Abstract
The coronavirus disease, called COVID-19, which is spreading fast worldwide since the end of 2019, and has become a global challenging pandemic. Until 27th May 2020, it caused more than 5.6 million individuals infected throughout the world and resulted in greater than 348,145 deaths. CT images-based classification technique has been tried to use the identification of COVID-19 with CT imaging by hospitals, which aims to minimize the possibility of virus transmission and alleviate the burden of clinicians and radiologists. Early diagnosis of COVID-19, which not only prevents the disease from spreading further but allows more reasonable allocation of limited medical resources. Therefore, CT images play an essential role in identifying cases of COVID-19 that are in great need of intensive clinical care. Unfortunately, the current public health emergency, which has caused great difficulties in collecting a large set of precise data for training neural networks. To tackle this challenge, our first thought is transfer learning, which is a technique that aims to transfer the knowledge from one or more source tasks to a target task when the latter has fewer training data. Since the training data is relatively limited, so a transfer learning-based DensNet-121 approach for the identification of COVID-19 is established. The proposed method is inspired by the precious work of predecessors such as CheXNet for identifying common Pneumonia, which was trained using the large Chest X-ray14 dataset, and the dataset contains 112,120 frontal chest X-rays of 14 different chest diseases (including Pneumonia) that are individually labeled and achieved good performance. Therefore, CheXNet as the pre-trained network was used for the target task (COVID-19 classification) by fine-tuning the network weights on the small-sized dataset in the target task. Finally, we evaluated our proposed method on the COVID-19-CT dataset. Experimentally, our method achieves state-of-the-art performance for the accuracy (ACC) and F1-score. The quantitative indicators show that the proposed method only uses a GPU can reach the best performance, up to 0.87 and 0.86, respectively, compared with some widely used and recent deep learning methods, which are helpful for COVID-19 diagnosis and patient triage. The codes used in this manuscript are publicly available on GitHub at (https://github.com/lichun0503/CT-Classification).
Keywords: Transfer learning, Classification, Small-sized samples learning, COVID-19 Pneumonia
1. Introduction
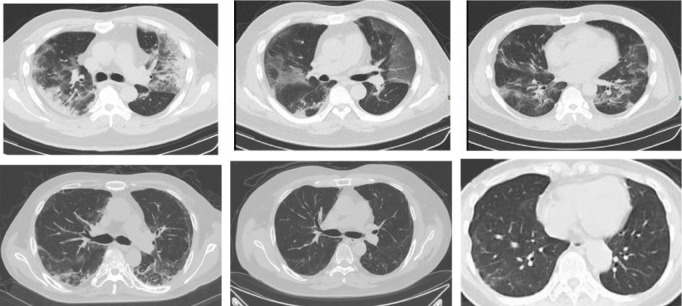
The coronavirus disease 2019 (COVID-19) is spreading fast worldwide since the end of 2019, and has become a global challenging pandemic. As shown in Fig. 1, until May 21, 2020, the COVID-19 has been affecting 213 countries and territories around the world and 2 international conveyances, 4,891,330 coronavirus cases, and 320,134 deaths are confirmed, reported by https://www.worldometers.info/coronavirus/. World Health Organization (WHO) has declared the COVID-19 as a global health emergency on January 30, 2020, [1], which poses a great threat to international human health. Because it is highly infectious and vaccine development is still in clinical trials, early diagnosis of COVID-19 has become more and more urgent. It not only prevents the disease from spreading further but allows a more reasonable allocation of limited medical resources. Currently, as the golden standard for COVID-19 diagnosis, the detection of the reverse transcription-polymerase chain reaction (RT-PCR) of the viral nucleic acid takes 4 to 6 h to obtain results. To me, as one of the testers who has personally experienced two nucleic acid tests, compared with COVID-19’s horrible speed of propagation, this is a long time. The chest computed tomography (CT) is useful for the clinical assistant diagnosis of COVID-19 [2], [3], [4], [5], [6], [7], [8], [9], [10], [11], however, since the terrible increase in the number of COVID-19 infections, there is a huge shortage of clinicians and radiologists, moreover, the routine use of CT puts a huge burden on the radiology department, meanwhile, there is a potential infection in CT equipment. As a result, many infected people cannot get a timely diagnosis, so they continue to infect others unconsciously. Therefore, it is favorably hunger for to researchers explore automated computer-aided diagnostic system classification methods for COVID-19 based CT images. About the examples of chest CT images with infection of COVID-19 and Non-COVID-19 as shown in Fig. 2.
Fig. 1.
As of May 21, 2020, the world’s statistics on the number of COVID-19 infections and deaths.
Fig. 2.
Examples of chest CT images with infection of COVID-19 (the first row) and non-infection of COVID-19 (the second row).
Nowadays, the COVID-19 pandemic threatens the health system worldwide and the health of people all over the world, if the diagnostic performance of CT images is improved, even if CT images cannot completely replace the nucleic acid detection method of COVID-19, CT images information can contribute to treatment decisions based on more accurate and reproducible data. Other benefits include reduced need for CT scans, which reduces delays, radiation levels, and costs. Consequently, in the field of medical image analysis, more and more imaging-based artificial intelligence diagnostic systems were developed to combat COVID-19. Those systems can be roughly divided into several categories: segmentation-based [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], classification-based [22], [23], [24], [25], [26], and follow-up and prognosis [27]. Those methods so that categorize the population through a computer-aided diagnosis system based on CT classification, which can reduce the medical burden of the hospital, make effective use of limited medical resources, and enable effective treatment of critically ill patients, thereby reducing the spread of disease. In terms of image-based disease diagnosis, some valuable works should be introduced, for example, the related works for multi-modal disease diagnosis are as follows [28], [29]. Additionally, Zhou et al. [30] proposed a novel hybrid-fusion network (Hi-Net) for multi-modal MR image synthesis, which learned mapping from multi-modal source images (i.e., existing modalities) to target images (i.e., missing modalities).
