Significance
HIV is a highly mutable pathogen that can mutate to evade vaccine-induced immune responses, thus negating the vaccine’s protective effects. If mutations that evade immune responses incur a fitness penalty for the virus, other mutations can evolve to partially compensate for fitness loss. Accounting for such effects, we designed a single long peptide immunogen comprised of parts of HIV proteins wherein mutations would be difficult to evolve without fitness penalties for the virus. This designed immunogen was expressed in adenovirus vectors, which are in clinical development (e.g., for COVID-19). Monkeys immunized with our vaccine exhibited strong T-cell responses directed toward regions contained in our immunogen. This suggests that further exploration of this vaccine for use in humans is warranted.
Keywords: subunit vaccines, HIV, fitness landscape
Abstract
An effective vaccine that can protect against HIV infection does not exist. A major reason why a vaccine is not available is the high mutability of the virus, which enables it to evolve mutations that can evade human immune responses. This challenge is exacerbated by the ability of the virus to evolve compensatory mutations that can partially restore the fitness cost of immune-evading mutations. Based on the fitness landscapes of HIV proteins that account for the effects of coupled mutations, we designed a single long peptide immunogen comprising parts of the HIV proteome wherein mutations are likely to be deleterious regardless of the sequence of the rest of the viral protein. This immunogen was then stably expressed in adenovirus vectors that are currently in clinical development. Macaques immunized with these vaccine constructs exhibited T-cell responses that were comparable in magnitude to animals immunized with adenovirus vectors with whole HIV protein inserts. Moreover, the T-cell responses in immunized macaques strongly targeted regions contained in our immunogen. These results suggest that further studies aimed toward using our vaccine construct for HIV prophylaxis and cure are warranted.
Over 30 y ago, HIV was shown to be the causative agent for AIDS (1, 2). The devastating effects of AIDS have been ameliorated by the development of effective antiviral therapies. However, current antiviral therapies are not curative, because they do not eradicate the virus. Infected individuals must stay on therapy for their lifetimes. Although new infections have declined substantially in recent years, over 1.5 million people were infected with HIV in 2019, with infection rates being especially high in parts of sub-Saharan Africa. An effective vaccine against HIV would likely end the long-standing global health challenge presented by HIV. Unfortunately, despite decades of research and enormous expense, such a vaccine does not exist.
A major barrier in the quest for a vaccine against HIV is its high mutability, which enables the virus to evade human immune responses. One approach that is being actively pursued to address this challenge is to develop vaccination strategies to elicit antibodies that are able to neutralize diverse HIV strains (3–5). These broadly neutralizing antibodies target relatively conserved epitopes on the trimeric HIV spike, such as the CD4 binding site (which binds to human receptors to enter host cells) (3–5).
T lymphocytes (T cells) can also be an important component of immune responses induced by a prophylactic or therapeutic vaccine (6–11). In the therapeutic setting, a potent T-cell response is essential. HIV is known to be able to evolve mutations that evade T-cell responses directed against peptide epitopes targeted by humans (12–15). An effective vaccine must elicit T-cell responses directed against epitopes wherein such escape mutations are unlikely to evolve because they severely impair the virus’ fitness, that is, its ability to replicate and propagate infection. One approach to identifying such epitopes is to examine sequences of HIV proteins derived from virus samples extracted from diverse patients and screen for epitopes with residues that exhibit low amino acid variability. A common way to quantify the variability is to compute the entropy corresponding to the extent of variation of amino acids at residues within epitopes. Epitopes containing residues with low entropy are relatively conserved, most likely because mutations therein incur a fitness penalty. One possible strategy to elicit potent vaccine-induced T-cell responses is to employ an immunogen composed of such “low entropy” epitopes in the HIV proteome, and such approaches are being pursued (16–21).
In order to evade T-cell responses, HIV is known to evolve compensatory mutations that partially restore the fitness cost associated with the primary escape mutation (12, 22, 23). Even if only a limited number of mutations can compensate for the fitness costs associated with mutations in a relatively conserved epitope, the high mutability and replication rate of HIV can enable the evolution of the compensatory mutations. This effect would limit the efficacy of the vaccination strategy outlined above. An additional challenge is that subunit vaccines that contain only parts of the viral proteome are difficult to deliver in a way that induces strong T-cell responses. In this paper, we take steps toward addressing these challenges, with the ultimate goal of developing vaccination strategies that can elicit potent T-cell responses in humans.
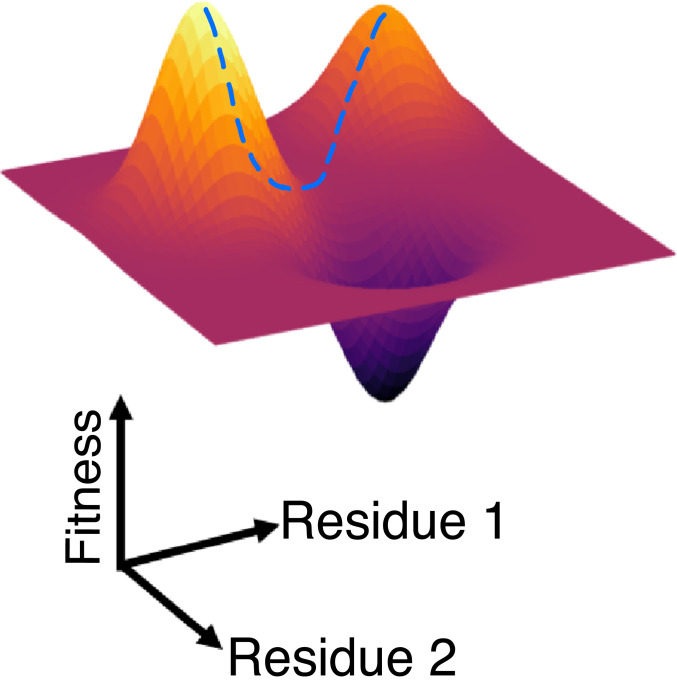
The mutational fitness landscape of a virus is defined by the way in which its ability to replicate and propagate infection in vivo depends on its protein sequences (Fig. 1). Previously, we determined the fitness landscape of HIV polyproteins by analyzing thousands of sequences using methods rooted in computer science and physics that explicitly accounted for the coupled effects of mutations at different protein residues (12, 24–29). This was accomplished in two steps. First, we used a physics-based learning algorithm to develop a model for the “prevalence landscape” of HIV polyproteins. By prevalence landscape, we mean a model that describes the probability of observing a virus with a particular sequence in circulation. Since the sequences used for the analyses were obtained from virus samples derived from patients, the information contained in the inferred model reflects the virus’ ability to replicate and propagate infection in humans. However, the observed sequence data are also influenced by the response of the circulating virus population to human immunity. So, the prevalence landscape may not reflect the intrinsic fitness landscape. We sought the intrinsic fitness landscape, as we wish to define targets for T-cell responses wherein mutations would severely impair the intrinsic ability of the virus to replicate and propagate infection. Therefore, as a second step, we studied the relationship between the prevalence and fitness landscapes given the evolutionary history of HIV (30). These studies suggested that, upon deconvoluting the effects of population-wide human immunity, the rank order of prevalence and fitness is statistically similar for HIV strains. We have previously described the principal biological reasons, as revealed by our analyses, for this relatively simple relationship between the prevalence and fitness landscapes of HIV (30, 31).
Fig. 1.
A cartoon illustrating the fitness landscape of a virus with only one protein with two residues. The horizontal plane represents the amino acid sequence space of the virus, and the relative fitness of strains is depicted as the height of the third dimension. The dotted line represents a potential compensatory pathway.
