Abstract
The extensively utilized tool to detect novel coronavirus (COVID-19) is a real-time polymerase chain reaction (RT-PCR). However, RT-PCR kits are costly and consume critical time, around 6 to 9 hours to classify the subjects as COVID-19(+) or COVID-19(-). Due to the less sensitivity of RT-PCR, it suffers from high false-negative results. To overcome these issues, many deep learning models have been implemented in the literature for the early-stage classification of suspected subjects. To handle the sensitivity issue associated with RT-PCR, chest CT scans are utilized to classify the suspected subjects as COVID-19 (+), tuberculosis, pneumonia, or healthy subjects. The extensive study on chest CT scans of COVID-19 (+) subjects reveals that there are some bilateral changes and unique patterns. But the manual analysis from chest CT scans is a tedious task. Therefore, an automated COVID-19 screening model is implemented by ensembling the deep transfer learning models such as Densely connected convolutional networks (DCCNs), ResNet152V2, and VGG16. Experimental results reveal that the proposed ensemble model outperforms the competitive models in terms of accuracy, f-measure, area under curve, sensitivity, and specificity.
Keywords: Deep learning, COVID-19, Chest CT, Transfer learning
Introduction
The SARS-CoV-2 known as novel coronavirus is a communicable virus that belongs to the brood of coronaviruses. SARS-CoV-2 is 80% similar to SARS-CoV-1 in terms of genetic sequence and both are originated from bats [1, 2]. The disease caused by SARS-CoV-2 is known as COVID-19. The first infected case of this virus was reported in the Wuhan city of China in December 2019. Thereafter, COVID-19 was declared a global pandemic on 11 March 2020 [3]. This virus has promptly spread in the whole world to date. As of 12 December 2020, more than 20 million persons have been infected and more than 1.6 million persons have died due to this deadly virus [4]. These numbers are continuously increased due to the possibility of a second wave of coronavirus. The resources such as ICU, ventilators, and protecting kits are becoming exhausted due to the increase in the number of COVID-19 patients. The main symptoms of COVID-19 are fever, cough, smell loss, fatigue, and shortness in breathing [5, 6]. In some cases, infected persons may be asymptomatic. Due to the contiguous nature of the virus, the early detection of COVID-19 is required. Nowadays, the infection of coronavirus is tested through Reverse Transcription-Polymerase Chain Reaction (RT-PCR). However, RT-PCR test is time-consuming and has high false-negative [7]. To resolve the above-mentioned problems, radiographic imaging techniques such as computed tomography(CT) and X-ray are used [8]. These techniques are used to quantify the disease severity in the lung infection. CT provides better sensitivity than the X-ray and RT-PCR [9]. The analysis of CT scans requires radiological experts. In this pandemic situation, a lot of burdens have been put on radiological experts for analysis. To alleviate this issue, deep learning techniques are utilized. The pre-trained deep learning models are used for automatic analysis of chest CT scan [10, 11]. The sensitivity and specificity of pre-trained deep learning models are still far from the optimal results. The manual analysis of CT scan takes a lot of time and error-prone.
The main contributions of this paper are three-fold as follows:
To handle the less sensitivity issue with RT-PCR, chest CTs images are utilized in this paper to classify the suspected subjects as COVID-19 (+), tuberculosis, pneumonia, or healthy subjects.
An automated COVID-19 screening model is implemented by ensembling the deep transfer learning models such as Densely connected convolutional networks (DCCNs), ResNet152V2, and VGG16.
The proposed ensemble model outperforms the competitive models in terms of various performance measures such as accuracy, f-measure, area under curve, sensitivity, and specificity.
The rest of this paper is organized as follows. Section 2 presents related works. Section 3 presents the proposed deep learning model. The experimental results and discussion are mentioned in Section 4. The concluding remarks are drawn in Section 5.
