Abstract
This study was conducted to explore whether trichostatin A-assisted epigenomic modulation (TSA-EM) can affect the expression of not only recombinant human α1,2-fucosyltransferase (rhα1,2-FT) and α-galactosidase A (rhα-Gal A) immune system enzymes but also Galα1→3Gal epitopes in ex vivo proliferating adult cutaneous fibroblast cells (ACFCs) derived from hFUT2×hGLA bi-transgenic pigs that had been produced for the needs of future xenotransplantation efforts. The ACFC lines were treated with 50 nM TSA for 24 h and then the expression profiles of rhα1,2-FT and rhα-Gal A enzymes were analyzed by Western blot and immunofluorescence. The expression profiles of the Galα1→3Gal epitope were determined by lectin blotting and lectin fluorescence. The ACFCs derived from non-transgenic (nTG) pigs were served as the negative (TSA−) and positive (TSA+) control groups. For both hFUT2×hGLA and nTG samples, the expression levels of α1,2-FT and α-Gal A proteins in TSA+ cells were more than twofold higher in comparison to TSA− cells. Moreover, a much lower expression of the Galα1→3Gal epitopes was shown in TSA− hFUT2×hGLA cells as compared to the TSA− nTG group. Interestingly, the levels of Galα1→3Gal expression in TSA-treated hFUT2×hGLA and nTG ACFCs were significantly higher than those noticed for their TSA-untreated counterparts. Summing up, ex vivo protection of effectively selected bi-transgenic ACFC lines, in which TSA-dependent epigenetic transformation triggered the enhancements in reprogrammability and subsequent expression of hFUT2 and hGLA transgenes and their corresponding transcripts, allows for cryopreservation of nuclear donor cells, nuclear-transferred female gametes, and resultant porcine cloned embryos. The latter can be used as a cryogenically conserved genetic resource of biological materials suitable for generation of bi-transgenic cloned offspring in pigs that is targeted at biomedical research in the field of cell/tissue xenotransplantation.
Keywords: pig; TSA-mediated epigenomic modulation; in vitro culture; bi-transgenic; adult cutaneous fibroblast cells; α1,2-fucosyltransferase; α-galactosidase A; Galα1→3Gal epitope; xenotransplantation
1. Introduction
Xenotransplantation seems to be a response to the contemporary shortage of organs for transplantation [1]. Choosing the right donor is difficult, but nowadays pigs are considered as the most suitable donors of tissues and organs for human patients with chronic or end-stage organ failure. Pigs share a large amount of similarities in organ anatomy and physiology; moreover, they share approximately 96% of their genetic identity with humans [2,3,4,5]. Unfortunately, the large phylogenetic distance between pig and human is the cause of the immunological barrier including immune response leading to hyperacute rejection (HAR), which currently makes pig-to-human of xenotransplantation unsuccessful [6]. The HAR is caused by the Galα1→3Gal epitope, which is present on the surface of porcine cells. The epitope Galα1→3Gal is formed by a reaction catalyzed by α1,3-galactosyltransferase (α1,3-GT), an enzyme encoded by the GGTA1 gene. The α1,3-GT enzyme catalyzes the galactose transfer reaction from UDP-Gal and its α1→3 glycosidic bond with glycoproteins or glycosphingolipids containing terminal Galβ1→4GlcNAc-R residues [7]. One of the strategies to remove the Galα1→3Gal epitope from the porcine cells involves combined transgenic expression of recombinant human α1,2-fucosyltransferase (rhα1,2-FT) and α-galactosidase A (rhα-Gal A) enzymes encoded by hFUT2 and hGLA genes, respectively [8]. Both endogenous porcine α1,3-galactosyltransferase and the introduced recombinant human α1,2-fucosyltransferase utilize N-acetyllactosamine. However, α1,2-FT is located in the intermediate compartment, and α1,3-GT in the trans part of the Golgi apparatus; therefore, α1,2-FT acts earlier than porcine endogenous α1,3-GT. As oligosaccharide moves through the Golgi apparatus it is first fucosylated by rhα1,2-FT, whereby it cannot accept the terminal galactose residue in subsequent reaction catalyzed by α1,3-GT [9]. In turn, the recombinant human α-galactosidase A cuts off the terminal D-galactose residues, and thus the Galα1→3Gal epitope becomes less xenoreactive [10]. Zeyland et al. [11] have shown the successful production of double-transgenic pigs with combined expression of α1,2-fucosyltransferase and α-galactosidase A. The above-mentioned investigators demonstrated that the co-expression of these two transgenes leads to a considerable reduction of the Galα1→3Gal epitope level on the surface of skin-derived fibroblast cells.
Thus far, various strategies of epigenomic modulation (epigenetic transformation) that are mediated by non-specific inhibitors of histone deacetylases (HDACi) and/or DNA methyltransferases (DNMTi) have been applied to improve reprogrammability of donor cell-inherited genome in somatic cell nuclear transfer (SCNT)-derived oocytes and resultant cloned embryos in different mammalian species [12,13,14,15,16]. Analogous methods of HDACi- and/or DNMTi-dependent epigenetic transformation have been adapted to enhance capabilities of parental genomes to be epigenetically reprogrammed in the in vitro fertilization (IVF)-derived embryos [17,18,19,20]. Such approaches have also been used either to expedite the epigenomic reprogramming and molecular dedifferentiation of adult somatic cells or mesenchymal stem cells (MSCs) into induced pluripotent stem cells (iPSCs) [21,22,23,24,25] or to facilitate the molecular rejuvenation of adult MSCs [21,26,27]. Moreover, the efforts that are aimed at the epigenomic modulation dependent on HDACi and/or DNMTi approved by the U.S. Food and Drug Administration (FDA) or by European Medicines Agency (EMA) have been undertaken to accomplish clinical treatments (i.e., epigenetic therapies) in oncologic patients and medical patients afflicted by neurodegenerative diseases or psychiatric disorders [28,29,30,31].
Taking into consideration the above-mentioned broad spectrum of practical applying exogenous epigenomic modulation, HDACi-assisted epigenetic transformation of in vitro-cultured somatic cells stemming from pigs genetically modified for the purposes of pig-to-human tissue xenotransplantation has not yet been reported. The scientific justification of such HDACi-dependently epigenomically modulating the genetically modified somatic cell lines is reflected in the finding that their treatment with non-selective HDACi designated as trichostatin A (TSA) can lead to improving reprogrammability of transgenes integrated with nuclear host genome. The molecular mechanism underlying improvements in the capabilities of the transgenes to be epigenetically reprogrammed in somatic cell nuclei encompasses direct enhancements in lysine moiety acetylation (i.e., hyperacetylation) processes of histones derived from chromatin nucleosomal cores associated with exon DNA sequences within promoters and/or enhancers of the transgenes that have been efficiently incorporated into host genome. TSA-dependent diminishments in histone deacetylation processes can indirectly trigger the enhancements in DNA cytosine residue demethylation (i.e., hypomethylation) reactions within promoter- and/or enhancer-related regions of the successfully integrated transgenes. These latter seem to evoke progressive onset and increase of transcriptional and translational activities of the incorporated transgenes and their mRNA transcript counterparts, respectively.