Currently, people actively explored the method of predicting whether COVID-19 is positive based on deep learning CT and achieved gratifying results [31], [32], [33], [34], [35], [36], [37]. Wang and Wong [32] developed a platform called COVID-Net for identification of COVID-19, and achieved good sensitivity. Sun et al. [22] leveraged a deep forest-based method to learn some high-level features from chest CT for the diagnosis of COVID-19. Di et al. [38] developed a hypergraph learning-based method for the identification of COVID-19 with CT imaging. He et al. [39] proposed a multi-task multi-instance learning framework to jointly assess the severity of the COVID-19 patients and segment the lung lobe in chest CT images.
Based on previous work and inspired by their early success, the purpose of this paper is to evaluate the feasibility of deep neural networks in COVID-19 assisted-diagnosis. Unfortunately, the current public health emergency, which has caused great difficulties in collecting a large set of precise data for training neural networks. Under the current grim circumstances, COVID-19 patients occupy a large degree of medical resources (clinicians and radiologists), therefore, they are unlikely to have time to collect and annotate a large number of COVID-19 CT scans. Consequently, one of the main concentration of this paper is to develop a deep learning-based method such that can obtain the desired diagnostic effect for COVID-19 from CT scans, in the case of limited training samples.
To this end, in this paper, we propose a novel transfer learning-based method with a small-size of medical image data for the classification between COVID-19 with Non-COVID-19. Transfer Learning (TL) is not a strange thing for human beings, which is the ability to learn by analogy. For example, If we have learned to ride a bicycle, learning to ride a motorcycle becomes very simple for us. If we have learned to play badminton, learning to play tennis is not that difficult. However, for computers, the TL is a technology that allows the existing model algorithms to be slightly adjusted to be applied to a new field or function. Regarding TL, the following researchers have done several valuable works, such as Long et al. [40] developed a conditional adversarial domain adaptation focused on discriminative information conveyed in the classifier predictions. Also, Wang et al. [41] presented visual domain adaptation with manifold embedded distribution alignment. Furthermore, Liu [42] developed an unsupervised approach for heterogeneous domain adaptation. According to the scenarios of TL, which can be split into three main categories: inductive (the source and target domains have different learning tasks), transductive (the source domain and target domain are different and the learning tasks are the same), and unsupervised TL (neither the source domain nor the target domain has labels). According to feature space, the TL can be split into two main categories: homogeneous and heterogeneous TL. Usually, in terms of homogeneous TL, the feature spaces of the data for the source and target domains are described by the same attributes, but space has the same dimension. This plays a crucial role in shortening the difference in the data distribution between domains when the data are transmitted cross-domain [43], [44]. Currently, several references address this problem. In contrast, in terms of heterogeneous TL, the feature spaces from the source and target domain are nonequivalent, moreover, they even are usually non-overlapping, also, the dimensions of the feature spaces may be different. Therefore, the key role of this approach is not only to deal with the gaps in knowledge transfer but to handle the differences in data distribution of cross-domain. Hence, this situation is more challenging, since few same modal or features can be shared. From the perspective of transforming the feature spaces, heterogeneous TL can be roughly divided into two categories: symmetric and asymmetric transformation [43], [44]. More currently, with the development of modern technology, TL has made new developments, such as deep transfer learning (DTL) and multi-source domain TL have received widespread attention. The knowledge can be effectively transferred by the deep neural network is called DTL [45]. Das et al. [46] leveraged the DTL-based approach for detection of COVID-19 infection in chest X-rays. For the multi-source domain TL, Lu et al. [47] presented new methods for merging fuzzy rules from multiple domains for regression tasks. These integrated methods are possible to open a door for future transfer learning.
From the perspective of the authenticity of the data, we use the publicly-available COVID-19-CT dataset, which contains 349 CT images with clinical findings of 216 COVID-19 patient cases, it is built by He et al. [23]. However, the deep neural network model is a data-hungry method. When training on a small-sized dataset, it is easy to produce the risk of overfitting. In typical transfer learning, researchers usually divide the data for transfer learning into two categories, one is source data and the other is target data. Source data refers to additional data and is not directly related to the task to be solved, while target data is data directly related to the task. Source data is often huge, and target data is often small. How to make good use of source data to help and even improve the performance of the model on target data is the issue to be considered for transfer learning. Furthermore, the purpose of transfer learning is using the knowledge for new and relevant tasks under the premise of obtaining certain additional data or the existence of an existing model. Additionally, transfer learning allows the domains, tasks, and distributions used in training and testing to be different. Based on the advantages of transfer learning and to make up for the data deficiency, we thought of transfer learning aims to build a good classifier use a small-sized dataset.
The main contributions of this paper are summarized three-fold as follows:
-
1.
A transfer learning-based framework is proposed for the identification of COVID-19 with limited CT imaging.
-
2.
The proposed method is evaluated on the publicly-available COVID19-CT dataset, which contains 349 CT images with clinical findings of 216 COVID-19 patient cases. Experimental results display that our method achieves good performance compared with several state-of-the-art methods.
-
3.
Through extensive experiments and comprehensive research aims to explore the consequences of transfer learning-based method for recognition of COVID-19 diagnosis and provides clinically interpretable saliency maps, which are useful for patient treatment and make the medical resources more effective and reasonable application.
The rest of the paper is organized as follows, in Section 2, we first present some related works for the diagnosis of COVID-19. Furthermore, we introduce the materials utilized in this study and the proposed transfer learning for recognition of COVID-19 in Section 3. After that, in Section 4, we describe the experimental settings, experimental results, the limitations of the current study, and present several future research directions. Finally, we summarize this paper in Section 5.
2. Related work
In this section, we briefly review the most relevant studies from the following three aspects: (1) prediction-based learning method for identification of COVID-19, (2) segmentation-based methods for identification of COVID-19 with CT imaging, and (3) classification-based diagnosis of COVID-19 patients.