We have tested predictions derived from the fitness landscape of HIV polyproteins in a number of different ways. Predictions of the fitness of mutant strains were tested positively against in vitro measurements of the ability of the virus to infect human cells and replicate (24–27). We also studied the evolution of escape mutations in a cohort of untreated patients infected with HIV. By combining evolutionary dynamics and the HIV fitness landscape with knowledge of the T-cell epitopes targeted by individuals in this patient cohort, we predicted the location and timing of escape mutations that arise in these individuals with high statistical accuracy (12). These results also highlighted the role of compensatory and antagonistic mutational pathways in the virus’ ability to evolve escape mutations that evade T-cell responses in humans. Epitopes commonly targeted by T-cell responses in elite controllers, or by broadly neutralizing antibodies, contain residues that our fitness landscape predicts to be multidimensionally conserved (12, 24–26, 28, 29)—that is, mutations therein incur a significant fitness cost regardless of amino acids at other residues of the proteins. In some instances, such as for Gag proteins and Env, we observed that the predicted multidimensional constraints in vulnerable regions of HIV proteins originate in structural constraints due to contacts in multiprotein assemblies or individual proteins (25, 28). Our fitness landscape was also predictive of escape mutations observed in protease for patients undergoing antiretroviral therapy (29).
Using the fitness landscape, we can identify epitopes containing residues that are multidimensionally conserved. That is, the fitness landscape allows us to define epitopes where mutations within the epitope incur a high fitness penalty, compensatory pathways are unlikely to be available, and/or mutations at other residues are linked antagonistically to mutations within the epitope. If such multidimensionally conserved epitopes are targeted by T cells, escape mutations that can evade the response are less likely to evolve. Vaccinating with immunogens containing such epitopes may therefore be an effective strategy for eliciting potent T-cell responses.
In past work, we identified some T-cell epitopes that are multidimensionally conserved (24, 28). More recently, based on structural analyses and experiments in mice, Gaiha et al. (32) defined epitopes that are likely to be mutationally vulnerable because they contain residues that interact with many others in protein structures (networked epitopes). McKay and coworkers (33) have recently used extended versions of methods that we first described in our quest for the HIV fitness landscape (28) to also define mutationally vulnerable epitopes that have overlaps with those reported by Gaiha et al. (32).
One practical challenge to deploying short peptide epitopes as immunogens is that they can be difficult to deliver in a way that elicits strong T-cell responses. We have developed nonreplicating adenovirus (Ad) vectors with whole HIV protein mosaic inserts, which have been shown to elicit strong and efficacious T-cell and antibody responses in macaques (34–37). A vaccine with an adenovirus serotype 26 (Ad26) vector and whole HIV protein inserts has also been shown to be safe and immunogenic in humans, and is now being studied in phase 2b and phase 3 clinical efficacy trials (37–43). An Ad26-Ebola virus vaccine has been licensed in Europe, and an Ad26-COVID-19 vaccine is in phase 3 clinical efficacy trials. In this study, we aimed to design a subunit vaccine using the Ad vector delivery platform and an immunogen composed of multidimensionally conserved epitopes identified by our fitness landscape.
For stable expression in Ad vectors, the inserted genes corresponding to the immunogen optimally lie within a length range longer than individual T-cell epitopes. Here we describe a computational algorithm that used the knowledge from our fitness landscape to design long peptide immunogens composed principally of multidimensionally conserved regions of the HIV proteome, and which are of appropriate length for use with Ad vectors. These immunogens were then stably expressed in Ad vectors. Immunization with our designed vaccine elicited strong T-cell responses in rhesus macaques. Our studies show that the designed vaccine is at least as immunogenic as Ad-vectored vaccines expressing whole HIV protein antigens. Moreover, as expected, the immune responses elicited by our designed vaccine construct were focused on the regions included in our immunogen. There is substantial clinical experience with Ad vectors, and proven manufacturing capacity exists. Furthermore, the designed immunogen is based on a fitness landscape of HIV proteins that has been tested against clinical and experimental data in the past (12, 24–27). Therefore, we conclude this paper by discussing the steps necessary to advance our subunit vaccine for use in humans, for both prophylaxis and a functional cure.
Results
The Immunogen Design Algorithm and the Designed Immunogen.
As noted earlier, in the past, we have analyzed thousands of sequences of HIV polyproteins to obtain corresponding mutational fitness landscapes which account for the coupled effects of different mutations. The sequence of a polyprotein of length N is denoted by a vector, s, as it is a list of amino acids at each of the N residues. Our model for the fitness of a strain with the polyprotein sequence, s, is denoted by f(s), and has the following form:
| [1] |
where is a parameter that quantifies the fitness cost of evolving a mutation to amino acid, α, at residue i ), with a large negative value corresponding to a large fitness cost. is a parameter that describes the fitness cost of evolving a double mutation with amino acids and at sites i and j, respectively. A positive value of corresponds to a pair of compensatory mutations; a negative value implies that, if the two corresponding mutations arise together, the fitness cost is larger than the independent effects of each individual mutation. Mutations are defined with respect to a reference sequence. The set of parameters, {, }, are inferred by fitting them to recapitulate the single and double mutation probabilities observed in the sequence data. This was accomplished using various methods (24, 25, 44).
Armed with the fitness landscape of HIV polyproteins, we embarked on developing an algorithm that could design single long peptide immunogens containing multidimensionally conserved epitopes that are 300 to 1,600 amino acids in length. Inserts in this range of length can potentially be stably expressed in Ad vectors.
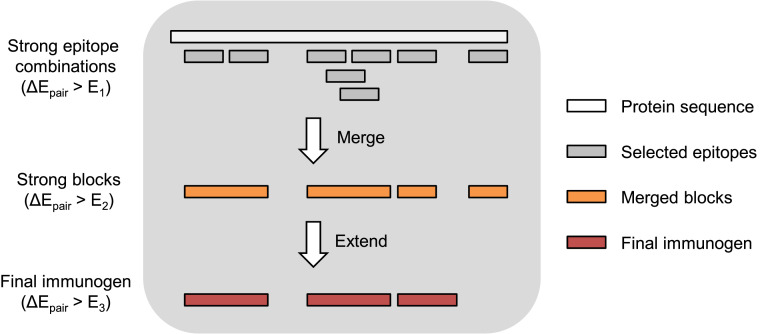
The algorithm that we developed is schematically depicted in Fig. 2. We first use the fitness landscape to compute the fitness cost of all double mutations in pairs of nonoverlapping epitopes, averaged over all possible sequences that may arise in the rest of the polyproteins (see Methods). Briefly, we fix the amino acids at a pair of residues in the two chosen epitopes. Then Eq. 1 is used to compute the fitness cost of the chosen pair of mutations. The fitness cost is then averaged over an ensemble of sequences generated according to the probabilities with which they are in circulation (Eq. 1) using a Monte Carlo procedure. This process is repeated for all pairs of amino acids that could arise at the two epitopes, and an average over all the fitness costs thus incurred is calculated. We refer to this as the “pairwise fitness cost” for a given pair of epitopes. The pairwise fitness cost has contributions from direct fitness effects of individual mutations, interactions between mutations in the two epitopes, and interactions between amino acid mutations in the epitopes and amino acids in the rest of the polyproteins’ sequence.
Fig. 2.
Schematic depiction of our immunogen design algorithm. First, we select a pair of epitopes where the fitness cost of mutations required to escape from both epitopes is high in all sequence backgrounds, and then we add additional epitopes sequentially as described in step 1 of the algorithm described in the text. Next, we bridge and merge together overlapping epitopes to form larger blocks, provided that the average pairwise fitness cost for mutations in epitopes in these larger blocks remains high (step 2 of the algorithm). Finally, we extend blocks to a minimum length to avoid introducing too many unnatural junctional epitopes that bridge between blocks when the blocks are concatenated into an immunogen (step 3 of the algorithm). Blocks that cannot be extended to this minimum length without significantly decreasing the pairwise fitness cost of mutations are eliminated.
