SUMMARY
Mammalian cells respond to insufficient oxygen through transcriptional regulators called hypoxia-inducible factors (HIFs). While transiently protective, prolonged HIF activity drives distinct pathological responses in different tissues. Using a model of chronic HIF1a accumulation in pluripotent stem cell-derived oligodendrocyte progenitors (OPCs), we demonstrate that HIF1a activates non-canonical targets to impair generation of oligodendrocytes from OPCs. HIF1a activated a unique set of genes in OPCs through interaction with the OPC-specific transcription factor OLIG2. Non-canonical targets, including Ascl2 and Dlx3, were sufficient to block differentiation through suppression of the oligodendrocyte regulator Sox10. Chemical screening revealed that inhibition of MEK/ERK signaling overcame the HIF1a-mediated block in oligodendrocyte generation by restoring Sox10 expression without impacting canonical HIF1a activity. MEK/ERK inhibition also drove oligodendrocyte formation in hypoxic regions of human oligocortical spheroids. This work defines mechanisms by which HIF1a impairs oligodendrocyte formation and establishes that cell-type-specific HIF1a targets perturb cell function in response to low oxygen.
Graphical Abstract
eTOC
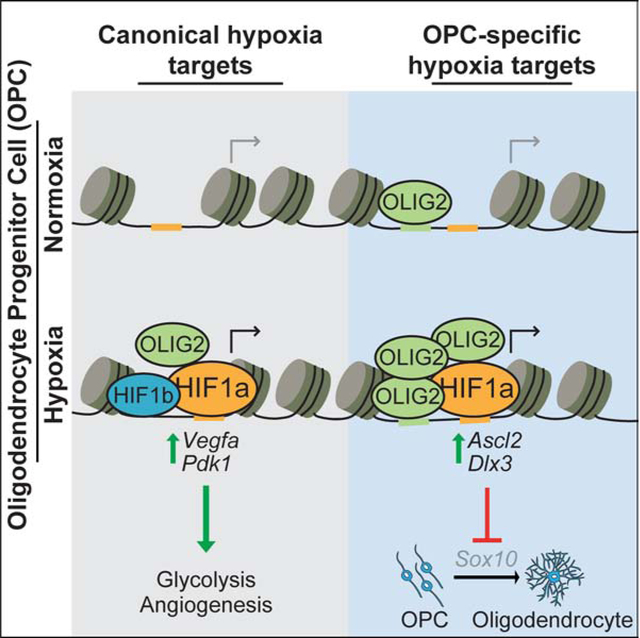
Hypoxia impairs the generation of oligodendrocytes and myelin in various neurological disorders. Allan and colleagues map the genome-wide binding profile of HIF1a in oligodendrocyte progenitor cells (OPCs). HIF1a activated non-canonical, cell-type-specific target genes in OPCs that converge to suppress Sox10 expression and block oligodendrocyte formation.
INTRODUCTION
The ability to sense and respond to fluctuations in oxygen levels is required to maintain homeostasis in every cell in the body (Kaelin and Ratcliffe, 2008; Semenza, 2012). Insufficient concentrations of molecular oxygen rapidly trigger an evolutionary conserved transcriptional response that enables cell survival in low oxygen by promoting anaerobic metabolism for energy production as well as angiogenesis and erythropoiesis to increase access to local oxygen. While this is initially protective, prolonged activation of this response leads to cellular dysfunction and disease in many tissues. For example, the response to chronic hypoxia blocks white matter formation in premature birth (Scafidi et al., 2014; Volpe, 2009; Volpe et al., 2011), promotes inflammation and insulin resistance in obesity (Lee et al., 2014), and impairs hematopoietic stem cell transplantation capacity (Takubo et al., 2010). This cellular dysfunction has largely been attributed to prolonged activation of the canonical response to low oxygen shared across all cell types; however, it is difficult to explain how activation of a conserved set of hypoxia signature genes can lead to such diverse cellular phenotypes. An alternative unexplored possibility is that cell-type-specific differences in chromatin landscape enable access to unique non-canonical targets, which could account for tissue-specific pathologies.
The response to low oxygen is mediated by hypoxia inducible factors (HIFs), a family of transcription factors that are stabilized under hypoxic conditions in all mammalian cells and are primarily thought to upregulate multiple pathways to adapt cells to low oxygen (Cassavaugh and Lounsbury, 2011; Choudhry and Harris, 2018; Kupferschmidt, 2019). HIFs are heterodimeric complexes consisting of an alpha and beta subunit. In the presence of oxygen, alpha subunits are hydroxylated by prolyl-hydroxylases, allowing for recognition and ubiquitination by von Hippel Lindau (VHL) (Ivan et al., 2001; Jaakkola et al., 2001), and rapid degradation by the proteasome. In low oxygen conditions, alpha subunits escape hydroxylation, avoid degradation, and translocate to the nucleus to pair with constitutive beta subunits and regulate gene expression (Cassavaugh and Lounsbury, 2011; Choudhry and Harris, 2018; Semenza, 2007). The HIF1a motif is present more than 1 million times in the genome; however, HIF1a binds to a small fraction of these sites suggesting that HIF1a binding is heavily regulated (Schodel et al., 2011; Smythies et al., 2019). Still, the determinants of HIF1a binding in each cell type and whether cell-type-specific targets are functional remain unknown.
The central nervous system (CNS) consumes 20% of the total body oxygen and white matter of the CNS is highly susceptible to hypoxic insults as seen in stroke, vascular dementia, respiratory distress syndromes, premature birth, and subsets of cerebral palsy (Hankey, 2017; Salmaso et al., 2014; Shindo et al., 2016; Volpe, 2009). In fact, chronic HIF1a activity is sufficient to block white matter development (Yuen et al., 2014). White matter of the CNS is formed by oligodendrocytes, which wrap neuronal axons in a lipid-rich protective sheath called myelin, allowing for rapid transmission of action potentials and maintenance of axonal integrity (Chang et al., 2016; Emery, 2010; Nave, 2010). Oligodendrocytes arise from oligodendrocyte progenitor cells (OPCs), which are prevalent in the developing and adult CNS, and HIF1a accumulation has been shown to be sufficient to impair oligodendrocyte formation from OPCs (Jablonska et al., 2016; van Tilborg et al., 2018; Yuen et al., 2014). However, the mechanism of the HIF1a-mediated block in oligodendrocyte formation from OPCs remains unclear. In this study, we use OPCs as an archetypal hypoxia-disease relevant cell type to define the mechanism by which chronic HIF1a drives cell dysfunction.
RESULTS
Knockout of Vhl models chronic HIF1a accumulation in OPCs
Defining the mechanisms by which HIF activity perturbs cell function is notoriously challenging as HIFs are rapidly degraded, in minutes, when cells are restored to normoxia. Because of this instability and the low abundance of HIFs, biochemical studies often require extraordinary numbers of cells, which is challenging for hypoxia disease relevant cell types outside of cancer cell lines. To explore the mechanisms underlying the HIF-mediated block in oligodendrocyte development from OPCs, we generated a cellular model of chronic HIF1a accumulation in mouse pluripotent stem cell-derived OPCs, which are uniquely scalable and amenable to genetic manipulation (Hubler et al., 2018; Lager et al., 2018; Najm et al., 2015; Najm et al., 2011). CRISPR-Cas9-mediated knockout of Vhl, a central component of the ubiquitin-proteasome system that degrades HIFs (Choudhry and Harris, 2018; Haase, 2009; Rechsteiner et al., 2011), in OPC cultures resulted in stable HIF1a protein accumulation and significant 9-fold and 18-fold activation of downstream hallmark HIF1a targets Vegfa and Bnip3, respectively, without affecting viability (Figures 1A, 1B, S1A, S1B, and S1C). The response of OPCs to Vhl knockout mirrored that of OPCs cultured in hypoxia at both the protein and RNA level (Figures 1C, 1D, and S1D). Vhl knockout OPCs were generated with two independent single guide RNAs targeting Vhl (sgVhl and sgVhl.2), each of which caused significant decreases in Vhl transcript and protein levels through Cas9-mediated insertion-deletions (in-dels) at the respective target sites compared to control (Cas9 expressing OPCs with no sgRNA) (Figures S1E–S1G). It is possible that loss of VHL stabilizes pathways independent of HIF1a, therefore we confirmed key findings from Vhl knockout OPCs in wild-type OPCs treated with hypoxia (1% O2), primary mouse OPCs exposed to hypoxia (1% O2), and human OPCs in pluripotent stem cell-derived oligocortical spheroids (Madhavan et al., 2018).
Figure 1. HIF1a Accumulation Impairs the Induction of Oligodendrocytes from OPCs.
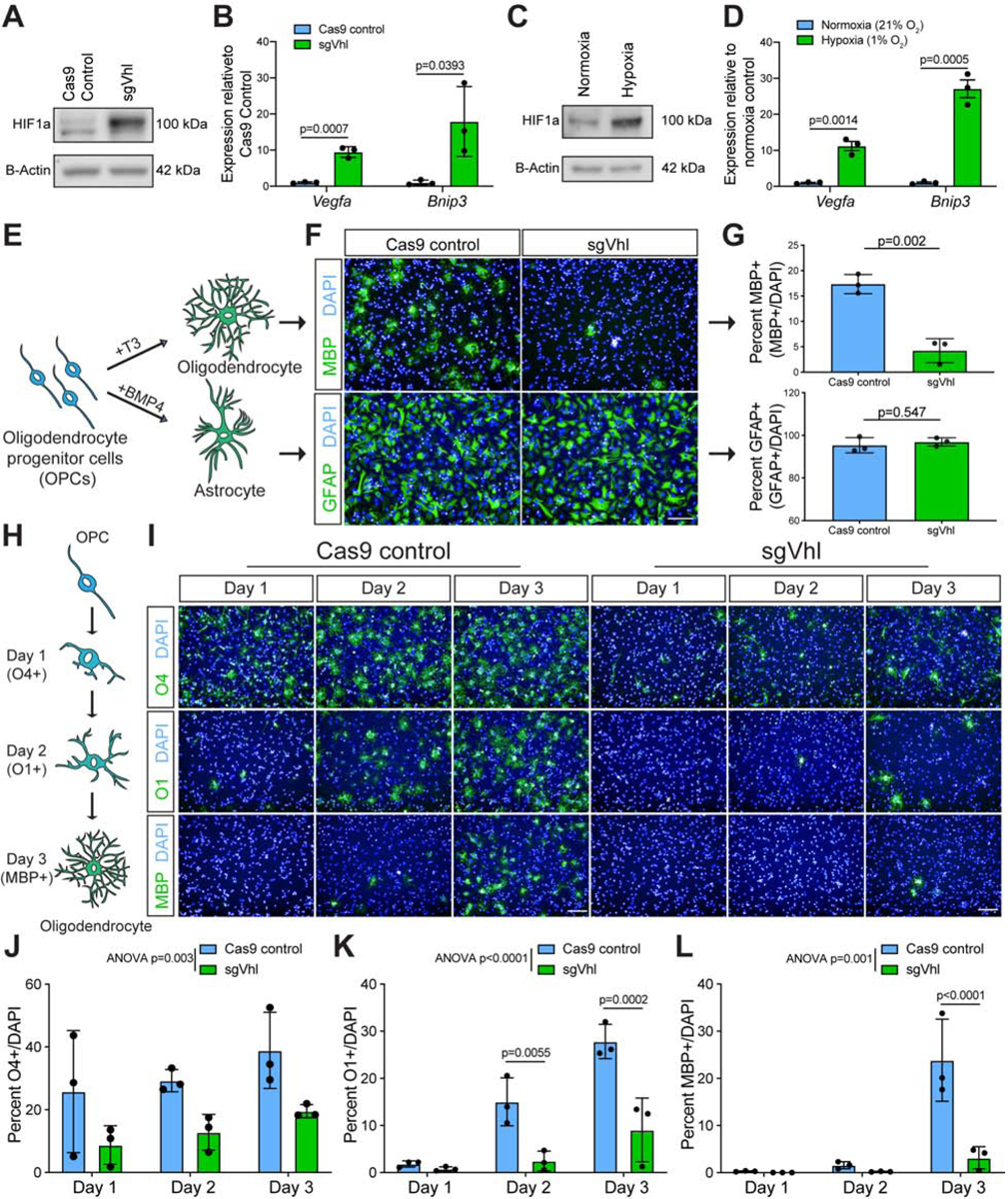
(A) Western blot of HIF1a in sgVhl and Cas9 control OPCs.