2.1. Prediction-based learning method
X-rays can provide effective information for the computer-assisted diagnosis of COVID-19. Several deep learning methods have been proposed for the prediction of CT images with COVID-19. For instance, Wang and Wong [32] proposed a deep network named COVID-Net, for the detection of COVID-19 cases from chest radiography images. This paper open-sourced a deep convolutional neural network designed to detect COVID-19 cases from chest X-ray images, and the chest radiographic image dataset used to train COVID-Net is named COVIDx and contains 5941 back-to-front chest radiographic images of 2839 patients from two open databases. The paper also analyzes how COVID-Net uses interpretable methods to make predictions, intending to gain an in-depth understanding of the key factors associated with COVID cases, which can help clinicians to perform better screening. Open data and codes help to further develop high-precision and practical deep learning solutions for the detection of COVID-19 cases. Di et al. [38] proposed an uncertainty vertex-weighted hypergraph learning method to identify COVID-19 using CT images.
2.2. Segmentation-based methods for identification of COVID-19 with CT imaging
Segmentation of the lung or lung lobe has been seen as a common pre-requisite procedure for COVID-19 identification performance. To improve the diagnostic accuracy of COVID-19, many scholars have done a lot of research on the segmentation of the lung or lung lobe. For example, to help detect infections early and assess the disease progression of COVID-19, He et al. [39] proposed a synergistic learning framework for automated severity assessment of COVID-19 in 3D CT images. Selvan et al. [48] used lung segmentation from chest X-rays using the variational data imputation method aimed at automated risk scoring of COVID-19 from CXRs. Oh, et al. [15] proposed a patch-based convolutional neural network approach for COVID-19 diagnosis by learning COVID-19 features on CXR using limited training data sets. Moreover, U-net [49], as a classic successful model for medical image segmentation, the related works of using U-net to predict COVID-19 are presented in [50], [51], [52], and several variants of U-Net have been applied to the diagnosis or severity assessment of COVID-19 [53], [54], [55], [56].
2.3. Classification-based diagnosis of COVID-19 patients
Sun et al. [22] developed a deep-forest-based identification method for COVID-19, they used the deep forest to learn the high-level representation of location-based-specific features, meanwhile, an adaptive feature selection operation is employed to reduce the redundancy of features. He et al. [23] developed sample-efficient deep learning methods to accurately diagnose COVID-19 from CT scans and claimed that they built the largest of publicly-available CT-dataset for COVID-19.
3. Material and method
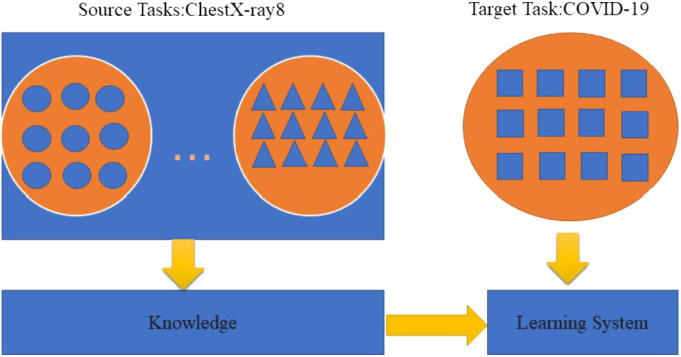
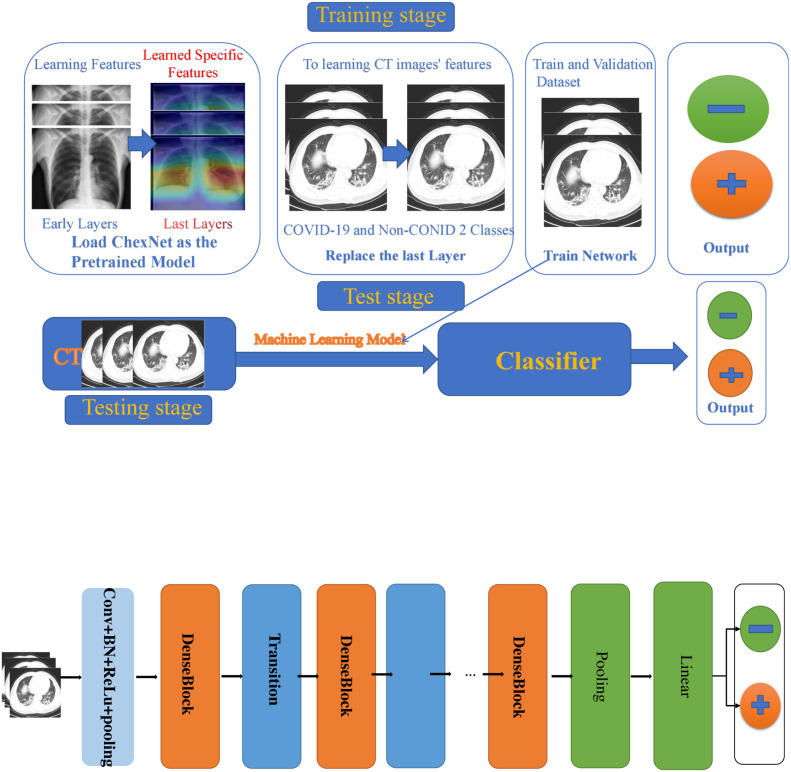
First of all, in this section, we will introduce the dataset used in our investigation. Furthermore, we present the implementation details for our proposed transfer learning for COVID-19 classification based on CT images. About the learning processing of transfer learning we can see from Fig. 3.
Fig. 3.
Learning processing of transfer learning.
