Abstract
Background
In 2004, in response to high levels of treatment failure associated with sulfadoxine-pyrimethamine (SP) resistance, Benin changed its first-line malaria treatment from SP to artemisinin-based combination therapy for treatment of uncomplicated Plasmodium falciparum malaria. Resistance to SP is conferred by accumulation of single nucleotide polymorphisms (SNPs) in P. falciparum genes involved in folate metabolism, dihydrofolate reductase (Pfdhfr) and dihydropteroate synthase (Pfdhps), targeted by pyrimethamine and sulfadoxine, respectively. Because SP is still used for intermittent preventive treatment in pregnant women (IPTp) and seasonal malaria chemoprevention (SMCP) in Benin, the prevalence of Pfdhfr and Pfdhps SNPs in P. falciparum isolates collected in 2017 were investigated.
Methods
This study was carried out in two sites where the transmission of P. falciparum malaria is hyper-endemic: Klouékanmey and Djougou. Blood samples were collected from 178 febrile children 6–59 months old with confirmed uncomplicated P. falciparum malaria and were genotyped for SNPs associated with SP resistance.
Results
The Pfdhfr triple mutant IRN (N51I, C59R, and S108N) was the most prevalent (84.6%) haplotype and was commonly found with the Pfdhps single mutant A437G (50.5%) or with the Pfdhps double mutant S436A and A437G (33.7%). The quintuple mutant, Pfdhfr IRN/Pfdhps GE (A437G and K540E), was rarely observed (0.8%). The A581G and A613S mutant alleles were found in 2.6 and 3.9% of isolates, respectively. Six isolates (3.9%) were shown to harbour a mutation at codon I431V, recently identified in West African parasites.
Conclusions
This study showed that Pfdhfr triple IRN mutants are near fixation in this population and that the highly sulfadoxine-resistant Pfdhps alleles are not widespread in Benin. These data support the continued use of SP for chemoprevention in these study sites, which should be complemented by periodic nationwide molecular surveillance to detect emergence of resistant genotypes.
Keywords: Drug resistance, Sulfadoxine Pyrimethamine, Pfdhfr, Pfdhps, Intermittent Preventive Treatment in Pregnant, Seasonal Malaria Chemoprevention, Pregnant women, Malaria
Background
Malaria is a major health problem in Benin and is the leading cause of mortality among children under 5 years of age and morbidity among adults. In 2019, the World Health Organization (WHO) reported an estimated four million malaria cases and over 7000 deaths in the country [1]. In 2004, Benin joined many other countries in Africa in changing their recommended first-line treatment of uncomplicated malaria to artemisinin-based combination therapy (ACT) [2] due to reported high treatment failure rates in children treated with sulfadoxine-pyrimethamine (SP) for uncomplicated malaria [3–5]. However, the WHO recommends the continued use of SP for intermittent preventive treatment in pregnant women (IPTp) [6], as well as for seasonal malaria chemoprevention (SMC), used in combination with amodiaquine (SP-AQ) for the latter indication, in countries with highly seasonal malaria transmission such as the Sahel region of sub-Saharan Africa [7].
Resistance to SP is conferred by accumulation of single nucleotide polymorphisms (SNPs) in two genes that code for enzymes involved in Plasmodium falciparum folate metabolism: P. falciparum dihydrofolate reductase (Pfdhfr) and P. falciparum dihydropteroate synthase (Pfdhps), which are targeted by pyrimethamine and sulfadoxine, respectively. At least five mutations in Pfdhfr confer resistance to pyrimethamine: C50R, N51I, C59R, S108N, and I164L (amino acid substitutions in bold face). Similarly, at least five SNPs in Pfdhps are involved in resistance to sulfadoxine: S436A/F, A437G, K540E, A581G, and A613S/T [8–12].
The combination of Pfdhfr triple mutant IRN with the Pfdhps double mutant GE results in a quintuple mutant, which has been shown to lead to clinical treatment failure of SP [13–15]. In general, these quintuple mutants are commonly found throughout East Africa, but rarely in West and Central Africa [16–18]. In contrast, the Pfdhfr triple mutant IRN with the Pfdhps A437G is often found in West Africa and is also associated with treatment failure, but to a lesser degree than the IRN plus GE quintuple mutants [16–21]. Studies have demonstrated that the efficacy of SP for IPTp is still acceptable even when a high prevalence of SP resistance markers exists, including the IRN plus GE quintuple mutants and the IRN plus G quadruple mutants [6, 22–25], justifying the continued use of SP for IPTp and SMC. However, the occurrence of additional Pfdhps mutations at codons A581G [19, 26–28] and A613S/T [27] or Pfdhfr I164L [29, 30] to the quintuple IRN plus GE mutant genotype was shown to lead to declines in SP’s IPTp efficacy [28] and protection in infants [31].