(B) qRT-PCR of Vegfa and Bnip3 in Cas9 control (in blue) and sgVhl (in green) OPCs. Data are presented as mean ± SEM from 3 biological replicates.
(C) Western blot of HIF1a in hypoxic (1% O2) and normoxic OPCs.
(D) qRT-PCR of Vegfa and Bnip3 in hypoxic (in green) and normoxic (in blue) OPCs. Data are presented as mean ± SEM from 3 biological replicates.
(E) Schematic of the two in vitro differentiation schemes directing OPCs to either oligodendrocytes or astrocytes.
(F) Representative images of Cas9 control and sgVhl oligodendrocytes (MBP+ in green) and astrocytes (GFAP+ in green). Nuclei are marked by DAPI (in blue). Scale bar, 100μm.
(G) Quantification of the percentage of oligodendrocytes (MBP+ cells / DAPI) and astrocytes (GFAP+ cells / DAPI) formed from sgVhl (in green) and Cas9 control OPCs (in blue). Data are presented as mean ± SD from 3 independent biological replicates. p-values were calculated using Student’s two-tailed t-test.
(H) Schematic illustrating acquisition of early (O4), intermediate (O1) and late (MBP) oligodendrocyte markers during in vitro oligodendrocyte differentiation.
(I) Representative images of early (O4+ in green), intermediate (O1+ in green) and late (MBP+ in green) oligodendrocytes during days 1, 2, and 3 of differentiation of Cas9 control and sgVhl OPCs. Nuclei are marked by DAPI (in blue). Scale bars, 100μm.
J-L) Quantification of the percentage of early O4+ (J), intermediate O1+ (K), and late MBP+ (L) oligodendrocytes in sgVhl (in green) and Cas9 control OPCs (in blue) at days 1, 2, and 3 of differentiation. Data are presented as mean ± SD from 3 independent biological replicates. p-values were calculated using two-way ANOVA (reported as ANOVA p=) for overall group differences and Sidak’s multiple comparisons test for individual timepoint differences.
p-values for qRT-PCRs were calculated using Student’s two tailed t-test.
See also Figure S1.
HIF1a accumulation specifically delays OPC differentiation into oligodendrocytes
HIF1a accumulation in OPCs is sufficient to impair oligodendrocyte formation (Yuen et al., 2014). To test whether HIF1a accumulation is a general or specific inhibitor of OPC differentiation, sgVhl OPCs were stimulated to form either astrocytes (Grinspan et al., 2000), or oligodendrocytes (Baas et al., 1997; Barres et al., 1994; Gao et al., 1998; Najm et al., 2011) (Figure 1E). This revealed a significant 4-fold reduction in oligodendrocyte formation by staining for myelin basic protein (MBP), a marker of mature oligodendrocytes, with no change in astrocyte formation by staining for glial fibrillary acidic protein (GFAP) in sgVhl OPCs compared to control (Figures 1E–1G, and S1H), which shows that the effects of HIF1a are cell type specific. Staining for early (O4+), intermediate (O1+), and late (MBP+) oligodendrocyte markers throughout the differentiation process demonstrated a significant and delayed acquisition of all oligodendrocyte markers in sgVhl and sgVhl.2 OPCs compared to control (Figures 1H–1L, and S1I–S1K). These data suggest that HIF1a accumulation specifically impairs early OPC differentiation, thereby delaying the formation of oligodendrocytes and ultimately myelin.
HIF1a upregulates unique target genes in OPCs
To delineate the gene targets of HIF1a in OPCs responsible for blocking oligodendrocyte development, chromatin-linked immunoprecipitation sequencing (ChIP-seq) was used to map its genome-wide chromatin binding profile. Utilizing 100 million control and sgVhl OPCs, we identified 503 high-stringent HIF1a peaks (FDR<0.001) in sgVhl OPCs with clear enrichment proximal to the annotated transcription start site (TSS), which agrees with HIF1a as a promoter centric transcription factor (Schodel et al., 2011; Smythies et al., 2019) (Figures 2A and S2A). HIF1a was enriched at canonical target genes Vegfa and Bnip3 and globally peaks were enriched for HIF motifs and motifs for ubiquitously expressed transcription factors that have been shown to interact with HIF1a including Sp1, c-Myc, and Bmal1 (Huang, 2008; Kaluz et al., 2003; Wu et al., 2017) (Figures S2A and S2B; Tables S1–S3). HIF1a was not found proximal to Sirt1, Wnt7a, or Wnt7b (Figures S2C–S2E), which have previously been suggested as putative HIF targets that impair oligodendrocyte formation in hypoxia-treated OPCs (Jablonska et al., 2016; Yuen et al., 2014). Moreover, sgVhl OPCs, wild type OPCs, and primary OPCs exposed to hypoxia exhibited no increase in expression of Sirt1, Wnt7a, or Wnt7b transcripts, suggesting that other targets are likely functioning to block oligodendrocyte formation (Figures S2F–S2I) (Zhang et al., 2020). Without HIF1a ChIP-seq data in OPCs, these previous studies focused on potential HIF1a target genes identified in other cell types, which overlooked that HIF1a might bind to unique cell-type-specific targets in OPCs that suppress oligodendrocyte formation.
Figure 2. HIF1a directly upregulates unique targets in OPCs.
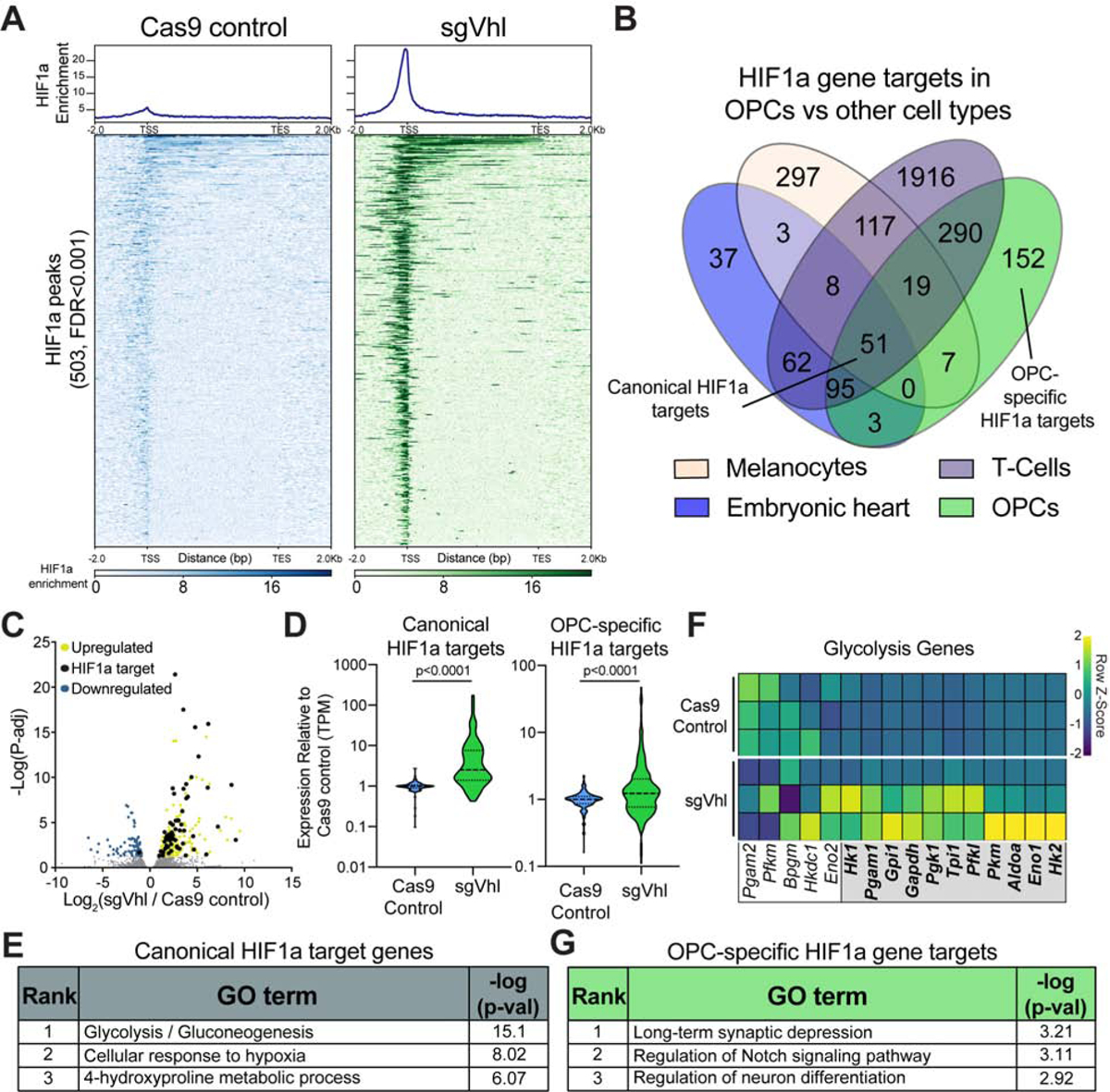
(A) Aggregate binding profile and heatmap of 503 HIF1a peaks (FDR<0.001) in sgVhl OPCs within 2Kb of the transcription start site (TSS) and transcription end site (TES) of the closest expressed gene in Cas9 control and sgVhl OPCs. See also Table S1 for full list of peaks.
(B) Venn diagram of direct HIF1a target genes in diverse mouse tissues overlapped with HIF1a targets in OPCs. See also Table S2 for full list of peaks.
(C) Volcano plot of genes that significantly increase and decrease in yellow and blue respectively (P-adj < 0.05) in sgVhl OPCs compared to Cas9 control OPCs with direct targets of HIF1a in black. Gray dots are genes not significantly different between conditions. Data are from 3 biological replicates.
(D) Violin plots of expression data (TPM) normalized to Cas9 control OPCs for canonical and OPC-specific HIF1a targets in Cas9 control (in blue) and sgVhl (in green) OPCs. Bold dashed line represents the median with the thin dashed lines representing the upper and lower quartiles. p-values were calculated using the Mann-Whitney test.
(E) Gene ontology (GO) analysis of shared canonical targets of HIF1a. Table shows the rank of the GO term along with −log(p-value).
(F) Heatmap of row normalized expression of glycolysis genes (TPM) between Cas9 control and sgVhl OPCs. Genes in the gray box are direct targets of HIF1a. Each row represents a biological replicate.
(G) Gene ontology (GO) analysis of OPC-specific HIF1a target genes. Table shows the rank of the GO term along with −log(p-value).
See also Figure S2.
To define the cell-type-specific targets of HIF1a in OPCs, we overlapped HIF1a targets in OPCs with HIF1a targets from the limited number of publicly available datasets derived from other mouse cell types including melanocytes (Loftus et al., 2017), T cells (Ciofani et al., 2012), and embryonic heart (Guimaraes-Camboa et al., 2015). This analysis identified 51 genes that were HIF1a targets across all 4 cell types (“canonical HIF1a targets”), 152 genes that were HIF1a targets only in OPCs based on these datasets (“OPC-specific HIF1a targets”) and 2250 genes that were specific to either heart, T cells, or melanocytes (“other tissue-specific HIF1a targets”) (Figure 2B).