3.1. Dataset description
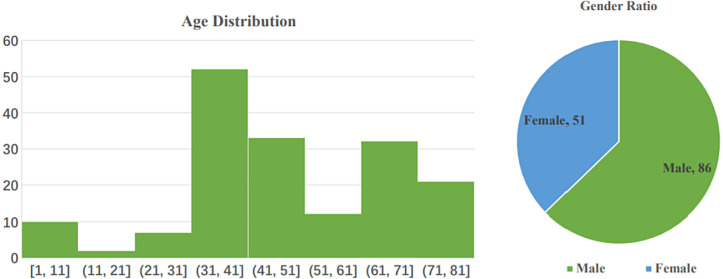
COVID-CT-Dataset, which is an open-sourced dataset provided by Zhao and Xie [57]. They claimed that it is the largest COVID-19-CT dataset that is publicly available to date. They collected 760 preprints about COVID-19 from bioRxiv3 and medRxiv2 from Jan 19th to Mar 25th. According to the original description of the database established by them, numerous of these preprints contain the cases of COVID-19 and the CT images in their report. The authors used the python package (PyMuPDF4) to extract the CT images from these preprints, after that, they manually selected all CT images, then determine whether the CT is positive. For each CT image, the authors collected the meta information extracted from the paper, such as patient age, gender, location, medical history, scan time, the severity of COVID-19, and radiology report [57]. Through various selections, they got 349 CT images labeled as being positive for COVID-19. The CT images have different sizes. The minimum, average, and maximum heights are 153, 491, and 1853. The minimum, average, and maximum widths are 124, 383, and 1485. These images are from 216 patient cases, For patients labeled with positive, 169 of them have age information and 137 of them have gender information [57]. From Fig. 4 we can learn the information on the age distribution and gender ratio of patients with COVID-19. Also, we can see those male patients are more than female ones. The CT images for training are extracted from papers. However, the test dataset and validation CT images are from hospital donations, and none of them is extracted from papers. The training data include the positive samples (the 349 COVID-19 CT images), and the negative samples (Non-COVID-19), the Non-COVID-19 CT images are collected by the authors from the MedPix5 database https://medpix.nlm.nih.gov/home, the LUNA6 dataset https://luna16.grand-challenge.org/, the radio media website https://radiopaedia.org/articles/covid-19-3, and PubMed Central (PMC) https://www.ncbi.nlm.nih.gov/pmc/, respectively.
Fig. 4.
(Left) Age distribution of COVID-19 patients. (Right) The gender ratio of COVID-19 patients. The ratio of male: female is 86: 51 [57].
To test the effectiveness of the training model, the validation set and the test set were built by the authors. The sources of original COVID-19 CTs are from the COVID-19 CT segmentation dataset http://medicalsegmentation.com/covid19, which contains 20 axial volumetric COVID-19 CT scans. The sources of original non-COVID-19 CTs are from the LUNA10 dataset https://luna16.grand-challenge.org/ and the radio media website https://radiopaedia.org/articles/covid-19-3, which contains 888 lung cancer CT scans from 888 patients and contains radiology images from 36559 patient cases, respectively. For detailed information about the database, please refer to [57].
3.2. Data preprocessing
Since the CT images have different sizes, therefore, they were resized to 224 × 224, we adopted the method of dataset splitting by the authors, namely, a training, validation, and a test set by patient IDs with a ratio of 0.6: 0.15: 0.25. Since the training data was limited, therefore, to avoid the overfitting problem, data augmentation strategies were employed including normalization processing random affine transformation, (mean=[0.485,0.456,0.406], std=[0.229,0.224,0.225]), the affine transformation was composed of rotation (0 to 10 angles), horizontal and vertical translations (0 to 10 angles).
3.3. Deep learning method
Recently, numerous works show that if a network performs satisfactory results, image-based classification also will have high performance on the COVID-19 task. Numerous network structures [58], [59], [60], [61], [62], [63], [64], [65] have been proposed for visual object recognition, and achieved gratifying performance. Both ResNets [58] and DensNets [59] architectures are successfully applied to in various image recognition tasks, and achieved outstanding performance.
Supervised and semi-supervised learning as the traditional machine learning algorithms aim to make predictions on the future data employing some models that are trained on previously collected labeled or unlabeled training data and assume that the labeled and distributions of unlabeled data are the same domains. Transfer learning, in contrast, allows the domains, tasks, and distributions used in training and testing to be different [66]. It is a technique that aims to transfer the knowledge from one or more source tasks to a target task when the latter has fewer training data. Transfer learning is only interested in the target task instead of source tasks. Consequently, a typical approach is to pre-train a deep learning algorithm, which is used for feature extraction on large datasets in the source tasks by fitting the human-annotated labels therein, then fine-tune this pre-trained network on the target task [67].
3.4. Predefined models
Our pre-defined models are as follows, ResNet34 [58], ResNet50 [58], resNet101 [58], DensNet121 [59], and the latest Network named ResNest [68]. We did a lot of training experiments on several pre-defined network frameworks, and finally chose the CheXNet [69] trained using the ChestX-ray14 dataset [70], which contains 112,120 frontal chest X-rays of 14 different chest diseases (including Pneumonia) that are individually labeled. CheXNet [69] is a 121-layer convolutional neural network. Its input is a chest X-ray, the output is the probability of Pneumonia and a heat map (heatmap), which is used to locate the image area that best indicates Pneumonia. Compare with other predefined network frameworks, CheXNet has done a lot of training on the large ChestX-ray14 dataset and achieved good performance. Consequently, we choose CheXNet as a pre-train model. About the framework of the proposed method, please refer to Fig. 5.
Fig. 5.
The modified CheXNet for diagnosing disease COVID-19.
3.5. Training and testing procedures
The COVID-19 classification deep network software was developed based on the Pytorh framework https://pytorch.org/. The network was trained for 100 epochs using SGD (stochastic gradient descent) with the momentum [71] of 0.9, a base learning rate of 0.05, and the weight-decay of 1e−5. Our classification network aims to discriminate against the COVID-19 and Non-COVID-19 from the lung CT images. So we adopted to extend DensNet121 to perform the COVID-19 classification task. And the training objective is as follow,
| (1) |
Since the Pneumonia detection can be seen as a binary classification problem, therefore, the binary cross-entropy loss function was exploited to compute the loss between predicted-labels and the ground-truth labels, where the input is a lung CT images and the output is a binary label indicating the presence or absence of Pneumonia. The formulation of the loss function is as follow,
| (2) |
here, is the probability that the network assigned to label .