Studies of molecular markers of SP resistance in Benin carried out between 2003 and 2012 in the north, south [32–34], and the coast [4, 35, 36] showed that the majority (> 90%) of parasites carry IRN Pfdhfr mutations. The most common mutation in Pfdhps was A437G (71.4%) [34]. A low prevalence of K540E (8.3%) was found [36] and no mutation was found at codon S436A in a study conducted between 2008 and 2010 [37]. However, recent data on molecular markers associated with SP resistance is lacking. This study investigated the prevalence of SNPs in Pfdhfr and Pfdhps in P. falciparum isolates collected from Benin in 2017.
Methods
Study population and sample collection
The samples utilized in this study were obtained from a therapeutic efficacy study (TES) of artemether-lumefantrine conducted by the National Malaria Control Programme (NMCP) in Benin in 2017 (results unpublished). The study was carried out in two NMCP sentinel sites, Klouékanmey and Djougou, where the transmission of P. falciparum malaria is hyper-endemic (Fig. 1). Criteria for inclusion included children 6–59 months old with monoinfection of P. falciparum, measured by microscopy, a parasite density between 2000 and 200,000 parasites/µl, axillary temperature of 37.5 °C or higher, and ability to take oral medication. Children with signs of severe illness and malnutrition were excluded. Enrolled patients were treated with a supervised 3-day course of artemether-lumefantrine and monitored for 28 days with weekly scheduled visits on days 7, 14, 21, and 28. Patients were also asked to return to the clinic if they became ill any other day during the follow-up period (unscheduled visits). Dried blood spots (DBS) were collected on Whatman grade 3 filter paper (GE Healthcare Life Sciences, Marlborough, USA) from enrolled patients on the day of enrollment (pre-treatment) and on the scheduled and unscheduled visits. Only samples from the day of enrollment were utilized in this study.
Fig. 1.
Location of study sites used for the therapeutic efficacy study, Benin, 2017. Benin map (shaded in gray) indicating the location of the two sentinel sites, Klouékanmey and Djougou (purple dots), in which samples were collected and used to determine the prevalence of sulfadoxine-pyrimethamine resistance markers in Plasmodium falciparum isolates
Sample processing andPfdhfrandPfdhpsmolecular analysis
Genomic DNA was isolated from the DBS using the QIAamp® blood mini kit (Qiagen Inc., CA, USA) per the manufacturer’s recommendations. The Pfdhfr and Pfdhps gene fragments were amplified by polymerase chain reaction (PCR) using previously published primers [38]. SNPs at Pfdhfr codons 50, 51, 59, 108, and 164 and Pfdhps codons 431, 436, 437, 540, 581, and 613 were investigated using Sanger sequencing as previously described [38]. The PCR products were precipitated in 70% ethanol to clean up dye terminators, rehydrated in 10 µl Hi-Di Formamide™, and sequenced using the Applied Biosystems 3130xl sequencer (Life Technologies, Grand Island, NY). Sequences were analysed using Geneious software (Biomatters, San Francisco, CA, USA). The 3D7 P. falciparum Pfdhps (Gene ID: 2,655,294) and Pfdhfr (Gene ID: 9,221,804) were used as reference sequences in the analysis.
Data management and analysis
Data were entered into a Microsoft Excel database and descriptive statistics such as percentage, mean, and range were reported as appropriate. The prevalence of different alleles and haplotypes in the Pfdhfr and Pfdhps genes were reported per site. The prevalence of the different alleles was reported as wild type (having only the wild type allele), mutant (having only the mutant allele), or mixed infection (having both wild type and mutant alleles).
Results
A total of 178 pre-treatment samples (85 from Klouékanmey and 93 from Djougou) were evaluated for molecular markers of resistance in the Pfdhfr and Pfdhps genes. The mean age of the patients was 33 (SD = 14) months and 62.1% were male. The geometric mean P. falciparum parasite density was 24,154 (95% CI 16,600–31,700); range: 2,081–200,000) parasites/µl.
Prevalence ofPfdhfr and Pfdhpsalleles (SNPs)
A total of 169 (94.9%) specimens were successfully sequenced for the Pfdhfr gene. Twelve samples (7.1%) had a mixed infection. No mutations were found at codons 50 or 164. Overall, a high prevalence of mutations was observed in codons N51I, (86.4%; 146), C59R, (89.9%; 152), and S108N, (94.7%; 160); an additional 4.1, 6.5, and 2.4% of samples, respectively, contained both mutant and wild-type alleles as part of a mixed infection, Table 1. For the Pfdhps gene, 153 (86.0%) samples were successfully sequenced and 21 of these (13.7%) had a mixed infection. Overall, a majority of the samples had the A437G mutation (94.8%; 145) followed by the S436A mutation (35.3%; 54), Table 1. All samples from Klouékanmey (100%; 68) had the A437G mutation compared to 95.3% (81) in Djougou, of which four (4.7%) were mixed alleles. The A581G and A613S alleles were observed in 2.6 and 3.9% of isolates, respectively, with an additional sample from Djougou having a 613A/S mixed infection. Six isolates (3.9%) were shown to harbour a mutation at codon I431V, three from Klouékanmey and three from Djougou, of which two were mixed with wildtype parasites, Table 1.