For all the reasons above, it is worth highlighting the fact that the effects of TSA-assisted epigenetic transformation of porcine double-transgenic fibroblast cell lines not only on the enzymatic expression levels of recombinant human α1,2-fucosyltransferase (α1,2-FT) and α-galactosidase A (α-Gal A) proteins but also on the semi-quantitative profiles of Galα1→3Gal antigenic determinants, i.e., epitopes, have been explored for the first time.
2. Results
2.1. Western Blot Analysis of the Relative Expression of Recombinant Human α1,2-Fucosyltransferase (rhα1,2-FT) and α-Galactosidase A (rhα-Gal A) Proteins
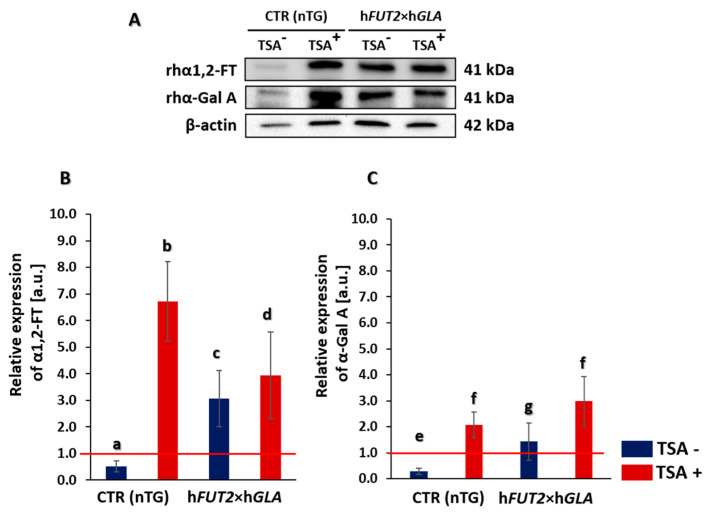
The adult cutaneous fibroblast cells (ACFCs) treated (TSA+) and non-treated with trichostatin A (TSA−) were derived from hFUT2×hGLA double-transgenic pigs and from non-transgenic pigs, which served as a control (CTR nTG). Western blot analysis of total protein samples showed the presence of recombinant human α1,2-fucosyltransferase (rhα1,2-FT) and α-galactosidase A (rhα-Gal A) proteins in all the transgenic samples (Figure 1A). A weak positive signal for both α1,2-FT and α-Gal A appeared in the TSA− control group, but it was found to be non-significant. A clear positive signal was observed for both rhα1,2-FT and rhα-Gal A in total protein samples derived from TSA+ cells. Signal intensities of analyzed proteins were normalized to β-actin, which was used as a loading control. The semi-quantitative analysis of Western blot confirmed our observation. Indeed, the relative expression of both analyzed proteins was significantly higher (at least p < 0.05) in TSA+ cells as compared to the corresponding samples derived from TSA− cells (Figure 1B). Interestingly, a clear positive signal was also identified for both α1,2-FT and α-Gal A proteins in TSA+ cells derived from non-transgenic pigs (CTR nTG). This result was confirmed both qualitatively and semi-quantitatively (at least p < 0.05) (Figure 1).
Figure 1.
Western blot analysis of the relative expression of human α1,2-fucosyltransferase (α1,2-FT) and α-galactosidase A (α-Gal A) proteins in porcine bi-transgenic and non-transgenic adult cutaneous fibroblast cells (ACFCs) undergoing or not undergoing treatment with trichostatin A (TSA+ and TSA−, respectively). Representative blots of the expression of α1,2-FT and α-Gal A proteins in the ACFC samples derived from epigenetically modulated (TSA+) and non-modulated (TSA−) cells. The samples of non-transgenic animals served as a control group (CTR nTG—panel (A)). β-Actin served as a loading control for all analyzed samples. The results of relative expression (in arbitrary units) of α1,2-FT and α-Gal A are shown in panels (B,C), respectively. Relative optical density (ROD) from three separate analyses of at least three animals for each variant is expressed as mean. Bar graphs show mean ± standard error of the mean (SEM). The red line is taken as the cut-off value 1.0. Statistics: One-way ANOVA and Tukey’s honestly significant difference (HSD) post hoc test. The bars marked with different letters differ significantly; values denoted as a-b, a-d, b-c, b-d, e-f: p < 0.01; a-c, c-d, e-g, f-g: p < 0.05.
2.2. Immunofluorescence Localization of Recombinant Human α1,2-Fucosyltransferase (rhα1,2-FT) and α-Galactosidase A (rhα-Gal A) in Trichostatin A-Treated and Untreated ACFCs
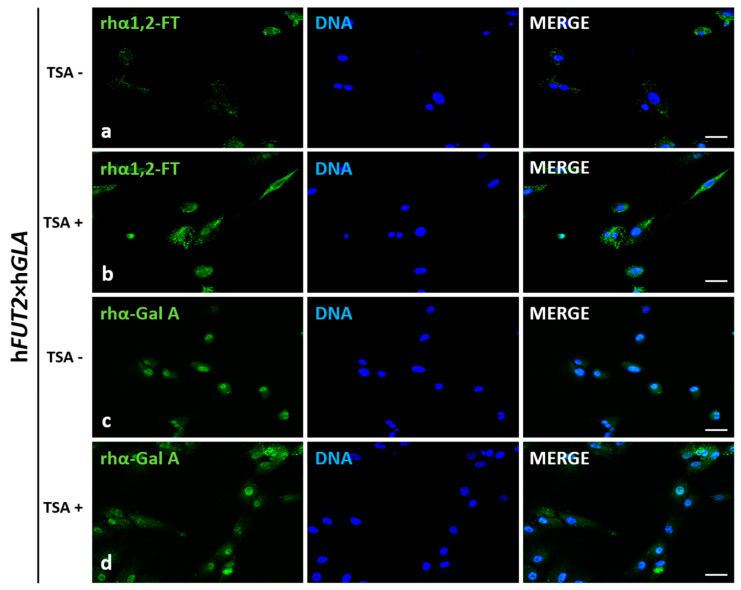
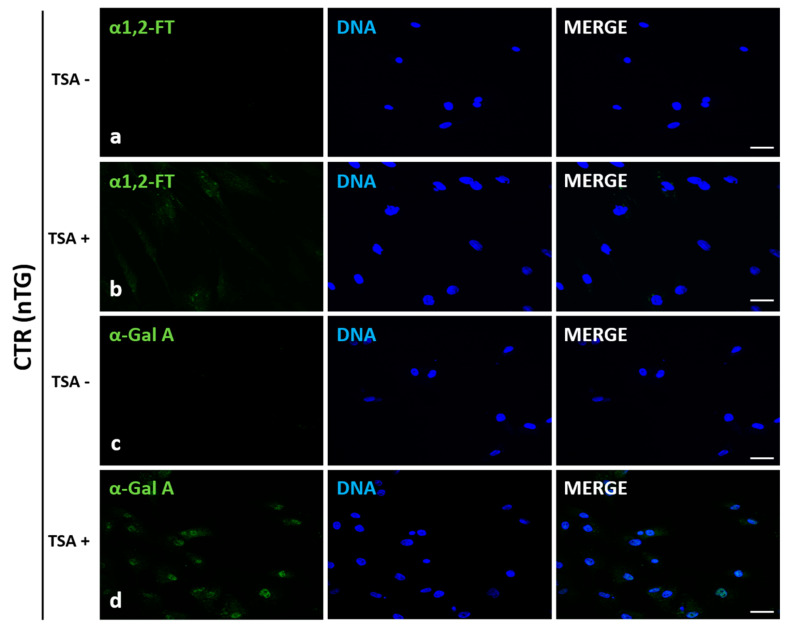
Localization of recombinant human α1,2-fucosyltransferase (rhα1,2-FT) and α-galactosidase A (rhα-Gal A) was examined by immunofluorescence staining of trichostatin A-exposed (TSA+) and non-exposed (TSA−) ACFCs derived from hFUT2×hGLA double-transgenic (Figure 2) and non-transgenic (Figure 3) pigs utilized as a control (CTR nTG). The positive immunofluorescence signal descended from rhα1,2-FT was mainly distributed in small perinuclear clusters in bi-transgenic cells. In turn, rhα-Gal A immunostaining was found to be more homogenous and highly/intensively detectable in whole cytosol of transgenic (hFUT2×hGLA) cells (Figure 2). Clearly stronger immunofluorescence signal was observed for both rhα1,2-FT and rhα-Gal A proteins in cells treated with trichostatin A (TSA+) (Figure 2b,d). Moreover, a weak immunofluorescence signal for both α1,2-FT and α-Gal A was also identified in non-transgenic (CTR nTG) TSA+ cells (Figure 3b,d). However, in the TSA− CTR nTG cells, the α1,2-FT was barely detectable by immunofluorescence, while any positive signal descended from α-galactosidase A was not identified (Figure 2).
