Abstract
Excessive alcohol intake is a well-known modifiable risk factor for many cancers. It is still unclear whether genetic variants or single nucleotide polymorphisms (SNPs) can modify alcohol intake’s impact on prostate cancer (PCa) aggressiveness. The objective is to test the alcohol–SNP interactions of the 7501 SNPs in the four pathways (angiogenesis, mitochondria, miRNA, and androgen metabolism-related pathways) associated with PCa aggressiveness. We evaluated the impacts of three excessive alcohol intake behaviors in 3306 PCa patients with European ancestry from the PCa Consortium. We tested the alcohol–SNP interactions using logistic models with the discovery-validation study design. All three excessive alcohol intake behaviors were not significantly associated with PCa aggressiveness. However, the interactions of excessive alcohol intake and three SNPs (rs13107662 [CAMK2D, p = 6.2 × 10−6], rs9907521 [PRKCA, p = 7.1 × 10−5], and rs11925452 [ROBO1, p = 8.2 × 10−4]) were significantly associated with PCa aggressiveness. These alcohol–SNP interactions revealed contrasting effects of excessive alcohol intake on PCa aggressiveness according to the genotypes in the identified SNPs. We identified PCa patients with the rs13107662 (CAMK2D) AA genotype, the rs11925452 (ROBO1) AA genotype, and the rs9907521 (PRKCA) AG genotype were more vulnerable to excessive alcohol intake for developing aggressive PCa. Our findings support that the impact of excessive alcohol intake on PCa aggressiveness was varied by the selected genetic profiles.
Keywords: alcohol intake, SNP interaction, prostate cancer
1. Introduction
Prostate cancer (PCa) is the most common cancer, and the second leading cause of cancer deaths among men in the United States [1] and the sixth cause of cancer-related deaths among men in highly developed regions worldwide [2]. It is commonly accepted that a complex interplay between genetic and environmental factors plays a role in cancer prognosis [3,4]. Both environmental and genetic factors play essential roles in PCa etiology [5,6,7]. Despite many research efforts, the major proportion of the familial risk of PCa remains unknown [8].
Alcohol consumption is considered one of the most important and modifiable environmental risk factors for human cancers, such as liver and colorectal cancers [9]. Alcohol intake is also one of the potentially modifiable factors associated with PCa aggressiveness and prognosis. Among adult Americans, 55.3% consumed alcohol, while 26.5% and 6.6% reported binge and heavy drinking, respectively, in the past month based on the 2018 National Survey on Drug Use and Health [10]. PCa patients have similar excessive alcohol intake compared with non-cancer individuals [11]. The heavy alcohol intake rate in 2019 is similar for PCa patients (4.2%) and non-cancer individuals (5.2%), and the frequent binge alcohol drinking is 4.9% and 4.2% for PCa patients and non-cancer individuals, respectively. Frequent binge drinking was defined as ≥4 binge drinking days (≥5 for males or ≥4 for females of alcohol drinks per day) per month in the past year [11]. Demoury et al. conducted a large population-based case-control study and found high beer intake was associated with PCa aggressiveness [12]. Additionally, a meta-analysis study revealed alcohol consumption to be associated with an increased risk of PCa aggressiveness [13]. However, associations between alcohol use and PCa aggressiveness remain inconclusive [14,15].
Genetic variation has been recognized as a risk factor for PCa aggressiveness. Approximately 40 single nucleotide polymorphisms (SNPs), inherited genetic variants, were suggested to be associated with PCa progression in genome-wide association studies (GWASs) [16]. Several SNPs in alcohol-metabolizing genes (ALDH1A2 and ALDH1B1) were associated with PCa specific mortality [17]. Studies on gene–environment interactions can help discover new environmental factors impacting diseases and identify novel genes and high-risk individuals (23–25). Genetic variants may influence alcohol’s impact on PCa aggressiveness. Several pathogenetic mechanisms for alcohol-induced carcinogenesis have been reported [18]. However, the effects of crosstalk between alcohol use and inherited genetic variants on PCa aggressiveness remain understudied.
Accumulating evidence suggests that interplay among angiogenesis, mitochondria, miRNA, and androgen metabolism-related pathways may play a critical role in PCa aggressiveness [19,20,21]. For crosstalk between angiogenesis and androgen, expression of androgen, epigenetic factors, and oxygen level in tumor micro-environment regulates angiogenesis, which leads to metastatic PCa [22]. Recurrent patients who had therapies targeting both angiogenesis and androgen resulted in increasing survival [23]. In addition, androgens influence angiogenesis in androgen-sensitive prostate tumors [24]. These findings suggested the relationship between androgen and angiogenesis and how they interact in aggressive PCa patients. Genes involved in the androgen metabolism pathway also lead to oncogenic metabolic phenotypes, such as mitochondrial respiration and cell proliferation in PCa cells. In addition, androgen repression in PCa cells decreases mitochondrial activity [25]. We and others have reported that miRNAs (such as miR-221, miR-222, and miR-155) are involved in the regulation of various aspects of angiogenesis [20] and progression of PCa [26]. Thus, this study aims to evaluate alcohol intake and explore the alcohol–SNP interactions of the SNPs in the four PCa-related pathways (angiogenesis, mitochondria, miRNA, and androgen metabolism-related pathways) associated with PCa aggressiveness.
2. Materials and Methods
2.1. Study Population
This study included 3306 PCa patients with European ancestry from the Collaborative Oncological Gene–Environment Study (COGS) in the Prostate Cancer Association Group to investigate Cancer Associated Alterations in the Genome (PRACTICAL) Consortium. In this study, PCa aggressiveness is defined as Gleason score ≥8, Prostate Specific Antigen (PSA) >100, distant disease stage at diagnosis, or death from PCa. Ethnic groups were determined based on ~37,000 uncorrelated markers that passed quality control, including ~1000 selected as ancestry informative markers. European ancestry was defined as ≥85% European component by multidimensional scaling using the three HapMap 2 populations [27]. We randomly assigned half of the participants to the discovery and validation set with a sample size of 1636 and 1670, respectively. The combined set is the sum of the discovery and validation set. There were seven study sites, and we combined the small sites based on geographic region into five integrated study sites (Table S1) for modeling. Details of the PRACTICAL Consortium study have been previously reported [27]. This project was approved by the Louisiana State University Health Sciences Center Institutional Review Boards (IRB #9338).
2.2. SNP Selection
This study included 7501 SNPs in the four PCa-related pathways (angiogenesis, mitochondria, miRNA, and androgen metabolism) from the PRACTICAL COGS project. The SNPs with a minor allele frequency (MAF) <0.05 and call rates <95% were excluded. For each SNP, we let a lowercase letter ‘a’ to denote the minor (low frequency) allele and an uppercase ‘A’ to denote the major allele. Each SNP potentially has three genotype categories: homozygous major type (‘AA’), heterozygous type (‘Aa’), and homozygous minor type (‘aa’). The additive mode treated an SNP as a continuous variable by counting the number of minor alleles (AA = 0, Aa = 1, and aa = 2). The comparison of Aa/aa versus AA was performed for the dominant mode, and the comparison of aa versus AA/Aa was made for the recessive mode.
2.3. Alcohol Intake Behaviors
We evaluated three excessive alcohol intake behaviors: heavy alcohol intake, heavy beer intake, and high ethanol intake. Heavy alcohol intake was defined as ≥2 times alcohol consumption per day [28]. Heavy beer intake was defined as ≥1 time beer consumption per day [29]. High ethanol intake was defined as intake ≥30 g of alcohol (similar to the recommended limit of two servings) per day [30].