Table 1.
Summary of alleles observed in Pfdhfr and Pfdhps genes
| Pfdhfr | Klouékanmey n = 79 (%) |
Djougou n = 90 (%) |
Overall n = 169 (%) |
|---|---|---|---|
| C50 | 79 (100) | 90 (100) | 169 (100) |
| 50R | 0 | 0 | 0 |
| 50 C/R | 0 | 0 | 0 |
| N51 | 3 (3.8) | 13 (14.4) | 16 (9.5) |
| 51I | 72 (91.1) | 74 (82.3) | 146 (86.4) |
| 51 N/I | 4 (5.1) | 3 (3.3) | 7 (4.1) |
| C59 | 1 (1.3) | 5 (5.6) | 6 (3.6) |
| 59R | 72 (91.1) | 80 (88.8) | 152 (89.9) |
| 59 C/R | 6 (7.6) | 5 (5.6) | 11 (6.5) |
| S108 | 0 | 5 (5.6) | 5 (3.0) |
| 108 N | 78 (98.7) | 82 (91.1) | 160 (94.7) |
| 108S/N | 1 (1.3) | 3 (3.3) | 4 (2.4) |
| I164 | 79 (100) | 90 (100) | 169 (100) |
| 164L | 0 | 0 | 0 |
| 164I/L | 0 | 0 | 0 |
| Pfdhps | Klouékanmey n = 68 (%) |
Djougou n = 85 (%) |
Overall n = 153 (%) |
|---|---|---|---|
| I431 | 65 (95.6) | 82 (96.4) | 147 (96.1) |
| 431V | 3 (4.4) | 1 (1.2) | 4 (2.6) |
| 431I/V | 0 | 2 (2.4) | 2 (1.3) |
| S436 | 41 (60.2) | 40 (47.1) | 81 (52.9) |
| 436A | 22 (32.4) | 32 (37.6) | 54 (35.3) |
| 436S/A | 5 (7.4) | 13 (15.3) | 18 (11.8) |
| A437 | 0 | 4 (4.7) | 4 (2.6) |
| 437G | 68 (100) | 77 (90.6) | 145 (94.8) |
| 437A/G | 0 | 4 (4.7) | 4 (2.6) |
| K540 | 66 (97.0) | 83 (97.6) | 149 (97.4) |
| 540E | 1 (1.5) | 1 (1.2) | 2 (1.3) |
| 540K/E | 1 (1.5) | 1 (1.2) | 2 (1.3) |
| A581 | 65 (95.6) | 84 (98.8) | 149 (97.4) |
| 581G | 3 (4.4) | 1 (1.2) | 4 (2.6) |
| 581A/G | 0 | 0 | 0 |
| A613 | 65 (95.6) | 81 (95.3) | 146 (95.4) |
| 613S | 3 (4.4) | 3 (3.5) | 6 (3.9) |
| 613A/S | 0 | 1 (1.2) | 1 (0.7) |
Bold letters denote mutant alleles
Observed haplotypes per gene
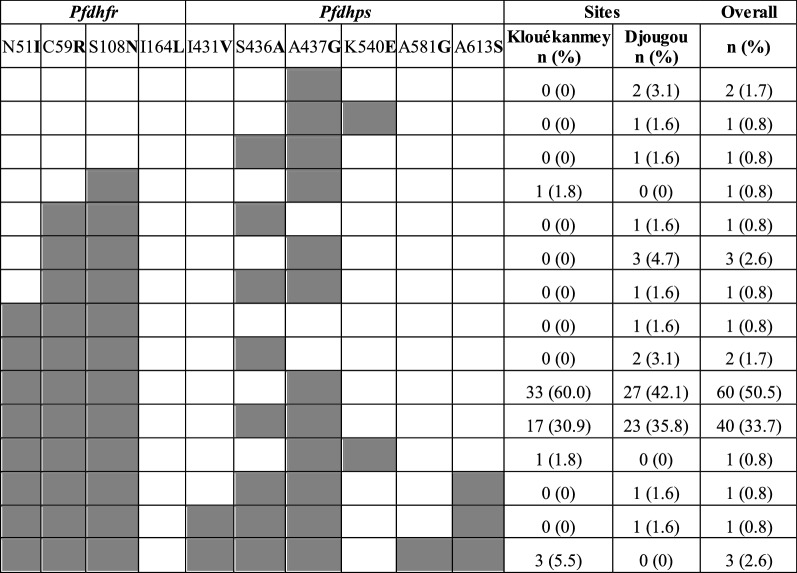
Table 2 summarizes the observed haplotypes for the Pfdhfr and Pfdhps genes. Haplotypes were constructed using codons C50R, N51I, C59R, S108N, and I164L in the Pfdhfr gene and I431V, S436A, A437G, K540E, A581G, and A613S in the Pfdhps gene. Mixed infections in the Pfdhfr (12) and Pfdhps (21) genes were excluded for the haplotype construction. The majority of parasites (84.6%, 143) harboured the triple mutant Pfdhfr CIRNI haplotype: 89.8% (71) in Klouékanmey and 79.9% (72) in Djougou. The most common Pfdhps haplotype was ISGKAA (49.6%; 76), followed by the double mutant Pfdhps IAGKAA (29.3%; 45). The I431V mutation was seen in combination with other Pfdhps mutations, with three isolates (2.0%) possessing S436A, A437G, A581G, and A613S, and one isolate (0.7%) possessing S436A, A437G, and A613S, Table 2.