Figure 2.
Immunofluorescence analysis of in vitro-cultured porcine ACFCs treated (TSA+) (b,d) and not treated with trichostatin A (TSA−) (a,c). Representative microphotographs of immunofluorescence localization of recombinant human α1,2-fucosyltransferase (rhα1,2-FT; a,b) and α-galactosidase A (rhα-Gal A; c,d) in ACFCs derived from double-transgenic pigs (hFUT2×hGLA). Immunofluorescent staining with Alexa Fluor 488-labelled secondary antibodies (green fluorescence) and 4′,6-diamidino-2-phenylindole (DAPI)-mediated counterstaining of cell nuclei (blue fluorescence). Scale bars represent 100 μm. Immunoreaction was performed on in vitro cultured porcine ACFCs from at least three pigs of each experimental group. The immunofluorescence signal stemming from rhα1,2-FT was distributed in the perinuclear region of all the analyzed cells from each variant. The recombinant human α-galactosidase A was located homogeneously in whole cytoplasm of all the analyzed cells. The TSA+ cells exhibited a more intense signal for both rhα1,2-FT and rhα-Gal A proteins (b,d) than their TSA− counterparts (a,c).
Figure 3.
Immunofluorescence analysis of in vitro cultured porcine ACFCs treated (TSA+) (b,d) and not treated with trichostatin A (TSA−) (a,c). Representative microphotographs of immunofluorescence localization of human α1,2-fucosyltransferase (α1,2-FT; a,b) and α-galactosidase A (α-Gal A; c,d) in ACFCs derived from non-transgenic pigs (CTR nTG). Immunofluorescent staining with Alexa Fluor 488-labelled secondary antibodies (green fluorescence) and 4′,6-diamidino-2-phenylindole (DAPI)-mediated counterstaining of cell nuclei (blue fluorescence). Scale bars represent 100 μm. Immunoreaction was performed on in vitro-cultured porcine ACFCs from at least three pigs of each experimental group. The TSA+ cells exhibited a more intense signal for both α1,2-FT and α-Gal A proteins in control group (CTR nTG) (b,d). No positive signal stemming from α1,2-FT and α-Gal A in the TSA− control (CTR nTG) was noticed (a,c).
2.3. Lectin Blotting Analysis of Galα1→3Gal Epitope Expression at the Protein Level in the In Vitro-Cultured Porcine Bi-Transgenic and Non-Transgenic ACFCs Treated and Not Treated with Trichostatin A
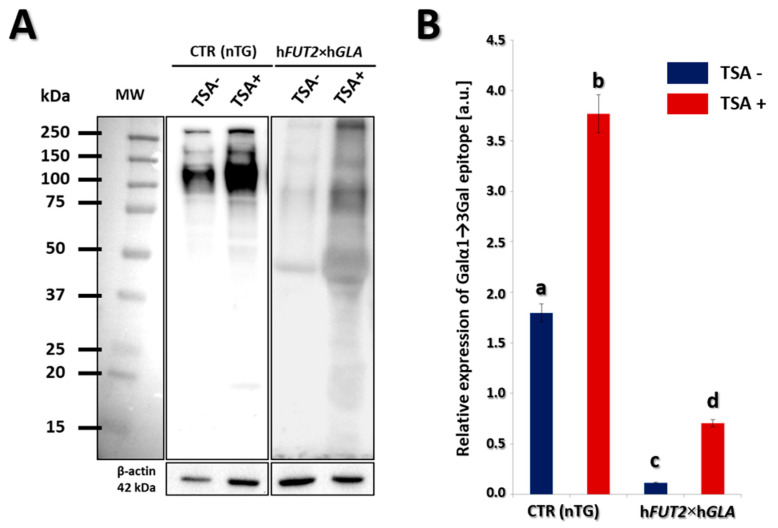
Lectin blot analysis was used to determine the expression profile of Galα1→3Gal epitopes at the total protein level using horseradish peroxidase (HRP)-conjugated lectin GS-IB4. The results of our study confirmed that significantly lower expression of Galα1→3Gal epitopes was identified in bi-transgenic fibroblast cells as compared to the control group. In turn, both in control (CTR nTG) and double-transgenic (hFUT2×hGLA) groups, expression of Galα1→3Gal epitopes was found to be significantly increased in the cell samples derived from trichostatin A (TSA)-treated ACFCs (TSA+; Figure 4A). β-Actin served as a loading control. The quantitative analysis of hFUT2×hGLA TSA confirmed these observations. The significantly lowest expression of Galα1→3Gal epitopes was detected in TSA-untreated (TSA−) ACFCs derived from hFUT2×hGLA double-transgenic pigs, transpiring to statistically differ from both CTR nTG TSA− (p < 0.01) and CTR nTG TSA+ groups (p < 0.01). The expression of Galα1→3Gal epitopes in ACFCs originating from hFUT2×hGLA TSA+ group was significantly higher (p < 0.05) than that noticed for cells stemming from hFUT2×hGLA TSA− group, but it was found to still be lower than that indicated in both CTR nTG groups (p < 0.01) (Figure 4B).