Related works
Mukherjee et al. [12] presented a lightweight CNN model for classification of COVID-19 infected persons. Chest CT-scan and X-ray were used to validate the proposed ensemble model and reported the accuracy of 96.28%. The performance of the proposed ensemble model can be further improved by reducing the computational time. Singh et al. [13] developed an automatic approach for chest CT-based COVID-19 classification. They used CNN model to categorize whether the person is infected with coronavirus or other viruses. A multi-objective function was designed by considering specificity and sensitivity. The proposed ensemble model outperforms the other competitive algorithms in terms of accuracy, sensitivity, specificity, and F-measure. Wang et al. [14] proposed a novel learning framework for the identification of COVID-19 patients. They redesigned COVID-Net by incorporate the modification in architecture and learning strategy. The proposed framework achieved improvement over COVID-Net by 12.6% in terms of accuracy. However, the performance of this framework can be enhanced by utilizing transfer learning. Javaheri et al. [15] implemented a CovidCTNet for detection of coronavirus infection in patients using CT scans. The performance of CovidCTNet was tested on 89,145 slices of all CT scans of 287 patients. The accuracy obtained from CovidCTNet was 90%. Mishra et al. [16] proposed a fusion approach that integrated the predictions of existing deep CNN models. The proposed approach was trained on the COVID-CT dataset and attained an accuracy of 86%. However, the proposed approach was only applied to axial slices. Ahuja et al. [17] implemented a three-phase deep learning model for the identification of COVID-19 patients using CT scans. Data augmentation was applied to the CT scans. Four pre-trained deep architectures namely ResNet18, ResNet50, ResNet101, and SqueezeNet were used for disease classification. They reported that the accuracy obtained from ResNet18 was 99.65%. Liu et al. [18] designed a lesion-attention deep neural network to predict the COVID-19 in chest CT scans. The weights of VGG16, ResNet18, ResNet50, DenseNet121, DenseNet169, and EfficientNetwere utilized in the proposed approach. The proposed approach was trained on 746 chest CT scans and attained a classification accuracy of 88.6%. Wang et al. [19] developed an automatic deep learning framework for discriminating coronavirus infected persons from CT scans. UNet was trained for lung segmentation. This pre-trained net was used to obtain the lung mask for CT scans. The proposed DeCoVNet was trained on 499 CT scans and achieved a classification accuracy of 90.1%. The main limitations of this model are inadequate use of temporal information and applicability on a small dataset. Harmon et al. [20] implemented the 3D anisotropic hybrid network architecture for the classification of corona infected persons. DenseNet121 was used to segment 3D lung regions from the chest CT scan. The sensitivity and specificity obtained from this model were 84% and 93%, respectively. However, the small dataset was used for validation process. Xu et al. [21] presented an automatic screening model for COVID-19 in CT scans. This model was tested on 618 CT scans and attained an accuracy of 86.7%. However, the manifestation of other pneumonia may be overlapped with COVID-19. Due to this, the performance of this model was greatly affected. Bai et al. [22] designed an artificial intelligence system to discriminate the COVID-19 from other pneumonia. EfficientNet B4 was used in this system for classification. This system was tested on the CT scans of 1186 patients and achieved an accuracy of 96%. However, the validation of the proposed approach was tested on a limited sample size. Han et al. [23] proposed an attention-based deep learning model for screening of COVID-19. An attention mechanism and deep multiple instance learning were utilized in this model. The classification accuracy of this model was 94.3%. Li et al. [24] developed a COVNet model for classification of coronavirus infected patients using CT scans. The pre-trained ResNet50 deep learning model was used in COVNet. COVNet was trained on 4352 chest CT scans. The sensitivity and specificity obtained from COVNet were 90% and 96%, respectively. Ni et al. [25] proposed a model that consists of 3D UNet and MVPNet. 3D UNet was used to extract the lesion in the given CT scan. 19,291 CT scans were used for experimentation. The classification accuracy obtained from this model was 94%. Xiao et al. [26] implemented a pre-trained ResNet34 to envisage the severity of COVID-19 using chest CT scans. 23,812 CT scans were used for experimentation. This model attained the accuracies of 97.4% and 81.9% in training and testing sets, respectively. The main limitations of this model are biases in the results due to the small sample size, lack of transparency, and interpretability. Pu et al. [27] developed an automatic approach to perceive and quantify the pneumonia regions of CT scans associated with COVID-19. UNet framework was used to segment lung boundary, which is further used to assess the progression of disease. They used both deep learning and computer vision techniques. The sensitivity and specificity obtained from this approach were 95%, and 84%, respectively. Jaiswal et al. [28] proposed an automatic tool for detection of COVID-19 using chest CT scans. The pretrainedDenseNet201 model was used to classify the patients as coronavirus infected or not. The features were extracted from DenseNet201model using the weights learned from the ImageNet dataset. The classification accuracy obtained from this model was 97%. The problem associated with the above-mentioned techniques can be resolved by developing a novel deep learning framework to classify and monitor the progression severity of COVID-19. Therefore, a novel deep learning framework is developed in this paper.