2.4. Statistical Analyses
The PCa patients’ alcohol intake and smoking behaviors by PCa aggressiveness (yes/no) and study sets (discovery, validation, and combined set) were summarized using descriptive statistics. We tested the 7501 candidate SNPs in the four target pathways. The selected SNPs, alcohol intake, and smoking behaviors associated with PCa aggressiveness were tested using the Chi-square test or Fisher’s exact test. In order to control for population substructure, principal component analysis was performed, with the first six principal components of population stratification, which was applied as suggested by the PRACTICAL study [27]. Logistic regressions were applied to evaluate alcohol intake associated with PCa aggressiveness adjusting for the study site, the first six principal components, and smoking status. For testing an alcohol–SNP interaction on PCa aggressiveness, the full interaction logistic model with one alcohol intake term, one SNP, and their interaction term was applied. For each SNP, three different inheritance modes (additive, dominant, and recessive) based on the minor allele were tested, and the results with the lowest p-value were selected.
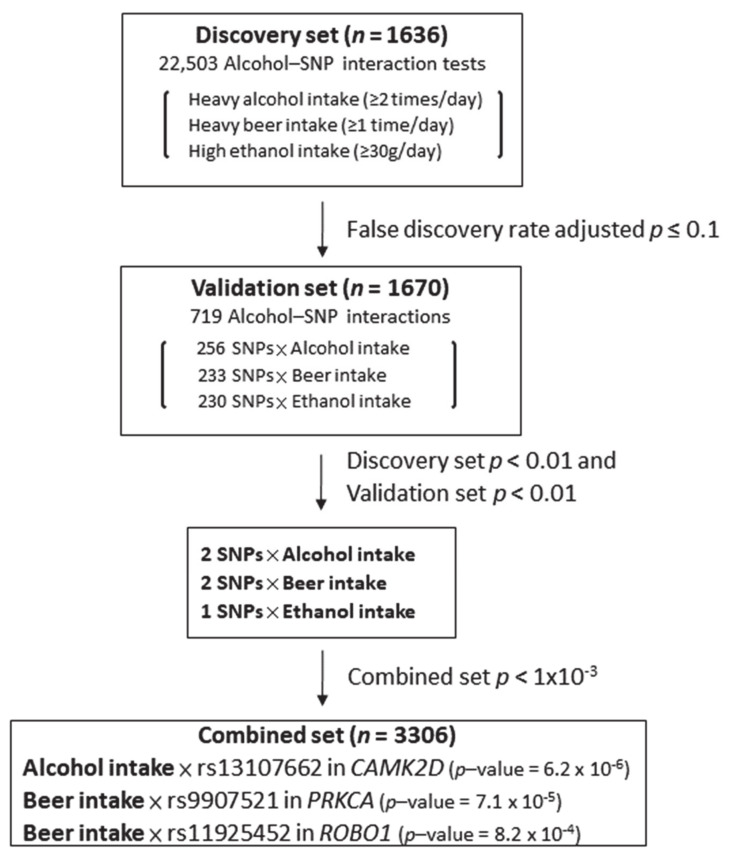
We applied the multi-stage approach (discovery and validation stage, Figure 1) to identify and verify significant alcohol–SNP interactions. In the discovery stage, we used the false discovery rate method to adjust for multiple comparisons [31]. First, alcohol–SNP interaction effect analyses associated with PCa aggressiveness were performed in the discovery set. Interactions with a false discovery rate adjusted p-value ≤ 0.10 in the discovery set were further tested in the validation set. Some interactions may have an opposite effect in the discovery and validation sets, so we retested the promising interactions (with p < 0.01 in both sets) in the combined set. We defined the significant and verified alcohol–SNP interactions with p < 0.01 in both discovery and validation sets and p < 0.001 in the combined set. For significant alcohol–SNP interactions, we performed subgroup analyses for evaluating associations between alcohol intake and PCa aggressiveness within the selected genotypes. We also reported the top alcohol–SNP interactions by expanding the p-value criteria in the validation set (p < 0.05). Smoking is a well-known environmental factor interacting with alcohol intake, so all models for testing alcohol effects, SNP individual effects, and alcohol–SNP interactions were adjusted for the study site, the first six principal components, and smoking. Data analyses were performed using SAS and SNPassoc R package.
Figure 1.
Process of identifying alcohol–single nucleotide polymorphism (SNP) interactions associated with prostate cancer aggressiveness. Note: p-values were based on the logistic model adjusting for study site, the first six principal components, and smoking status.
3. Results
The prevalence of PCa aggressiveness was 17.7%, 17.5%, and 17.9% in the combined, discovery, and validation sets, respectively. In the combined set (Table 1), 29.6%, 20.0%, and 28.2% of PCa patients reported heavy alcohol intake, heavy beer intake, and high ethanol intake, respectively, and 10% of them were current smokers. As shown in Table 1 and Table S2, there were no significant associations between these alcohol intake factors and PCa aggressiveness in the combined, discovery, and validation sets. The prevalence of PCa aggressiveness was the same (20.5%) in the combined set for those with and without heavy alcohol intake. The prevalence of PCa aggressiveness was similar (17.4–20.2%) regardless of beer and ethanol intake status. In contrast, smoking status was significantly associated with PCa aggressiveness in the validation set (p = 0.04), but not in the discovery and combined set. In the validation set, current smokers had a higher prevalence of PCa aggressiveness than non-smokers and former smokers (24.7% vs. 16.1% and 17.8%). Similarly, heavy alcohol intake, heavy beer intake, and high ethanol intake showed no significant associations with PCa aggressiveness in the discovery, validation, and combined set after adjusting for study site, the first six principal components, and smoking status (Table S2).
Table 1.
Alcohol intake and smoking status associated with prostate cancer (PCa) aggressiveness by study sets.
| Combined (n = 3306) PCa Aggressiveness (17.7%) |
Discovery (n = 1636) PCa Aggressiveness (17.5%) |
Validation (n = 1670) PCa Aggressiveness (17.9%) |
|||||
|---|---|---|---|---|---|---|---|
| Factors | Total N (%) |
Yes N (%) |
No N (%) |
Yes N (%) |
No N (%) |
Yes N (%) |
No N (%) |
| Heavy alcohol intake (≥2 times/day) | |||||||
| No Yes |
1066 (70.4) 448 (29.6) |
218 (20.5) 92 (20.5) |
848 (79.5) 356 (79.5) |
114 (21.8) 46 (20.8) |
410 (78.2) 175 (79.2) |
104 (19.2) 46 (20.3) |
438 (80.8) 181 (79.7) |
| Heavy beer intake (≥1 time/day) | |||||||
| No Yes |
1690 (80.0) 423 (20.0) |
341 (20.2) 75 (17.7) |
1349 (79.8) 348 (82.3) |
167 (20.2) 35 (16.6) |
659 (79.8) 176 (83.4) |
174 (20.1) 40 (18.9) |
690 (79.9) 172 (81.1) |
| High ethanol intake (≥30 g/day) | |||||||
| No Yes |
2375 (71.8) 931 (28.2) |
414 (17.4) 171 (18.4) |
1961 (82.6) 760 (81.6) |
205 (17.3) 81 (17.9) |
978 (82.7) 372 (82.1) |
209 (17.5) 90 (18.8) |
983 (82.5) 388 (81.2) |
| Smoking status 1 | |||||||
| Non-smoker Former smoker Current smoker |
1263 (38.6) 1679 (51.4) 327 (10.0) |
218 (17.3) 291 (17.3) 71 (21.7) |
1045 (82.7) 1388 (82.7) 256 (78.3) |
116 (18.4) 138 (16.8) 31 (18.8) |
515 (81.6) 683 (83.2) 134 (81.2) |
102 (16.1) 153 (17.8) 40 (24.7) |
530 (83.9) 705 (82.2) 122 (75.3) |
1p = 0.040 for smoking vs. PCa aggressiveness in the validation set; other factors associated with PCa aggressiveness were not significant (p-value > 0.05).
