Significance
Plant cells are surrounded by an extracellular matrix known as the cell wall. We have analyzed the contribution of the Arabidopsis cell wall to disease resistance to pathogens with different parasitic styles. Here, we demonstrate that plant cell walls are determinants of immune responses since modification of their composition in a set of Arabidopsis cell wall mutants has an impact on their disease resistance and fitness phenotypes. In these genotypes, we identified specific correlations between the amounts of specific wall carbohydrate epitopes and disease resistance/fitness phenotypes through mathematical analyses. These data support the relevant and specific function of plant cell wall composition in plant immune responses and provide the basis for using wall traits in crop breeding programs.
Keywords: cell wall, disease resistance, immunity, fitness, glycomics
Abstract
Plant cell walls are complex structures subject to dynamic remodeling in response to developmental and environmental cues and play essential functions in disease resistance responses. We tested the specific contribution of plant cell walls to immunity by determining the susceptibility of a set of Arabidopsis cell wall mutants (cwm) to pathogens with different parasitic styles: a vascular bacterium, a necrotrophic fungus, and a biotrophic oomycete. Remarkably, most cwm mutants tested (29/34; 85.3%) showed alterations in their resistance responses to at least one of these pathogens in comparison to wild-type plants, illustrating the relevance of wall composition in determining disease-resistance phenotypes. We found that the enhanced resistance of cwm plants to the necrotrophic and vascular pathogens negatively impacted cwm fitness traits, such as biomass and seed yield. Enhanced resistance of cwm plants is not only mediated by canonical immune pathways, like those modulated by phytohormones or microbe-associated molecular patterns, which are not deregulated in the cwm tested. Pectin-enriched wall fractions isolated from cwm plants triggered immune responses in wild-type plants, suggesting that wall-mediated defensive pathways might contribute to cwm resistance. Cell walls of cwm plants show a high diversity of composition alterations as revealed by glycome profiling that detect specific wall carbohydrate moieties. Mathematical analysis of glycome profiling data identified correlations between the amounts of specific wall carbohydrate moieties and disease resistance phenotypes of cwm plants. These data support the relevant and specific function of plant wall composition in plant immune response modulation and in balancing disease resistance/development trade-offs.
Plants are under continuous pathogen threats that might compromise their survival and reproduction. To cope with these threats, plants have evolved a plethora of resistance mechanisms, which are either constitutively expressed or induced after pathogen attack (1–4). One common resistance mechanism to all plant cells is the presence of a cell wall that shields plants from pathogen invasion. The cell wall acts first as a passive barrier that pathogens have to hydrolyze by secreting cell wall–degrading enzymes for infection progression but also functions as a reservoir of antimicrobial compounds (5–7). Plant cell walls are also a source of carbohydrate moieties that are released during wall degradation and could act as damage-associated molecular patterns (DAMPs) triggering plant immune responses upon their perception by plant pattern recognition receptors (PRRs) (6–12). Plant walls are complex and dynamic structures that consist of a primary wall composed of carbohydrate-based polymers—cellulose, pectic polysaccharides (homogalacturonan, rhamnogalacturonan (RGI), and RGII), hemicelluloses (xyloglucan and xylans) and minor polysaccharides—and of structural glycoproteins (13). In addition, to reinforce their structure, some plant cells deposit a secondary wall that is mainly composed of cellulose, hemicelluloses (mostly xylans), and lignin (14, 15). The biosynthesis, transport, deposition, remodeling, and turnover of cell walls, along with the regulation of these processes, involve ∼10% of genes encoded in plants genomes (16, 17).
Modifications of cell wall composition and structure occur during plant development but also upon plant exposure to environmental stresses (e.g., drought or pathogen attack) or treatments with chemicals disrupting wall biosynthesis (e.g., isoxaben). These wall modifications have a direct effect on cell wall integrity (CWI) and can initiate molecular adaptive mechanisms, such as cell wall composition remodeling and defensive responses activation (12, 18–21). CWI alteration also occurs in plants impaired in or overexpressing cell wall–related genes. Some of these plants/mutants show altered disease-resistance phenotypes that were initially associated with the misadaptation of pathogens to overcome the modified wall structures of these genotypes (5, 7, 22–27). However, activation of defensive pathways takes place in the majority of these mutants/overexpressing lines with wall alterations (5, 7, 22–27). For instance, impairment of cellulose synthesis for secondary cell walls by inactivating cellulose synthase subunits, as it occurs in Arabidopsis thaliana irregular xylem mutants (irx1, irx3, and irx5), or for primary cell walls, as it occurs in Arabidopsis isoxaben–resistant (ixr1), also known as constitutive expression ofvegetative storage protein 1 (cev1) mutant, results in constitutive activation of some canonical defensive responses and enhanced resistance to different pathogens. For example, irx1-6 shows enhanced resistance to the necrotrophic fungus Plectosphaerella cucumerina (Pc) and the vascular bacterium Ralstonia pseudosolanacearum (Rp) (formerly Ralstonia solanacearum (28–31)). Similarly, alteration of the biosynthesis and/or structure of wall pectins (e.g., degree of methyl-esterification) can also affect pathogen resistance (8, 32–37). Moreover, modification of glucuronoxylans and xyloglucans structure also has impacts on disease resistance, as it occurs in the Arabidopsis de-etiolated 3 (det3) mutant that shows enhanced resistance to Pc (38, 39) or in agb1 mutant (impaired in Gβ subunit of the heterotrimeric G protein), which has reduced xylose content and shows enhanced susceptibility to several pathogens, including Pc, the biotrophic oomycete Hyaloperonospora arabidopsidis (Hpa), and the hemibiotrophic bacterium Pseudomonas syringae (40–43). Also, modification of the degree of acetylation of wall polysaccharides and of lignin composition affect disease resistance, growth, and adaptation to environmental changes of plants (12, 44–46). Interestingly, cell wall modification can also result in contrasting disease resistance effects as illustrated by the arr6-3 mutant that shows enhanced resistance to Pc but is highly susceptible to Rp (20).
Alteration of CWI can initiate the release of DAMPs that regulate plant immune responses in a similar way to those triggered by microbe-associated molecular patterns (MAMPs) (6, 24, 47). Despite the diversity of glycan structures of plant cell walls, only a limited number of wall-associated DAMPs have been identified so far, including some oligosaccharides structures derived from β-1,3-glucan (callose), cellulose, xyloglucan, mannan, homogalacturonan, and arabinoxylan polysaccharides of plant cell walls (6, 8–10, 12, 47–51). Modification of CWI also leads to developmental phenotype alterations (e.g., reduced plant size and biomass or fertility), indicating that the cell wall contributes to plant fitness (5, 19, 52, 53). Notwithstanding the evidence of the roles of plant cell walls in immunity and fitness, correlations between variations in cell wall carbohydrate moiety composition and specific phenotypes have not been described until recently (12, 41, 54).
We have investigated the specific contribution of plant cell wall to disease resistance by testing the susceptibility to three different pathogens of a large set of Arabidopsis cell wall mutants (cwm; n = 34). We found that a significant proportion of these mutants (29 of 34; 85.3%) showed altered disease-resistance phenotypes, supporting a more relevant function of such extracellular layer in plant immunity than currently considered. Here, we demonstrate, combining mathematical analyses and glycome profiling, that the content of specific wall glycan moieties in cwm plants correlates with some of their disease resistance and fitness phenotypes, providing a link between plant cell wall composition and plant development/immunity phenotypes.
Results
Arabidopsis Cell Wall Composition Specifically Contributes to Disease- Resistance Responses.
