SUMMARY
Bacterial type II toxin-antitoxin (TA) modules encode a toxic protein that downregulates metabolism and a specific antitoxin that binds and inhibits the toxin during normal growth. In non-typeable Haemophilus influenzae, a common cause of infections in humans, the vapXD locus was found to constitute a functional TA module and contribute to pathogenicity; however, the mode of action of VapD and the mechanism of inhibition by the VapX antitoxin remain unknown. Here, we report the structure of the intact H. influenzae VapXD complex, revealing an unusual 2:1 TA molecular stoichiometry where a Cas2-like homodimer of VapD binds a single VapX antitoxin. VapX consists of an oligonucleotide/oligosaccharide-binding domain that docks into an asymmetrical cavity on the toxin dimer. Structures of isolated VapD further reveal how a symmetrical toxin homodimer adapts to interacting with an asymmetrical antitoxin and suggest how a primordial TA system evolved to become part of CRISPR-Cas immunity systems.
In Brief
In this paper, Bertelsen et al. present the three-dimensional structure of a toxic protein complex from the pathogenic bacterium Haemophilus influenzae, which causes a range of diseases in humans. The authors demonstrate that the toxin, which makes the bacterium more virulent, is inactivated by an antitoxin of unusual structure.
Graphical Abstract

INTRODUCTION
Bacterial type II toxin-antitoxin (TA) modules encode two small protein components: a stable toxin capable of downregulating cellular metabolism and an antitoxin that specifically binds and inhibits the toxin (Harms et al., 2018). Antitoxins are metabolically and structurally unstable and must thus be continuously replenished in order to maintain toxin inhibition. This principle forms the basis for plasmid maintenance via post-segregational killing, in which cells that do not inherit a TA-containing plasmid fail to survive because they no longer express the antitoxin, and abortive infection that increases bacterial survival during phage infection (Harms et al., 2018). Most type II antitoxins are composed of two distinct domains: an N-terminal DNA-binding domain that regulates transcription from the TA locus through direct interaction with the promoter, and an intrinsically unstructured C-terminal region responsible for toxin binding and inhibition (Harms et al., 2018). During nutritional or oxidative stress, the antitoxins are specifically degraded by cellular proteases such as Lon, thus providing a metabolic cue to toxin activation (Page and Peti, 2016). Once activated, type II toxins typically act as nucleases that modulate gene expression through degradation of specific, stable RNAs, including tRNA, rRNA, and mRNA (Goeders and Van Melderen, 2014). By targeting central cellular processes that affect bacterial growth and cell division, the toxins thus generate a fitness advantage for their hosts by allowing them to quickly adapt their growth rate to environmental changes (LeRoux et al., 2020; Page and Peti, 2016).
Non-typeable (non-encapsulated) Haemophilus influenzae (NTHi) is a human pathogen and common cause of infections such as recurrent middle ear infection (otitis media) and sinusitis in children and severe lower respiratory tract infections (pneumonia) in patients with chronic obstructive pulmonary disease (Murphy, 2003). The NTHi genome contains four TA operons, toxAvapA, vapBC-1, vapBC-2, and vapXD, of which vapBC-1 and vapBC-2 are canonical and well-characterized vapBC-type TA modules containing a PIN-domain RNase toxin (VapC) (Daines et al., 2007; Harms et al., 2018; Molinaro et al., 2019). The vapXD locus encodes a non-canonical TA system, which was linked to the survival and virulence of NTHi in an animal model system and demonstrated to act as a bona fide TA module in vivo (Daines et al., 2004; Ren et al., 2012) (Figure 1A). The VapD toxin was shown to be active as a homodimer in vivo, but very little is known about the structure and function of the expressed VapXD protein complex (Ren et al., 2012). A structure exists of a homologous VapD protein from Helicobacter pylori, which is remarkable because it is expressed from a locus that does not appear to encode an antitoxin, and the protein thus apparently is not part of a canonical TA system (Kwon et al., 2012). The H. pylori structure revealed a clear structural similarity to Cas2, which is required for spacer integration during CRISPR-Cas adaptation (Nunez et al., 2014). This observation as well as comparative sequence studies have fueled speculations that the bacterial CRISPR-Cas immunity systems might have in fact evolved from a primordial vapXD-type TA system (Makarova et al., 2012).
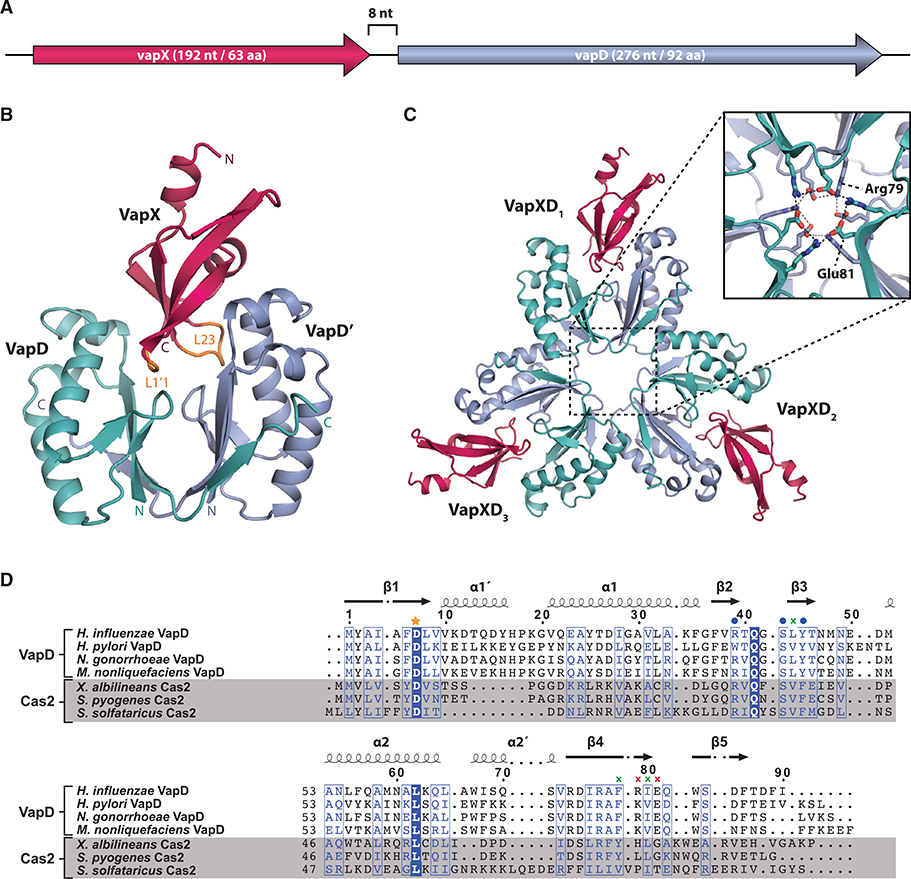
Figure 1. The Structure of H. influenzae VapXD.
(A) The genetic organization of the H. influenzae vapXD operon. The vapX gene (192 nt/63 aa) precedes the vapD gene (276 nt/92 aa) spaced by 8 nt.
(B) Overall structure of the VapXD2 heterotrimer composed of a toxin of two VapD molecules (sea green/light blue) and one VapX antitoxin molecule (red).
(C) Symmetry expansion of the crystal structure showing the ring-shaped higher-order structure composed of three VapXD2 heterotrimers. The inset shows the conserved residues, Arg79 and Glu81, that stabilize the assembly of the ring structure through electrostatic interactions.