To test whether SNPs in the four pathways influenced the associations between excessive alcohol intake impact and PCa aggressiveness, we evaluated a total of 7501 alcohol–SNP interaction tests for each of the alcohol outcomes in the discovery set. As shown in Figure 1, there were 719 alcohol–SNP interactions (256 for heavy alcohol intake, 233 for heavy beer intake, and 230 for high ethanol intake) with a false discovery rate adjusted p-value ≤ 0.10 in the discovery set. These interactions were retested in the validation set. There were five alcohol–SNP interactions with a p < 0.01 in both the discovery and validation set (two for heavy alcohol intake, two for heavy beer intake, and one for high ethanol intake). Two of them had an opposite effect in the discovery and validation set, so these two interactions became insignificant in the combined set. Although none of the alcohol–SNP interactions reached the Bonferroni significance level (p < 2.2 × 10−6 = 0.05/22503), the top three alcohol–SNP interactions were validated and had p < 0.001 in the combined set: alcohol–rs13107662 (p = 6.2 × 10−6), beer–rs9907521 (p = 7.1 × 10−5), and beer–rs11925452 (p = 8.2 × 10−4).
As shown in Table 2, the interaction of rs13107662 (CAMK2D) and heavy alcohol intake was significantly associated with PCa aggressiveness (p = 6.2 × 10−6 in the combined, p = 2.1 × 10−4 discovery, and p = 0.003 validation set). The interaction between heavy beer intake and rs9907521 (PRKCA) was significantly associated with PCa aggressiveness (p = 7.1 × 10−5 in the combined set). In addition, the interaction of heavy beer intake and rs11925452 (ROBO1) was significantly associated with PCa aggressiveness (p = 8.2 × 10−4 in the combined set). For these three alcohol–SNP interactions, all of them selected the additive mode, which means the impact of alcohol intake on PCa aggressiveness varied by the number of the minor allele. For the individual SNP effects for the three SNPs involved in these interactions, only rs13107662 in CAMK2D was significantly associated with PCa aggressiveness (p = 0.003 in the combined set, Table 3). The PCa patients with the rs13107662 GG genotype had a higher chance of PCa aggressiveness than those with the AA or AG genotypes (30.0%, 19.8%, and 18.5% for GG, AA, and AG genotypes, respectively). The other two SNPs (rs9907521 and rs11925452) did not have significant individual SNP effects associated with PCa aggressiveness (p = 0.640 and 0.514 in the combined set, respectively).
Table 2.
Three verified alcohol–single nucleotide polymorphism (SNP) interactions associated with prostate cancer aggressiveness.
| Alcohol-SNP Interaction | Combined | Discovery | Validation | |||
|---|---|---|---|---|---|---|
| p-Value 1 | Mode 2 | p-Value 1 | Mode 2 | p-Value 1 | Mode 2 | |
| Heavy alcohol Intake–rs13107662 (CAMK2D) | 6.2 × 10−6 | Add | 2.1 × 10−4 | Add | 0.003 | Rec 4 |
| Heavy beer Intake–rs9907521 (PRKCA) | 7.1 × 10−5 | Add | 0.005 | Add | 0.005 | Add |
| Heavy beer Intake–rs11925452 (ROBO1) | 8.2 × 10−4 | Add | 0.003 | Rec 3 | 0.002 | Add |
1p-value of alcohol–SNP interactions were based on logistic regression adjusted for study site, first six principal components, and smoking status. 2 Add: additive, Rec: recessive mode; 3 p = 0.073 for the additive mode; 4 p = 0.006 for the additive mode.
Table 3.
SNP individual effects for SNPs involved with alcohol–SNP interactions associated with prostate cancer (PCa) aggressiveness.