Figure 4.
Lectin blotting analysis of Galα1→3Gal epitope expression at the protein level in the in vitro cultured porcine transgenic and non-transgenic ACFCs treated (TSA+) and not treated with trichostatin A (TSA−). (A) Representative lectin blots of the expression of Galα1→3Gal epitope in TSA− and TSA+ in vitro-cultured ACFCs derived from non-transgenic (CTR nTG) control and double-transgenic (hFUT2×hGLA) pigs. MW indicates the molecular weight of protein standards (Precision Plus Protein Dual Color Standards, Bio-Rad). Each band represents a glycosylated protein containing the Galα1→3Gal epitope. β-Actin served as a loading control for all analyzed samples. (B) The semi-quantitative analysis of Galα1→3Gal epitope relative expression (in arbitrary units). Relative optical density (ROD) from three separate analyses of at least three animals for each variant is expressed as mean. Graph bar shows mean ± SEM. Statistics: One-way ANOVA and Tukey’s HSD post hoc test. The bars marked with different letters differ significantly. Values denoted as a-b, a-c, a-d, b-c, b-d: p < 0.01; c-d: p < 0.05. The relative expression of Galα1→3Gal epitope was significantly lower in the ACFCs stemming from hFUT2×hGLA bi-transgenic pigs as compared to the control (CTR nTG) non-transgenic animals. It is also noteworthy to highlight the fact that the TSA− cell samples derived from hFUT2×hGLA double-transgenic pigs were characterized by the remarkably lowest expression of Galα1→3Gal epitopes. However, expression of Galα1→3Gal epitopes was significantly higher in TSA+ cell samples compared to that identified for their TSA− counterparts derived from both transgenic and non-transgenic pigs.
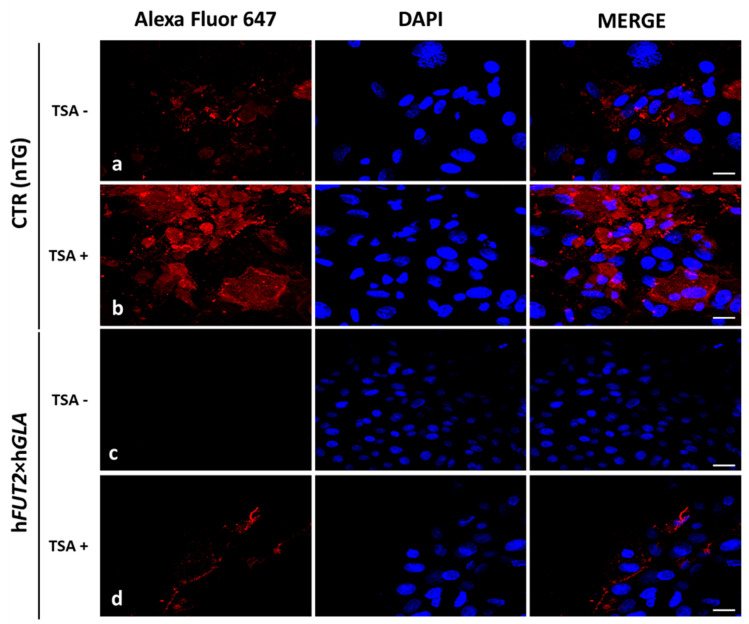
2.4. Identification of the Expression Profiles of Galα1→3 Gal Epitope by Fluorescently Labelled Lectin GS-IB4
For identifying the expression profiles of Galα1→3 Gal epitope, we used lectin GS-IB4 labelled with Alexa Fluor 647 (Figure 5). The lectin GS-IB4 strongly stained Galα1→3 Gal antigenic determinants in trichostatin A-treated (TSA+) ACFCs originating from the CTR nTG group (Figure 5b). In contrast, TSA+ and TSA- ACFCs stemming from hFUT2×hGLA double-transgenic pigs displayed a much lower Galα1→3Gal fluorescence intensity as compared to both TSA+ and TSA− control groups. However, in hFUT2×hGLA group, we observed stronger Galα1→3Gal fluorescence intensity in TSA+ ACFCs compared to their TSA− counterparts (Figure 5c,d).
Figure 5.
Lectin fluorescence analysis of Galα1→3Gal epitope expression in the in vitro-cultured porcine transgenic and non-transgenic ACFCs treated (TSA+) (b,d) and not treated with trichostatin A (TSA−) (a,c). Representative microphotographs of lectin GS-IB4 labelled sections derived from non-transgenic control (CTR nTG; a,b) and double-transgenic (hFUT2×hGLA; c,d) pigs. The lectin fluorescence analysis was performed with the use of Alexa Fluor 647-conjugated lectin GS-IB4 (red fluorescence) and 4′,6-diamidino-2-phenylindole (DAPI)-mediated counterstaining of cell nuclei (blue fluorescence). Scale bars represent 50 μm.
3. Discussion
The hyperacute rejection is the main obstacle in pig-to-human xenotransplantation. To overcome the immune response, genetically modified pigs must be generated. Research around the world shows different ways of genetic modification and their combinations. These include generating homozygous pigs lacking the gene for α1,3-galactosyltransferase for the purpose of depletion of anti-pig antibodies, removal of the Galα1→3Gal epitope using enzymes, and engineering pigs transgenic for certain graft-protective proteins [32,33,34,35].
