SUMMARY
Recent advances in single particle cryogenic electron microscopy (cryo-EM) have enabled the structural determination of numerous protein assemblies at high resolution, yielding unprecedented insights into their function. However, despite its extraordinary capabilities, cryo-EM remains time-consuming and resource-intensive. It is therefore beneficial to have a means for rapidly assessing and optimizing the quality of samples prior to lengthy cryo-EM analyses. To do this, we have developed a native mass spectrometry (nMS) platform that provides rapid feedback on sample quality and highly streamlined biochemical screening. Because nMS enables accurate mass analysis of protein complexes, it is well-suited for routine evaluation of the composition, integrity, and homogeneity of samples prior to their plunge-freezing on EM grids. We demonstrate the utility of our nMS-based platform for facilitating cryo-EM studies using structural characterizations of exemplar bacterial transcription complexes as well as the replication-transcription assembly from the SARS-CoV-2 virus that is responsible for the COVID-19 pandemic.
Graphical Abstract

eTOC blurb
A major bottleneck in single-particle cryo-EM involves sample preparation and assessment of sample stability and homogeneity. Olinares et. al. have developed a time-saving native mass spectrometry-based platform that provides rapid feedback on sample quality and enables highly streamlined biochemical screening for optimal sample conditions prior to cryo-EM analysis.
INTRODUCTION
The central players in most cellular events are multi-subunit protein complexes with highly coordinated components (Alberts, 1998). To understand how these complexes work, it is valuable to elucidate their molecular architectures and capture the repertoires of conformational changes that they undergo in performing their function. Among the currently available structural biology methodologies, single particle cryogenic electron microscopy (cryo-EM) is generating near-atomic resolution structures of macromolecular assemblies at an ever-increasing pace, ushering in an exciting new era in structural biology (Callaway, 2015; Cheng, 2018; Kuhlbrandt, 2014; Nogales and Scheres, 2015; Ognjenovic et al., 2019). Technological advances in cryo-EM, particularly in image detection and data processing, are enabling the structural determination of conformationally heterogeneous protein assemblies, including reversible and intermediate states that have previously proved largely intractable (Frank, 2002; Murata and Wolf, 2018; Nogales and Scheres, 2015; Wu and Lander, 2020).
The single particle cryo-EM workflow (Cheng et al., 2015; Costa et al., 2017; Lyumkis, 2019) involves blotting or spraying the protein sample onto an EM grid followed by immediate plunge-freezing into liquid ethane to preserve the protein complexes in a frozen hydrated state. The resulting specimen is imaged with a transmission electron microscope that records pictures of millions of particles in various orientations. The individual particles are picked, sorted, and aligned by shape and orientation. The resulting 2D and 3D classifications are used to reconstruct and refine three-dimensional structure(s) of potentially multiple conformations of the target complex.
Despite its spectacular capabilities, cryo-EM analysis comes with a high price tag and demands considerable time investment (Hand, 2020). Access to appropriate electron microscopes, which are often shared resources among multiple laboratories or institutes, can involve long waiting periods. After acquiring vast amounts of data, the subsequent image classification, reconstruction, and refinement steps require substantial computing capacity and processing time. To maximize the use of all this effort, time and resources, it is critical to prepare high-quality samples with an efficient means of checking sample quality (Lyumkis, 2019; Passmore and Russo, 2016; Takizawa et al., 2017). Single particle cryo-EM capitalizes on averaging large numbers of similar particles to improve the signal-to-noise ratio and structural resolution. Obtaining high quality structures ultimately depends on collecting a sufficiently large number of individual particle images that originate from the undissociated target protein complex in the desired assembly state(s) with negligible degradation products or contaminating proteins. Hence, it is highly desirable to have available a rapid and reliable method to assess sample stability and homogeneity and to screen for the presence of the desired constituents, stoichiometry, and assembly state(s).
Standard methods for initially evaluating sample quality during purification and reconstitution of the target protein complex include SDS-PAGE, native gel electrophoresis and size exclusion chromatography (SEC). These sizing techniques can provide information on sample composition and homogeneity but at low mass accuracy and resolution. Negative stain EM is also employed to evaluate particle size and distribution in the reconstituted sample; however, specimen preparation can introduce artifacts in certain cases (Lyumkis, 2019) and correlating particle size distribution with the presence of a completely assembled target complex can yield ambiguous results. Differential scanning fluorimetry and thermal melting assays (Boivin et al., 2013; Chari et al., 2015) are automated, high-throughput screening methods that correlate unfolding transitions of protein complexes with increasing temperature; these yield readouts of global sample stability that are sometimes difficult to interpret for multi-component assemblies, particularly in pinpointing which components are labile or unstable in the conditions being tested. Mass photometry (MP) is a relatively fast, single-particle mass measurement method based on light scattering of molecular assemblies that is useful for assessing sample heterogeneity; however, with ~20 kDa mass resolution and 2% mass accuracy, it might not fully resolve relevant protein modifications or bound small molecules, cofactors, and ligands (Sonn-Segev et al., 2020; Young et al., 2018).
Native mass spectrometry (nMS) enables direct mass measurement of intact noncovalent macromolecular assemblies upon gas-phase transfer from nondenaturing solution conditions (Heck, 2008; Hernandez and Robinson, 2007; Leney and Heck, 2017; Loo, 1997). By maintaining the quaternary structure of protein assemblies, nMS has provided key complementary structural information for hybrid and integrative structural studies involving cryo-EM, including complex composition, subunit stoichiometry, inter-subunit connectivity, and assembly dynamics (Abbas et al., 2020; Casanal et al., 2017; Chase et al., 2018; Geiger et al., 2010; Kuhlen et al., 2018; Liko et al., 2016; Lorenzen et al., 2007; Martinez-Rucobo et al., 2015; Snijder et al., 2017). In addition, high-resolution nMS can resolve and identify relevant post-translational modifications as well as bound ligands, cofactors, substrates or stabilizing lipids that are useful in assigning previously unknown EM densities during structure reconstruction and refinement (Liko et al., 2016).
Here we describe an nMS platform that readily integrates into the earlier steps of cryo-EM workflows to generate critical information on sample stability and homogeneity. In addition, for characterizing protein assemblies that contain labile components or are dynamic, the nMS platform enables screening for optimal biochemical conditions that promote and preserve the assembly state(s) of the target protein complex. To illustrate the utility of our nMS-based platform, we describe three structural biology projects involving the bacterial transcription machinery that contains the DNA-dependent RNA polymerase (DdRp) at various stages of the transcription cycle (Chen et al., 2020a; Kang et al., 2020; Kang et al., 2017), and a fourth project involving the coupling of a helicase and RNA-dependent RNA polymerase (RdRp) in SARS-CoV-2 (Chen et al., 2020b). Most of these examples were initiated with starting conditions that did not yield the desired target assemblies for cryo-EM, shortcomings that if not recognized prior to EM data collection and analysis would have led to a considerable waste of valuable EM resources and investigators’ time and effort. However, with the aid of iterative nMS-based screening, it proved possible to rapidly establish optimized sample conditions that eventually yielded cryo-EM structures at near-atomic resolution. The high-resolving capability of nMS also proved useful in revealing sample heterogeneities, including those that differed by just a single nucleotide mass. Overall, we find that our nMS platform provides an enormously time-saving strategy that enables streamlined biochemical screening and routine assessment of sample quality in solution prior to cryo-EM analyses.
RESULTS
The native MS platform
Our nMS-based diagnostic and screening platform integrates into the earlier steps of the cryo-EM workflow by rapidly assessing which components and conditions are optimal in preparing samples for blotting or spraying onto EM grids and subsequent plunge-freezing. The method can be used to interrogate samples that range from those obtained from the initial steps along the preparation/purification of each protein or oligonucleotide component all the way to the reconstitution of the desired target macromolecular assembly. Using accurate mass readouts of the relevant constituents and assemblies, nMS provides feedback on whether or not the sample prepared under a particular condition contains the desired target protein complex containing all the anticipated components, often including bound cofactors. It also provides a detailed assessment of the presence of degradation products, contaminant proteins, cloning artifacts, or unwanted modifications. The rapid feedback facilitates iterative testing and optimization to select the best condition(s) for preparing high-quality cryo-EM samples.
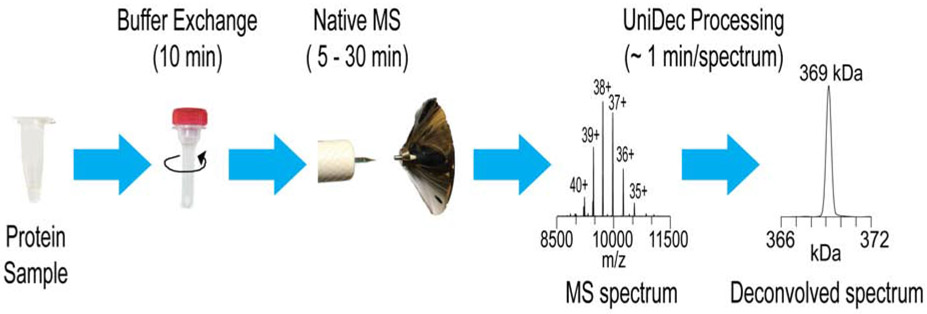
Figure 1 shows the schematic for nMS analysis of each sample or sample condition. First, the protein samples are buffer exchanged into an nMS-compatible solution that is volatile and maintained in the physiological pH range. We have previously optimized this step to facilitate rapid, multiplexed buffer exchange using microspin desalting columns with low sample volumes (<13 μL) to minimize sample consumption (Olinares et al., 2016). Multiple protein samples can be buffer exchanged in parallel with a multi-slot centrifuge. Ammonium acetate (100 to 500 mM) is our typical nMS buffer of choice. The starting amount for each protein or nucleic acid component required for nMS screening depends on: (1) the component stoichiometry, assembly state, and stability of the target protein complex, (2) the sample concentration range that yields good MS peak signals, and (3) the number of iterative experiments involved. The typical electrospray sample concentration for our nMS analyses ranges from 0.5 – 10 μM, which is in the same range of concentrations that is generally used for cryo-EM experiments. Usually, 30 – 50 μL each of the protein and nucleic acids components at 5 – 20 μM concentration is sufficient for several MS optimization experiments. The protein components can be buffer exchanged at these higher protein concentrations (up to 20 μM in the samples analyzed here) to allow further sample mixing, reconstitution, or dilution to the desired concentration prior to nMS analysis.
Figure 1. nMS-based screening platform.
nMS analysis workflow for each sample with the typical time duration involved for each step. The target complex can be reconstituted before or after the buffer-exchange step prior to nMS analysis.