To determine functional targets of HIF1a in OPCs, we performed RNA-seq of sgVhl and control OPCs. Overlapping transcripts that significantly changed between sgVhl and control OPCs (P-adj<0.05) with direct targets of HIF1a revealed that HIF1a directly bound to 61 significantly increased genes and only 1 significantly decreased gene (Figures 2C and S2J; Table S1), consistent with the role of HIF1a as a transcriptional activator (Dengler et al., 2014; Guimaraes-Camboa et al., 2015). Genes that were both significantly upregulated (P-adj<0.05) and not called as top targets of HIF1a (at a stringent threshold FDR<0.001) were enriched for immune and hypoxia pathways suggesting that these genes are weaker direct targets of HIF1a or upregulated indirectly in the context of constitutive HIF accumulation (Figure S2J). Both shared canonical and OPC-specific HIF1a target genes collectively increased in expression in sgVhl OPCs compared to control, with canonical targets induced to a greater extent than non-canonical targets (Figures 2D, S2K, and S2L). However, both of these gene sets showed a significantly greater increase in expression compared to the “other tissue-specific” HIF1a targets in sgVhl OPCs (Figure S2K).
Gene ontology (GO) analysis of canonical HIF1a target genes showed enrichment for metabolic and hypoxia pathways, which agrees with HIF1a’s role to promote glycolysis in a majority of cell types (Figure 2E) (Choudhry and Harris, 2018; Majmundar et al., 2010; Miska et al., 2019; Nagao et al., 2019). In fact, more than half of the enzymes in the glycolysis pathway were direct HIF1a targets in OPCs, and both sgVhl and sgVhl.2 OPCs exhibited a 2-fold increase in levels of the glycolysis byproduct, lactate, compared to control OPCs (Figures 2F and S2M).
Interestingly, GO analysis for cell-type-specific HIF1a targets demonstrated enrichment for pathways separate from the canonical hypoxic response and related to the tissue of origin (Figure S2N). In particular, GO analysis for OPC-specific HIF1a targets showed enrichment for neural development and differentiation pathways (Figure 2G). This demonstrates that HIF1a regulates genes critical for the canonical response to low oxygen as well as separate cell-type-specific targets.
Chromatin accessibility and cell-type-specific transcription factors define non-canonical HIF1a targets
How HIF1a is able to bind uniquely in a cell-type-specific manner remains unknown. To better understand the determinants of the cell type specificity of HIF1a binding profiles, we compared the open and active chromatin landscape in OPCs at canonical, non-canonical OPC-specific, and “other tissue-specific” HIF1a peaks (Figure S3A; Table S2). ChIP-seq for the active chromatin mark H3K27Ac (Creyghton et al., 2010) demonstrated a significant gain of H3K27Ac in sgVhl OPCs compared to control (FDR<0.1), and these regions were enriched for HIF motifs (Figures S3B and S3C). Shared canonical and OPC-specific HIF1a peaks exhibited a greater enrichment for HIF1a, H3K27Ac, and open chromatin (defined by ATAC-seq regions in non-transduced OPCs) compared to “other tissue-specific” HIF1a sites (Figure 3A and S3D). This agrees with previous findings that HIF1a preferentially binds open and active chromatin and that the level of HIF1a binding correlates with the magnitude of expression of gene targets in hypoxia (Smythies et al., 2019; Xia and Kung, 2009). However, there was a subset of “other tissue-specific” HIF1a peaks enriched for open and active chromatin that lacked HIF1a binding in OPCs, suggesting that chromatin accessibility and activity were not the sole predictors of HIF1a binding. HIF1b, a common binding partner with HIF1a, was more strongly enriched at canonical HIF1a targets compared to OPC-specific targets, which further suggests that other interacting transcription factors could define cell-type-specific targets of HIF1a (Figure S3D).
Figure 3. Chromatin Accessibility and Cell-Type-Specific Transcription Factors Define Non-Canonical HIF1a Targets.
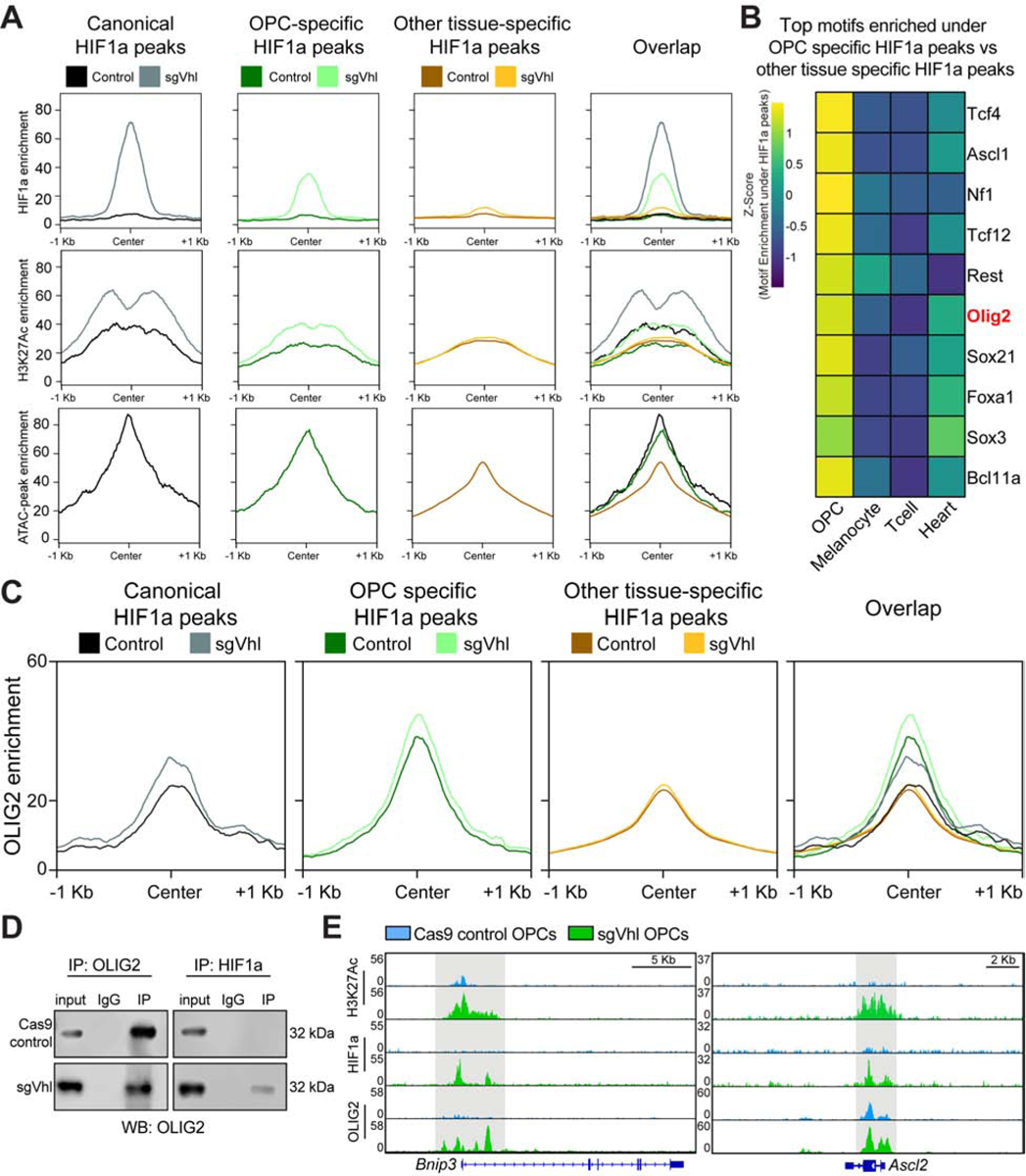
(A) Aggregate plots of HIF1a and H3K27ac enrichment in Cas9 control and sgVhl OPCs and open chromatin enrichment in non-transduced OPCs at shared canonical HIF1a peaks, OPC-specific HIF1a peaks, and “other tissue-specific” HIF1a peaks.
(B) Heatmap of the enrichment of the top 10 transcription factor motifs under non-canonical HIF1a peaks in OPCs compared to other tissue types. Olig2 (highlighted in red) is a dominant transcription factor of the oligodendrocyte lineage.
(C) Aggregate plots of OLIG2 enrichment in Cas9 control and sgVhl OPCs at canonical HIF1a peaks, OPC-specific HIF1a peaks, and “other tissue-specific” HIF1a peaks.
D) Western blot of OLIG2 from total cell lysate (input), IgG control lysate, and lysate from pulldown (IP) of either OLIG2 (on left) or HIF1a (on right) in Cas9 control (top) and sgVhl OPCs (bottom). See also figures S3H and S3I for reciprocal co-IP and replicate co-IP in sgVhl OPCs.
(E) Genome browser view of H3K27Ac, HIF1a, and OLIG2 ChIP-seq in Cas9 control (blue) and sgVhl (green) OPCs at Bnip3 and Ascl2. Scale bars, 5 Kb and 2 Kb respectively.
See also Figure S3.
Motif enrichment analysis under cell-type-specific HIF1a peaks demonstrated an enrichment for lineage defining transcription factors such as the Mitf family in melanocytes (Levy et al., 2006), Nkx2 family in embryonic heart (Bartlett et al., 2010), and basic-helix-loop helix (bHLH) motifs in OPCs (Figure S3E; Table S3). Specifically, the motif for Olig2, a lineage-defining bHLH transcription factor in OPCs (Yu et al., 2013), was highly enriched (in the top 5% of motifs) under OPC-specific HIF1a peaks, whereas the Olig2 motif was not enriched under any other tissue-specific HIF1a peaks (Figure 3B, S3E; Table S3). ChIP-seq for OLIG2 in control and sgVhl OPCs validated the enrichment for OLIG2 at OPC-specific HIF1a peak loci compared to canonical and “other tissue-specific” peaks (Figure 3C, S3F, S3G; Table S4). Furthermore, both canonical and non-canonical HIF1a peaks demonstrated an increase in OLIG2 binding in sgVhl OPCs compared to control suggesting that HIF1a and OLIG2 physically interact (Figure 3C and S3G). Co-immunoprecipitation (co-IP) of OLIG2 and HIF1a validated this physical interaction specifically in sgVhl OPCs compared to Cas9 control OPCs (Figure 3D, S3H, and S3I). Given this physical interaction between HIF1a and OLIG2, we observe an appearance of OLIG2 at canonical HIF1a target genes in sgVhl OPCs, such as Bnip3 (Figure 3E, S3J, S3K; Table S4). Non-canonical HIF1a gene targets, such as Ascl2, are already enriched for OLIG2 in control OPCs and display increased OLIG2 binding in sgVhl OPCs suggesting OLIG2 also interacts with HIF1a at these loci (Figure 3E). Collectively, these data demonstrate that, outside of canonical HIF1a targets, lineage-defining transcription factors, such as OLIG2, generate a permissible chromatin landscape to drive HIF1a to unique non-canonical targets in diverse cell types.
OPC-specific targets of HIF1a suppress Sox10 expression and oligodendrocyte formation
To understand whether OPC-specific targets are sufficient to impair oligodendrocyte formation, we first examined whether HIF1a induction impacts the expression of critical lineage transcription factors and lineage markers in OPCs. This revealed a selective reduction in Sox10, a transcription factor required for formation of oligodendrocytes from OPCs (Stolt et al., 2004; Stolt et al., 2002), and downstream SOX10 target genes Plp1 and Pdgfra, whereas other lineage transcription factors, such as Olig2 and Nkx2–2, remained unchanged (Figures 4A, S4A, and S4B). Of note, HIF1a did not bind proximal to any master regulators of oligodendrocyte development; however, the proximal promoter region of Sox10 showed a significant reduction of H3K27Ac in sgVhl OPCs (Figure 4B). The decrease in expression of Sox10 in sgVhl OPCs was validated at the mRNA and protein level as well as by reduction in expression of downstream SOX10 target genes Plp1 and Pdgfra in sgVhl OPCs and primary OPCs treated with hypoxia (1% O2) (Figures 4C, 4D, and S4C–E). These data suggest that HIF1a indirectly represses Sox10 through activation of other gene targets to block oligodendrocyte development.
