Siglec-7, however, was complicated by the expression of CD43 on NK cells as well as on target cells. NK cells are reported to express a glycoform of CD43 dense in core 2 O-glycans that is distinct from the core 1 glycosylated molecule we identify as a Siglec-7 ligand (60). It was therefore challenging to interpret whether synaptic accumulation reflected mobilization of target cell CD43, NK cell CD43, or both.
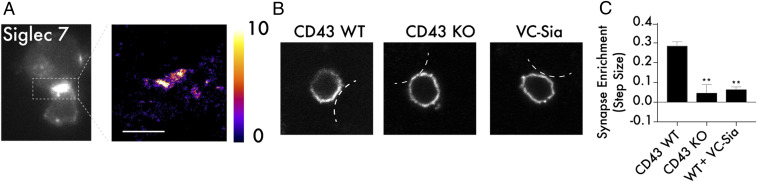
Fig. 5.
CD43 drives Siglec-7 recruitment to the immunological synapse. (A) Primary NK cells and K562 cells were cocultured at a 2:1 ratio, fixed in 4% paraformaldehyde, and stained with a fluorescent antibody against Siglec-7. Super-resolution microscopy was performed to assess recruitment of Siglec-7 to the immune synapse. The scale bar corresponds to 2 microns and the color bar corresponds to number of localizations per bin. (B) Target cell expression of CD43 is required for recruitment of Siglec-7 to the immune synapse. Primary NK cells and K562 cells that were either CD43 WT, CD43 KO, or treated with VC-Sia at 100 nM for 30 min were prepared as in A, and normal resolution confocal microscopy was used to capture the distribution of Siglec-7 along the NK cell membrane at NK cell/target cell contact points. For simplicity, the membrane of the target cell is indicated with white lines in each image. (C) A synaptic enrichment algorithm was used to quantitate the level of Siglec-7 synaptic recruitment over a series of images. A value above 0 indicates higher abundance inside the synapse than outside, and a value below 0 indicates lower abundance inside the synapse than outside. n = 3, and error bars indicate SEM. **P < 0.01.
We therefore developed a quantitative confocal microscopy approach to study the specific role of tumor cell CD43 in mediating Siglec-7 recruitment to the immune synapse. For this, an intensity trace of the Siglec-7 signal along the NK cell membrane as a measurement dimension was recorded, starting from the opposite side of the synapse, as this point could be identified easily by eye. Using this strategy, the density of Siglec-7 at the synaptic contact area was measured for a series of cells expressing CD43 (Fig. 5B and SI Appendix, Fig. S9). The majority of cells imaged in a given sample exhibited significant polarization of Siglec-7 toward the immune synapse under these conditions (SI Appendix, Figs. S9 and S10). When the same experiment was repeated with a CD43 KO cell line, polarization of Siglec-7 no longer occurred consistently, with cells just as likely to display higher density of Siglec-7 outside the synapse as inside it (Fig. 5 B and C and SI Appendix, Figs. S9 and S10). Indeed, KO of the CD43 protein created the same reduction in synaptic polarization seen following treatment with a sialidase enzyme, which nonspecifically cleaves sialic acid off the cell surface (Fig. 5 B and C and SI Appendix, Figs. S9 and S10).
These data indicate a role for CD43 in physiological recruitment of Siglec-7 to cell–cell contact points. Therefore, it follows that CD43 KO should enhance the killing of cancer cell lines by NK cells. As expected, coculture of NK cells with CD43 KO K562 cells led to substantially higher cell killing than with WT cells (Fig. 6A). Lentiviral transduction of WT CD43 into CD43 KO cells eliminated this hypersensitivity to NK cell lysis, indicating that sensitization to NK cell killing is a specific effect of CD43 KO (SI Appendix, Fig. S11). Notably, when both cell lines were pretreated with a sialidase enzyme, this difference in cellular sensitivity was abolished, indicating that hypersensitivity of CD43 KO cells is produced specifically by elimination of glycan ligands presented in the extracellular domain and not by some other function of the protein (Fig. 6A).
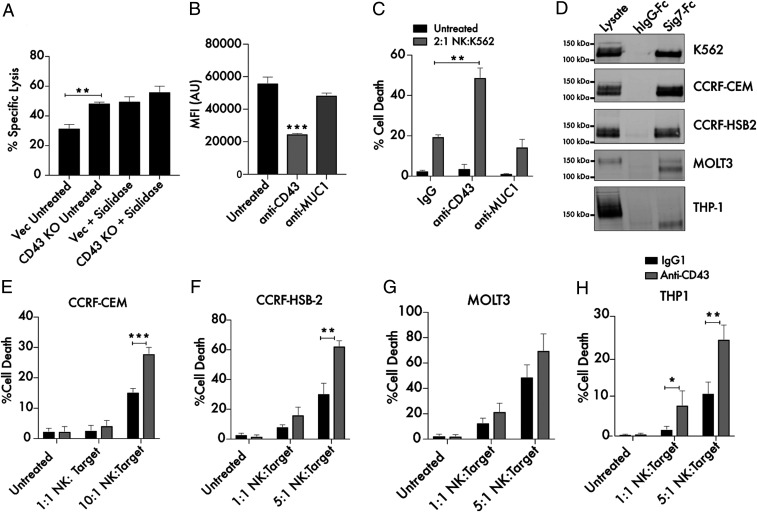
Fig. 6.