As a good starting point for determining nMS solution conditions, we match the ammonium acetate concentration and pH with the range of ionic strength and buffer pH used in the original purification, storage, or reconstitution of the protein samples (Hernandez and Robinson, 2007). In addition, we found that adding a small amount of Tween-20 (0.001 – 0.01 %v/v), a non-ionic detergent that is commonly used for surface passivation, into the nMS solution enables maximal sample recovery by preventing adsorptive losses during buffer exchange without introducing significant chemical background or signal interference during nMS analysis (Olinares et al., 2016). For example, if the protein sample is well-behaved in 20 mM Tris, 200 mM NaCl, pH 7.5, we would initially buffer exchange it into 200 mM ammonium acetate, 0.01% Tween-20, pH 7.5. The ionic strength and/or pH can be subsequently varied during the screening process, if deemed necessary.
Prior to nMS characterization of the reconstituted macromolecular assemblies, the purified protein components are usually analyzed separately by nMS to check if the individual components have the correct masses based on their sequences. This mass information can be valuable for assessing (1) sample purity, (2) the efficacy of protein purification protocols (e.g., correct protein expression and complete affinity tag removal during protease cleavage), and (3) the presence of post-translational modifications (e.g., truncations or loss of N-terminal methionine) and noncovalently bound small molecules (e.g., Zn2+ cofactors). In addition, we can verify the masses of any DNA and/or RNA components using the same nMS workflow for protein analysis in positive mode with minor modifications in the MS parameters (see STAR Methods and Figures S1, S2, and S4). Here, nucleic acid samples were analyzed at 1 – 5 μM concentration in 500 mM ammonium acetate, 0.01% Tween-20. The nucleic acid construct with the highest mass analyzed in the present work was a 60-kDa, 98-bp duplex DNA (Figure S1).
For our nMS experiments, we use a commercial Exactive Plus EMR, an Orbitrap-based mass analyzer that is configured for measuring the masses of macromolecular assemblies at high resolving power and high sensitivity (Rose et al., 2012). The samples are manually introduced into the instrument at nanoflow rates (typically 20 – 50 nL/min) with a modified static nanospray source that enables easy access and on-the-fly troubleshooting of the electrospray (Olinares and Chait, 2020). Sample loading, source setup and nanospray initiation generally take ~ 2 min. Mass spectra are acquired efficiently with 100 scans within 1 – 1.5 min depending on the ion injection time, number of microscans, and instrument resolution settings, which impact the overall duty cycle. We used the same set of nMS parameters for optimal desolvation and ion transmission of all the target protein complexes analyzed in this study (see STAR Methods).
The resulting raw data from nMS analyses are mass-to-charge (m/z) spectra. The detected species are multiply charged and thus register multiple m/z peaks on the spectrum (e.g., peaks for charge-state z = 40+, 39+,…35+ in Figure 1). To obtain the actual mass values for each species present, the charge-state series are then processed and converted into zero-charge state spectra (also called deconvolved spectra) showing a single mass peak for each observed species (e.g., a peak for the 369-kDa protein complex in Figure 1). For spectrum processing, we use the fast and freely available Bayesian-based deconvolution software UniDec that is part of the MetaUniDec software suite (Marty et al., 2015; Reid et al., 2019) (see STAR Methods for parameters used). Each MS spectrum takes ~1 min to deconvolve, enabling us to process raw nMS spectra during or immediately after acquisition to yield mass information that guides the selection of subsequent screening conditions.
Example 1: Promoter melting pathway in bacterial transcription initiation
We have applied our nMS screening platform to facilitate in-depth structural characterization of the bacterial transcription cycle, a highly coordinated multi-step process wherein genetic information encoded in DNA is transcribed into RNA. In bacteria, a single catalytically active DdRp synthesizes all cellular RNA (Feklistov et al., 2014). The ~400-kDa bacterial DdRp core enzyme (termed here as E) consists of five protein subunits (α2ββ’ω). During the early stage of transcription initiation, sigma factor 70 (σ70), associates with the DdRp core to form the holoenzyme (Eσ70), which is then capable of interacting with promoter DNA containing specific recognition sequences (elements) upstream from the transcription start site (Feklistov et al., 2014; Gruber and Gross, 2003). The resulting promoter-bound Eσ70 complex is called the closed complex (RPc), referring to the initial state of the duplex DNA. Without using ATP as external energy source, the Eσ70 complex in the RPc “melts” a specific area of the duplex DNA and separates the two DNA strands (generating the transcription bubble), and positions the single-stranded DNA template into the active site to form the open complex (RPo) (Bae et al., 2015; Ruff et al., 2015; Saecker et al., 2011; Zuo and Steitz, 2015). This promoter melting process has been challenging to characterize in structural detail because the RPc isomerizes rapidly through several intermediate states to form the stable, and often irreversible, RPo. Hence, our main goals were to reconstitute a stable RPc and visualize the conformational changes that occur during promoter DNA melting prior to reaching the RPo state.
Our initial efforts focused on assembling RPc by reconstituting Eσ70 complexes from E. coli with the well-characterized rrnB P1 promoter DNA (Figure S1), a ribosomal RNA promoter that forms an unstable RPo (relative to most E. coli promoters) that is in rapid equilibrium with earlier intermediates (Gourse et al., 2018; Rutherford et al., 2009). To stabilize and capture these intermediates, we added a transcription factor called TraR, which inhibits transcription of ribosomal RNAs and ribosomal proteins during nutrient starvation or stringent response (Frost et al., 1994; Gopalkrishnan et al., 2017; Maneewannakul and Ippen-Ihler, 1993). DNA footprinting and transcription assays indicated the presence of transcriptionally active Eσ70 complexes together with the rrnB P1 promoter (Gopalkrishnan et al., 2017). Negative stain EM imaging of the TraR-Eσ70 sample with rrnB P1 DNA showed particles with size dimensions consistent with the intact TraR-Eσ70 protein complex. These results led us to commit the considerable time and effort of performing cryo-EM analysis only to discover that the particle densities obtained only corresponded to TraR-Eσ70 complexes with no promoter DNA bound (Figure 2C) (Chen et al., 2019). At this point, we postulated that nMS might be a much more efficient and faster assay to determine the integrity of such complexes, prior to the time-consuming and labor-intensive single particle cryo-EM structure determination.
Figure 2. nMS-based screening to obtain intact RPc complexes containing Eσ70, transcription factor TraR, and promoter DNA.
(A) Screening for RPc with rrnB P1 promoter DNA. The conditions that were tested include DNA concentration, addition of transcription factor Fis, and incubation with glutaraldehyde (GA) that can stabilize complex formation. Crosses indicate that the expected complex containing DNA was not observed. (B) Screening for RPc with rpsT P2 promoter DNA. Checks indicate that the expected complex containing DNA was observed. The constructs tested include the wild-type and T-7A mutant sequences as well as fully complementary and partially melted promoters. (C) Representative single particle cryo-EM structures for transcription complexes with rrnB P1 (no DNA-bound), wild-type rpsT P2 and T-7A rpsT P2* promoters (PDB IDs: 6n57, 6psq, and 6pst, respectively) (Chen et al., 2020a; Chen et al., 2019).
We thus tested the efficacy of our nMS platform to assay sample integrity. After buffer exchange into ammonium acetate, we incubated the pre-assembled TraR-Eσ70 complex with the rrnB P1 promoter DNA. Subsequent nMS analysis showed an intact 471-kDa TraR-Eσ70 complex at 1:1 stoichiometry but without bound DNA (Figure 2A), consistent with the cryo-EM results. Informed by previous biochemical and functional studies, we rapidly screened a variety of conditions using our nMS platform. These conditions included (1) increases in DNA concentration, (2) addition of FIS, a protein that binds rrnB P1 and stabilizes complexes with the DdRp (Bokal et al., 1997), and (3) glutaraldehyde (GA) stabilization based on a previous successful cryo-EM study with another transcription complex (Kang et al., 2017). However, none of these conditions yielded the target DNA-bound TraR-Eσ70 complexes (Figure 2A).
Given that we were unable to identify any DNA-containing transcription complexes either from cryo-EM or nMS, we decided to test the binding of a different promoter, the rpsT P2 promoter, which controls transcription of the gene encoding ribosomal protein S20. Like rrnB P1, rpsT P2 is inhibited by TraR during the stringent response; however, DdRp complexes with rpsT P2 are more stable than those with the rrnB P1 promoter (Chen et al., 2019; Gopalkrishnan et al., 2017). In this trial, nMS analysis of TraR-Eσ70 and the 65-bp rpsT P2 duplex promoter yielded complete DNA-bound transcription complexes (Figure 2B). Encouraged by this positive nMS result using the wild-type fully double-stranded rpsT P2 promoter, we engineered and tested three additional sequence variants (Figure S1 and Table S1). The first was a variant with a T-7A mutation, which yielded DNA footprinting results suggesting stabilization of early promoter melting intermediates (Chen et al., 2020a). The second contained a partially melted bubble that was introduced into the wild type using two non-complementary base pairs in the −10 element region. The third contained this partially melted bubble introduced into the T-7A mutant (we call this promoter variant rpsT P2*). Our nMS assay indicated that the T-7A mutant promoter bound less well to the TraR-Eσ70 complex than the wild-type duplex, as expected (Figure 2B). However, introduction of a partial bubble with two unpaired base pairs at the upstream edge of the bubble on both wild-type and mutant sequences yielded fully assembled TraR-Eσ70-promoter DNA complexes (Figure 2B).
Guided by these nMS screening results, we prepared cryo-EM grids for both the TraR-Eσ70 plus WT rpsT P2 duplex DNA and the TraR-Eσ70 plus rpsT P2* DNA. From the resulting single particle cryo-EM structures, we were able to capture and discern several distinct structural intermediates that revealed a detailed stepwise view of how promoters are melted by Eσ70 (Chen et al., 2020a). Representative cryo-EM structures are shown in Figure 2C, with the bound promoter DNA still in closed (top) or partially melted (bottom) form (Chen et al., 2020a).
Example 2: Arrest of bacterial transcription elongation by a phage protein
Coliphages (viruses that infect E. coli) hijack and reprogram the host’s transcription machinery to transcribe their viral genomes. Multiple coliphage strains that infect one bacterial host devise strategies to outcompete rival strains for cellular resources. In particular, the HK022 prophage blocks superinfection of a rival coliphage λ by producing a 13-kDa protein called Nun that specifically inhibits transcription of λ DNA (Hung and Gottesman, 1995; Robert et al., 1987; Robledo et al., 1991). We sought to determine the structural basis for transcription elongation arrest by the HK022 Nun protein.