Proposed ensemble deep transfer learning model
This section discusses the proposed ensemble model for screening of suspected patients.
Motivation
Recently many researchers have started working on the diagnosis of suspected COVID-19 infected persons using deep transfer learning models. These models have shown significantly better results, especially for early-stage classification at rapid speed as compared to RT-PCR. However, these models are still suffer from the over-fitting issue. Additionally, some models are effective for COVID-19 diagnosis but not for similar diseases such as tuberculosis and pneumonia. Therefore, in this paper, an ensemble DCCNs is designed for diagnosis the suspected objects into four classes. These four classes are COVID-19(+), tuberculosis, pneumonia, and healthy subjects.
Ensemble DCCNs
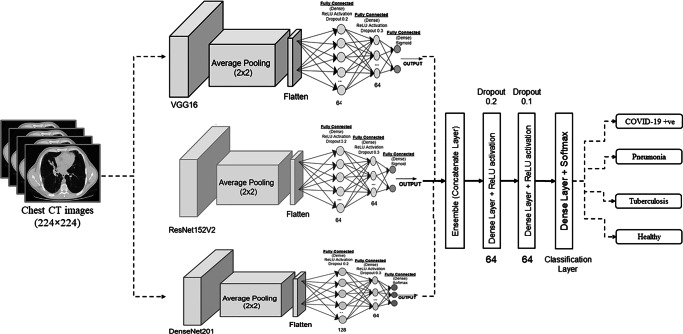
In this paper, ResNet152V2 [29], DenseNet201 [30], and VGG16 [31] models are considered to build the ensemble model. As known in prior ensembling of models [32, 33] can obtain more efficient results. It can boost feature extraction and accuracy of the used models [34]. Figure 1 shows the ensemble DCCNs. 64 neurons are used for initial dense layer [35, 36]. A fine-tuned pre-trained transfer learning model is used with many layers to extract the features. A softmax activation function is utilized. The models are trained for 10 epochs with a batch size of 10. During the tuning of initial attributes, fully connected layers [37, 38] with 64 neurons along with dropout of 0.3 and 0.2, respectively are used. To overcome the over-fitting issue, regularization [39, 40] is also done by considering the concept of early stopping. lr = 0.00001 is used as learning rate.
Fig. 1.
Proposed ensemble densely connected convolutional neural networks
Layers of DCCNs
A.
Pooling Layers
Pooling layers are utilized to achieve multi-scale analysis. These layers can also minimize the size of input data for feature reduction. Average and Max pooling layers are extensively used in CNNs. An average pooling layer (La) is utilized in this paper and can be computed as [41]:
| 1 |
Here, |di| and i define cardinal number and pooling region, respectively. d represents the activation set in i and can be computed as:
| 2 |
where j ∈ pooling region (i).