We average over different sequence backgrounds because we wish to make a single vaccine construct for all individuals. Because of HIV’s high mutability, the virus that is present in different individuals will exhibit different mutations. We aim to avoid targeting epitopes where escape mutations may incur a high fitness cost in some sequence backgrounds, but where compensatory mutations in other backgrounds can lower the fitness cost for escape. Averaging the fitness cost over viable sequence backgrounds enables us to identify regions where mutations will incur a large fitness penalty independent of the sequence background in which the mutation has to evolve.
Calculating the pairwise fitness cost allows us to determine a rank-ordered list of epitope pairs; the highest ranked pair corresponds to the one wherein mutations in the two epitopes would be most deleterious for the virus regardless of the rest of the protein sequence. If the highly ranked pairs of epitopes are targeted by T cells, the virus would likely be cornered between being killed by this immune response or evolving unviable mutations, regardless of the amino acids at other residues of the proteins. Below, we will also refer to an “average pairwise fitness cost” of the immunogen, which is simply the average of the “pairwise fitness cost” over all pairs of nonoverlapping epitopes in the immunogen.
Our algorithm proceeds in four steps for each polyprotein.
-
1)
Seed: Begin the immunogen by finding the best pair of 11-mer epitopes in a polyprotein with pairwise fitness cost greater than a threshold . The value that we have used for was based on studies of HIV evolution in vivo, where we found that escape was unlikely to occur in epitopes when the fitness cost of potential escape mutations exceeded (12). Selecting from the remaining epitopes in the protein, add the epitope with the highest average pairwise fitness cost when paired with the epitopes already in the immunogen. Repeat this selection and addition step until the average pairwise fitness cost of the new epitope, averaged over all pairs of epitopes in the immunogen, falls below .
-
2)
Bridge and merge: The output of step 1 is a list of blocks of variable length that are either noncontiguous or overlapping by <10 residues. Because we assume putative epitopes are 11-mers, if two blocks overlapped by 10 residues, then they could simply be merged into one block without changing the included epitopes. To bridge noncontiguous blocks, we consider the intervening amino acid segments between all successive blocks. Such a segment is added to the immunogen if the epitopes so included do not reduce the average pairwise fitness cost below a threshold . This threshold value was chosen to be smaller than , thus allowing us to form larger blocks, but was still large enough that escape would still be suppressed. Lengthening blocks is important due to the lower bound of 300 amino acids required for stable expression in Ad vectors. To merge successive overlapping blocks, we perform a similar procedure for the epitopes that would be included by combining the two blocks. To illustrate this simply, let us consider 4-mer epitopes instead of 11-mers. For example, the amino acid sequence ACDEFG contains three 4-mers: ACDE, CDEF, and DEFG. The epitopes ACDE and DEFG overlap by two residues, but, if they are concatenated as ACDEDEFG, then the CDEF epitope is not included. However, all three epitopes can be included with the immunogen sequence ACDEFG. If including the CDEF epitope keeps the average pairwise fitness cost above , then it will be included, and the immunogen sequence will be ACDEFG. Otherwise, if the CDEF epitope does not synergize well with the rest of the immunogen, then the immunogen sequence will be ACDEDEFG.
-
3)
Extend or reject: After step 2, some of the blocks may still be very short. If concatenated together with other blocks, these would introduce unnatural junctional epitopes that contain some residues from one block and some from another. Such an epitope would not be processed and presented on cells infected with HIV, as the junction introduced by connecting two short blocks does not exist in its proteome. Targeting an unnatural junctional epitope with a vaccine-induced T-cell response is of no value. Therefore, we wish to minimize the number of unnatural junctional epitopes in our immunogen. Blocks shorter than 31-mers contain more unnatural than natural epitopes. This is because 21 overlapping 11-mers fit completely within a 31-mer block, and each junction between blocks introduces ten 11-mer junctional epitopes. For the short blocks still under consideration, all 31-mers that contain them are considered. We include these 31-mers in the immunogen as long as the average pairwise fitness cost of the new epitopes with the existing epitopes in the immunogen exceeds a threshold , which was selected following a similar rationale to the threshold. The blocks which cannot be extended this way due to poor synergy are removed from the immunogen. This leads to deletion of some multidimensionally conserved regions from our immunogen.
-
4)
Concatenate: The remaining unconnected blocks are concatenated. The blocks can be concatenated in different orders. We concatenated the blocks in their native 5′-to-3′ order as well as a shuffled variation wherein the blocks are joined in a different order. Compared to the 5′ to 3′ immunogen, the shuffled immunogen thus contains a different set of unnatural junctional epitopes, but the same mutationally vulnerable epitopes. Thus, upon immunization with both immunogens, T-cell responses to the natural epitopes are expected to be more likely than undesired responses to unnatural junctional epitopes.
Note that a lower threshold corresponds to a more lenient criterion for including a region of the viral proteome in the immunogen. As noted above, in past work (12), we found that the virus was unable to evolve escape mutations for long times if the fitness penalties as per our fitness landscape were sufficiently large. The threshold values that we used were guided by these data (12). The specific values used for the fitness penalty thresholds are E1 = 8.5, E2 = 7.5, and E3 = 7.0 (for definition of E, see Eq. 1 and Methods). For Pol proteins, we use a threshold that is more stringent than for the other proteins (in particular, ) because it is not as immunogenic, and so we wish to include only the regions that contain residues where mutations are more deleterious for the virus’ fitness. We did not test the effects of lowering these thresholds substantially, because this would lead to the inclusion of regions where escape mutations would be likely to evolve within a short time.
Our designed immunogen following this procedure is 551 amino acids in length. The amino acid and nucleotide sequences of the two immunogens we designed are shown in Tables 1 and 2, respectively. We will refer to the immunogen arranged in its 5′-to-3′ order as “fit1” and the shuffled immunogen as “fit2.” As shown (SI Appendix, Fig. S1), the designed immunogen contains many regions that are known to be immunogenic in macaques (45, 46), as well as human epitopes in the CTL A-list (47). As described in Discussion, moving our immunogen forward to human studies will also require us to assess the immunodominance of different epitopes contained in our immunogen for people with diverse human leukocyte antigen (HLA) types.
Table 1.
