Abstract
Bioluminescence imaging with luciferase-luciferin pairs is commonly used for monitoring biological processes in cells and whole organisms. Traditional bioluminescent probes are limited in scope, though, as they cannot be easily distinguished in biological environments, precluding efforts to visualize multicellular processes. Additionally, many luciferase-luciferin pairs emit light that is poorly tissue penetrant, hindering efforts to visualize targets in deep tissues. To address these issues, we synthesized a set of π-extended luciferins that were predicted to be red-shifted luminophores. The scaffolds were designed to be rotationally labile such that they produced light only when paired with luciferases capable of enforcing planarity. A luciferin comprising an intramolecular “lock” was identified as a viable light-emitting probe. Native luciferases were unable to efficiently process the analog, but a complementary luciferase was identified via Rosetta-guided enzyme design. The unique enzyme-substrate pair is red-shifted compared to well known bioluminescent tools. The probe set is also orthogonal to other luciferase-luciferin probes and can be used for multicomponent imaging. Four substrate-resolved luciferases were imaged in a single session. Collectively, this work provides the first example of Rosetta-guided design in engineering bioluminescent tools and expands the scope of orthogonal imaging probes.
Graphical Abstract
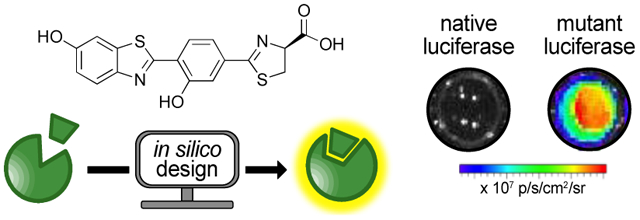
INTRODUCTION
Bioluminescence imaging (BLI) is among the most sensitive and versatile platforms for visualizing cells and biological processes in vivo.1 The core of this technology is a light-emitting reaction involving luciferase enzymes and luciferin small molecules. One of the most popular bioluminescent pairs comprises firefly luciferase (Fluc) and its substrate, d-luciferin (d-luc, Figure 1a). Imaging with Fluc and d-luc affords excellent signal-to-noise ratios in cells and tissues, as these materials do not emit large numbers of endogenous photons.2 BLI also does not require excitation light for photon production, making it well suited for serial imaging. Indeed, the Fluc/d-luc reaction has been routinely employed to examine biological processes over time, including gene expression and cell proliferation.3–4
Figure 1. Orthogonal bioluminescence imaging with engineered luciferases and chemically modified luciferins.
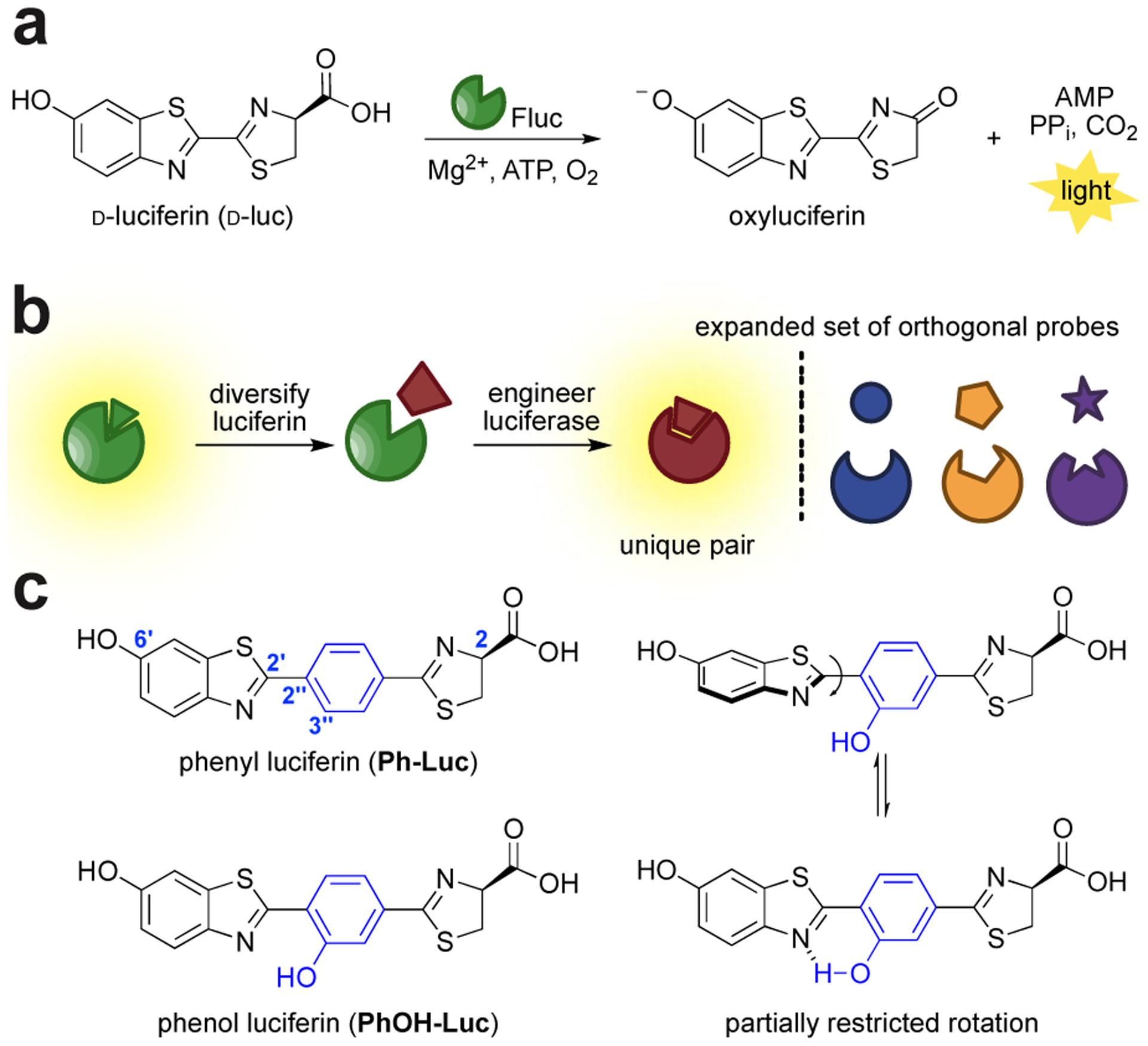
(a) Luciferase-mediated oxidation of luciferin produces a photon of light. (b) Schematic of parallel engineering to generate orthogonal luciferase-luciferin pairs. (c) Two π-extended luciferins with red-shifted emission. The analogs were designed to be more flexible at the C2′-C2′′ junction. The conformation of PhOH-Luc is partially restricted via intramolecular hydrogen bonding.
While popular, bioluminescence has been historically limited to monitoring one or two features at a time.5–6 Visualizing collections of cells or multicomponent processes is exceedingly difficult due, in part, to a lack of distinguishable probes. Numerous luciferases from the insect family are available, but most use the same substrate (d-luc) for light production and are difficult to discriminate in tissue.7–8 Efforts to develop more spectrally resolved probes have been successful, but in most cases, the wavelengths cannot be easily resolved in complex tissues or whole animals.9–12
In contrast to spectral resolution, bioluminescent reporters can be readily distinguished based on substrate usage. Luciferases that use unique luciferins are widely used in dual reporter assays.13–14 For example, Fluc (with d-luc) and Renilla luciferase (with coelenterazine) are routinely used in tandem to monitor differential gene expression.13–14 Additional targets, in theory, could be simultaneously visualized using other naturally occurring luciferases. However, most of these enzymes are not well suited for imaging applications based on suboptimal photon penetrance or limited substrate bioavailability.15–17 The discovery of alternative light-emitting probes18 has also not kept pace with the demand for more bioluminescent tools.
To address the need for more substrate-resolved (i.e., orthogonal) enzymes, we and others are identifying artificial luciferases that can selectively process chemically distinct luciferins.19–22 Our starting point has been the Fluc/d-luc pair, an attractive native bioluminescent system for in vivo work.23–25 We have synthesized several d-luc analogs comprising steric or electronic modifications that perturb photon outputs with Fluc.6 Concomitant engineering of the luciferase active site has yielded complementary enzymes for the modified probes (Figure 1b). Dozens of substrate-selective pairs have been identified via this method, but limitations remain. Many of the pairs exhibit suboptimal emission profiles, with photons being absorbed by surrounding tissue. The molecular diversity of the substrates has also been limited in scope, complicating the search for additional orthogonal luciferases. Identifying triplet, quadruplet, or higher order sets of bioluminescent probes requires more unique luciferins.