Figure 4. OPC-specific HIF1a Targets Ascl2 and Dlx3 Suppress Sox10 and Impair Oligodendrocyte Formation.
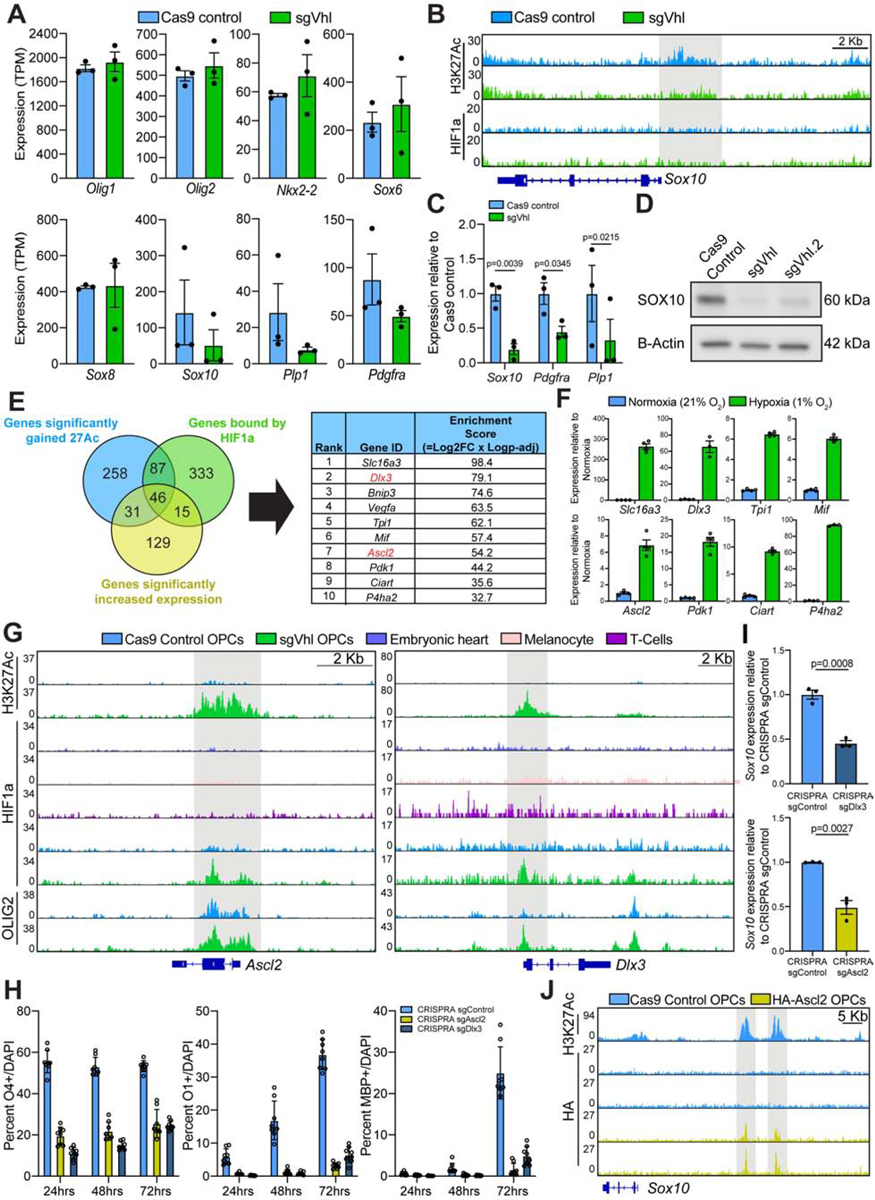
(A) Quantification of the normalized expression (TPM) of oligodendrocyte lineage markers in Cas9 control (in blue) and sgVhl (in green) OPCs. Data represent mean ± SEM from 3 independent RNA-seq replicates.
(B) Genome browser view of H3K27ac and HIF1a ChIP-seq in Cas9 control (in blue) and sgVhl (in green) OPCs at the locus for Sox10. The gray bar highlights significant H3K27Ac enrichment at the Sox10 promoter in control cells (−log10(q value) = −5.6) that is absent in Vhl knockout OPCs. Scale bar, 2Kb.
(C) qRT-PCR of Sox10 and downstream SOX10 target genes Pdgfra and Plp1 in sgVhl (in green) and Cas9 control (in blue) OPCs. Data are presented as mean ± SEM from 3 biological replicates.
(D) Western blot for SOX10 in sgVhl, Vhl.2 OPCs, and Cas9 control OPCs. Data represent results from a single biological replicate.
(E) Venn diagram overlapping genes with proximal HIF1a binding (in green), significant gains in H3K27ac (in blue), and significantly increased expression (in purple) in OPCs. The top 10 genes were then ranked and listed by their enrichment score, which is the product of the Log2(Fold change in gene expression) and −Log(P-adj). See also Table S1 for the full target list.
(F) qRT-PCR of 8 top HIF1a target genes (see Figure 1D for Vegfa and Bnip3) in hypoxic (1% O2, in green) and normoxic (21% O2, in blue) OPCs. Data are presented as mean ± SEM from 3–4 technical replicates.
(G) Genome browser view of HIF1a ChIP-seq at Ascl2 and Dlx3 in sgVhl OPCs (in green), Cas9 control OPCs (in blue), and other diverse cell types. H3K27ac and OLIG2 enrichment are also shown in sgVhl and Cas9 control OPCs. The gray bars highlight HIF1a accumulation in sgVhl OPCs. Scale bars, 2Kb.
(H) Quantification of the percentage of early O4+, intermediate O1+, and late MBP+ oligodendrocytes from sgAscl2 (in yellow), sgDlx3 (in dark blue) and sgControl (in light blue) CRISPRA OPCs at day 1, 2 and 3 of differentiation. Data are presented as mean ± SD of 6–8 technical replicates (individual wells) per condition.
(I) qRT-PCR of Sox10 in sgAscl2 (in yellow), sgDlx3 (in dark blue), and sgControl (in light blue) CRISPRA OPCs. Data are presented as mean ± SEM from 3 biological replicates.
(J) Genome browser view of HA ChIP-seq upstream of Sox10 in Cas9 control (in blue) and HA-Ascl2 (in yellow) OPCs. The gray bars highlight enhancers for Sox10 defined by H3K27Ac ChIP-seq in Cas9 Control OPCs. Scale bar, 5 Kb.
p-values for qRT-PCRs were calculated using Student’s two-tailed t-test.
See also Figure S4.
To define the top functional targets of HIF1a in OPCs, we overlapped direct HIF1a targets (FDR<0.001) with genes that exhibited both increased transcription (P-adj<0.05) and increased H3K27Ac (FDR<0.1) in sgVhl OPCs compared to control (Figure 2C, 4E; Table S1). Hits were independently validated in OPCs treated with hypoxia (1% O2), which showed that all of our top 10 HIF1a targets were significantly upregulated compared to normoxia (Figures 1D and 4F). Out of the top 10 targets of HIF1a in OPCs, Ascl2 and Dlx3 were the only non-canonical, OPC-specific HIF1a targets (Figure 4E; Table S2). Both are transcription factors that regulate differentiation of somatic stem cells in the periphery and are not normally expressed by any cell type in the mouse CNS (Tabula Muris et al., 2018; Zhang et al., 2014), which is reflected by the lack of active chromatin at Ascl2 and Dlx3 in control OPCs (Figures 4G and S4F). However, OLIG2 binds to both of these loci in Cas9 control OPCs which agrees with our finding that OLIG2 is enriched at non-canonical, OPC-specific HIF1a target genes (Figure 4G). Both targets display robust HIF1a peaks proximal to their promoters specifically in sgVhl OPCs compared to heart, melanocytes and T cells along with gained H3K27Ac and OLIG2 binding in sgVhl OPCs compared to control OPCs (Figure 4G). Ascl2 and Dlx3 both accumulate at the protein level in sgVhl OPCs and are induced in primary OPCs treated with hypoxia compared to normoxic controls (Figures S4G–S4I). In fact, ASCL2 was induced in vivo in the brains of mouse pups reared in chronic hypoxia (10% O2) and this correlated with a decrease in SOX10 and white matter proteins MBP and MAG compared to normoxia reared controls (Figures S4J and S4K).
Ectopic expression of Dlx3 and Ascl2 in OPCs using CRISPR activation (CRISPRA) technology was sufficient to impair the acquisition of early (O4+), intermediate (O1+), and late (MBP+) oligodendrocyte markers across the course of differentiation (Figure 4H). Moreover, ectopic expression of Dlx3 and Ascl2 led to a significant reduction in Sox10 expression (Figures 4I and S4L), whereas activation of canonical HIF1a targets, Slc16a3, Bnip3, and Vegfa, did not (Figures S4L and S4M). ASCL2 is known to impair somatic stem cell differentiation and can act as a repressor at critical differentiation genes (Murata et al., 2020; Schuijers et al., 2015; Wang et al., 2017), therefore we hypothesized that ASCL2 could directly bind to upstream regulatory regions of Sox10 (Yu et al., 2013). ChIP-seq for HA-tagged ASCL2 in OPCs revealed that ASCL2 is able to directly bind upstream enhancers of Sox10 and reduce expression of Sox10 compared to control OPCs (Figure 4J, S4N–S4P).
While Ascl2, Dlx3, and possibly other non-canonical OPC-specific HIF1a targets are each sufficient to decrease Sox10 expression and impair oligodendrocyte formation, it was not clear if these targets were individually necessary for the functional effect. RNAi-mediated knockdown of Ascl2 or Dlx3 in sgVhl OPCs did not rescue oligodendrocyte formation (Figures S4Q and S4R) suggesting that non-canonical HIF1a targets collectively or redundantly converge to suppress Sox10 expression and impair oligodendrocyte formation from OPCs.
Chemical inhibition of MEK/ERK increases oligodendrocyte formation from sgVhl OPCs
To identify potential pathways that could overcome the differentiation block imposed by HIF1a, we screened a library of 1753 bioactive compounds for the ability to increase the formation of MBP+ oligodendrocytes relative to DMSO treated sgVhl OPCs (Figures 5A and S5A–S5D; Table S5). Compounds that were non-toxic (fold change in total cell number > 0.7 relative to DMSO treated sgVhl OPCs at the primary screening dose of 3μM) and enhanced the number and percentage of MBP+ oligodendrocytes (fold change > 3 relative to DMSO treated sgVhl OPCs) were considered primary hits (Figure S5E). MEK inhibitors were enriched among the primary hits and, as a class, demonstrated a significant increase in oligodendrocyte formation compared to all other non-toxic compounds tested in the primary screen (Figures 5B, 5C, and S5F; Table S5). Interestingly, drugs previously identified to enhance oligodendrocyte formation, such as miconazole, clemastine, and tamoxifen (Gonzalez et al., 2016; Mei et al., 2014; Najm et al., 2015), were not enriched as hits in this screen, highlighting the ability of this screen to identify context-specific modulators of the differentiation block imposed by HIF1a (Figure 5A and S5B).
Figure 5. Chemical Inhibition of MEK/ERK Increases Oligodendrocyte Formation from sgVhl OPCs.