B.
Softmax and fully connected layer
The fully connected layer comes up with full connections with all the neurons. Its input is multiplied with its weight matrix to obtain the multiplicative result. Additionally, softmax is used as an activation function. Fully connected layers utilize conditional probability to achieve its functionality (more details please see [41]). Softmax (Pty(C,b)) defines probability of a subject s belong to given class C. It can be computed as:
| 3 |
Here, Pty(C) defines class probability. C shows total number of classes. Pty(C) defines probability of given C.
Equation (3) can be rewritten as:
| 4 |
where
| 5 |
C. Dropout
Dropouts are utilized to regularize CNNs by randomly placing the outgoing neurons at hidden layers to 0 during the model building process. These neurons have no effect on back-propagation during the model building stage. It can prevent over-fitting issue [42].
D. Rectified linear units
Rectified linear units (ReLU) can be computed as:
| 6 |
ReLU takes lesser time as compared to the sigmoidal functions. It can efficiently handle gradient-based training and poor performance with CNNs due to the widespread saturation. It can provide faster convergence of stochastic gradient descent [41].
Performance analysis
To evaluate the performance of the proposed ensemble model, the experiments are carried on MATLAB 2020b software with 64-bit 8-core, and 32GB RAM. The competitive models are also implemented on the same platform and dataset.
Dataset
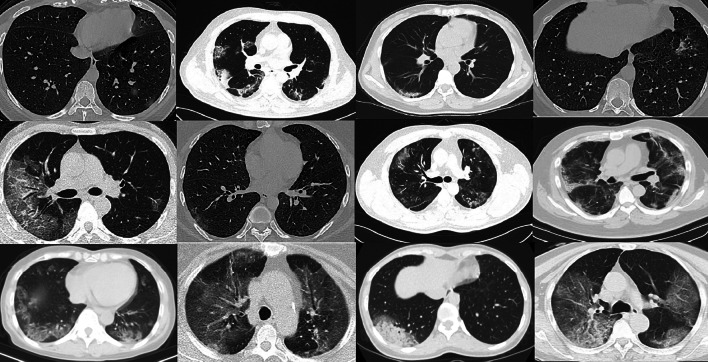
Four class chest-CT scanned images are collected from various sources such as COVID-19 [43], Pneumonia [44, 45], and Tuberculosis [13, 46]. We have collected group of chest CT scanned images as 2373 images of COVID-19 infected patients, 2890 pneumonia infected patients, 3193 tuberculosis images and 3038 healthy subjects. Figure 2 shows a view of chest-CT scanned images dataset. Out of this 65% fraction of total images are utilized for training purpose and remaining are used for testing purpose.
Fig. 2.
A view of chest-CT scanned images dataset
Comparative analyses
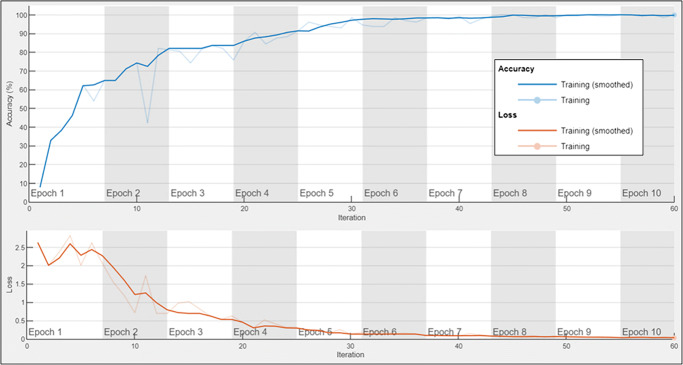
Figure 3 shows the accuracy and loss analysis of the proposed ensemble model on training dataset. The binary cross-entropy is used as a loss function. It clearly shows that the proposed ensemble model convergence to the optimal results after 33 iterations and 6th epoch. The proposed ensemble model convergence at a good speed.