To achieve an expanded set of orthogonal tools for in vivo imaging, we were attracted to scaffolds with extended chromophores. Luciferins with elongated π-systems can potentially provide more red photons (>650 nm).26–30 Such wavelengths are desirable for imaging in animals, as they are less absorbed by hemoglobin and more readily escape tissue.31–33 Tissue-penetrant photons are produced in the canonical Fluc/d-luc reaction, but they comprise only a small portion of the emission spectrum (λmax ~600 nm in vivo34). More red-shifted bioluminescent tools could thus provide added sensitivity in imaging experiments. Indeed, probes with λmax ~650 nm exhibit dramatically improved signal-to-noise ratios in tissues and animals.30,35 Red-shifted analogs of d-luc have been developed by replacing the aromatic core with aminostyrenyl-26,35 or aminonaphthyl-based heterocycles.36 These substrates produced near-infrared bioluminescence with Fluc, but the signals were weak, likely due to inefficient turn over or poor quantum yields. Improved photon outputs were achieved, though, via further engineering of the luciferase.
We surmised that probes with extended conjugation could also provide access to unique chemical space for orthogonal substrate development. Here we report the design and synthesis of a unique class of π-extended analogs. These probes were designed to be rotationally labile. Complementary luciferases that could enforce light-emitting geometries were engineered using a combination of computational and semi-rational approaches. A novel luciferase-luciferin pair comprising a π-extended analog was identified. This probe set emitted more red light than Fluc/d-luc. The engineered pair was also readily integrated into the current bioluminescence toolkit, enabling multicomponent imaging.
RESULTS AND DISCUSSION
Designing and synthesizing π-extended luciferins.
Altering the structure of the luciferin emitter can provide unique bioluminescent colors. Indeed, a variety of π-extended luciferins have been produced in recent years that exhibit red-shifted emission.3 Most are suboptimal substrates for Fluc, though, and produce lower levels of light.12,37 Some of the analogs also comprise rigid π-architectures, which can be more prone to non-specific oxidation.38–39 To create more robust probes and restore bioluminescence activity, we envisioned engineering luciferases to accept flexible, red-emitting luciferins. π-Extended luciferins are also structurally distinct from other orthogonal substrates, providing a unique avenue for accessing additional probes for multicomponent imaging.
We were initially drawn to Ph-Luc, an analog comprising an intervening phenyl moiety within the d-luc core (Ph-Luc, Figure 1c). Computational modeling suggested that a fully planarized Ph-Luc could emit >800 nm light, which is desirable for imaging in tissues and whole organisms (Figure S1a). The benzothiazole-phenyl junction (C2′-C2′′) was predicted to have a rotational barrier of ~6 kcal/mol based on DFT calculations (Figure S1b). These data suggested that Ph-Luc could reside in different conformers in the luciferase active site. The majority of the rotational states would likely be non-emissive, due to the interrupted push-pull system.40 This conformational flexibility could also provide a molecular switch for substrate specificity. Only luciferases capable of enforcing a planar geometry should produce light, making them suitable candidates for orthogonal probe development.
Evolving an enzyme to control discrete chromophore conformation was a perceived challenge as only a handful of successes have been reported.41–42 To mitigate risk, we designed a complementary analog, PhOH-Luc, with partially restricted rotation. PhOH-Luc comprises a phenol moiety that can engage in an intramolecular hydrogen bond that modulates C2′-C2′′ rotation.43 The introduction of the hydroxyl group increased the predicted rotational barrier (Figure S1b). The attenuated rotational flexibility could potentially provide a more accessible planarized π-system. Like Ph-Luc, the phenolic scaffold was predicted to exhibit red-shifted emission (Figure S1a). Both molecules were also structurally distinct from most d-luc analogs synthesized to date. Such unique architectures could potentially expedite the search for orthogonal pairs.
We envisioned accessing Ph-Luc and PhOH-Luc from a common brominated synthon. Late stage Suzuki-Miyaura coupling and D-cysteine condensation (Scheme 1) would provide both analogs. Brominated intermediate 1 was accessed in two steps from commercial materials (Scheme S1). Coupling this fragment with pinacol boronate 2 forged the key biaryl C-C bond en route to Ph-Luc. Benzonitrile product 3 was ultimately condensed with D-cysteine to provide Ph-Luc in good yield. PhOH-Luc was similarly prepared. Earlier attempts to isolate PhOH-Luc from a more streamlined route (via a late-stage double demethylation, Scheme S2) were unsuccessful.
Scheme 1.
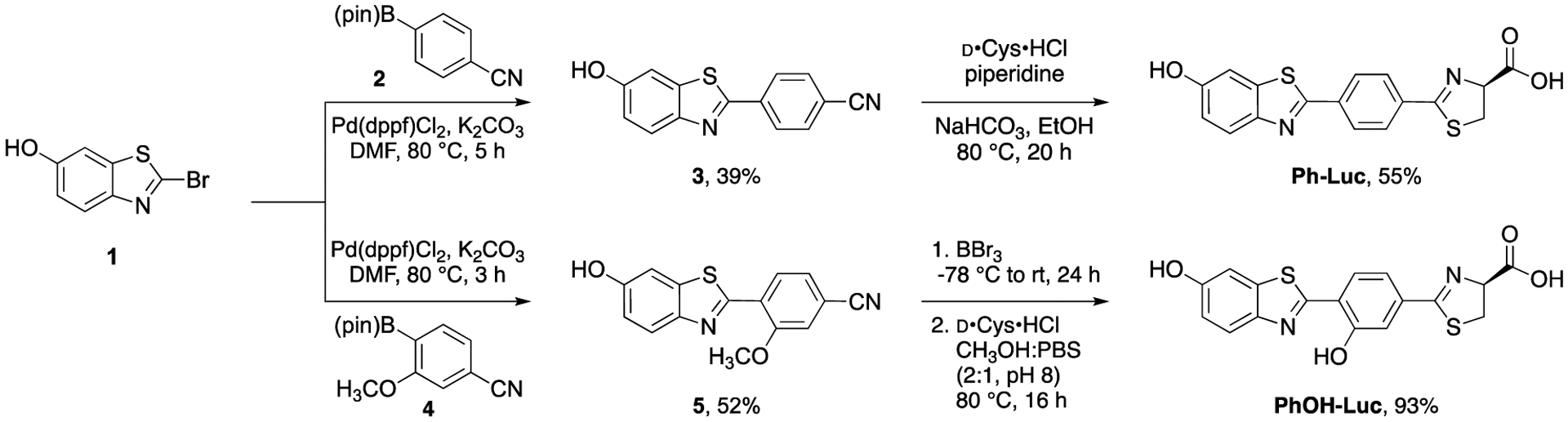
Synthesis of π-extended luciferins via a modular Suzuki-Miyaura coupling.
With the π-extended chromophores in hand, we evaluated their propensity to form planar structures via fluorescence spectroscopy. The relative emission profiles of Ph-Luc and PhOH-Luc would inform on whether intramolecular H-bonding was an effective rotational lock. Measurements were performed with the luciferin scaffolds instead of the oxyluciferin products. These latter molecules are notoriously difficult to isolate and analyze,44–45 and the key conformational parameter (rotation about C2-C2”) could be directly assayed using the parental luciferins. Ph-Luc exhibited an emission maximum (λem) of 442 nm (Figure S2a). This wavelength was characteristic of an isolated benzothiazole chromophore, suggesting a twisted excited-state geometry. By contrast, PhOH-Luc exhibited a λem of 529 nm. This red-shifted emission suggested that PhOH-Luc is likely more conjugated in the excited state compared to Ph-Luc. Methylated analogs further revealed that the pendant hydroxyl group was key to confining the chromophore in a planar geometry (Figure S2b, Scheme S3). Ph-Luc exhibited a second emission peak at 595 nm in water, which was not observed for PhOH-Luc (Figure S2a). This emission is likely due to a solvent-mediated intermolecular proton transfer at the 6’-OH, which is well documented for d-luc and related chromophores.46 Solvent-mediated proton transfer was less likely for PhOH-Luc due to competing intramolecular processes (Figures S2–3).
Analyzing bioluminescent light emission with π-extended luciferins
We next evaluated the bioluminescent properties of the π-extended analogs with Fluc. Surprisingly, no steady state light emission was observed for either luciferin when incubated with recombinant luciferase (Figure S4). This result was in contrast with analogs comprising intervening vinyl units, which were previously reported as robust emitters.35 It is possible that Fluc processed the π-extended analogs, but only extremely low levels of light were produced. In any case, the photon output was incompatible with screening. Since Ph-Luc and PhOH-Luc were already “dark” with Fluc, they would be excellent candidates for orthogonal probes. Identifying complementary enzymes was expected to be difficult, though, due to Fluc inactivity. Luciferases can be readily evolved to process unnatural luciferin analogs.19,30,36 However, evolving new enzymatic functions requires a starting point (i.e., an enzyme with some basal level of activity).47 Indeed, initial screens of small Fluc mutant libraries did not reveal any functional hits with the π-extended analogs (Figure S5).