Concatenated amino acid sequence for two different versions of the immunogen
| Version | Amino acid sequence |
| Immunogen:5-3 (fit1) | VWASRELERFAVNPGLLETSEGCRQILGQLQQAISPRTLNAWVKVVEEKAFSPEVIPMFSALSEGATPQDLNTMLNTVGGHQAAMQMLKETINEEAAEWDRLHPVHAGPIAPGQMREPRGSDIAGTTSTLQEQIGWMTNNPPIPVGEIYKRWIILGLNKIVRMYSPTSILDIRQGPKEPFRDYVDRFYKTLRAEQASQEVKNWMTETLLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPEALLDTGADDTVLEEMNLPGRWKPKMIGGIGGFIKVTPDKKHQKEPPFLWMGYELHPDKWTVQPIVLPEKDSWTVNDIQKLVGKLNWASQIYMENRWQVMIVWQVDRMRIRTWKSLVKHHMYIDAKLVITTYWGLHTGERDWHLGQGVSIEWRKFLGFLGAAGSTMGAASITLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARSLCLFSYHRLRDLLLIVTRIVELLGRRGWEANADCAWLEAQEEEEVGFPVRPQVPLRPMTYKYSQKRQDILDLWVYHTQGYFPDWQNYTPGPG |
| Shuffled immunogen (fit2) | YSQKRQDILDLWVYHTQGYFPDWQNYTPGPGQAISPRTLNAWVKVVEEKAFSPEVIPMFSALSEGATPQDLNTMLNTVGGHQAAMQMLKETINEEAAEWDRLHPVHAGPIAPGQMREPRGSDIAGTTSTLQEQIGWMTNNPPIPVGEIYKRWIILGLNKIVRMYSPTSILDIRQGPKEPFRDYVDRFYKTLRAEQASQEVKNWMTETLLVQNANPDCKTILKALGPAATLEEMMTACQGVGGPTPDKKHQKEPPFLWMGYELHPDKWTVQPIVLPEKDSWTVNDIQKLVGKLNWASQIYDAKLVITTYWGLHTGERDWHLGQGVSIEWRKSLCLFSYHRLRDLLLIVTRIVELLGRRGWEAEALLDTGADDTVLEEMNLPGRWKPKMIGGIGGFIKVMENRWQVMIVWQVDRMRIRTWKSLVKHHMYIFLGFLGAAGSTMGAASITLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARNADCAWLEAQEEEEVGFPVRPQVPLRPMTYKVWASRELERFAVNPGLLETSEGCRQILGQLQ |
Table 2.
Concatenated nucleotide sequence for two different versions of the immunogen
| Version | Nucleotide sequence |
| Immunogen:5-3 (fit1) | ATGGTCTGGGCCAGCAGAGAGCTGGAAAGATTCGCCGTGAATCCCGGCCTGCTGGAAACCTCTGAGGGCTGCAGACAGATCCTGGGACAGCTGCAGCAGGCCATCTCTCCCAGAACACTGAACGCCTGGGTCAAAGTGGTGGAAGAGAAGGCTTTCAGCCCCGAAGTGATCCCCATGTTCAGCGCCCTTTCTGAGGGCGCCACACCTCAGGACCTGAACACCATGCTGAATACCGTTGGCGGACACCAGGCCGCCATGCAGATGCTGAAAGAGACAATCAACGAAGAGGCCGCCGAGTGGGATAGACTGCACCCTGTTCATGCCGGACCTATCGCTCCAGGCCAGATGAGAGAGCCTAGAGGCTCTGATATCGCCGGCACCACCAGCACACTGCAAGAGCAGATCGGCTGGATGACCAACAATCCTCCTATTCCTGTGGGCGAGATCTACAAGCGGTGGATCATCCTGGGCCTGAACAAGATCGTGCGGATGTACAGCCCCACCAGCATCCTGGATATCCGGCAGGGACCCAAAGAGCCCTTCAGAGACTACGTGGACCGGTTCTACAAGACCCTGAGAGCCGAGCAGGCCAGCCAAGAAGTGAAGAACTGGATGACAGAGACACTGCTGGTGCAGAACGCCAATCCTGACTGCAAGACCATCCTGAAGGCCCTGGGACCTGCCGCCACACTGGAAGAAATGATGACCGCCTGTCAAGGCGTTGGCGGCCCTGAAGCTTTGCTGGATACAGGCGCCGATGACACCGTGCTGGAAGAGATGAATCTGCCTGGCCGGTGGAAGCCCAAGATGATCGGAGGAATCGGCGGCTTCATCAAAGTGACCCCTGACAAGAAGCACCAGAAAGAACCACCTTTCCTGTGGATGGGCTACGAGCTGCACCCCGATAAGTGGACCGTGCAGCCTATTGTGCTGCCCGAGAAGGATAGCTGGACCGTGAACGACATCCAGAAACTCGTGGGCAAGCTGAATTGGGCCAGCCAGATCTACATGGAAAACCGGTGGCAAGTGATGATCGTGTGGCAGGTCGACCGGATGCGGATCAGAACCTGGAAGTCCCTGGTCAAGCACCACATGTACATCGACGCCAAGCTGGTCATCACCACCTACTGGGGACTGCACACCGGCGAGAGAGATTGGCATCTTGGACAGGGCGTGTCAATCGAGTGGCGGAAGTTCCTGGGCTTTCTGGGAGCCGCCGGATCTACAATGGGAGCTGCCAGCATCACCCTGACAGTGCAGGCTAGACAGCTGCTGAGCGGAATCGTGCAGCAGCAGAACAACCTGCTGAGAGCCATTGAGGCCCAGCAGCATCTCCTGCAGCTGACAGTGTGGGGCATCAAGCAGCTCCAGGCTAGAAGCCTGTGCCTGTTCAGCTACCACAGACTGAGGGACCTGCTGCTGATCGTGACCCGGATTGTGGAACTGCTGGGAAGAAGAGGCTGGGAAGCCAATGCCGATTGCGCCTGGCTGGAAGCTCAAGAGGAAGAGGAAGTCGGCTTCCCCGTCAGACCTCAGGTGCCACTCAGACCCATGACCTACAAGTACAGCCAGAAGCGGCAGGACATCCTGGACCTGTGGGTGTACCACACACAGGGCTACTTCCCCGACTGGCAGAACTACACACCTGGACCAGGC |
| Shuffled immunogen (fit2) | ATGTACAGCCAGAAGCGGCAGGACATCCTGGACCTGTGGGTGTACCACACACAGGGCTACTTCCCCGACTGGCAGAACTACACACCTGGACCAGGACAGGCCATCTCTCCCAGAACACTGAACGCCTGGGTCAAAGTGGTGGAAGAGAAGGCTTTCAGCCCCGAAGTGATCCCCATGTTCAGCGCCCTTTCTGAGGGCGCCACACCTCAGGACCTGAACACCATGCTGAATACCGTTGGCGGACACCAGGCCGCCATGCAGATGCTGAAAGAGACAATCAACGAAGAGGCCGCCGAGTGGGACAGACTGCATCCTGTTCATGCCGGACCTATCGCTCCCGGCCAGATGAGAGAACCTAGAGGCTCTGATATCGCCGGCACCACCAGCACACTGCAAGAGCAGATCGGCTGGATGACCAACAATCCTCCTATTCCTGTGGGCGAGATCTACAAGCGGTGGATCATCCTGGGCCTGAACAAGATCGTGCGGATGTACTCCCCTACCAGCATCCTGGATATCCGGCAGGGCCCCAAAGAGCCCTTCAGAGACTACGTGGACCGGTTCTACAAGACCCTGAGAGCCGAGCAGGCCAGCCAAGAAGTGAAGAACTGGATGACAGAGACACTGCTGGTGCAGAACGCCAATCCTGACTGCAAGACCATCCTGAAGGCCCTGGGACCTGCCGCCACACTGGAAGAAATGATGACCGCCTGTCAAGGCGTCGGCGGACCCACACCTGATAAGAAGCACCAGAAAGAACCACCGTTCCTGTGGATGGGCTACGAGCTGCACCCTGACAAGTGGACCGTGCAGCCTATTGTGCTGCCCGAGAAGGATAGCTGGACCGTGAACGACATCCAGAAACTCGTGGGCAAGCTGAACTGGGCCAGCCAGATCTACGATGCCAAGCTGGTCATCACCACCTACTGGGGACTGCACACCGGCGAGAGAGATTGGCATCTTGGACAGGGCGTGTCCATCGAGTGGCGGAAGTCCCTGTGCCTGTTCAGCTACCACAGACTGAGGGACCTGCTGCTGATCGTGACCCGGATTGTGGAACTGCTGGGAAGAAGAGGCTGGGAAGCCGAGGCTCTGCTTGATACAGGCGCCGATGATACCGTGCTGGAAGAGATGAACCTGCCTGGCAGATGGAAGCCCAAGATGATCGGCGGCATCGGCGGATTCATCAAAGTCATGGAAAACCGGTGGCAAGTGATGATCGTGTGGCAGGTCGACCGGATGCGGATCAGAACCTGGAAGTCTCTGGTCAAGCACCACATGTATATCTTTCTGGGATTCCTGGGCGCTGCCGGCTCTACAATGGGAGCCGCTTCTATCACCCTGACTGTGCAGGCTAGACAGCTGCTGAGCGGAATCGTGCAGCAGCAGAACAACCTGCTGAGAGCCATTGAGGCCCAGCAGCATCTCCTGCAGCTGACAGTGTGGGGCATCAAGCAGCTCCAGGCCAGAAATGCCGATTGCGCCTGGCTGGAAGCTCAAGAGGAAGAGGAAGTCGGCTTTCCCGTCAGACCTCAGGTGCCACTGAGGCCTATGACCTACAAAGTGTGGGCCAGCAGAGAGCTGGAAAGATTCGCCGTGAATCCCGGCCTGCTGGAAACCTCTGAGGGCTGCAGACAGATCCTGGGGCAGCTGCAG |
The Designed Immunogens Are Stably Expressed in Adenovirus Vectors and Are Immunogenic in Macaques.