Targeting CD43 enhances killing of leukemia cells by primary NK cells. (A) Primary NK cells and K562 Vec (empty vector transfected) or CD43 KO target cells were cocultured for 4 h at a 2:1 ratio and subjected to flow cytometry using live cell (CellTrace Far Red) and dead cell (Sytox Green) stains to quantitate specific lysis of the target cells. The experiment was also performed following treatment of both Vec and KO target cells with 2 μM endotoxin-free sialidase from Salmonella Typhimurium. (B) A total of 104 K562 cells were treated with 10 μg/mL anti-CD43 (MEM-59) or anti-Muc1 (VU4H5) antibodies for 30 min, incubated with 100 ng/mL Siglec-7–Fc/Alexa Fluor 488, and subjected to flow cytometry. MFI indicates mean fluorescence intensity, and AU indicates arbitrary units. (C) K562 cells were treated for 30 min with 10 μg/mL isotype control, anti-CD43, and anti-MUC1 antibodies. Cells were then cocultured with primary NK cells at the indicated ratio as in A. The “Untreated” condition refers to target cells that have not been incubated with NK cells. (D) Lysates from the indicated leukemia and lymphoma cell lines were passed over Siglec-7–Fc functionalized beads. Bound proteins were eluted from the beads and subjected to Western blot to assess the presence of a Siglec-7–binding CD43 glycoform. Beads conjugated to huFc were used as a control. A total of 25 μg protein was loaded in the input lysate, while 500 μg protein was used for each IP. (E) CCRF-CEM cells, (F) CCRF-HSB2 cells, (G) MOLT3 cells, and (H) THP-1 cells were treated for 30 min with 10 μg/mL isotype control or anti-CD43 mouse IgG1 antibody and cocultured with primary NK cells at the indicated ratios as in A. n = 3 in all cases. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars indicate SEM.
Given the enhancement of NK cell activity we observed toward CD43 KO cells, we wondered whether it would be possible to selectively enhance immune cell killing activity by treatment with CD43-blocking antibodies. As Siglec-7 was found to bind a defined epitope at the N terminus of CD43, we sought to identify antibodies that would specifically occlude interactions with this Siglec-7–binding domain. Indeed, we found that a commercially available anti-CD43 antibody (MEM-59 clone), which binds the N-terminal region of CD43, could block binding of recombinant Siglec-7–Fc to the surface of K562 cells (Fig. 6B) (61). An antibody targeted against MUC1, another heavily glycosylated cell-surface protein also expressed in K562s (SI Appendix, Fig. S12), did not block Siglec-7–Fc binding (Fig. 6B) (35). Pretreatment of K562 cells with the same anti-CD43 antibody was also found to enhance NK cell lysis of K562 cells. Treatment with a MUC1 antibody (also a mouse IgG1 isotype), conversely, had no effect on NK cell cytotoxic activity (Fig. 6C). These results indicate that enhancement of NK cell activity results specifically from CD43 blockade and not from antibody-dependent cellular cytotoxicity. This finding is consistent with prior work showing that mouse IgG1 antibodies exhibit low cross-reactivity with the CD16 family of activating human Fc receptors expressed on NK cells (62, 63). Enhanced NK cell activity was still observed following pretreatment of NK cells with an antibody blocking the CD16 Fc receptor, confirming this finding (SI Appendix, Fig. S13). Anti-CD43 Fab fragments lacking an intact Fc domain were also generated and similarly potentiated NK cell killing activity (SI Appendix, Fig. S14). These results suggest that selective blockade of glyco-immune checkpoint ligands like CD43 can enhance anticancer activity of cytotoxic immune cells.
In order to assess the broader implications of these findings, we identified a series of leukemia and lymphoma cell lines of various cytological origin and tested for expression of Siglec-7–binding CD43 by pulldown assay (Fig. 6D). All cell lines tested exhibited some expression of the previously observed 125 kDa CD43 glycoform that interacted with Siglec-7 (hereafter referred to as CD43Sig7-Lig). For some cell lines (K562, CCRF-CEM, and CCRF–HSB-2), this glycoform was the most abundant CD43 species observed in the cell lysate. In other cell lines (MOLT3 and THP-1 cells) the Siglec-7–binding glycoform represented a minority of total cellular CD43, with the most abundant CD43 species running at a higher MW. These differences demonstrate the heterogeneity of CD43 glycosylation across different cell types and further illustrate that Siglec-7 selectively binds one glycoform of CD43. Notably, cell lines that exhibited higher expression of CD43Sig7-Lig by pulldown assay were also found to have higher expression of Siglec-7 ligands by cell-surface staining (SI Appendix, Fig. S15).