In the current study, we investigated the effects of epigenetic modulation by trichostatin A on overexpression of recombinant human α1,2-fucosyltransferase and α-galactosidase A, and on the amount of Galα1→3Gal antigen in the porcine ACFCs derived from bi-transgenic and non-transgenic lines. Since genetically modified pigs were produced to avoid hyperacute rejection [12,36,37], we discuss this aspect in relation to an adult cutaneous fibroblast cell (ACFC)-based in vitro model. Our Western blot and immunofluorescence analyses revealed the significant enhancements in the expression of both recombinant human α1,2-fucosyltransferase and α-galactosidase A proteins in the epigenomically modulated ACFCs (TSA+) derived from hFUT2×hGLA bi-transgenic pigs as compared to their TSA− cell counterparts. Moreover, immunofluorescence staining with antibodies against human α1,2-fucosyltransferase and α-galactosidase A provided evidence that the epigenetic modulation with TSA led to increase in the expression α1,2-fucosyltransferase and α-galactosidase A proteins in ACFCs. The study by Jia et al. [38] confirmed that α-galactosidase alone reduced the expression of Galα1→3Gal antigenic determinants by 78%, while the co-expression of both α1,2-FT and α-Gal A diminished the quantitative profile of these epitopes to a negligible level on the surface of SV40 immortalized aortic porcine endothelial cells. In turn, the previous investigation by Wiater et al. [39] has proven that co-expression of recombinant human α-galactosidase A and α1,2-fucosyltransferase decreased the expression of Galα1→3Gal antigenic determinants by 62% (on the basis of histochemistry) and 47% (on the basis of blotting), respectively. However, in the previously mentioned studies, relatively low expression of rhα1,2-FT and rhα-Gal A was demonstrated, which could be the reason for the lack of complete silencing the expression of Galα1→3Gal epitopes. Therefore, the use of epigenomic modulation of the cells to increase expression of these proteins seems to be inevitable. It is also worth noting that the impacts of TSA-dependent epigenetic transformation of porcine bi-transgenic ACFCs not only on the biocatalytic expression levels of recombinant human α1,2-fucosyltransferase (rhα1,2-FT) and α-galactosidase A (rhα-Gal A) proteins but also on the semi-quantitative profiles of Galα1→3Gal epitopes have been investigated for the first time. Indeed, in the current study, we showed by Western blot analyses that TSA-mediated epigenomic modulation of ACFCs led to increase in the relative expression of both rhα1,2-FT and rhα-Gal A by 28.32% and 108.96%, respectively. In our present study, we demonstrated by semi-quantitative (lectinblotting) and qualitative (lectin fluorescence) methods that the expression of Galα1→3Gal antigenic determinants in porcine ACFCs was significantly declined in cells derived from hFUT2×hGLA double-transgenic pigs. The efficiency of Galα1→3Gal epitope reduction in cells not treated with TSA remained at the level of 93.63%, while in TSA+ cells, at the level of 81%. Moreover, Western blot analyses showed that the relative expression of Galα1→3Gal epitopes increased by 109.83% in non-transgenic (CTR nTG) TSA+ cells compared to the TSA− cells. Thus, these results provided proof that trichostatin A-assisted epigenetic transformation affects not only expression profiles of rhα1,2-FT and rhα-Gal A proteins but also relative abundance of Galα1→3Gal antigenic determinants in ACFCs.
To sum up, we have shown that the overexpression of both recombinant human α1,2-fucosyltransferase and α-galactosidase A in a porcine hFUT2×hGLA bi-transgenic ACFCs model significantly reduced but did not eliminate the Galα1→3Gal epitope from the surface of cells. Moreover, we have shown that the treatment of ACFCs with non-selective HDACi designated as trichostatin A (TSA) can lead to improvement in the reprogrammability of transgenes integrated with nuclear host genome, but possibly also the host’s own genes. Therefore, taking into account cell/tissue xenotransplantation, it is clear that the most effective way to eliminate Galα1→3Gal epitopes is by producing homozygous α-1,3-galactosyltransferase knock-out (GTKO) pigs lacking the gene for α1,3-galactosyltransferase.
4. Materials and Methods
4.1. In Vitro Culture and Trichostatin A-Mediated Epigenomic Modulation of Fibroblast Cells
The adult cutaneous fibroblast cell (ACFC) lines were established according to the protocols described in our previous studies [40,41,42]. In our current investigation, ACFCs derived from hFUT2×hGLA bi-transgenic pigs (n = 3) were utilized [11]. The ACFCs stemming from non-transgenic pigs were served as a control group (CTR nTG). All animal procedures that were used in the study by Zeyland et al. [11] were conducted in accordance with the European Directive 2010/63/EU and approved by the Second Local Ethics Committee in Kraków, Poland (Permission 1181/2015 from 21st May 2015). All ACFC lines were cultured in Dulbecco’s Modified Eagle’s Medium/Nutrient Ham’s Mixture F-12 (DMEM/F-12) (1:1) (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 15% fetal bovine serum (FBS; Sigma-Aldrich) and 1% penicillin/streptomycin cocktail (Sigma-Aldrich) in CO2 incubator under the stabilized conditions as follows: temperature of +38.5 °C, 5% CO2, and relative humidity (RH) of air atmosphere ranging from 90 to 95%. For Western and lectin blot analyses, cells were cultured in T-25 flasks up to 2–3 passage, but for immunofluorescence, the cells at the second passage were seeded onto sterile coverslips in 6-well plates. When the ex vivo-expanded ACFC lines reached approximately 85% of confluence, their epigenomic modulation was initiated by adding 50 nM of trichostatin A (TSA; Sigma-Aldrich) to the culture medium. Both bi-transgenic and non-transgenic fibroblast cells were epigenetically transformed by exposure to TSA for 24 h.
4.2. Total Protein Extraction and Western/Lectin Blot Analyses
Total protein was extracted from harvested ACFCs by using radioimmunoprecipitation assay lysis buffer (RIPA buffer, Thermo Fisher Scientific, Waltham, MA, USA) containing 1% of proteinase inhibitor cocktail (RIPA+PI; Bioshop Inc., Burlington, VT, Canada). After TSA treatment, the cells were washed with phosphate-buffered saline (PBS; Biomed, Lublin, Poland) and then 300 μL of RIPA+PI was added per flask, and cells were harvested with cell scrapers. The samples were subsequently sonicated and centrifuged at 13,200 rpm for 15 min at +4 °C and supernatant collected. Protein concentration was estimated with microassay DC Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA) using bovine serum albumin (BSA) as a standard. Protein samples were stored at −80 °C for further analyses.
For SDS-PAGE, protein samples were diluted in 2× Laemmli Sample Buffer (Bio-Rad Laboratories, Hercules, CA, USA) containing β-mercaptoethanol and denaturated at 99.5 °C per 5 min. Electrophoresis was performed with 5% stacking and 10% resolving polyacrylamide gels. Each lane was loaded with 20 μg of protein. Then, proteins were electro-transferred onto poly(vinylidene fluoride) (PVDF) membrane (Immobilon-P; Merck, Darmstadt, Germany) at constant amperage 250 mA for 120 min.
For immunoblotting, membranes after several washes in Tris-buffered saline (TBS) were blocked for 1 h in 5% non-fat milk in TBST (Tris-buffered saline with 0.1% v/v Tween20; Bioshop Inc., Burlington, VT, Canada). Then, membranes were washed several times in TBST and incubated overnight at +4 °C with the following primary antibodies: against human α1,2-fucosyltransferase (diluted 1:1000 in TBST; rabbit polyclonal antibodies, ab198712, Abcam, Cambridge, UK) and human α-galactosidase A (diluted 1:1000 in TBST; rabbit polyclonal antibodies, PA5-27349, ThermoFisher Scientific, Waltham, MA, USA). β-Actin was served as a loading control protein (diluted 1:2000 in TBST; mouse monoclonal antibodies, ab8224, Abcam). Then, membranes were washed several times in TBST and incubated with goat anti-rabbit or goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (ThermoFisher Scientific, Waltham, MA, USA) at a dilution 1:6000 in TBST for 1 h at room temperature.
For lectinblotting, membranes were blocked for 30 min in 1% BSA (Bioshop Inc., Burlington, Canada) in TBST. Then, membranes were washed three times in Dulbecco’s phosphate-buffered saline (DPBS) containing Ca2+/Mg2+ ions (Gibco, ThermoFisher Scientific, Waltham, MA, USA) followed by TBS. In the next step, membranes were incubated overnight at +4 °C with lectin GS-IB4 labelled with HRP (L5391, Sigma-Aldrich) diluted 1:2000 in DPBS. Finally, membranes were washed in TBS buffer.
