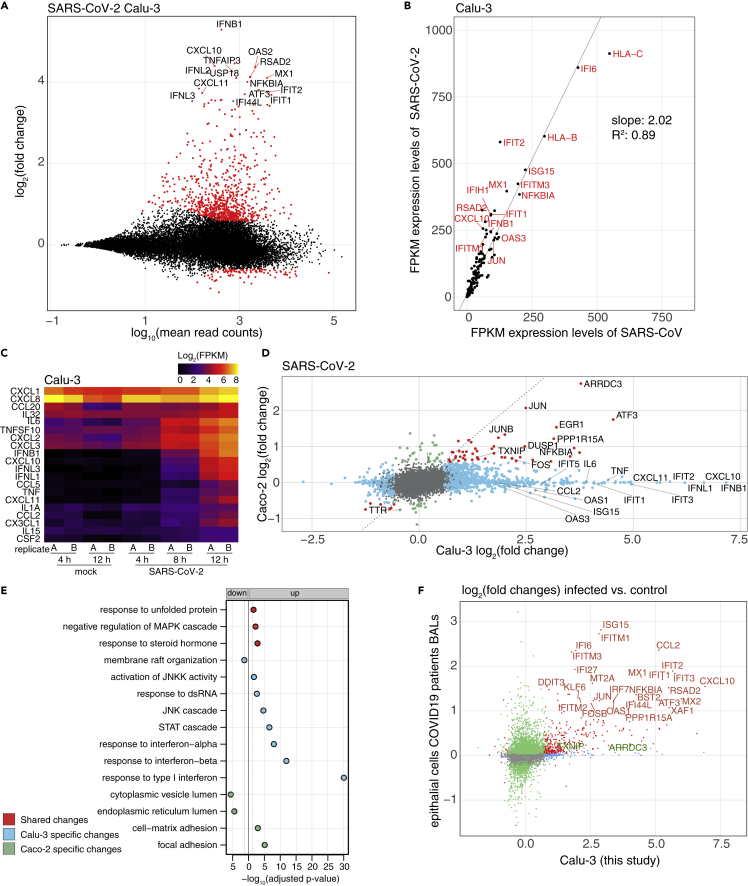
Figure 2.
Dissection of the transcriptional response to SARS-CoV/-2 infection
(A) Mean average vs. log2 fold change plot of differentially expressed (DE) genes at the mRNA level of Calu-3 cells infected with SARS-CoV-2 (12 hpi). X axis depicts log10 of mean counts associated with a gene. Y axis depicts mRNA log2 fold changes. Significant differentially expressed genes (red dots) are defined as those with an absolute log2 fold change of greater than 0.58 and a Benjamini-Hochberg corrected adjusted p value > 0.05. Selected outliers are labeled.
(B) Expression values (FPKM) of genes significantly induced in cells infected with either virus at 12 hpi in Calu-3 cells. Values represent averages of the two replicates.
(C) Expression values (FPKM) of differentially expressed cytokine genes in SARS-CoV-2 infected Calu-3 cells. Values of individual replicates are log2 transformed and represented as a heatmap.
(D) log2-transformed fold changes of SARS-CoV-2 infected Calu-3 cells at 12 hpi vs. mock (horizontal axis) and Caco-2 cells 12 hpi vs. mock (vertical axis). Genes exhibiting significant changes in both cell lines are shown in red, significant changes only in Calu-3 cells in light blue, only in Caco-2 cells in light green. All other genes are shown in gray. Selected genes with significant fold changes are labeled.
(E) Gene Ontology (GO) terms in a gene set enrichment analysis of cell lines infected by SARS-CoV-2. In blue are indicated enriched GO terms from differentially expressed genes in Calu-3 cells, in green from Caco-2 cells and in red from both cell lines. Adjusted p values were -log10-transformed. GO terms from downregulated genes are shown to the left, those from upregulated genes on the right of the solid line. The dotted line represents the cutoff value (p = 0.1).
(F) Log2-transformed fold changes of SARS-CoV-2 infected Calu-3 cells of this study at 12 hpi (horizontal axis) and epithelial cells from severe COVID19 patients from Liao et al. (2020) (vertical axis). Genes exhibiting significant changes in both cells are shown in red, significant changes only in Calu-3 cells in light blue, only in patient cells in light green. All other genes are shown in gray. Selected genes with significant fold changes are labeled in red, and additional genes mentioned in the text (TXNIP and ARRDC3) in green.