We finally tested whether blockade of CD43Sig7-Lig would enhance activity of primary NK cells toward this broad range of leukemia cell lines. Across this panel of cancer cell lines, we found that treatment with an anti-CD43 antibody consistently increased tumor cell lysis by human NK cells (Fig. 6 D–H). Enhancement of activity was proportional to the levels of CD43Sig7-Lig expressed on the target cell line. These findings imply that CD43Sig7-Lig is therefore a bona fide inhibitory immune molecule that suppresses the detection and killing of a range of leukemia cell types by cytotoxic immune cells. CD43’s interaction with Siglec-7 can be selectively blocked using currently available CD43-binding agents, a finding with possible implications for therapeutic development.
Discussion
The emergence of Siglecs as targets for cancer immune therapy creates a strong motivation to identify biological ligands for these receptors. Here, we used a cell-based genetic screening approach to characterize the determinants of Siglec-binding interactions on live cells. We found that Siglec-7, but not Siglec-9, exhibits significant selective binding to a glycoform of CD43. Further experiments identified a Siglec-7–binding motif present at the N terminus of CD43 that is densely glycosylated with disialyl core 1 epitopes. The presence of this single protein was required to mediate inhibition of immune activity by Siglec-7. The precise molecular interactions that underlie this unusual specificity will be an interesting topic for further structural biology studies. The determinants of Siglec-7 binding to CD43 are reminiscent of those that structure the binding of the immune adhesion receptor P-selectin to its ligand PSGL-1 (22). Both GBP–glycan interactions are mediated by a discrete glycan motif presented in the context of a specific polypeptide backbone (22). This work, along with other more recent studies identifying biological ligands for Siglecs (such as Siglec-10/CD24), suggests that binding of many of these receptors to cancer cells may be mediated by association with specific glycoforms of cell-surface protein ligands (7).
The finding that the CD43 protein is a primary determinant of Siglec-7 ligand expression on cancer cell lines has important implications for therapeutic development. As our screen was conducted in the K562 (CML) genetic background, these findings primarily have relevance for leukemia- and lymphoma-targeted therapeutics. Indeed, clinical studies have linked CD43 expression to adverse patient outcomes specifically in the context of diffuse large B cell lymphoma (64). The discovery that CD43 acts as a ligand for immune checkpoint receptors like Siglec-7 could be related to these findings. It should be noted that the primacy of CD43 as a ligand for Siglec-7 on these cancer types does not preclude other structures playing a similar role in different cell models. The clustered glycopeptide motif defined here as the Siglec-7 binding epitope, for example, could also be expressed on other mucin-type glycoproteins bearing homologous regions of clustered glycosylation. Indeed, our studies indicated that Siglec-7 ligand expression is significantly reduced by CD43 KO but almost completely ablated by treatment with an O-glycoprotein specific protease (SI Appendix, Fig. S1). These results imply that other mucins could potentially carry the Siglec-7–binding motif found at the N terminus of CD43 (35).
In the future, it will also be interesting to study how blockade of these glyco-immune checkpoints affects tumor growth in vivo. Such studies will have to be carefully designed, as the lack of direct homology between murine and human Siglecs means that insights derived from human cell studies are often difficult to straightforwardly translate into mouse model systems (27). While the closest murine homolog of Siglec-7 is understood to be mouse Siglec-E, it is unclear whether the selective binding relationship we describe between Siglec-7 and human CD43 will exist for Siglec-E and murine CD43 (65). If not, further study of this immune checkpoint in vivo may require extensive new work to generate immune-competent humanized mouse models.
While our primary intention in designing this study was the investigation of Siglec-7 and -9, our findings also have relevance to the large body of work that has attempted to define CD43’s immunological function. A glycoform of CD43 expressing disialyl core 1 ligands has been found on the surface of naive T cells (66). Indeed, we have found that Siglec-7 efficiently interacts with T cell CD43 using the pulldown assays we have previously described (SI Appendix, Fig. S16). Monocyte CD43, which is known to be differently glycosylated with hexasaccharide glycans, does not interact with Siglec-7 (66) (SI Appendix, Fig. S16). These data hint at the possibility that Siglec-7/CD43 binding may have evolved in humans as a mechanism of endogenous immune regulation. Just as Siglec-7 possesses a canonical ITIM, CD43 also has an intracellular signaling domain that becomes hyperphosphorylated during T cell activation (40). The discovery of a specific interaction between these two proteins implies rich possibilities for regulatory crosstalk between the innate and adaptive immune systems, with particular applicability to understanding interactions of CD43-expressing T cells with Siglec-expressing antigen-presenting cells (67). These topics could also have therapeutic relevance, especially in light of the recent successful development of recombinant Siglec ligands as anti-inflammatory therapeutics (68).
Finally, these studies represent application of genomic CRISPR screening to defining a functional glyco-immune receptor-ligand pair. CRISPR screening is, in some ways, the ideal tool for investigating such interactions, which are driven by polygenetic contributions from glycosyltransferase enzymes and cell-surface proteins that act in concert to create complex binding epitopes. By rapidly profiling the function of thousands of genes in a single biological experiment, genome-wide CRISPR screening allows dissection of the genetic basis of such binding events at an unprecedented level of detail. We envision many future applications of this approach to uncover new ligands and biological functions for the huge number of glycan-binding receptors that play important roles in human physiology and disease.