Identifying a starting point via RosettaDesign
The lack of robust emission in our initial screens suggested that a major redesign of the luciferase active site was needed. To identify enzymes that could provide more photons with the π-extended analogs, we took cues from previous efforts to engineer lipoic acid ligase (LplA).48 LplA catalyzes the adenylation of a dithiolane-containing lipid and its subsequent attachment to target peptides. Initial attempts to modify this enzyme to accept an elongated structure (resorufin) were challenging, as LplA did not exhibit any activity towards the molecule. Rosetta-guided enzyme design was ultimately used to engineer the LplA active site to accept unnatural substrates. Beyond this example, Rosetta software has enabled the de novo design of other challenging enzyme active sites49 and protein interfaces.50–51
RosettaDesign has also been used to craft enzymes capable of binding distinct rotamers.52 In a key example, Schultz and coworkers identified an engineered aminoacyl-tRNA synthetase capable of “locking” a flexible, biphenyl amino acid into a flat geometry. We aimed to use the platform to similarly identify Fluc mutants that could maximize packing interactions between a π-extended luciferin and surrounding amino acids. Ideally, the mutations would provide luciferases with active sites that not only facilitate light emission, but also restrict substrate geometry. Because the mutants would be designed to complement the π-extended scaffolds, they would also be less likely to accept other luciferin analogs and, thus, be orthogonal.
To identify luciferase variants that could stabilize the extended π-systems in a planar conformation, the RosettaMatch53 and RosettaDesign54 algorithms were used (see Supplementary Note for detailed protocols). The modified luciferins were first generated in silico and docked into the luciferase enzyme (PDB ID: 4g36) using the RosettaMatch protocol. The RosettaDesign algorithm was then used to optimize the residues surrounding the extended substrates. Inspection of the calculation outputs indicated that many mutations suggested by Rosetta were likely necessary to accommodate the excess bulk of the new luciferin substrates. These residues served as a guide for the development of luciferase libraries that were generated as described below.
Mutants from the Rosetta calculations were ranked by shape complementarity between the designed residues and luciferin analogs. We developed a mutant library targeting the top residues predicted by Rosetta (Figure 2a). This library was prepared via targeted combinatorial codon mutagenesis (Figure S6).55 Based on a small sample, ~3–4 sites were mutated in each library member. The library was screened for light emission with Ph-Luc and PhOH-Luc via a two-tiered approach (Figure S7).19 In brief, the library was introduced into bacteria, and the transformants were plated on agar plates embedded with one of the analogs. Light-emitting colonies were identified and picked for a secondary screen. Fluc was included in the second screen, and any mutants that exhibited light emission on par or greater than that of Fluc were considered “hits.”
Figure 2. Searching for an evolutionary starting point for π-extended luciferins.
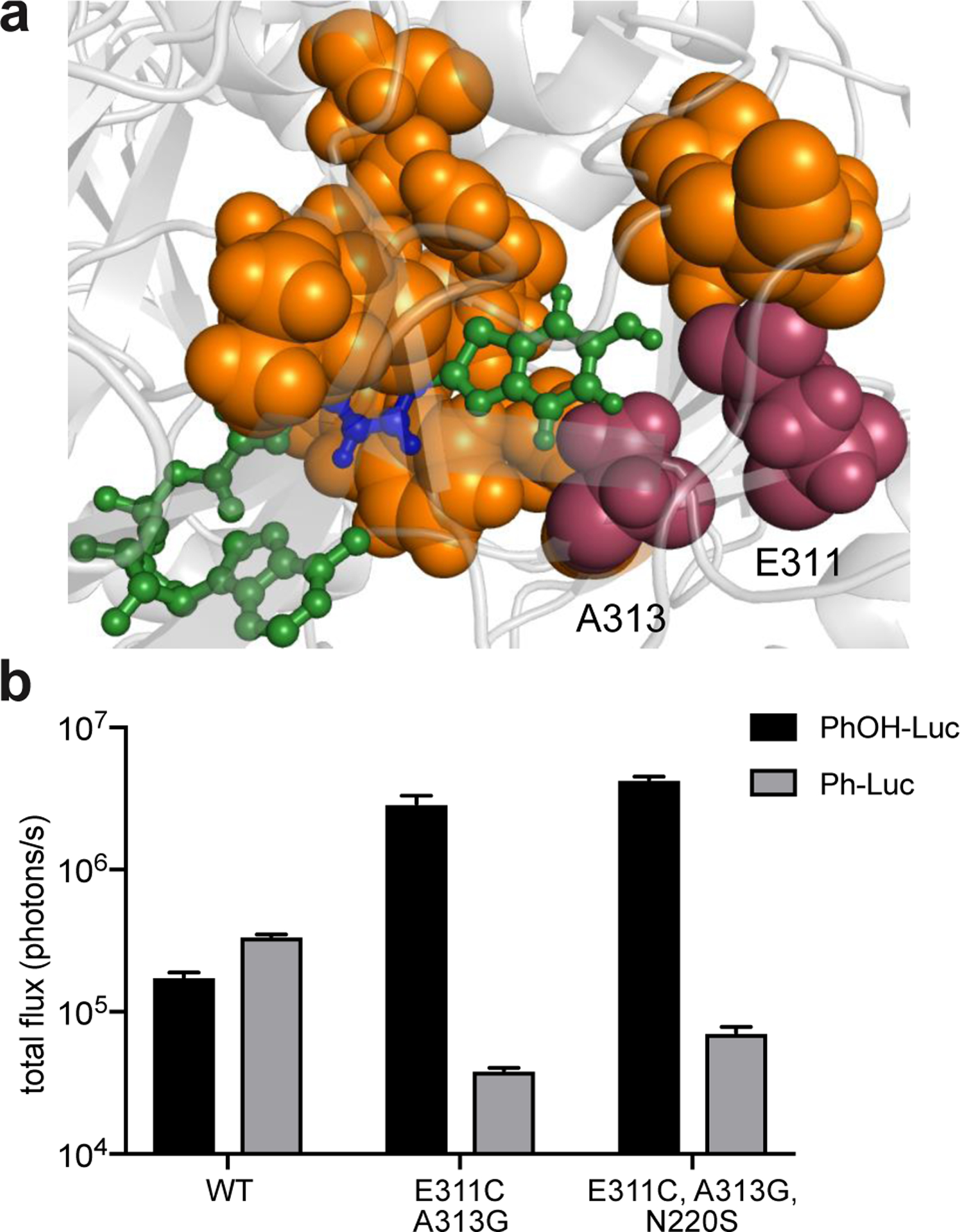
(a) Fluc docked with a π-extended luciferin was generated using existing crystal structure data (PDB: 4G36) and Rosetta. Residues targeted for library design are highlighted in orange. Key residues Glu311 and Ala313 are highlighted in magenta. (b) Two “hit” mutants identified from screening the Rosetta library. Bacteria expressing mutant enzymes were lysed and treated with π-extended analogs (250 μM). Emission intensities are provided as total photon flux values. Error bar represent the standard error of the mean for n = 3 experiments.
Gratifyingly, we were able to identify a functional mutant (G1) that exhibited improved photon outputs compared to Fluc with PhOH-Luc (Figure 2b). This mutant appeared to be selective toward the partially rigidified substrate, as it did not emit light with Ph-Luc. No functional mutant was identified from further screening the Rosetta library with Ph-Luc. The lack of hits for this analog suggested that it could take multiple mutations (>4) to remodel the luciferase active site to compensate for the substrate’s flexibility. Introducing more diversity in the Rosetta library could potentially reveal a functional variant for Ph-Luc. Since we had a starting point with PhOH-Luc, though, we decided to focus on this analog.
Characterization of the Rosetta “hits”
Sequence analyses revealed that the brightest “hit” (G1) for PhOH-Luc comprised two mutations, E311C and A313G. Both residues are part of a β-strand that sculpts the binding pocket for luciferin intermediates.12,56 Glu311 is known to participate in a hydrogen bond network involving multiple water molecules and active site side chains. Disruption of this network could provide space to accommodate the π-extended luciferin. Less is known about direct interactions involving Ala313, though mutations at this position could likely alter the conformation of the luciferase active site. Further screening revealed a mutant (G2) comprising S220N that was also functional with PhOH-Luc. Ser220 is distal from the luciferin-binding site and was not targeted in the original library. S220N likely conferred additional thermal stability.
We were able to recapitulate light emission with PhOH-Luc and recombinant G2 (Figure 3a). Signal was also detected when PhOH-Luc was incubated with a large (1000-fold) excess of Fluc. This latter result corroborated our previous steady state measurements, where photon output was found to be too low for screening. With G2, we observed a significant increase in substrate turnover, which is likely the basis for improved photon output. These results suggest that disruption of the water cluster (via the E311C mutation) promotes substrate turnover. A similar outcome was observed in recent efforts to engineer an insect luciferase to process a naphthyl modified luciferin.36
Figure 3. Biochemical analyses of lead mutant.