For both Western blotting and lectin blotting, protein bands were detected by chemiluminescence using Clarity Western ECL Blotting Substrate (Bio-Rad Laboratories, Hercules, CA, USA) and visualized with the ChemiDoc XRS+ Imaging System (Bio-Rad Laboratories, Hercules, CA, USA). Protein bands were quantified using the Image Lab 2.0 Software (Bio-Rad Laboratories, Hercules, CA, USA). Semi-quantitative analysis was performed for 3 separately repeated experiments for each control and experimental group and normalized on reference protein (i.e., β-actin)-related signal. Each analysis was calculated as follows:
| (1) |
Subsequently, the results encompassing the relative expression of the analyzed proteins (i.e., rhα1,2-FT and rhα-Gal A enzymes) were shown as a mean ± standard error of the mean (SEM).
4.3. Immunofluorescence Staining
The ACFCs after TSA treatment were washed with sterile PBS and fixed with 4% paraformaldehyde for 10 min at room temperature (RT). After several washes in PBS, cells were blocked in 5% normal goat serum (NGS) in PBST (PBS containing 0.1% Triton X-100) for 30 min. Cells were then incubated overnight at +4 °C in humidified chamber with the following primary antibodies (the same as those for Western blot): against human α1,2-fucosyltransferase (diluted 1:150 in PBST) and human α-galactosidase (diluted 1:200 in PBST). In the next step, cells were washed several times in PBST and treated with goat anti-rabbit Alexa Fluor 488-conjugated secondary antibodies (diluted 1:300 in PBST; ThermoFisher Scientific, Waltham, MA, USA) for 1 h at room temperature. After final washes, cells were mounted in Fluoroshield with 4′,6-diamidino-2-phenylindole (DAPI) mounting medium (F6057, Sigma-Aldrich). Fluorescently labelled ACFCs were examined as described in Section “4.5. Confocal microscope analyses”.
4.4. Lectin Fluorescence
For intracellular localization of Galα1→3Gal epitopes and consequent semi-quantitative comparison of their expression profiles between TSA-treated and untreated dermal fibroblast cells originating from hFUT2×hGLA and non-transgenic (CTR nTG) pigs, we used lectin from Griffonia simplicifolia (GS-IB4) conjugated with Alexa Fluor 647 fluorescent dye (I32450, Molecular Probes, Invitrogen, ThermoFisher Scientific, Waltham, MA, USA). The ACFCs after TSA treatment were washed with sterile PBS and fixed with 4% paraformaldehyde for 10 min at RT. After several washes in PBS, cells were blocked in 1% BSA in PBST for 1 h. Cells were subsequently rinsed thrice in PBS and treated with lectin GS-IB4 diluted 1:200 in DPBS at +4 °C overnight in a dark humidified chamber. After final washes, cells were mounted in Fluoroshield with DAPI and coverslipped. Fluorescently labelled ACFCs were examined as described in Section “4.5. Confocal microscope analyses”.
4.5. Confocal Microscope Analyses
Fluorescently labelled cells were examined by the confocal microscope Olympus FluoView 1200 on inverted stand IX83 (Olympus, Tokyo, Japan). Forty-times magnification objective (NA = 0.95) was used, and diode laser (473 nm), diode laser (635 nm), and diode laser (405 nm) were applied to excite green (Alexa Fluor 488), far-red (Alexa Fluor 647), and blue (DAPI) fluorescence, respectively.
4.6. Statistical Analysis
For each TSA+ or TSA− cell derived from genetically modified and non-modified pigs and for all analyses, we performed three repeats. Quantitative data were expressed as the mean ± standard error of the mean (SEM) and examined using the Shapiro–Wilk W test for normality. Comparisons between the appropriate means were performed by one-way analysis of variance (ANOVA), followed by Tukey’s honestly significant difference (HSD) post hoc test for multiple ranges. All statistical analyses were accomplished using Statistica 13 Software (StatSoft Inc., Tulsa, OK, USA). Statistical significance was marked by letters at the appropriate charts. The bars marked with different letters differ significantly.
5. Conclusions
The ex situ conservation of efficiently selected (desirable) double-transgenic adult cutaneous fibroblast cell (ACFC) lines, in which TSA-mediated epigenomic modulation has resulted in the improved reprogrammability and thereby increased transcriptional and translational activities of hFUT2 and hGLA transgenes and their counterpart mRNA transcripts, enables us to cryogenically preserve reservoirs of nuclear donor somatic cells, nuclear-transferred oocytes reconstructed with somatic cells, and corresponding cloned pig embryos. These biorepositories of somatic cell lines, nuclear-transferred female gametes, and resultant embryos can provide a source of cryopreserved biological materials that are reliable and feasible for research targeted at producing bi-transgenic somatic cell nuclear transfer (SCNT)-derived piglets for the purposes of pig-to-human cell and/or tissue xenograft transplantation.
It is noteworthy to highlight that irrespective of the mechanism, by which TSA-mediated epigenomic modulation of ACFCs brings about increase in the relative abundance for Galα1→3Gal epitopes, the extent of Galα1→3Gal expression decreased remarkably in both TSA-treated and untreated hFUT2×hGLA bi-transgenic ACFCs displaying overabundance of rhα1,2-FT and rhα-Gal A enzymes as compared to their non-transgenic counterparts.
Finally, further detailed research is required to focus on not only transcriptomic and proteomic profiling but also recognizing the molecular mechanisms underlying TSA-mediated enhancement of the relative abundance for Galα1→3Gal epitopes in porcine hFUT2×hGLA double-transgenic ACFCs and subsequently their potential use for procedures of cloning pigs by SCNT.