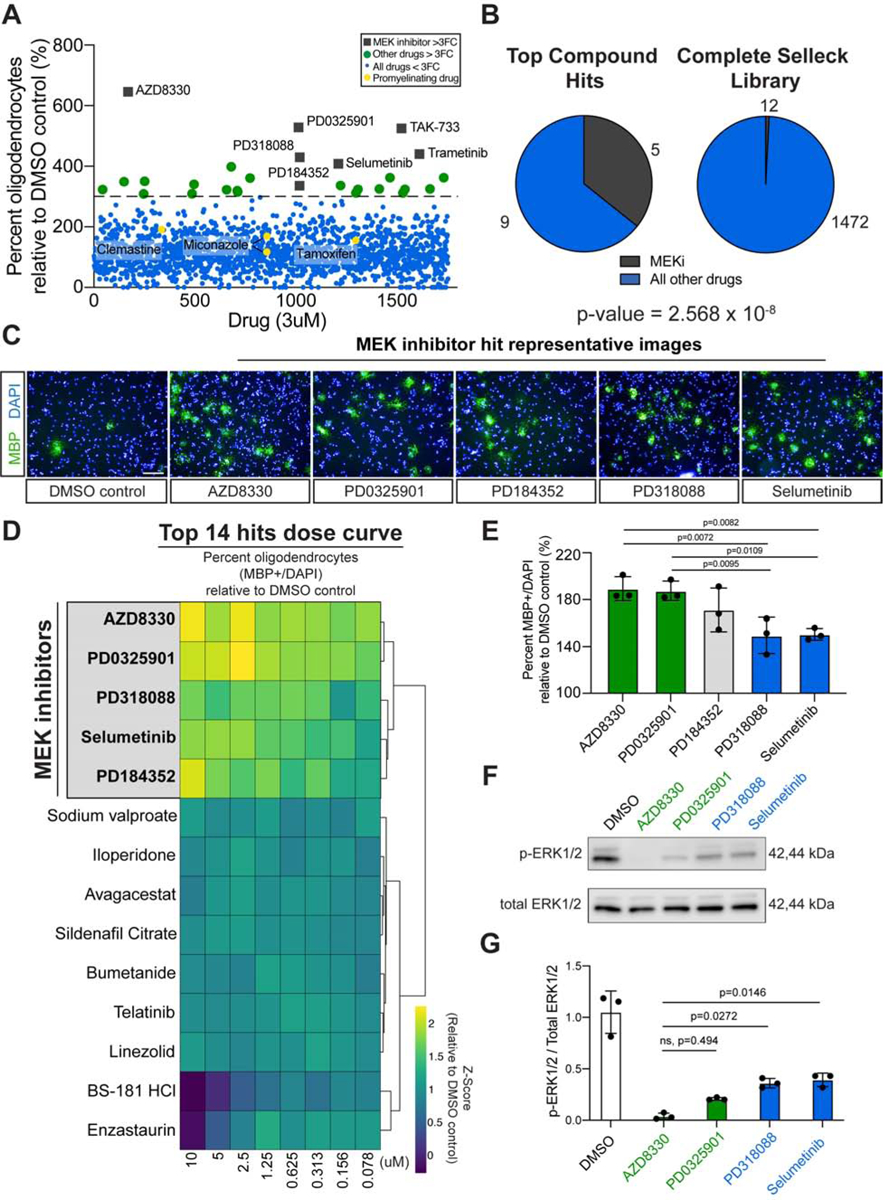
(A) Primary bioactives library screen showing the effect of 1753 molecules on percentage of oligodendrocytes (MBP+ cells/ total DAPI) formed by sgVhl OPCs relative to DMSO treated sgVhl OPCs. The dotted line represents a 3-fold increase in oligodendrocyte formation and compounds that clear this threshold are indicated as green dots. MEK inhibitors are highlighted as gray boxes and pro-myelinating drugs are highlighted as yellow dots. See also Table S5. (B) Pie charts of the number of MEK inhibitors (in dark gray) and other compounds classes (in blue) within top hits compared to their prevalence in the non-toxic compounds of the Selleck library. p-value was calculated using hypergeometric analysis. (C) Representative immunocytochemistry images of oligodendrocytes (MBP+ in green) from the primary drug screen of the top 5 MEK inhibitor hits along with the DMSO negative control. Nuclei are marked by DAPI (in blue). Scale bars, 100μm.
(D) Heatmap showing the row normalized fold change in the percentage (MBP+ / DAPI) of oligodendrocytes relative to DMSO treated sgVhl OPCs of an 8-point dose curve of the primary hits. The heatmap rows are sorted by unsupervised hierarchical clustering with columns in order from high (10μM) to low dose (78nM). MEK inhibitors are highlighted in gray and bolded. Data are presented as the mean from 3 separate dose curve plates. See also Table S6.
(E) Averaging all 8 doses shows the ability of each MEK inhibitor to increase the formation of oligodendrocytes (MBP+/DAPI) relative to DMSO treated sgVhl OPCs. Green and blue columns represent the most and least effective compounds respectively. Data are presented as the mean ± SD from 3 separate dose curve plates. p-values were calculated using a one-way ANOVA with Tukey’s multiple comparisons test.
(F) Representative western blot for phosphorylated ERK1/2 (p-ERK1/2) relative to total ERK1/2 of sgVhl OPCs incubated with 100nM of indicated MEK inhibitors for 30 minutes.
(G) Quantification of the ratio of p-ERK1/2 to total ERK1/2 for the most (in green) and least (in blue) effective MEK inhibitors relative to DMSO treated controls. Data are presented as mean ± SD from 3 biological replicates. p-values were calculated using one-way ANOVA with Dunnett’s multiple comparisons test.
See also Figure S5.
To identify compounds that were effective across a range of doses, we performed an 8-point dose curve from 78nM to 10μM of the top 14 hits. Performing unbiased hierarchical clustering of the results revealed that all 5 MEK inhibitors clustered together and led to a pronounced increase in oligodendrocyte formation relative to DMSO treated sgVhl OPCs, with AZD8330 and PD0325091 significantly outperforming PD318088 and Selumetinib while PD184352 was not significantly different from the other 4 MEK inhibitors (Figures 5D, 5E, and S5G; Table S6). The ability of the top two MEK inhibitors to impact oligodendrocyte formation correlated with their on-target IC50 values for MEK1 and MEK2 as well as their on-target ability to reduce ERK1/2 phosphorylation in sgVhl OPCs (Figures 5F, 5G, and S5H). Performance of an 8-point dose curve consisting of 12 drugs that each inhibit a potential downstream target of MEK revealed that ERK1/2 inhibitors led to the greatest increase in oligodendrocyte formation compared to the other classes of drugs tested (Shaul and Seger, 2007; Yohe et al., 2018) (Figures S5I and S5J). Compounds targeting the closely related MEK5 pathway did not increase oligodendrocyte formation from sgVhl OPCs and AZD8330, the most potent MEK inhibitor, did not impair ERK5 phosphorylation in sgVhl OPCs (Figures S5K and S5L). Collectively, these results demonstrate that chemical inhibition of MEK/ERK acts as a node of intervention to restore the formation of oligodendrocytes in the context of HIF1a accumulation.
MEK/ERK inhibition drives Sox10 expression without changing HIF1a activity
We next asked whether MEK inhibition increased differentiation of sgVhl OPCs by modulating HIF1a or its direct downstream targets. Treating OPCs with 300nM AZD8330, our most potent MEK inhibitor (Figures 5F and 5G), for 14 hours did not alter nuclear accumulation of HIF1a (Figures 6A–6C). Furthermore, downstream non-canonical HIF1a targets Ascl2 and Dlx3 did not increase activation of MEK/ERK signaling (Figure S6A). These data suggest that MEK inhibitor treatment does not directly counter HIF1a or downstream HIF1a target activity, but rather circumvents the effect of HIF1a accumulation by increasing oligodendrocyte differentiation despite persistent HIF signaling.
Figure 6. MEK Inhibitors Restore Sox10 Expression Independent of HIF1a Activity in OPCs.
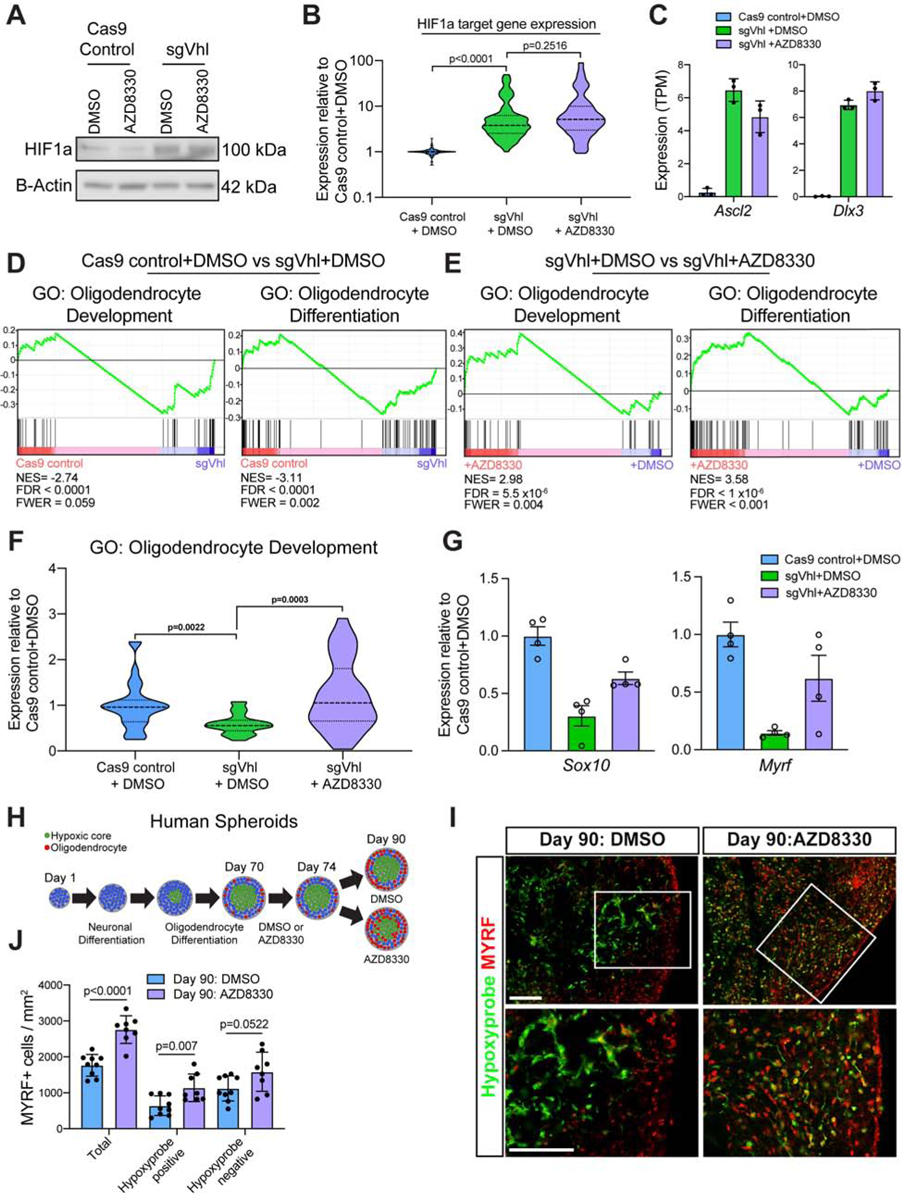
(A) Western blot for HIF1a in sgVhl OPCs treated with AZD8330 or DMSO. Data represent results from a single biological replicate.
(B) Violin plot of normalized expression (TPM values normalized to Cas9 control+DMSO) of direct functional targets of HIF1a (see Figure 2B) in Cas9 control+DMSO (in blue), sgVhl + DMSO (in green), and sgVhl + AZD8330 (in purple) OPCs.