Table 2.
Summary of haplotypes observed in Pfdhfr and Pfdhps genes
| Klouékanmey n (%) | Djougou n (%) | Overall n (%) | |
|---|---|---|---|
| Pfdhfr | |||
| CIRNI | 71 (89.8) | 72 (79.9) | 143 (84.6) |
| CNRNI | 1 (1.3) | 7 (7.8) | 8 (4.7) |
| CNCNC | 1 (1.3) | 0 | 1 (0.6) |
| CNCSI | 0 | 5 (5.6) | 5 (3.0) |
| Mixed-infection | 6 (7.6) | 6 (6.7) | 12 (7.1) |
| Pfdhps | |||
| ISGKAA | 39 (57.4) | 37 (43.5) | 76 (49.6) |
| IAGKAA | 19 (27.9) | 26 (30.6) | 45 (29.3) |
| IAAKAA | 0 | 3 (3.5) | 3 (2.0) |
| VAGKGS | 3 (4.4) | 0 | 3 (2.0) |
| ISGEAA | 1 (1.5) | 1 (1.2) | 2 (1.3) |
| VAGKAS | 0 | 1 (1.2) | 1 (0.7) |
| IAGKAS | 0 | 1 (1.2) | 1 (0.7) |
| ISAKAA | 0 | 1 (1.2) | 1 (0.7) |
| Mixed-infection | 6 (8.8) | 15 (17.6) | 21 (13.7) |
Bold letters denote mutant alleles
Combined Pfdhfr/Pfdhps haplotypes
Combined Pfdhfr and Pfdhps haplotypes were constructed using 119 samples that were successfully sequenced at each of the codons investigated, Table 3. Mixed infections at any of the codons were excluded. The N51I/C59R/S108N/A437G haplotype was found in 106 (89.1%) of the samples. Of these 106, 45 also contained the S436A mutation (N51I/C59R/S108N/S436A/A437G) and one contained the K540E mutation (N51I/C59R/S108N/A437G/K540E). Another two samples contained neither the A437G nor the K540E mutation but contained the S436A mutation (N51I/C59R/S108N/S436A). In the samples with the N51I/C59R/S108N/S436A/A437G haplotype, the A613S mutation was found in five samples, three of which also contained the A581G mutation.
Table 3.
Summary of combined Pfdhfr and Pfdhps haplotypes. Shaded boxes and bold letters denote mutant alleles. Key haplotypes associated with SP resistance include: the quadruple haplotypes, N51I, C59R, S108N plus S436A or A437G; the quintuple haplotypes, N51I, C59R, S108N plus A437G and K540E; or N51I, C59R, S108N plus S436A and A437G
Discussion
Results from this study demonstrate that the prevalence of the Pfdhfr IRN triple mutant is very high, implying these mutants are well established in this region, similar to observations made previously in Benin and in many African countries [16, 18, 39, 40]. In contrast, the prevalence of multiple mutations in the Pfdhps gene was low, with the majority of parasites having only a single mutation at codon A437G and 29.3% of the parasites with a double mutant (S436A/A437G), as commonly observed in West Africa [18, 21, 41, 42]. Only a handful of isolates had mutations at codons K540E (2.6%), A581G (2.6%), and A613S (4.6%). A low prevalence or complete absence of these mutations was also observed in other studies in Benin [36, 37]. Several studies have shown the prevalence of these mutations in West Africa is very low compared to East Africa, (reviewed in [16, 18]). This is especially the case for the K540E mutation, which has a prevalence greater than 10% in many countries in East Africa but is rarely reported in West Africa [16, 18]. A significant increase in the prevalence of the mutations at codons A581G and A613S was observed in Nigeria [27, 43], demonstrating the emergence of these mutations. However, their prevalence, even in Benin, is well below the WHO thresholds for consideration of changes in the use of IPTp (> 95% for K540E and > 10 % for A581G) [44].