Methods
Cell Culture.
K562, CCRF-CEM, CCRF–HSB-2, MOLT-3, and THP-1 cells (acquired from the American Type Culture Collection) were all cultured at low passage in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% fetal bovine serum (FBS; Sigma) and 1% Penicillin/Streptomycin (Gibco) at 5% CO2 and 37 °C. HEK293T cells were all cultured at low passage in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS (Sigma) and 1% Penicillin/Streptomycin (Gibco) at 5% CO2 and 37 °C. Primary NK cells were cultured following isolation in RPMI supplemented with 10% heat-inactivated FBS (Sigma) at 5% CO2 and 37 °C. Methods for peripheral blood mononuclear cell (PBMC) isolation and immune cell subtype isolation are detailed in SI Appendix.
Genome-Wide CRISPRi Screening.
K562-dCas9–KRAB cells were transduced with the genome-wide CRISPRi library, stained with Siglec-7–Fc/Siglec-9–Fc, and sorted so as to isolate a population of cells exhibiting at least a 10-fold decrease in Siglec-Fc staining. Libraries were subsequently amplified from genomic DNA of both sorted and unsorted samples. Illumina sequencing was then performed to obtain sgRNA read counts for every guide in each sample. Detailed protocols for all these steps are described in SI Appendix.
Generation of CD43 KO, CD43-myc, and CD43 Knock-In Lines.
CD43 KO was performed by lentiviral transduction of a LentiCRISPR-v2 vector (69) encoding the relevant sgRNA sequence and Cas9, followed by selection and sorting for CD43 KO cells. CD43-myc and CD43 knock-in lines were generated by lentiviral transduction of CD43 cDNA sequences. Detailed experimental protocols are included in SI Appendix.
Lectin Staining.
Cells were pelleted by centrifugation and washed once with phosphate-buffered saline (PBS). Siglec-Fc precomplexes with anti-huIgG–Alexa Fluor 488 were prepared at either 1 μg/mL or 100 ng/mL concentration in FACS buffer (PBS containing 0.5% BSA). SNA-biotin and MAL-II-biotin lectins (Vector Labs B-1305-2 and B-1265-1) were precomplexed with Alexa Fluor 488-Streptavidin (Thermo Fisher Scientific) at 1 μg/mL in FACS buffer. Staining was performed for 30 min on ice at 1 × 106 cells/mL. Cells were subsequently washed twice with cold PBS and analyzed by flow cytometry on a BD Accuri C6 instrument. Gating on forward scatter/side scatter was used to identify intact cells. Where indicated, antibody treatments prior to lectin staining were performed for 30 min at 37 °C using 10 μg/mL anti-CD43 (MEM-59 clone, Abcam, ab9088) or 10 μg/mL anti-MUC1 (VU4H5 clone, Cell Signaling).
Siglec Pulldown for Western Blot.
Cells were lysed in ice cold 0.1% Nonidet P-40 containing 1 mM ethylenediaminetetraacetic acid (EDTA) and 1× Halt Protease Inhibitor mixture (Thermo Fisher Scientific) and rotated at 4 °C for 1 h. The insoluble fraction was then cleared by centrifugation at 18,000 × g for 15 min. Protein concentration was determined by bicinchoninic acid (BCA) assay, and lysates were diluted further in lysis buffer to 500 μL with a final protein concentration of 1 μg/mL. In parallel, 5 μg Siglec-Fc was mixed with 50 μL Protein G Dynabeads (Thermo Fisher Scientific) in 250 μL PBS and rotated at room temperature for 1 h. Beads were isolated with a magnet and washed once with PBS. Cell lysates were then added to Siglec-coated beads at rotated at 4 °C for at least 1 h. Where indicated, digestions were performed with 100 nM sialidase from Vibrio Cholerae (VC-Sialidase), 100 ng/mL StcE, 1 μL β1-3,4–galactosidase (New England Biolabs P0746S) in a total volume of 30 μL, and 1 μL S. pneumoniae O-glycosidase (Sigma G1163) in a total volume of 30 μL for 1 h at 37 °C. VC-Sialidase was expressed and purified as previously reported (14). Following pulldown, flow-through was removed and beads were washed four times with ice cold 0.1% Nonidet P-40. Western blot analysis was performed as described in SI Appendix.
Siglec Pulldown for Proteomics.
A total of 10 × 106 K562 cells were lysed and diluted as described above. Lysate was split into two 500-μL aliquots; one aliquot was left untreated, while the other was treated with 100 nM VC-Sialidase for 1 h. In parallel, 5 μg Siglec-Fc was mixed with 50 μL Protein G Dynabeads in 250 μL PBS and rotated at room temperature for 1 h. Beads were isolated with a magnet and washed once with PBS. Treated and untreated cell lysates were then added to the beads and incubated overnight at 4 °C with rotation. Following pulldown, flow-through was removed and beads were washed twice with ice cold 0.1% Nonidet P-40 and twice with ice cold 50 mM ammonium bicarbonate. Trypsin (1 μg, Promega) and GluC (1 μg, Progema) were then added to beads in 100 μL 50 mM ammonium bicarbonate, and beads were incubated overnight with shaking at 37 °C. The following day, digestion was acidified by adding formic acid (FA) to beads to a final concentration of 2%, followed by shaking at 37 °C for 30 min. Supernatant was then removed; beads were washed once with 100 μL 2% FA, and this wash was pooled with the original supernatant. The digested sample was dried down in a speedvac and resuspended in 10 μL 0.1% FA. Half of the sample (5 μL) was analyzed by MS/MS according to methods detailed in SI Appendix.