(C) Quantification of normalized expression (TPM) for both Ascl2 and Dlx3 in Cas9 control+DMSO (in blue), sgVhl+DMSO (in green) and sgVhl+AZD8330 (in purple) OPCs. Data represent mean ± SD from 3 independent RNA-seq replicates.
(D) Gene set enrichment analysis (GSEA) analysis of gene program changes in sgVhl compared to Cas9 control OPCs demonstrates a significant reduction (FDR<0.001) in GO terms for Oligodendrocyte Development (normalized enrichment score/NES = −2.74) and Oligodendrocyte Differentiation (NES = −3.11).
(E) GSEA analysis of gene program changes in sgVhl + AZD8330 compared to sgVhl + DMSO OPCs demonstrates a significant enrichment (FDR<0.001) in GO terms for Oligodendrocyte Development (normalized enrichment score/NES = 2.98) and Oligodendrocyte Differentiation (NES = 3.58).
(F) Violin plot showing normalized expression (TPM values normalized to Cas9 control + DMSO OPCs) of genes in the GO term Oligodendrocyte Development (GO:0014003) that decrease (FC<0.75) in sgVhl + DMSO OPCs (in green) relative to Cas9 control + DMSO OPCs (in blue) as well as sgVhl OPCs following treatment with AZD8330.
(G) qRT-PCR of Sox10 and Myrf in Cas9 control+DMSO (in blue), sgVhl+DMSO (in green) and sgVhl+AZD8330 (in purple) OPCs. Data are presented as mean ± SEM from 4 technical replicates (individual wells).
(H) Schematic of human brain oligocortical spheroids treated at days in vitro (DIV) 70 with either DMSO or AZD8330 for 4 days. At day 90, organoids were incubated with hypoxyprobe, fixed, and sectioned for immunohistochemistry.
(I) Representative immunohistochemistry images for oligodendrocytes (MYRF+ in red) and hypoxic regions (hypoxyprobe in green) of DIV 90 oligocortical spheroids at low and high magnification that were treated from DIV 70–74 with DMSO or AZD8330. Scale bars, 100μM.
(J) Quantification of oligodendrocytes (MYRF+ / mm2) in the whole oligocortical spheroid (total), hypoxic region of the spheroid (hypoxyprobe positive), and normoxic region of the spheroid (hypoxyprobe negative) in DIV 90 spheroids that had been treated with DMSO or AZD8330 from DIV 70–74. Data represent mean ± SD from 8–9 individual spheroids. p-values were calculated using Student’s two-tailed t-test.
AZD8330 was used at a dose of 300nM for all experiments and p-values for violin plots were calculated using the Kruskal Wallis One-Way ANOVA with Dunn’s multiple comparisons test.
See also Figure S6.
Gene set enrichment analysis (GSEA) revealed that AZD8330 treatment of sgVhl OPCs led to an enrichment for “Oligodendrocyte Differentiation” and “Oligodendrocyte Development” pathways, which were normally depleted in sgVhl OPCs compared to control (Figures 6D and 6E). Further supporting this, AZD8330 treatment of sgVhl OPCs led to a significant increase in the subset of genes within the GO term “Oligodendrocyte Development, GO:0014003” that were normally decreased in sgVhl OPCs compared to control (fold change of sgVhl to control OPCs <0.75), such as Sox10 and Myrf (Figures 6F and 6G). We validated the AZD8330-mediated increase in expression of both of these transcription factors, which are critical for oligodendrocyte differentiation, by qPCR (Figure 6G). To confirm that this was a function of impaired MEK/ERK signaling, treatment of sgVhl OPCs with ERK1/2 inhibitors AZD0364 and SCH772984 led to a similar increase in Sox10 expression compared to DMSO (Figure S6B).
Given that MEK inhibitors do not change HIF1a translocation to the nucleus or downstream HIF1a target gene expression, it is unlikely that the MEK inhibitor mediated increase in Sox10 expression is specific to sgVhl OPCs. In agreement with this point, Sox10 expression increases in Cas9 control OPCs treated with 300nM AZD8330 for 14 hours (Figure S6C and S6D). However, MEK inhibitors lead to a pronounced increase oligodendrocyte formation in sgVhl OPCs with minimal impact on the oligodendrocyte formation from control OPCs, highlighting the context specific block imposed by HIF1a (Figure S6E and S6F). Collectively, these data suggest that the reduction of Sox10 expression by cell-type-specific HIF1a targets is critical for the HIF-mediated block in oligodendrocyte differentiation, such that restoration of Sox10 expression without altering canonical HIF function in sgVhl OPCs restores oligodendrocyte formation.
MEK/ERK inhibition drives oligodendrocyte formation in hypoxic regions of human oligocortical spheroids
To evaluate the effect of MEK inhibition in hypoxic human OPCs, we leveraged a previously established method of generating human myelinating cortical spheroids from pluripotent stem cells (Madhavan et al., 2018). The interior of human brain spheroids is hypoxic (Brawner et al., 2017; Giandomenico and Lancaster, 2017), and we hypothesized that these hypoxic regions would inhibit SOX10 expression and oligodendrocyte formation. Immunohistochemistry using hypoxyprobe to define hypoxic regions of the spheroid (Pogue et al., 2001) demonstrated that SOX10 expression is significantly impaired in hypoxic regions of the spheroid (Figures S6G and S6H). We next tested whether hypoxic regions of the oligocortical spheroid were inhibitory to oligodendrocyte formation and whether this could be overcome using a MEK inhibitor. To test this, we treated oligocortical spheroids with either DMSO or 300nM AZD8330 for 4 days starting at 70 days in vitro, immediately following induction of oligodendrocytes. At day 90, spheroids were treated with hypoxyprobe and harvested for analysis (Figure 6H). Immunohistochemistry for oligodendrocytes (MYRF+ cells) and hypoxic regions (defined by hypoxyprobe staining) demonstrated a significant 2.8-fold reduction in the number of MYRF+ oligodendrocytes in hypoxic regions compared to normoxic regions within the spheroids (Figures 6I, 6J and S6H). MEK/ERK inhibition with AZD8330 treatment led to a significant 1.8-fold increase in the number of oligodendrocytes within hypoxic regions of the spheroid (Figures 6I, 6J and S6I). Collectively these results show that oxygen tensions shape oligodendrocyte development and that MEK inhibition circumvents the hypoxia-mediated inhibition of oligodendrocyte formation in 3D models of human brain development.
DISCUSSION
Cells are equipped to translate external cues from the environment into internal signals that ultimately alter transcriptional programs. Molecular oxygen is crucial to support energy production of the cell, and low oxygen upregulates a rapid and conserved transcriptional response mediated largely by HIF transcription factors in all mammalian cells. HIF1a promotes an adaptive response by upregulating oxygen-independent metabolism and increasing blood vessel formation; however, chronic accumulation of HIF1a negatively impacts the function of almost every organ system (Kullmann et al., 2020; Lee et al., 2019; Menendez-Montes et al., 2016; Takubo et al., 2010).
Here, we profiled the genome-wide functional targets of HIF1a in OPCs and found that HIF1a not only binds to canonical hypoxia response genes that are shared across multiple cell types, but also activates a unique set of non-canonical genes in a cell-type-specific manner. In the context of the brain, these non-canonical HIF1a targets that impair oligodendrocyte formation could have implications in the numerous hypoxia driven pathologies of white matter such as diffuse white matter injury of prematurity (Salmaso et al., 2014; van Tilborg et al., 2018), white matter stroke in adults (Hankey, 2017; Marin and Carmichael, 2018) and multiple sclerosis (Ding et al., 2020; Graumann et al., 2003; Zeis et al., 2008). More broadly, we suggest that non-canonical HIF1a targets in diverse cell types impact a variety of cell-type-specific functions, such as oligodendrocyte differentiation, heart morphogenesis, and T cell activation. This concept has previously been overlooked as many studies have focused on canonical targets of HIF1a or those that were discovered in immortalized cell lines, which may behave differently in response to HIF1a accumulation. Wnt7a and Wnt7b, for example, were identified as direct HIF1a targets in mouse fibroblasts and extrapolated to be direct targets in mouse OPCs (Yuen et al., 2014). However, we demonstrate that Wnt7 ligands are not direct HIF1a targets in OPCs, a finding corroborated by a recent study demonstrating that Wnt7 ligands are targets of HIF1a in astrocytes, but not in OPCs (Zhang et al., 2020). This highlights that the response to hypoxia is not universal across all cell types and that the cell-type-specific response to HIF1a induction, such as Wnt signaling in astrocytes or downregulation of Sox10 in OPCs, have critical phenotypic consequences (Zhang et al., 2020).
The same transcription factor can bind to different gene targets in different cell types through interaction with transcriptional machinery unique to each cell type (Mullen et al., 2011; Trompouki et al., 2011). HIF1a binding has been shown to depend on the openness and activation status of chromatin; however, we and others show that chromatin accessibility is not the sole predictor of HIF1a binding (Schodel et al., 2011; Smythies et al., 2019; Xia and Kung, 2009). Our results highlight that HIF1a binds more strongly and induces a greater increase in canonical hypoxia response genes compared to non-canonical cell-type-specific genes, which are enriched for open chromatin as well as motifs for lineage-defining transcription factors. This implies that chronic HIF1a upregulation could allow for sufficient accumulation of non-canonical HIF1a targets that then impair development and function in a cell-type-specific manner. These non-canonical targets could represent either a pathological “off-target” effect of sustained HIF1a accumulation or a normal cell-type-specific response to molecular oxygen levels that is coopted in the context of hypoxia disease.
To cast a wide net to uncover pathways that circumvent the disruption of oligodendrocyte formation imposed by HIF, we performed a chemical-genetics screen revealing that MEK/ERK inhibition enhanced oligodendrocyte programs. This adds to the ongoing debate of the functionality of MEK/ERK signaling in the oligodendrocyte lineage. Previous studies have shown that MEK/ERK signaling can promote myelination (Ishii et al., 2012; Yang et al., 2016), while others demonstrate that inhibition of MEK/ERK signaling enhances OPC differentiation to oligodendrocytes (Newbern et al., 2011; Suo et al., 2019). We believe that this discrepancy owes to a potential stage-specific function of MEK/ERK such that this pathway impairs the transition from OPC to immature oligodendrocyte but then promotes mature oligodendrocyte formation and myelination. However, the mechanism of how inhibition of MEK/ERK promotes early oligodendrocyte programs remains elusive. One possibility could be that impairing ERK2 phosphorylation leads to transcriptional disinhibition at Sox10 and other key development genes, as has been shown in cancer and mouse embryonic stem cells (Tee et al., 2014; Yohe et al., 2018). MEK inhibitors have also been shown to directly upregulate SOX10 translocation to the nucleus, which would then drive downstream oligodendrocyte gene programs (Fufa et al., 2019). Continued dissection of the roles of MEK/ERK signaling during the process of differentiation would provide critical insight to how this pathway plays a bidirectional role regulating oligodendrocyte formation from OPCs.
Overall, this work advances our conceptual understanding of the tissue specific response to chronic HIF accumulation and how oxygen tensions regulate tissue physiology and pathology.
Limitations of Study
While we show that Ascl2 and Dlx3 individually are not necessary for the HIF1a-mediated block in differentiation, it is possible that inactivation of combinations of Ascl2, Dlx3, and other non-canonical targets might elucidate a core set of non-canonical HIF1a targets that are necessary to impair oligodendrocyte formation. The mechanism by which MEK/ERK signaling regulates Sox10 expression remains unknown and will benefit from further study with a particular focus on developmental stage-specific effects. Due to the number of cells required in this study, most experiments were limited to in vitro cultured mouse OPCs. Given that hypoxia also impairs oligodendrocyte formation in vivo in rodents and in vitro in human oligocortical spheroid tissue, it will be important for future studies to validate the effects of non-canonical HIF1a targets in these more therapeutically relevant contexts. Finally, it will be exciting to understand if cell-type specific non-canonical HIF1a targets are the key drivers of hypoxia-mediated dysfunction in other cell types outside of OPCs.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for materials, datasets, and protocols should be directed to and will be fulfilled by the Lead Contact, Paul Tesar (paul.tesar@case.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
All datasets generated in this study have been deposited in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under SuperSeries accession codeGEO: GSE143474 with subseries for RNA-seq (GSE143473), ChIP-seq (GSE143472), and ATAC-seq (GSE155366).