(D) Structure-based sequence alignment of four VapD homologs and three Cas2 homologs, H. influenzae VapD (PDB: 6ZN8, UniProt: P71351, this work), H. pylori VapD (HP0315, PDB: 3UI3, UniProt: 19909) (Kwon et al., 2012), Neisseria gonorrhoeae VapD (UniProt: Q51003), Moraxella nonliquefaciens VapD (UniProt: A0A1B8QQI3), Xanthomonas albilineans Cas2 (PDB: 5H1O, UniProt: D2UG58) (Ka et al., 2017), Streptococcus pyogenes Cas2 (PDB: 4QR0, UniProt: Q99YS8) (Ka et al., 2014), and S. solfataricus Cas2 (PDB: 2I8E, UniProt: Q97YC2) (Beloglazova et al., 2008). Strictly conserved residues are shown on a blue background, and similar residues are in blue letters and boxed. Secondary structure elements as observed for the NTHi VapD structure are indicated on top of the alignment and labeled as in Figure 2A. The conserved Asp7 involved in catalysis is marked with an orange star. Arg39, Ser43, and Tyr45 located near the active site are marked with blue spheres; hydrophobic residues Leu44, Phe78, and Ile80 involved in dimerization are marked with green crosses; and the charged residues Arg79 and Glu81 involved in dimerization and formation of the ring shape are marked with red crosses. The sequences were aligned using PROMALS 3D (Pei and Grishin, 2014) and printed using ESPript 3.0 (Robert and Gouet, 2014).
See also Figures S1 and S2 and Table S1.
Most characterized Cas2-type proteins display nonspecific endonuclease activity, with some preferring single-stranded RNA (Beloglazova et al., 2008) and others double- or single-stranded DNA (Ka et al., 2014; Nam et al., 2012). The activity has been found to depend on divalent cations (Mg2+ or Mn2+) as well as a strictly conserved aspartic acid residue, which was proposed to be involved in coordination of the ion in the active site (Beloglazova et al., 2008; Ka et al., 2014, 2017; Nam et al., 2012). Surprisingly, however, the RNase activity of Cas2 does not appear to be required for its role in CRISPR-Cas adaptation, where the protein functions as a spacer of dimeric Cas1 subunits, which in turn catalyze the dsDNA cleavage reaction required for spacer acquisition (Nunez et al., 2014). H. pylori VapD appears to be active solely on single-stranded RNA, thus suggesting that VapD orthologs, when part of a TA system, function as RNases, similar to RelE, MazF, and VapC, but independent of cations (Kwon et al., 2012; Pedersen et al., 2003; Zhang et al., 2003). Despite this, mutation of the strictly conserved aspartic acid residue abolishes VapD activity, suggesting that the protein contains an active site similar to those of the Cas2 proteins, but possibly uses a different catalytic mechanism (Kwon et al., 2012).
Understanding the mechanistic details underlying toxin inhibition and activity in the VapXD TA system is highly relevant to understanding their influence on pathogenicity in H. influenzae and could prove useful for drug development against infectious diseases. Moreover, insights into the VapXD system could support the proposed evolutionary links between CRISPR-Cas and the TA modules. Here, we provide structural insights into the architecture of the intact VapXD TA complex and toxin activation. The crystal structure of the NTHi VapXD complex confirms the similarity to Cas2 and reveals an unusual 2:1 TA complex in which an otherwise symmetrical toxin homodimer adapts to the binding of an asymmetrical antitoxin. Surprisingly, the VapX antitoxin contains neither an intrinsically disordered region nor a DNA-binding domain, but consists of a single oligonucleotide/oligosaccharide-binding (OB) fold, which intercalates between monomers of the toxin dimer. Our results thus shed light on the mechanism of toxin inhibition in this unique TA system and suggest that transcriptional regulation is markedly different from canonical type II TA systems.
RESULTS
The Structure of the VapXD Complex Reveals an Unusual 2:1 Stoichiometry
To understand the molecular basis for VapD inhibition by its cognate VapX antitoxin, we determined the structure of the NTHi VapXD protein complex by X-ray crystallography. Briefly, selenomethionine-labeled VapXD complex was produced by overexpression of the natural, full-length operon, modified to include a C-terminal His-tag on the VapD toxin, in the methionine auxotroph Escherichia coli strain B834(DE3) growing in minimal medium. The labeled protein was purified to homogeneity and crystallized in the cubic space group I4132, and the structure was determined to 3.2 Å using Se single-wavelength anomalous diffraction phasing, followed by manual model building and refinement to a final Rwork = 25% and Rfree = 29% (see STAR Methods and Table 1 for details). The relatively high crystallographic R factors are likely due to an unusually high solvent content (77.5%, VM = 5.46 Å3/Da) as well as the presence of pseudo-symmetry in the cubic space group resulting in extended solvent channels with weak unexplained and repetitive density, pathologies that could not be accounted for using regular refinement protocols. Modeling of this spurious density did not improve refinement. Nevertheless, the electron density is generally well defined and the model is nearly complete, with a few terminal residues missing in each chain likely due to flexibility (Figure S1A). The structure consists of two VapXD2 heterotrimers in the crystallographic asymmetric unit, each consisting of a VapD homodimer (2•92 aa) and a single VapX antitoxin (63 aa) molecule, which is located in a cleft formed between the two VapD subunits (Figure 1B). Although the VapD toxin is known to be active as a dimer, such a 2:1 molecular stoichiometry is highly unusual for type II TA systems, for which the toxins usually always associate 1:1 with their cognate antitoxin proteins (Page and Peti, 2016).
Table 1.
Crystallographic Data Collection and Refinement Statistics
| PDB Entry | VapXD (Selenomethionine) 6ZN8 | VapD (Wild Type) 6ZI0 | VapD (D7N) 6ZI1 |
|---|---|---|---|
| Data Collection | |||
| Wavelength (Å) | 0.979 | 0.98021 | 0.98021 |
| Resolution range | 46.88–3.21 (3.33–3.21)a | 85.88–2.50 (2.56–2.50) | 42.19–2.20(2.26–2.20) |
| Space group | I4132 | P6322 | P321 |
| Unit cell dimensions | |||
| a, b, c (Å) | 256.75, 256.75, 256.75 | 99.168, 99.168, 44.486 | 84.380, 84.380, 49.476 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 120 |
| Total no. of reflections | 172,895 (17,802) | 49,715(3,673) | 34,356 (2,279) |
| Unique reflections | 23,771 (2,337) | 4,800 (344) | 10,306 (736) |
| Multiplicity | 7.3 (7.6) | 10.3(10.7) | 3.3 (3.1) |
| Completeness (%) | 99.7 (99.7) | 99.9(100.0) | 97.2 (97.9) |
| Rmerge | 0.169 (1.37) | 0.212 (1.581) | 0.060 (0.928) |
| I/σ (I) | 9.2 (1.3) | 9.5 (1.3) | 12.1 (1.3) |
| CC1/2 | 0.99 (0.53) | 0.996 (0.55) | 0.999(0.61) |
| Refinement | |||
| Average B factor (Å2) | 100.7 | 76.2 | 58.9 |
| No. of reflections | 23,774 (2,337) | 4,560 (326) | 9,282 (668) |
| No. of reflections (free) | 2,351 (209) | 241 (18) | 1,028 (68) |
| Rwork (%) | 25.8 (34.9) | 22.0 (28.4) | 21.4(38.6) |
| Rfree (%) | 29.9 (39.6) | 26.4 (28.9) | 26.7 (40.9) |
| No. of atoms | |||
| Protein | 3,939 | 722 | 1,333 |
| Solvent | 0 | 55 | 88 |
| Ligand | 0 | 0 | 0 |
| RMSD bonds (Å) | 0.010 | 0.008 | 0.007 |
| RMSD angles (°) | 1.11 | 1.74 | 1.48 |
| Ramachandran statistics | |||
| favored (%) | 93.7 | 91.6 | 84.8 |
| Allowed (%) | 6.3 | 7.2 | 14.6 |
| Generously allowed (%) | – | 1.2 | 0.7 |
| Outliers (%) | 0.0 | 0.0 | 0.0 |
Numbers in parentheses indicate values for the outermost resolution shell.