Large-Scale Isolation of CD43.
A total of 500 × 108 K562 CD43-myc–FLAG cells were pelleted at 600 × g, washed once with PBS, and resuspended in 10 mL Mammalian Protein Extraction Reagent (M-PER) (Thermo Fisher Scientific) with 1 mM EDTA and 1× Halt Protease Inhibitor mixture. To facilitate cell lysis, the cell suspension was sonicated with six 10-s pulses (output 2.0) using a Branson Sonicator Microtip and rotated for at least 1 h at 4 °C. Insoluble material was then pelleted by centrifugation at 18,000 × g for 15 min. Lysate was heated at 95 °C for 5 min to denature protein and maximally expose myc-tag epitope for binding and then placed back on ice and allowed to cool to 4 °C. A packed volume anti-cMyc agarose resin (Pierce, 500 μL) was then added to the lysate and rotated overnight at 4 °C. The resin was subsequently loaded onto a fritted 30 mL column. Lysate was removed, and resin was subsequently washed with 300 mL cold PBS containing 0.1% Tween-20 detergent (PBST), followed by 100 mL cold PBS (without detergent). To elute bound CD43-myc–FLAG, resin was resuspended twice in 500 μL 50 mM NaOH and allowed to stand for 1 min; eluate was then collected and neutralized by addition 50 μL 1 M Tris, pH 7.5. Protein was then buffer exchanged into PBS by repeatedly running eluate through 500-μL-volume Amicon columns with a 10 kDa MW cutoff (Sigma UFC501096). Samples were concentrated to <50 μL final volume and immediately processed for downstream analysis (see below). Purity of protein was confirmed by sodium dodecyl sulfate polyacrylamide gel electrophoresis and silver stain. Glycomic analysis of CD43 was performed as described in SI Appendix.
Glyco-Site Mapping of CD43.
CD43-myc–FLAG was isolated as described above. Sample eluate (50 μL) was treated with 100 nM VC-Sialidase, 1 μg trypsin and 1 μg GluC (Promega) at 37 °C overnight, with shaking. Reduction and alkylation were not performed as the CD43 protein sequence does not contain cysteines. Sample was acidified by addition of FA to 2%, followed by shaking at 37 °C for 30 min. Samples were dried down in the speedvac and subsequently resuspended in 10 μL MS-grade H2O (Pierce). Cleanup was performed using a C18 ZipTip resin (EMD Millipore). Desalted samples were subsequently dried down in the speedvac. MS was performed as described in SI Appendix.
Super-Resolution Cell-Surface Imaging.
Chambered 1.5-mm borosilicate slides (Thermo Fisher Scientific) were coated for 1 h with 20 μg/mL Fibronectin (Sigma) in PBS for 1 h at 37 °C. Slides were subsequently washed once with PBS and allowed to air dry at room temperature. A total of 1 × 105 K562 cells/well were subsequently plated in 250 μL media and allowed to adhere overnight. Media was removed from slides, and 250 μL 4% electron microscopy-grade paraformaldehyde (Fisher) was added directly to each well. Fixation was performed for 20 min. Cells were subsequently washed with PBS twice for 10 min each with gentle agitation. Cells were stained for 1 h at 4 °C with 1 μg/mL Sig7-Fc or Sig-9–Fc/anti-hIgG–Alexa Fluor 647 precomplex (prepared as above) and 1 μg/mL anti-CD43 antibody (Abcam) conjugated to a CF568 dye (conjugation performed using Biotium Mix-n-Stain Antibody Labeling Kit) for 1 h at 4 °C with protection from ambient light. Cells were then washed twice for 5 min each with PBS. Single molecule localization microscopy was performed as described in SI Appendix.
NK Cell Killing Assays.
For all NK cell killing assays, cells were isolated as described in SI Appendix. Target cells were harvested by centrifugation and resuspended in serum-free RPMI containing 5 μM CellTrace Far Red at 5 × 105 cells/mL. Cells were then incubated for 30 min at 37 °C. Following staining, cells were spun down, washed once with complete media, and resuspended in complete media. Cell concentration was then requantitated, and cells were diluted to a concentration of 1 × 105 cells/mL in complete media containing 100 nM Sytox Green. The cell suspension (100 μL) was then aliquoted into a flat bottom 96-well plate. Separately, NK cells were diluted to various cell concentrations to generate the indicated ratios of effector:target cells in complete media containing 100 nM Sytox Green. Then, 100 μL/well of these cell suspensions were mixed with the target cell suspensions to generate a 200-μL total volume. Where indicated, target cell populations were treated with either isotype mouse IgG1 (BioRad), mouse IgG1 anti-CD43 (MEM-59 clone, Thermo Fisher Scientific or Abcam), or mouse IgG1 anti-MUC1 (VU4H5 clone, Cell Signaling), or corresponding Fab fragments for 30 min. Antibodies were subsequently washed out by spinning cells down in 96-well plates for 5 min at 600 × g. For CD16 blocking experiments, NK cells were treated with 10 μg/mL purified mouse IgG1 anti-human CD16 (BioLegend, 3G8 clone) prior to coculture. For sialidase treatment experiments, target cells were treated for 30 minutes with 2 μM endotoxin-free sialidase from Salmonella Typhimurium, which was expressed and endotoxin-purified as previously reported (16). Killing was allowed to proceed at 37 °C for 4 h, and target cell death was quantitated by flow cytometry as described in SI Appendix.