Our pre-defined models (PM) are as follows, PM={ResNet34, ResNet50, ResNet101, Resnest, DenseNet}, PM refers to a set of pre-defined models, then each model in PM is fine-tuned on the COVID-19 dataset , refers to images of input, the size of each is 224 × 224, refers to the corresponding labels (COVID-19, Non-COVID-19), the base learning rate is 0.005, which is used to updates the weights of our model, where the . the dataset consists of three parts, namely, the training set , validate set , and test set , the batch-size of the training set is 8, then by iteratively optimizing (fine-tuning) the networks to reduce the objective loss function:
| (3) |
where refer to the deep model that predicts class for input and the given weight . Let layer be a convolutional layer, then we can compute the input layer as follows:
| (4) |
where , refer to a bias matrix, weight matrix, respectively. After that, the activation function as follows can be applied to the convolutional layer:
| (5) |
The common activation functions are sigmoid, tanh, rectified linear units (), and so on. In our method, we use function as the activation function, namely,
| (6) |
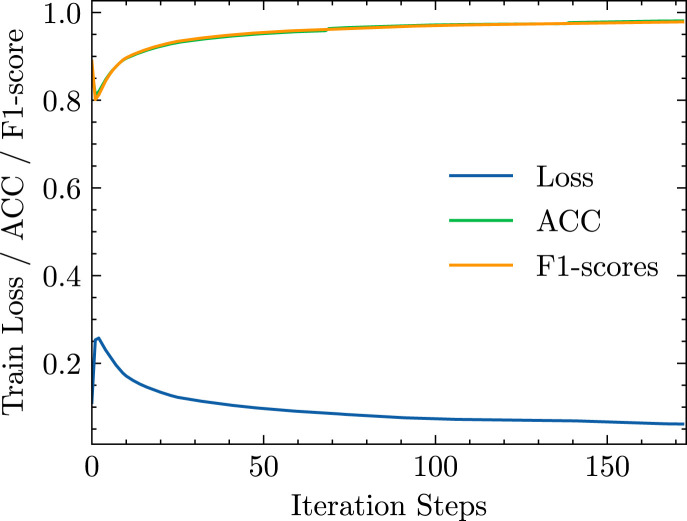
During the procedure of testing, data augmentation strategies were not used, and we test the model one time after training 1 epoch. After that, the predicted probabilities of all COVID-19 cases and the corresponding true labels were accumulated for statistical analysis. The loss function training history, ACC, and F1-score are shown in Fig. 6.
Fig. 6.
The history of training loss, ACC, and F1-score.
About the transfer parameters from the pre-train model, since the original CheXNet is a multi-classification task, it contains 14 categories, which are Atelectasis, Cardiomegaly, Effusion, Infiltration, Mass, Nodule, Pneumonia, Pneumothorax, Consolidation, Edema, Emphysema, Fibrosis, Pleural Thickening, and Hernia, respectively, they replaced the final fully-connected layer of Densenet-121 with a single output unit, and then they applied the nonlinear sigmoid function to output an image containing the probability of pneumonia. As COVID-19 is a two-class problem, we change the neurons in the output layer to two neurons, and the weights of the neurons are initialized by Kaiming’s initialization [72].
Table 1.
Dataset split.
| Class | Train | Val | Test | |
|---|---|---|---|---|
| COVID | 130 | 32 | 54 | |
| Patients | Non-COVID | 105 | 24 | 42 |
| COVID | 191 | 60 | 98 | |
| Image | Non-COVID | 234 | 58 | 105 |
4. Experiments
In this section, we will describe more details about experimental setup and results while testing the performance of our method.
4.1. Evaluation metrics
In our experiments, five criteria are employed to evaluate the COVID-19 identification performance, and the definitions are shown as follows. We use five types of commonly used metrics to evaluate the classification performance achieved by different methods in the classification task.
-
1.
Accuracy (ACC): ACC measures the proportion of samples that are correctly classified. .
-
2.
Recall: The Recall measures the proportion of actual positives that are correctly identified as such. This reflecting the misdiagnose proportion, and it is a critical evaluation metric in medical diagnostic applications. .
-
3.
Precision: Precision measures the proportion of detected positives that are truly positive. .
-
4.
F1-score: F1-score measures the harmonic mean of precision and recall. . where , , , and above represent True Positive, True Negative, False Positive, and False Negative, respectively.
-
5.
AUC: which is the area under the receiver operating characteristic curve showing how false positive rate increases as true positive rate increases. For all five metrics, the higher, the better.
4.2. Experimental settings
We did the experiments on the open-sourced dataset, which is named COVID-CT-Dataset. It consists of 349 COVID-19 CTs and 397 Non-COVID-19 CTs. Since the CT images have different sizes. Therefore, we resized the CT images to 224 × 224. For the division of the data set, we adapt to the division of the authors. The dataset contains three parts: a training set, a validation set, and a test set by patient IDs with a ratio of 0.6: 0.15: 0.25. About the method division for the dataset by the authors please refer to Table 1. About the data split information for the patient ID of Non-COVID-19, the number of train-id, test-id, and validate-id is 105, 42, and 24, respectively. Also, the data split information for the patient ID of COVID-19, the number of train-id, test-id, and validate-id is 191, 98, and 60, respectively. For more detail please refer to [57] and https://github.com/UCSD-AI4H/COVID-CT/blob/master/Data-split/.
We trained the deep neural network models on our equipped with a GeForce RTX 1080 super GPU. The deep network software was developed based on the Pytorh framework. In the procedures of training, the binary cross-entropy loss function and SGD optimization function with momentum were used, respectively. Batch normalization is used through all models, and the training BathSize is applied through all models.
4.3. Classification performance
We evaluate the COVID-19 vs. Non-COVID-19 classifications on the COVID-CT-Dataset. The detection results of COVID-19 were presented and analyzed using ACC and F1-score, and AUC. Because Precision and Recall are already reflected in the computing formula of F1-score, it is omitted here. Specifically, ACC and F1-score, and AUC are some critical indicators that are used to estimate the deep learning algorithm, for medical analysis. Table 2 shows the quantitative results (i.e. ACC, F1-score, and AUC) achieved by different methods. From Table 2, we also can observe that our deep classification model archives the best performance in terms of ACC, F1-score. However, it is worth noting that our AUC value is lower than SEDLCD model, it is also possible to be due to the number of GPUs and computer configuration, which caused our AUC to be lower than the SEDLCD (4 GPUs required).