Fig. 3.
Accuracy and loss analysis of the proposed ensemble model on training dataset
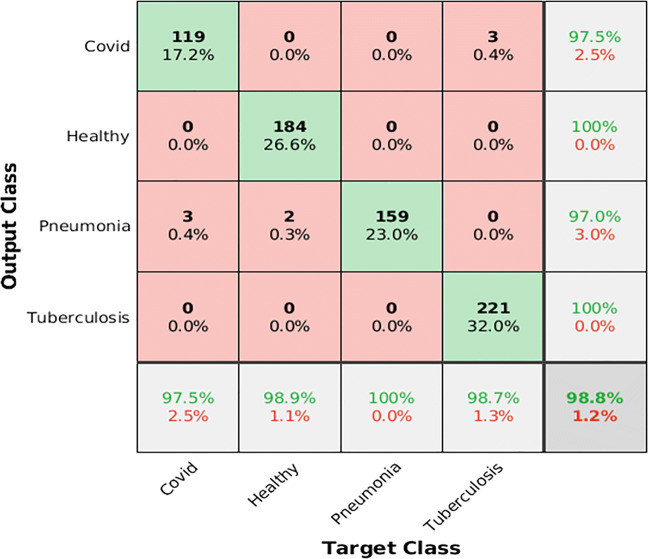
Figure 4 shows the confusion matrix of the obtained classification results from the proposed ensemble model on the testing dataset. The proposed ensemble model achieves an average accuracy of 99.2% on a training dataset. Thus, the proposed ensemble model provide significantly better screening results for suspected subjects as it does not much suffer from the overfitting issue.
Fig. 4.
Confusion matrix of the proposed ensemble model on the testing dataset
Table 1 shows the comparative analysis among the proposed and the competitive deep transfer learning models in terms of accuracy, f-measure, area under curve, sensitivity, and specificity. Bold values indicate the best results. The proposed ensemble model outperforms competitive models. The proposed ensemble model achieves good performance as compared to the competitive models in terms of accuracy, f-measure, area under curve, sensitivity, and specificity by 1.2738%, 1.3274%, 1.8372%, 1.283%, and 1.8382%, respectively.
Table 1.
Testing analysis (in %) of the proposed automated screening model
| Model | Accuracy | Area under curve | F-measure | Sensitivity | Specificity |
|---|---|---|---|---|---|
| CNN | 97.2818 | 97.1372 | 97.2912 | 98.1837 | 96.9382 |
| VGG16 | 97.9821 | 97.8324 | 97.8372 | 98.1949 | 96.9827 |
| ResNet152V2 | 98.1392 | 98.2831 | 97.8292 | 98.78929 | 97.8821 |
| Alexnet | 98.2939 | 98.3928 | 97.9394 | 98.8929 | 97.3928 |
| DenseNet201 | 98.4822 | 98.2932 | 97.9282 | 99.2833 | 97.2939 |
| Inceptionnet V3 | 98.2839 | 98.1382 | 98.1933 | 98.3283 | 98.1839 |
| Proposed | 98.8392 | 98.2893 | 98.3083 | 98.8372 | 98.8293 |
Table 2 shows the performance analysis of the proposed automated screening model over the existing models. The results reveals that the proposed ensemble model outperforms the existing models. Additionally, most of the existing work has focused on binary class or three-class classification chest CT scans dataset. Therefore, besides accuracy, the proposed ensemble model can handle a four-class problem in an efficient manner.
Table 2.