NK Cell Immune Synapse Imaging.
Chambered 1.5-mm borosilicate slides (Thermo Fisher Scientific) were coated for 1 h with 20 μg/mL Fibronectin (Sigma) in PBS for 1 h at 37 °C. Slides were subsequently washed once with PBS and allowed to air dry at room temperature. A total of 1 × 105 K562 cells/well were subsequently plated in 250 μL media and allowed to adhere overnight. The next day, NK cells were isolated from whole blood PBMCs as described above and resuspended in PBS at 8 × 105 cells/mL. Media was removed from slides, and 250 μL/well of this NK cell suspension was added to achieve an effector:target ratio of 2:1. Slides were then incubated at 37 °C for 30 min in PBS. Following incubation, 16% EM-grade paraformaldehyde (Fisher) was added directly to each well to a final concentration of 4% in each well. Fixation was performed for 20 min. Cells were subsequently washed with PBS twice for 10 min each with gentle agitation. Cells were then stained with 1 μg/mL anti–Siglec-7 antibody (BioLegend, Clone S7.7) conjugated to a CF568 dye (conjugation performed as described above) and/or with 1 μg/mL anti-CD43-Alexa Fluor 647 (Thermo Fisher Scientific) in FACS Buffer for 1 h at 4 °C with protection from ambient light. Cells were then washed twice for 5 min with PBS under gentle agitation. Confocal microscopy was performed on a Nikon A1R confocal microscope equipped with a Plan Fluor 60× oil immersion 1.30-numerical aperture objective. This instrument was equipped with a 561-nm green laser and a 639-nm red laser for CF568 and Alexa Fluor 647 excitation, respectively. Quantitative image analysis was performed as described in SI Appendix.
Statistical Analysis.
In all cases, a standard Student’s two-tailed t test was applied as a standard method for determining statistical significance. Further statistical details are included in figure legends. No statistical method was used to determine sample size. No data exclusions were performed. All data were confirmed with multiple biological replicates as indicated in the figure legends.
Supplementary Material
Acknowledgments
S.W. is supported by a Banting Postdoctoral Fellowship from the Canadian Institutes of Health Research. Work was broadly supported by NIH grants R01CA227942 and U01 CA226051-01A1 to C.R.B. and by R35GM118067 to W.E.M. We thank the Complex Carbohydrates Research Consortium, in particular Parastoo Azadi and Asif Shajahan, for assistance with glycomics analysis. Glycomics analysis was supported in part by NIH grant 1S10OD018530 to Parastoo Azadi. N.M.R. was funded through an NIH Predoctoral to Postdoctoral Transition Award (Grant K00 CA21245403). S.A.M. was supported by a National Institute of General Medical Sciences F32 Postdoctoral Fellowship (F32-GM126663-01). K.P. was supported by a US NSF Graduate Research Fellowship, a Stanford Graduate Fellowship, and the Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program. B.A.H.S. was supported by a Predoctoral Fellowship from the NIH (F30CA232541) and the Stanford School of Medicine Medical Scientist Training Program. M.A.G. was supported by the NSF Graduate Research Fellowship and the Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program. Human CRISPRi-v2 was a gift from Jonathan Weissman (Addgene identification 83969). LentiCRISPR v2 was a gift from Feng Zhang (Addgene identification 52961).
Footnotes
Competing interest statement: C.R.B. is a cofounder and Scientific Advisory Board member of Palleon Pharmaceuticals, Enable Biosciences, Redwood Bioscience (a subsidiary of Catalent), and InterVenn Biosciences, and a member of the Board of Directors of Eli Lilly and Company. S.W., B.A.H.S., and C.R.B. are co-inventors on a patent application related to this work held by Stanford University (PCT/US2020/041603).