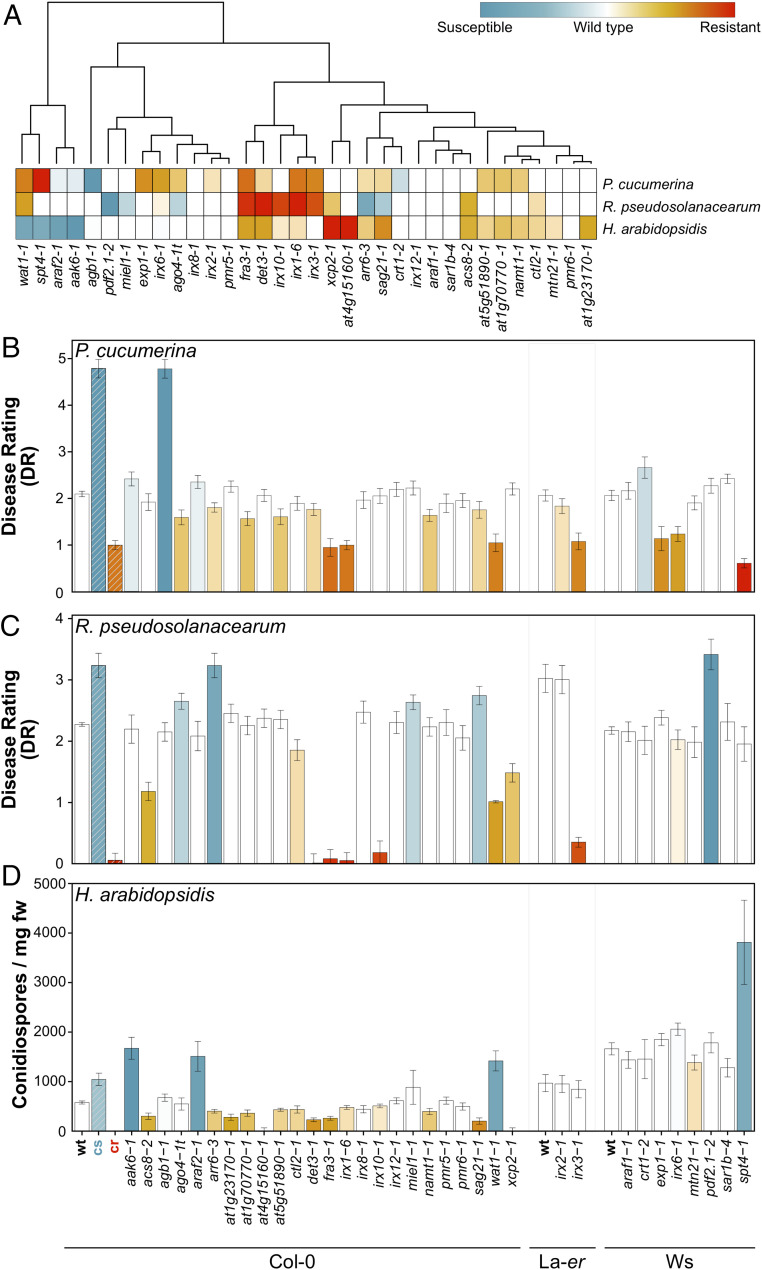
To determine the specific function of plant cell wall in immunity, we selected two large sets of Arabidopsis cwm and tested their resistance to three pathogens with different parasitic styles: the necrotrophic fungus Pc, the vascular bacterium Rp, and the biotrophic oomycete Hpa. The first set of cwm included 18 previously described mutants, such as irregular xylem (irx1-6, irx2-1, irx3-1, irx6-1, irx8-1, irx10-1, and irx12-1), powdery mildew resistance (pmr5-1 and pmr6-1) mutants, and det3-1, fra3-1, wat1-1, arr6-3, agb1-1, exp1-1, araf1-1, araf2-1, and ctl2-1 lines (SI Appendix, Figs. S1 and S2). The second cwm set was composed of 16 transfer DNA (T-DNA) insertional mutants, which were impaired in orthologs of Zinnia elegans genes differentially expressed during xylogenesis, a process involving secondary wall biosynthesis (SI Appendix, Fig. S1) (22). The majority of these mutations (e.g., those in T-DNA insertional mutants) resulted in loss-of-function mutants, as expression of the impaired genes was not detected by RT-PCR or led to truncated proteins, but hypomorphic alleles were also included in the analyses (SI Appendix, Figs. S1 and S2). We checked the resistance phenotypes of these 34 cwm lines and their wild-type counterparts (Col-0, Ws, or La-er) upon infection with Pc, Rp, or Hpa either by evaluating plants macroscopic disease symptoms caused by Rp and Pc and assigning Disease Rating values (DR) or by determining Hpa sporangiophores formation on plant leaves and conidiospore production by these sporangiophores per plant fresh weight. In all these disease-resistance analyses, susceptible and resistant control genotypes (mainly for Col-0 background) were included for comparison as follows: 1) irx1-6 as control of resistance (cr) for Pc and Rp, and La-er and Col-0 wild-type ecotypes as cr (gene for gene resistance) for Hpa (Col-0 and La-er/Ws, mutant backgrounds, respectively); 2) controls of susceptibility (cs) for Col-0 genotypes were agb1-1 for Pc, arr6-3 for Rp, and NahG plants, defective in salicylic acid (SA) pathway, for Hpa; and 3) cs of Hpa for Ws and La-er genotypes were eds1-1 (Ws) and eds1-2 (Col-0) alleles, respectively, which are impaired in the gene for gene resistance (20, 29, 31, 39, 55‒56). In addition, irx6-1 (Ws) and irx3-1 (La-er) were included as cr of Pc and Pc/Rp for Ws and La-er, respectively (20).
We found that 29 of the 34 cwm lines tested (85.3%) showed, in comparison to wild-type plants, altered resistance responses (mainly enhanced resistance) to at least one of these pathogens: 20 of 34 mutants to Pc (58.8%), 19 of 34 to Hpa (55.9%), and 15 of 34 mutants to Rp (44.2%; Fig. 1 and SI Appendix, Fig. S3). Cluster analyses of these phenotypes identified some specific groups of cwm mutants with similar disease-resistance phenotypes but also a high diversity of disease-resistance phenotypes illustrated by mutants with unique phenotypes (Fig. 1A). Remarkably, 15 of the 34 mutants showed enhanced resistance to more than 1 pathogen and 3 mutants to all 3 (fra3-1 and det3-1 and the previously characterized irx1-6) (29, 31) (Fig. 1 and SI Appendix, Fig. S3). For Pc, several mutants (e.g., det3-1, fra3-1, at1g70770-1, ago4-1t, sag21-1, irx2-1, at5g51890, or arr6-3) showed lower DR than wild-type plants and enhanced resistance, whereas several mutants showed higher DR than wild-type plants and enhanced susceptibility (e.g., xcp2-1, crt1-1, araf2-1, and akk6-2), but the levels of disease resistance values were weaker than those of cr (irx1-6/irx3-1/irx6-1) and cs (agb1-1) controls, respectively (Fig. 1B and SI Appendix, Fig. S3) (29, 40). Pc resistance phenotypes were further validated by infection of a representative set of additional alleles of some mutants (SI Appendix, Fig. S4). In the analysis of resistance to Rs, we identified 4 mutants (pdf2.1-2, ago4-t, sag21-1, and miel1-1) that showed, like arr6-3, enhanced susceptibility and more severe disease symptoms and DR than wild-type plants and 10 mutants showing lower DR and enhanced resistance (det3-1, xcp2-1, irx10-1, fra3-1, wat1-1, ctl2-1, irx1-6, acs8-2, irx6-1, and irx3-1) than their corresponding wild-type plants (Fig. 1C and SI Appendix, Fig. S3). Except for fra3-1 and irx10-1, the enhanced resistance of these 10 mutants was weaker than that of the previously characterized irx1-6, irx3-1, or wat1-1 partially resistant genotypes, whereas only pdf2.1-2 plants were as susceptible as the recently described hypersusceptible arr6-3 plants (20, 29, 57; Fig. 1C and SI Appendix, Fig. S3). Notably, we found 14 mutants with enhanced resistance to the biotroph Hpa (e.g., det3-1, xcp2-1, at1g23170-1, fra3-1, at1g70770-1, acs8-2, at4g15160-1, namt1-1, sag21-1, at5g518-1, ctl1-1, irx1-6, at5g51890-1, and arr6-3), showing two of them (at4g15160-1 and xcp2-1) a reduction in conidiospore production similar to that of cr control, whereas four lines (wat1-1, at3g47510-1, irx6-1, and spt4-1) were more susceptible than wild-type plants to this oomycete, but their susceptibility was weaker than that of NahG or agb1-1 (Col-0), eds1-1 (Ws), or eds1-2 (La-er) included as cs (Fig. 1D and SI Appendix, Fig. S3). All these susceptible genotypes included as controls developed a significantly higher number of sporangiophores in their leaves/cotyledons than the corresponding wild-type plants, the cr genotypes, or cwm lines showing enhanced resistance (SI Appendix, Fig. S5), further supporting their disease-resistance phenotype classification based on conidiospore production. These data pointed to a relevant function of the cell wall composition in resistance to different types of pathogens.
Fig. 1.
Arabidopsis cell wall mutants show alterations of their disease-resistance phenotypes in comparison to wild-type plants. (A) Clustering of disease-resistance phenotypes of Arabidopsis cell wall mutants to P. cucumerina (Pc), R. pseudosolanacearum (Rp), and H. arabidopsidis (Hpa). Clusters were computed using Euclidean distances using disease-resistance indexes relative to wild-type (wt) plants (DR for Pc and Rp; number of conidiospores per milligram of rosette fresh weight (mg fw) for Hpa). The color-coding of the corresponding columns/squares indicates the level of the resistance phenotype, from susceptible (blue) to resistant (red), that have been established for each pathogen tested. Colored squares/columns indicate values of significant differences compared with wt values (ANOVA nonbalanced analysis and Dunnett’s test, P ≤ 0.05). (B) DR (average ± SD) of cell wall mutants and wt plants (Col-0, La-er, and Ws backgrounds; n > 10) at 7 dpi with the necrotrophic fungus Pc. DR varies from 0 (noninfected plants) to 5 (dead plants). The irx1-6 and agb1-1 mutants were included as cr and cs, respectively. (C) DR (average ± SD) of wt and mutants (n > 10) at 8 dpi with bacterium Rp. DR varies between 0 (no symptoms) and 4 (dead plants). irx1-6 and arr6-3 mutants were included as cr and cs, respectively. (D) Number of conidiospore/milligram fresh weight in wt and mutant plants (average ± SD; n > 20) at 7 dpi with the oomycete Hpa. La-er and Col-0 wild-type ecotypes were included as cr for Col-0 and La-er/Ws mutant backgrounds, respectively, and NahG plants (Col-0), eds1-1 (Ws), and eds1-2 (Col-0) alleles were used as cs for Col-0, Ws, and La-er mutant backgrounds, respectively (SI Appendix, Fig. S3). Data in B–D are from one representative experiment of the three performed that gave similar results. References and details of cwm mutants are listed in SI Appendix, Figs. S1 and S2, and the DR and conidiospore/mg fw values (average ± SD) are shown in SI Appendix, Fig. S3.
Enhanced Resistance of cwm Plants to Pc and Rp Negatively Impacts Plant Fitness.