| KERRYPNX | Combine PCa Aggressiveness (17.7%) |
Discovery PCa Aggressiveness (17.5%) |
Validation PCa Aggressiveness (17.9%) |
||||
|---|---|---|---|---|---|---|---|
| SNP (Gene, Major > Minor Allele, MAF) 1 | Total N (%) |
Yes N (%) |
No N (%) |
Yes N (%) |
No N (%) |
Yes N (%) |
No N (%) |
| rs13107662 (CAMK2D, A > G, 34.5%) 2 | |||||||
| AA | 657 (43.4) | 130 (19.8) | 527 (80.2) | 74 (21.8) | 266 (78.2) | 56 (17.7) | 261 (82.3) |
| AG | 670 (44.3) | 124 (18.5) | 546 (81.5) | 56 (18.3) | 250 (81.7) | 68 (18.7) | 296 (81.3) |
| GG | 187 (12.4) | 56 (30.0) | 131 (70.1) | 30 (30.3) | 69 (69.7) | 26 (29.6) | 62 (70.5) |
| p = 0.003 | p = 0.046 | p = 0.046 | |||||
| rs9907521 (PRKCA, A > G, 7.0%) 2 | |||||||
| AA | 1824 (86.5) | 362 (19.9) | 1462 (80.2) | 172 (19.1) | 727 (80.9) | 190 (20.5) | 735 (79.5) |
| AG | 273 (13.0) | 51 (18.7) | 222 (81.3) | 27 (21.1) | 101 (78.9) | 24 (16.6) | 121 (83.5) |
| GG | 11 (0.5) | 3 (27.3) | 8 (72.7) | 3 (33.3) | 6 (66.7) | 0 | 2 (100) |
| p = 0.640 | p = 0.456 | p = 0.561 | |||||
| rs11925452 (ROBO1, G > A, 21.1%) 2 | |||||||
| GG | 1312 (62.1) | 267 (20.4) | 1045 (79.7) | 130 (20.4) | 507 (79.6) | 137 (20.3) | 538 (79.7) |
| AG | 710 (33.6) | 130 (18.3) | 580 (81.7) | 61 (17.1) | 295 (82.9) | 69 (19.5) | 285 (80.5) |
| AA | 90 ( 4.3) | 19 (21.1) | 71 (78.9) | 11 (25.0) | 33 (75.0) | 9 (17.4) | 38 (82.6) |
| AA | |||||||
| AA | |||||||
| AA | |||||||
| p = 0.514 | p = 0.274 | p = 0.889 | |||||
1 MAF: minor allele frequency. 2 p-values were based on Fisher’s exact test.
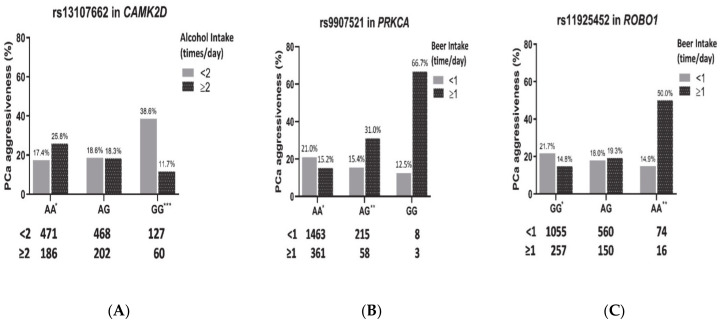
For these three significant alcohol–SNP interactions, we performed subgroup analyses to evaluate the effects of alcohol intake on PCa aggressiveness within each genotype. For rs13107662 in CAMK2D (A > G representing for major “A” allele and minor “G” allele), heavy alcohol intake associated with PCa aggressiveness was significant for PCa patients with the AA and GG genotypes (Figure 2A). For 43.4% PCa patients with the rs13107662 AA genotype (Table 3), those with heavy alcohol intake had a higher chance of PCa aggressiveness (25.8%) compared with those without heavy alcohol intake (17.4%, p = 0.015). For PCa patients with the rs13107662 GG genotype, PCa aggressiveness prevalence was 38.6% and 11.7% for low and heavy alcohol intake, respectively (p = 1.8 × 10−4). Adjusting for the potential confounding factors (study site, the first six principal components, and smoking status), these effects of heavy alcohol intake remained significant. As shown in Figure 3 and Table S3, heavy alcohol intake had a significant risk effect on PCa aggressiveness (odds ratio (OR) = 1.83, p = 0.008 in the combined set) for the rs13107662 AA genotype group, but a significantly protective effect on PCa aggressiveness (OR = 0.2, p = 0.002) for those with the rs13107662 GG genotype group. Similar trends of this alcohol–SNP interaction can be observed for both the discovery and validation set.
Figure 2.
Alcohol intake impact on prostate cancer aggressiveness by the selected genotype profile. (A) Heavy alcohol intake impact by genotypes of rs13107662 in CAMK2D; (B) heavy beer intake impact by genotypes of rs9907521 in PRKCA; (C) heavy beer intake impact by genotypes of rs11925452 in ROBO1. Note: *: p < 0.05, **: p < 0.01; Logistic models regression adjusted for study site, the six principal components, and smoking status. For a sample size <100 (rs13107662 GG for the discovery and validation set, rs11925452 AA for all sets, and rs9907521GG for all sets), unadjusted results were reported. The order of the genotypes was based on homozygous major, heterozygous, and homozygous minor types.
Figure 3.
Prevalence of prostate cancer aggressiveness by the selected SNPs and alcohol intake behaviors. (A) Interaction of heavy alcohol intake and rs13107662 in CAMK2D; (B) interaction of heavy beer intake and rs9907521 in PRKCA; (C) interaction of heavy beer intake rs11925452 in ROBO1. Note: *: p < 0.05, **: p < 0.01, ***: p < 0.001.
For rs9907521 in PRKCA (Figure 2B), heavy beer intake had a significant risk effect on PCa aggressiveness for those with the AG genotype, but a significantly protective effect for those with the AA genotype. For the rs9907521 common AA genotype, PCa aggressiveness prevalence was 21.0% and 15.2% for those with low and heavy beer intake, respectively (p = 0.014). For the PCa patients with the rs9907521 AG genotype, PCa aggressiveness prevalence was significantly higher for those with heavy beer intake than those without (31.0% vs. 15.4%, p = 0.007). Beer intake effects remained significant after adjusting for the selected factors. For rs9907521 in PRKCA (Figure 3 and Table S4), patients with heavy beer intake were more likely to have PCa aggressiveness in the AG genotype group (OR = 2.71, p = 0.006 in the combined set), but less likely to have PCa aggressiveness in the AA genotype group (OR = 0.71, p = 0.036 in the combined set).
For rs11925452 in ROBO1 (G > A, Figure 2C), heavy beer intake was significantly associated with PCa aggressiveness for those with the GG and AA genotypes. For 62.1% of PCa patients with the rs11925452 GG genotype (Table 3), heavy beer intake had a protective effect on PCa aggressiveness. In PCa patients with the rs11925452 AA genotype, those with heavy beer intake had a higher chance of PCa aggressiveness than those with low beer intake (50.0% vs. 14.9%, p = 0.004). The opposite effect was observed in the GG genotype. The prevalence of PCa aggressiveness was 14.8% and 21.7% for those with heavy and low beer intake, respectively (p = 0.014), for those with the rs11925452 GG genotype. Similarly, these beer effects were validated and remained significant after adjusting other factors (Figure 3). Heavy beer intake had a significant risk effect on PCa aggressiveness for PCa patients with the rs11925452 AA genotype (OR = 5.73, p = 0.004 in the combined set), but had a significant protective effect for those with the GG genotype (OR = 0.64, p = 0.023 in the combined set, Table S5) after adjusting for study site, the first six principal components, and smoking status. In addition to these top three pairs, there were five extra alcohol-SNP pairs by expanding the p-value criterion as p < 0.05 in the validation set (Table S6). These five alcohol–SNP interactions were beer–rs5745616 (HGF, p = 2.0 × 10−4), ethanol–rs2050143 (PDGFB, p = 2.0 × 10−4), beer–rs4744514 (SYK, p = 3.0 × 10−4), alcohol–rs11226159 (PDGFD, p = 5.0 × 10−4), and alcohol–rs10933175 (COL4A3, p = 8.0 × 10−4).
4. Discussion
This study examined the alcohol–SNP interactions in the four pathways (angiogenesis, mitochondria, miRNA, and androgen metabolism) associated with PCa aggressiveness. Our results showed that excessive alcohol intake factors (≥2 times alcohol intake per day, ≥1 time beer intake per day, and ≥30 g of alcohol per day) were not associated with PCa aggressiveness. However, the interactions of excessive alcohol intake and three SNPs (rs13107662 in CAMK2D, rs9907521 in PRKCA, and rs11925452 in ROBO1) had a significant effect on PCa aggressiveness. Among these three SNPs, only the rs13107662 had a significant individual effect associated with aggressive PCa (Table 3). These significant alcohol–SNP interactions suggested that excessive alcohol intake significantly impacts PCa aggressiveness on the specific genetic sub-groups in CAMK2D, PRKCA, and ROBO1. For all three identified SNPs, excessive alcohol intake had a significant risk effect for one genotype, but had a protective impact on another genotype within the same SNP. Using rs13107662 in CAMK2D as an example, high alcohol intake (≥2 times alcohol intake per day) was not significantly associated with PCa aggressiveness, but this alcohol effect varied by rs13107662 genotype status. High alcohol intake had a risk effect on PCa aggressiveness for PCa patients with the rs13107662 AA genotype, but had a protective effect for those with the GG genotype.
The literature supports the biological functions of our three identified alcohol–SNP interactions. Studies reported that CAMK2D, PRKCA, and ROBO1 were associated with both alcohol intake and PCa. CAMK2D is part of a larger family of CAMKII, a key component in the common five pathways involved in alcohol, cocaine, opioids, and nicotine addiction [32]. CAMKII has been associated with increased frequency of alcohol consumption and drinking patterns [33]. Gene expression of CAMK2D is differentially expressed in cocaine-addicted individuals and controls [34]. In addition, one CAMK2D genetic variant (rs3815072) is significantly associated with pathological gambling [35]. CAMK2D is also overexpressed with ethanol exposure in rats [36]. In addition, gene expression of CAMK2D, enriched in the cell cycle and the calcium signaling pathway, is associated with PCa metastasis [37]. It has been shown that an elevated calcium concentration may facilitate the metastasis of PCa by bone remodeling and the Akt signaling pathway [38]. In addition, CAMK2D appeared to be regulated by miRNA-30, known as a tumor suppressor miRNA. miRNA-30 inhibits cell migration and invasion and is generally under-expressed in PCa tissues [39]. Although there is no report on a role of CAMK2D SNP in PCa, two SNPs were investigated as a biomarker for other cancers. rs13107662 was found to be associated with ovarian cancer risk [40], and rs10023113 was found as a prognostic marker for survival of lung cancer patients [41].