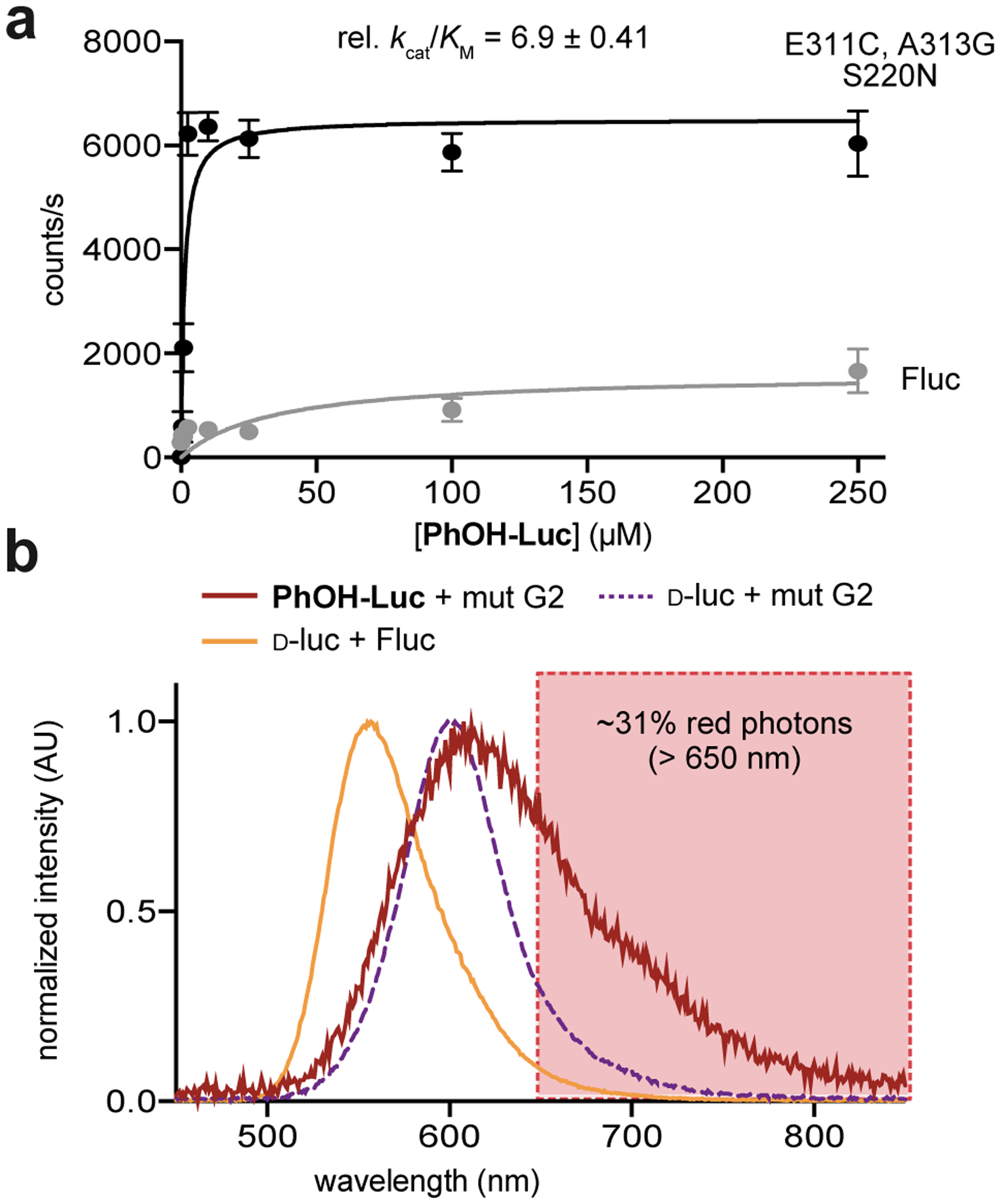
(a) Kinetic studies revealed that improved activity was likely due to increased substrate turnover. Kinetic parameters are apparent values, determined via measurements of initial rates of light emission over a range of substrate concentrations. Values were normalized to emission with Fluc. Error bars represent the standard error of the mean for n = 3 experiments. (b) Bioluminescence spectrum for PhOH-Luc with recombinant G2. Photons in the 650–850 nm region accounted for ~31% of the total emission.
With a light-emitting mutant in hand, we next evaluated its bioluminescence spectrum. Compared to Fluc and d-luc, G2 and PhOH-Luc exhibited a red-shifted λmax. The overall photon output was weak, but ~31% of the photons were > 650 nm (Figures 3b and S8). A similar red shift was observed when G2 was incubated with the native substrate, d-luc, suggesting that the change might not be substrate specific. However, an additional “shoulder” at ~700 nm was observed in the PhOH-Luc spectrum. The bimodal emission suggested the presence of two distinct light-emitting species, perhaps indicative of a flexible luminophore.57 Docking analyses predicted that PhOH-Luc adopts a twisted conformation in the binding pocket (Figure S9). Confirming the structure of the exact light emitter experimentally proved challenging, though. PhOH-Luc comprises an O-2-benzothiazolyl phenol unit. This chromophore is known to undergo an excited state intramolecular proton transfer to afford a keto tautomer (Figure S3).58 The keto tautomer could be another contributor to bimodal emission.
Evolving a panel of brighter and specific luciferases
We proceeded to optimize the Rosetta “hit” (G2) via conventional mutagenesis performed over multiple generations. We targeted residues known to modulate substrate binding (highlighted in Figure 4a). Arg218 influences the local structure of the d-luc binding site.59 Residues 314–316 are located in close proximity to the aryl-benzothiazole junction. Arg337 and Ser286 are also involved in the same hydrogen-bonding network as Glu311.60–61 Moreover, mutations at all of these positions have been shown to perturb d-luc binding, modulating both substrate turnover and emission color.10,62 Libraries targeting the sites were prepared. Screening with PhOH-Luc revealed a panel of functional mutants, but only modest gains in photon output were achieved (Figure 4b). Two enzymes comprising all beneficial mutations were also prepared. This mutant exhibited no significant improvement in activity when compared to the brightest hit (Figure S10), suggesting that a local maximum was reached in the evolutionary landscape.
Figure 4. Improving light emission of G2 via iterative screening.
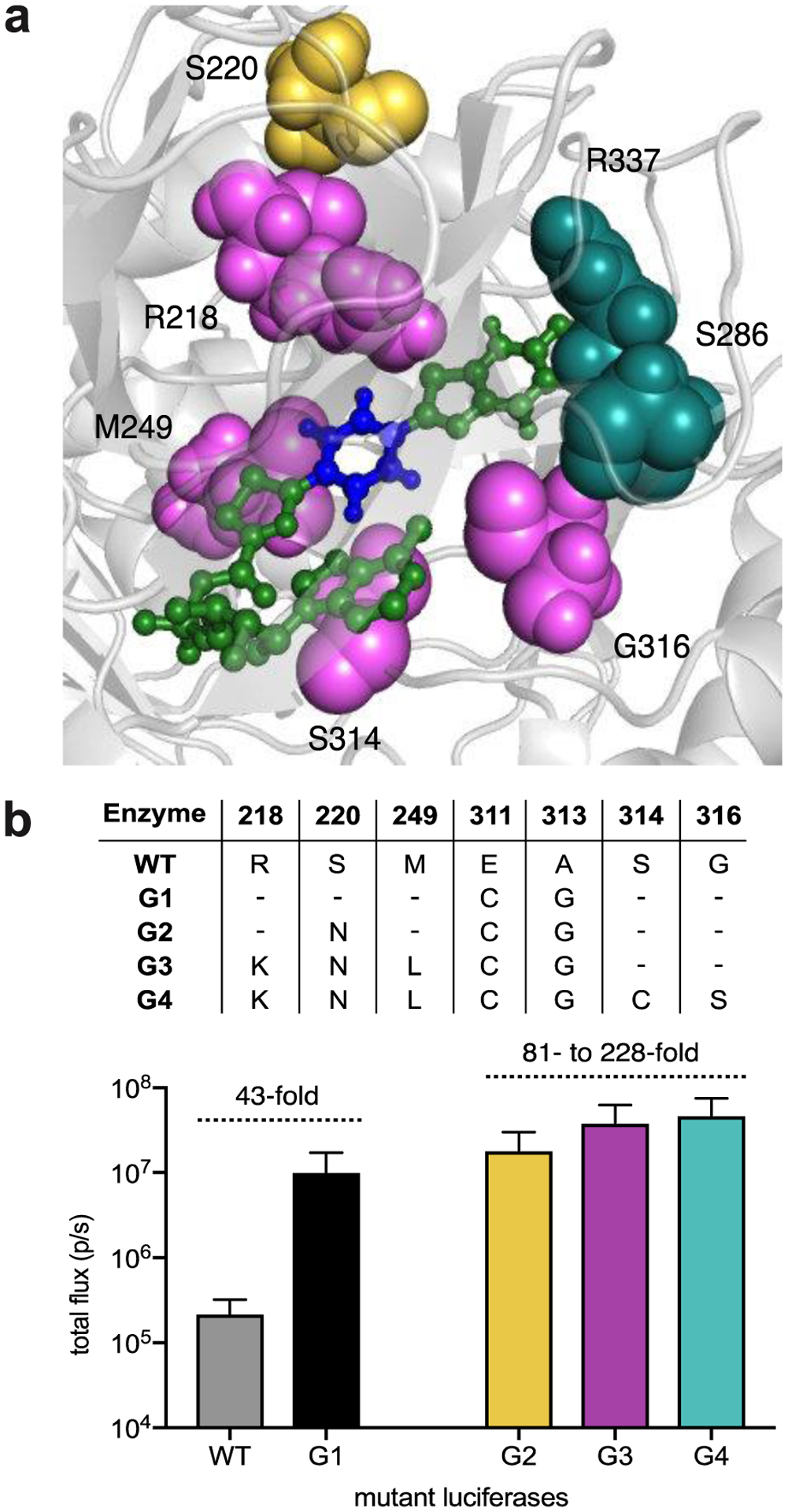
(a) Residues targeted for subsequent mutagenesis (highlighted using the docked structure generated from initial Rosetta design). These residues were selected based on their ability to modulate substrate binding. (b) Light emission of PhOH-Luc with the top performing mutants identified from each round of mutagenesis. Bacteria expressing mutant enzymes were lysed and treated with PhOH-Luc (250 μM). Emission intensities are plotted as total photon flux values. Error bars represent the standard error of the mean for n = 3 experiments.