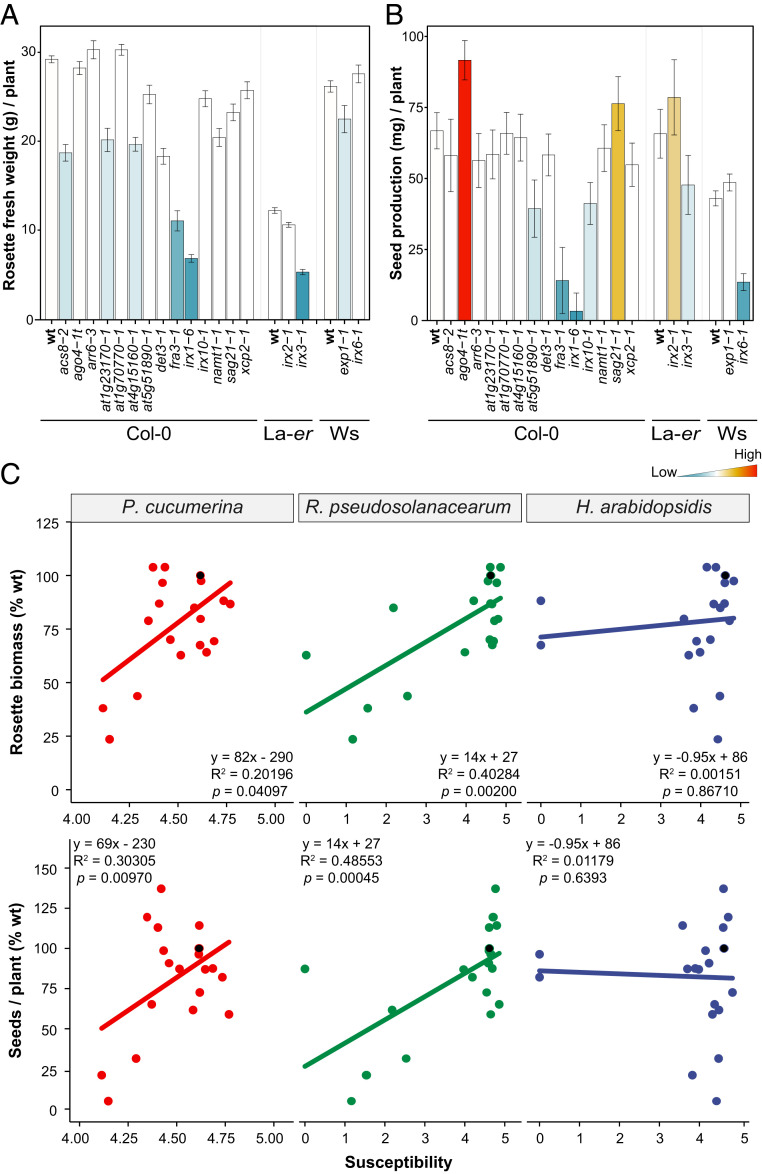
The overall developmental phenotypes (e.g., rosette size and leaf architecture) of the majority of cwm tested did not differ significantly from those of wild-type plants. This is in contrast with the previously described dwarf/altered phenotypes of irx1-6, irx3-1, fra3-1, or det3-1 mutants and that found here for at5g51890-1 (SI Appendix, Fig. S6 and references in SI Appendix, Fig. S1). Since disease resistance/developmental growth trade-offs have been described in Arabidopsis (4, 58, 59), we selected 18 cwm mutants from representative clusters of resistance phenotypes (Fig. 1A) and different ecotype backgrounds, and we measured vegetative (rosette biomass) and reproductive (seed production) traits related to fitness under growth conditions with no limitation of nutrients and water and no infection. Rosette biomass (fresh weight) of 4-wk-old plants was, in comparison to wild-type plants, reduced (between 30 and 80%) in 9 out of 18 cwm tested (det3-1, irx1-6, at4g15160-1, acs8-2, namt1-1, at1g23170-1, fra3-1, irx6-1, and irx3-1), and no significant increase in rosette biomass was observed in any of cwm lines (Fig. 2A). Seed production at the end of the reproductive cycle was significantly reduced, in comparison to wild-type plants, in six mutants (irx10-1, irx1-6, at5g51890-1, fra3-1, irx3-1, and irx6-1) and notably increased in two cwm lines (ago4-t1 and sag21-1) (Fig. 2B). Both fitness traits were negatively affected only in three mutants, irx1-6, fra3-1, and irx3-1, as described previously (29, 53), suggesting that these two traits are decoupled.
Fig. 2.
Arabidopsis cell wall mutants show associated resistance/fitness trade-offs. (A) Rosette fresh weight biomass (average g/plant ± SD) of 4-wk-old mutants and wild-type (wt) plants (Col-0, La-er, and Ws backgrounds). (B) Seed yield (average milligram/plant ± SD) of wt plants and mutants at the end of reproductive cycle. Data are the average of 10 plants. The column color indicates significant differences compared with wt values (ANOVA nonbalanced analysis and Dunnett’s test, P ≤ 0.05), with higher and lower values than wt indicated in red and blue, respectively. This is one representative experiment of the three performed that gave similar results. (C) Correlation analysis between biotic stress susceptibility to pathogens (Pc, Rp, and Hpa) and fitness parameters (seed yield and rosette biomass) of 18 cwm mutants and wt plants (Col-0, La-er, and Ws backgrounds). The average response information of each genotype (dot in the graph) is expressed in relation to that of the reference wt plant (black dot; value of 100% at the y-axes). Disease resistance susceptibility ratios were log-transformed, and accordingly, x-axes range from 0 (lower susceptibility) to 5 (greater susceptibility), with the wt plants situated at 4.72 = ln (1 + 100). A linear model was fitted for each combination and correlations determined. Fitted equations, R-squares, and P values are indicated in the insets of the graphs. The x-axes of the figures involving Pc are enlarged in the 4 to 5 range for better visualization.
We next determined in this subset of 18 cwm mutants if their fitness alteration was associated with their resistance/susceptibility phenotypes. Correlation analyses were performed after conversion of DR and fitness data to percentage susceptibility ratios (with respect to each ecotype’s wild-type value), followed by least-squares (LS) means estimation. A negative correlation was found in cwm plants between both rosette biomass and seed production and resistance to Pc (P = 0.04097 and R2 = 020196 for seed yield and P = 0.0097 and R2 = 0.30305 for biomass) and Rp (P = 0.002 and R2 = 0.40284 for seeds yield and P = 0.00045 and R2 = 0.48553 for biomass) (Fig. 2C). In contrast, a negative association was not found between the resistance phenotype to Hpa of cwm plants and their seeds yield (P = 0.8671 and R2 = 0.00151) or biomass (P = 0.6393 and R2 = 0.01179) (Fig. 2C). These results indicated that trade-offs between resistance to Pc/Rp and plant development exist. Of note, we did not find, among the cwm mutants with enhanced resistance, any with higher seed yield or rosette biomass than wild-type plants (Fig. 2C), indicating that cwm defensive responses associated to CWI alteration are costly for plant development.
Associations between resistance to Pc or Rp and tolerance to abiotic stresses (e.g., drought, desiccation, and salinity) have been reported (29, 54). Accordingly, we quantified the tolerance to desiccation (survival percentage rate upon rewatering after desiccation) of the 18 cwm genotypes. We found that six of them (det3-1, irx1-6, fra3-1, irx3-1, irx10-1, and irx2-1) were more tolerant to desiccation than the wild-type plants (SI Appendix, Fig. S7A). Of note, a positive correlation was found between desiccation tolerance of cwm plants and disease resistance to either Pc (P = 0.03141 and R2 = 0.22128) or Rp (P = 7.196 × 10−6 and R2 = 0.66227) but not to Hpa (P = 0.5254 and R2 = 0.02155; SI Appendix, Fig. S7B). These results are in line with previous findings indicating that resistance to Pc/Rp and desiccation tolerance could be linked traits (29, 57).
Enhanced Resistance Phenotypes of cwm Plants Are Associated with Different Alterations of Their Cell Wall Compositions.