Software used to analyze the datasets are freely or commercially available and detailed in the Key Resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD140a-APC | eBioscience | Cat#17-1401-81; RRID:AB_529482 |
| NG2-AF488 | Millipore | Cat#AB5320A4; RRID:AB_11203143 |
| OLIG2 | Proteintech | Cat#13999-1-AP; RRID:AB_2157541 |
| OLIG2 | Millipore | Cat#AB9610; RRID:AB_570666 |
| OLIG2 | Millipore | Cat#MABN50; RRID:AB_10807410 |
| MBP | Abcam | Cat#ab7349; RRID:AB_305869 |
| O1 | CCF Hybridoma Core Facility | N/A |
| O4 | CCF Hybridoma Core Facility | N/A |
| ASCL2 | Millipore | Cat#MAB4417; RRID:AB_10562654 |
| GFAP | Agilent/Dako | Cat#Z033401-2 |
| Donkey anti-rat AF488 | Thermo Fisher Scientific | Cat#A-21208; RRID:AB_2535794 |
| Donkey anti-mouse AF488 | Thermo Fisher Scientific | Cat# A-21202; RRID:AB_141607 |
| Donkey anti-mouse AF647 | Thermo Fisher Scientific | Cat#A31571; RRID:AB_162542 |
| Donkey anti-rabbit AF647 | Thermo Fisher Scientific | Cat#A31573; RRID:AB_2536183 |
| HIF1a | Abcam | Cat#Ab2185; RRID:AB_302883 |
| Phospho-p44/42. (phosphorylated-ERK1/2) | Cell Signaling Technologies | Cat#9101S; RRID:AB_331646 |
| P44/42 (total ERK1/2) | Cell Signaling Technologies | Cat#9102S; RRID:AB_330744 |
| Phospho-ERK5 (Thr218/Tyr220) | Cell Signaling Technologies | Cat#3371S; RRID:AB_2140424 |
| ERK5 (total) | Cell Signaling Technologies | Cat#3372S; RRID:AB_330491 |
| DLX3 | Abcam | Cat#Ab178428 |
| SOX10 | R&D Systems | Cat#AF2864; RRID:AB_442208 |
| B-Actin peroxidase | Sigma Aldrich | Cat#A3854; RRID:AB_262011 |
| VHL | BD Biosciences | Cat#556347; RRID:AB_396376 |
| MBP | Abcam | Cat#ab7349; RRID:AB_305869 |
| MBP | BioLegend | Cat#808401; RRID:AB_2564741 |
| MAG | Thermo Fisher Scientific | Cat#346200; RRID:AB_2533179 |
| H3K27Ac | Abcam | Cat#Ab4729; RRID:AB_2118291 |
| MyRF | Gift from Michael Wegner | |
| HA | Cell Signaling Technologies | Cat#3724S; RRID:AB_1549585 |
| Normal Rabbit IgG | Cell Signaling Technologies | Cat#2729S; RRID:AB_1031062 |
| SOX6 | abcam | Cat#30455; RRID:AB_1143033 |
| A2B5 | Millipore | Cat#MAB312; RRID:AB_94709 |
| NKX2-2 | DSHB | Cat#74.5A5; RRID:AB_531794 |
| Bacterial and Virus Strains | ||
| Stbl3 E.Coli | ThermoFisher | Cat#C737303 |
| Lenti-X shots for lenti-virus generation | Clonetech | Cat# 631276 |
| Biological Samples | ||
| MCH forebrain mouse protein lysate samples | Gift from Brian Popko | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N2 Max | R&D Systems | Cat#AR009 |
| B27 | Thermo Fisher Scientific | Cat#12587010 |
| bFGF | R&D Systems | Cat#23-3FB-01M |
| PDGFA | Thermo Fisher Scientific | Cat#221-AA-050 |
| DMEM/F12 | Thermo Fisher | Cat#11320-033 |
| DMEM | Thermo Fisher | Cat# 11960-044 |
| Fetal Bovine Serum (FBS) | Fisher | Cat#A3160402 |
| MEM Non-essential amino acids | Thermo Fisher | Cat#11140-050 |
| Glutamax | Thermo Fisher | Cat#35050061 |
| 2-Mercaptoethanol | Sigma | Cat#M3148 |
| Poly-L-ornithine | Sigma Aldrich | Cat#P3655-50MG |
| Laminin | Sigma Aldrich | Cat#L2020-1MG |
| Noggin | R&D Systems | Cat#3344NG050 |
| IGF-1 | R&D Systems | Cat#291G1200 |
| cyclic AMP | Sigma Aldrich | Cat#D0260-100MG |
| NT3 | Thermo Fisher Scientific | Cat#267N3025 |
| T3 (Thyroid hormone) | Sigma Aldrich | Cat#T-6397 |
| Hb-EGF | R&D Systems | Cat#259-HE-050 |
| CNTF | R&D Systems | Cat#557-NT-010 |
| BMP4 | R&D Systems | Cat#314-BP-050 |
| CloneR | Stem Cell Technologies | Cat#5889 |
| LDN193189 | Sigma | Cat#SML0559 |
| SB-43152 | Sigma | Cat#S4317 |
| Normal donkey serum | Jackson ImmunoResearch | Cat#017-000-121 |
| Paraformaldehyde | Electron microscopy sciences | Cat#15710 |
| Triton X-100 | Sigma Aldrich | Cat#1086431000 |
| DAPI | Sigma Aldrich | Cat#D8417 |
| Puromycin | Thermo Fisher Scientific | Cat#A1113802 |
| Blasticidin | Thermo Fisher Scientific | Cat#A1113903 |
| Hygromycin | Thermo Fisher Scientific | Cat#10687010 |
| SCH772984 | Selleck Chemicals | Cat#S7101 |
| AZD0364 | Selleck Chemicals | Cat#S8708 |
| VX-11e | Selleck Chemicals | Cat#S7709 |
| BIX02188 | Selleck Chemicals | Cat#S1530 |
| BIX02189 | Selleck Chemicals | Cat#S1531 |
| RIPA Buffer | Sigma | Cat# R0278 |
| Protease and Phosphatase Inhibitor | Thermo Fisher Scientific | Cat#78441 |
| NuPAGE Bis-Tris gels | Thermo Fisher Scientific | Cat#NP0335BOX |
| PVDF membranes | Thermo Fisher Scientific | Cat#LC2002 |
| TRIzol | Ambion | Cat#15596-026 |
| Protein G magnetic DynaBeads | Thermo Fisher Scientific | Cat#10004D |
| Proteinase K | Thermo Fisher | Cat#AM2546 |
| Ampure XP beads | Aline Biosciences | Cat#C-1003-5 |
| Hypoxyprobe-1 | Hypoxyprobe Inc | Cat#HP1-100Kit |
| Penicillin-streptomycin | ThermoFisher | Cat#15070063 |
| Bioactive Compound Library-I | Selleck Chemicals | Cat#1700 |
| Critical Commercial Assays | ||
| QIAquick Gel Extraction Kit | Qiagen | Cat#28115 |
| Qiagen Plasmid Maxi Kit (25) | Qiagen | Cat#12163 |
| KAPA HiFi HotStart ReadyMix | KAPA Biosystems | Cat#kk2602 |
| KAPA Sybr Fast qPCR kit | KAPA Biosystems | Cat# KK4605 |
| Taqman gene expression master mix | Thermo Fisher | Cat#4369016 |
| NEBNext® Library Quant Kit for Illumina® | New England 641 Biolabs | Cat#E7630 |
| Protein Assay Kit | Bio-Rad | Cat#5000001 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| iScript cDNA synthesis kit | Bio-Rad | Cat#1708891 |
| NEBNext Poly(A) mRNA Magnetic Isolation Module | NEB | Cat#E7490L |
| NEBNext Ultra RNA Library Prep Kit for Illumina | NEB | Cat#E7530L |
| truChIP Chromatin Shearing Kit with Formaldehyde | Covaris | Cat#520154 |
| Hypoxyprobe Kit | Hypoxyprobe Inc | Cat#HP1-100Kit |
| ON-TARGETplus Mouse siRNA smartpool: Ascl2 | Horizon/Dharmacon | Cat#L-047262-01-0005 |
| ON-TARGETplus Mouse siRNA smartpool: Dlx3 | Horizon/Dharmacon | Cat#L-041957-01-0005 |
| siGENOME Mouse siRNA pool: Non-Targeting Control #2 | Horizon/Dharmacon | Cat#D-001206-14-05 5 nmol |
| Basic NucleofectorTM Kit for Primary Mammalian Glial Cells | Lonza | Cat# VPI-1006 |
| Qiagen MinElute PCR purification Kit | Qiagen | Cat#28004 |
| MTS assay kit | abcam | Cat# ab197010 |
| Deposited Data | ||
| RNA-seq datasets | This paper | GSE143474; GSE143473 (subseries) |
| ChIP-seq datasets | This paper | GSE143474; GSE143472 (subseries) |
| ATAC-seq datasets | This paper | GSE143474; GSE155366 (subseries) |
| Experimental Models: Cell Lines | ||
| Lenti-X 293T Cell line | Takara Biosciences | Cat#632180 |
| Epiblast stem cell (EpiSC) derived OPCs | Najm et al. 2011 | N/A |
| Human iPSCs (H7) to derive Oligocortical spheroids | NIH hESC registry | NIHhESC-10-0061; RRID:CV-CL_9772 |
| Experimental Models: Organisms/Strains | ||
| Mouse, Male: 129S/SvEv E3.5 for EpiSC isolation | Jackson Laboratory | RRID:MGI:3050593 |
| Mouse, mixed sex, postnatal day 2 and 7: C57BL/6J | Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primer: sgVHL Forward: TCCCTACACGACGCTCTTCCGATCTCTCTCAGGTCATCTTCTGCAACC | This manuscript | IDT |
| Primer: sgVHL Reverse: AGTTCAGACGTGTGCTCTTCCGATCTGACAAGATGCTCGGGGTCGG | This manuscript | IDT |
| Primer: sgVHL.2 Forward: TCCCTACACGACGCTCTTCCGATCTAATAAACAGGTGCCATGCCC | This manuscript | IDT |
| Primer: sgVHL.2 Reverse: AGTTCAGACGTGTGCTCTTCCGATCTAGATTGACTATTAACCTGGCAATG | This manuscript | IDT |
| sgRNAs | See methods | IDT |
| Taqman probes | See methods | IDT |
| Recombinant DNA | ||
| CRISPRv2 constructs | Sanjana et al., 2014 | Addgene plasmid #52961; RRID:Addgene_52961 |
| lentiSAMv2 constructs | Joung et al., 2017 | Addgene plasmid #75112; RRID:Addgene_75112 |
| Activation helper | Joung et al., 2017 | Addgene plasmid #89308; RRID:Addgene_89308 |
| N-terminus tagged 3x HA Ascl2 construct | Genecopoeia | Cat# EX-Mm03774-Lv117 |
| Software and Algorithms | ||
| Columbus Image Data Storage and Analysis System | Perkin Elmer | www.perkinelmer.com/Product/columbus |
| Outknocker | Schmid-Burgk et al., 2014 | http://www.outknocker.org/outknocker2.htm |
| Graphpad Prism | Graphpad Software | https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
| Image Studio Acquisition software | LI-COR | LI-COR Image Studio Software; RRID:SCR_015795 |
| Salmon 0.14.1 | Patro et al. 2017 | https://github.com/COMBINE-lab/salmon |
| DESEQ2 | Love et al. 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html; RRID:SCR_015687 |
| Metascape | Zhou et al. 2019 | http://metascape.org/gp/index.html#/main/step1; RRID:SCR_016620 |
| GSEA | Subramanian et al. 2005 | https://www.gsea-msigdb.org/gsea/index.jsp; RRID:SCR_003199 |
| Trim Galore! Version 0.4.1 | Krueger, F. 2015 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; RRID:SCR_011847 |
| Bowtie2 Version 2.3.2 | Langmead and Slazberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml; RRID:SCR_005476 |
| MACs Version 2.1.1 | Feng et al. 2012 | http://liulab.dfci.harvard.edu/MACS/; RRID:SCR_013291 |
| Integrative Genomics Viewer | Broad Institute | http://www.broadinstitute.org/igv/; RRID:SCR_011793 |
| Galaxy | Afgan et al. 2018 | http://galaxyproject.org/; RRID:SCR_006281 |
| Diffbind software | Ross-Innes et al. 2012 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html; RRID:SCR_012918 |
| HOMERv4.11.1 | Heinz et al., 2010 | http://homer.ucsd.edu/; RRID:SCR_010881 |