The photon outputs from PhOH-Luc and engineered mutants are sufficient for biological imaging. The emission values (~1.7–3.0% of Fluc/d-luc emission in lysate, Figure S11) are on par with other designer bioluminescent probes that have been used for cell and animal imaging (Figure S11).19 We further found that PhOH-Luc could be used to visualize HEK293 cells expressing mutant luciferases (Figure S12).
While mutants that provided greater light outputs with PhOH-Luc were not identified, mutants that could selectively process this analog over other luciferins were found. The G4 mutants comprised R218K, M249L, and S347G. R218K and M249L have been previously found to preclude binding of C4’-modified luciferins, setting the stage for orthogonality.20,63 Likewise, S347G was previously found to exclude C7’-modified analogs. Enzyme hits from later generations (G4 and G5) were also not functional with Ph-Luc. This result was surprising, given the structural similarity shared among the extended scaffolds. It is possible that the hydroxy group of PhOH-Luc might participate in additional hydrogen bonding with the enzyme, facilitating binding or turnover (Figure S9b).
Searching for orthogonal luciferase-luciferin pairs
Given the unique structural properties of PhOH-Luc, we reasoned that the probe was likely also orthogonal to existing luciferin analogs. Indeed, mutants identified from the PhOH-Luc screens were minimally reactive towards luciferins with steric or electronic modifications (Figure S13). Analogously, PhOH-Luc was not emissive with luciferases evolved for different analogs. To further demonstrate the preference between PhOH-Luc and its complementary luciferase, we screened the π-extended analog against a panel of 209 previously published luciferase mutants (Figure S14).20 A cross-comparison algorithm predicted that PhOH-Luc and G3-G5 mutants would be orthogonal to 23 other engineered luciferase-luciferin pairs. These data highlight the value of structural diversity in achieving orthogonality.
We also reasoned that the π-extended scaffold could be used in tandem with Fluc/d-luc, as PhOH-Luc is minimally active with Fluc. Differentiating an engineered luciferase from Fluc has been historically challenging—most synthetic analogs react with Fluc and most mutants react with d-luc.6,64 Moreover, bioluminescent pairs that are truly orthogonal to Fluc/d-luc typically require long imaging times.65 The signal acquired from one luciferase-luciferin pair must clear prior to administration of the second luciferin, a process that can take hours-to-days. To reduce imaging time, we recently developed a platform that relies on rapid substrate unmixing.65 Orthogonal probes can be applied and imaged sequentially with minimal delay. The method requires collections of enzymes with unique substrate preferences. The probes must also differ in emission intensity, enabling signals to be “layered” in, beginning with the dimmest probe first. We hypothesized that PhOH-Luc/G4 would be amenable for rapid, multicomponent imaging as this pair is both substrate- and intensity- resolved from other well-known bioluminescent tools. For example, PhOH-Luc is used preferentially by mutant G4, and this pair is ~100-fold dimmer from Fluc/d-luc (Figure 5a). Substrate resolution ensures that mutant G4 will be illuminated when PhOH-Luc is first added to a mixture of the two luciferases. Signal from Fluc/d-luc can then be directly layered on top of the PhOH-Luc/G4 emission, without the need for signal clearance. Since the brighter probes are added last, the residual signal from the dimmer first pair becomes part of the background. This layering effect can be coupled with substrate-unmixing algorithms to deconvolute both sets of signals.65–66
Figure 5. Rapid, two-component imaging with a π-extended luciferin.
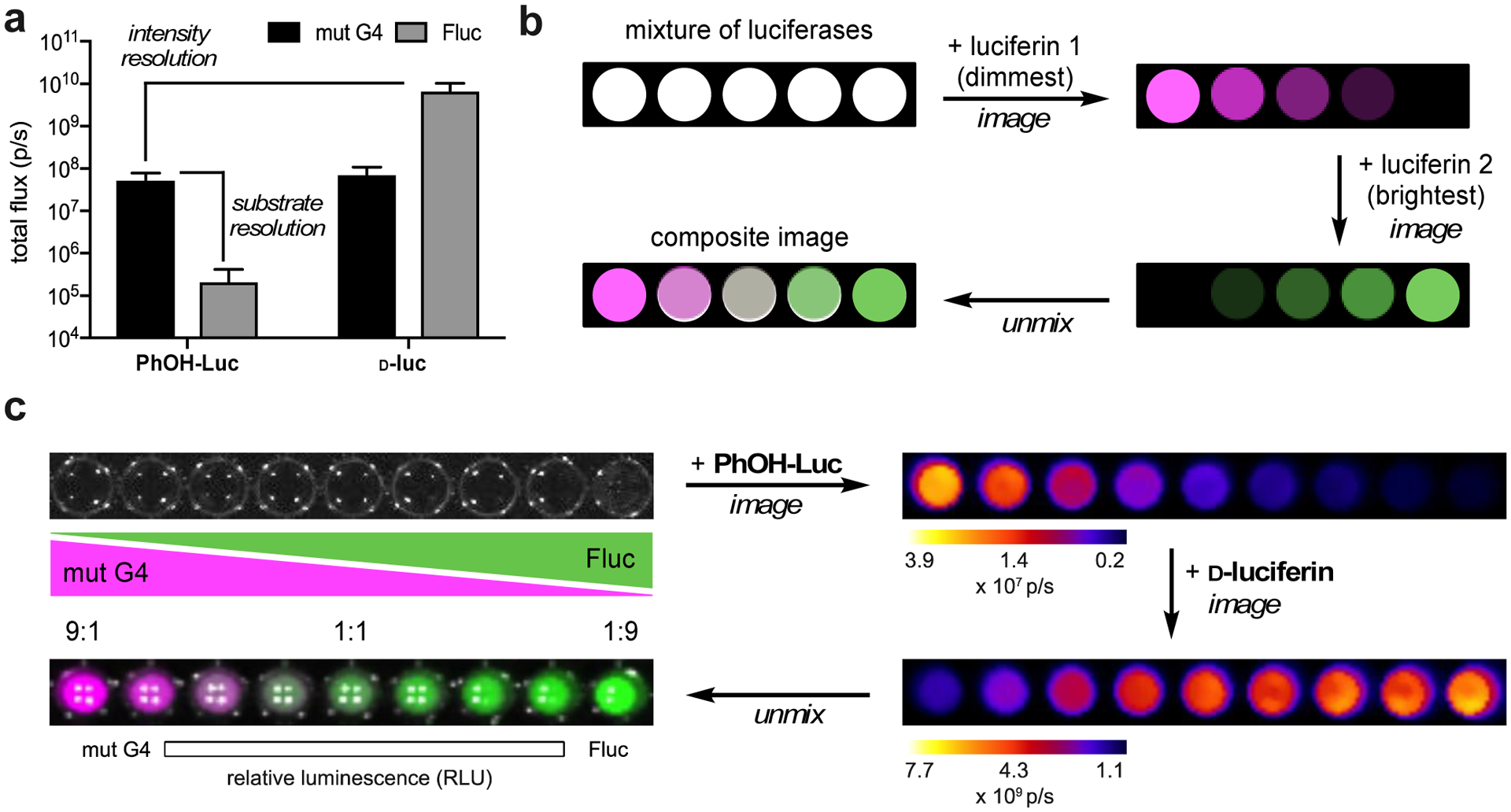
(a) G4/PhOH-Luc are both substrate- and intensity-resolved from Fluc/d-luc. (b) Schematic for resolving multiple luciferases via substrate unmixing. Luciferin analogs were administered sequentially with minimal delay time. Bioluminescent signals generated from each addition were deconvoluted via linear unmixing. (c) A model two-component assay. Gradients of G4 and Fluc (expressed in bacterial lysate) were plated in a 96-well plate. PhOH-Luc (250 μM) was administered, followed by d-luc (100 μM). Images were acquired after each addition. The raw data were stacked and unmixed. The processed image is false colored.