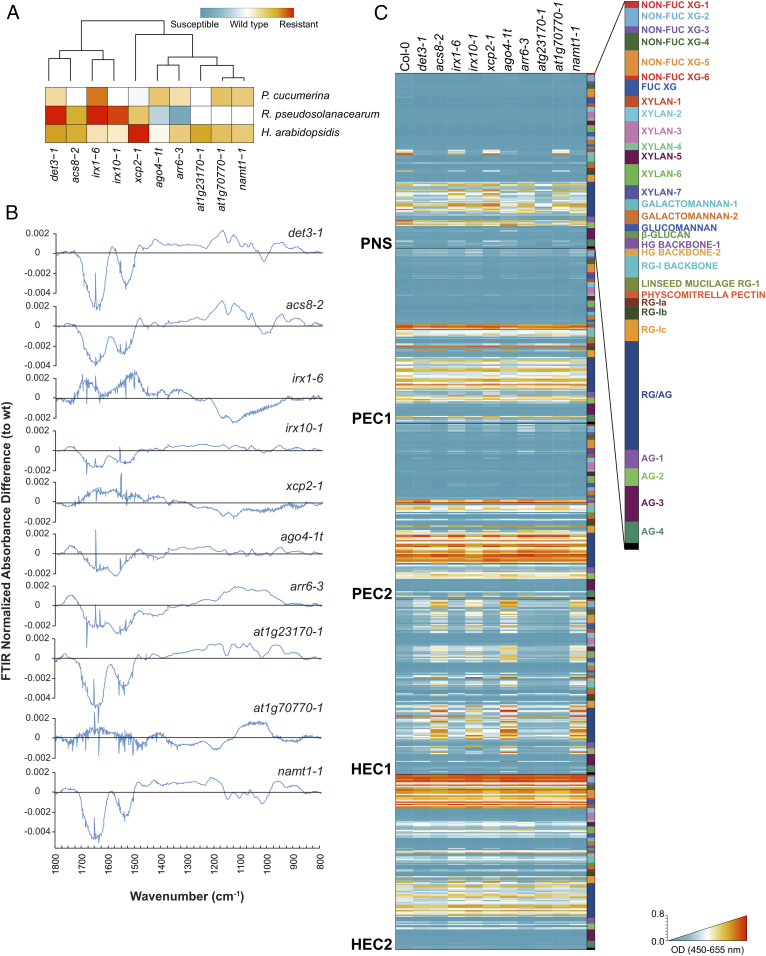
We next determined the putative correlations between the observed resistance phenotypes of cwm plants and their wall composition (e.g., cellulose, neutral sugars, and uronic acid content). Of the subset of 18 mutants used in trade-off analyses, only 9 have been previously characterized as cell wall mutants (SI Appendix, Fig. S1), whereas 9 were putative wall mutants (22). We found, in comparison to wild-type plants, differences in the composition of the walls of the majority of this subset of 18 mutants; at1g23170-1, at1g70770-1, xcp2-1 ago4-1t, and irx6-1 had reduced and acs8-2 had increased levels of cellulose; det3-1 and irx1-6 possessed less pectic uronic acids; and noncrystalline neutral sugars levels were increased in arr6-3 and decreased in at1g70770 and acs8-2 (SI Appendix, Fig. S8). These data confirmed that the majority of nine putative cwm initially selected showed wall alterations. Since these biochemical characterizations of the cell wall composition of cwm plants were not very precise, we narrowed down the collection to a set of 10 mutants, representing six different clusters with different resistance phenotypes, and performed a deeper cell wall profiling (Fig. 3A). We subjected mutants and wild-type purified cell walls to Fourier-Transform InfraRed (FTIR) spectroscopy that can assign wall polymers and functional groups to different wavenumbers of the FTIR spectra (60). Differential FTIR spectra obtained after digital subtraction of the wild-type values from the mutants showed clearly that xcp2-1, namt2-1, acs8-2, at1g70770-1, at1g23170-1, and ago4-1t were cell wall mutants with biochemical alterations that differ from those observed in the previously characterized det3-1, irx1-6, irx10-1, and arr6-3 wall mutants (Fig. 3B; 21, 28). Since some of the wavenumbers of the differential FTIR spectra were associated to lignin components (wavenumbers at 1,515, 1,630, and 1,720 cm−1), we determined total lignin content, and we found that it was altered in four mutants (det3-1, irx1-6, namt1-1, and at1g23170-1), further supporting that these genotypes were mainly affected in secondary wall composition (SI Appendix, Fig. S9).
Fig. 3.
Cell wall analyses by FTIR spectroscopy and glycome profiling of a core set of Arabidopsis cell wall mutants. (A) Selection of a core set of representative mutants with different levels of disease resistance to Pc, Rp, and Hpa. Clusters were computed by Euclidean distances using disease-resistance indexes relative to wild-type (wt) plants. (B) Cell wall FTIR difference spectra of mutants and wt plants (Col-0). The black line indicates wt values, and values over this line are differential FTIR spectra in the mutants tested. (C) Heatmaps of glycome profiling of cell wall extracts (PNS, PEC1, PEC2, HEC1, and HEC2) of cwm and wt (Col-0) rosette leaves of 25-d-old plants (see Dataset S1 for details). Heatmaps depict antibody binding strength based on optical density (OD) indicated as a color gradient ranging from blue (no binding) to red (strongest binding). The list of monoclonal antibodies used for glycome profiling of each fraction and wall structures recognized by them are indicated (Right) (see Dataset S1 for details). Data represent average values of two independent experiments (n > 10).
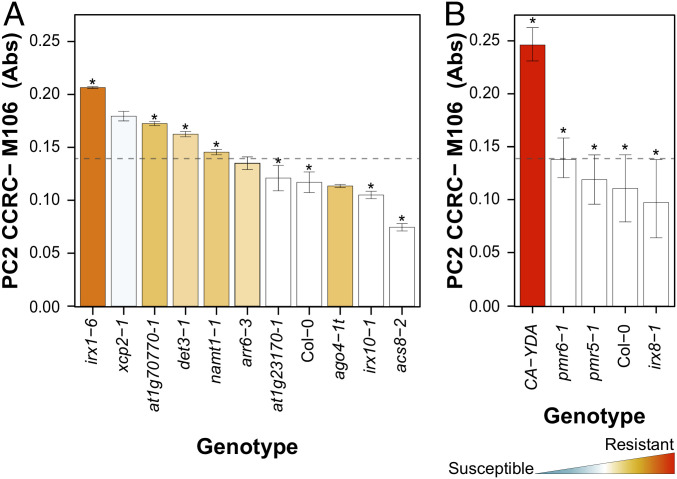
These classical cell wall analyses were complemented with an in-depth characterization of cwm wall composition by glycome profiling using a collection of 155 glycan-directed antibodies, recognizing diverse cell wall substructures (61, 62). These analyses were carried out on the following five sequential wall extracts obtained from rosettes of each of plant genotypes: protein and neutral sugars (PNS), two pectin (PEC1 and PEC2), and two hemicellulose (HEC1 and HEC2) wall extracts, which are known to be enriched in different glycans (61, 62). Glycome profiling confirmed that all of the selected mutants showed significant differences in the abundances of some wall glycan epitopes in comparison to wild-type plants (Fig. 3C and Dataset S1) and also corroborated the diversity of wall compositions in the selected wall mutants. The relative abundances of some specific wall epitopes in the PEC1 and PEC2 glycome profiles (e.g., fucosylated-xyloglucan) showed opposite patterns in the resistant cwm mutants in comparison to those showing wild-type or hypersusceptible phenotypes, suggesting some kind of correlation between specific wall epitopes and disease resistance (Dataset S1). To test this hypothesis, we performed a model analysis to uncover and generalize the potential relationships between each mutant’s glycomic data (that act as independent or explanatory variables) and disease resistance to Pc, Rp, or Hpa as response variables. We used a nonparametric Classification and Regression Tree (CRT) methodology for these analyses, which provides simple and interpretable classification models with almost no statistical assumptions (SI Appendix, Fig. S10A and Materials and Methods). CRT identified a set of antibodies whose reaction values explained, with an estimated cross-validation accuracy between 83.43% and 84.34% (in 10 cwm and wild-type genotypes), the resistance phenotypes of cwm plants (SI Appendix, Fig. S10B and Table S1). For example, the abundance of fucosylated-xyloglucans (recognized by CCRC-M106 antibody) correlated with the level of resistance to Pc and explained the response phenotypes of 8 out of 11 genotypes tested (Fig. 4A). Similarly, CCR5-M5 (detecting a yet undefined RGI epitope) correlated with the resistance to Rp (8 out of 10 cwm genotypes), and CCRC-M174 (detecting galactomannan) and CCRC-M106 (detecting fucosylated-xyloglucans) explained the resistance to Hpa (8 out of 10 cwm genotypes) (SI Appendix, Figs. S10B and S11). Additional carbohydrate moieties may also contribute to explain a mutant’s disease-resistance phenotypes but with lower accuracy values (SI Appendix, Table S1). To further validate the association of fucosylated-xyloglucan (CCRC-M106) with the Pc disease-resistance phenotype, we performed glycomic analyses with selected antibodies on three additional mutants (pmr5-1, pmr6-1, and irx8-1), with disease resistance to Pc similar to that of wild-type plants (Col-0; Fig. 1A), and on CA-YDA plants that overexpress the constitutive active YODA MAP3K and show enhanced resistance to Pc and additional pathogens (63). As predicted by the model, walls of CA-YDA plants, but not those of pmr5-1, pmr6-1, and irx8-1, showed an enhanced accumulation of the fucosylated-xyloglucan epitope recognized by CCRC-M106 in comparison to wild-type plant cell walls (Fig. 4B).
Fig. 4.
CRT analyses correlate wall composition and disease-resistance phenotypes of the Arabidopsis cell wall mutants. (A) Biological validation of CRT model for resistance to Pc with cell wall mutants from six different clusters (Fig. 3A). The absolute value (average ± SD) of the epitope signal detected by CCR-M106 antibody is shown. Columns are colored according to the resistance level of the corresponding mutant, from red (resistant) to blue (susceptible), in comparison with wild-type (wt) level of resistance (white column; ANOVA nonbalanced analysis, Dunnett’s test, P ≤ 0.05). The scale used is the same of that in Figs. 1 and 3A. The absorbance cutoff value for considering a mutant as resistant or susceptible/wt phenotype, as determined by CRT, is indicated by the dotted lines. The mutant genotypes which follow the CRT model are marked with an asterisk. (B) Biological validation of the CRT model with pmr5-1, pmr6-1, and irx8-1 mutants that do not show enhanced resistance to Pc and CA-YDA plants that show enhanced resistance to the fungus. All these mutants follow the absorbance cutoff value predicted by the CRT model, and, accordingly, they are marked with an asterisk.