Upon transcription bubble formation (RPo state), the bound sigma factor dissociates and the DdRp translocates along the template DNA and synthesizes RNA in the presence of nucleotide substrates in a transcription elongation complex (TEC). In order to assemble TECs that promote Nun association, we used a modified nucleic acid scaffold (NAS) comprising a DNA:RNA hybrid (Figure 3A), that was previously shown to recapitulate Nun-mediated transcription arrest in vitro (Vitiello et al., 2014). The pre-translocated RNA used for this analysis is a 10-mer unit that spans the whole 10-nt bubble and has an identical sequence to the post-translocated, 9-mer RNA but with an additional cytosine base at the 3’-end (Figure 3A). We thus aimed to determine whether a pre-translocated or post-translocated transcript was the better choice for structural studies of Nun-TEC. Our nMS analyses showed that the 384-kDa TECs can be successfully reconstituted with either pre- or post-translocated RNA (Figure 3B). Upon incubation of both TECs with the Nun protein, we observed a Nun-TEC assembly at 1:1 stoichiometry (Figure 3B and Table S1).
Figure 3. nMS-based screening for Nun-bound TECs.
(A) Nucleic acid scaffold (NAS) sequences used in assembling the TEC. (B) nMS analyses of TEC containing pre- or post-translocated RNA transcript with or without incubation with the Nun protein. The pre-translocated RNA has an additional nucleotide at its 3’-end (cytosine with mass of 305 Da) compared to the post-translocated version. (C) Zoom-in of spectra from (B) highlighting the peaks for the TEC and TEC/Nun assemblies. (D) EM structure of the Nun-bound TEC containing a post-translocated RNA transcript in the NAS (PDB ID: 6alg) (Kang et al., 2017).
Although samples containing pre- or post-translocated RNA both yielded assembled complexes, closer inspection of the deconvolved spectra (Figure 3C and Table S1) revealed differences in sample homogeneity. Doublet peaks were observed for the TEC and Nun-TEC assemblies containing the 10-mer, pre-translocated transcript. The main peaks (about 80% relative intensity in each pair of peaks) matched the masses of complexes with the 10-mer RNA. A lower-intensity peak that was 305 Da lower in mass than the main peak corresponded to complexes that lost the cytosine nucleotide (305 Da) at the 3’-end, yielding 9-mer RNA-containing assemblies. This finding indicated that a population of the TEC complex harboring the pre-translocated RNA had undergone intrinsic nucleolytic cleavage wherein the DdRp hydrolyzes the phosphodiester bond linking the penultimate and the 3’-end nucleotides. This terminal RNA cleavage was also observed in Nun-free, TEC-only samples (Figure 3B) indicating that it had occurred prior to the addition of Nun.
In contrast to the situation observed for TECs containing pre-translocated RNA, we did not observe RNA cleavage with TECs containing the post-translocated RNA (Figure 3C), indicating that these samples are stable and homogeneous relative to RNA content and translocation state (only post-translocated register). For our structural study, we thus reconstituted a TEC/Nun sample with NAS containing a post-translocated RNA transcript that yielded a high-resolution Nun-TEC structure (Kang et. al., 2017). From the structure (Figure 3D), it was seen that Nun wedges into the TEC active site cleft forming an extensive interaction network that explains how Nun essentially 'crosslinks' the nucleic acids to the DdRp, preventing translocation necessary for transcript elongation (Kang et al., 2017).
Example 3: Transcription termination of stalled bacterial elongation complexes by the Mfd translocase
E. coli Mfd is an ATP-dependent transcription-repair coupling factor (TRCF) that couples transcription and DNA repair by recognizing and disassembling TECs that are stalled at damaged DNA sites (Bockrath et al., 1987; Witkin, 1966). Mfd is a 132-kDa multi-domain protein that can translocate on DNA, associate with stalled TECs, disassemble TECs to terminate transcription, and recruit the nucleotide-excision repair machinery (Selby and Sancar, 1993; Selby et al., 1991; Witkin, 1966).
We focused our nMS screening on variations of the nucleic acid scaffold (NAS) sequences used to assemble a nucleoprotein complex mimicking a stalled TEC that can be processed by Mfd (Komissarova et al., 2003; Park et al., 2002; Park and Roberts, 2006; Vvedenskaya et al., 2014; Zhang et al., 2012). As outlined in Figure S2, we systematically modified the NAS constructs by varying the DNA sequences upstream of the bubble, the DNA sequence complementarity, and the length of the RNA transcript (21-mer or 20-mer for pre- or post-translocated versions, respectively). We also screened Mfd-TEC formation for various TECs mixed with Mfd that had been purified from two different preparations (Figure S3D).
In summary, with the five different NAS sequences screened, we found one NAS sequence (NAS 3) and a specific Mfd protein preparation that generated the highest amount of intact Mfd-TEC complexes with no sample heterogeneity from RNA transcript cleavage, no dissociated TECs, and no dimeric Mfd (Figure 4A, Figure S3 and Data S1). We therefore prepared EM grids using the optimal composition and condition inferred from our nMS screening and analyzed these by single particle cryo-EM. Using image classification approaches, we captured distinct Mfd-TEC complexes (a representative structure is shown in Figure 4B) that depict the extensive remodeling of Mfd and the upstream duplex DNA upon TEC engagement (Kang et al., 2020).
Figure 4. nMS-based screening to capture intact Mfd-TEC assemblies.
(A) nMS analyses of TECs containing various nucleic acid scaffolds (NAS) incubated with Mfd. The check mark indicates the condition which generated the highest amount of intact Mfd-TEC complexes that did not show evidence for dissociation or transcript cleavage. Refer to Figure S2, Figure S3, and Data S1 for more details. (B) Representative single particle cryo-EM structure of the Mfd-bound TEC containing NAS 3 (PDB ID: 6x5q) (Kang et al., 2020).
Example 4: Coupling of the helicase and RdRp for replication and transcription in SARS-CoV-2
SARS-CoV-2 is the pathogenic agent responsible for the current globally devastating Coronavirus Disease 2019 (COVID-19) pandemic (Wu et al., 2020; Zhou et al., 2020). The SARS-CoV-2 RNA genome is replicated and transcribed by the RdRp holoenzyme (holo-RdRp) comprising nsp7, nsp8, and nsp12 (Kirchdoerfer and Ward, 2019; Subissi et al., 2014) and is the target for antiviral drugs such as remdesivir (Agostini et al., 2018; Yin et al., 2020). The holo-RdRp has been shown to coordinate with a number of essential co-factors (Snijder et al., 2016; Sola et al., 2015), including nsp13, a superfamily 1B (SF1B) helicase that can unwind DNA or RNA in an NTP-dependent manner (Ivanov and Ziebuhr, 2004; Lee et al., 2010; Seybert et al., 2000a; Seybert et al., 2000b; Tanner et al., 2003). Several studies suggest that nsp13 and nsp12 can associate (Adedeji et al., 2012; Jia et al., 2019), but a stable complex had not been reconstituted nor structurally characterized. We thus aimed to assemble the holo-RdRP with an RNA scaffold, forming the so-called replication-transcription complex (RTC), with bound nsp13 to obtain information on the structure and function of the helicase in viral replication and transcription.
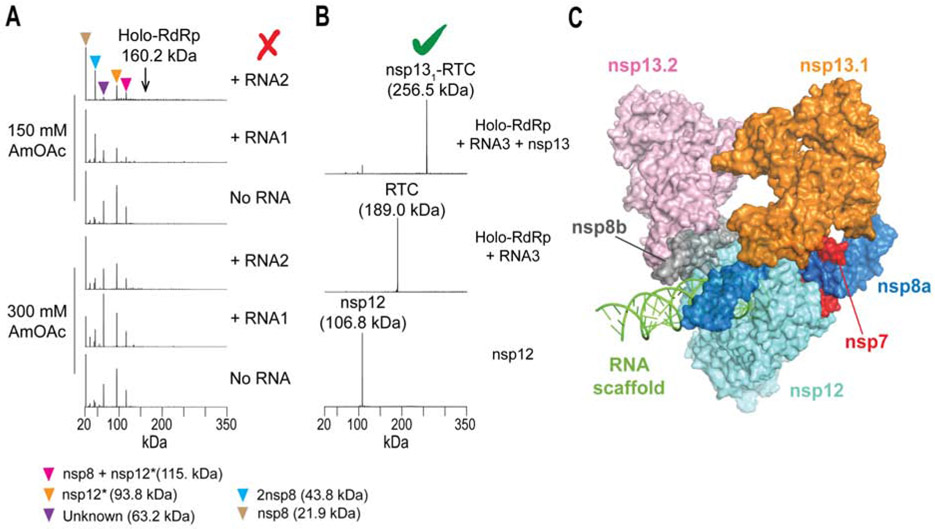
The SARS-CoV-2 nsp7/nsp8 (co-expressed), nsp12, and nsp13 genes were cloned separately in expression plasmids using coding sequences optimized for recombinant production in E. coli (Chen et al., 2020b). Prior to nMS analysis, we assembled the RTC by incubation of purified nsp7/nsp8 and nsp12 at 3:1 ratio, performed SEC to remove excess nsp7/8, and added the RNA scaffold. We used nMS to screen two different RNA scaffolds (Figure S4A) incubated in either 150 mM or 300 mM ammonium acetate. nMS results showed no holo-RdRp nor RTC peaks detected in any of the tested conditions (Figure 5A). Instead, we observed peaks for subcomplexes, individual subunits, a truncated nsp12 (measured mass was lower by 13 kDa, see Table S2) as well as an unknown 63-kDa species. These results showed that the sample was heterogeneous and did not contain the properly assembled complex. The main issue was that the purified, N-terminal tagged nsp12 was missing its C-terminal domain (thumb region), which harbors critical interaction surfaces for holo-RdRp assembly and binding to substrate RNA (Table S2). We thus replaced the codon-optimized gene construct for nsp12 with the cDNA obtained by reverse-transcription of SARS-CoV-2 RNA propagated from Vero E6 cells (Chen et al., 2020b). nMS analysis of the purified nsp12 expressed from the new construct showed the correct mass for nsp12 (Figure 5B and Table S2). With the expression issue resolved, we then proceeded to put together the RTC using a longer primer-template RNA scaffold (RNA3, Figure S4), finally obtaining a fully assembled RTC (Figure 5B) with a stoichiometry of nsp7:2nsp8:nsp12:RNA, consistent with recently determined cryo-EM structures of the SARS-CoV RTC (Hillen et al., 2020; Kirchdoerfer and Ward, 2019; Wang et al., 2020; Yin et al., 2020). Subsequent addition of nsp13 helicase to the RTC sample yielded samples with an nMS peak corresponding to a fully reconstituted nsp13-RTC at 1:1 stoichiometry (Figure 5B). These optimal nMS sample preparation conditions were then used in preparing specimens for cryo-EM analysis, which yielded high-resolution structures for the helicase-bound RTC (Figure 5C, (Chen et al., 2020b)).