VapXD2 Heterotrimers Associate into Higher-Order Structures
Symmetry expansion of the crystal structure reveals that three VapXD2 heterotrimers come together to form a larger, ring-shaped structure, which is stabilized by the conserved charged residues Arg79 and Glu81 that form a circle of alternating positive and negative charges at the center (Figures 1C, inset, and 1D). Protein Interfaces, Surfaces, and Assemblies (PISA) anal-ysis (Krissinel and Henrick, 2007) suggests several possible higher-order assemblies, including the VapXD2 heterotrimer and VapX3D6 (the ring), as well as VapX6D12 (two stacked rings), and even VapX12D24 (four stacked rings, Figure S1B), all with favorable calculated free energies of dissociation (Table S1), suggesting that they all potentially could be stable and, in theory, biologically relevant. To investigate which of these species is most prevalent in solution, we performed gel-filtration chroma-tography, dynamic light scattering (DLS), and protein cross-linking. During gel-filtration chromatography, the VapXD complex elutes with an apparent molecular weight of 150 kDa (Figure S2A), which is significantly larger than expected for a single VapXD2 trimer (32.3 kDa) and closer to the expected mass of the ring structure (3•32.3 = 96.9 kDa). The presence of higher-order assemblies is further supported by glutaraldehyde cross-linking in vitro, which reveals several species with molecular masses up to around 90 kDa (Figure S2B). Finally, we measured the DLS from a solution containing purified VapXD, which provides an overall average size (not mass) estimate (Figure S2C). This revealed a largely monodisperse solution, with a single species having an average hydrodynamic diameter of 10.7 ± 0.1 nm. Calculation of theoretical hydrodynamic diameters of the various assemblies suggested by PISA using HYDROPRO (Ortega et al., 2011) shows that the measured size fits both one and two stacked rings, which have calculated hydrodynamic diameters of 8.1 and 9.8 nm, respectively. Together, these results support that the VapXD complex forms complexes of higher order than a single VapXD2 heterotrimer in solution, and most likely in the form of trimers of trimers (rings), and possibly stacked, as suggested by PISA and DLS. Moreover, we note that ring stacking is coaxial, which in theory would allow binding of longer macromolecules through the central hole (Figure S1B). Further studies will be necessary to determine the stoichiometry of these assemblies in vivo as well as their biological relevance.
The NTHi VapD Toxin Dimer Is Structurally Similar to Cas2
The VapD toxin contains a modified ferredoxin fold (β1α1β2-β3α2β4), where each of the two α helices has been split into two shorter helices connected by short loops, resulting in a β1α1′α1β2β3α2α2′β4 topology (Figure 2A). In the VapXD complex, two VapD monomers form a butterfly-shaped dimer with a deep cleft between them. This dimerization is primarily mediated by a fifth C-terminal β strand (β5) outside the core ferredoxin fold, via formation of an extended β sheet with the corresponding region of the other VapD monomer (Figures 2A and 2B). The structure of NTHi VapD is highly similar to that of H. pylori VapD, both at the sequence and at the structural level (40% sequence identity, 68% sequence similarity, root-mean-square deviation [RMSD] Cα atoms 0.98 Å, Figure 2C) (Kwon et al., 2012). In NTHi VapD, β2 and β3 are very short (only two residues each), and β2 is slightly distorted from an ideal β-strand conformation. This contributes to the dimerization interface by providing shape complementarity at the bottom of the cleft. Interestingly, the sequence of β4 is well conserved among VapD homologs (Figure 1D), suggesting that a similar mode of dimerization/cleft is likely to be found for these and that side chains could be functionally important. Indeed, the conserved hydrophobic side chains of Leu44, Phe78, and Ile80 appear to form a hydrophobic core that further contributes to the stability of the dimer. In addition, the conserved charged residues Arg79 and Glu81, which are involved in formation of the ring-shaped higher-order structure, also interact across monomers at the bottom of the dimer (Figure S3A).
Figure 2. The VapD Toxin Is Similar to Cas2 and GhoS.
(A) The NTHi VapD monomer with secondary structure elements colored individually and labeled.
(B) Dimerization interface of the VapD homodimer with monomers in different colors and the backbone of residues 75–85 shown as sticks to illustrate the hydrogen bonds stabilizing the dimer.
(C) Structural alignment of H. pylori VapD (PDB: 3UI3, red, RMSD Cα atoms 0.98 Å) and E. coli Cas2 (PDB: 5DS4, green, RMSD Cα atoms 3.9 Å) on the NTHi VapD dimer (blue) (Kwon et al., 2012; Nuñez et al., 2015). In both cases, the alignment was based on the complete dimers and used the VapD dimer from the VapXD complex. Differences are shown with bright green (Cas2) and dark blue (VapD), respectively. The conserved acidic residues (Asp7 in NTHi VapD, Asp65 in H. pylori VapD, and Glu9 in Cas2) are shown with sticks.
(D) Left, structural alignment of the type V antitoxin, GhoS, from the E. coli GhoST TA system (orange and red, PDB: 2LLZ) (Wang et al., 2012), with an NTHi VapD monomer (blue), with structural differences shown in red (GhoS) and dark blue (VapD). Right, close up of the active-site region with relevant amino acids marked.
See also Figure S3.
The VapD dimer is structurally similar to Cas2 as found within the Cas1-Cas2 complex required for spacer cleavage during the CRISPR-Cas adaptation stage in many microorganisms (Beloglazova et al., 2008; Kwon et al., 2012; Nunez et al., 2014). The NTHi VapD monomer superposes with Sulfolobus solfataricus Cas2 (PDB: 2I8E) with an overall RMSD of Cα atoms of 2.6 Å. Interestingly, alignment of complete dimers is significantly worse (5.5 Å), suggesting that mostly the monomer fold, and less so dimer interaction, is conserved between the TA systems and CRISPR-Cas. Comparison of the structures shows that the charged Arg79/Glu81 interaction is unique to VapD, while the as2 dimer does have a cluster of hydrophobic residues between the monomers similar to VapD. Compared with E. coli Cas2, which has been structurally characterized in complex with Cas1 and DNA, NTHi VapD has two short, additional α helices (α1′ and α2′) at the top, but lacks a β hairpin at the C terminus, which is responsible for Cas1 interaction in the Cas1-Cas2 complex (Figure 2C) (Nunez et al., 2014). Moreover, the additional helices in VapD, which are involved in VapX antitoxin binding, would overlap with DNA in the Cas1-Cas2-DNA structure, explaining their absence in Cas2 (Nunez et al., 2015).
Mutational studies of S. solfataricus Cas2 and H. pylori VapD have mapped the nuclease active site to the cleft between the two monomers (Beloglazova et al., 2008; Kwon et al., 2012). In particular, Asp10 was found to be crucial for the activity of S. solfataricus Cas2 (Beloglazova et al., 2008). The two equivalent Asp10 residues are adjacent to each other in the cleft between the molecules of the corresponding Cas2 dimer, and have been proposed to coordinate a Mg2+ ion, which in turn could prepare a water molecule for a nucleophilic attack on the phosphate backbone of RNA and/or DNA (Beloglazova et al., 2008). This acidic residue is conserved among both Cas2 and VapD homologs and corresponds to Glu9 in E. coli Cas2 and Asp7 of NTHi VapD (Figures 2C and 1D). In the structure of NTHi VapD bound to its antitoxin, the two Asp7 residues are located 11.9 Å apart (as opposed to 6.5 Å in S. solfataricus Cas2), which is clearly too far for coordination of a single Mg2+ ion (Figure S3B). It thus seems more likely that, if divalent cations are involved in catalysis, one is bound in each site in a symmetrical fashion.
VapD Is Structurally Similar to the Antitoxin of the Type V TA System, GhoST
The GhoST system from E. coli is the only known type V TA system, which is defined by the fact that the antitoxin (GhoS) is an RNase that cleaves the toxin mRNA under normal conditions, thus preventing its expression and activity (Wang et al., 2012). Curiously, the structure of GhoS (Figure 2D) revealed a ferredoxin-like fold similar to that of S. solfataricus Cas2, including the conserved catalytic residues (Wang et al., 2012). The monomer of NTHi VapD superposes well with E. coli GhoS antitoxin with an RMSD of the Cα atoms of 2.4 Å (Figure 2D). When the two structures are aligned, the conserved catalytic residue Asp7 in VapD superimposes with Asp17 in GhoS, which is oriented into the cleft of the dimer, suggesting the two residues could have the same function (Figure 2D, inset). However, GhoS does not contain the extra C-terminal β strand and does not form a dimer (Wang et al., 2012). Instead, GhoS has a small β hairpin near its C terminus, which precludes the dimer interaction (Figure 2D). Interestingly, the GhoS antitoxin is specific for its substrate, the mRNA of its cognate toxin (Wang et al., 2012). Mutational studies of five conserved residues revealed that Arg30 and Phe57 are crucial for the activity of GhoS in vitro, and furthermore that Arg30 is important for the activity in vivo (Wang et al., 2012), but none of these residues are conserved in VapD. In summary, it is likely that VapD and GhoS derived from the same, original ferredoxin-like fold, possibly with similar reaction mechanisms involving an Asp, and later diverged in terms of oligomeric state and substrate specificity.