Performance analysis of the proposed automated screening model over the existing models
| Model | Accuracy(in %) |
|---|---|
| Lightweight CNN [12] | 96.28% |
| COVID-Net [14] | 12.6% |
| CovidCTNet [15] | 90% |
| CNN [16] | 86% |
| Three-phase deep learning [17] | 99.65% |
| DenseNet169 [18] | 88.6% |
| DeCoVNet [19] | 90.1% |
| DenseNet121 [20] | 93% |
| EfficientNet B4 [22] | 96% |
| Attention-based deep learning [23] | 94.3% |
| COVNet [24] | 96% |
| 3D UNet [25] | 94% |
| ResNet34 [26] | 81.9% |
| UNet [27] | 84% |
| DenseNet201 [28] | 97% |
| Proposed | 98.83% |
Conclusion
In this paper, an ensemble deep learning model was proposed for COVID-19 classification in chest CT scan images. The proposed ensemble model utilized the three well-known models namely DCCNs, ResNet152V2 and VGG16. The proposed ensemble model is able to handle the sensitivity issue that is associated with RT-PCR. The proposed ensemble model has been tested on large chest CT dataset and compared with fifteen competitive models. Experimental results reveal that the proposed ensemble model outperforms the existing models in terms of accuracy, f-measure, area under curve, sensitivity, and specificity by 1.2738%, 1.3274%, 1.8372%, 1.283%, and 1.8382%, respectively.
Footnotes
This article belongs to the Topical Collection: Artificial Intelligence Applications for COVID-19, Detection, Control, Prediction, and Diagnosis
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hui DS, Azhar EI, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C, et al. The continuing 2019-ncov epidemic threat of novel coronaviruses to global healththe latest 2019 novel coronavirus outbreak in wuhan, china. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothan HA, Byrareddy SN (2020) The epidemiology and pathogenesis of coronavirus disease (covid-19) outbreak. J Autoimmun:102433 [DOI] [PMC free article] [PubMed]
- 3.Zhu J, Shen B, Abbasi A, Hoshmand-Kochi M, Li H, Duong TQ. Deep transfer learning artificial intelligence accurately stages covid-19 lung disease severity on portable chest radiographs. PloS one. 2020;15(7):e0236621. doi: 10.1371/journal.pone.0236621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ting DSW, Carin L, Dzau V, Wong TY. Digital technology and covid-19. Nat Med. 2020;26(4):459–461. doi: 10.1038/s41591-020-0824-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozsahin I, Sekeroglu B, Musa MS, Mustapha MT, UzunOzsahin D (2020) Review on diagnosis of covid-19 from chest ct images using artificial intelligence. Computational and Mathematical Methods in Medicine 2020 [DOI] [PMC free article] [PubMed]
- 6.Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R (2020) Severe acute respiratory syndrome coronavirus 2 (sars-cov-2) and corona virus disease-2019 (covid-19): the epidemic and the challenges. Int J Antimicr Agents:105924 [DOI] [PMC free article] [PubMed]
- 7.Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, Fan Y, Zheng C (2020) Radiological findings from 81 patients with covid-19 pneumonia in wuhan, china: a descriptive study. The Lancet Infectious Diseases [DOI] [PMC free article] [PubMed]
- 8.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L (2020) Correlation of chest ct and rt-pcr testing in coronavirus disease 2019 (covid-19) in china: a report of 1014 cases. Radiology:200642 [DOI] [PMC free article] [PubMed]
- 9.Fang Y, Zhang H, Xie J, Lin M, Ying L, Pang P, Ji W (2020) Sensitivity of chest ct for covid-19: comparison to rt-pcr. Radiology:200432 [DOI] [PMC free article] [PubMed]