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015024118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Waldman A. D., Fritz J. M., Lenardo M. J., A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seidel J. A., Otsuka A., Kabashima K., Anti-PD-1 and anti-CTLA-4 therapies in cancer: Mechanisms of action, efficacy, and limitations. Front. Oncol. 8, 86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang J., et al. , Trial watch: The clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug Discov. 17, 854–855 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Wolchok J. D., et al. , Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 377, 1345–1356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muñoz-Wolf N., Lavelle E. C., Innate immune receptors. Methods Mol. Biol. 1417, 1–43 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Kawai T., Akira S., The role of pattern-recognition receptors in innate immunity: Update on toll-like receptors. Nat. Immunol. 11, 373–384 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Barkal A. A., et al. , CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature 572, 392–396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J., et al. , Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 25, 656–666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jandus C., et al. , Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Invest. 124, 1810–1820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown G. D., Willment J. A., Whitehead L., C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 18, 374–389 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Macauley M. S., Crocker P. R., Paulson J. C., Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bornhöfft K. F., Goldammer T., Rebl A., Galuska S. P., Siglecs: A journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev. Comp. Immunol. 86, 219–231 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Beatson R., et al. , The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat. Immunol. 17, 1273–1281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao H., Woods E. C., Vukojicic P., Bertozzi C. R., Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc. Natl. Acad. Sci. U.S.A. 113, 10304–10309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Büll C., et al. , Targeted delivery of a sialic acid-blocking glycomimetic to cancer cells inhibits metastatic spread. ACS Nano 9, 733–745 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Gray M. A., et al. , Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat. Chem. Biol. 16, 1376–1384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majeti R., et al. , CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bochner B. S., et al. , Glycan array screening reveals a candidate ligand for Siglec-8. J. Biol. Chem. 280, 4307–4312 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Gieseke F., et al. , Siglec-7 tetramers characterize B-cell subpopulations and leukemic blasts. Eur. J. Immunol. 42, 2176–2186 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Narimatsu Y., et al. , An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol. Cell 75, 394–407.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godula K., Bertozzi C. R., Density variant glycan microarray for evaluating cross-linking of mucin-like glycoconjugates by lectins. J. Am. Chem. Soc. 134, 15732–15742 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sako D., et al. , A sulfated peptide segment at the amino terminus of PSGL-1 is critical for P-selectin binding. Cell 83, 323–331 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Marcelo F., et al. , Identification of a secondary binding site in human macrophage galactose-type lectin by microarray studies: Implications for the molecular recognition of its ligands. J. Biol. Chem. 294, 1300–1311 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudak J. E., Canham S. M., Bertozzi C. R., Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 10, 69–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanczak M. A., et al. , Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J. Clin. Invest. 128, 4912–4923 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horlbeck M. A., et al. , Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duan S., Paulson J. C., Siglecs as immune cell checkpoints in disease. Annu. Rev. Immunol. 38, 365–395 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Isaji T., et al. , An oncogenic protein Golgi phosphoprotein 3 up-regulates cell migration via sialylation. J. Biol. Chem. 289, 20694–20705 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith R. D., Lupashin V. V., Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 343, 2024–2031 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pang S. H. M., et al. , PU.1 is required for the developmental progression of multipotent progenitors to common lymphoid progenitors. Front. Immunol. 9, 1264 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gomes C., et al. , Expression of ST3GAL4 leads to SLe(x) expression and induces c-Met activation and an invasive phenotype in gastric carcinoma cells. PLoS One 8, e66737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicoll G., et al. , Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via siglec-7-dependent and -independent mechanisms. Eur. J. Immunol. 33, 1642–1648 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto N., et al. , The ceramide moiety of disialoganglioside (GD3) is essential for GD3 recognition by the sialic acid-binding lectin SIGLEC7 on the cell surface. J. Biol. Chem. 294, 10833–10845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitazume-Kawaguchi S., Kabata S., Arita M., Differential biosynthesis of polysialic or disialic acid Structure by ST8Sia II and ST8Sia IV. J. Biol. Chem. 276, 15696–15703 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Malaker S. A., et al. , The mucin-selective protease StcE enables molecular and functional analysis of human cancer-associated mucins. Proc. Natl. Acad. Sci. U.S.A. 116, 7278–7287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magistretti P. J., et al. , Gangliosides: Treatment avenues in neurodegenerative disease. Front. Neurol. 10, 859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An H. J., Froehlich J. W., Lebrilla C. B., Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 13, 421–426 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenstein Y., Santana A., Pedraza-Alva G., CD43, a molecule with multiple functions. Immunol. Res. 20, 89–99 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Tuccillo F. M., et al. , Cancer-associated CD43 glycoforms as target of immunotherapy. Mol. Cancer Ther. 13, 752–762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mody P. D., et al. , Signaling through CD43 regulates CD4 T-cell trafficking. Blood 110, 2974–2982 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Velázquez F., et al. , CD43 functions as an E-selectin ligand for Th17 cells in vitro and is required for rolling on the vascular endothelium and Th17 cell recruitment during inflammation in vivo. J. Immunol. 196, 1305–1316 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manjunath N., Correa M., Ardman M., Ardman B., Negative regulation of T-cell adhesion and activation by CD43. Nature 377, 535–538 (1995). [DOI] [PubMed] [Google Scholar]