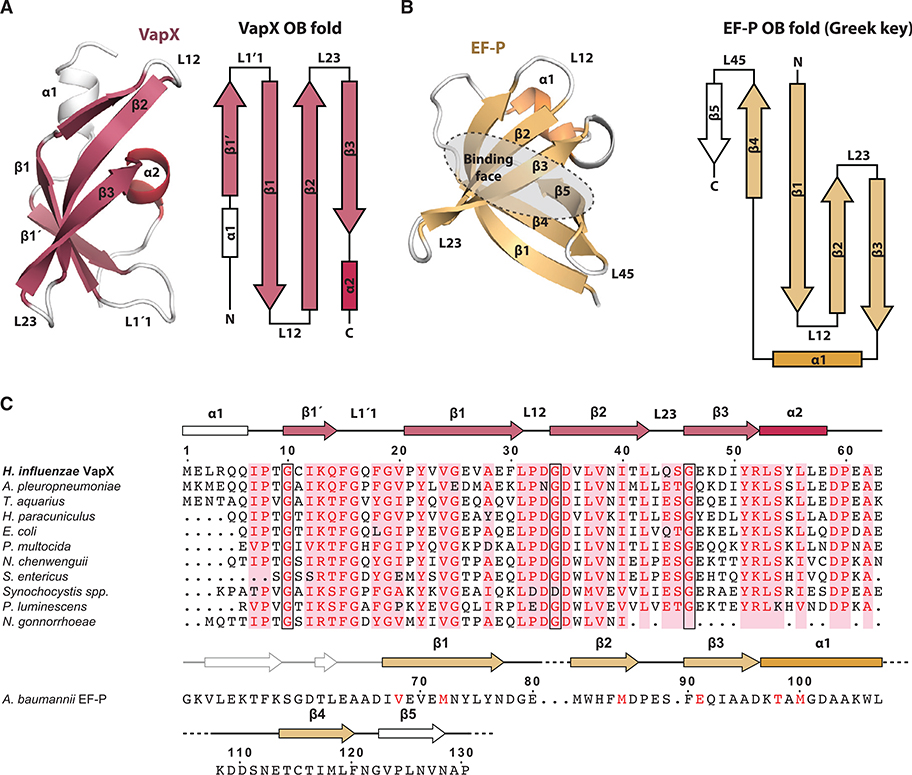
The VapX Antitoxin Contains an OB Fold with Swapped Architecture
VapX is a small, monomeric, and single-domain antitoxin with a compact α(β)4α fold consisting of an N-terminal region composed of a loop and an α helix linked to a core fold of four highly curved and antiparallel β sheets connected by loops and, finally, a C-terminal region also composed by an α helix and a loop (Figure 3A). VapX structurally resembles the OB domain proteins, of which translation elongation factor P is an example (EF-P; Z score 6.1, RMSD Cα atoms 1.8 Å, Figure 3B). VapX superimposes well with EF-P; however, closer inspection reveals that its topology is different in that it contains a swapped OB fold in which the β4 strand, which runs alongside the β1 in the canonical OB fold and forms a classical Greek key motif, is replaced by an N-terminal β strand (β1′, Figure 3A). The OB domain is found ubiquitously in all kingdoms of life, despite a general lack of sequence similarity (Murzin, 1993), and is usually involved in binding RNA, ssDNA, oligosaccharides, or other proteins (Arcus, 2002). Regardless of binding target, OB-fold proteins tend to use the same face for interactions, a region that is bounded by several loop regions (Figure 3B). Specifically, binding of single-stranded nucleic acids is accomplished through aromatic stacking, hydrogen bonding, and polar interactions by side chains located between β strands 1 and 2 (L12) and between β strands 4 and 5 (L45). Together, these loops generate a cleft that runs across the surface of the domain, perpendicular to the β strands, with a fixed polarity in nucleic acid binding running 5′ to 3′ from β4 and β5 toward β2 (Theobald et al., 2003). Interestingly, the OB-fold binding face is conserved in VapX and located on the flat side of the VapXD complex. However, due to the swapped OB-fold topology, the L45 loop is structurally replaced by the loop between the extra N-terminal β strand (β1′) and β1 (L1′1), while L12 is in the canonical location (Figure 3A). Consistently, a sequence comparison does not show any pattern of sequence similarity between VapX and the canonical OB-fold proteins (Figure 3C). In summary, we conclude that VapX is a highly unusual antitoxin in that it contains neither a well-known DNA-binding domain nor an intrinsically disordered region used for toxin inhibition.
Figure 3. VapX has an OB Fold.
(A) The structure of NTHi VapX (left) and topology (right). The colored parts of the diagrams represent the canonical elements of the OB fold.
(B) Structure of the Acinetobacter baumannii EF-P OB fold with conserved secondary structure in gold (PDB: 5J3B, left, J. Abendroth et al., unpublished) and corresponding topology (right). EF-P has a classical Greek key topology consisting of a three-stranded, antiparallel sheet followed by a helix and β4 hydrogen bonding to β1. The conserved binding face of the OB fold is indicated in gray.
(C) Sequence alignment of NTHi VapX with homologs from other bacteria as well as A. baumannii EF-P (bottom). The alignment shows the secondary structure on top of the sequences, with the sequence of EF-P wrapped around to show the structural homology between β1′ in VapX and β4 in EF-P. Conserved residues are shown in red and the conserved glycine residues at the beginning of some of the β-strands in black boxes.
Binding of VapX Introduces Asymmetry in the VapD Dimer
Probably the most unusual feature of the VapXD TA system is that a single antitoxin (VapX) binds between two monomers of a toxin (VapD) homodimer. This interaction is largely driven by charged and polar interactions, with VapX being mostly negatively charged and VapD being positively charged (Figure S3C). Moreover, the 2:1 molecular interaction between toxin and antitoxin proteins allows us to understand how an asymmetrical domain interacts with a symmetrical dimer. Interestingly, the β strands and loops of the VapX OB fold take a pseudo-two-fold symmetrical conformation, which allows them to bind in similar ways into the two VapD subunits. In this interaction, the loop connecting β1′ and β1 (L1′1) and that connecting β2 and β3 (L23) in VapX form two legs that are able to interact with each of the two VapD monomers (Figure 4). The two loops interact with some of the same residues (Arg39, Tyr25, Tyr45, and Gln22) in the two VapD monomers, but the details of the interactions differ to accommodate the lack of symmetry in VapX. Also, there is no sign of an internal pseudo-symmetry such as a palindromic protein sequence in VapX (Figure 3C). In VapD chain A, the L1′1 loop of VapX thus inserts into a small cleft between α1 and the conserved loop that connects β2 and β3 of the toxin. Here, Arg39 of VapD forms several hydrogen bonds to the backbone carbonyl groups of Phe18 and Gly19 in VapX (Figure 4A). In the other VapD monomer (chain B), the L23 loop of VapX instead binds on top of the conserved β2-β3 loop of VapD, farther away from α1 (Figure 4B). Here, both Arg39 and Ser43 form hydrogen bonds to the carbonyl group of VapX Gln44. Moreover, there is an interaction from VapD Tyr25, via the VapX Gln44 side chain, to VapD Asp7, which could directly affect activity.
Figure 4. The VapD Homodimer Adapts to Binding the Asymmetrical VapX.
(A) Interactions between VapD (chain A) and L23 of VapX with relevant residues labeled.