Ultimately, the combination of mutations in the Pfdhfr and Pfdhps genes is one of several factors that determines a parasite’s response to SP. Marked regional differences in the Pfdhfr and Pfdhps genotypes have been observed across Africa [16, 18]. The quadruple (N51I, C59R, S108N plus A437G) mutants are widespread in West Africa, while the quintuple mutants (N51I, C59R, S108N plus A437G, 540E) and sextuple mutants (addition of Pfdhps A581G and A613T/S or Pfdhfr I164L on the quintuple background) predominate in East Africa [16, 18–20, 38, 45, 46]. In keeping with these observations and with a study conducted in Benin between 2008 and 2010 [37], the majority of isolates in this study were quadruple (N51I, C59R, S108N plus A437G ) mutants. The sustained high prevalence of these quadruple mutant parasites is likely due to persistent SP drug pressure from its continual use for IPTp and SMC in Benin. The quintuple (N51I, C59R, S108N plus A437G and K540E) mutant was observed only in one isolate in Klouékanmey. The minority of isolates in this study with the A581G and A613S mutations were found in the absence of the K540E mutation. These results demonstrate that genotypes conferring a high level of SP resistance have not fully emerged in these study sites, providing support for the continued use of SP for IPTp and SMC in Benin.
A few isolates (2.6 %) in this study possessed the Pfdhps I431V mutation, which was first described in travelers from Nigeria identified in the UK [47]. A recent study conducted in Nigeria demonstrated an increase in the prevalence of this mutation from 0–6.5 % between 2003 and 2008, and as high as 46 % in 2010 in Enugu, Nigeria [43]. It was also found in pregnant women from other sites in Nigeria such as Epe and Ibeju-Lekki [42]. Furthermore, the I431V mutation was seen in isolates from pregnant women in Cameroon and Ghanaian travellers [48, 49], suggesting this mutation is emerging in the region. Interestingly, to date, the I431V mutation has not been observed in other parts of Africa. In concordance with previous studies [42, 49], this mutation was observed in combination with other Pfdhps mutations (S436A/A437G/A581G/A613S), suggesting this mutation may have occurred only in the presence of the other Pfdhps mutations. The implications of this combination of Pfdhps mutations remain unclear. While some propose these mutations may disrupt the binding of sulfadoxine to the Pfdhps active site [43], additional studies are needed to fully support this notion and to understand the mechanisms involved. Therefore, it is worthwhile to continue monitoring the prevalence of the I431V mutation, along with other mutations, in this region.
Limitations of this study include the fact that the samples were obtained from a TES conducted in only two sites in Benin, and therefore the results obtained may not be generalizable to other regions or sites. Moreover, the sample size used was small; additional study sites are recommended for future studies.
Conclusions
The results from this study indicate that the highly sulfadoxine resistant Pfdhps alleles are not widespread in Benin, supporting the current policy of using SP for IPTp and SMC in Benin. However, given the continued use of this drug and limited alternative options, frequent monitoring of SP resistance markers in order to inform IPTp and SMC policies in Benin remains important.
Acknowledgements
The authors would like to thank the National Malaria Control Programme of Benin, all study team members, and children’s parents and relatives.
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Authors’ contributions
AK, ESH, AS, RS, MP, and FD designed the study; SS and AA performed all laboratory work; SSS and NWL analysed and presented the data. SSS, ESH, VW and NL drafted the paper, all authors reviewed and approved the final manuscript. All authors read and approved the final manuscript.
Funding
The study was funded by the U.S. President’s Malaria Initiative.
Availability of data and materials
The full anonymized clinical dataset will be uploaded to Worldwide Antimalarial Resistance Network and WHO repositories upon request and after publication.
Ethics approval and consent to participate
Written informed consent was obtained from each study participant before study participation. The ethical approvals were granted by the National Ethics Committee for Health Research (CNERS) in Benin, IRB#06860,1/20/2018. Laboratory analysis was performed with a Benin laboratory trainee at the Centers for Disease Control and Prevention (CDC) as part of a technical training program [50]. The work performed at CDC did not constitute engagement in human subjects research (CDC protocol #2017 − 141).
Consent for publication
Authors gave their consent for publication.