Similar CRT analyses were then performed with the fitness parameters (biomass and seed yield, acting as dependent variables) of these 10 cwm mutants and wild-type plants. Of note, we found a relationship between the reaction signal of some antibodies recognizing some particular carbohydrate moieties and these fitness traits, which explained between 87.31% and 87.62% of the phenotypes; CCRC-M22 (selective for a six-linked β-galactan epitope in RGI and arabinogalactan) explained biomass phenotypes, and the levels of epitope detected by CCRC-M175 (galactomannans) and CCRC-M170 (acetylated mannans) correlated with seed yield (SI Appendix, Figs. S10B and S12 and Table S1). Similarly, we found a relationship between tolerance to desiccation and the epitope recognized by the JIM101 antibody (detecting an RGI epitope) that explained 83.16% of the phenotypes (SI Appendix, Table S1). Together, these data suggest that the cell wall composition of Arabidopsis is a determinant of plant developmental phenotypes and resistance/tolerance to biotic and abiotic stresses.
Disease-Resistance Responses of Cell Wall Mutants Is Not Associated with the Differential Regulation of Canonical Defensive Pathways.
The molecular defensive mechanisms underlying the enhanced resistance/susceptibility of the cwm lines were further investigated by qRT-PCR determination of the expression of defense genes in noninfected and Pc-inoculated plants (1-d post inoculation [dpi]). The tested genes are either up-regulated by MAMPs (e.g., WRKY33, PHI1, CYP81F2, and PAD3), CWI alteration (At1g51890; 64), or defensive phytohormones (PR1, LOX3, PR4, LTP3, and PDF1.2: gene markers of SA, ethylene [ET], jasmonic acid [JA], abscisic acid, and ET plus JA, respectively). We clustered the expression levels of these genes in cwm with their resistance phenotypes to identify potential correlations, and only a significant cluster between expression of LTP3 in noninoculated irx1-6 plants and disease resistance to Pc was found (SI Appendix, Fig. S13A), as previously described (29). In Pc-inoculated plants, only one cluster was found associated to PR1 expression, but it did not explain the cwm resistance phenotypes to any pathogen, since it includes two mutants and wild-type plants (SI Appendix, Fig. S13B). These data indicated that a constitutive expression or enhanced up-regulation upon infection of phytohormone-, or MAMP-triggered- or CWI-related genes might not explain the cwm-enhanced resistance or susceptibility phenotypes observed.
We have recently shown that pectin wall fractions (PEC1 and PEC2) of Col-0 and cell wall mutant arr6-3 (Fig. 1C) contain potential glycan-derived DAMPs that regulate immune responses when applied to Col-0 wild-type plants (20). Further biochemical subfractionation and characterization of arr6-3 PEC1 has led to the identification of an arabinoxylan pentasaccharide (33-α-l-arabinofuranosyl-xylotetraose) as a novel active DAMP, triggering immune responses such as Ca+2 burst and mitogen-activated protein kinases (MPKs) phosphorylation in Col-0 wild-type plants (51). Similar immune responses triggered by elicitor activities have been described in wall extracts of additional Arabidopsis wall mutants (12, 44). Given these previous data, we investigated and found that the PEC1 and PEC2 wall extracts from cwm plants, like those of arr6-3 included as control, triggered early immune responses, such as Ca+2 bursts, upon their application to Arabidopsis Col-0AEQ lines expressing the apoaequorin Ca+2 sensor protein (35S::Apoaequorincyt) (20, 47; SI Appendix, Fig. S14). To further determine whether the signaling mechanisms regulating cwm PEC1-mediated Ca+2 bursts were similar to those triggered by other MAMPs/DAMPs, we generated an agb1-2AEQ line and tested Ca+2 bursts upon PEC1 treatment, as agb1-2 is impaired in immune responses triggered by several MAMPs such as flg22, elf18, and chitin (65, 66). Col-0AEQ and agb1-2 AEQ lines treated either with PEC1 from Col-0 or the most active cwm fractions (SI Appendix, Fig. S14) showed similar Ca+2 bursts, which contrast with the reduced Ca+2 burst of agb1-2AEQ line treated with flg22 in comparison to Col-0AEQ (SI Appendix, Fig. S15A), indicating that PEC1-triggered immunity does not require the immune regulator AGB1. To further characterize PEC1-mediated immunity, we tested the activity of Col-0 and cwm PEC-1 in triggering MPK phosphorylation in Col-0 and in bak1-5 mutant that is impaired in BAK1 coreceptor required for several immune responses (67). We found that PEC1 from cwm genotypes trigger MPK phosphorylation, which was, in general, higher than that of PEC1 from Col-0, and we observed that MPK phosphorylation was not impaired in PEC1-treated bak1-5 plants, which was different from the significant reduction in MPK phosphorylation observed in bak1-5 treated with flg22 when compared with Col-0–treated plants (SI Appendix, Fig. S15B). These data suggest that these wall extracts might contain additional DAMPs or increased amounts of DAMPs, in comparison to wild-type wall extracts that might regulate Arabidopsis immune responses through signaling pathways that do not seem to involve the immune regulator AGB1 and the BAK1 coreceptor. The immune activity of these cell wall fractions (e.g., PEC1) might contribute to and explain the disease-resistance phenotypes of the Arabidopsis cell wall mutants tested.
Discussion
Plant cell walls are important components of both preexisting and inducible plant defense mechanisms against pathogen infection (5, 6, 18, 19). Accordingly, modifications of cell wall composition and structure in some mutants or transgenic lines have been demonstrated to result in the alteration of their resistance phenotypes to different pathogens, including hemibiotrophic (e.g., P. syringae; 8, 29, 68) and vascular (e.g., Rp; 29, 40, 57) bacteria and necrotrophic (e.g., Pc and Botrytis cinerea; 21, 29–32, 36, 39, 42, 57, 69) or biotrophic fungi (e.g., Erysiphe sp. 28; 70–74). For example, 15 of the wall mutants analyzed in this study have been previously described to show differential disease resistance phenotypes to one or two pathogens (e.g., irx1-6 and irx3-1), and in a few cases, like irx1-6, agb1-1, and arr6-3 used as controls in the analyses, their resistance to three or more pathogens have been determined (Fig. 1 and SI Appendix, Fig. S1). Despite these previous data, specific correlations between wall composition/structure and the resistance phenotypes and/or immune responses of these plant genotypes have not been described.
Here, we have determined the contribution of Arabidopsis cell walls to disease-resistance responses against three pathogens with very different parasitic styles by selecting a large set of cell wall mutants that includes well-characterized and putative wall mutants (22). Given the different molecular bases of Arabidopsis resistance to the three pathogens tested (Pc, Rp, and Hpa; 20, 29, 39, 40, 57, 75) and the putative diversity of cell wall alterations in the mutants screened, we initially anticipated that we could obtain a global view of Arabidopsis cell wall contribution to resistance. Notably, we found that 85.3% of the cell wall mutants tested (29 of 34) showed, in comparison to wild-type plants, differential phenotypes (enhanced resistance, mainly, or susceptibility in a few cases) to at least one of the three pathogens tested. Of note, we have identified different clusters containing one or several mutants with specific phenotypes (e.g., from enhanced resistance to the three pathogens to specific resistance to one pathogen) (Fig. 1A). These data support the diverse and significant functions of cell walls on plant disease resistance responses to vascular and necrotrophic pathogens, as previously described (20, 29, 57). Our data also identified a contribution of cell walls to plant resistance to biotrophic oomycetes, such as Hpa (Fig. 1C and SI Appendix, Figs. S3 and S5), which is in line with the described wall function in Arabidopsis resistance to biotrophic fungi causing powdery mildew diseases (70–73, 75). The proportion of mutants with differential disease resistance phenotypes identified in this screening is several orders of magnitude higher than the expected proportion that would be obtained in blind, unbiased disease-resistance screenings using T-DNA or chemically mutagenized plant populations. It can be anticipated that a similar proportion of genotypes to that obtained here would be found if a biased screening was performed with Arabidopsis mutants impaired in known components of key defensive pathways, such as phytohormone signaling or MAMP-triggered immunity (76, 77). Therefore, the data obtained here with the set of cwm plants strongly supports the relevant function of cell walls in plant immunity and disease resistance to different pathogens.
The genetic and molecular basis of Arabidopsis resistance to the three pathogens analyzed here differ significantly: 1) plant resistance to Hpa mainly depends on activation of effector-triggered immunity (ETI) and of the SA pathway (see resistance and susceptible controls in Fig. 1D and SI Appendix, Figs. S3 and S5); 2) disease resistance to Rp is mediated by ET, and just a few examples of ETI responses have been described; and 3) Arabidopsis resistance to necrotrophic fungi, including Pc, has been shown to depend on hormones signaling (mainly ET and JA, but also SA) and on the synthesis of tryptophan-derived metabolites (such as camalexin and indole glucosinolates), and few examples of ETI-mediated resistance have been described so far (78, 79). This lack of source of resistance genes triggering ETI to control Rp and necrotrophic fungi, such as Pc or B. cinerea, might explain the strong incidence of the diseases caused by these two types of pathogens in crops and the associated yield losses since breeding programs have not been effective in selecting traits conferring enhanced resistant to these pathogens (80, 81). This contrasts with the effectiveness of breeding programs in controlling biotrophic pathogens, such as oomycetes (e.g., Hpa) causing downy mildews or fungi (e.g., Erysiphe sp.) causing powdery mildew diseases (79, 82). Our data indicate that CWI disruption could be, initially, an effective strategy in the control of diseases caused by necrotrophic and vascular pathogens and, therefore, that a genomic-assisted breeding selection of CWI-associated traits could be used in breeding programs, as suggested previously for other crop traits, such as biomass digestibility (83). However, modification of cell wall composition and structure usually results in alterations of plant developmental phenotypes (e.g., reduced plant size, biomass, or fertility) that impact fitness (19, 84). In line with these previous data, we describe here a negative correlation between fitness parameters, such as rosette biomass and seed production, of the cell wall mutants tested and their enhanced resistance to vascular and necrotrophic pathogens (e.g., Rp and Pc) (Fig. 2C). These trade-offs associated to increased resistance to these pathogens are also probably hampering the selection of crop traits conferring improved resistance. In contrast, we have not found in the genotypes tested associated trade-offs to the enhanced resistance to the biotroph Hpa (Fig. 2C), indicating that some wall-associated traits identified here might be of interest for improving resistance to biotrophic pathogens.