Figure 5. nMS-based screening to reconstitute the SARS-CoV-2 helicase-RTC.
(A) Screening for the viral holo-RdRp with varying RNA scaffolds and ammonium acetate (AmOAc) concentrations. (B) nMS results after optimizing expression of the correct nsp12 subunit and subsequent reconstitution of RTC using a new RNA scaffold (RNA3, Figure S4A). Incubation of the assembled RTC with the nsp13 helicase yielded a single peak corresponding to nsp131-RTC. (C) cryo-EM structure of the nsp13-bound RTC (PDB ID: 6xez) (Chen et al., 2020b).
Cryo-EM analyses showed a distribution of three major structural classes, namely nsp132-RTC (67%) shown in Figure 5C, nsp131-RTC (20%), and dimers of nsp132-RTC [(nsp132-RTC)2] (13%) (Chen et al., 2020b). In contrast, we only observed nsp131-RTC during nMS screening (Figure 5B), indicating that one of the bound nsp13 moieties likely dissociated at some point during the nMS analysis. Note, however, that from the structure of nsp132-RTC, one nsp13 helicase subunit (designated as nsp13.1) interacts more extensively with the three RdRp subunits and the substrate RNA and is more stably associated to RTC than the other bound helicase (nsp13.2), which only interacts with one nsp8 and nsp13.1 (Figure 5C). In addition, nsp12 and nsp13 are initially expressed as part of a single viral polyprotein before subsequent processing (de Groot et al., 2012) and are presumably present at equimolar concentrations. Overall, the structure of nsp13-RTC captured notable structural features and revealed possible roles for nsp13 in viral replication and transcription (Chen et al., 2020b).
DISCUSSION
We have described an nMS platform that provides critical feedback on sample quality and is especially useful for assaying samples prior to cryo-EM analyses. There are several advantages in integrating this nMS platform into cryo-EM workflows. First, the relatively high speed in nMS analysis enables rapid feedback. Each sample interrogation takes ~20 min from buffer exchange to production of a deconvolved spectrum (Figure 1). This throughput allows us to screen 15 – 20 different sample conditions (including replicates) per day, which has proved sufficient for our screening pipeline. Overall, the nMS timeline enables optimization and iterative screening on a scale of hours or days compared to iterations performed through cryo-EM analyses, which can take several weeks. Ongoing developments such as automated online buffer exchange coupled to nMS analysis, albeit performed at high flow rates (100 μL/min) with commensurately higher rates of sample consumption, point the way to further increases in sample throughput (VanAernum et al., 2020). Second, the sensitivity of the nMS workflow involves minimal sample consumption and analysis at concentration ranges that match the working concentrations and volumes used for cryo-EM analyses. These improvements in overall sensitivity are brought about by (1) ensuring minimal sample losses during buffer exchange, (2) electrospraying low sample volumes at nanoflow rates and (3) the exceptional desolvation efficiencies attained from analysis with the current generation of commercial nMS instrumentation (here, Thermo Exactive Plus EMR) (Hernandez and Robinson, 2007; Olinares et al., 2016; Olinares and Chait, 2020; Rose et al., 2012). Third, compared to other sizing and mass measurement techniques, nMS generates accurate and high-resolution mass measurements. Current commercially available nMS platforms (e.g., Orbitrap-based and Q-TOF analyzers) can generally resolve the charge-states of intact protein assemblies (up to 800 kDa), subcomplexes, and individual components as well as nucleic acids. In addition to instrument settings, the main factors that limit achieving high resolution nMS measurements originate from inefficient desolvation of the protein complexes and inherent microheterogeneities in the target protein species (Lossl et al., 2014). The typical mass accuracies observed with the nMS platform used here ranged from 0.003 – 0.03% for most samples (Tables S1 and S2) given the instrument resolution setting of 17,500 at m/z 200. The observed mass deviations for the RPc samples (DNA-containing transcription assemblies) were higher (~0.2%) mainly due to data acquisition at lower resolution setting (8,750 at m/z 200) as well as peak broadening from adduction in promoter DNA-bound complexes and incomplete desolvation. Overall, our high-resolution nMS measurements yielded accurate masses and revealed heterogeneities involving small mass modifications. For example, we captured the 305-Da mass difference due to cleavage of one terminal nucleotide from TECs containing pre-translocated RNA transcripts (Figures 3C and S3B). We confirmed the masses of the nucleic acid components prior to reconstituting the transcription complexes. We also verified that the nsp12 and nsp13 components of the SARS-CoV-2 nsp13-RTC contained the expected number of Zn2+ cofactors, consistent with what was observed in the previous cryo-EM structures (Table S2). In addition, the subunit stoichiometries, bound small molecules and post-translational modifications (Tables S1 and S2) extracted simultaneously from the same nMS data acquired during the screening have been extremely useful in downstream EM density map assignment and structure reconstruction.
The success of the nMS screening depends on the synergistic work and open communication between the analytical/mass spectrometry and structural biology researchers involved with the project. To ensure effective selection of test conditions for iterative screening, optimal nMS sample preparation and accurate interpretation of the resulting mass measurements, the following information should be provided for each sample: (1) accurate sequences of the protein and nucleic acid components including affinity tags and known modifications, (2) composition of the buffer used for sample storage and/or preparation, (3) sample concentration, (4) basic scheme of how the sample was purified and initial assessments of sample purity (e.g., SDS-PAGE gel), and (5) additional biochemical information such as bound cofactors, substrates and/or ligands.
On occasion, as in the SARS-Cov-2 replication-transcription complex example, the detailed correlation between the nMS and cryo-EM results is not perfect – likely because of the differential stabilities of complexes in these two physically different techniques. As previously noted, the typical composition of the buffers used for preparing samples for cryo-EM include salts, specific metal ions, reducing agents, detergents and other additives that are incompatible with nMS analysis as these nonvolatile components suppress electrospray ionization, increase chemical noise, and cause extensive adduct formation and peak broadening (Hernandez and Robinson, 2007). Although the relative ionic strength and pH are maintained as the samples are buffer exchanged into an nMS-compatible solution such as ammonium acetate, issues in protein complex stability and solubility can still arise in some cases. Potential instabilities in the protein assembly can also occur during the electrospray process and the transition from the solution phase into the gas phase during nMS analysis. However, overall, we have found an excellent correlation between observing assembled transcription complexes from the nMS screening and subsequently obtaining an analytically useful number of intact particles to produce high-resolution cryo-EM structures. As single particle cryo-EM is becoming an increasingly dominant structural technique, we anticipate routine application and integration of our platform in cryo-EM workflows, particularly for characterizing protein assemblies that are dynamic, conformationally heterogeneous and have transiently interacting components.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests on resources and datasets should be directed to the lead contact, Brian T. Chait (chait@rockefeller.edu).
Materials Availability
This study did not generate any new and unique reagents.
Data and Code Availability
The nMS RAW files generated in this work are available at http://dx.doi.org/10.17632/nbyz8j4j34.1.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
The DdRp core complex (α2ββ’ω), σ70, TraR, and Mfd are proteins from E. coli. Nun is a protein from HK022 virus. nsp7, nsp8, nsp12, and nsp13 are proteins from the SARS-CoV-2 virus.
METHOD DETAILS
Protein expression and purification
Transcription initiation complex with TraR-Eσ70
The E. coli DdRp was expressed and purified as previously described (Chen et al., 2019). A pET-based plasmid overexpressing each subunit of the DdRp (full-length α, β, ω) as well as β’-PPX-His10 (PPX; PreScission protease cleavage site, LEVLFQGP) was co-transformed with a pACYCDuet-1 plasmid containing E. coli rpoZ (encoding ω) into E. coli BL21(DE3) (Novagen). Protein expression was induced with 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) for 4 h at 30°C. Cells were harvested and lysed with a French Press (Avestin) at 4°C. Lysate was precipitated using polyethyleneimine [PEI, 10% (w/v), pH 8.0, Acros Organics]. Pellets were washed and DdRp was eluted. The PEI elutions were precipitated with ammonium sulfate. Pellets were harvested, resuspended and loaded on to HiTrap immobilized metal affinity chromatography (IMAC) HP column (GE Healthcare Life Sciences) for purification. Bound DdRp was washed on column, eluted and dialyzed. The dialyzed DdRp was loaded onto a Biorex-70 column (Bio-Rad) for purification by ion exchange (IEX) chromatography. Eluted DdRp was concentrated, then loaded onto a HiLoad 26/600 Superdex 200 column (GE Healthcare Life Sciences) for purification by size exclusion chromatography (SEC). The purified DdRp sample was supplemented with glycerol to a final concentration of 20% (v/v), flash frozen in liquid N2, and stored at −80°C.
E. coli σ70 and TraR were expressed and purified as previously described (Chen et al., 2019). The corresponding plasmids encoding for His10-SUMO-σ70 and His10-SUMO-TraR plasmid were transformed separately into E. coli BL21(DE3) (Novagen). Protein expression of His10-SUMO-σ70 was induced with 1 mM IPTG for 1 h at 30°C. Protein expression for His10-SUMO-TraR was induced with 1 mM IPTG for 3 h at 37°C. The subsequent purification protocol for each protein involved similar steps. Briefly, cells were harvested and lysed with a French Press (Avestin) at 4°C. Lysate was loaded onto a HiTrap IMAC HP column (GE Healthcare Life Sciences) for purification. Eluted proteins were cleaved with SUMO protease (Thermo Fisher Scientific) to remove the His10-SUMO-tag followed by dialysis. The cleaved sample was further purified on a HiTrap IMAC HP column (GE Healthcare Life Sciences). The fractions containing the tagless protein were collected from the flowthrough, concentrated and loaded onto a HiLoad 16/60 Superdex 200 for SEC. The purified σ70 sample was supplemented with glycerol to a final concentration of 20% (v/v), flash-frozen in liquid N2, and stored at −80° C. The purified TraR sample was concentrated by centrifugal filtration, flash-frozen in liquid N2, and stored at −80°C.