Competing interests
The authors have no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO . World malaria report 2019. Geneva: World Health Organization; 2019. [Google Scholar]
- 2.Zinsou C, Cherifath AB. The malaria testing and treatment landscape in Benin. Malar J. 2017;16:174. doi: 10.1186/s12936-017-1808-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubouy A, Fievet N, Bertin G, Sagbo JC, Kossou H, Kinde-Gazard D, et al. Dramatically decreased therapeutic efficacy of chloroquine and sulfadoxine‐pyrimethamine, but not mefloquine, in southern Benin. Trop Med Int Health. 2007;12:886–94. doi: 10.1111/j.1365-3156.2007.01859.x. [DOI] [PubMed] [Google Scholar]
- 4.Nahum A, Erhart A, Gazard D, Agbowai C, Van Overmeir C, van Loen H, et al. Adding artesunate to sulphadoxine-pyrimethamine greatly improves the treatment efficacy in children with uncomplicated falciparum malaria on the coast of Benin, West Africa. Malar J. 2007;6:170. doi: 10.1186/1475-2875-6-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faucher JF, Aubouy A, Adeothy A, Cottrell G, Doritchamou J, Gourmel B, et al. Comparison of sulfadoxine-pyrimethamine, unsupervised artemether‐lumefantrine, and unsupervised artesunate‐amodiaquine fixed‐dose formulation for uncomplicated Plasmodium falciparum malaria in Benin: a randomized effectiveness noninferiority trial. J Infect Dis. 2009;200:57–65. doi: 10.1086/599378. [DOI] [PubMed] [Google Scholar]
- 6.WHO . Technical expert group meeting on intermittent preventive treatment in pregnancy (IPTp) Geneva: World Health Organization; 2007. [Google Scholar]
- 7.WHO . Seasonal malaria chemoprevention with sulfadoxine–pyrimethamine plus amodiaquine in children: a field guide. Geneva: World Health Organization; 2013. [Google Scholar]
- 8.Brooks DR, Wang P, Read M, Watkins WM, Sims PF, Hyde JE. Sequence variation of the hydroxymethyldihydropterin pyrophosphokinase: dihydropteroate synthase gene in lines of the human malaria parasite, Plasmodium falciparum, with differing resistance to sulfadoxine. Eur J Biochem. 1994;224:397–405. doi: 10.1111/j.1432-1033.1994.00397.x. [DOI] [PubMed] [Google Scholar]
- 9.Cowman AF, Morry MJ, Biggs BA, Cross G, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:9109–13. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci USA. 1988;85:9114–8. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Triglia T, Cowman AF. Primary structure and expression of the dihydropteroate synthetase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1994;91:7149–53. doi: 10.1073/pnas.91.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev. 2005;57:117–45. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- 13.Omar S, Adagu I, Gump D, Ndaru N, Warhurst D. Plasmodium falciparum in Kenya: high prevalence of drug-resistance-associated polymorphisms in hospital admissions with severe malaria in an epidemic area. Ann Trop Med Parasitol. 2001;95:661–9. doi: 10.1080/00034983.2001.11813683. [DOI] [PubMed] [Google Scholar]
- 14.Staedke SG, Sendagire H, Lamola S, Kamya MR, Dorsey G, Rosenthal PJ. Relationship between age, molecular markers, and response to sulphadoxine–pyrimethamine treatment in Kampala, Uganda. Trop Med Int Health. 2004;9:624–9. doi: 10.1111/j.1365-3156.2004.01239.x. [DOI] [PubMed] [Google Scholar]
- 15.Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, et al. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–8. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- 16.Naidoo I, Roper C. Mapping ‘partially resistant’,‘fully resistant’, and ‘super resistant’malaria. Trends Parasitol. 2013;29:505–15. doi: 10.1016/j.pt.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Ruizendaal E, Tahita MC, Traoré-Coulibaly M, Tinto H, Schallig HD, Mens PF. Presence of quintuple dhfr N51, C59, S108–dhps A437, K540 mutations in Plasmodium falciparum isolates from pregnant women and the general population in Nanoro, Burkina Faso. Mol Biochem Parasitol. 2017;217:13–5. doi: 10.1016/j.molbiopara.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 18.Sridaran S, McClintock SK, Syphard LM, Herman KM, Barnwell JW, Udhayakumar V. Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar J. 2010;9:247. doi: 10.1186/1475-2875-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gesase S, Gosling RD, Hashim R, Ord R, Naidoo I, Madebe R, et al. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One. 2009;4:e4569. doi: 10.1371/journal.pone.0004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juma DW, Omondi AA, Ingasia L, Opot B, Cheruiyot A, Yeda R, et al. Trends in drug resistance codons in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in Kenyan parasites from 2008 to 2012. Malar J. 2014;13:250. doi: 10.1186/1475-2875-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearce RJ, Pota H, Evehe M-SB, Bâ E-H, Mombo-Ngoma G, Malisa AL, et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gies S, Coulibaly SO, Ouattara FT, D’Alessandro U. Individual efficacy of intermittent preventive treatment with sulfadoxine–pyrimethamine in primi-and secundigravidae in rural Burkina Faso: impact on parasitaemia, anaemia and birth weight. Trop Med Int Health. 2009;14:174–82. doi: 10.1111/j.1365-3156.2008.02215.x. [DOI] [PubMed] [Google Scholar]