(B) Interactions between VapD (chain B) and L1′1 of VapX with relevant residues labeled.
(C) Interactions between VapD helix α1′ (chain A) and VapX. These interactions are not found in chain B due to the asymmetry of VapX.
(D) Structural alignment of VapD chain B (in gray) onto VapD chain A (blue), including its interaction with VapX to show how the symmetrical dimer adapts to the binding of an asymmetrical molecule.
Whereas VapX interacts differently with the conserved β2-β3 loops of the two VapD molecules, its interactions with the N-terminal ends of α1 mirror each other and induce only slight differences in the conformation of the two VapD monomers in this region (Figures 4A and 4B). In one VapD monomer, the side chain of Gln22 forms a hydrogen bond with Asp59 of VapX, while in the other monomer it interacts with Thr41. The nearby hydrophobic residues, Val21 and Tyr25, are buried by hydrophobic residues of VapX for both monomers of VapD, further stabilizing the interaction. Finally, there is an interaction between the dedicated α1′ helix of VapD chain A and VapX, which is tilted and comes closer to VapD at the top of the trimer. Several hydrogen bonds bind the helix to β3 of VapX, mainly using backbone atoms as well as the side chains of Arg73 and Gln14 on VapD (Figure 4C).
In summary, the analysis of the VapD-VapX interaction demonstrates an interesting example of how a symmetrical homodimer adapts to binding of a folded, asymmetrical partner protein, in this case primarily facilitated via the β2-β3 loop in VapD. The structure also sheds new light on the diversity of the classic OB fold as a protein-protein interaction domain. The two different modes of interaction induce different conformations in the conserved β2-β3 loop of the two VapD monomers, which can be seen upon superposition of the two VapD monomers, including both orientations of VapX (Figure 4D). Interestingly, this loop was also found in two different conformations and with increased B factors compared with the rest of the model in the H. pylori VapD dimer (Kwon et al., 2012). In both cases, however, and in line with what is observed for most type II TA complexes, the putative active-site region around Asp7 of VapD is sterically blocked by interaction of the antitoxin.
Isolated VapD Has a Closed Active-Site Cleft
To investigate the conformation of the activated VapD toxin as well as any changes induced by VapX binding, we determined the crystal structures of isolated VapD and a D7N active-site mutant. Surprisingly, expression of the wild-type (WT) toxin did not affect cell growth despite earlier results showing that VapD expression in E. coli causes growth arrest, suggesting that the effects of the toxin in this heterologous host are strain dependent or too subtle to observe during large-scale growth (Daines et al., 2004). Isolated VapD elutes with an apparent MW of 50 kDa during gel-filtration chromatography, indicating that it also forms a higher-order structure, possibly a tetramer, in the absence of VapX (data not shown). The WT protein was crystallized in the hexagonal space group, P6322, and the structure was refined to 2.5 Å, while the D7N mutant crystals could be best modeled in P321 and diffracted to 2.2 Å (see STAR Methods and Table 1). In both cases, a VapD monomer was used to determine the structure by molecular replacement. The structures were refined to R = 22.0%/Rfree = 26.4% (WT) and R = 21.4%/Rfree = 26.7% (D7N) and are complete except for a few residues in the C terminus and, in the case of the D7N mutant, the extended region 10–23. Symmetry expansion of the structures reveals that the VapD dimer and the ring-shaped trimer (of dimers) are present in both cases in the absence of VapX (Figures S3D and S4A). Moreover, since the structure of the isolated WT VapD toxin contains only a single molecule in the crystallographic asymmetric unit, it is perfectly two-fold symmetrical, and thus, VapD forms a completely symmetrical homodimer (or trimer of dimers) in the absence of VapX. The D7N mutant structure contains a pseudo-symmetrical dimer in the asymmetric unit, with very few differences between the monomers.
Structural alignment of isolated VapD with the VapX-bound form reveals differences at the top of the cleft between VapD monomers (Leu8-Thr26 of the β1α1′α1 motif), where the long α1 helix has straightened and become extended by one turn, while the shorter α1′ has unfolded completely into a loop that points upward and toward the other VapD monomer (Figure 5A). The overall effect of this conformational change is that the binding cleft between the monomers of the dimer is closed by contacts between the loops on either side, with Tyr16 and Pro18 from one monomer coming into close contact with the corresponding residues from the other monomer (Figure 5B). This hydrophobic interaction is stabilized by a hydrogen bond from His17 to Asp12 of the same monomer on each side. Furthermore, the β2-β3 loop that forms the bottom of the cleft takes a slightly different turn compared with the VapX-bound form, with the side chains pointing more inward packing more closely together. This also affects the putative catalytic residue, Asp7, which points more down into the cleft in the isolated structure (Figure 5A). However, the distance between these residues is even longer than in the VapX-bound structure (14.5 Å), supporting that they do not need to come close during catalysis and perhaps are part of separate ion-binding sites. Interestingly, the isolated VapD dimer has a highly electropositive channel running through the closed cleft, which is compatible with binding of negatively charged nucleic acids (Figure S4B). This potential is generated by the combined positive charge from several basic arginine residues, Arg39, Arg73, and Arg76, likely in combination with backbone amino groups. Together, these observations are thus compatible with a potential for nucleic acid binding at the cleft region. This area is also the most conserved part of the structure, again supporting that it constitutes an important site (Figure S4C).
Figure 5. The Structure of the Isolated WT and D7N VapD Toxins.
(A) Overview of the structure of the isolated VapD dimer (green) and comparison to the VapX-bound conformation (gray). The conserved Asp7 is shown and the arrows indicate the closure of the lobes surrounding the active-site cleft upon loss of the VapX antitoxin.
(B) Details of the interactions at the top of the isolated VapD dimer where the lobes meet. The hydrophobic interaction area involving Tyr16 and Pro18 is shown in gray.
(C) Structural alignment of the isolated VapD D7N structure (in two shades of blue) on isolated WT VapD (gray). The inset shows the conformation of the mutated residue (D7) as well as nearby residues in the β2-β3 loop (in blue, D7N only). In VapD D7N, there is a hydrogen bond between Asn7 and the carbonyl oxygen of Thr41 of the opposing VapD monomer (dashed line).
See also Figure S4.
In the VapD D7N mutant, the loop connecting β1 and the first α helix (α1, Val10-Gln23) is not visible by electron density. Since the missing part exactly covers the region that differs between the VapX-bound and the isolated form of VapD, the structure is therefore compatible with both the open and the closed conformation (Figure 5C). This suggests that this region of the structure is highly flexible in the absence of VapX and thus might adjust to accommodate substrate during binding. Interestingly, the unit cell dimensions and optimal space group differ between the WT and the D7N mutant structures, suggesting a slightly different packing (Figure S4B). The side chain of the mutated residue (D7N) points in the same direction as the WT but slightly more into the cleft than the corresponding residue of WT VapD, now forming a hydrogen bond to the carbonyl atom of Thr40 in the other VapD monomer, which is absent in the WT structure due to the more retracted position of Asp7 (Figure 5C, inset).
DISCUSSION
In this paper, we present the structure of a TA complex of the VapXD type, revealing a highly unusual stoichiometry of interaction, in which one antitoxin protein binds and inhibits a toxin dimer. Binding of the antitoxin directly to the active site of the toxin is a common feature of type II TA complexes, and by far the most common molecular stoichiometry is 1:1, where each toxin molecule is inhibited by a cognate antitoxin (Page and Peti, 2016). There are examples of 2:1 toxin:antitoxin interactions among the VapBC TA modules, where extended C-terminal tails of the VapB antitoxins adopt a linear structure and use pseudo-palindromic protein sequences to inhibit both active sites of a nearby VapC toxin dimer (Bendtsen et al., 2017). However, in these cases, there is always a second VapB antitoxin present, thus preserving the overall 1:1 molecular stoichiometry of the complex, even though one antitoxin is not used to inhibit a toxin directly. The interaction observed between VapD and VapX is more similar to the gyrase poison TA system, CcdBA, where an extended structure of the antitoxin CcdA interacts with both molecules of a symmetrical CcdB toxin dimer (De Jonge et al., 2009). However, common to nearly all TA interactions is that the toxin-interacting part of the antitoxin forms an extended (and intrinsically disordered) structure. VapXD is thus unique in that a single, folded-domain antitoxin is able to bind both molecules of a toxin dimer. The compact structure of VapX thus raises the question of how toxin activation takes place in the VapXD system. For many TA systems, a stress-induced cellular protease like Lon or Clp is believed to be involved in targeted degradation of the antitoxin via the disordered region (Muthuramalingam et al., 2016). Given that VapX consists of a compact single-domain OB fold, this suggests that perhaps another, unknown, mechanism is involved in activation.