- 10.Tayarani-N M-H (2020) Applications of artificial intelligence in battling against covid-19: A literature review. Chaos, Solitons Fractals:110338 [DOI] [PMC free article] [PubMed]
- 11.Girdhar A, Kapur H, Kumar V, Kaur M, Singh D, Damasevicius R (2020) Effect of covid-19 outbreak on urban health and environment. Air Quality, Atmosph Health:1–9 [DOI] [PMC free article] [PubMed]
- 12.Mukherjee H, Ghosh S, Dhar A, Obaidullah SM, Santosh KC, Roy K (2020) Deep neural network to detect covid-19: one architecture for both ct scans and chest x-rays. Appl Intell:1–13 [DOI] [PMC free article] [PubMed]
- 13.Singh D, Kumar V, Kaur M (2020) Classification of covid-19 patients from chest ct images using multi-objective differential evolution–based convolutional neural networks. Eur J Clin Microbiol Infect Diseas:1–11 [DOI] [PMC free article] [PubMed]
- 14.Wang Z, Liu Q, Dou Q. Contrastive cross-site learning with redesigned net for covid-19 ct classification. IEEE J Biomed Health Inf. 2020;24(10):2806–2813. doi: 10.1109/JBHI.2020.3023246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javaheri T, Homayounfar M, Amoozgar Z, Reiazi R, Homayounieh F, Abbas E, Laali A, Radmard AR, Gharib MH, Mousavi S AJ et al (2020) Covidctnet: An open-source deep learning approach to identify covid-19 using ct image. arXiv:2005.03059 [DOI] [PMC free article] [PubMed]
- 16.Mishra AK, Das SK, Roy P, Bandyopadhyay S (2020) Identifying covid19 from chest ct images: A deep convolutional neural networks based approach. J Healthcare Eng 2020 [DOI] [PMC free article] [PubMed]
- 17.Ahuja S, Panigrahi BK, Dey N, Rajinikanth V, Gandhi TK (2020) Deep transfer learning-based automated detection of covid-19 from lung ct scan slices [DOI] [PMC free article] [PubMed]
- 18.Liu B, Gao X, He M, Liu L, Yin G (2020) A fast online covid-19 diagnostic system with chest ct scans. In: Proceedings of KDD, vol 2020
- 19.Wang X, Deng X, Fu Q, Zhou Q, Feng J, Ma H, Liu W, Zheng C (2020) A weakly-supervised framework for covid-19 classification and lesion localization from chest ct. IEEE Trans Med Imaging [DOI] [PubMed]
- 20.Harmon SA, Sanford TH, Xu S, Turkbey EB, Roth H, Xu Z, Yang D, Myronenko A, Anderson V, Amalou A, et al. Artificial intelligence for the detection of covid-19 pneumonia on chest ct using multinational datasets. Nat Commun. 2020;11(1):1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Jiang X, Ma C, Du P, Li X, Lv S, Yu L, Ni Q, Chen Y, Su J et al (2020) A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering [DOI] [PMC free article] [PubMed]
- 22.Bai HX, Wang R, Xiong Z, Hsieh B, Chang K, Halsey K, Tran T ML, Choi JW, Wang D-C, Shi L-B et al (2020) Ai augmentation of radiologist performance in distinguishing covid-19 from pneumonia of other etiology on chest ct. Radiology:201491 [DOI] [PMC free article] [PubMed]
- 23.Han Z, Wei B, Hong Y, Li T, Cong J, Zhu X, Wei H, Zhang W (2020) Accurate screening of covid-19 using attention based deep 3d multiple instance learning. IEEE Trans Med Imaging [DOI] [PubMed]
- 24.Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q et al (2020) Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary ct: evaluation of the diagnostic accuracy. Radiology 296(2) [DOI] [PMC free article] [PubMed]
- 25.Ni Q, Sun ZY, Qi L, Chen W, Yang Y, Wang L, Zhang X, Yang L, Fang Y, Xing Z, et al. A deep learning approach to characterize 2019 coronavirus disease (covid-19) pneumonia in chest ct images. Euro Radiol. 2020;30(12):6517–6527. doi: 10.1007/s00330-020-07044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao L-, Li P, Sun F, Zhang Y, Xu C, Zhu H, Cai F-Q, He Y-L, Zhang W-F, Ma S-C, et al. Development and validation of a deep learning-based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Front Bioengin Biotechnol. 2020;8:898. doi: 10.3389/fbioe.2020.00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pu J, Leader JK, Bandos A, Ke S, Wang J, Shi J, Du P, Guo Y, Wenzel SE, Fuhrman CR et al (2020) Automated quantification of covid-19 severity and progression using chest ct images. Eur Radiol:1–11 [DOI] [PMC free article] [PubMed]