- 23.Hill J, Kayentao K, Toure M, Diarwara S, Bruce J, Smedley J, et al. Effectiveness of antenatal clinics to deliver intermittent preventive treatment and insecticide treated nets for the control of malaria in pregnancy in Mali: a household survey. PLoS ONE. 2014;9:e92102. doi: 10.1371/journal.pone.0092102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hommerich L, Von Oertzen C, Bedu-Addo G, Holmberg V, Acquah PA, Eggelte TA, et al. Decline of placental malaria in southern Ghana after the implementation of intermittent preventive treatment in pregnancy. Malar J. 2007;6:144. doi: 10.1186/1475-2875-6-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mpogoro FJ, Matovelo D, Dosani A, Ngallaba S, Mugono M, Mazigo HD. Uptake of intermittent preventive treatment with sulphadoxine-pyrimethamine for malaria during pregnancy and pregnancy outcomes: a cross-sectional study in Geita district, North-Western Tanzania. Malar J. 2014;13:455. doi: 10.1186/1475-2875-13-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alifrangis M, Lusingu JP, Mmbando B, Dalgaard MB, Vestergaard LS, Ishengoma D, et al. Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the Plasmodium falciparum dihydropteroate synthase gene. Am J Trop Med Hyg. 2009;80:523–7. doi: 10.4269/ajtmh.2009.80.523. [DOI] [PubMed] [Google Scholar]
- 27.Spalding MD, Eyase FL, Akala HM, Bedno SA, Prigge ST, Coldren RL, et al. Increased prevalence of the pfdhfr/phdhps quintuple mutant and rapid emergence of pfdhps resistance mutations at codons 581 and 613 in Kisumu, Kenya. Malar J. 2010;9:338. doi: 10.1186/1475-2875-9-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrington W, Mutabingwa T, Muehlenbachs A, Sorensen B, Bolla M, Fried M, et al. Competitive facilitation of drug-resistant Plasmodium falciparum malaria parasites in pregnant women who receive preventive treatment. Proc Natl Acad Sci USA. 2009;106:9027–32. doi: 10.1073/pnas.0901415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynch C, Pearce R, Pota H, Cox J, Abeku TA, Rwakimari J, et al. Emergence of a dhfr mutation conferring high-level drug resistance in Plasmodium falciparum populations from southwest Uganda. J Infect Dis. 2008;197:1598–604. doi: 10.1086/587845. [DOI] [PubMed] [Google Scholar]
- 30.Karema C, Imwong M, Fanello CI, Stepniewska K, Uwimana A, Nakeesathit S, et al. Molecular correlates of high-level antifolate resistance in Rwandan children with Plasmodium falciparum malaria. Antimicrob Agents Chemother. 2010;54:477–83. doi: 10.1128/AAC.00498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosling RD, Gesase S, Mosha JF, Carneiro I, Hashim R, Lemnge M, et al. Protective efficacy and safety of three antimalarial regimens for intermittent preventive treatment for malaria in infants: a randomised, double-blind, placebo-controlled trial. Lancet. 2009;374:1521–32. doi: 10.1016/S0140-6736(09)60997-1. [DOI] [PubMed] [Google Scholar]
- 32.Ogouyèmi-Hounto A, Ndam NT, Fadégnon G, Azagnandji C, Bello M, Moussiliou A, et al. Low prevalence of the molecular markers of Plasmodium falciparum resistance to chloroquine and sulphadoxine/pyrimethamine in asymptomatic children in Northern Benin. Malar J. 2013;12:413. doi: 10.1186/1475-2875-12-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahlström S, Aubouy A, Maïga-Ascofaré O, Faucher J-F, Wakpo A, Ezinmègnon S, et al. Plasmodium falciparum polymorphisms associated with ex vivo drug susceptibility and clinical effectiveness of artemisinin-based combination therapies in Benin. Antimicrob Agents Chemother. 