For quantitative evaluation, as shown in Table 2, both conventional learning models and the proposed transfer learning methods achieve considerable improvement with the learned latent representation in terms of all two metrics. Table 2 also demonstrates the classification performance of the proposed method and compared methods. Table 2 shows that the proposed method reaches the best accuracy performance, up to 0.87. In terms of F1-score, our method also achieves the best performance, up to 0.86. However, it is worth noting that our AUC value is lower than sample-efficient deep learning for COVID-19 diagnosis based on CT scans (SEDLCD) model, it is also possible to be due to the number of GPUs and computer configuration, which caused our AUC to be lower than the SEDLCD (4 GPUs required).
Table 2.
Performance of COVID-29 vs Non-COVID-19 classification achieved by the historical models and our proposal method, refer to the mean and standard deviation, respectively.
| Method | ACC (%) | F1-score (%) | AUC (%) |
|---|---|---|---|
| SVM [73] | 0.61 ± 1.37 | 0.60 ± 1.69 | 0.62 ± 0.31 |
| LR [74] | 0.70 ± 1.67 | 0.68 ± 0.75 | 0.75 ± 1.21 |
| DANN [75] | 0.77 ± 1.27 | 0.78 ± 0.51 | 0.75 ± 0.52 |
| CheXNet [69] | 0.85 ± 1.64 | 0.84 ± 1.64 | 0.75 ± 0.78 |
| AFS-DF [22] | 0.86 ± 1.04 | 0.86 ± 0.91 | 0.87 ± 0.91 |
| SEDLCD [23] | 0.86 ± 0.00 | 0.85 ± 0.00 | 0.94 ± 0.00 |
| Ours | 0.87 ± 1.13 | 0.86 ± 0.83 | 0.75 ± 1.39 |
4.4. Limitations, challenging, and future work
There are still several limitations to our current study. Firstly, network design and optimizer may be further improved. For example, Laplacian smoothing-gradient descent [76] was not utilized to optimize the network. Secondly, although the data used in this study came from the large of publicly-available CT-dataset for COVID-19, because deep learning belongs to the data-hungry method, so a few hundred images are not enough for it, to be further cross-validated in the more publicly-available dataset. Thirdly, it is worth noting that our AUC value is lower than SEDLCD model, it is also possible be due to the number of GPUs and computer configuration, which caused our AUC to be lower than the SEDLCD (4 GPUs required). Moreover, we still face difficulties when trying to capture the features of COVID-19 and Non-COVID-19, since data dependence is one of the most serious problems in deep learning, insufficient training data is an inevitable problem in some special fields, especially in the early stage of the COVID-19 outbreak. Although TL relaxes the assumption that training data must be independent and identically distributed with test data, however, in the training stage, since insufficient data such that over-fitting is easy to occur, resulting in high accuracy in the training stage, and there is a certain gap in the accuracy of the test and the training phase. Data distribution is another problem, in future work, we will combine TL and model-driven methods to reduce over-fitting, thereby reducing the gap between training accuracy and test refinement, and improving the performance of the model. Also, more features (patient age, gender, location, medical history, scan time, the severity of COVID-19) will be considered in our proposed method aiming to improve further performance. Furthermore, such as the Laplacian smoothing optimizer [76] also will be leveraged to speed up network optimization, shorten training time, and improve classification accuracy.
5. Conclusion
In this paper, we propose a transfer learning-based deep method that aims to identify COVID-19 from Non-COVID-19 using relatively small-sized CT images. We confront one of the challenging issues that have great difficulties in collecting a large set of precise data for training neural networks. Our proposed method employs the characteristic of transfer learning that transfers the knowledge from one or more source tasks to a target task when the latter has fewer training data. CheXNet as the pre-trained network was used for the target task (COVID-19 classification) by fine-tuning the network weights on the small-sized dataset in the target task. Experimental results on a real COVID-19 CT image dataset demonstrate that our method achieves good performance in severity assessment of COVID-19 patients, compared with several state-of-the-art methods, except AUC.
CRediT authorship contribution statement
Chun Li: Conceptualization, Methodology, Software. Yunyun Yang: Writing - original draft, Validation. Hui Liang: Writing - review & editing. Boying Wu: Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
The code (and data) in this article has been certified as Reproducible by Code Ocean:https://help.codeocean.com/en/articles/1120151-code-ocean-s-verification-process-for-computational-reproducibility. More information on the Reproducibility Badge Initiative isavailable at www.elsevier.com/locate/knosys.