Mapping of the electrostatic surface potential reveals that VapX has an overall electronegative surface that binds to a positively charged cleft of VapD. The OB fold of the VapX antitoxin is a well-known nucleic acid-binding fold; however, the overall charge of VapX suggests that this might not be the case. It thus appears that the VapX OB fold evolved specifically to bind a protein instead of nucleic acids (Arcus, 2002). OB domains involved in protein-protein interactions are found in some bacterial enterotoxins, such as the superantigen SEC-B from Staphylococcus aureus, which uses an OB fold to bind the T cell receptor:major histocompatibility complex on the surface of immune cells (Rodstrom et al., 2014). Despite an overall negative surface charge, the binding region of VapX does contain several residues that potentially could be used for nucleic acid interaction, including acidic (Asp33, Asp35, Glu 47, and Asp49), basic (Arg52), and aromatic (Phe18 and Tyr55) residues. Several of these residues are conserved and, especially those in the loop between β1 and β2 (L12), appear not to interact with VapD and could thus have other functions. We hope that future studies will reveal if VapX has a separate role that involves DNA/RNA binding aside from its interaction with VapD. If VapX does not bind DNA, this is contrary to what is observed for most type II TA modules, where the antitoxin functions as a repressor of transcription from the TA locus. This also raises the question of how the transcription of the vapXD locus is controlled; it is possible that it is regulated by a global transactivator protein (such as Fis), as has been shown for the vapBC-1 module (Cline et al., 2012). However, this would also have to be confirmed experimentally.
VapD is structurally similar to Cas2, which we know is active as a nuclease. Some RNase activity of VapD has been shown in vitro, but we do not know the specificity of the reaction (Ren et al., 2012). We know that a conserved residue (Asp7 in NTHi VapD) is important, but its role in catalysis remains enigmatic. Moreover, reports are conflicting when it comes to the requirement for divalent cations such as Mg2+ and Mn2+, which are required for the function of many nucleases. The consensus is that while Cas2 requires the presence of such ions (Beloglazova et al., 2008; Ka et al., 2014, 2017; Nam et al., 2012), VapD-type proteins do not (Kwon et al., 2012; Pedersen et al., 2003; Zhang et al., 2003), which is surprising given their obvious structural homology. For Cas2, it has been suggested that there is hinge motion involving the loop region between β4 and β5, which would reduce the distance between the aspartates to become compatible with Mg2+ binding (Ka et al., 2014, 2017; Nam et al., 2012). However, these conclusions are based on separate experiments, and careful parallel experiments using identical conditions would be needed to confirm if this is really the case. Finally, the concept of a toxin dimer acting as a nuclease involves the question of whether there are one or two active sites, a question that is also pertinent for understanding the VapC toxins and still not resolved. To better understand the catalytic mechanism of VapD, a structure with RNA substrate bound is needed. Such a structure would be highly valuable and would also give insights into Cas2 RNase activity, as there currently is no structure of Cas2 with RNA substrate bound.
The VapXD and VapD structures also shed light on the (yet) speculative evolutionary connections between CRISPR-Cas and the TA systems (Makarova et al., 2012). According to the proposed model, a primordial VapXD-type TA system became associated with an ancestral transposable DNA element containing cas genes (a “casposon”), which was later incorporated next to a cas cassette to allow for adaptive immunity via incorporation of novel genetic elements in the nearby CRISPR arrays (Koonin and Krupovic, 2015). According to this hypothesis, the VapD fold and nuclease evolved to become the Cas2 protein, which is known today for its role in the Cas1-Cas2 universal CRISPR-Cas adaptation module (Koonin and Krupovic, 2015). Curiously, Cas2 appears to have kept its nuclease activity despite no apparent need for this during cleavage of protospacers during CRISPR-Cas adaptation (Nunez et al., 2014; Nuñez et al., 2015). Studying the VapD-type nucleases thus provides a unique glimpse into a world that predates CRISPR-Cas and can perhaps offer explanations as to why this is the case. Comparison of VapD and Cas2, however, offers some insight into how the transformation from a TA toxin to a CRISPR Cas protein took place. We know that Cas2 lacks the small α1′ helix, which sits on top of the VapD dimer and is involved in recognition of VapX. This functionality is not required in Cas2, which has likely meant that the helix was lost over time. Moreover, Cas2 contains an additional β hairpin at its C terminus, which we know is involved in CRISPR-specific functions (Cas1 binding) (Nunez et al., 2014). This element is not present in VapD and could thus represent a later addition to the protein as it evolved to become part of the Cas1-Cas2 complex. In light of this, it is intriguing that natural variations of VapD have been found in some strains of NTHi that lack residues 30–77 (Daines et al., 2004). This would remove a large part of the core Cas2 fold and connect the long α1 with β4, thus making it difficult, from a structural perspective, to understand how the protein can remain folded (Figure S4D). Nevertheless, all natural deletions are in-frame, and the resulting proteins have been shown to interact with full-length VapD in vivo. It is thus possible that this represents an extra twist on the VapXD TA system and its relevance in the pathogenicity of H. influenzae.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Lead contact: Ditlev Egeskov Brodersen (deb@mbg.au.dk).
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact.
Materials Availability
There is no restriction on materials generated for this study and first reported here. They are available from the lead author upon reasonable request.
Data and Code Availability
Atomic coordinates and structure factors have been deposited to the protein database (PDB:6ZN8, VapXD; 6ZI0, VapD(wt); 6ZI1, VapD(D7N)). All other data are available from the lead author upon reasonable request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Microbes
E. coli B834 DE3 cells were cultured at 20°C to 37°C in Se-Met Complete media. E. coli BL21 cells were cultured at 20°C to 37°C in LB media.
METHOD DETAILS
Protein Purification
The plasmid pDD791 expressing the wt VapXD complex from NTHi strain 86–028NP C-terminally His-tagged on VapD, was transformed into the Met auxotroph E. coli strain B834 (DE3) (Merck) for seleno-methionine (Se-Met) incorporation using Se-Met Complete medium (Molecular Dimensions). Cells were grown at 37°C to OD600 = 0.6–0.8, followed by induction with 0.1 mM IPTG overnight at 20°C. Cell pellets were resuspended in a lysis buffer (50 mM Tris-HCl, pH=8, 300 mM NaCl) followed by sonication and clarification by centrifugation at 14,000 rpm for 45 min. Cell extract containing the VapX:VapD-His6 complex was loaded onto a 1 ml His-Trap column (GE Healthcare). After binding, a multi-step wash/elution was performed using a buffer identical to the lysis buffer but including 10, 20, 40, 80, 150, and 300 mM imidazole, respectively. Fractions containing most protein complex were further purified on a 1 ml Source 15Q column (GE Healthcare) running in 50 mM Tris-HCl, pH=8.5, 5 mM β-mercaptoethanol (BME) and eluted with a gradient into 1 M NaCl. Final separation was achieved on a Superdex 200 10/300 GL column (GE Healthcare), equilibrated in 20 mM Tris-HCl, pH 7.5, 300 mM NaCl, and 5 mM BME. Peak fractions were concentrated to 12 mg/ml prior to crystallisation.