Our results show that PCa patients with heavy beer intake are more likely to have PCa aggressiveness in the rs9907521 AG genotype in PRKCA. Although the risk effect of heavy beer intake was not significant for those with the rs9907521 GG genotype because of the small sample size (n = 13), the difference in PCa aggressiveness prevalence for those with and without heavy beer intake (66.7% vs. 12.5%) is the largest among all genotypes for these three SNPs in Figure 2. PRKCA (protein kinase C alpha) plays an essential role in many different cellular processes, such as cell adhesion and cell transformation. PRKCA has been associated with alcohol dependence and early onset of alcohol dependence for European Americans and African Americans in the GWAS [42,43]. Four SNPs in PRKCA (rs17688881, rs721429, rs7217618, and rs8077110) are associated with alcohol dependence and brain activations [44]. PRKCA is also a GWAS identified gene significantly associated with food addiction [45]. Knockout studies in mice suggest that PRKCA activity regulates cancer, and PRKCA was overexpressed in PCa cells [46]. No study evaluated a role of PRKCA SNPs associated with PCa aggressiveness. One study reported that rs11079651 in PRKCA was associated with pancreatic cancer risk [47]. A preclinical study showed PRKCA expression was significantly decreased in rats exposed to ethanol compared with those exposed to either water or saccharin [48]. In addition, expression of PRKCA was changed after ethanol exposure in liver-derived cells [49].
ROBO1 was shown to be involved with both PCa and alcohol intake. ROBO1, a member of the roundabout (ROBO) immunoglobulin superfamily of protein, plays a role in cell migration [50,51] and acts as a tumor suppressor gene [52,53]. ROBO1 expression was negatively associated with prognosis for PCa risk/metastasis [52,54], breast cancer [53,55], and colorectal cancer [56]. The ROBO/SLIT signaling pathway acts as a critical regulator for tumor developmental and pathological processes for several cancers, including PCa [57]. ROBO1 acts like a natural inhibitor of metastasis; therefore, ROBO1 is considered as a potential cancer therapeutic target [58]. We previously reported several SNP–SNP interactions between R0BO1 and MMP16 associated PCa aggressiveness, and the biological function of the ROBO1–MMP16 interactions was supported by gene expression results [21,59]. ROBO1 is one gene enriched in the axon guidance pathway, significantly associated with alcoholism and alcohol addiction [60].
We also identified five additional alcohol–SNP interaction pairs associated with PCa aggressiveness by expanding the p-value criteria to p < 0.05 in the validation set. They are rs5745616 (HGF), rs2050143 (PDGFB), rs4744514 (SYK), rs11226159 (PDGFD), and rs10933175 (COL4A3). Previous studies indicated that HGF significantly increases the proliferation and invasion of prostate tumor cells [61,62,63]. In addition, HGF leads to recover from alcohol-induced fatty liver by participation in lipid metabolism [64]. Platelet-derived growth factor F (PDGF) signaling plays an important role in cancer risk and progression. The PDGF isoforms are composed of four different genes, PDGFA, PDGFB, PDGFC, and PDGFD [65]. These genes regulate cell proliferation, transformation, apoptosis, invasion, angiogenesis, and metastasis [66,67,68]. Duan et al. reported that two SNPs of PDGFB are associated with pancreatic cancer risk [69]. SYK is required for angiogenesis and lymphangiogenesis during embryonic development [70,71]. The role of SYK in human cancers is not completely established. SYK expression inversely correlates with tumor growth and metastasis in breast cancer [72,73]. In addition, binge alcohol intake could induce SYK activation in an animal study [74], and SYK was associated with the development of alcoholic liver disease and liver fibrosis [75]. In addition, expression of COL4A3 was associated with tumor size, higher grade, metastasis, and invasion in several malignancies [76,77,78].
We are aware of the limitations of this study. First, our results were generated from PCa patients with European ancestry, so these findings may not be applied in other racial groups. Second, genetic variations and alcohol interactions may reveal the complexity of aggressiveness of prostate cancer. However, these interactions may not be applied in other cancers. Third, this study did not have detailed beverage types (such as red wine, white wine, and spirits) and may have recall bias of the self-report alcohol intake behaviors. Finally, the biological role of identified SNPs in alcohol metabolism is not known. Therefore, basic research on the function of these genetic variants in alcohol metabolism is warranted. Our study findings can be used to identify high-risk genetic groups of PCa aggressiveness. For men with risky alleles, special attention to alcohol intervention for decreasing alcohol consumption should be considered to prevent PCa progression. Future research with a large sample size and information of various alcohol beverage types will be needed to further validate the identified effects of alcohol–SNP interactions on PCa aggressiveness. In summary, our findings suggest that the impact of alcohol intake on PCa aggressiveness may not be universal in PCa patients owing to the effects of genetic heterogeneity, such as CAMK2D, PRKCA, and ROBO1. These alcohol–SNP interactions may explain the inconclusive results of alcohol’s impact on PCa aggressive when genetic profiles are not considered. These results suggested that excessive alcohol intake significantly impacts PCa aggressiveness in the specific genetic subgroups.
Acknowledgments
We thank our anonymous reviewers for their valuable comments, which have led to many improvements to this article. This study was supported by the National Cancer Institute (R21CA202417, PI: Lin, HY). Kenneth R. Muir was supported by the National Institute for Health Research (NIHR) Manchester Biomedical Research Centre.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/10/3/553/s1, Table S1: Study site information; Table S2: Alcohol intake associated with prostate cancer (PCa) aggressiveness by study set; Table S3: Impact of alcohol intake on prostate cancer aggressiveness for the sub-groups with different genotypes of rs13107662 in CAMK2D; Table S4: Impact of alcohol intake on prostate cancer aggressiveness for the sub-groups with different genotypes of rs9907521 in PRKCA; Table S5: Impact of alcohol intake on prostate cancer aggressiveness for the sub-groups with different genotypes of rs11925452 in ROBO1; Table S6. Eight alcohol–SNP interactions associated with prostate cancer aggressiveness based on liberal significance criteria.
Author Contributions
H.-Y.L. and X.W. conducted data analyses. H.-Y.L., X.W., Y.-H.K., T.-S.T., and J.Y.P. were mainly responsible for the study design and drafted the paper’s initial version. Z.F. and P.E.M. were part of the writing group and were primarily responsible for the result interpretation. All other co-authors were responsible for cohort-level data collection, cohort-level data analysis, and critical reviews of this paper. All authors approved the final version of the paper submitted to the journal. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Cancer Institute (R21CA202417, PI: Hui-Yi Lin).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board of Louisiana State University Health Sciences Center (protocol code: IRB-9338). (24 May 2016).
Informed Consent Statement
Not applicable.
Data Availability Statement
Restrictions apply to the availability of the data used in this project. Data were obtained from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome consortium (PRACTICAL Consortium, http://practical.icr.ac.uk/blog/) and are available with the permission of PRACTICAL Consortium.