A model assay was used to evaluate the utility of PhOH-Luc/G4 for rapid, sequential imaging. Bacteria expressing either mutant G4 or Fluc were lysed and plated in a gradient fashion (Figure 5b). PhOH-luc was added the wells, followed by d-luc, and images were collected for each substrate. Image acquisition was completed within 10–15 minutes, a significant improvement over conventional protocols requiring hours or days. The resulting data were then combined and processed using a linear unmixing algorithm.65 The analysis provides a false-colored image of the wells with signal-intensity correlated to the light output of unique enzyme-substrate pairs (e.g. red representing signal from G4/PhOH-Luc and green representing Fluc/d-luc pair). Excitingly, the concentration gradient mirrored the color distribution in the processed image (Figure 5c). The unmixing algorithm also removed residual signal from G4/d-luc, indicating that perfect substrate orthogonality is not necessary. Rapid, two-component imaging was also possible when PhOH-Luc and its associated mutants were paired with other engineered bioluminescent probes, including Pecan/4′Br-Luc and Akaluc/AkaLumine (Figure S15).30,65 This result demonstrated that PhOH-Luc and mutants can be readily used in combination with existing bioluminescent tools for fast multicomponent imaging in vitro.
Multicomponent imaging often requires resolving more than two probes. Thus, we set out to identify expanded collections of unique enzyme-substrate pairs. We reasoned that the unique architecture of PhOH-Luc could expedite the search for triplets, quadruplets and higher order sets. Because PhOH-Luc was already orthogonal to most existing analogs, a triplet set could be readily achieved by combining PhOH-Luc and its preferred mutant with a known orthogonal pair. Indeed, mutant G4 was readily differentiated from two mutant luciferases that process sterically modified luciferins (Figure S16), providing an orthogonal triplet. A similar approach was used to identify a bioluminescent quartet. Mutant G4/PhOH-Luc was combined with a triplet set of orthogonal probes previously reported by our lab.20 Light emission patterns unique to each enzyme/substrate pair were observed (Figure 6a). Image processing was again used to recognize and deconvolute the unique analog signatures. The enzyme-substrate pairs were readily differentiated as shown in the false-colored image (Figure 6b). To our knowledge, this is the first example of imaging four bioluminescent reporters via substrate resolution. It should be noted that rapid compound administration and signal unmixing was not possible in the triplet and quadruple imaging examples. The overall emission intensities of the enzyme-substrate pairs are similar, precluding the “layering-in” of signals. Bioluminescence from the first pair must clear before the second can be imaged. Further engineering of the probes—to achieve varying degrees of light output (and thus rapid imaging)—is underway.
Figure 6. Quadruple bioluminescence imaging.
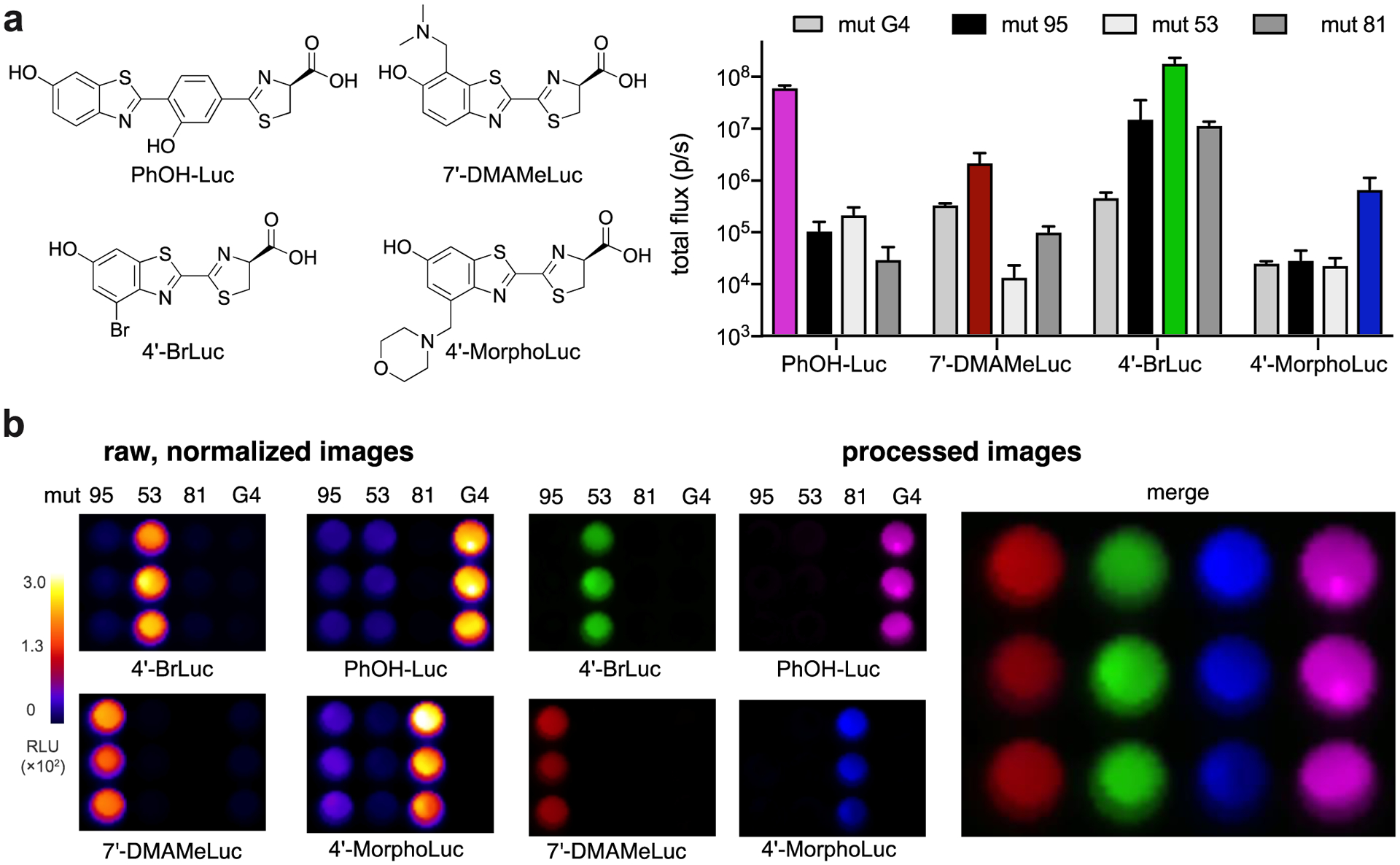
(a) Mutant luciferases were expressed in bacteria. Comparable levels of expression were observed (Figure S17). Cells were lysed, then distributed across four 96-well plates (in a 3×4 matrix). Saturating doses of the corresponding luciferin analogs were administered ([PhOH-Luc] = 250 μM, [7′-DMAMeLuc] = 250 μM, [4′-BrLuc] = 100 μM, [4′-MorphoLuc] = 250 μM). Images were acquired, and the raw emission data were stacked and unmixed. (b) Unmixed images from (a) depicting a quartet of unique luciferase-luciferin pairs. Emission intensities are plotted as total photon flux values. Error bars represent the standard error of the mean for n = 3 experiments.
CONCLUSIONS
We developed novel luciferin analogs with extended π-conjugation for multicomponent imaging. These substrates not only provide unique molecular architectures for orthogonal substrate design, but also exhibit red-shifted emission. Like many other modified luciferin analogs, the π-extended probes were poor substrates with the native luciferase (Fluc). Efforts to improve enzyme activity by rational, site-directed mutagenesis were not successful. Thus, we turned to computationally guided active site remodeling using RosettaDesign. Rosetta software was used to reshape the luciferase active site and identify enzymes tolerant of longer scaffolds. Screening of a combinatorial library provided a mutant with ~100-fold improved activity. Further engineering provided enzymes that could potentially enforce substrate and were substrate selective. The unique luciferase-luciferin pairs identified with the π-extended molecules were also orthogonal to previously published probes. The added structural diversity enabled orthogonal triplet and quartet imaging to be readily achieved. Such expanded bioluminescence capabilities can enable rapid, multicomponent imaging. While the exact nature of the π-extended light emitters remains unknown, the analogs could serve as templates for next-generation scaffolds. More diverse luciferins are necessary to grow the bioluminescent toolkit. Future work will also focus on evolving mutants that can process π-extended luciferins more efficiently and provide even more red-shifted light.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the U.S. National Institutes of Health (R01 GM107630 to J.A.P.). Z.Y. was supported by the National Science Foundation via the BEST IGERT (DGE-1144901) program and a Graduate Research Fellowship (DGE-1321846). B.S.Z. was supported by a GAANN fellowship. Some experiments were performed at the Laser Spectroscopy labs (LSL) at UCI. We thank Colin Rathbun and Caroline Brennan for assisting with image processing, Krysten Jones and Sierra Williams for assisting with analog screening, along with other members of the Prescher lab for helpful discussions. We also thank members of the Weiss, Overman, Jarvo, Rychnovsky and Martin laboratories for providing equipment and reagents.