The designed immunogens were expressed in E1/E3-deleted, replication-incompetent Ad26 and Ad5 vectors, essentially as described previously (48). In this study, four rhesus macaques (Macaca mulatta) were first immunized by the intramuscular route with 1011 viral particles (vp) of Ad26 expressing the “fit2” immunogen, and two macaques served as positive controls, as they were immunized with 1011 vp Ad26 expressing whole HIV Gag/Pol/Env inserts (45). It is important to note that previous studies in rhesus macaques have demonstrated the immunogenicity of the Ad vectors expressing whole inserts; strong T-cell responses distributed across HIV proteins are elicited (34–37). Table 3 shows the immunization schedule for both groups of macaques. The four macaques that received our designed immunogen were last boosted at 48 wk postprime with 1011 vp of Ad5 expressing the “fit1” immunogen. The vector used to boost the two macaques in the control group at this time point was Ad5HVR48. Ad5HVR48 and Ad5 are 99% identical and comparably immunogenic (49), and so, due to vector availability, Ad5HVR48 was used rather than Ad5. The use of the Ad5 vector for the final boost allowed us to study the effect of a heterologous vector on the magnitude of T-cell responses elicited by our designed vaccine.
Table 3.
Immunization schedule for animals immunized with the designed vaccine and those immunized with whole HIV protein antigens
| Week | Animals immunized with designed vaccine | Animals immunized with whole HIV proteins |
| 0 | Ad26-Fit2 | Ad26-Env/Gag/Pol |
| 12 | Ad26-Fit1 | Ad26-Env/Gag/Pol |
| 24 | RhAd66-Fit2 | Ad26-Env/Gag/Pol |
| 48 | Ad5-Fit1 | Ad5HVR48-Env/Gag/Pol |
The vector is shown before the antigen. Week 0 is the prime, and the following weeks are boosts.
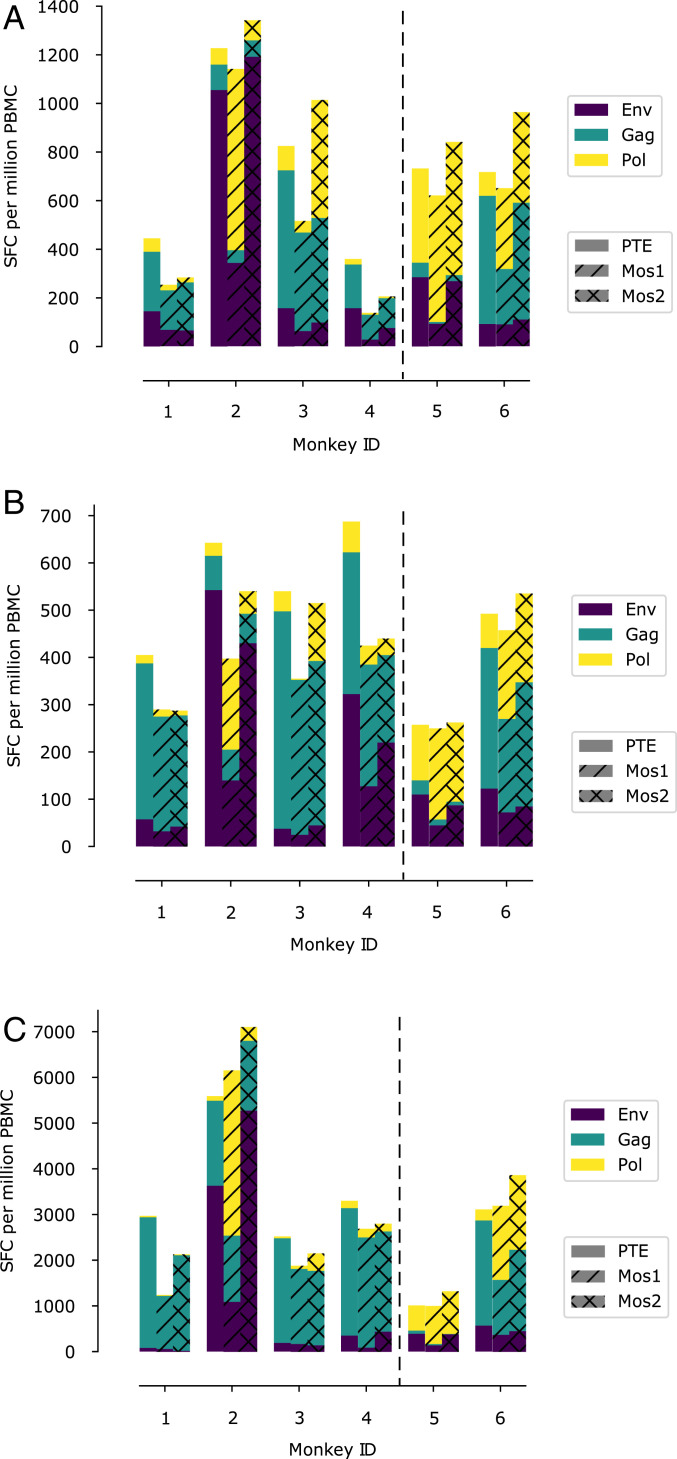
Vaccine-elicited cellular immune responses were assessed by interferon (IFN)-γ-enzyme linked immune absorbent spot (ELISPOT) assays. The raw data from the immunization studies are provided (SI Appendix, Tables S1–S22). Fig. 3 shows the magnitude of the ELISPOT responses directed against different HIV proteins that were elicited in both groups of macaques at week 4 after priming, at week 20, and at week 50 (2 wk after the final boost). Animals immunized with our designed immunogen elicited ELISPOT responses that were comparable in magnitude to those immunized with whole protein antigens. After priming with Ad26-Fit2, the next two boosts did not increase responses much, but the final Ad5-Fit1 boost increased cellular immune responses substantially. These data point to the potency of a heterologous vector for delivery of the designed immunogens.
Fig. 3.
Bar graphs showing immunogenicity to various peptide pools for macaques 4 wk after priming (A), 20 wk after priming (B), and 50 wk after priming (C). The first four macaques (1 to 4) were immunized with the immunogen designed by us (Table 1). The last two macaques (5 and 6) were immunized with standard whole protein immunogens. All immunogens were vectored by adenovirus serotypes. The ordinates show the results of ELISPOT assays as the number of spot-forming cells (SFC) per million peripheral blood mononuclear cells (PBMC). The proportion of the response directed toward Gag, Env, and Pol are depicted by the bars colored green, purple, and yellow, respectively. Three different peptide pools (see Methods) were used for the ELISPOT assays: PTE (plane bars), Mos1 (hatched bars), and Mos2 (cross-hatched bars).