Abbreviations
| ACFCs | Adult cutaneous fibroblast cells |
| DNMTi | Inhibitors of DNA methyltransferases |
| HAR | Hyperacute rejection |
| HDACi | Inhibitors of histone deacetylases |
| HRP | Horseradish peroxidase |
| iPSCs | Induced pluripotent stem cells |
| MSCs | Mesenchymal stem cells |
| rhα1,2-FT | Recombinant human α1,2-fucosyltransferase |
| rhα-Gal A | Recombinant human α-galactosidase A |
| SCNT | Somatic cell nuclear transfer |
| TSA | Trichostatin A |
| α1,3GT | α1,3-galactosyltransferase |
Author Contributions
Conceptualization, M.S. (Marcin Samiec) and J.W.; analysis of data and their interpretation, J.W., M.S. (Marcin Samiec), and D.L.; performance of experiments and preparation of results, J.W., M.S. (Marcin Samiec), and M.S. (Maria Skrzyszowska); writing the article, J.W. and M.S. (Marcin Samiec); supervision and funding acquisition, M.S. (Marcin Samiec); graphic and photographic documentation, J.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Ministry of Science and Higher Education in Poland as a statutory activity No. 04-19-05-00.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Niemann H.P. The production of multi-transgenic pigs: Update and perspectives for xenotransplantation. Transgenic Res. 2016;25:361–374. doi: 10.1007/s11248-016-9934-8. [DOI] [PubMed] [Google Scholar]
- 2.Cooper D.K., Gollackner B., Sachs D.H. Will the pig solve the transplantation backlog? Annu. Rev. Med. 2002;53:133–147. doi: 10.1146/annurev.med.53.082901.103900. [DOI] [PubMed] [Google Scholar]
- 3.Galili U., Shohet S.B., Kobrin E., Stults C.L., Macher B.A. Man, apes, and Old World monkeys differ from other mammals in the expression of alpha-galactosyl epitopes on nucleated cells. J. Biol. Chem. 1988;263:17755–17762. doi: 10.1016/S0021-9258(19)77900-9. [DOI] [PubMed] [Google Scholar]
- 4.Hryhorowicz M., Zeyland J., Słomski R., Lipiński D. Genetically modified pigs as organ donors for xenotransplantation. Mol. Biotechnol. 2017;59:435–444. doi: 10.1007/s12033-017-0024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whyte J.J., Prather R.S. Genetic modifications of pigs for medicine and agriculture. Mol. Reprod. Dev. 2011;78:879–891. doi: 10.1002/mrd.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper D.K.C., Ekser B., Tector A.J. Immunobiological barriers to xenotransplantation. Int. J. Surg. 2015;23:211–216. doi: 10.1016/j.ijsu.2015.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanken W.M., Van den Eijnden D.H. Biosynthesis of terminal Gal α1→3Galβ1→4GlcNAc-R oligosaccharide sequences on glycoconjugates. Purification and acceptor specificity of a UDP-Gal:N-acetyllactosaminide α1→3-galactosyltransferase from calf thymus. J. Biol. Chem. 1985;260:12927–12934. doi: 10.1016/S0021-9258(17)38814-2. [DOI] [PubMed] [Google Scholar]
- 8.Osman N., McKenzie I.F., Ostenried K., Ioannou Y.A., Desnick R.J., Sandrin M.S. Combined transgenic expression of alpha-galactosidase and alpha1,2-fucosyltransferase leads to optimal reduction in the major xenoepitope Galalpha(1,3)Gal. Proc. Natl. Acad. Sci. USA. 1997;94:14677–14682. doi: 10.1073/pnas.94.26.14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartel-Schenk S., Minnifield N., Reutter W., Hanski C., Bauer C., Morré D.J. Distribution of glycosyltransferases among Golgi apparatus subfractions from liver and hepatomas of the rat. Biochim. Biophys. Acta. 1991;1115:108–122. doi: 10.1016/0304-4165(91)90019-D. [DOI] [PubMed] [Google Scholar]
- 10.Luo Y., Wen J., Luo C., Cummings R.D., Cooper D.K.C. Pig xenogeneic antigen modification with green coffee bean α-galactosidase. Xenotransplantation. 1999;6:238–248. doi: 10.1034/j.1399-3089.1999.00035.x. [DOI] [PubMed] [Google Scholar]
- 11.Zeyland J., Woźniak A., Gawrońska B., Juzwa W., Jura J., Nowak A., Słomski R., Smorąg Z., Szalata M., Mazurek U., et al. Double transgenic pigs with combined expression of human α1,2-fucosyltransferase and α-galactosidase designed to avoid hyperacute xenograft rejection. Arch. Immunol. Ther. Exp. (Warsz.) 2014;62:411–422. doi: 10.1007/s00005-014-0280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huan Y., Wang H., Wu Z., Zhang J., Zhu J., Liu Z., He H. Epigenetic modification of cloned embryos improves Nanog reprogramming in pigs. Cell. Reprogram. 2015;17:191–198. doi: 10.1089/cell.2014.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samiec M., Skrzyszowska M. Intrinsic and extrinsic molecular determinants or modulators for epigenetic remodeling and reprogramming of somatic cell-derived genome in mammalian nuclear-transferred oocytes and resultant embryos. Pol. J. Vet. Sci. 2018;21:217–227. doi: 10.24425/119040. [DOI] [PubMed] [Google Scholar]
- 14.Saini M., Selokar N.L., Agrawal H., Singla S.K., Chauhan M.S., Manik R.S., Palta P. Treatment of donor cells and reconstructed embryos with a combination of trichostatin-A and 5-aza-2′-deoxycytidine improves the developmental competence and quality of buffalo embryos produced by handmade cloning and alters their epigenetic status and gene expression. Cell. Reprogram. 2017;19:208–215. doi: 10.1089/cell.2016.0061. [DOI] [PubMed] [Google Scholar]
- 15.Opiela J., Samiec M., Romanek J. In vitro development and cytological quality of inter-species (porcine→bovine) cloned embryos are affected by trichostatin A-dependent epigenomic modulation of adult mesenchymal stem cells. Theriogenology. 2017;97:27–33. doi: 10.1016/j.theriogenology.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 16.Silva C.G.D., Martins C.F., Bessler H.C., da Fonseca Neto Á.M., Cardoso T.C., Franco M.M., Mendonça A.D.S., Leme L.O., Borges J.R.J., Malaquias J.V., et al. Use of trichostatin A alters the expression of HDAC3 and KAT2 and improves in vitro development of bovine embryos cloned using less methylated mesenchymal stem cells. Reprod. Domest. Anim. 2019;54:289–299. doi: 10.1111/rda.13360. [DOI] [PubMed] [Google Scholar]
- 17.Oliveira C.S., Saraiva N.Z., Cruz M.H., Mazeti B., Oliveira L.Z., Lopes F.L., Garcia J.M. HDAC inhibition decreases XIST expression on female IVP bovine blastocysts. Reproduction. 2013;145:9–17. doi: 10.1530/REP-11-0343. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda S., Tatemizo A., Iwamoto D., Taniguchi S., Hoshino Y., Amano T., Matsumoto K., Hosoi Y., Iritani A., Saeki K. Enhancement of histone acetylation by trichostatin A during in vitro fertilization of bovine oocytes affects cell number of the inner cell mass of the resulting blastocysts. Zygote. 2009;17:209–215. doi: 10.1017/S0967199409005279. [DOI] [PubMed] [Google Scholar]
- 19.Laguna-Barraza R., Sánchez-Calabuig M.J., Gutiérrez-Adán A., Rizos D., Pérez-Cerezales S. Effects of the HDAC inhibitor scriptaid on the in vitro development of bovine embryos and on imprinting gene expression levels. Theriogenology. 2018;110:79–85. doi: 10.1016/j.theriogenology.2017.12.043. [DOI] [PubMed] [Google Scholar]