Conflicts of Interest
All authors disclosed no potential conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J. Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Brennan P. Gene-environment interaction and aetiology of cancer: What does it mean and how can we measure it? Carcinogenesis. 2002;23:381–387. doi: 10.1093/carcin/23.3.381. [DOI] [PubMed] [Google Scholar]
- 4.Simonds N.I., Ghazarian A.A., Pimentel C.B., Schully S.D., Ellison G.L., Gillanders E.M., Mechanic L.E. Review of the Gene-Environment Interaction Literature in Cancer: What Do We Know? Genet. Epidemiol. 2016;40:356–365. doi: 10.1002/gepi.21967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizos C., Papassava M., Golias C., Charalabopoulos K. Alcohol consumption and prostate cancer: A mini review. Exp. Oncol. 2010;32:66–70. [PubMed] [Google Scholar]
- 6.Perdana N.R., Mochtar C.A., Umbas R., Hamid A.R. The Risk Factors of Prostate Cancer and Its Prevention: A Literature Review. Acta Med. Indones. 2016;48:228–238. [PubMed] [Google Scholar]
- 7.Gronberg H. Prostate cancer epidemiology. Lancet. 2003;361:859–864. doi: 10.1016/S0140-6736(03)12713-4. [DOI] [PubMed] [Google Scholar]
- 8.Van den Broeck T., Joniau S., Clinckemalie L., Helsen C., Prekovic S., Spans L., Tosco L., Van Poppel H., Claessens F. The role of single nucleotide polymorphisms in predicting prostate cancer risk and therapeutic decision making. Biomed. Res. Int. 2014;2014:627510. doi: 10.1155/2014/627510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boffetta P., Hashibe M. Alcohol and cancer. Lancet Oncol. 2006;7:149–156. doi: 10.1016/S1470-2045(06)70577-0. [DOI] [PubMed] [Google Scholar]
- 10.Substance Abuse and Mental Health Services Administration . Key Substance Use and Mental Health Indicators in the United States: Results from the 2018 National Survey on Drug Use and Health. Substance Abuse and Mental Health Services Administration; Rockville, MD, USA: 2019. ((HHS Publication No. PEP19-5068, NSDUH Series H-54)). [Google Scholar]
- 11.Lin H.Y., Fisher P., Harris D., Tseng T.S. Alcohol intake patterns for cancer and non-cancer individuals: A population study. Transl. Cancer Res. 2019;8:S334–S345. doi: 10.21037/tcr.2019.06.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demoury C., Karakiewicz P., Parent M.E. Association between lifetime alcohol consumption and prostate cancer risk: A case-control study in Montreal, Canada. Cancer Epidemiol. 2016;45:11–17. doi: 10.1016/j.canep.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J., Stockwell T., Roemer A., Chikritzhs T. Is alcohol consumption a risk factor for prostate cancer? A systematic review and meta-analysis. BMC Cancer. 2016;16:845. doi: 10.1186/s12885-016-2891-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickerman B.A., Markt S.C., Koskenvuo M., Pukkala E., Mucci L.A., Kaprio J. Alcohol intake, drinking patterns, and prostate cancer risk and mortality: A 30-year prospective cohort study of Finnish twins. Cancer Causes Control. 2016;27:1049–1058. doi: 10.1007/s10552-016-0778-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohrmann S., Linseisen J., Key T.J., Jensen M.K., Overvad K., Johnsen N.F., Tjonneland A., Kaaks R., Bergmann M.M., Weikert C., et al. Alcohol consumption and the risk for prostate cancer in the European Prospective Investigation into Cancer and Nutrition. Cancer Epidemiol. Biomark. Prev. 2008;17:1282–1287. doi: 10.1158/1055-9965.EPI-07-2888. [DOI] [PubMed] [Google Scholar]
- 16.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J., et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brunner C., Davies N.M., Martin R.M., Eeles R., Easton D., Kote-Jarai Z., Al Olama A.A., Benlloch S., Muir K., Giles G., et al. Alcohol consumption and prostate cancer incidence and progression: A Mendelian randomisation study. Int. J. Cancer. 2017;140:75–85. doi: 10.1002/ijc.30436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seitz H.K., Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer. 2007;7:599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 19.Fukumori T., Oka N., Takenaka Y., Nangia-Makker P., Elsamman E., Kasai T., Shono M., Kanayama H.O., Ellerhorst J., Lotan R., et al. Galectin-3 regulates mitochondrial stability and antiapoptotic function in response to anticancer drug in prostate cancer. Cancer Res. 2006;66:3114–3119. doi: 10.1158/0008-5472.CAN-05-3750. [DOI] [PubMed] [Google Scholar]
- 20.Poliseno L., Tuccoli A., Mariani L., Evangelista M., Citti L., Woods K., Mercatanti A., Hammond S., Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 21.Lin H.Y., Amankwah E.K., Tseng T.S., Qu X., Chen D.T., Park J.Y. SNP-SNP Interaction Network in Angiogenesis Genes Associated with Prostate Cancer Aggressiveness. PLoS ONE. 2013;8:e59688. doi: 10.1371/journal.pone.0059688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eisermann K., Fraizer G. The Androgen Receptor and VEGF: Mechanisms of Androgen-Regulated Angiogenesis in Prostate Cancer. Cancers. 2017;9:32. doi: 10.3390/cancers9040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKay R.R., Zurita A.J., Werner L., Bruce J.Y., Carducci M.A., Stein M.N., Heath E.I., Hussain A., Tran H.T., Sweeney C.J., et al. A Randomized Phase II Trial of Short-Course Androgen Deprivation Therapy With or Without Bevacizumab for Patients With Recurrent Prostate Cancer After Definitive Local Therapy. J. Clin. Oncol. 2016;34:1913–1920. doi: 10.1200/JCO.2015.65.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boddy J.L., Fox S.B., Han C., Campo L., Turley H., Kanga S., Malone P.R., Harris A.L. The androgen receptor is significantly associated with vascular endothelial growth factor and hypoxia sensing via hypoxia-inducible factors HIF-1a, HIF-2a, and the prolyl hydroxylases in human prostate cancer. Clin. Cancer Res. 2005;11:7658–7663. doi: 10.1158/1078-0432.CCR-05-0460. [DOI] [PubMed] [Google Scholar]
- 25.Audet-Walsh E., Yee T., McGuirk S., Vernier M., Ouellet C., St-Pierre J., Giguere V. Androgen-Dependent Repression of ERRgamma Reprograms Metabolism in Prostate Cancer. Cancer Res. 2017;77:378–389. doi: 10.1158/0008-5472.CAN-16-1204. [DOI] [PubMed] [Google Scholar]
- 26.Amankwah E.K., Anegbe E., Park H., Pow-Sang J., Hakam A., Park J.Y. miR-21, miR-221 and miR-222 expression and prostate cancer recurrence among obese and non-obese cases. Asian J. Androl. 2013;15:226–230. doi: 10.1038/aja.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eeles R.A., Olama A.A., Benlloch S., Saunders E.J., Leongamornlert D.A., Tymrakiewicz M., Ghoussaini M., Luccarini C., Dennis J., Jugurnauth-Little S., et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat. Genet. 2013;45:385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention (CDC), Glossary: Drinking Status. [(accessed on 23 January 2021)]; Available online: https://www.cdc.gov/nchs/nhis/alcohol/alcohol_glossary.htm.