References
- 1.Sohrabi C., Alsafi Z., O’Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World Health Organization declares global emergency: A review of the 2019 novel Coronavirus (COVID-19) Int. J. Surg. (London, England) 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical 2019-nCoV pneumonia: Relationship to negative RT-PCR testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W.-J., Yang Y., Fayad Z., Jacobi A., Li K., Li S., Shan H. CT imaging features of 2019 novel Coronavirus (2019-nCoV) Radiology. 2020 doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li M., Lei P., Zeng B., Li Z., Yu P., Fan B., Wang C., Li Z., Zhou J., Hu S., et al. Coronavirus disease (COVID-19): spectrum of CT findings and temporal progression of the disease. Acad. Radiol. 2020 doi: 10.1016/j.acra.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020 doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li W., Cui H., Li K., Fang Y., Li S. Chest computed tomography in children with COVID-19 respiratory infection. Pediatric Radiol. 2020:1–4. doi: 10.1007/s00247-020-04656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai K., Tabata S., Ikeda M., Noguchi S., Kitagawa Y., Matuoka M., Miyoshi K., Tarumoto N., Sakai J., Ito T., et al. Clinical evaluation of an immunochromatographic IgM/IgG antibody assay and chest computed tomography for the diagnosis of COVID-19. J. Clin. Virol. 2020 doi: 10.1016/j.jcv.2020.104393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen R., Chen J., Meng Q.-t. Chest computed tomography images of early Coronavirus disease (COVID-19) Canad. J. Anaesthesia. 2020:1. doi: 10.1007/s12630-020-01625-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salehi S., Abedi A., Radmard A.R., Sorouri M., Gholamrezanezhad A. Chest computed tomography manifestation of Coronavirus disease 2019 (COVID-19) in patients with cardiothoracic conditions. J. Thorac. Imaging. 2020 doi: 10.1097/RTI.0000000000000531. [DOI] [PubMed] [Google Scholar]
- 10.Huang E.P.-C., Sung C.-W., Chen C.-H., Fan C.-Y., Lai P.-C., Huang Y.-T. Can computed tomography be a primary tool for COVID-19 detection? Evidence appraisal through meta-analysis. Crit. Care. 2020;24:1–3. doi: 10.1186/s13054-020-02908-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nie S., Han S., Ouyang H., Zhang Z. Coronavirus disease 2019-related dyspnea cases difficult to interpret using chest computed tomography. Respir. Med. 2020 doi: 10.1016/j.rmed.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., Xu Z., Zheng Z., et al. AI-Assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. MedRxiv. 2020 doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi X., Jiang Z., Yu Q., Shao C., Zhang H., Yue H., Ma B., Wang Y., Liu C., Meng X., et al. Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study. MedRxiv. 2020 doi: 10.21037/atm-20-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., Shi F., Gao Y., Liu J., Zhang J., et al. 2020. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. arXiv preprint arXiv:2005.03832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh Y., Park S., Ye J.C. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 16.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 17.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., Shen J., Shao L. 2020. Inf-net: Automatic COVID-19 lung infection segmentation from CT scans. arXiv preprint arXiv:2004.14133. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y.-H., Gao S.-H., Mei J., Xu J., Fan D.-P., Zhao C.-W., Cheng M.-M. 2020. JCS: An explainable COVID-19 diagnosis system by joint classification and segmentation. arXiv preprint arXiv:2004.07054. [DOI] [PubMed] [Google Scholar]
- 19.Muggeo V., Sottile G., Porcu M. 2020. Modelling COVID-19 outbreak: segmented regression to assess lockdown effectiveness. [Google Scholar]
- 20.Amyar A., Modzelewski R., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19: Classification and segmentation. medRxiv. 2020 doi: 10.1016/j.compbiomed.2020.104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alom M.Z., Rahman M., Nasrin M.S., Taha T.M., Asari V.K. 2020. COVID_MTNet: COVID-19 detection with multi-task deep learning approaches. arXiv preprint arXiv:2004.03747. [Google Scholar]
- 22.Sun L., Mo Z., Yan F., Xia L., Shan F., Ding Z., Shao W., Shi F., Yuan H., Jiang H., et al. 2020. Adaptive feature selection guided deep forest for COVID-19 classification with chest CT. arXiv preprint arXiv:2005.03264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E., Xie P. Sample-efficient deep learning for COVID-19 diagnosis based on CT scans. MedRxiv. 2020 [Google Scholar]
- 24.Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., Jiang B. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395(10228) doi: 10.1016/S0140-6736(20)30558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K., Fang Y., Li W., Pan C., Qin P., Zhong Y., Liu X., Huang M., Liao Y., Li S. CT image visual quantitative evaluation and clinical classification of Coronavirus disease (COVID-19) Eur. Radiol. 2020:1–10. doi: 10.1007/s00330-020-06817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi F., Xia L., Shan F., Wu D., Wei Y., Yuan H., Jiang H., Gao Y., Sui H., Shen D. 2020. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. arXiv preprint arXiv:2003.09860. [DOI] [PubMed] [Google Scholar]
- 27.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 28.Zhou T., Thung K.-H., Zhu X., Shen D. Effective feature learning and fusion of multimodality data using stage-wise deep neural network for dementia diagnosis. Hum. Brain Mapp. 2019;40(3):1001–1016. doi: 10.1002/hbm.24428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou T., Liu M., Thung K.-H., Shen D. Latent representation learning for Alzheimer’s disease diagnosis with incomplete multi-modality neuroimaging and genetic data. IEEE Trans. Med. Imaging. 2019;38(10):2411–2422. doi: 10.1109/TMI.2019.2913158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou T., Fu H., Chen G., Shen J., Shao L. Hi-net: hybrid-fusion network for multi-modal MR image synthesis. IEEE Trans. Med. Imaging. 2020 doi: 10.1109/TMI.2020.2975344. [DOI] [PubMed] [Google Scholar]
- 31.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Zhang H., Ji W., Bernheim A., Siegel E. 2020. Rapid AI development cycle for the Coronavirus (COVID-19) pandemic: Initial results for automated detection & patient monitoring using deep learning CT image analysis. arXiv preprint arXiv:2003.05037. [Google Scholar]
- 32.Wang L., Wong A. 2020. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narin A., Kaya C., Pamuk Z. 2020. Automatic detection of Coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. Covidx-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. arXiv preprint arXiv:2003.11055. [Google Scholar]
- 35.Apostolopoulos I.D., Aznaouridis S.I., Tzani M.A. Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. J. Med. Biol. Eng. 2020:1. doi: 10.1007/s40846-020-00529-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farooq M., Hafeez A. 2020. COVID-Resnet: A deep learning framework for screening of COVID-19 from radiographs. arXiv preprint arXiv:2003.14395. [Google Scholar]
- 37.Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. 2020. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. arXiv preprint arXiv:2004.02696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di D., Shi F., Yan F., Xia L., Mo Z., Ding Z., Shan F., Li S., Wei Y., Shao Y., et al. 2020. Hypergraph learning for identification of COVID-19 with CT imaging. arXiv preprint arXiv:2005.04043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., Shi F., Gao Y., Liu J., Zhang J., et al. 2020. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. arXiv preprint arXiv:2005.03832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long M., Cao Z., Wang J., Jordan M.I. NeurIPS. 2018. Conditional adversarial domain adaptation. [Google Scholar]
- 41.J. Wang, W. Feng, Y. Chen, H. Yu, M. Huang, P.S. Yu, Visual domain adaptation with manifold embedded distribution alignment, in: Proceedings of the 26th ACM International Conference on Multimedia, 2018.