Nun and TEC
The ΔαCTD E. coli DdRp was expressed and purified as previously described (Twist et al., 2011). Briefly, the pVS10 plasmid harboring genes encoding for DdRp α-X234-241, β, β’, and ω subunits and pACYCDuet-1_Ec_rpoZ plasmid containing ω subunit gene were transformed into an engineered E.coli strain BL21(DE3)T-X234-241H. Recombinant protein expression was induced with 1 mM IPTG for 4 h at 30°C. Cells were harvested and lysed with a French Press (Avestin) at 4°C. The clarified cell lysate was precipitated using PEI. Pellets were washed and DdRp was eluted. The PEI elutions were then precipitated with ammonium sulfate. Pellets were harvested, resuspended and loaded on to HiTrap IMAC HP column (GE Healthcare Life Sciences) for purification. The eluted sample was dialyzed and incubated overnight with PreScission protease (GE Healthcare). Afterward, a subtractive IMAC chromatographic step removed uncleaved DdRp and the cleaved aCTD-(His)10 fusion protein. The flowthrough from the column that contained DdRp was dialysed and purified by loading onto a Bio-Rex 70 column (Bio-Rad). The pooled fractions were then applied onto a HiLoad 26/600 Superdex200 gel filtration column (GE Healthcare) for SEC. The peak fractions containing DdRp was supplemented with glycerol to a final concentration of 20% (v/v), flash-frozen in liquid N2, and stored at −80° C.
The full-length HK022 Nun protein was expressed and purified as previously described (Kang et al., 2017). The construct encoding for HK022 Nun was cloned into the pET21d vector (Novagen) without any tag and transformed into the E. coli BL21-AI (arabinose inducible) strain (Invitrogen). HK022 Nun was expressed via an autoinduction system due to its high toxicity to cells (Studier, 2005). The transformed cells were inoculated into non-inducing media and grown overnight. The overnight culture was added to inducing media and grown for 16 h at 37°C. Cells were harvested and lysed with a French Press (Avestin) at 4°C. The clarified lysate was then loaded onto a HiTrap SP column (GE Healthcare Life Sciences) and the eluted proteins were subsequently loaded onto a MonoS column (GE Healthcare Life Sciences). The resulting pooled fractions were then purified on a HiLoad Superdex75 column (GE Healthcare Life Sciences) by SEC. The peak fractions containing Nun was supplemented with glycerol to a final concentration of 20% (v/v), flash-frozen in liquid N2, and stored at −80° C.
Mfd-TEC
The E. coli DdRp (full-length subunits) used here was similar to the DdRp used in the structural determination of transcription initiation assemblies (see above) and was purified as previously described (Chen et al., 2019).
The E. coli Mfd was expressed and purified as previously described (Deaconescu and Darst, 2005). A pET28a-based plasmid overexpressing N-terminal His6-tagged E. coli Mfd was transformed into Rosetta(DE3)pLysS cells (Novagen). Protein expression was induced with 1 mM IPTG for 4 h at 30°C. Cells were harvested and lysed with a French Press (Avestin) at 4°C. The clarified lysate was loaded onto a HiTrap IMAC HP column (GE Healthcare Life Sciences) for purification. The elutions were dialyzed and loaded onto a HiTrap Heparin HP (GE Healthcare Life Sciences) column. The protein was eluted and further purified by SEC using a HiLoad 16/600 Superdex200 (GE Healthcare Life Sciences). The purified Mfd sample was supplemented with glycerol to 20% (v/v), flash frozen in liquid N2, and stored at −80°C.
SARS-CoV-2 RTC and nsp13 helicase
For the initial set of RTC purification experiments, the nsp12 and nsp7/8 proteins were co-expressed and purified (Chen et al., 2020b). The coding sequences for nsp12 and nsp7/8 were codon-optimized for E. coli expression. The pET28a-based plasmid encoding for His10-SUMO-nsp12 and the pCDFduet plasmid expressing His6-PPX-nsp7/8 were co-transformed into E. coli BL21 (DE3) cells (Novagen). Protein expression was induced with IPTG (0.1 mM final) and 10 μM ZnCl2 for 16 h at 16°C. Cells were harvested and lysed with a continuous-flow French Press (Avestin) at 4°C. The clarified lysate was loaded onto a HiTrap Heparin column (GE Healthcare Life Sciences). The eluted samples were then pooled and loaded onto a HisTrap HP column (GE Healthcare Life Sciences). The fractions containing the target proteins were dialyzed overnight in the presence of PPX to release the His6-tag from nsp7. After the dialysis step, the Ulp1 SUMO protease was added to cleave the His10-SUMO tag from nsp12. The sample was again passed through the HisTrap HP column and the flow-through was collected, concentrated, and loaded onto a Superdex 200 Hiload 16/600 (GE Healthcare Life Sciences) for purification by SEC. The fractions containing nsp12, nsp7, and nsp8 were pooled, supplemented with glycerol to 20% (v/v), flash frozen in liquid N2, and stored at −80°C.
nMS analysis of the RTC prepared from the initial procedure showed the presence of only truncated nsp12 and unassembled RTC indicating issues with expressing the full-length nsp12. For subsequent optimization of RTC purification (Chen et al., 2020b), nsp12 and nsp7/8 were purified separately and then reconstituted to form the RTC. Instead of using the E. coli codon-optimized sequence for nsp12, the SARS-CoV-2 RNA was used as the source and was obtained from the supernatant of propagated Vero E6 cells provided by B.R. tenOever (Blanco-Melo et al., 2020). The sequence encoding nsp12 was reverse transcribed into cDNA and the ORF1a/1b programmed ribosomal frameshift that naturally occurs during ORF1b translation was corrected to express the nsp12 open reading frame without any frameshift. The SARs-CoV-2 nsp12 coding sequence was subsequently cloned into a modified pRSFDuet-1 vector (Novagen) bearing an N-terminal His6-SUMO-tag.
For production and purification of the optimized nsp12, the pRSFDuet-1 plasmid expressing His6-SUMO-nsp12 was transformed into Eco BL21-CodonPlus(DE3)-RP (Agilent). Protein expression was induced with IPTG (0.1 mM final) for 16 h at 16°C. Cells were harvested and lysed with a continuous-flow French Press (Avestin) at 4°C. The clarified lysate was loaded onto a HisTrap HP column (GE Healthcare Life Sciences) and the eluted samples were dialyzed overnight in the presence of His6-Ulp1 SUMO protease to cleave the His6-SUMO tag. Cleaved nsp12 was again passed through the HisTrap HP column and the flow-through was collected, concentrated, and loaded onto a Superdex 200 Hiload 16/600 (GE Healthcare Life Sciences) for purification by SEC. The purified nsp12 sample was supplemented with glycerol to 20% (v/v), flash frozen in liquid N2, and stored at −80°C.
nsp7 and nsp 8 were expressed and purified as previously described (Chen et al., 2020b). For nsp7/nsp8 expression, the pCDFduet plasmid expressing His6-PPX-nsp7/8 was transformed into E. coli BL21 (DE3) cells. Protein expression was induced with IPTG (0.1 mM final) for 14 h at 16°C. Cells were harvested and lysed with a continuous-flow French Press (Avestin) at 4°C. The clarified lysate was loaded onto a HisTrap HP column (GE Healthcare Life Sciences) and the eluted samples were dialyzed overnight in the presence of His6-PPX to cleave the His6-tag. Cleaved nsp7/8 was passed through the HisTrap HP column and the flow-through was collected, concentrated, and loaded onto a Superdex 75 Hiload 16/600 (GE Healthcare Life Sciences) for SEC purification. The purified nsp7/8 sample was supplemented with glycerol to 20% (v/v), flash frozen in liquid N2, and stored at −80°C.
The nsp13 helicase was expressed and purified as previously described (Chen et al., 2020b). The pet28 plasmid expressing His6-PPX-nsp13 was transformed into E. coli Rosetta (DE3) (Novagen). Protein expression was induced with IPTG (0.2 mM final) for 17 h at 16°C. Cells were harvested and lysed with a continuous flow French Press (Avestin) at 4°C. The clarified lysate was loaded onto a HisTrap HP column (GE Healthcare Life Sciences) and the eluted samples were dialyzed overnight in the presence of His6-PPX to cleave the His6-tag. Cleaved nsp7/8 was passed through the HisTrap HP column and the flow-through was collected, concentrated, and loaded onto a Superdex 200 Hiload 16/600 (GE Healthcare Life Sciences) for SEC purification. The purified nsp13 sample was supplemented with glycerol to 20% (v/v), flash frozen in liquid N2, and stored at −80°C.
Sample preparation and screening conditions for nMS analysis
For nMS sample preparation, all buffer exchange steps were performed using Zeba microspin desalting columns (Thermo Fisher Scientific) with a 40-kDa MWCO. This ten-minute buffer exchange protocol involves storage buffer removal and four washing/equilibration steps—each step involving one-minute centrifuge spins at 4°C—followed by the actual two-minute buffer exchange (Olinares et al., 2016; Olinares and Chait, 2020).
As a general protocol in preparing the transcription initiation samples (Chen et al., 2020a), the DdRp holoenzyme (Eσ70) was assembled by incubating the DdRp core and σ70 (1:1.3 molar ratio) at RT for 10 min. TraR was then added at five-fold molar excess to an aliquot of pre-assembled Eσ70 and incubated at RT for 10 min. E and Eσ70 samples were concentrated using Amicon Ultra 0.5-mL centrifugal filters (EMD Millipore) with a 100 kDa molecular weight cutoff (MWCO) to remove unbound TraR. The samples were buffer-exchanged into 150 mM ammonium acetate, pH 7.5, 0.01% Tween-20. Promoter DNA constructs were desalted into HPLC-grade H2O. Prior to reconstitution, the concentrations of the protein complex post-buffer exchange and the DNA components were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific). To assemble the protein-DNA complexes, the promoter DNA was mixed at 1-1.3x ratio with the buffer-exchanged protein sample (typically at 3 μM) and incubated at RT for 10 min prior to nMS characterization. The ammonium acetate concentration was also varied from 75 – 300 mM to test the effect of ionic strength on sample stability.