- 20.Kong P., Yin M., Chen D., Li S., Li Y., Xing F., Jiang M., Fang Z., Lyu Q., Chen X. Effects of the histone deacetylase inhibitor ‘Scriptaid’ on the developmental competence of mouse embryos generated through round spermatid injection. Hum. Reprod. 2017;32:76–87. doi: 10.1093/humrep/dew290. [DOI] [PubMed] [Google Scholar]
- 21.Chen X., Zhai Y., Yu D., Cui J., Hu J.F., Li W. Valproic acid enhances iPSC induction from human bone marrow-derived cells through the suppression of reprogramming-induced senescence. J. Cell. Physiol. 2016;231:1719–1727. doi: 10.1002/jcp.25270. [DOI] [PubMed] [Google Scholar]
- 22.Zhai Y., Chen X., Yu D., Li T., Cui J., Wang G., Hu J.F., Li W. Histone deacetylase inhibitor valproic acid promotes the induction of pluripotency in mouse fibroblasts by suppressing reprogramming-induced senescence stress. Exp. Cell Res. 2015;337:61–67. doi: 10.1016/j.yexcr.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Chen G., Guo Y., Li C., Li S., Wan X. Small molecules that promote self-renewal of stem cells and somatic cell reprogramming. Stem Cell Rev. Rep. 2020;16:511–523. doi: 10.1007/s12015-020-09965-w. [DOI] [PubMed] [Google Scholar]
- 24.Mahapatra P.S., Singh R., Kumar K., Sahoo N.R., Agarwal P., Mili B., Das K., Sarkar M., Bhanja S.K., Das B.C., et al. Valproic acid assisted reprogramming of fibroblasts for generation of pluripotent stem cells in buffalo (Bubalus bubalis) Int. J. Dev. Biol. 2017;61:81–88. doi: 10.1387/ijdb.160006sb. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Madoz J.R., San Jose-Eneriz E., Rabal O., Zapata-Linares N., Miranda E., Rodriguez S., Porciuncula A., Vilas-Zornoza A., Garate L., Segura V., et al. Reversible dual inhibitor against G9a and DNMT1 improves human iPSC derivation enhancing MET and facilitating transcription factor engagement to the genome. PLoS ONE. 2017;12:e0190275. doi: 10.1371/journal.pone.0190275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han B., Li J., Li Z., Guo L., Wang S., Liu P., Wu Y. Trichostatin A stabilizes the expression of pluripotent genes in human mesenchymal stem cells during ex vivo expansion. PLoS ONE. 2013;8:e81781. doi: 10.1371/journal.pone.0081781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samiec M., Romanek J., Lipiński D., Opiela J. Expression of pluripotency-related genes is highly dependent on trichostatin A-assisted epigenomic modulation of porcine mesenchymal stem cells analysed for apoptosis and subsequently used for generating cloned embryos. Anim. Sci. J. 2019;90:1127–1141. doi: 10.1111/asj.13260. [DOI] [PubMed] [Google Scholar]
- 28.McClure J.J., Li X., Chou C.J. Advances and challenges of HDAC inhibitors in cancer therapeutics. Adv. Cancer Res. 2018;138:183–211. doi: 10.1016/bs.acr.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Sermer D., Pasqualucci L., Wendel H.G., Melnick A., Younes A. Emerging epigenetic-modulating therapies in lymphoma. Nat. Rev. Clin. Oncol. 2019;16:494–507. doi: 10.1038/s41571-019-0190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon S., Eom G.H. HDAC and HDAC inhibitor: From cancer to cardiovascular diseases. Chonnam Med. J. 2016;52:1–11. doi: 10.4068/cmj.2016.52.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalin J.H., Butler K.V., Kozikowski A.P. Creating zinc monkey wrenches in the treatment of epigenetic disorders. Curr. Opin. Chem. Biol. 2009;13:263–271. doi: 10.1016/j.cbpa.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Pierson R.N., III Antibody-mediated xenograft injury: Mechanisms and protective strategies. Transpl. Immunol. 2009;21:65–69. doi: 10.1016/j.trim.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weiss E.H., Lilienfeld B.G., Müller S., Müller E., Herbach N., Kessler B., Wanke R., Schwinzer R., Seebach J.D., Wolf E., et al. HLA-E/human b2-microglobulin transgenic pigs: Protection against xenogeneic human anti-pig natural killer cell cytotoxicity. Transplantation. 2009;87:35–43. doi: 10.1097/TP.0b013e318191c784. [DOI] [PubMed] [Google Scholar]
- 34.Crew M.D., Cannon M.J., Phanavanh B., Garcia-Borges C.N. An HLA-E single trimer inhibits human NK cell reactivity towards porcine cells. Mol. Immunol. 2005;42:1205–1214. doi: 10.1016/j.molimm.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Forte P., Baumann B.C., Weiss E.H., Seebach J.D. HLA-E expression on porcine cells: Protection from human NK cytotoxicity depends on peptide loading. Am. J. Transplant. 2005;5:2085–2093. doi: 10.1111/j.1600-6143.2005.00987.x. [DOI] [PubMed] [Google Scholar]
- 36.Lipiński D., Jura J., Zeyland J., Juzwa W., Mały E., Kalak R., Bochenek M., Pławski A., Szalata M., Smorąg Z., et al. Production of transgenic pigs expressing human a1,2-fucosyltransferase to avoid humoral xenograft rejection. Med. Weter. 2010;66:316–322. [Google Scholar]
- 37.Zeyland J., Gawrońska B., Juzwa W., Jura J., Nowak A., Słomski R., Smorąg Z., Szalata M., Woźniak A., Lipiński D. Transgenic pigs designed to express human α-galactosidase to avoid humoral xenograft rejection. J. Appl. Genet. 2013;54:293–303. doi: 10.1007/s13353-013-0156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia Y., Ren H., Gao X., Ji S., Yang J., Liu Z., Li S., Zhang Y. Expression of human alpha-galactosidase and alpha1,2-fucosyltransferase genes modifies the cell surface Galalpha1,3Gal antigen and confers resistance to human serum-mediated cytolysis. Chinese Med. Sci. J. (Chung-kuo i hsueh k’o hsueh tsa chih) 2004;19:31–37. [PubMed] [Google Scholar]
- 39.Wiater J., Karasiński J., Słomski R., Smorąg Z., Wartalski K., Gajda B., Jura J., Romek M. The effect of recombinant human alpha-1,2-fucosyltransferase and alpha-galactosidase A on the reduction of alpha-gal expression in the liver of transgenic pigs. Folia Biol. (Krakow) 2020;68:121–133. doi: 10.3409/fb_68-4.14. [DOI] [Google Scholar]
- 40.Samiec M., Skrzyszowska M., Opiela J. Creation of cloned pig embryos using contact-inhibited or serum-starved fibroblast cells analysed intra vitam for apoptosis occurrence. Ann. Anim. Sci. 2013;13:275–293. doi: 10.2478/aoas-2013-0009. [DOI] [Google Scholar]
- 41.Samiec M., Skrzyszowska M. The use of different methods of oocyte activation for generation of porcine fibroblast cell nuclear-transferred embryos. Ann. Anim. Sci. 2010;10:399–411. [Google Scholar]
- 42.Samiec M., Skrzyszowska M. Roscovitine is a novel agent that can be used for the activation of porcine oocytes reconstructed with adult cutaneous or fetal fibroblast cell nuclei. Theriogenology. 2012;78:1855–1867. doi: 10.1016/j.theriogenology.2012.06.029. [DOI] [PubMed] [Google Scholar]