Plant cell walls (primary and secondary) are complex and dynamic structures composed mainly of carbohydrate-based polymers of differing monosaccharide and glycosyl-linkage compositions (13). Among the genotypes included in our analysis, we selected 18 previously described wall mutants showing a great diversity of wall alterations, such as reduction/alteration of the content/decorations of cellulose (irx1-6, irx3-1, irx6-1, irx2-1, or ctl2), xylan (irx10-1), glucuronoxylan (irx8-1), pectin (pmr6-1, pmr5-1, arr6-3, and irx8-1), xyloglucans (agb1-1), or lignin (irx12-1) (see SI Appendix, Fig. S1 for references), or impairment in glycan transport or in muro biosynthesis of wall components (e.g., ctrl1-1, det3-1, wat1-1, and fra3-1) (see SI Appendix, Fig. S1 for references). We also tested 16 putative cell wall mutants (SI Appendix, Figs. S1 and S3), including some that have been recently characterized as wall mutants (e.g., araf2-1 and araf1-1 impaired in arabinan-containing pectins or xcp2-1 and arr6-3), and seven mutants whose wall alterations have been demonstrated for the first time here (Fig. 3, SI Appendix, Figs. S8 and S9, and Dataset S1). These data corroborate that the majority of genotypes initially selected are bona fide Arabidopsis wall mutants. Cell wall modifications identified by FTIR spectroscopy or biochemical analyses in the mutants from the six phenotypic clusters selected (Fig. 3A and SI Appendix, Figs. S8 and S9) were not precise enough to find specific associations between wall composition and disease-resistance phenotypes. Chemically extracted cell wall fractions (e.g., PEC1 and PEC2) contain mixtures of carbohydrate moieties derived from various polymer classes and can be enriched in certain carbohydrates detectable by glycome profiling. We show here that glycome profiling analysis of these extracts provides a more precise picture of wall modifications impacting disease resistance.
Our data show that mathematical modeling by CRT of glycome profiling of plant genotypes provides detailed and biologically consistent links between cell wall composition and disease resistance/fitness phenotypes, as it has been previously reported for the determination of cell wall digestibility of plant genotype biomass (12, 85, 86). The CRT algorithm used here allows for both the identification of variables (cell wall components recognized by some antibodies) and the definition of cut-points on these variables, separating mathematically in different branches and nodes the genotypes belonging to different phenotypic classes (best, equal, or worse than wild-type phenotypes). Since CRT is based on binary branching, it obtains more pure or homogenous nodes (in terms of their class composition) in contrast to other supervised classification methods (e.g., linear discriminant analysis, logistic regression, random forest, or classification trees). Using CRT, we have identified significant epitope associations explaining as much as 84.34% of the disease-resistance phenotypes tested (Fig. 4B and SI Appendix, Fig. S11B and Table S1). Remarkably, the abundance of fucosylated-xyloglucan (detected by CCRC-M106), RGIa (CCRC-M5), and galactomannan (CCRC-M174)/fucosylated-xyloglucan (CCRC-M106) in the cell walls of the mutants correlated with the level of resistance to Pc, Rp, and Hpa, respectively (Fig. 4 A and B and SI Appendix, Figs. S11 and S12). The relevance of xyloglucan and xylose content in Arabidopsis disease resistance to Pc has been previously described (39) and is further validated here by the content of wall epitope recognized by the CCRC-M106 antibody, which is enhanced in the Pc-resistant CA-YDA plants (Fig. 4B; 60). In contrast, galactomannan and RGIa/fucosylated-xyloglucan contribution to Rp and Hpa resistance, respectively, have not been previously described. Notably, we have also demonstrated here that the level of a six-linked β-galactan epitope present in RGI/arabinogalactans (CCRC-M22) and of acetylated-mannan/galactomannan (CCRC-M170/CCRC-M175) correlated with rosette biomass and seed production, respectively, indicating that wall composition can also determine plant fitness (SI Appendix, Fig. S13). These data are in accordance with previous results showing that high-density quantitative glycan microarrays, used in conjunction with association mapping, can detect pertinent variations related to plant cell wall genetics (12, 85, 86). Since the specificity of the carbohydrate moieties, recognized by some of the antibodies identified here, has not been fully established yet, in contrast to other antibodies of the glycomics collection (62), it cannot be excluded that disease resistance/fitness traits could be associated with other types of wall epitopes than those described here.
Various hypotheses have been put forward to explain why modification of cell wall composition often appears to enhance rather than reduce plant disease resistance (7, 12, 87). These hypotheses include strategies to avoid plant wall breakdown by microbial cell wall–degrading enzymes either by reshuffling (“masking”) wall composition or by releasing inhibitor proteins targeting microbial enzymes, but also the activation of immune responses upon recognition by PRRs of released elicitor-active molecules (DAMPs) from incorrectly assembled plant cell walls (20, 51). Our data support that some particular immune pathways are differentially regulated in some of the mutants, but their expression patterns do not explain their disease-resistance phenotypes (SI Appendix, Fig. S13). Notably, we also show that cell wall fractions of some cwm plants trigger immune responses, suggesting that they might contain additional DAMPs or enhanced levels of DAMPs in comparison to wild-type fractions (SI Appendix, Fig. S14), as it has been described recently to occur in other Arabidopsis wall mutants (12, 20, 44, 51). In this regard, our approach pointed to a role of fucosylated-xyloglucans and galactomannans in plant disease resistance, and, interestingly, recent reports have proposed these β-1, 4-linked components of hemicelluloses as potential plant DAMPs (49, 88). Although these reports do not allow us to narrow down the type of xyloglucans and galactomannans that our methodology has found to be involved in immunity, the fact that such structures can trigger defense responses in plants is at least promising. Of note, immune responses activated by the cwm wall fractions, such as PEC1, do not seem to require key regulators of canonical immune responses mediated by MAMPs, such as AGB1 and BAK1 (SI Appendix, Fig. S15), suggesting that novel signaling mechanisms and molecular components might be involved in the activation of immune responses activated by plant cell walls glycans. A growing number of plant cell wall–associated DAMPs have been identified so far; however, the mechanisms involved in their perception by plant PRRs are poorly characterized. Notably, several of these plant cell wall DAMPs trigger enhanced disease resistance responses when applied exogenously to Arabidopsis and crops (49, 51, 88). In line with these previous data, DAMP-triggered immunity, together with the canonical immune pathways that might be constitutively expressed or primed for stronger activation upon pathogen infection in some of the cell wall mutants analyzed, would contribute to regulating their immune responses and disease-resistance phenotypes. The characterization of these cwm defensive responses, and the wall DAMPs and plant PRRs involved in their activation, deserves further attention to understand these novel wall-associated immune responses.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis genotypes used in this study and oligonucleotides used for T-DNA insertional mutant characterization are listed in SI Appendix, Fig. S1 and Table S2. For plants used in Rp assays, seeds were germinated on Murashige and Skoog (MS) medium and then grown in Jiffy pots (www.jiffygroup.com) in a chamber at 22 °C, with a 9-h light period and a light intensity of 200 μmol ⋅ m−2 ⋅ sec−1 and 50% relative humidity. Plants used in Pc and Hpa disease resistance and fitness experiments were grown on soil in a growth chamber as described (20). Plant rosette biomass was determined on 4-wk-old plants (n = 10), and seeds were harvested at 8 wk after plants (n = 10) completed their vegetative cycle. Experiments were repeated at least three times with similar results. Genotyping of T-DNA insertional mutants was performed by PCR amplification of DNA extracted from mutant leaves following established protocols (20) and the oligonucleotides indicated in SI Appendix, Table S2. The Simple Sequence Length Polymorphisms (SSLP) markers nga119 or nga151 were used to confirm Ws background of the mutant tested.
Pathogen Growth Conditions and Plant Infections.
Pc Brigitte Mauch-Mani (BMM) strain and Hpa (isolates Noco2, Emwa1, and Cala) were grown as described (20). Rp (strains GMI1000 and RD15) were grown at 28 °C on Bacto-Agar (15 mg/mL) and glucose (5 mg/mL) medium. For Pc infection, 3-wk-old plants (n > 10) were sprayed with a suspension spore (4 × 106 spores/mL) of virulent Pc BMM isolate, progression of the infection was followed by visual evaluation of DR at different dpi, and the average DR, from 0 to 5, was scored as follows: 0 = no symptoms, 1 = plant with some necrotic spots, 2 = one or two necrotic leaves, 3 = three or more leaves showing necrosis, 4 = more than half of the plant showing profuse necrosis, and 5 = decayed/dead plant (39). For Hpa assays, 12-d-old plants (n > 20) were sprayed with a conidiospore suspension (2 × 104 spores/mL) of virulent isolates (Noco2, Emwa1, and Cala for plants in Col-0, Ws, and La-er backgrounds, respectively). Then, plants were incubated under short day conditions (10-h illumination) for 7 d, and the aerial parts of all plants were harvested and shaken in water, released conidiospores counted, and the average per milligram plant fresh weight determined (47). For Rp infections, roots of 4-wk-old plants (n > 10) were dipped into a bacterial suspension (5 × 107 cfu/mL) of virulent strains GMI1000 (for Col-0 and La-er) or RD15 (for Ws). Following inoculation, plants were transferred to a growth chamber under the following conditions: 12 h photoperiod, 27 °C, and 80% relative humidity. The average DR was scored in leaves as follows: 0 = no symptoms, 1 = 25% wilted leaves, 2 = 50% wilted leaves, 3 = 75% wilted leaves, and 4 = 100% wilted leaves (dead plant; 74). All pathogen resistance assays were repeated at least three times, and in all these experiments, susceptible and resistant control genotypes were included for comparisons (Fig. 1 and SI Appendix, Figs. S3 and S4).