Additional screening conditions were tested with the TraR-Eσ70 and rrnB P1 promoter DNA including addition of another transcription factor FIS and glutaraldehyde (GA) stabilization. The pre-assembled Eσ70 was mixed with five-fold excess TraR, followed by an equivalent amount of promoter DNA, and then FIS protein (1:1:1 Eσ70:DNA:FIS) with each step involving a 10-min incubation at RT. The resulting sample was buffer-exchanged into 150 mM ammonium acetate, pH 7.5, 0.01% Tween-20 for nMS analysis. For GA treatment, Eσ70, TraR and promoter DNA were mixed as above. Then, the sample was incubated with 0.1 mM (0.001%) or 0.2 mM GA (0.0025%) for 10 min at RT and quenched with 100 mM Tris pH 8 prior to buffer exchange into 150 mM ammonium acetate, pH 7.5, 0.01% Tween-20 and subsequent nMS analysis.
For the Nun/TEC (Kang et al., 2017) and Mfd/TEC (Kang et al., 2020) samples, the TEC was assembled by mixing the DdRp core complex with the annealed template DNA:RNA hybrid at a molar ratio of 1:1.3 and incubated for 15 min at RT. Non-template DNA was then added and incubated for 10 min. To form Nun-TECs, Nun was added to the TEC sample at 3- to 3.5-fold molar excess. To reconstitute Mfd-TEC assemblies, 7 μM TEC was incubated with 3.5 μM Mfd (2:1 molar ratio) in buffer containing 20 mM Tris, 150 mM KCl, 5 mM DTT, 10 mM MgCl2, and 2 mM ATP for 1 min at RT. The resulting samples were immediately buffer-exchanged into 500 mM ammonium acetate, pH 7.5, 0.01% Tween-20 before nMS analysis.
For the replication-transcription assemblies from SARS-CoV-2 (Chen et al., 2020b), the initial set of RTC purification involved co-expression of the nsp12 and nsp7/8 subunits that did not require further reconstitution after purification. For the next set of experiments, the optimized nsp12 (codon sequence obtained from RT-PCR of SARS-CoV-2 RNA from propagated Vero E6 cells) and nsp7/8 proteins were purified separately and then reconstituted to assemble the RTC. The purified nsp12 and nsp7/8 were concentrated by centrifugal filtration (Amicon), mixed in a 1:3 molar ratio and dialyzed into 20 mM HEPES-NaOH pH 8.0, 300 mM NaCl, 10 mM MgCl2, 2 mM DTT for 20 min at RT. The annealed RNA scaffold was added to the dialyzed nsp7/8/12 mixture and incubated for 15 min at RT. The sample was buffer exchanged into S6 buffer (20 mM HEPES-NaOH, pH 8.0, 150 mM K-acetate, 10 mM MgCl2, 2 mM DTT) using Zeba spin desalting columns. After buffer exchange, the sample was further incubated for 20 min at 30°C and then purified over a Superose 6 Increase 10/300 GL column (GE Healthcare Life Sciences) in S6 buffer. The peak corresponding to the RTC was pooled and concentrated by centrifugal filtration (Amicon). The reconstituted RNA-bound holo-RdRp (RTC) and the purified nsp13 were buffer exchanged separately into 150 mM ammonium acetate, pH 7.5, 0.01% Tween-20 using Zeba microspin desalting columns with a 40-kDa MWCO (Thermo Scientific). The buffer-exchanged samples were mixed yielding a final concentration of 4 μM RTC and 5 μM nsp13, and then incubated for 5 min at RT prior to nMS characterization.
nMS analysis
To perform the nMS analysis for the reconstituted transcription assemblies, 2 – 3 μL of the sample was loaded into a gold-coated quartz emitter that was prepared in-house and then electrosprayed into an Exactive Plus EMR instrument (Thermo Fisher Scientific) with a modified static nanospray source—see (Olinares and Chait, 2020) for details. For the modified source, we removed the plastic dome on the Nanoflex Ion Source that houses the emitter holder to enable easy access to the emitter tip during nMS analysis and replaced the syringe pump with a hand vacuum pump to apply backpressure during electrospray. Making the emitter more accessible facilitated efficient emitter tip opening (by lightly touching it with a fine gel-loading tip) to initiate the electrospray while the high voltage is already turned on, and streamlined troubleshooting of the spray when there is suspected buildup or clogging (Olinares and Chait, 2020).
The following set of nMS parameters worked well for all the protein complexes (mass > 200 kDa) analyzed in this study. These parameters can be set and saved as an MS tune file that can be automatically loaded when performing the nMS analysis. The parameters include: spray voltage, 1.2 – 1.3 kV; capillary temperature, 125 – 150 °C; in-source dissociation (ISD), 10 V; S-lens RF level, 200; instrument resolution setting, 8,750 (for the transcription initiation samples) or 17,500 (all the other samples) at m/z of 200; AGC target, 1 x 106; maximum injection time, 200 ms; number of microscans, 5; injection flatapole, 8 V; interflatapole, 4 V; bent flatapole, 4 V; high energy collision dissociation (HCD), 180 – 200 V; ultrahigh vacuum (UHV) pressure, 6 – 7 × 10−10 mbar; and total number of scans, at least 100. Mass calibration in positive EMR mode was performed using cesium iodide.
To analyze component proteins, subcomplexes or protein assemblies (mass < 200 kDa), we generally change the transmission parameters to injection flatapole, 8 V; interflatapole, 7 V; bent flatapole, 5 – 6 V. The following activation parameters can be varied accordingly: ISD voltage (0 – 10 V), HCD voltage (50 – 150 V) and trapping gas pressure in the collision cell (UHV pressure: 3 – 7 × 10−10 mbar).
For characterization of nucleic acids, the samples were desalted or buffer-exchanged into 500 mM ammonium acetate, pH 7.5, 0.01% Tween-20. Samples were then diluted to 1 – 5 μM into the same ammonium acetate solution before nMS analysis. The nMS parameters include: spray voltage, 1.2 – 1.3 kV; capillary temperature, 150 °C; ISD, 10 V; S-lens RF level, 200; instrument resolution setting, 17,500 at m/z of 200; AGC target, 1 x 106; maximum injection time, 200 ms; number of microscans, 5; injection flatapole, 8 V; interflatapole, 7 V; bent flatapole, 5 – 6 V; HCD, 60 – 90 V; UHV pressure, 6 – 7 × 10−10 mbar; and total number of scans, at least 100.
Data Processing
For data processing, the acquired nMS spectra were visualized using Thermo Xcalibur Qual Browser (versions 3.0.63 and 4.2.47). MS spectra deconvolution was performed either manually or using the UniDec software versions 3.2 and 4.1 (Marty et al., 2015; Reid et al., 2019). The general UniDec settings used include: No background subtraction, charge range of 1 – 60, sample mass every 1 Da and smooth charge state distributions setting ON. To focus on the relevant assemblies, deconvolution was performed for peaks within specific m/z ranges: 9,500 – 14,000 Th for the transcription initiation assemblies; 8,000 – 12,000 Th for the Nun-TEC assemblies; 6,000 – 12,000 Th for the Mfd-TEC assemblies; and 2,000 – 12,000 Th for the viral RdRp samples. The deconvolved spectra obtained from UniDec were re-plotted using the m/z software (Proteometrics LLC) or Adobe Illustrator v.24.0.1. Experimental masses obtained by manual computation were reported in Tables S1 and S2 as the average mass ± standard deviation (SD) across all the calculated mass values within each observed charge state series (n ≥ 4, where n is the number of charge-state peaks per series). Mass accuracies were calculated as the % difference between the measured and expected masses relative to the expected mass.
Structure Visualization
The relevant cryo-EM structures were obtained from the Protein Databank (https://www.rcsb.org/). The structures were visualized and presented using the PyMOL Molecular Graphics System v.2.4.0 (Schrodinger, LLC).