Table 4 shows the evolution of the T-cell responses in animals immunized with our designed vaccine. As time ensued, the proportion of responses directed toward Gag increased in all four outbred macaques, with the extent of this increase depending upon the specific peptide pools used for assaying T-cell responses.
Table 4.
Evolution of the CTL response in animals immunized with the designed vaccine as determined by assays with three different standard peptide pools
| Animal number and time point | GAG:ENV:POL, % | ||
| PTE | MOS1 | MOS2 | |
| Macaque 1 (4 wk) | 55:33:12 | 64:27:9 | 70:23:7 |
| Macaque 1 (20 wk) | 81:14:4 | 84:11:5 | 82:15:3 |
| Macaque 1 (50 wk) | 96:3:1 | 93:6:2 | 98:1:1 |
| Macaque 2 (4 wk) | 9:86:5 | 5:30:65 | 5:89:6 |
| Macaque 2 (20 wk) | 11:84:4 | 16:35:48 | 12:80:9 |
| Macaque 2 (50 wk) | 33:65:2 | 24:18:59 | 22:74:4 |
| Macaque 3 (4 wk) | 69:19:12 | 79:12:9 | 42:10:48 |
| Macaque 3 (20 wk) | 85:7:8 | 92:7:1 | 67:9:24 |
| Macaque 3 (50 wk) | 91:8:2 | 87:10:4 | 75:7:18 |
| Macaque 4 (4 wk) | 50:44:6 | 75:20:5 | 60:37:4 |
| Macaque 4 (20 wk) | 44:47:9 | 61:30:9 | 42:50:8 |
| Macaque 4 (50 wk) | 85:11:5 | 89:4:7 | 78:16:6 |
Analyses of the ELISPOT data using peptide subpools (raw data in SI Appendix, Tables S17–S22) (37, 45) provided more granular information on the regions of HIV proteins targeted by the T-cell responses in macaques immunized with our designed vaccine (Table 5). The regions targeted by these T-cell responses are contained in the subunit immunogen that was used (compare Tables 5 and 6). In three out of the four macaques (animals 1, 3, and 4) immunized with our designed immunogen, the T-cell responses elicited at 50 wk after priming were largely focused on regions broadly distributed across Gag, and the responses were strong (Table 5). We note that previous studies have shown that Gag-focused responses correlate with better virus control and lower viral loads in humans and macaques (35, 46, 50). The subpool data for the fourth macaque (animal 2) show that this animal mounted strong immune responses to subpools containing both regions of Gag and Env contained in our immunogen. When examined using pooled peptides of the mosaic 1 type, in this animal, strong T-cell responses targeting regions of Pol that contain our immunogen were also elicited (3,560 spot-forming cells per million peripheral blood mononuclear cells). Thus, in the four outbred macaques immunized with our designed Ad-vectored vaccine, strong Gag-focused T-cell responses were elicited in three animals, and, in the fourth animal, strong and broader Gag-Pol-Env directed responses were detected. The reason for the different responses in animal 2 is not clear, but such variability is common in outbred nonhuman primates. It is encouraging to see that strong T-cell responses directed against mutationally vulnerable regions (especially in Gag) were elicited in all animals.
Table 5.
Strong T-cell responses against regions contained in our designed immunogen are detected by ELISPOT assays in all four immunized animals (data shown are for 50 wk after the prime)
| Region | Animal 1 | Animal 2 | Animal 3 | Animal 4 |
| Gag 1 to 52 | 3,250 | |||
| Gag 160 to 212 | 540 | 1,460 | 1,750 | 2,590 |
| Gag 204 to 253 | 720 | |||
| Gag 241 to 294 | 380 | 400 | ||
| Env 487 to 540 | 3,830 |
The first column lists the HXB2 positions of the regions in the PTE peptide subpools that elicit strong ELISPOT responses (greater than 350 spot-forming cells [SFC] per million peripheral blood mononuclear cells). Columns 2 to 5 list the strength of the response detected in each immunized animal after the last boost. As described in the main text, in assays with pooled peptides, animal 2 also developed a response to a region in Pol that is contained in our immunogen.
Table 6.
Regions in Gag, Env, and Pol contained in the immunogen
| HIV polyprotein | Region included in immunogen |
| Gag | 35 to 65, 145 to 356 |
| Env | 519 to 564, 762 to 792 |
| Pol | 77 to 112, 371 to 426 |
For reference to data in Table 5, the regions in Gag, Env, and Pol contained in the immunogen (HXB2 numbering) are noted. The immunogen also contained regions in Vif and Nef that were not substantially targeted by the immunized animals.
Taken together, these data show that, when delivered using Ad vectors, the immunogens designed using our algorithm that contain multidimensionally conserved regions of the HIV proteome are at least as immunogenic as well-studied whole protein antigens. Furthermore, the responses elicited by our designed immunogen were focused on multidimensionally conserved regions of the HIV proteome in outbred macaques of diverse genotypes.
Discussion
Despite over 30 y of effort, neither a prophylactic or therapeutic vaccine is available for HIV. A major barrier in this quest has been the high mutability of the virus. Major efforts to overcome this challenge are focused on eliciting broadly neutralizing antibodies by vaccination (3–5). But T-cell responses can also be important components of effective vaccine-induced immune responses for prophylaxis or therapy (6–11). Potent T-cell responses are essential for therapeutic vaccination, which could potentially either eradicate the virus or achieve a functional cure by keeping viral loads to undetectable levels in vaccinated individuals. Indeed, the ability of elite controllers of HIV, cohorts of patients who maintain undetectable or very low viral loads, is correlated with particular types of T-cell responses that may contribute to controlling viral loads to low levels (8, 51, 52).
Because of its high mutability and high replication capacity, HIV is known to evolve mutations that can escape T-cell responses. This challenge could be addressed if vaccine-induced T-cell responses targeted epitopes wherein mutations incurred a large fitness penalty. Using low entropy of amino acid variation as a metric of the fitness cost of evolving mutations, an immunogen containing parts of the HIV proteome, tHIVconsvX, was designed by Hanke and coworkers (16) and studied in mice. However, HIV can evolve compensatory mutations that partially restore the fitness costs incurred by making escape mutations at targeted epitopes (12, 22, 23). This may limit the efficacy of the strategy of designing immunogens based on consideration of epitope entropy alone (as discussed in the Introduction).
In past work, we have developed a fitness landscape for HIV polyproteins that accounts for the coupled effects of mutations (12, 24–27, 30). Availability of the fitness landscape enables identification of multidimensionally conserved epitopes, that is, those wherein mutations incur a high fitness cost regardless of the rest of the polyprotein sequence. Based on this knowledge, and an algorithm developed in this paper, we designed single long peptide immunogens composed principally of multidimensionally conserved epitopes. These long peptide immunogens are suitable for delivery using the Ad-based delivery platform for which there is considerable clinical experience (37–43). Indeed, Ad26 vectors expressing whole “mosaic” Env, Gag, and Pol immunogens are currently in phase 2b and phase 3 vaccine clinical trials. We immunized macaques with our designed immunogens using Ad vectors and showed that our vaccine constructs are comparably immunogenic as whole protein antigens. Moreover, these responses are directed against multidimensionally conserved regions of the HIV proteome in a group of outbred macaques. These data set the stage for advancing our vaccine concept to human studies.