Funding Sources
The authors declare no competing financial interests.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Experimental details on luciferin synthesis, Rosetta analysis, library development, analog screens, and bioluminescence imaging are included.
REFERENCES
- 1.Paley MA; Prescher JA Bioluminescence: a versatile technique for imaging cellular and molecular features. MedChemComm 2014, 5, 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prescher JA; Contag CH Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol 2010, 14, 80–89. [DOI] [PubMed] [Google Scholar]
- 3.Yeh H-W; Ai H-W Development and applications of bioluminescent and chemiluminescent reporters and biosensors. Annu. Rev. Anal. Chem 2019, 12, 129–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thorne N; Inglese J; Auld DS Illuminating insights into firefly luciferase and other bioluminescent reporters used in chemical biology. Chem. Biol 2010, 17, 646–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yao Z; Zhang BS; Prescher JA Advances in bioluminescence imaging: new probes from old recipes. Curr. Opin. Chem. Biol 2018, 45, 148–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams SJ; Prescher JA Building biological flashlights: Orthogonal luciferases and luciferins for in vivo imaging. Acc. Chem. Res 2019, 52, 3039–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaskova ZM; Tsarkova AS; Yampolsky IV 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev 2016, 45, 6048–6077. [DOI] [PubMed] [Google Scholar]
- 8.Rathbun CM; Prescher JA Bioluminescent probes for imaging biology beyond the culture dish. Biochemistry 2017, 56, 5178–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mezzanotte L; Que I; Kaijzel E; Branchini B; Roda A; Löwik C Sensitive dual color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS ONE 2011, 6, e19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branchini BR; Ablamsky DM; Rosenman JM; Uzasci L; Southworth TL; Zimmer M Synergistic mutations produce blue-shifted bioluminescence in firefly luciferase. Biochemistry 2007, 46, 13847–13855. [DOI] [PubMed] [Google Scholar]
- 11.Branchini BR; Ablamsky DM; Murtiashaw MH; Uzasci L; Fraga H; Southworth TL Thermostable red and green light-producing firefly luciferase mutants for bioluminescent reporter applications. Anal. Biochem 2007, 361, 253–262. [DOI] [PubMed] [Google Scholar]
- 12.Stowe CL; Burley TA; Allan H; Vinci M; Kramer-Marek G; Ciobota DM; Parkinson GN; Southworth TL; Agliardi G; Hotblack A; Lythgoe MF; Branchini BR; Kalber TL; Anderson JC; Pule MA Near-infrared dual bioluminescence imaging in mouse models of cancer using infraluciferin. eLife 2019, 8, e45801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhaumik S; Lewis XZ; Gambhir SS Optical imaging of Renilla luciferase, synthetic Renilla luciferase, and firefly luciferase reporter gene expression in living mice. J. Biomed. Opt 2004, 9, 578–586. [DOI] [PubMed] [Google Scholar]
- 14.Fan F; Wood KV Bioluminescent assays for high-throughput screening. ASSAY Drug Dev. Techn 2007, 5, 127–136. [DOI] [PubMed] [Google Scholar]
- 15.Kotlobay AA; Sarkisyan KS; Mokrushina YA; Marcet-Houben M; Serebrovskaya EO; Markina NM; Somermeyer LG; Gorokhovatsky AY; Vvedensky A; Purtov KV; Petushkov VN; Rodionova NS; Chepurnyh TV; Fakhranurova LI; Guglya EB; Ziganshin R; Tsarkova AS; Kaskova ZM; Shender V; Abakumov M; Abakumova TO; Povolotskaya IS; Eroshkin FM; Zaraisky AG; Mishin AS; Dolgov SV; Mitiouchkina TY; Kopantzev EP; Waldenmaier HE; Oliveira AG; Oba Y; Barsova E; Bogdanova EA; Gabaldón T; Stevani CV; Lukyanov S; Smirnov IV; Gitelson JI; Kondrashov FA; Yampolsky IV Genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. U. S. A 2018, 115, 12728–12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsarkova AS; Kaskova ZM; Yampolsky IV A tale of two luciferins: fungal and earthworm new bioluminescent systems. Acc. Chem. Res 2016, 49, 2372–2380. [DOI] [PubMed] [Google Scholar]
- 17.Kim SB; Nishihara R; Citterio D; Suzuki K Fabrication of a new lineage of artificial luciferases from natural luciferase pools. ACS Comb. Sci 2017, 19, 594–599. [DOI] [PubMed] [Google Scholar]
- 18.Hananya N; Shabat D Recent advances and challenges in luminescent Imaging: bright outlook for chemiluminescence of dioxetanes in water. ACS Cent. Sci 2019, 5, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones KA; Porterfield WB; Rathbun CM; McCutcheon DC; Paley MA; Prescher JA Orthogonal luciferase–luciferin pairs for bioluminescence imaging. J. Am. Chem. Soc 2017, 139, 2351–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathbun CM; Porterfield WB; Jones KA; Sagoe MJ; Reyes MR; Hua CT; Prescher JA Parallel screening for rapid identification of orthogonal bioluminescent tools. ACS Cent. Sci 2017, 3, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mofford DM; Reddy GR; Miller SC Aminoluciferins extend firefly luciferase bioluminescence into the near-infrared and can be preferred substrates over d-luciferin. J. Am. Chem. Soc 2014, 136, 13277–13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams ST; Mofford DM; Reddy GSKK; Miller SC Firefly luciferase mutants allow substrate-selective bioluminescence imaging in the Mouse Brain. Angew. Chem. Int. Ed 2016, 55, 4943–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Contag CH; Spilman SD; Contag PR; Oshiro M; Eames B; Dennery P; Stevenson DK; Benaron DA Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem. Photobiol 1997, 66, 523–531. [DOI] [PubMed] [Google Scholar]
- 24.Berger F; Paulmurugan R; Bhaumik S; Gambhir SS Uptake kinetics and biodistribution of 14C-d-luciferin—a radiolabeled substrate for the firefly luciferase catalyzed bioluminescence reaction: impact on bioluminescence based reporter gene imaging. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 2275–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H; Doyle TC; Coquoz O; Kalish F; Rice BW; Contag CH Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J. Biomed. Opt 2005, 10, 41210. [DOI] [PubMed] [Google Scholar]
- 26.Iwano S; Obata R; Miura C; Kiyama M; Hama K; Nakamura M; Amano Y; Kojima S; Hirano T; Maki S; Niwa H Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light. Tetrahedron 2013, 69, 3847–3856. [Google Scholar]
- 27.Miura C; Kiyama M; Iwano S; Ito K; Obata R; Hirano T; Maki S; Niwa H Synthesis and luminescence properties of biphenyl-type firefly luciferin analogs with a new, near-infrared light-emitting bioluminophore. Tetrahedron 2013, 69, 9726–9734. [Google Scholar]
- 28.Kitada N; Saitoh T; Ikeda Y; Iwano S; Obata R; Niwa H; Hirano T; Miyawaki A; Suzuki K; Nishiyama S; Maki SA Toward bioluminescence in the near-infrared region: Tuning the emission wavelength of firefly luciferin analogues by allyl substitution. Tetrahedron Lett 2018, 59, 1087–1090. [Google Scholar]
- 29.Jathoul AP; Grounds H; Anderson JC; Pule MA A dual-color far-red to near-infrared firefly luciferin analogue designed for multiparametric bioluminescence imaging. Angew. Chem. Int. Ed 2014, 53, 13059–13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwano S; Sugiyama M; Hama H; Watakabe A; Hasegawa N; Kuchimaru T; Tanaka KZ; Takahashi M; Ishida Y; Hata J; Shimozono S; Namiki K; Fukano T; Kiyama M; Okano H; Kizaka-Kondoh S; McHugh TJ; Yamamori T; Hioki H; Maki S; Miyawaki A Single-cell bioluminescence imaging of deep tissue in freely moving animals. Science 2018, 359, 935–939. [DOI] [PubMed] [Google Scholar]
- 31.Weissleder R; Ntziachristos V Shedding light onto live molecular targets. Nat. Med 2003, 9, 123–128. [DOI] [PubMed] [Google Scholar]
- 32.Hilderbrand SA; Weissleder R Near-infrared fluorescence: application to in vivo molecular imaging. Curr. Opin. Chem. Biol 2010, 14, 71–79. [DOI] [PubMed] [Google Scholar]
- 33.Dawson JB; Barker DJ; Ellis DJ; Cotterill JA; Grassam E; Fisher GW; Feather JW A theoretical and experimental study of light absorption and scattering by in vivo skin. Phys. Med. Biol 1980, 25, 695–709. [DOI] [PubMed] [Google Scholar]