For expression of isolated VapD, the wt vapD gene from NTHi strain 86–028NP was inserted into pET24b encoding a C-terminal Leu-Glu-His6 affinity tag. E. coli BL21 (DE3) transformed with this plasmid were grown to OD600 = 0.6–0.8 in Luria-Bertani (LB) medium including kanamycin (0.05 mg/mL) at 37°C in a shaking incubator. Expression was induced with 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and growth continued overnight at 20°C before harvesting cells at 6000 rpm. Cells were resuspended in 50 mM Tris-HCl, pH=8.0, 300 mM NaCl supplemented with 0.2 mg/mL lysozyme, incubated 30 minutes on ice and were lysed by sonication. After removal of cell debris, the supernatant was applied to a pre-packed 1 mL HisTrap FF (GE Healthcare) column, washed in 20–30 column volumes (CV) of lysis buffer +10 mM imidazole before elution using lysis buffer + 300 mM imidazole. Peak fractions were further purified on a 1 mL Source 15Q column (GE Healthcare Life Sciences) equilibrated in 50 mM Tris-HCl, pH=8.5, 50 mM NaCl, 5 mM BME by gradient salt elution up to 700 mM NaCl. Finally, the pooled and concentrated fractions were applied to a Superdex200 increase 10/300 column (GE Healthcare) pre-equilibrated in 20 mM Tris-HCl, pH=8.0, 150 mM NaCl, 5 mM BME running at 0.4 mL/min. The peak fractions were analysed on a 4–20 % SDS-PAGE and pooled for crystallisation trials and biochemical experiments. The column was calibrated with standard proteins of known mass. The VapD D7N site-directed mutant was synthesised by Integrated DNA Technologies (Coralville, IA, USA) and inserted into pET24b encoding a C-terminal Leu-Glu-His6 affinity tag (pDD943) and expressed and purified in a similar way, except pH=7.5 was used in the final gel filtration buffer. Both protein samples were concentrated to 6–7 mg/mL using a Vivaspin 500 mini spin column (Sartorius) before crystallisation.
Structure Determination
The NTHi VapXD complex was crystallised using the sitting-drop vapor-diffusion method at 19°C. 96-well crystallisation plates (Hampton research) were dispensed using a Mosquito automated crystallisation robot (TTP labtech). Protein crystals belonging to the cubic space group I4132 appeared in 1.6 M ammonium sulphate, 0.1 M MES pH=6.5, and 10 % v/v 1,4-dioxane. Individual crystals were transferred to a drop of the corresponding mother liquor supplemented with 25% (v/v) glycerol and immediately flash-cooled in liquid nitrogen. A complete Se-Met SAD data set was collected at the ID29 beamline of ESRF and phases obtained using SHELXD/hkl2map pipeline (de Sanctis et al., 2012; Sheldrick, 2010). The structure was built manually in Coot (Emsley et al., 2010) based on the initial model output by hkl2map and iteratively refined using Phenix.refine (Adams et al., 2010). R factors were significantly improved by optimising bulk solvent mask parameters. Ramachandran outliers and clashes were corrected by hand in Coot, and Ramachandran restraints were used in the final rounds of refinement to avoid some Ramachandran outliers from reappearing.
For isolated VapD (wt), crystals were obtained in 1 M LiCl, 0.1 M citrate, pH=4.0, 20% PEG 6000 while crystals of the VapD (D7N) mutant appeared in 1.8 M ammonium sulphate, 50 mM Na cacodylate, pH=6.0. In both cases, the crystals grew at 19°C and were frozen in mother liquor supplemented with 20% PEG 400 before data collection (360 degrees at 0.1° per frame) at 100 K at beamline P13 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany) (Cianci et al., 2017). Data were processed in XDS through XDSGUI (Kabsch, 2010) and the structures were determined by molecular replacement in Phenix.phaser (McCoy et al., 2007) using a monomer of VapD from the VapXD as a search model. Initial models were built using Phenix.autobuild (Terwilliger et al., 2008) and the structures were iteratively refined in Phenix.refine (Adams et al., 2010) with manual rebuilding in Coot (Emsley et al., 2010). Both crystal forms suffer from various crystallographic irregularities and pseudo-symmetries likely due to the higher order symmetry of the VapD dimer and ring structure. In both cases, automated software initially suggested the high-symmetry space group P6322, which was also used for the VapD (wt) structure. For the higher resolution structure of VapD (D7N), the map in this space group revealed overlaps between symmetry mates and the structure was therefore expanded to P321 to accommodate slightly different structures of some of the side chains of the two monomers in the crystallographic asymmetric unit. This structure thus has a non-crystallographic symmetry (NCS) two-fold axis parallel to the two-fold axis in P6322 but is modelled as an NCS.
Analytical Gel Filtration and Cross-Linking
Peak fractions from a Source 15Q column were pooled and concentrated to app. 6 mg/mL using a Vivaspin 6 spin filter (Sartorius) with a 5 kDa molecular mass cut-off. During concentration, the buffer was exchanged to 20 mM Hepes, pH=7.5,150 mM NaCl, 5 mM BME. 400 μL of the protein sample was loaded on a Superdex 200 increase 10/300 GL column (GE Healthcare), which had been pre-equilibrated in the same buffer. The protein was eluted at 0.4 mL/min while measuring OD280. The elution volume (Ve) from the Superdex 200 increase 10/300 GL column was used to calculate the apparent molecular mass of the sample based on a calibration run with the Gel Filtration Calibration Kit HMW (GE Healthcare). Peak fractions from the analytical SEC run were concentrated to 4 mg/ml, and a reaction mixture containing 63 μL of VapXD complex and 7 μL of 0.5% or 1% glutaraldehyde was prepared. At the time points 5, 10, 20, 40, 60, and 120 min, 11 μL aliquots were removed, and the reaction quenched with 4 μL of 2 M Tris, pH=8.0. Samples were analysed on precast protein gels (Bio Rad).
Dynamic Light Scattering (DLS)
The hydrodynamic diameter of the VapXD complex in solution was measured by DLA using a Zetasizer μV (Malvern) cuvette setup. For this, the concentration of purified VapXD complex was first adjusted to 1.5 mg/mL, and DLS was measured at 5°C. After an equilibration time of 120 s, three consecutive measurements were made, each consisting of 13 runs.
Calculation of Theoretical Hydrodynamic Diameter
The program HYDROPRO version 10.0 (Ortega et al., 2011) was used to predict the translational diffusion coefficient D, of the different suggested assemblies of VapXD. The viscosity of the solvent (water), was set to 0.0015 N s/m2 and the temperature was set to 278 K, to mimic the conditions in the DLS measurements. The predicted translational diffusion coefficients (D) were then used to calculate the hydrodynamic diameters (Dhyd) by the Stokes-Einstein equation, Dhyd = kT/3πηD, where k is Boltzmann’s constant, T is the absolute temperature, and η is the viscosity. The same values for temperature and viscosity as used in the HYDROPRO program were used for this calculation.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli B834 (DE3) | Merck | N/A |
| E. coli BL21 (DE3) | Novagene | Cat#69450 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| His-tagged H. influenzae VapXD complex | This paper | N/A |
| His-tagged H. influenzae VapD | This paper | N/A |
| His-tagged H. influenzae VapD (D7N) | This paper | N/A |
| Deposited Data | ||
| Crystal structure of the H. influenzae VapXD complex | This paper | PDB:6ZN8 |
| Crystal structure of H. influenzae VapD | This paper | PDB:6ZI0 |
| Crystal structure of H. influenzae VapD (D7N) | This paper | PDB:6ZI1 |
| Structure of SSO1404, a predicted DNA repair-associated protein from Sulfolobus solfataricus P2 | (Beloglazova et al., 2008) | PDB:2I8E |
| Structural and Biochemical Characterization of HP0315 from Helicobacter pylori as a VapD Protein with an Endoribonuclease Activity | (Kwon et al., 2012) | PDB:3UI3 |
| CRISPR-associated protein | (Ka et al., 2017) | PDB:5H1O |
| Crystal structure of Streptococcus pyogenes Cas2 at pH 5.6 | (Ka et al., 2014) | PDB:4QR0 |
| Crystal structure the Escherichia coli Cas1-Cas2 complex bound to protospacer DNA | (Nuñez et al., 2015) | PDB:5DS4 |
| GhoS (YjdK) monomer | (Wang et al., 2012) | PDB:2LLZ |
| Structure of translation elongation factor P from Acinetobacter baumannii | (J. Abendroth et al., unpublished) | PDB:5J3B |
| Recombinant DNA | ||
| pDD791 (pET24b-VapXD-His) | (Ren et al., 2012) | N/A |
| pDD1205 (pET24b-VapD-His) | This paper | N/A |
| pDD943 (pET24b-VapD(D7N)-His) | This paper | N/A |
| Software and Algorithms | ||
| XDS/XDSGUI | (Kabsch, 2010) | Download |
| SHELXD/hkl2map | (de Sanctis et al., 2012; Sheldrick, 2010). | Download |
| Phenix | (Adams et al., 2010) | Download |
| Coot | (Emsley et al., 2010) | Download |
| HYDROPRO | (Ortega et al., 2011) | Download |
Highlights.