We note that the immunogens we have designed have both similarities to and differences from tHIVconsvX, which was designed based on sequence entropy considerations; 33.3% of residues in tHIVconsvX are contained in our immunogen, and 52.1% of residues in our immunogen are contained in tHIVconsvX. So, our vaccine is a different construct that needs to be independently explored, as should other approaches to immunogen design (32). Several steps must be taken to further advance the use of our vaccine.
Given that the Ad vectors are known to be safe in humans, the next step to advance the vaccine construct that we have described in this paper may be to carry out a study in humans in the therapeutic setting. Here the goal would be to use our designed vaccine to either eradicate the virus from vaccinated patients or control viral load to very low or undetectable levels without therapy. Elite controllers achieve the latter goal, and the situation is tantamount to a functional cure. Recent work suggests that such potent cytotoxic T lymphocte (CTL) responses may enable elite controllers to eliminate infected cells in which viral DNA is integrated in euchromatin (53). These results encourage the use of designed therapeutic vaccines like ours as a route to a potential functional cure of HIV. A study to assess the efficacy of our designed vaccine for achieving a functional cure could be carried out in a treatment interruption clinical trial (54) with a small cohort of patients currently under antiretroviral therapy. Such a study could be carried out after, or in parallel with, a simian human immunodeficiency virus challenge study in a large cohort of macaques.
Several challenges have to be confronted in order to achieve the goal of a functional cure by therapeutic vaccination. Our designed immunogen contains mutationally vulnerable consensus epitopes. In infected persons, the predominant strains in their viral reservoir might have already evolved escape mutations at some of these epitopes. For such epitopes, the vaccine-induced CTL response would not be useful. This challenge can potentially be overcome by taking the following steps. First, we need to determine the epitopes that a patient’s T cells are likely to have targeted. Immunodominance patterns are strongly influenced by the HLA type of the patient, and so this must be established. Given the HLA type, the immunodominance pattern of the epitopes that have likely been targeted needs to be determined. This is a challenging problem, but data exist on immunodominance patterns for HIV infection. Furthermore, recently, by training a model to data from HIV-infected patients, we have made progress in being able to predict immunodominance hierarchies, given HLA type (55). Given the epitopes that have likely been immunodominantly targeted, we can use our fitness landscape to determine the likelihood that escape mutations may have evolved in the dominant epitopes. Alternatively, this knowledge could also be obtained by sequencing the viral reservoir in the patient. If escape mutations have evolved in the most dominant epitope, using a variant of the method developed in ref. 55, one would ask whether the remaining epitopes that are likely to be immunodominant and wherein escape mutations have not evolved are contained in our immunogen. If they are, then the patient is a candidate for therapeutic vaccination. Not all patients will be good candidates. Finally, we may also explore the use of mosaic versions of our designed immunogen (45).
In conclusion, we have designed a subunit vaccine containing an immunogen with mutationally vulnerable multidimensionally conserved regions of the HIV proteome. This vaccine elicits strong T-cell responses in macaques. We believe that further studies exploring the use of this vaccine concept for prophylaxis or therapy in humans are warranted.
Methods
Determination of the Pairwise Fitness Cost.
Let us illustrate the approach for calculating an average fitness cost by first considering a single epitope. Following Eq. 1, let denote a sequence, and denote the corresponding “energy.” We use the word “energy” for E by analogy with statistical mechanics, where Eq. 1 describes a Boltzmann probability and E appears as the energy of the system. Similar language has been used when similar models have been used to study protein sequences (56). The value of the energy correlates negatively with the fitness of the viral strain with sequence (24–26). The full sequence can be divided into two parts: , the region containing the epitope of interest, and , which contains the rest of the protein.
To average over the possible sequence backgrounds, in which the epitope, , might appear, the energy/fitness cost of realizable mutations at different points in the epitope, given all possible sequence backgrounds, can be computed. This is accomplished using a standard Monte Carlo procedure in which various possible sequences, , are sampled according to their prevalence probability (Eq. 1). The average fitness cost for evolving mutations at the epitope under consideration can then be computed as follows. First, the region containing the epitope is fixed to be equal to that of the targeted epitope, . The average energy difference between a mutant and the unmutated epitope is
| [2] |
Eq. 2 is evaluated using standard Monte Carlo sampling techniques (57). The estimated fitness cost of evolving escape mutations in the epitope is then
| [3] |
This average is used for computing the average fitness cost of mutations in order to put the most weight on more probable escape routes. The method used to obtain average fitness cost is illustrated using a single epitope for simplicity. It is easily generalized for calculations carried out with a pair of epitopes following the steps described in Results.
Immunization.
Six outbred Indian-origin adult male and female rhesus macaques (M. mulatta), 3 y to 8 y old, were randomly allocated to two groups. All animals were housed at Alpha Genesis, Inc. Animals received 1011 vp Ad26, RhAd66, and Ad5 (or Ad5HVR48) vectors expressing the “fit1,” “fit2,” or whole mosaic Env/Gag/Pol immunogens by the intramuscular route without adjuvant. All animal studies were conducted in compliance with all relevant local, state, and federal regulations and were approved by the Bioqual Institutional Animal Care and Use Committee.
ELISPOT and Peptide Pools.
ELISPOT plates were coated with mouse anti-human IFN-γ monoclonal antibody from BD Pharmingen at a concentration of 5 µg per well overnight at 4 °C. Plates were washed with Dulbecco's phosphate-buffered saline containing 0.25% Tween 20, and blocked with R10 media (Roswell Park Memorial Institute with 11% fetal bovine serum and 1.1% penicillin−streptomycin) for 1 h at 37 °C. Peptide pools contained 15 amino acid peptides overlapping by 11 amino acids that spanned the mosaic 1 (Mos1), mosaic 2 (Mos2), or potential T-cell epitope (PTE) Env, Gag, and Pol protein sequences. Peptide pools were prepared at a concentration of 2 µg per well, and 200,000 cells per well were added. The peptides and cells were incubated for 18 h to 24 h at 37 °C. All steps following this incubation were performed at room temperature. The plates were washed with Coulter buffer and incubated for 2 h with Rabbit polyclonal anti-human IFN-γ Biotin from U-Cytech (1 µg/mL). The plates are washed a second time and incubated for 2 h with Streptavidin−alkaline phosphatase antibody from Southern Biotechnology (1 µg/mL). The final wash was followed by the addition of Nitro blue Tetrazolium Chloride/5-bromo-4-chloro-3-indolyl phosphate p-toluidine salt (NBT/BCIP chromogen) substrate solution for 7 min. The chromogen was discarded, and the plates were washed with water and dried in a dim place for 24 h. Plates were scanned and counted on a Cellular Technologies Limited Immunospot Analyzer.
Supplementary Material
Acknowledgments
We are very grateful to Darrell Irvine and Bruce Walker for fruitful discussions about this work. This research was funded primarily by the Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard. D.K.M. was also supported by an NSF Graduate Research Fellowship, and A.G. was supported by NSF Grant PHY 2026995. D.H.B. was also supported by NIH Grants AI129797, AI124377, AI128751, and AI126603.
Footnotes
Competing interest statement: The authors have filed an application for a patent on the immunogen.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022496118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or SI Appendix.