QUANTIFICATION AND STATISTICAL ANALYSIS
nMS spectra deconvolution was performed either manually or using the UniDec software versions 3.2 and 4.1 (Marty et al., 2015; Reid et al., 2019). The experimental masses obtained by manual computation were reported in Tables S1 and S2 as the average mass ± standard deviation (SD) across all the calculated mass values within each charge-state series (n ≥ 4, where n is the number of charge-state peaks per series). Mass accuracies were calculated as the % difference between the measured and expected masses relative to the expected mass.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli BL21(DE3) | Novagen/EMD Millipore | N/A |
| E. coli BL21(DE3)T-X234-241H | Twist et al., 2011 | N/A |
| E. coli BL2-AI | Invitrogen | Cat# C6070-03 |
| E. coli Rosetta (DE3) pLysS | Novagen | N/A |
| E. coli BL21-CodonPlus | Agilent | Cat# 230280 |
| E. coli Rosetta (DE3) | Novagen | Cat# 70954-3 |
| SARS-CoV-2 infected Vero E6 cells | Blanco-Melo et al., 2020 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ammonium Acetate, TraceSELECT | Sigma-Aldrich | Cat#373432-100G-F; CAS: 631-61-8 |
| Tween-20 Surfact-Amps (20% v/v) Detergent Solution | Thermo Scientific | Cat#28320; CAS: 9005-64-5 |
| Polyethyleneimine | Fisher Scientific | Cat#AC178572500; CAS: 9002-98-6 |
| Glutaraldehyde (50% v/v) solution | Sigma Aldrich | Cat#G7651-10ML; CAS: 111-30-8 |
| E. coli DdRp core (full-length subunits) | Chen et al., 2019; Kang et al., 2020 | N/A |
| E. coli σ70 | Chen et al., 2019 | N/A |
| E. coli TraR | Chen et al., 2019 | N/A |
| E. coli DdRp core (ΔCTD for α subunit) | Twist et al., 2011 | N/A |
| HK022 Nun | Kang et al., 2017 | N/A |
| E. coli Mfd | Deaconescu and Darst, 2005; Kang et al., 2020 | N/A |
| SARS-CoV-2 nsp12 | Chen et al., 2020b | N/A |
| SARS-CoV-2 nsp7/8 | Chen et al., 2020b | N/A |
| SARS-CoV-2 nsp13 | Chen et al., 2020b | N/A |
| Deposited Data | ||
| native MS RAW files | This paper; Mendeley Data | http://dx.doi.org/10.17632/nbyz8j4j34.1 |
| Oligonucleotides | ||
| The promoters used for the screening of transcription initiation complexes (DNA sequences shown in Figure S1A) | Integrated DNA Technologies | N/A |
| Template DNA for TEC/Nun assembly: 5’-GGGTATTCGCCGTGTACCTCTCC-3’ |
Integrated DNA Technologies | N/A |
| Non-template DNA for TEC/Nun assembly: 5’-GGGCTACCTCTCCATGACGGCGAATACCC-3’ |
Integrated DNA Technologies | N/A |
| Pre-translocated RNA for TEC/Nun assembly: 5’-GGAGAGGUAC-3’ |
Dharmacon | N/A |
| Post-translocated RNA for TEC/Nun assembly: 5’-GGAGAGGUA-3’ |
Dharmacon | N/A |
| The template and non-template DNA constructs used for the screening of Mfd-TEC (DNA sequences shown in Figure S2A) | Integrated DNA Technologies | N/A |
| The RNA transcripts used for the screening of Mfd-TEC (RNA sequences shown in Figure S2A) | Dharmacon | N/A |
| The RNA constructs used for the nsp13-RTC screening (Figure S4A) | Horizon Discovery Ltd./Dharmacon | N/A |
| Recombinant DNA | ||
| pACYCDuet-1_Ec_rpoZ | Twist et al., 2011 | N/A |
| pEcrpoABC(-XH)Z | Twist et al., 2011 | N/A |
| pET28a | EMD Millipore | N/A |
| pET28a-His10-SUMO rpoD | Chen et. al., 2017 | N/A |
| pET28a-His10-SUMO traR | Chen et al., 2019 | pRLG15142 |
| pVS10 plasmid harboring genes encoding for E.coli RNAP α-X234-241, β, β’, and ω | Twist et al., 2011 | N/A |
| pET21d plasmid harboring HK022 Nun | Kang et al., 2017 | N/A |
| pET21d | Novagen | N/A |
| pAD6 (pET28a plasmid harboring Mfd) | Deaconescu et. al., 2005 | N/A |
| pET28a-His10-SUMO nsp12 | Chen et al., 2020b | N/A |
| pCDFDuet-1 | Novagen | Cat#71340-3 |
| pCDFDuet-1-His6-SARS-CoV-2-nsp7_nsp8 | Chen et al., 2020b | N/A |
| pRSFDuet-1 | Novagen | Cat#71341-3 |
| pRSFDuet-1-His6-SUMO-SARS-CoV-2-nsp12 | Chen et al., 2020b | N/A |
| pET28-His6-SARS-CoV-2-nsp13 | GenScript | N/A |
| Software and Algorithms | ||
| UniDec version 3.2 & 4.1 | Marty et al., 2015; Reid et al., 2019 | https://github.com/michaelmarty/UniDec/releases |
| Qual Browser Thermo Xcalibur version 3.0.63 & 4.2.47 | Thermo Fisher Scientific Inc. | Thermo Scientific MS instruments |
| m/z- Knexus edition | Proteometrics, LLC | N/A |
| PyMOL Molecular Graphics System | Schrödinger, LLC | http://www.pymol.org |
| Other | ||
| cOmplete Protease Inhibitor Cocktail | Roche | Cat#11697498001 |
| Bio-Rex 70 cation exchange resin, analytical grade, 100-200 mesh | Bio-Rad | Cat#1425842 |
| HiLoad 26/600 Superdex 200 pg | GE Healthcare Life Sciences | Cat#28989336 |
| HiTrap IMAC HP | GE Healthcare Life Sciences | Cat#17092003 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare Life Sciences | Cat#GE28-9893-3 |
| HiLoad 16/600 Superdex 75 pg | GE Healthcare Life Sciences | Cat#GE28-9893-33 |
| HisTrap HP | GE Healthcare Life Sciences | Cat#GE17-5248-02 |
| HiTrap Heparin HP | GE Healthcare Life Sciences | Cat#GE17-0407-03 |
| HiTrapSP | GE Healthcare Life Sciences | Cat#17115201 |
| MonoS 10/100 GL | GE Healthcare Life Sciences | Cat#17516901 |
| Superose 6 Increase 10/300 GL column | GE Healthcare Life Sciences | Cat#GE29-0915-96 |
| Zeba Micro Spin Desalting Columns, 40K MWCO | Thermo Pierce | Cat#87765 |
| Amicon Ultra-0.5 Centrifugal Filter Unit with Ultracel-100 membrane, 100 kDa MWCO | EMD Millipore | Cat#UFC510096 |
| Quartz tubing without filament: 1.0mm O.D., 0.70mm I.D., 10cm length | Sutter | Cat#Q100-70-10 |
HIGHLIGHTS.
A native MS-based diagnostic and screening platform for cryo-EM samples
Provides rapid assessment of sample stability and homogeneity
Enables iterative biochemical screening for optimizing sample conditions for cryo-EM
Applied to structural studies of transcription complexes from bacteria and SARS-CoV-2
Acknowledgements.
We are grateful to the members of the Chait and the Darst/Campbell labs for helpful discussions. We also thank Michael Marty from the University of Arizona for the development of the freely available UniDec software. This work is supported by the Pels Foundation to The Rockefeller University, NIH grants P41 GM109824 and P41 GM103314 to BTC, R35 GM118130 to SAD, and R01 GM114450 to EAC. Access to the cryo-EM microscopes and support was through The Rockefeller University Evelyn Gruss Lipper Cryo-EM Resource Center and at The Simons Electron Microscopy Center (SEMC), National Resource for Automated Molecular Microscopy (NRAMM), and National Center for CryoEM Access and Training (NCCAT) at the NYSBC, supported by NIH NIGMS (P41 GM103310), NYSTAR, the Simons Foundation (SF349247), the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24 GM129539) and NY State Assembly Majority.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests. The authors declare no competing interests.
REFERENCES
- Abbas YM, Wu D, Bueler SA, Robinson CV, and Rubinstein JL (2020). Structure of V-ATPase from the mammalian brain. Science 367, 1240–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adedeji AO, Marchand B, Te Velthuis AJ, Snijder EJ, Weiss S, Eoff RL, Singh K, and Sarafianos SG (2012). Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One 7, e36521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, et al. (2018). Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 9, e00221–00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B (1998). The cell as a collection of protein machines: Preparing the next generation of molecular biologists. Cell 92, 291–294. [DOI] [PubMed] [Google Scholar]
- Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, and Darst SA (2015). Structure of a bacterial RNA polymerase holoenzyme open promoter complex. Elife 4, e08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, Jordan TX, Oishi K, Panis M, Sachs D, et al. (2020). Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 181, 1036–1045 e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockrath R, Barlow A, and Engstrom J (1987). Mutation frequency decline in Escherichia coli B/r after mutagenesis with ethyl methanesulfonate. Mutation Research/DNA Repair Reports 183, 241–247. [DOI] [PubMed] [Google Scholar]
- Boivin S, Kozak S, and Meijers R (2013). Optimization of protein purification and characterization using Thermofluor screens. Protein Expr Purif 91, 192–206. [DOI] [PubMed] [Google Scholar]
- Bokal AJ, Ross W, Gaal T, Johnson RC, and Gourse RL (1997). Molecular anatomy of a transcription activation patch: FIS-RNA polymerase interactions at the Escherichia coli rrnB P1 promoter. EMBO J 16, 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E (2015). The revolution will not be crystallized: a new method sweeps through structural biology. Nature 525, 172–174. [DOI] [PubMed] [Google Scholar]
- Casanal A, Kumar A, Hill CH, Easter AD, Emsley P, Degliesposti G, Gordiyenko Y, Santhanam B, Wolf J, Wiederhold K, et al. (2017). Architecture of eukaryotic mRNA 3'-end processing machinery. Science 358, 1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A, Haselbach D, Kirves JM, Ohmer J, Paknia E, Fischer N, Ganichkin O, Moller V, Frye JJ, Petzold G, et al. (2015). ProteoPlex: stability optimization of macromolecular complexes by sparse-matrix screening of chemical space. Nat Methods 12, 859–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase J, Catalano A, Noble AJ, Eng ET, Olinares PD, Molloy K, Pakotiprapha D, Samuels M, Chait B, des Georges A, et al. (2018). Mechanisms of opening and closing of the bacterial replicative helicase. Elife 7, e41140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiu C, Gopalkrishnan S, Chen AY, Olinares PDB, Saecker RM, Winkelman JT, Maloney MF, Chait BT, Ross W, et al. (2020a). Stepwise Promoter Melting by Bacterial RNA Polymerase. Mol Cell 78, 275–288 e276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Gopalkrishnan S, Chiu C, Chen AY, Campbell EA, Gourse RL, Ross W, and Darst SA (2019). E. coli TraR allosterically regulates transcription initiation by altering RNA polymerase conformation. Elife 8, e49375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Malone B, Llewellyn E, Grasso M, Shelton PMM, Olinares PDB, Maruthi K, Eng ET, Vatandaslar H, Chait B, et al. (2020b). Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell, 30941–30947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y (2018). Single-particle cryo-EM-How did it get here and where will it go. Science 361, 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Grigorieff N, Penczek PA, and Walz T (2015). A primer to single-particle cryo-electron microscopy. Cell 161, 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TRD, Ignatiou A, and Orlova EV (2017). Structural Analysis of Protein Complexes by Cryo Electron Microscopy. Methods Mol Biol 1615, 377–413. [DOI] [PubMed] [Google Scholar]
- de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJM, Talbot PJ, et al. (2012). Family Coronaviridae In Virus taxonomy Ninth report of the international committee on taxonomy of viruses, King AMQ, Adams MJ, Carstens EB, and Lefkowitz EJ, eds. (Amsterdam: Elsevier Academic Press; ), pp. 806–828. [Google Scholar]
- Deaconescu AM, and Darst SA (2005). Crystallization and preliminary structure determination of Escherichia coli Mfd, the transcription-repair coupling factor. Acta Crystallogr Sect F Struct Biol Cryst Commun 61, 1062–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklistov A, Sharon BD, Darst SA, and Gross CA (2014). Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 68, 357–376. [DOI] [PubMed] [Google Scholar]
- Frank J (2002). Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Biomol Struct 31, 303–319. [DOI] [PubMed] [Google Scholar]
- Frost LS, Ippen-Ihler K, and Skurray RA (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol Rev 58, 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, and Cramer P (2010). RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell 39, 583–594. [DOI] [PubMed] [Google Scholar]
- Gopalkrishnan S, Ross W, Chen AY, and Gourse RL (2017). TraR directly regulates transcription initiation by mimicking the combined effects of the global regulators DksA and ppGpp. Proc Natl Acad Sci U S A 114, E5539–E5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Chen AY, Gopalkrishnan S, Sanchez-Vazquez P, Myers A, and Ross W (2018). Transcriptional Responses to ppGpp and DksA. Annu Rev Microbiol 72, 163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, and Gross CA (2003). Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol 57, 441–466. [DOI] [PubMed] [Google Scholar]