- 28.Jaiswal A, Gianchandani N, Singh D, Kumar V, Kaur M (2020) Classification of the covid-19 infected patients using densenet201 based deep transfer learning. J Biomol Struct Dyn:1–8 [DOI] [PubMed]
- 29.He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
- 30.Huang G, Liu Z, Van DerMaaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
- 31.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
- 32.Siwek K, Osowski S, Szupiluk R. Ensemble neural network approach for accurate load forecasting in a power system. Int J Appl Math Comput Sci. 2009;19(2):303–315. doi: 10.2478/v10006-009-0026-2. [DOI] [Google Scholar]
- 33.Islam J, Zhang Y. Brain mri analysis for alzheimer’s disease diagnosis using an ensemble system of deep convolutional neural networks. Brain Inf. 2018;5(2):2. doi: 10.1186/s40708-018-0080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alshazly H, Linse C, Barth E, Martinetz T. Ensembles of deep learning models and transfer learning for ear recognition. Sensors. 2019;19(19):4139. doi: 10.3390/s19194139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh S, Shivakumara P, Roy P, Pal U, Lu T. Graphology based handwritten character analysis for human behaviour identification. CAAI Trans Intell Technol. 2020;5(1):55–65. doi: 10.1049/trit.2019.0051. [DOI] [Google Scholar]
- 36.Wiens T. Engine speed reduction for hydraulic machinery using predictive algorithms. Int J Hydromechatron. 2019;2(1):16–31. doi: 10.1504/IJHM.2019.098949. [DOI] [Google Scholar]
- 37.Gupta B, Tiwari M, Lamba SS. Visibility improvement and mass segmentation of mammogram images using quantile separated histogram equalisation with local contrast enhancement. CAAI Trans Intell Technol. 2019;4(2):73–79. doi: 10.1049/trit.2018.1006. [DOI] [Google Scholar]
- 38.Osterland S, Weber J. Analytical analysis of single-stage pressure relief valves. Int J Hydromechatron. 2019;2(1):32–53. doi: 10.1504/IJHM.2019.098951. [DOI] [Google Scholar]
- 39.Basavegowda HS, Dagnew G. Deep learning approach for microarray cancer data classification. CAAI Trans Intell Technol. 2020;5(1):22–33. doi: 10.1049/trit.2019.0028. [DOI] [Google Scholar]
- 40.Wang R, Yu H, Wang G, Zhang G, Wang W. Study on the dynamic and static characteristics of gas static thrust bearing with micro-hole restrictors. Int J Hydromechatron. 2019;2(3):189–202. doi: 10.1504/IJHM.2019.102893. [DOI] [Google Scholar]
- 41.Wang S-H, Phillips P, Sui Y, Liu B, Yang M, Cheng H. Classification of alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J Med Syst. 2018;42(5):85. doi: 10.1007/s10916-018-0932-7. [DOI] [PubMed] [Google Scholar]
- 42.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]
- 43.Zhao J, Zhang Y, He X, Xie P (2020) Covid-ct-dataset: a ct scan dataset about covid-19. arXiv:2003.13865
- 44.Challenge RPD (2018) Radiological society of north america
- 45.Pathak Y, Shukla PK, Arya KV (2020) Deep bidirectional classification model for covid-19 disease infected patients. IEEE/ACM Transactions on Computational Biology and Bioinformatics [DOI] [PubMed]
- 46.Li L, Huang H, Jin X (2018) Ae-cnn classification of pulmonary tuberculosis based on ct images. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME). IEEE, pp 39–42