References
- 1.Barre-Sinoussi F., et al. , Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Popovic M., Sarngadharan M. G., Read E., Gallo R. C., Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science 224, 497–500 (1984). [DOI] [PubMed] [Google Scholar]
- 3.Sok D., Burton D. R., Recent progress in broadly neutralizing antibodies to HIV. Nat. Immunol. 19, 1179–1188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwong P. D., Mascola J. R., Nabel G. J., Broadly neutralizing antibodies and the search for an HIV-1 vaccine: The end of the beginning. Nat. Rev. Immunol. 13, 693–701 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Escolano A., Dosenovic P., Nussenzweig M. C., Progress toward active or passive HIV-1 vaccination. J. Exp. Med. 214, 3–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker B., McMichael A., The T-cell response to HIV. Cold Spring Harb. Perspect. Med. 2, 1–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng K., et al. , Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins D. R., Gaiha G. D., Walker B. D., CD8+ T cells in HIV control, cure and prevention. Nat. Rev. Immunol. 20, 471–482 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen S. G., et al. , A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci. Transl. Med. 11, eaaw2607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Früh K., Picker L., CD8+ T cell programming by cytomegalovirus vectors: Applications in prophylactic and therapeutic vaccination. Curr. Opin. Immunol. 47, 52–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen S. G., et al. , Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351, 714–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barton J. P., et al. , Relative rate and location of intra-host HIV evolution to evade cellular immunity are predictable. Nat. Commun. 7, 11660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMichael A. J., Borrow P., Tomaras G. D., Goonetilleke N., Haynes B. F., The immune response during acute HIV-1 infection: Clues for vaccine development. Nat. Rev. Immunol. 10, 11–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips R. E., et al. , Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354, 453–459 (1991). [DOI] [PubMed] [Google Scholar]
- 15.Feeney M. E., et al. , Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78, 8927–8930 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ondondo B., et al. , Novel conserved-region T-cell mosaic vaccine with high global HIV-1 coverage is recognized by protective responses in untreated infection. Mol. Ther. 24, 832–842 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borthwick N., et al. , Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol. Ther. 22, 464–475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Létourneau S., et al. , Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2, e984 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyo N., et al. , Efficient induction of T cells against conserved HIV-1 regions by mosaic vaccines delivered as self-amplifying mRNA. Mol. Ther. Methods Clin. Dev. 12, 32–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakoshi H., et al. , CD8+ T cells specific for conserved, cross-reactive Gag epitopes with strong ability to suppress HIV-1 replication. Retrovirology 15, 46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borthwick N., et al. , Novel, in-natural-infection subdominant HIV-1 CD8+ T-cell epitopes revealed in human recipients of conserved-region T-cell vaccines. PLoS One 12, e0176418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinez-Picado J., et al. , Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80, 3617–3623 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brockman M. A., et al. , Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81, 12608–12618 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferguson A. L., et al. , Translating HIV sequences into quantitative fitness landscapes predicts viral vulnerabilities for rational immunogen design. Immunity 38, 606–617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louie R. H. Y., Kaczorowski K. J., Barton J. P., Chakraborty A. K., McKay M. R., Fitness landscape of the human immunodeficiency virus envelope protein that is targeted by antibodies. Proc. Natl. Acad. Sci. U.S.A. 115, E564–E573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mann J. K., et al. , The fitness landscape of HIV-1 gag: Advanced modeling approaches and validation of model predictions by in vitro testing. PLOS Comput. Biol. 10, e1003776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barton J. P., et al. , Modelling and in vitro testing of the HIV-1 Nef fitness landscape. Virus Evol. 5, vez029 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahirel V., et al. , Coordinate linkage of HIV evolution reveals regions of immunological vulnerability. Proc. Natl. Acad. Sci. U.S.A. 108, 11530–11535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butler T. C., Barton J. P., Kardar M., Chakraborty A. K., Identification of drug resistance mutations in HIV from constraints on natural evolution. Phys. Rev. E 93, 022412 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Shekhar K., et al. , Spin models inferred from patient-derived viral sequence data faithfully describe HIV fitness landscapes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 88, 062705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakraborty A. K., Barton J. P., Rational design of vaccine targets and strategies for HIV: A crossroad of statistical physics, biology, and medicine. Rep. Prog. Phys. 80, 032601 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Gaiha G. D., et al. , Structural topology defines protective CD8+ T cell epitopes in the HIV proteome. Science 364, 480–484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed S. F., Quadeer A. A., Morales-Jimenez D., McKay M. R., Sub-dominant principal components inform new vaccine targets for HIV Gag. Bioinformatics 35, 3884–3889 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Barouch D. H., Picker L. J., Novel vaccine vectors for HIV-1. Nat. Rev. Microbiol. 12, 765–771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barouch D. H., et al. , Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482, 89–93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borducchi E. N., et al. , Ad26/MVA therapeutic vaccination with TLR7 stimulation in SIV-infected rhesus monkeys. Nature 540, 284–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barouch D. H., et al. , Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19). Lancet 392, 232–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barouch D. H., et al. , Characterization of humoral and cellular immune responses elicited by a recombinant adenovirus serotype 26 HIV-1 Env vaccine in healthy adults (IPCAVD 001). J. Infect. Dis. 207, 248–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baden L. R., et al. , First-in-human evaluation of the safety and immunogenicity of a recombinant adenovirus serotype 26 HIV-1 Env vaccine (IPCAVD 001). J. Infect. Dis. 207, 240–247 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baden L. R.et al.; B003-IPCAVD004-HVTN091 Study Group , Assessment of the safety and immunogenicity of 2 novel vaccine platforms for HIV-1 prevention: A randomized trial. Ann. Intern. Med. 164, 313–322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baden L. R., et al. , Induction of HIV-1-specific mucosal immune responses following intramuscular recombinant adenovirus serotype 26 HIV-1 vaccination of humans. J. Infect. Dis. 211, 518–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephenson K. E., et al. , First-in-human randomized controlled trial of an oral, replicating adenovirus 26 vector vaccine for HIV-1. PLoS One 13, e0205139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colby D. J., et al. , Safety and immunogenicity of Ad26 and MVA vaccines in acutely treated HIV and effect on viral rebound after antiretroviral therapy interruption. Nat. Med. 26, 498–501 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Barton J. P., De Leonardis E., Coucke A., Cocco S., ACE: Adaptive cluster expansion for maximum entropy graphical model inference. Bioinformatics 32, 3089–3097 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Barouch D. H., et al. , Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 16, 319–323 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barouch D. H., et al. , Protective efficacy of a global HIV-1 mosaic vaccine against heterologous SHIV challenges in rhesus monkeys. Cell 155, 531–539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Los Alamos National Laboratory , Data from "Best-defined CTL/CD8+ epitope summary." HIV Molecular Immunology Database. https://www.hiv.lanl.gov/content/immunology/tables/optimal_ctl_summary.html. Accessed 2 January 2017.
- 48.Abbink P., et al. , Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J. Virol. 81, 4654–4663 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts D. M., et al. , Hexon-chimaeric adenovirus serotype 5 vectors circumvent pre-existing anti-vector immunity. Nature 441, 239–243 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Jia M., et al. , Preferential CTL targeting of Gag is associated with relative viral control in long-term surviving HIV-1 infected former plasma donors from China. Cell Res. 22, 903–914 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deeks S. G., Walker B. D., Human immunodeficiency virus controllers: Mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity 27, 406–416 (2007). [DOI] [PubMed] [Google Scholar]
- 52.The International HIV Controllers Study , The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330, 1551–1557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang C., et al. , Distinct viral reservoirs in individuals with spontaneous control of HIV-1. Nature 585, 261–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Julg B., et al. , Recommendations for analytical antiretroviral treatment interruptions in HIV research trials-report of a consensus meeting. Lancet HIV 6, e259–e268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao A., et al. , Predicting the immunogenicity of T cell epitopes: From HIV to SARS-CoV-2. bioRxiv [Preprint] (2020). 10.1101/2020.05.14.095885) (Accessed 25 October 2020). [DOI]
- 56.Morcos F., et al. , Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc. Natl. Acad. Sci. U.S.A. 108, E1293–E1301 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frenkel D., Smit B., Understanding Molecular Simulation: From Algorithms to Applications (Elsevier, 2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.