- 42.Liu F., Zhang G., Lu J. Heterogeneous domain adaptation: An unsupervised approach. IEEE Trans. Neural Netw. Learn. Syst. 2020;31:5588–5602. doi: 10.1109/TNNLS.2020.2973293. [DOI] [PubMed] [Google Scholar]
- 43.Day O., Khoshgoftaar T.M. A survey on heterogeneous transfer learning. J. Big Data. 2017;4(1):29. [Google Scholar]
- 44.Weiss K., Khoshgoftaar T.M., Wang D. A survey of transfer learning. J. Big data. 2016;3(1):9. [Google Scholar]
- 45.Tan C., Sun F., Kong T., Zhang W., Yang C., Liu C. International Conference on Artificial Neural Networks. Springer; 2018. A survey on deep transfer learning; pp. 270–279. [Google Scholar]
- 46.Das N.N., Kumar N., Kaur M., Kumar V., Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. Irbm. 2020 doi: 10.1016/j.irbm.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu J., Zuo H., Zhang G. Fuzzy multiple-source transfer learning. IEEE Trans. Fuzzy Syst. 2019;28(12):3418–3431. [Google Scholar]
- 48.Selvan R., Dam E.B., Rischel S., Sheng K., Nielsen M., Pai A. 2020. Lung segmentation from chest X-rays using variational data imputation. arXiv preprint arXiv:2005.10052. [Google Scholar]
- 49.Ronneberger O., Fischer P., Brox T. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 50.Qi X., Jiang Z., Yu Q., Shao C., Zhang H., Yue H., Ma B., Wang Y., Liu C., Meng X., et al. Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study. medRxiv. 2020 doi: 10.21037/atm-20-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv. 2020 [Google Scholar]
- 52.Huang L., Han R., Ai T., Yu P., Kang H., Tao Q., Xia L. Serial quantitative chest CT assessment of COVID-19: deep-learning approach. Radiol.: Cardiothoracic Imaging. 2020;2(2) doi: 10.1148/ryct.2020200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shan+ F., Gao+ Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. 2020. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655. [Google Scholar]
- 54.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., Xu Z., Zheng Z., et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. medRxiv. 2020 doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. Unet++: A nested u-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milletari F., Navab N., Ahmadi S.-A. 2016 Fourth International Conference on 3D Vision, 3DV. IEEE; 2016. V-net: Fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- 57.Zhao J., Zhang Y., He X., Xie P. 2020. COVID-CT-Dataset: a CT scan dataset about COVID-19. arXiv preprint arXiv:2003.13865. [Google Scholar]
- 58.K. He, X. Zhang, S. Ren, J. Sun, Deep residual learning for image recognition, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016, pp. 770–778.
- 59.G. Huang, Z. Liu, L. Van Der Maaten, K.Q. Weinberger, Densely connected convolutional networks, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 4700–4708.
- 60.C. Ledig, L. Theis, F. Huszár, J. Caballero, A. Cunningham, A. Acosta, A. Aitken, A. Tejani, J. Totz, Z. Wang, et al. Photo-realistic single image super-resolution using a generative adversarial network, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 4681–4690.
- 61.Simonyan K., Zisserman A. 2014. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. [Google Scholar]
- 62.Srivastava R.K., Greff K., Schmidhuber J. 2015. Highway networks. arXiv preprint arXiv:1505.00387. [Google Scholar]
- 63.Mazumder P., Singh P., Namboodiri V. ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing, ICASSP. IEEE; 2020. Cpwc: Contextual point wise convolution for object recognition; pp. 4152–4156. [Google Scholar]
- 64.Thermos S., Papadopoulos G.T., Daras P., Potamianos G. Deep sensorimotor learning for RGB-D object recognition. Comput. Vis. Image Underst. 2020;190 [Google Scholar]
- 65.Coquin D., Boukezzoula R., Benoit A., Nguyen T.L. Assistance via IoT networking cameras and evidence theory for 3D object instance recognition: Application for the NAO humanoid robot. Internet of Things. 2020;9 [Google Scholar]
- 66.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2009;22(10):1345–1359. [Google Scholar]
- 67.R. Girshick, J. Donahue, T. Darrell, J. Malik, Rich feature hierarchies for accurate object detection and semantic segmentation, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2014, pp. 580–587.
- 68.Zhang H., Wu C., Zhang Z., Zhu Y., Zhang Z., Lin H., Sun Y., He T., Mueller J., Manmatha R., et al. 2020. Resnest: Split-attention networks. arXiv preprint arXiv:2004.08955. [Google Scholar]
- 69.Rajpurkar P., Irvin J., Zhu K., Yang B., Mehta H., Duan T., Ding D., Bagul A., Langlotz C., Shpanskaya K., et al. 2017. Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint arXiv:1711.05225. [Google Scholar]
- 70.X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri, R.M. Summers, Chestx-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2017, pp. 2097–2106.
- 71.Qian N. On the momentum term in gradient descent learning algorithms. Neural Netw. 1999;12(1):145–151. doi: 10.1016/s0893-6080(98)00116-6. [DOI] [PubMed] [Google Scholar]
- 72.K. He, X. Zhang, S. Ren, J. Sun, Delving deep into rectifiers: Surpassing human-level performance on imagenet classification, in: Proceedings of the IEEE International Conference on Computer Vision, 2015, pp. 1026–1034.
- 73.Gunn S.R., et al. Support vector machines for classification and regression. ISIS Tech. Rep. 1998;14(1):5–16. [Google Scholar]
- 74.Liaw A., Wiener M., et al. Classification and regression by random forest. R News. 2002;2(3):18–22. [Google Scholar]
- 75.Ganin Y., Ustinova E., Ajakan H., Germain P., Larochelle H., Laviolette F., Marchand M., Lempitsky V. Domain-adversarial training of neural networks. J. Mach. Learn. Res. 2016;17(1):2030–2096. [Google Scholar]
- 76.Osher S., Wang B., Yin P., Luo X., Barekat F., Pham M., Lin A. 2018. Laplacian smoothing gradient descent. arXiv preprint arXiv:1806.06317. [Google Scholar]