- 43.van den Berg T. K., et al. , Cutting edge: CD43 functions as a T cell counterreceptor for the macrophage adhesion receptor sialoadhesin (Siglec-1). J. Immunol. 166, 3637–3640 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Fuhlbrigge R. C., King S. L., Sackstein R., Kupper T. S., CD43 is a ligand for E-selectin on CLA+ human T cells. Blood 107, 1421–1426 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibuya N., et al. , Fractionation of sialylated oligosaccharides, glycopeptides, and glycoproteins on immobilized elderberry (Sambucus nigra L.) bark lectin. Arch. Biochem. Biophys. 254, 1–8 (1987). [DOI] [PubMed] [Google Scholar]
- 46.Knibbs R. N., Goldstein I. J., Ratcliffe R. M., Shibuya N., Characterization of the carbohydrate binding specificity of the leukoagglutinating lectin from Maackia amurensis. Comparison with other sialic acid-specific lectins. J. Biol. Chem. 266, 83–88 (1991). [PubMed] [Google Scholar]
- 47.Wang Y., et al. , Cosmc is an essential chaperone for correct protein O-glycosylation. Proc. Natl. Acad. Sci. U.S.A. 107, 9228–9233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurosawa N., Kojima N., Inoue M., Hamamoto T., Tsuji S., Cloning and expression of Gal beta 1,3GalNAc-specific GalNAc alpha 2,6-sialyltransferase. J. Biol. Chem. 269, 19048–19053 (1994). [PubMed] [Google Scholar]
- 49.Marcos N. T., et al. , Role of the human ST6GalNAc-I and ST6GalNAc-II in the synthesis of the cancer-associated sialyl-Tn antigen. Cancer Res. 64, 7050–7057 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Vyas A. A., Blixt O., Paulson J. C., Schnaar R. L., Potent glycan inhibitors of myelin-associated glycoprotein enhance axon outgrowth in vitro. J. Biol. Chem. 280, 16305–16310 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitagawa H., Paulson J. C., Cloning of a novel alpha 2,3-sialyltransferase that sialylates glycoprotein and glycolipid carbohydrate groups. J. Biol. Chem. 269, 1394–1401 (1994). [PubMed] [Google Scholar]
- 52.Priatel J. J., et al. , The ST3Gal-I sialyltransferase controls CD8+ T lymphocyte homeostasis by modulating O-glycan biosynthesis. Immunity 12, 273–283 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Dimitroff C. J., Galectin-binding O-glycosylations as regulators of malignancy. Cancer Res. 75, 3195–3202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Z. L., et al. , Direct fluorescent glycan labeling with recombinant sialyltransferases. Glycobiology 29, 750–754 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Z. L., Huang X., Burton A. J., Swift K. A. D., Probing sialoglycans on fetal bovine fetuin with azido-sugars using glycosyltransferases. Glycobiology 26, 329–334 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Moody A. M., et al. , Sialic acid capping of CD8beta core 1-O-glycans controls thymocyte-major histocompatibility complex class I interaction. J. Biol. Chem. 278, 7240–7246 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Alphey M. S., Attrill H., Crocker P. R., van Aalten D. M. F., High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J. Biol. Chem. 278, 3372–3377 (2003). [DOI] [PubMed] [Google Scholar]
- 58.Cohen M., Varki A., Modulation of glycan recognition by clustered saccharide patches. Int. Rev. Cell Mol. Biol. 308, 75–125 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Falco M., et al. , Identification and molecular cloning of p75/AIRM1, a novel member of the sialoadhesin family that functions as an inhibitory receptor in human natural killer cells. J. Exp. Med. 190, 793–802 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aguado E., Santamaría M., Gallego M. D., Peña J., Molina I. J., Functional expression of CD43 on human natural killer cells. J. Leukoc. Biol. 66, 923–929 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Alvarado M., Klassen C., Cerny J., Horejsí V., Schmidt R. E., MEM-59 monoclonal antibody detects a CD43 epitope involved in lymphocyte activation. Eur. J. Immunol. 25, 1051–1055 (1995). [DOI] [PubMed] [Google Scholar]
- 62.Petricevic B., et al. , Trastuzumab mediates antibody-dependent cell-mediated cytotoxicity and phagocytosis to the same extent in both adjuvant and metastatic HER2/neu breast cancer patients. J. Transl. Med. 11, 307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Temming A. R., et al. , Cross-reactivity of mouse IgG subclasses to human Fc gamma receptors: Antibody deglycosylation only eliminates IgG2b binding. Mol. Immunol. 127, 79–86 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Ma X. B., Zheng Y., Yuan H. P., Jiang J., Wang Y. P., CD43 expression in diffuse large B-cell lymphoma, not otherwise specified: CD43 is a marker of adverse prognosis. Hum. Pathol. 46, 593–599 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Crocker P. R., Paulson J. C., Varki A., Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Brown T. J., et al. , Characterization of a CD43/leukosialin-mediated pathway for inducing apoptosis in human T-lymphoblastoid cells. J. Biol. Chem. 271, 27686–27695 (1996). [DOI] [PubMed] [Google Scholar]
- 67.Ding Y., et al. , The lectin Siglec-G inhibits dendritic cell cross-presentation by impairing MHC class I-peptide complex formation. Nat. Immunol. 17, 1167–1175 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Tian R.-R., et al. , CD24Fc protects against viral pneumonia in simian immunodeficiency virus-infected Chinese rhesus monkeys. Cell. Mol. Immunol. 17, 887–888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.