2014;58:1–10. doi: 10.1128/AAC.01790-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogouyèmi-Hounto A, Ndam NT, Gazard DK, d’Almeida S, Koussihoude L, Ollo E, et al. Prevalence of the molecular marker of Plasmodium falciparum resistance to chloroquine and sulphadoxine/pyrimethamine in Benin seven years after the change of malaria treatment policy. Malar J. 2013;12:147. doi: 10.1186/1475-2875-12-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertin G, Briand V, Bonaventure D, Carrieu A, Massougbodji A, Cot M, et al. Molecular markers of resistance to sulphadoxine-pyrimethamine during intermittent preventive treatment of pregnant women in Benin. Malar J. 2011;10:196. doi: 10.1186/1475-2875-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nahum A, Erhart A, Ahounou D, Bonou D, Van Overmeir C, Menten J, et al. Extended high efficacy of the combination sulphadoxine-pyrimethamine with artesunate in children with uncomplicated falciparum malaria on the Benin coast, West Africa. Malar J. 2009;8:37. doi: 10.1186/1475-2875-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moussiliou A, De Tove YS-S, Doritchamou J, Luty AJ, Massougbodji A, Alifrangis M, et al. High rates of parasite recrudescence following intermittent preventive treatment with sulphadoxine-pyrimethamine during pregnancy in Benin. Malar J. 2013;12:195. doi: 10.1186/1475-2875-12-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCollum AM, Poe AC, Hamel M, Huber C, Zhou Z, Shi YP, et al. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–97. doi: 10.1086/504687. [DOI] [PubMed] [Google Scholar]
- 39.Maiga H, Lasry E, Diarra M, Sagara I, Bamadio A, Traore A, et al. Seasonal malaria chemoprevention with sulphadoxine-pyrimethamine and amodiaquine selects Pfdhfr-dhps quintuple mutant genotype in Mali. PLoS ONE. 2016;11:e0162718. doi: 10.1371/journal.pone.0162718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fagbemi KA, Adebusuyi SA, Nderu D, Adedokun SA, Pallerla SR, Amoo AO, et al. Analysis of sulphadoxine–pyrimethamine resistance-associated mutations in Plasmodium falciparum isolates obtained from asymptomatic pregnant women in Ogun State, Southwest Nigeria. Infect Genet Evol. 2020;85:104503. doi: 10.1016/j.meegid.2020.104503. [DOI] [PubMed] [Google Scholar]
- 41.Fall B, Pascual A, Sarr FD, Wurtz N, Richard V, Baret E, et al. Plasmodium falciparum susceptibility to anti-malarial drugs in Dakar, Senegal, in 2010: an ex vivo and drug resistance molecular markers study. Malar J. 2013;12:107. doi: 10.1186/1475-2875-12-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quan H, Igbasi U, Oyibo W, Omilabu S, Chen S-B, Shen H-M, et al. High multiple mutations of Plasmodium falciparum-resistant genotypes to sulphadoxine-pyrimethamine in Lagos, Nigeria. Infect Dis Poverty. 2020;9:91. doi: 10.1186/s40249-020-00712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oguike MC, Falade CO, Shu E, Enato IG, Watila I, Baba ES, et al. Molecular determinants of sulfadoxine-pyrimethamine resistance in Plasmodium falciparum in Nigeria and the regional emergence of dhps 431V. Int J Parasitol Drugs Drug Resist. 2016;6:220–9. doi: 10.1016/j.ijpddr.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.WHO. Evidence Review Group on intermittent preventive treatment (IPT) of malaria in pregnancy. Geneva, World Health Organization. 2020. https://www.who.int/malaria/mpac/mpac_sep13_erg_ipt_malaria_pregnancy_report.pdf?ua=1.
- 45.Harrington WE, Mutabingwa TK, Kabyemela E, Fried M, Duffy PE. Intermittent treatment to prevent pregnancy malaria does not confer benefit in an area of widespread drug resistance. Clin Infect Dis. 2011;53:224–30. doi: 10.1093/cid/cir376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearce RJ, Pota H, Evehe M-SB, Bâ E-H, Mombo-Ngoma G, Malisa AL, et al. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med. 2009;6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutherland CJ, Fifer H, Pearce RJ, bin Reza F, Nicholas M, Haustein T, et al. Novel pfdhps haplotypes among imported cases of Plasmodium falciparum malaria in the United Kingdom. Antimicrob Agents Chemother. 2009;53:3405–10. doi: 10.1128/AAC.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chauvin P, Menard S, Iriart X, Nsango SE, Tchioffo MT, Abate L, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in pregnant women in Yaoundé, Cameroon: emergence of highly resistant pfdhfr/pfdhps alleles. J Antimicrob Chemother. 2015;70:2566–71. doi: 10.1093/jac/dkv160. [DOI] [PubMed] [Google Scholar]
- 49.Bansal D, Bharti PK, Acharya A, Abdelraheem MH, Patel P, Elmalik A, et al. Molecular surveillance of putative drug resistance markers of antifolate and artemisinin among imported Plasmodium falciparum in Qatar. Pathog Glob Health. 2019;113:158–66. doi: 10.1080/20477724.2019.1639018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halsey ES, Venkatesan M, Plucinski MM, Talundzic E, Lucchi NW, Zhou Z, et al. Capacity development through the US President’s malaria initiative–supported antimalarial resistance monitoring in Africa network. Emerg Infect Dis. 2017;23:53. doi: 10.3201/eid2313.170366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The full anonymized clinical dataset will be uploaded to Worldwide Antimalarial Resistance Network and WHO repositories upon request and after publication.