- 29.Papa N.P., MacInnis R.J., Jayasekara H., English D.R., Bolton D., Davis I.D., Lawrentschuk N., Millar J.L., Pedersen J., Severi G., et al. Total and beverage-specific alcohol intake and the risk of aggressive prostate cancer: A case-control study. Prostate Cancer Prostatic Dis. 2017;20:305–310. doi: 10.1038/pcan.2017.12. [DOI] [PubMed] [Google Scholar]
- 30.Downer M.K., Kenfield S.A., Stampfer M.J., Wilson K.M., Dickerman B.A., Giovannucci E.L., Rimm E.B., Wang M., Mucci L.A., Willett W.C., et al. Alcohol Intake and Risk of Lethal Prostate Cancer in the Health Professionals Follow-Up Study. J. Clin. Oncol. 2019;37:1499–1511. doi: 10.1200/JCO.18.02462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storey J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B. 2002;64:479–498. doi: 10.1111/1467-9868.00346. [DOI] [Google Scholar]
- 32.Chen P., Liang J., Wang Z., Zhou X., Chen L., Li M., Xie D., Hu Z., Shen H., Wang H. Association of common PALB2 polymorphisms with breast cancer risk: A case-control study. Clin. Cancer Res. 2008;14:5931–5937. doi: 10.1158/1078-0432.CCR-08-0429. [DOI] [PubMed] [Google Scholar]
- 33.Morisot N., Ron D. Alcohol-dependent molecular adaptations of the NMDA receptor system. Genes Brain Behav. 2017;16:139–148. doi: 10.1111/gbb.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z., Yuan Q., Mash D.C., Goldman D. Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc. Natl. Acad. Sci. USA. 2011;108:6626–6631. doi: 10.1073/pnas.1018514108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lobo D.S., Aleksandrova L., Knight J., Casey D.M., el-Guebaly N., Nobrega J.N., Kennedy J.L. Addiction-related genes in gambling disorders: New insights from parallel human and pre-clinical models. Mol. Psychiatry. 2015;20:1002–1010. doi: 10.1038/mp.2014.113. [DOI] [PubMed] [Google Scholar]
- 36.Heshmati E., Shirpoor A., Kheradmand F., Alizadeh M., Gharalari F.H. Chronic ethanol increases calcium/calmodulin-dependent protein kinaseIIdelta gene expression and decreases monoamine oxidase amount in rat heart muscles: Rescue effect of Zingiber officinale (ginger) extract. Anatol. J. Cardiol. 2018;19:19–26. doi: 10.14744/AnatolJCardiol.2017.8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X., Yao X., Qin C., Luo P., Zhang J. Investigation of the molecular mechanisms underlying metastasis in prostate cancer by gene expression profiling. Exp. Ther. Med. 2016;12:925–932. doi: 10.3892/etm.2016.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao J., Schneider A., Datta N.S., McCauley L.K. Extracellular calcium as a candidate mediator of prostate cancer skeletal metastasis. Cancer Res. 2006;66:9065–9073. doi: 10.1158/0008-5472.CAN-06-0317. [DOI] [PubMed] [Google Scholar]
- 39.Kao C.J., Martiniez A., Shi X.B., Yang J., Evans C.P., Dobi A., deVere White R.W., Kung H.J. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene. 2014;33:2495–2503. doi: 10.1038/onc.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Permuth-Wey J., Chen Y.A., Tsai Y.Y., Chen Z., Qu X., Lancaster J.M., Stockwell H., Dagne G., Iversen E., Risch H., et al. Inherited variants in mitochondrial biogenesis genes may influence epithelial ovarian cancer risk. Cancer Epidemiol. Biomark. Prev. 2011;20:1131–1145. doi: 10.1158/1055-9965.EPI-10-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang S., Pan Y., Wang Y., Hu L., Cao S., Chu M., Dai J., Shu Y., Xu L., Chen J., et al. Genome-wide association study of survival in early-stage non-small cell lung cancer. Ann. Surg. Oncol. 2015;22:630–635. doi: 10.1245/s10434-014-3983-0. [DOI] [PubMed] [Google Scholar]
- 42.Edenberg H.J., Koller D.L., Xuei X., Wetherill L., McClintick J.N., Almasy L., Bierut L.J., Bucholz K.K., Goate A., Aliev F., et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Treutlein J., Cichon S., Ridinger M., Wodarz N., Soyka M., Zill P., Maier W., Moessner R., Gaebel W., Dahmen N., et al. Genome-wide association study of alcohol dependence. Arch. Gen. Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Hutchison K.E., Calhoun V.D., Claus E.D., Turner J.A., Sui J., Liu J. CREB-BDNF pathway influences alcohol cue-elicited activation in drinkers. Hum. Brain Mapp. 2015;36:3007–3019. doi: 10.1002/hbm.22824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornelis M.C., Flint A., Field A.E., Kraft P., Han J., Rimm E.B., van Dam R.M. A genome-wide investigation of food addiction. Obesity. 2016;24:1336–1341. doi: 10.1002/oby.21476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konopatskaya O., Poole A.W. Protein kinase Calpha: Disease regulator and therapeutic target. Trends Pharmacol. Sci. 2010;31:8–14. doi: 10.1016/j.tips.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu X., Qian D., Liu H., Abbruzzese J.L., Luo S., Walsh K.M., Wei Q. Genetic variants of the peroxisome proliferator-activated receptor (PPAR) signaling pathway genes and risk of pancreatic cancer. Mol. Carcinog. 2020;59:930–939. doi: 10.1002/mc.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodd Z.A., Kimpel M.W., Edenberg H.J., Bell R.L., Strother W.N., McClintick J.N., Carr L.G., Liang T., McBride W.J. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol. Biochem. Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pochareddy S., Edenberg H.J. Chronic alcohol exposure alters gene expression in HepG2 cells. Alcohol. Clin. Exp. Res. 2012;36:1021–1033. doi: 10.1111/j.1530-0277.2011.01677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonda Y., Andrews W.D., Tabata H., Namba T., Parnavelas J.G., Nakajima K., Kohsaka S., Hanashima C., Uchino S. Robo1 regulates the migration and laminar distribution of upper-layer pyramidal neurons of the cerebral cortex. Cereb. Cortex. 2013;23:1495–1508. doi: 10.1093/cercor/bhs141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khusial P.R., Vadla B., Krishnan H., Ramlall T.F., Shen Y., Ichikawa H., Geng J.G., Goldberg G.S. Src activates Abl to augment Robo1 expression in order to promote tumor cell migration. Oncotarget. 2010;1:198–209. doi: 10.18632/oncotarget.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parray A., Siddique H.R., Kuriger J.K., Mishra S.K., Rhim J.S., Nelson H.H., Aburatani H., Konety B.R., Koochekpour S., Saleem M. ROBO1, a tumor suppressor and critical molecular barrier for localized tumor cells to acquire invasive phenotype: Study in African-American and Caucasian prostate cancer models. Int. J. Cancer. 2014;135:2493–2506. doi: 10.1002/ijc.28919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dallol A., Forgacs E., Martinez A., Sekido Y., Walker R., Kishida T., Rabbitts P., Maher E.R., Minna J.D., Latif F. Tumour specific promoter region methylation of the human homologue of the Drosophila Roundabout gene DUTT1 (ROBO1) in human cancers. Oncogene. 2002;21:3020–3028. doi: 10.1038/sj.onc.1205421. [DOI] [PubMed] [Google Scholar]