- Hand E (2020). ‘We need a people’s cryo-EM.’ Scientists hope to bring revolutionary microscope to the masses. Science 367, 354–358.31974231 [Google Scholar]
- Heck AJ (2008). Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods 5, 927–933. [DOI] [PubMed] [Google Scholar]
- Hernandez H, and Robinson CV (2007). Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc 2, 715–726. [DOI] [PubMed] [Google Scholar]
- Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, and Cramer P (2020). Structure of replicating SARS-CoV-2 polymerase. Nature 584, 154–156. [DOI] [PubMed] [Google Scholar]
- Hung SC, and Gottesman ME (1995). Phage HK022 Nun protein arrests transcription on phage lambda DNA in vitro and competes with the phage lambda N antitermination protein. J Mol Biol 247, 428–442. [DOI] [PubMed] [Google Scholar]
- Ivanov KA, and Ziebuhr J (2004). Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5'-triphosphatase activities. J Virol 78, 7833–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Yan L, Ren Z, Wu L, Wang J, Guo J, Zheng L, Ming Z, Zhang L, Lou Z, et al. (2019). Delicate structural coordination of the Severe Acute Respiratory Syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res 47, 6538–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Llewellyn E, Chen J, Olinares PDB, Brewer J, Chait BT, Campbell EA, and Darst SA (2020). Structural basis for transcription complex disruption by the Mfd translocase. bioRxiv, 10.1101/2020.1108.1112.248500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JY, Olinares PD, Chen J, Campbell EA, Mustaev A, Chait BT, Gottesman ME, and Darst SA (2017). Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. Elife 6, e25478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, and Ward AB (2019). Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 10, 2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Kireeva ML, Becker J, Sidorenkov I, and Kashlev M (2003). Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol 371, 233–251. [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W (2014). Biochemistry. The resolution revolution. Science 343, 1443–1444. [DOI] [PubMed] [Google Scholar]
- Kuhlen L, Abrusci P, Johnson S, Gault J, Deme J, Caesar J, Dietsche T, Mebrhatu MT, Ganief T, Macek B, et al. (2018). Structure of the core of the type III secretion system export apparatus. Nat Struct Mol Biol 25, 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NR, Kwon HM, Park K, Oh S, Jeong YJ, and Kim DE (2010). Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsP13. Nucleic Acids Res 38, 7626–7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leney AC, and Heck AJ (2017). Native Mass Spectrometry: What is in the Name? J Am Soc Mass Spectrom 28, 5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liko I, Allison TM, Hopper JT, and Robinson CV (2016). Mass spectrometry guided structural biology. Curr Opin Struct Biol 40, 136–144. [DOI] [PubMed] [Google Scholar]
- Loo JA (1997). Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrometry Reviews 16, 1–23. [DOI] [PubMed] [Google Scholar]
- Lorenzen K, Vannini A, Cramer P, and Heck AJ (2007). Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure 15, 1237–1245. [DOI] [PubMed] [Google Scholar]
- Lossl P, Snijder J, and Heck AJ (2014). Boundaries of mass resolution in native mass spectrometry. J Am Soc Mass Spectrom 25, 906–917. [DOI] [PubMed] [Google Scholar]
- Lyumkis D (2019). Challenges and opportunities in cryo-EM single-particle analysis. J Biol Chem 294, 5181–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneewannakul K, and Ippen-Ihler K (1993). Construction and analysis of F plasmid traR, trbJ, and trbH mutants. J Bacteriol 175, 1528–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rucobo FW, Kohler R, van de Waterbeemd M, Heck AJ, Hemann M, Herzog F, Stark H, and Cramer P (2015). Molecular Basis of Transcription-Coupled Pre-mRNA Capping. Mol Cell 58, 1079–1089. [DOI] [PubMed] [Google Scholar]
- Marty MT, Baldwin AJ, Marklund EG, Hochberg GK, Benesch JL, and Robinson CV (2015). Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal Chem 87, 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, and Wolf M (2018). Cryo-electron microscopy for structural analysis of dynamic biological macromolecules. Biochim Biophys Acta Gen Subj 1862, 324–334. [DOI] [PubMed] [Google Scholar]
- Nogales E, and Scheres SH (2015). Cryo-EM: A Unique Tool for the Visualization of Macromolecular Complexity. Mol Cell 58, 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ognjenovic J, Grisshammer R, and Subramaniam S (2019). Frontiers in Cryo Electron Microscopy of Complex Macromolecular Assemblies. Annu Rev Biomed Eng 21, 395–415. [DOI] [PubMed] [Google Scholar]
- Olinares PD, Dunn AD, Padovan JC, Fernandez-Martinez J, Rout MP, and Chait BT (2016). A Robust Workflow for Native Mass Spectrometric Analysis of Affinity-Isolated Endogenous Protein Assemblies. Anal Chem 88, 2799–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PDB, and Chait BT (2020). Native Mass Spectrometry Analysis of Affinity-Captured Endogenous Yeast RNA Exosome Complexes. Methods Mol Biol 2062, 357–382. [DOI] [PubMed] [Google Scholar]
- Park JS, Marr MT, and Roberts JW (2002). E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109, 757–767. [DOI] [PubMed] [Google Scholar]
- Park JS, and Roberts JW (2006). Role of DNA bubble rewinding in enzymatic transcription termination. Proc Natl Acad Sci U S A 103, 4870–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, and Russo CJ (2016). Specimen Preparation for High-Resolution Cryo-EM. Methods Enzymol 579, 51–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DJ, Diesing JM, Miller MA, Perry SM, Wales JA, Montfort WR, and Marty MT (2019). MetaUniDec: High-Throughput Deconvolution of Native Mass Spectra. J Am Soc Mass Spectrom 30, 118–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J, Sloan SB, Weisberg RA, Gottesman ME, Robledo R, and Harbrecht D (1987). The remarkable specificity of a new transcription termination factor suggests that the mechanisms of termination and antitermination are similar. Cell 51, 483–492. [DOI] [PubMed] [Google Scholar]
- Robledo R, Atkinson BL, and Gottesman ME (1991). Escherichia coli mutations that block transcription termination by phage HK022 Nun protein. Journal of Molecular Biology 220, 613–619. [DOI] [PubMed] [Google Scholar]
- Rose RJ, Damoc E, Denisov E, Makarov A, and Heck AJ (2012). High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods 9, 1084–1086. [DOI] [PubMed] [Google Scholar]
- Ruff EF, Record MT Jr., and Artsimovitch I (2015). Initial events in bacterial transcription initiation. Biomolecules 5, 1035–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford ST, Villers CL, Lee JH, Ross W, and Gourse RL (2009). Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev 23, 236–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saecker RM, Record MT Jr., and Dehaseth PL (2011). Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412, 754–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby CP, and Sancar A (1993). Molecular mechanism of transcription-repair coupling. Science 260, 53–58. [DOI] [PubMed] [Google Scholar]
- Selby CP, Witkin EM, and Sancar A (1991). Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc Natl Acad Sci U S A 88, 11574–11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A, Hegyi A, Siddell SG, and Ziebuhr J (2000a). The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5'-to-3' polarity. RNA 6, 1056–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A, van Dinten LC, Snijder EJ, and Ziebuhr J (2000b). Biochemical characterization of the equine arteritis virus helicase suggests a close functional relationship between arterivirus and coronavirus helicases. J Virol 74, 9586–9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Decroly E, and Ziebuhr J (2016). The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv Virus Res 96, 59–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J, Schuller JM, Wiegard A, Lossl P, Schmelling N, Axmann IM, Plitzko JM, Forster F, and Heck AJ (2017). Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science 355, 1181–1184. [DOI] [PubMed] [Google Scholar]
- Sola I, Almazan F, Zuniga S, and Enjuanes L (2015). Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu Rev Virol 2, 265–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonn-Segev A, Belacic K, Bodrug T, Young G, VanderLinden RT, Schulman BA, Schimpf J, Friedrich T, Dip PV, Schwartz TU, et al. (2020). Quantifying the heterogeneity of macromolecular machines by mass photometry. Nat Commun 11, 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW (2005). Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41, 207–234. [DOI] [PubMed] [Google Scholar]
- Subissi L, Posthuma CC, Collet A, Zevenhoven-Dobbe JC, Gorbalenya AE, Decroly E, Snijder EJ, Canard B, and Imbert I (2014). One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A 111, E3900–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa Y, Binshtein E, Erwin AL, Pyburn TM, Mittendorf KF, and Ohi MD (2017). While the revolution will not be crystallized, biochemistry reigns supreme. Protein Sci 26, 69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JA, Watt RM, Chai YB, Lu LY, Lin MC, Peiris JS, Poon LL, Kung HF, and Huang JD (2003). The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases. J Biol Chem 278, 39578–39582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twist KA, Husnain SI, Franke JD, Jain D, Campbell EA, Nickels BE, Thomas MS, Darst SA, and Westblade LF (2011). A novel method for the production of in vivo-assembled, recombinant Escherichia coli RNA polymerase lacking the alpha C-terminal domain. Protein Sci 20, 986–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanAernum ZL, Busch F, Jones BJ, Jia M, Chen Z, Boyken SE, Sahasrabuddhe A, Baker D, and Wysocki VH (2020). Rapid online buffer exchange for screening of proteins, protein complexes and cell lysates by native mass spectrometry. Nat Protoc 15, 1132–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello CL, Kireeva ML, Lubkowska L, Kashlev M, and Gottesman M (2014). Coliphage HK022 Nun protein inhibits RNA polymerase translocation. Proc Natl Acad Sci U S A 111, E2368–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, Ebright RH, and Nickels BE (2014). Interactions between RNA polymerase and the "core recognition element" counteract pausing. Science 344, 1285–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wu J, Wang H, Gao Y, Liu Q, Mu A, Ji W, Yan L, Zhu Y, Zhu C, et al. (2020). Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 182, 417–428 e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin EM (1966). Radiation-Induced Mutations and Their Repair. Science 152, 1345–1353. [DOI] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. (2020). A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, and Lander GC (2020). Present and Emerging Methodologies in Cryo-EM Single-Particle Analysis. Biophys J S0006-3495, 30677–30679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Mao C, Luan X, Shen DD, Shen Q, Su H, Wang X, Zhou F, Zhao W, Gao M, et al. (2020). Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 368, 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G, Hundt N, Cole D, Fineberg A, Andrecka J, Tyler A, Olerinyova A, Ansari A, Marklund EG, Collier MP, et al. (2018). Quantitative mass imaging of single biological macromolecules. Science 360, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, and Ebright RH (2012). Structural basis of transcription initiation. Science 338, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Y, and Steitz TA (2015). Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol Cell 58, 534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nMS RAW files generated in this work are available at http://dx.doi.org/10.17632/nbyz8j4j34.1.