Plant Cell Wall Purification, Fractionation, and Analyses.
Cell wall alcohol insoluble residues (AIR) were prepared from 25-d-old Arabidopsis plants according to ref. 89, and noncellulosic fraction, uronic acid, and crystalline cellulose and lignin contents were determined as previously described (63, 90). FTIR spectroscopy determination was done with discs prepared from mixtures of purified AIR and KBr (1:100, w:w) using a Graseby-Specac press. FTIR spectra were recorded and analyzed as described (91). Lignin-like material was quantified by the Klason gravimetric method with minor modifications (92). AIR fractions were subjected to sequential chemical extraction with increasingly harsh reagents in order to isolate fractions enriched in various cell wall components as previously described: PNS fraction, pectic fractions (PEC1 and PEC2), and hemicellulosic fractions (HEC1 and HEC2) (20, 89). Glycome profiling of the cell wall fractions was carried out by enzyme-linked immunosorbent assay (ELISA) using a toolkit of plant cell wall–directed monoclonal antibodies as previously described (see SI Appendix; 61, 62). Monoclonal antibodies are annotated in the database at glycomics.ccrc.uga.edu/wall2/antibodies/antibodyHome.html, and specific links to the antibodies are included in Dataset S1.
Gene Expression Analyses.
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) from Arabidopsis wild-type plants and mutants (SI Appendix, Fig. S2) and from mock-treated or Pc BMM–inoculated and rosettes (n > 25) at 1 dpi (four biological replicates), as reported previously (39). Quantitative real-time PCR amplification or RT-PCR detection were carried out as previously described (47). Oligonucleotides used for gene expression are detailed on SI Appendix, Table S3. The expression levels of each gene, relative to UBC21 (AT5G25760) expression, were determined using the Pfaffl method (93).
Clustering and Statistical Analyses.
Heatmaps and cluster aggrupation (Figs. 1A and 3A and SI Appendix, Fig. S13) were calculated using “ggplots” R package version 3.0.3. Clusters in Figs. 1A and 3A were computed using Euclidean distances using disease resistance indexes relative to wild-type plants (DR for Pc and Rp; the number of conidiospores per milligram of rosette fresh weight for Hpa). Clusters in SI Appendix, Fig. S13 were computed using Euclidean distances for absolute gene expression levels and disease indexes.
ANOVA models were fitted for each of the response variable (resistance to Pc, Rp, Ha, biomass and seed yield, and desiccation tolerance) (Figs. 1 and 2 A and B and SI Appendix, Fig. S7A) and each ecotype (Col-0, Ws, or La-er). LS means of these models were then obtained, providing a single estimation of the average response level (e.g., mean DR for both Pc and Rp, conidiospores/milligram plant fresh weight for Ha, seed yield in milligram and rosette fresh weight in milligram, and survival rate after desiccation) for each genotype. Afterward, correlation analyses (SI Appendix, Fig. S16) between biotic resistance and fitness features/desiccation were obtained by determining the ratio of each genotype LS mean to that of the corresponding wild-type ecotype for each response variable (e.g., percentage susceptibility levels with respect to wild-type plants). A logarithmic model was fitted for each combination of the biotic susceptibility ratios with the fitness and abiotic susceptibility ratios to analyze their correlations (see Fig. 2C and SI Appendix, Fig. S7B for the fitted equations, R-squares, and P values). For more details, see SI Appendix, Supplementary Material and Methods.
CRT predictive classification model (SI Appendix, Fig. S11), correlating wall composition with disease resistance and fitness phenotypes, was done by performing, first, a paired comparison analysis to assign Arabidopsis wild-type and cwm mutant genotypes into a class (e.g., a categorical valuation), which represents its status compared to wild-type plants with a similar performance (class equal), significantly better, or significantly worse than wild-type ones. The CRT method was then applied to link this class status to glycomics data. To avoid overfitting the data, the tree growing process of each CRT model was limited to a single binary branching to select a single antibody and its optimal cutoff point. The actual predictive capability or accuracy of the resulting classification tree models is evaluated as the percentage of correctly classified genotypes obtained through a 10-fold cross-validation process, replicated 100 times. The correlation and paired comparison analyses were implemented using the SAS software (glm and corr procedures), while the CRT classification model fitting and validation were implemented using Python (scikit-learn library: Data Set 2_CRTPythonscript, or see link: https://github.com/tinguarorg/PNAS_CellWall.git). See SI Appendix, Supplementary Material and Methods for further details.
Supplementary Material
Acknowledgments
This work was supported by the Spanish Ministry of Economy and Competitiveness (MINECO) grants BIO2015-64077-R and RTI2018-096975-B-I00 of Spanish Ministry of Science, Innovation, and Universities (MICIU) to A.M. and by the French National Agency for Research Grant ANR-07-GPLA-014 to D.G. This work has been also financially supported by the Severo Ochoa Program for Centers of Excellence in R&D from the Agencia Estatal de Investigación of Spain (Grant SEV-2016-0672 (2017-2021) to the Centro de Biotecnología y Genómica de Plantas). In the frame of this program, H.M. was a postdoctoral fellow. H.M. was also supported by an Individual Fellowship grant (SignWALLINg-624721) from the European Union. E.M. was a Juan de la Cierva Postdoctoral Fellow from MINECO, and L.B. was a Formacion Personal Investigador fellow of MICIU. The generation of the CCRC-series of plant cell glycan-directed monoclonal antibodies used in this work was supported by the US NSF (DBI-0421683 and IOS 0923992) to M.G.H. We thank Yves Marco and Philippe Ranocha for their help with the genotyping and selection of cwm mutants and the disease resistance screening with Rp. We thank the Molina laboratory members for useful discussion and comments on the manuscript, Javier Paz-Ares (Centro Nacional de Biotecnología, Spain) for their fresh ideas and interpretations about our challenging data, and Fernando García-Arenal (CBGP, Spain) for critical reading of the manuscript and suggestions.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010243118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Couto D., Zipfel C., Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Pieterse C. M. J., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C. M., Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Shigenaga A. M., Berens M. L., Tsuda K., Argueso C. T., Towards engineering of hormonal crosstalk in plant immunity. Curr. Opin. Plant Biol. 38, 164–172 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Denancé N., Sánchez-Vallet A., Goffner D., Molina A., Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4, 155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miedes E., Vanholme R., Boerjan W., Molina A., The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 5, 358 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacete L., Mélida H., Miedes E., Molina A., Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 93, 614–636 (2018). [DOI] [PubMed] [Google Scholar]
- 7.De Lorenzo G., Ferrari S., Cervone F., Okun E., Extracellular DAMPs in plants and mammals: Immunity, tissue damage and repair. Trends Immunol. 39, 937–950 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Bethke G., et al. , Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 28, 537–556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Souza C. A., et al. , Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol. 173, 2383–2398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voxeur A., et al. , Oligogalacturonide production upon Arabidopsis thaliana-Botrytis cinerea interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 19743–19752 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelsdorf T., et al. , Functional characterization of genes mediating cell wall metabolism and responses to plant cell wall integrity impairment. BMC Plant Biol. 19, 320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallego-Giraldo L., et al. , ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 1 (ADPG1) releases latent defense signals in stems with reduced lignin content. Proc. Natl. Acad. Sci. U.S.A. 117, 3281–3290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zablackis E., Huang J., Müller B., Darvill A. G., Albersheim P., Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 107, 1129–1138 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosgrove D. J., Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Sarkar P., Bosneaga E., Auer M., Plant cell walls throughout evolution: Towards a molecular understanding of their design principles. J. Exp. Bot. 60, 3615–3635 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Somerville C., et al. , Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211 (2004). [DOI] [PubMed] [Google Scholar]
- 17.McCann M. C., Carpita N. C., Designing the deconstruction of plant cell walls. Curr. Opin. Plant Biol. 11, 314–320 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Wolf S., Hématy K., Höfte H., Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 63, 381–407 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Vaahtera L., Schulz J., Hamann T., Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 5, 924–932 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Bacete L., et al. , Arabidopsis response regulator 6 (ARR6) modulates plant cell-wall composition and disease resistance. Mol. Plant Microbe Interact. 33, 767–780 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Bacete L., Hamann T., The role of mechanoperception in plant cell wall integrity maintenance. Plants 9, 574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pesquet E., et al. , Novel markers of xylogenesis in zinnia are differentially regulated by auxin and cytokinin. Plant Physiol. 139, 1821–1839 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellincampi D., Cervone F., Lionetti V., Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 5, 228 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malinovsky F. G., Fangel J. U., Willats W. G., The role of the cell wall in plant immunity. Front. Plant Sci. 5, 178 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kesten C., Menna A., Sánchez-Rodríguez C., Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 40, 106–113 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Nafisi M., Fimognari L., Sakuragi Y., Interplays between the cell wall and phytohormones in interaction between plants and necrotrophic pathogens. Phytochemistry 112, 63–71 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Houston K., Tucker M. R., Chowdhury J., Shirley N., Little A., The plant cell wall: A complex and dynamic structure as revealed by the responses of genes under stress conditions. Front. Plant Sci. 7, 984 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ellis C., Karafyllidis I., Wasternack C., Turner J. G., The Arabidopsis mutant cev1 links cell wall signaling to jasmonate and ethylene responses. Plant Cell 14, 1557–1566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernández-Blanco C., et al. , Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19, 890–903 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramírez V., et al. , MYB46 modulates disease susceptibility to Botrytis cinerea in Arabidopsis. Plant Physiol. 