- 54.Latil A., Chene L., Cochant-Priollet B., Mangin P., Fournier G., Berthon P., Cussenot O. Quantification of expression of netrins, slits and their receptors in human prostate tumors. Int. J. Cancer. 2003;103:306–315. doi: 10.1002/ijc.10821. [DOI] [PubMed] [Google Scholar]
- 55.Bhattacharya R., Mukherjee N., Dasgupta H., Islam M.S., Alam N., Roy A., Das P., Roychoudhury S., Panda C.K. Frequent alterations of SLIT2-ROBO1-CDC42 signalling pathway in breast cancer: Clinicopathological correlation. J. Genet. 2016;95:551–563. doi: 10.1007/s12041-016-0678-2. [DOI] [PubMed] [Google Scholar]
- 56.Grone J., Doebler O., Loddenkemper C., Hotz B., Buhr H.J., Bhargava S. Robo1/Robo4: Differential expression of angiogenic markers in colorectal cancer. Oncol. Rep. 2006;15:1437–1443. doi: 10.3892/or.15.6.1437. [DOI] [PubMed] [Google Scholar]
- 57.Choi Y.J., Yoo N.J., Lee S.H. Down-regulation of ROBO2 expression in prostate cancers. Pathol. Oncol. Res. 2014;20:517–519. doi: 10.1007/s12253-013-9722-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koohini Z., Koohini Z., Teimourian S. Slit/Robo Signaling Pathway in Cancer; a New Stand PoInt. for Cancer Treatment. Pathol. Oncol. Res. 2019;25:1285–1293. doi: 10.1007/s12253-018-00568-y. [DOI] [PubMed] [Google Scholar]
- 59.Lin H.Y., Cheng C.H., Chen D.T., Chen Y.A., Park J.Y. Coexpression and expression quantitative trait loci analyses of the angiogenesis gene-gene interaction network in prostate cancer. Transl. Cancer Res. 2016;5:S951–S963. doi: 10.21037/tcr.2016.10.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li C., Li Y., Xu J., Lv J., Ma Y., Shao T., Gong B., Tan R., Xiao Y., Li X. Disease-driven detection of differential inherited SNP modules from SNP network. Gene. 2011;489:119–129. doi: 10.1016/j.gene.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 61.Humphrey P.A., Zhu X., Zarnegar R., Swanson P.E., Ratliff T.L., Vollmer R.T., Day M.L. Hepatocyte growth factor and its receptor (c-MET) in prostatic carcinoma. Am. J. Pathol. 1995;147:386–396. [PMC free article] [PubMed] [Google Scholar]
- 62.Kasai S., Sugimura K., Matsumoto K., Nishi N., Kishimoto T., Nakamura T. Hepatocyte growth factor is a paracrine regulator of rat prostate epithelial growth. Biochem. Biophys. Res. Commun. 1996;228:646–652. doi: 10.1006/bbrc.1996.1710. [DOI] [PubMed] [Google Scholar]
- 63.Pisters L.L., Troncoso P., Zhau H.E., Li W., von Eschenbach A.C., Chung L.W. c-met proto-oncogene expression in benign and malignant human prostate tissues. J. Urol. 1995;154:293–298. doi: 10.1016/S0022-5347(01)67297-5. [DOI] [PubMed] [Google Scholar]
- 64.Tahara M., Matsumoto K., Nukiwa T., Nakamura T. Hepatocyte growth factor leads to recovery from alcohol-induced fatty liver in rats. J. Clin. Investig. 1999;103:313–320. doi: 10.1172/JCI4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andrae J., Gallini R., Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilhelm A., Aldridge V., Haldar D., Naylor A.J., Weston C.J., Hedegaard D., Garg A., Fear J., Reynolds G.M., Croft A.P., et al. CD248/endosialin critically regulates hepatic stellate cell proliferation during chronic liver injury via a PDGF-regulated mechanism. Gut. 2016;65:1175–1185. doi: 10.1136/gutjnl-2014-308325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/S0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 68.Hoch R.V., Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–4784. doi: 10.1242/dev.00721. [DOI] [PubMed] [Google Scholar]
- 69.Duan B., Hu J., Liu H., Wang Y., Li H., Liu S., Xie J., Owzar K., Abbruzzese J., Hurwitz H., et al. Genetic variants in the platelet-derived growth factor subunit B gene associated with pancreatic cancer risk. Int. J. Cancer. 2018;142:1322–1331. doi: 10.1002/ijc.31171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abtahian F., Guerriero A., Sebzda E., Lu M.M., Zhou R., Mocsai A., Myers E.E., Huang B., Jackson D.G., Ferrari V.A., et al. Regulation of blood and lymphatic vascular separation by signaling proteins SLP-76 and Syk. Science. 2003;299:247–251. doi: 10.1126/science.1079477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coopman P.J., Mueller S.C. The Syk tyrosine kinase: A new negative regulator in tumor growth and progression. Cancer Lett. 2006;241:159–173. doi: 10.1016/j.canlet.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Toyama T., Iwase H., Yamashita H., Hara Y., Omoto Y., Sugiura H., Zhang Z., Fujii Y. Reduced expression of the Syk gene is correlated with poor prognosis in human breast cancer. Cancer Lett. 2003;189:97–102. doi: 10.1016/S0304-3835(02)00463-9. [DOI] [PubMed] [Google Scholar]
- 73.Coopman P.J., Do M.T., Barth M., Bowden E.T., Hayes A.J., Basyuk E., Blancato J.K., Vezza P.R., McLeskey S.W., Mangeat P.H., et al. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–747. doi: 10.1038/35021086. [DOI] [PubMed] [Google Scholar]
- 74.Bukong T.N., Iracheta-Vellve A., Gyongyosi B., Ambade A., Catalano D., Kodys K., Szabo G. Therapeutic Benefits of Spleen Tyrosine Kinase Inhibitor Administration on Binge Drinking-Induced Alcoholic Liver Injury, Steatosis, and Inflammation in Mice. Alcohol. Clin. Exp. Res. 2016;40:1524–1530. doi: 10.1111/acer.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qu C., Zheng D., Li S., Liu Y., Lidofsky A., Holmes J.A., Chen J., He L., Wei L., Liao Y., et al. Tyrosine kinase SYK is a potential therapeutic target for liver fibrosis. Hepatology. 2018;68:1125–1139. doi: 10.1002/hep.29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Georgiou G.K., Igglezou M., Sainis I., Vareli K., Batsis H., Briasoulis E., Fatouros M. Impact of breast cancer surgery on angiogenesis circulating biomarkers: A prospective longitudinal study. World J. Surg. Oncol. 2013;11:213. doi: 10.1186/1477-7819-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kauppila S., Saarela J., Stenback F., Risteli J., Kauppila A., Risteli L. Expression of mRNAs for type I and type III procollagens in serous ovarian cystadenomas and cystadenocarcinomas. Am. J. Pathol. 1996;148:539–548. [PMC free article] [PubMed] [Google Scholar]
- 78.Liang Y., Lv Z., Huang G., Qin J., Li H., Nong F., Wen B. Prognostic significance of abnormal matrix collagen remodeling in colorectal cancer based on histologic and bioinformatics analysis. Oncol. Rep. 2020;44:1671–1685. doi: 10.3892/or.2020.7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Restrictions apply to the availability of the data used in this project. Data were obtained from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome consortium (PRACTICAL Consortium, http://practical.icr.ac.uk/blog/) and are available with the permission of PRACTICAL Consortium.