- 34.Rice BW; Contag CH The importance of being red. Nat. Biotechnol 2009, 27, 624–625. [DOI] [PubMed] [Google Scholar]
- 35.Kuchimaru T; Iwano S; Kiyama M; Mitsumata S; Kadonosono T; Niwa H; Maki S; Kizaka-Kondoh S A luciferin analogue generating near-infrared bioluminescence achieves highly sensitive deep-tissue imaging. Nat. Commun 2016, 7, 11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall MP; Woodroofe CC; Wood MG; Que I; van’t Root M; Ridwan Y; Shi C; Kirkland TA; Encell LP; Wood KV; Löwik C; Mezzanotte L Click beetle luciferase mutant and near infrared naphthyl-luciferins for improved bioluminescence imaging. Nat. Commun 2018, 9, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson JC; Grounds H; Jathoul AP; Murray JAH; Pacman SJ; Tisi L Convergent synthesis and optical properties of near-infrared emitting bioluminescent infra-luciferins. RSC Adv. 2017, 7, 3975–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stepanyuk GA; Unch J; Malikova NP; Markova SV; Lee J; Vysotski ES Coelenterazine-v ligated to Ca2+-triggered coelenterazine-binding protein is a stable and efficient substrate of the red-shifted mutant of Renilla muelleri luciferase. Anal. Bioanal. Chem 2010, 398, 1809–1817. [DOI] [PubMed] [Google Scholar]
- 39.Gorka AP; Nani RR; Zhu J; Mackem S; Schnermann MJ A near-IR uncaging strategy based on cyanine photochemistry. J. Am. Chem. Soc 2014, 136, 14153–14159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Upadhyayula S; Nuñez V; Espinoza EM; Larsen JM; Bao D; Shi D; Mac JT; Anvari B; Vullev VI Photoinduced dynamics of a cyanine dye: parallel pathways of non-radiative deactivation involving multiple excited-state twisted transients. Chem. Sci 2015, 6, 2237–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baloban M; Shcherbakova DM; Pletnev S; Pletnev VZ; Lagarias JC; Verkhusha VV Designing brighter near-infrared fluorescent proteins: insights from structural and biochemical studies. Chem. Sci 2017, 8, 4546–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shcherbakova DM; Baloban M; Emelyanov AV; Brenowitz M; Guo P; Verkhusha VV Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun 2016, 7, 12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang M; Yu R; Xu K; Ye S; Kuang S; Zhu X; Wan Y An arch-bridge-type fluorophore for bridging the gap between aggregation-caused quenching (ACQ) and aggregation-induced emission (AIE). Chem. Sci 2016, 7, 4485–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maltsev OV; Nath NK; Naumov P; Hintermann L Why is Firefly Oxyluciferin a Notoriously Labile Substance? Angew. Chem. Int. Ed 2014, 53, 847–850. [DOI] [PubMed] [Google Scholar]
- 45.Naumov P; Ozawa Y; Ohkubo K; Fukuzumi S Structure and spectroscopy of oxyluciferin, the light emitter of the firefly bioluminesence. J. Am. Chem. Soc 2009, 131, 11590–11605. [DOI] [PubMed] [Google Scholar]
- 46.Erez Y; Huppert D Excited-state intermolecular proton transfer of the firefly’s chromophore d-luciferin. J. Phys. Chem. A 2010, 114, 8075–8082. [DOI] [PubMed] [Google Scholar]
- 47.Brustad EM; Arnold FH Optimizing non-natural protein function with directed evolution. Curr. Opin. Chem. Biol 2011, 15, 201–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu DS; Nivón LG; Richter F; Goldman PJ; Deerinck TJ; Yao JZ; Richardson D; Phipps WS; Ye AZ; Ellisman MH; Drennan CL; Baker D; Ting AY Computational design of a red fluorophore ligase for site-specific protein labeling in living cells. Proc. Natl. Acad. Sci. U. S. A 2014, 111, E4551–E4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J; Liu Y; Liu Y; Zheng S; Wang X; Zhao J; Yang F; Zhang G; Wang C; Chen PR Time-resolved protein activation by proximal decaging in living systems. Nature 2019, 569, 509–513. [DOI] [PubMed] [Google Scholar]
- 50.Chen Z; Boyken SE; Jia M; Busch F; Flores-Solis D; Bick MJ; Lu P; VanAernum ZL; Sahasrabuddhe A; Langan RA; Bermeo S; Brunette TJ; Mulligan VK; Carter LP; DiMaio F; Sgourakis NG; Wysocki VH; Baker D Programmable design of orthogonal protein heterodimers. Nature 2019, 565, 106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dou J; Vorobieva AA; Sheffler W; Doyle LA; Park H; Bick MJ; Mao B; Foight GW; Lee MY; Gagnon LA; Carter L; Sankaran B; Ovchinnikov S; Marcos E; Huang P-S; Vaughan JC; Stoddard BL; Baker D De novo design of a fluorescence-activating β-barrel. Nature 2018, 561, 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson AD; Mills JH; Song Y; Nasertorabi F; Han GW; Baker D; Stevens RC; Schultz PG Trapping a transition state in a computationally designed protein bottle. Science 2015, 347, 863–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tinberg CE; Khare SD; Dou J; Doyle L; Nelson JW; Schena A; Jankowski W; Kalodimos CG; Johnsson K; Stoddard BL; Baker D Computational design of ligand-binding proteins with high affinity and selectivity. Nature 2013, 501, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanghellini A; Jiang L; Wollacott AM; Cheng G; Meiler J; Althoff EA; Röthlisberger D; Baker D New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006, 15, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Belsare KD; Andorfer MC; Cardenas FS; Chael JR; Park HJ; Lewis JC A simple combinatorial codon mutagenesis method for targeted protein engineering. ACS Synth. Biol 2017, 6, 416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Branchini BR; Southworth TL; Fontaine DM; Murtiashaw MH; McGurk A; Talukder MH; Qureshi R; Yetil D; Sundlov JA; Gulick AM Cloning of the orange light-producing luciferase from Photinus scintillans—a new proposal on how bioluminescence color is determined. Photochem. Photobiol 2017, 93, 479–485. [DOI] [PubMed] [Google Scholar]
- 57.Hu R; Feng J; Hu D; Wang S; Li S; Li Y; Yang G A rapid aqueous fluoride ion sensor with dual output modes. Angew. Chem. Int. Ed 2010, 49, 4915–4918. [DOI] [PubMed] [Google Scholar]
- 58.Chen C-L; Chen Y-T; Demchenko AP; Chou P-T Amino proton donors in excited-state intramolecular proton-transfer reactions. Nat. Rev. Chem 2018, 2, 131–143. [Google Scholar]
- 59.Branchini BR; Southworth TL; Murtiashaw MH; Boije H; Fleet SE A mutagenesis study of the putative luciferin binding site residues of firefly luciferase. Biochemistry 2003, 42, 10429–10436. [DOI] [PubMed] [Google Scholar]
- 60.Viviani VR; Simões A; Bevilaqua VR; Gabriel GVM; Arnoldi FGC; Hirano T Glu311 and Arg337 stabilize a closed active-site conformation and provide a critical catalytic base and countercation for green bioluminescence in beetle Luciferases. Biochemistry 2016, 55, 4764–4776. [DOI] [PubMed] [Google Scholar]
- 61.Branchini BR; Fontaine DM; Southworth TL; Huta BP; Racela A; Patel KD; Gulick AM Mutagenesis and structural studies reveal the basis for the activity and stability properties that distinguish the Photinus luciferases scintillans and pyralis. Biochemistry 2019, 58, 4293–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viviani VR; Amaral DT; Neves DR; Simões A; Arnoldi FGC The luciferin binding site residues C/T311 (S314) influence the bioluminescence color of beetle luciferases through main-chain Interaction with oxyluciferin phenolate. Biochemistry 2013, 52, 19–27. [DOI] [PubMed] [Google Scholar]
- 63.Liu MD; Warner EA; Morrissey CE; Fick CW; Wu TS; Ornelas MY; Ochoa GV; Zhang BS; Rathbun CM; Porterfield WB; Prescher JA; Leconte AM Statistical coupling analysis-guided library design for the discovery of mutant luciferases. Biochemistry 2018, 57, 663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miller SC; Mofford DM; Adams ST Lessons learned from luminous luciferins and latent luciferases. ACS Chem. Biol 2018, 13, 1734–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rathbun CM; Ionkina AA; Yao Z; Jones KA; Porterfield WB; Prescher JA Rapid multicomponent bioluminescence imaging via substrate unmixing. bioRxiv 2019, 811026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gammon ST; Leevy WM; Gross S; Gokel GW; Piwnica-Worms D Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal. Chem 2006, 78, 1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