155, 1920–1935 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escudero V., et al. , Alteration of cell wall xylan acetylation triggers defense responses that counterbalance the immune deficiencies of plants impaired in the β-subunit of the heterotrimeric G-protein. Plant J. 92, 386–399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lionetti V., et al. , Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea. Plant Physiol. 143, 1871–1880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferrari S., et al. , Transgenic expression of a fungal endo-polygalacturonase increases plant resistance to pathogens and reduces auxin sensitivity. Plant Physiol. 146, 669–681 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raiola A., et al. , Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol. Plant Microbe Interact. 24, 432–440 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Volpi C., et al. , The ectopic expression of a pectin methyl esterase inhibitor increases pectin methyl esterification and limits fungal diseases in wheat. Mol. Plant Microbe Interact. 24, 1012–1019 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Lionetti V., Raiola A., Cervone F., Bellincampi D., Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Mol. Plant Pathol. 15, 265–274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lionetti V., et al. , Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol. 173, 1844–1863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers L. A., et al. , Comparison of lignin deposition in three ectopic lignification mutants. New Phytol. 168, 123–140 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Delgado-Cerezo M., et al. , Arabidopsis heterotrimeric G-protein regulates cell wall defense and resistance to necrotrophic fungi. Mol. Plant 5, 98–114 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Llorente F., Alonso-Blanco C., Sánchez-Rodriguez C., Jorda L., Molina A., ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43, 165–180 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Trusov Y., Jordá L., Molina A., Botella J. R., “G proteins and plant innate immunity” in Integrated G Proteins Signaling in Plants, Yalovsky S., Baluška F., Jones A., Eds. (Springer, 2010), pp. 221–250. [Google Scholar]
- 42.Klopffleisch K., et al. , Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7, 532 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres M. A., Morales J., Sánchez-Rodríguez C., Molina A., Dangl J. L., Functional interplay between Arabidopsis NADPH oxidases and heterotrimeric G protein. Mol. Plant Microbe Interact. 26, 686–694 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Gallego-Giraldo L., et al. , Elicitors and defense gene induction in plants with altered lignin compositions. New Phytol. 219, 1235–1251 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Gille S., Pauly M., O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 3, 12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawar P. M. A., et al. , Expression of fungal acetyl xylan esterase in Arabidopsis thaliana improves saccharification of stem lignocellulose. Plant Biotechnol. J. 14, 387–397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mélida H., et al. , Non-branched β-1,3-glucan oligosaccharides trigger immune responses in Arabidopsis. Plant J. 93, 34–49 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Aziz A., et al. , Elicitor and resistance-inducing activities of beta-1,4 cellodextrins in grapevine, comparison with beta-1,3 glucans and alpha-1,4 oligogalacturonides. J. Exp. Bot. 58, 1463–1472 (2007). [DOI] [PubMed] [Google Scholar]
- 49.Claverie J., et al. , The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front. Plant Sci. 9, 1725 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hahn M. G., Darvill A. G., Albersheim P., Host-pathogen interactions. XIX. The endogenous elicitor, a fragment of a plant cell wall polysaccharide that elicits phytoalexin accumulation in soybeans. Plant Physiol. 68, 1161–1169 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mélida H., et al. , Arabinoxylan-oligosaccharides act as damage associated molecular patterns in plants regulating disease resistance. Front. Plant Sci. 11, 1210 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown D. M., Zeef L. A., Ellis J., Goodacre R., Turner S. R., Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Persson S., et al. , The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity. Plant Cell 19, 237–255 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood I. P., et al. , Carbohydrate microarrays and their use for the identification of molecular markers for plant cell wall composition. Proc. Natl. Acad. Sci. U.S.A. 114, 6860–6865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lawton K., et al. , Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant Microbe Interact. 8, 863–870 (1995). [DOI] [PubMed] [Google Scholar]
- 56.Parker J. E., et al. , Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8, 2033–2046 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Denancé N., et al. , Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73, 225–239 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Lozano-Durán R., Zipfel C., Trade-off between growth and immunity: Role of brassinosteroids. Trends Plant Sci. 20, 12–19 (2015). [DOI] [PubMed] [Google Scholar]
- 59.van Butselaar T., Van den Ackerveken G., Salicylic acid steers the growth-immunity tradeoff. Trends Plant Sci. 25, 566–576 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Alonso-Simón A., et al. , The use of FTIR spectroscopy to monitor modifications in plant cell wall architecture caused by cellulose biosynthesis inhibitors. Plant Signal. Behav. 6, 1104–1110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pattathil S., et al. , A comprehensive toolkit of plant cell wall glycan-directed monoclonal antibodies. Plant Physiol. 153, 514–525 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruprecht C., et al. , A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiol. 175, 1094–1104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sopeña-Torres S., et al. , YODA MAP3K kinase regulates plant immune responses conferring broad-spectrum disease resistance. New Phytol. 218, 661–680 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Van der Does D., et al. , The Arabidopsis leucine-rich repeat receptor kinase MIK2/LRR-KISS connects cell wall integrity sensing, root growth and response to abiotic and biotic stresses. PLoS Genet. 13, e1006832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang X., et al. , Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. eLife 5, e13568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J., et al. , Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. Plant Physiol. 161, 2146–2158 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schwessinger B., et al. , Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang H., Hong Y., Huang L., Li D., Song F., Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Sci. Rep. 6, 30251 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pogorelko G., et al. , Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiol. 162, 9–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jacobs A. K., et al. , An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15, 2503–2513 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishimura M. T., et al. , Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972 (2003). [DOI] [PubMed] [Google Scholar]
- 72.Vogel J. P., Raab T. K., Schiff C., Somerville S. C., PMR6, a pectate lyase-like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vogel J. P., Raab T. K., Somerville C. R., Somerville S. C., Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 40, 968–978 (2004). [DOI] [PubMed] [Google Scholar]
- 74.Deslandes L., et al. , Genetic characterization of RRS1, a recessive locus in Arabidopsis thaliana that confers resistance to the bacterial soilborne pathogen Ralstonia solanacearum. Mol. Plant Microbe Interact. 11, 659–667 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Engelsdorf T., et al. , Cell wall composition and penetration resistance against the fungal pathogen Colletotrichum higginsianum are affected by impaired starch turnover in Arabidopsis mutants. J. Exp. Bot. 68, 701–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sato M., et al. , Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog. 6, e1001011 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tsuda K., Sato M., Glazebrook J., Cohen J. D., Katagiri F., Interplay between MAMP-triggered and SA-mediated defense responses. Plant J. 53, 763–775 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Stotz H. U., Mitrousia G. K., de Wit P. J., Fitt B. D., Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 19, 491–500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vleeshouwers V. G., Oliver R. P., Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 27, 196–206 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Staal J., Kaliff M., Dewaele E., Persson M., Dixelius C., RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 55, 188–200 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Mengiste T., Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294 (2012). [DOI] [PubMed] [Google Scholar]
- 82.Spanu P. D., Panstruga R., Editorial: Biotrophic plant-microbe interactions. Front. Plant Sci. 8, 192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu Z., et al. , Genetic loci simultaneously controlling lignin monomers and biomass digestibility of rice straw. Sci. Rep. 8, 3636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonawitz N. D., Chapple C., Can genetic engineering of lignin deposition be accomplished without an unacceptable yield penalty? Curr. Opin. Biotechnol. 24, 336–343 (2013). [DOI] [PubMed] [Google Scholar]
- 85.Biswal A. K., et al. , Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat. Biotechnol. 36, 249–257 (2018). [DOI] [PubMed] [Google Scholar]
- 86.da Costa R. M. F., et al. , Desirable plant cell wall traits for higher-quality miscanthus lignocellulosic biomass. Biotechnol. Biofuels 12, 85 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin Q., et al. , Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J. 34, 327–338 (2003). [DOI] [PubMed] [Google Scholar]
- 88.Zang H., et al. , Mannan oligosaccharides trigger multiple defence responses in rice and tobacco as a novel danger-associated molecular pattern. Mol. Plant Pathol. 20, 1067–1079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bacete L., et al. , “Characterization of plant cell wall damage-associated molecular patterns regulating immune responses” in Plant Pattern Recognition Receptors, Shan L., He P., Eds. (Springer, Methods in Molecular Biology, 2017), pp. 13–23. [DOI] [PubMed] [Google Scholar]
- 90.Escudero V., et al. , Mitogen-activated protein kinase phosphatase 1 (MKP1) negatively regulates the production of reactive oxygen species during Arabidopsis immune responses. Mol. Plant Microbe Interact. 32, 464–478 (2019). [DOI] [PubMed] [Google Scholar]
- 91.Mélida H., et al. , Novel type II cell wall architecture in dichlobenil-habituated maize calluses. Planta 229, 617–631 (2009). [DOI] [PubMed] [Google Scholar]
- 92.Mélida H., et al. , Ectopic lignification in primary cellulose-deficient cell walls of maize cell suspension cultures. J. Integr. Plant Biol. 57, 357–372 (2015). [DOI] [PubMed] [Google Scholar]
- 93.Pfaffl M. W., A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.