The H. influenzae VapXD toxin-antitoxin complex has an unusual 2:1 stoichiometry
The protein complex forms a circular higher-order structure
The VapD toxin forms a dimer with a central active-site cleft
The VapX antitoxin has an oligonucleotide/oligosaccharide-binding domain
ACKNOWLEDGMENTS
This work was funded by the Danish National Research Foundation’s Centre for Bacterial Stress Response and Persistence (BASP, DNRF 120), the Novo Nordisk Foundation, and US National Institute on Deafness and Other Communication Disorders grant DC014756. The authors are indebted to beamline personnel at P13, EMBL-Hamburg, Germany, and ID29 at ESRF, France, for help and advice during data collection and Lan Bich Van for invaluable help with protein purification and crystallization.
Footnotes
DECLARATIONS OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.str.2020.10.002.
Quantification and Statistical Analysis
XDS/XDSGUI (Kabsch, 2010) were used for reduction of diffraction data, and the SHELXD/hkl2map pipeline for phase calculation for the structure of Se-Met VapXD (de Sanctis et al., 2012; Sheldrick, 2010). For VapD (wt) and VapD (D7N), Phenix.phaser (McCoy et al., 2007) we used for molecular replacement. Structure models were built using Phenix.autobuild (Terwilliger et al., 2008) and Coot (Emsley et al., 2010) and refined in Phenix.refine (Adams et al., 2010) with manual rebuilding in Coot (Emsley et al., 2010). All crystallographic data statistics are available in Table 1.
The program HYDROPRO version 10.0 (Ortega et al., 2011) was used to predict the translational diffusion coefficient D for VapXD.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus V (2002). OB-fold domains: a snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol 12, 794–801. [DOI] [PubMed] [Google Scholar]
- Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, and Ben-Tal N (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44, W344–W350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, et al. (2008). A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem 283, 20361–20371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen KL, Xu K, Luckmann M, Winther KS, Shah SA, Pedersen CNS, and Brodersen DE (2017). Toxin inhibition in C. crescentus VapBC1 is mediated by a flexible pseudo-palindromic protein motif and modulated by DNA binding. Nucleic Acids Res 45, 2875–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianci M, Bourenkov G, Pompidor G, Karpics I, Kallio J, Bento I, Roessle M, Cipriani F, Fiedler S, and Schneider TR (2017). P13, the EMBL macromolecular crystallography beamline at the low-emittance PETRA III ring for high- and low-energy phasing with variable beam focusing. J. Synchrotron Radiat 24, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline SD, Saleem S, and Daines DA (2012). Regulation of the vapBC-1 toxin-antitoxin locus in nontypeable Haemophilus influenzae. PLoS One 7, e32199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines DA, Jarisch J, and Smith AL (2004). Identification and characterization of a nontypeable Haemophilus influenzae putative toxin-antitoxin locus. BMC Microbiol 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daines DA, Wu MH, and Yuan SY (2007). VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J. Bacteriol 189, 5041–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, Wyns L, De Greve H, and Loris R (2009). Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol. Cell 35, 154–163. [DOI] [PubMed] [Google Scholar]
- de Sanctis D, Beteva A, Caserotto H, Dobias F, Gabadinho J, Giraud T, Gobbo A, Guijarro M, Lentini M, Lavault B, et al. (2012). ID29: a high-intensity highly automated ESRF beamline for macromolecular crystallography experiments exploiting anomalous scattering. J. Synchrotron Radiat 19, 455–461. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeders N, and Van Melderen L (2014). Toxin-antitoxin systems as multilevel interaction systems. Toxins (Basel) 6, 304–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Brodersen DE, Mitarai N, and Gerdes K (2018). Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol. Cell 70, 768–784. [DOI] [PubMed] [Google Scholar]
- Ka D, Hong S, Jeong U, Jeong M, Suh N, Suh JY, and Bae E (2017). Structural and dynamic insights into the role of conformational switching in the nuclease activity of the Xanthomonas albilineans Cas2 in CRISPR-mediated adaptive immunity. Struct. Dyn 4, 054701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka D, Kim D, Baek G, and Bae E (2014). Structural and functional characterization of Streptococcus pyogenes Cas2 protein under different pH conditions. Biochem. Biophys. Res. Commun 451, 152–157. [DOI] [PubMed] [Google Scholar]
- Kabsch W (2010). Xds. Acta Crystallogr. D Biol. Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, and Krupovic M (2015). Evolution of adaptive immunity from transposable elements combined with innate immune systems. Nat. Rev. Genet 16, 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Kwon AR, Kim JH, Park SJ, Lee KY, Min YH, Im H, Lee I, Lee KY, and Lee BJ (2012). Structural and biochemical characterization of HP0315 from Helicobacter pylori as a VapD protein with an endoribonuclease activity. Nucleic Acids Res 40, 4216–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoux M, Culviner PH, Liu YJ, Littlehale ML, and Laub MT (2020). Stress can induce transcription of toxin-antitoxin systems without activating toxin. Mol. Cell. 79, 280–292.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Anantharaman V, Aravind L, and Koonin EV (2012). Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct 7, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinaro AL, Kashipathy MM, Lovell S, Battaile KP, Coussens NP, Shen M, and Daines DA (2019). Crystal structure of VapBC-1 from nontypeable Haemophilus influenzae and the effect of PIN domain mutations on survival during infection. J. Bacteriol 201, 10.1128/JB.00026-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TF (2003). Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis 16, 129–134. [DOI] [PubMed] [Google Scholar]
- Murzin AG (1993). OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. EMBO J 12, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuramalingam M, White JC, and Bourne CR (2016). Toxin-antitoxin modules are pliable switches activated by multiple protease pathways. Toxins (Basel) 8, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Ding F, Haitjema C, Huang Q, DeLisa MP, and Ke A (2012). Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J. Biol. Chem 287, 35943–35952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Harrington LB, Kranzusch PJ, Engelman AN, and Doudna JA (2015). Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, and Doudna JA (2014). Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat. Struct. Mol. Biol 21, 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez JK, Lee ASY, Engelman A, and Doudna JA (2015). Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega A, Amoros D, and Garcia de la Torre J (2011). Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J 101, 892–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page R, and Peti W (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol 12, 208–214. [DOI] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, and Ehrenberg M (2003). The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140. [DOI] [PubMed] [Google Scholar]
- Pei J, and Grishin NV (2014). PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol. Biol 1079, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Walker AN, and Daines DA (2012). Toxin-antitoxin loci vapBC-1 and vapXD contribute to survival and virulence in nontypeable Haemophilus influenzae. BMC Microbiol 12, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodstrom KE, Elbing K, and Lindkvist-Petersson K (2014). Structure of the superantigen staphylococcal enterotoxin B in complex with TCR and peptide-MHC demonstrates absence of TCR-peptide contacts. J. Immunol 193, 1998–2004. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (2010). Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr 66, 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, and Adams PD (2008). Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr 64, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, and Wuttke DS (2003). Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct 32, 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lord DM, Cheng HY, Osbourne DO, Hong SH, Sanchez-Torres V, Quiroga C, Zheng K, Herrmann T, Peti W, et al. (2012). A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol 8, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, and Inouye M (2003). MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited to the protein database (PDB:6ZN8, VapXD; 6ZI0, VapD(wt); 6ZI1, VapD(D7N)). All other data are available from the lead author upon reasonable request.