| BEDTools Version 2.25.0 | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2; RRID:SCR_006646 |
| Deeptools Version 3.3.1 | Ramirez et al., 2016 | https://deeptools.readthedocs.io/en/develop/; RRID:SCR_016366 |
| Microsoft Excel | Microsoft | 2016 |
| Illustrator CC | Adobe | 2020 |
| Other | ||
| C-Chamber | BioSpherix | Cat#C-274 |
| 96-well plate | ThermoFisher | Cat#167008 |
| 384-well PDL-coated plate | PerkinElmer | Cat#6057500 |
| Operetta High Content Imaging and Analysis System | PerkinElmer | N/A |
| EL406 Microplate Washer Dispenser | BioTek | N/A |
| 5μl dispense cassette | BioTek | N/A |
| Janus automated workstation | PerkinElmer | N/A |
| Odyssey Fc Imaging System | LI-COR | N/A |
| 7300 real-time PCR system | Applied Biosystems | N/A |
| 5973 Mass Spectrometer | Agilent | N/A |
| Hamamatsu Nanozoomer S60 Slide Scanner | Hamamatsu | C13210-01 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | ThermoFisher | Cat#34580 |
| Halt Protease and Phosphatase Inhibitor Cocktail, EDTA-free (100x) | ThermoFisher | Cat#78441 |
| Nextera Tn5 Transposase | Illumina | Cat#FC-121-1030 |
| Covaris S2 Focused Ultrasonicator | Covaris | Cat#17015 |
| Amaxa Nucleofector 2b | Lonza | Cat#AAB-1001 |
| E12.5 heart HIF1a ChIP-Seq | Guimaraes-Camboa et al., 2015 | GSM1500750, GSM1500751, GSM1500749 |
| Melanocyte HIF1a ChIP-Seq | Loftus et al., 2017 | GSM2305570, GSM2305571, GSM2305572 |
| Th17 T-cell HIF1a ChIP-Seq | Ciofani et al., 2012 | GSM1004819, GSM1004991, GSM1004820, GSM1004993 |
| Mus musculus mm10 genome build | NCBI; Mouse Genome Sequencing Consotium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Culture of 293T Cells
293T cells (Takara Bioscience, 632180) were used for lentiviral production according to the vendor’s instructions and were cultured in the following medium: DMEM (Thermo Fisher, 11960–044) supplemented with 10% FBS (Fisher, A3160402), 1x MEM Non-essential amino acids (Thermo Fisher, 11140–050), 1x Glutamax (Thermo Fisher, 35050061), and 0.1mM 2-Mercaptoethanol (Sigma, M3148).
Pluripotent stem cell-derived OPC culture
Mouse OPCs were generated from epiblast stem cells (EpiSCs) as previously described except SHH was not used for maintenance or differentiation of OPCs (Najm et al., 2011). Mouse protocols were approved by Case Western Reserve University School of Medicine’s Institutional Animal Care and Use Committee (IACUC). In brief, epiblast stem cells (EpiSCs) were isolated from 129S/SvEv male embryos (E3.5; The Jackson Laboratory), pushed to form neural rosettes, and then rosettes were passaged into nunclon plates coated with poly-L-ornithine (Sigma, P3655–50MG) and laminin (Sigma, L2020–1MG) in OPC growth media consisting of DMEM/F12 supplemented with N2 Max (R&D Systems, AR009), B27 (Thermo Fisher, 12587010), 20ng/mL bFGF (R&D Systems, 23–3FB-01M), and 20ng/mL PDGFA (R&D Systems, 221-AA). Media was changed every 48 hours and cultures were maintained in 37°C with 5% CO2. After 4 passages these EpiSC derived OPCs were sorted to purity by fluorescence activated cell sorting using conjugated CD140a-APC (eBioscience, 17-1401-81; 1:80) and NG2-AF488 (Millipore, AB5320A4; 1:100) antibodies.
Primary mouse OPC culture
Primary mouse OPCs were derived using two methods. In the first method, cerebral cortices were harvested from postnatal day 2 (P2) C57BL/6J pups and dissociated using a Tumor Dissociation Kit (Miltenyi). Cells were then filtered through a 70 μm filter, washed in DMEM/F12, and plated on poly-ornithine (Sigma, P3655–50MG) and laminin (Sigma, L2020–1MG) coated plates to be expanded, passaged, and used in experiments. The second method follows the immunopanning protocol from P7 C57BL/6J mice (Barres et al., 1992). Primary in vivo derived OPCs using either method were pooled from multiple pups such that they are a combination of male and female cells. Similar to EpiSC-derived OPCs, primary OPCs were grown on Poly-L-ornithine and laminin coated flasks in OPC growth media. Media was changed every 48 hours.
Mouse OPC differentiation to oligodendrocytes and astrocytes
For oligodendrocyte generation, OPCs were seeded at either 40,000 cells per well (96-well plate, Fisher, 167008) or 15,000 cells per well (384-well PDL-coated cell carrier plates, PerkinElmer, 6057500) on plates coated with PO and laminin. Oligodendrocyte differentiation media consisted of DMEM/F12 supplemented with N2 Max, B27, 100ng/mL noggin (R&D, 3344NG050), 100ng/mL IGF-1 (R&D, 291G1200), 10uM cyclic AMP (Sigma, D0260–100MG), 10ng/mL NT3 (R&D, 267N3025) and 40ng/mL T3 (Thyroid hormone, Sigma, T-6397). Cells were analyzed after 3 days unless otherwise noted.
For OPC differentiation to astrocytes, 30,000 OPCs were plated per well in a 96 well plate (Fisher, 167008) containing astrocyte differentiation media described in (Liddelow et al., 2017). This media consisted of a 1:1 (v/v) mixture of neurobasal media and high glucose DMEM supplemented with sodium pyruvate, glutamax, N2 Max, and N-acetyl-cysteine and with growth factors including 20ng/mL bFGF, 5ng/mL Hb-EGF (R&D, 259-HE-050), 10ng/mL CNTF (R&D, 557-NT-010), and 10ng/mL BMP4 (R&D, 314-BP-050) for 3 days.
Human oligocortical spheroid production
Human embryonic stem cells (line H7, WiCell) were grown in mTesR1 media and oligocortical spheroids were generated with minor modifications to the protocol previously described (Madhavan et al., 2018). Briefly, in the first step of generating oligocortical spheroids, CloneR (Stem Cell Technologies, 5889) was used instead of Y-27632 and Dorsomorphin was replaced with 150nM LDN193189 (Sigma, SML0559). Spheroids were treated with 150nM LDN193189 and 10μM SB-43152 (Sigma, S4317) for the first 6 days followed 20ng/ml FGF-2 (R&D Systems, 233-FB-25/CF) and 20ng/ml EGF (R&D Systems, 236-EG-200) from day 7 to 25.
This was followed by 10ng/ml BDNF (R&D Systems, 248-BD) and 20ng/ml NT-3 (R&D Systems, 267-N3) treatment every other day between days 27 and 40. For OPC development and oligodendrocyte differentiation cultures 10ng/ml PDGF-AA (R&D Systems, 221-AA) and 10ng/ml IGF (R&D Systems, 291-GF-200) were added to cultures every other day between days 51 and 60 an 40n/ml T3 (Sigma, T6397) every other day between days 61 and 70. Spheroids were treated every other day with vehicle DMSO or 300nM MEKi between days 70 and 74 and harvested on day 90. Spheroids were treated with 200μM Hypoxyprobe-1 two hours prior to harvesting for IHC (pimonidazole, Hypoxyprobe Inc, Burlington MA, HP1–100Kit).
Tissue from mild chronic hypoxia (MCH) mice
MCH is a well described model of Diffuse White Matter Injury (DWMI) (Clayton et al., 2017a; Clayton et al., 2017b; Fancy et al., 2011; Scafidi et al., 2014; Yuen et al., 2014) and protein samples were generously provided by Brian Popko. In brief, postnatal day 3 (P3) C57Bl6 pups were placed into a BioSpherix chamber maintained at 10+/− 0.5% O2 by displacement of nitrogen until P11. Animals were then quickly sacrificed by CO2 asphyxiation followed by decapitation and the frontal cortex was isolated as this region has been shown to contain subcortical white matter that is susceptible to hypoxia-induced DWMI (Clayton et al., 2017b; Jablonska et al., 2012; Sanjana et al., 2014; Yuen et al., 2014).
METHOD DETAILS
In vitro hypoxia experiments
OPCs were plated in OPC growth media and then placed into a 2 shelf C-Chamber from BioSpherix (C-274). Oxygen tension was controlled using the ProOx 110 from BioSpherix such that nitrogen gas would flush out oxygen to maintain the chamber at the desired oxygen level. The subchamber was set at 1% O2 and cells were cultured for 48 hours unless otherwise noted. Hypoxia treated cells were then rapidly lysed for RNA or protein to minimize degradation of HIFs upon exposure to room air. The normoxic controls were cultured concurrently in the same cell culture incubator containing the BioSpherix C-chamber.
Immunocytochemistry
For antigens requiring live staining (O1 and O4), antibodies were diluted in N2B27 base media supplemented with 10% Donkey Serum (v/v) (Jackson ImmunoResearch, 017-000-121) and then added to cells for 18 minutes at 37°C. Cells were then fixed in cold 4% PFA (Electron microscopy sciences) for 18 minutes at room temperature, washed with PBS, and permeabilized and blocked in blocking solution containing 0.1% Triton X-100 in PBS supplemented with 10% normal donkey serum (v/v) for 30 minutes at room temperature. Primary antibodies were diluted in blocking solution and incubated with samples overnight at 4°C. Primary antibodies used included anti-OLIG2 (1.2μg/mL, Proteintech, 12999–1-AP), anti-MBP (1:100, Abcam, ab7349), anti-O1 (1:50, CCF Hybridoma Core), anti-O4 (1:100, CCF Hybridoma Core), anti-ASCL2 (1:10, EMD Millipore, MAB4417), anti-GFAP (1:5000, Dako, Z033401–2), anti-SOX6 (1:2000, abcam, 30455), anti-A2B5 (2μg/mL, Millipore, MAB312), and anti-NKX2–2 (1:200, DSHB, 74.5A5). The next day, cells were rinsed with PBS and incubated in blocking solution for one hour with the appropriate secondary antibody conjugated to an Alexa-Fluor (4μg/mL, Thermo Fisher) along with the nuclear stain DAPI (Sigma, 1ug/mL).
High content imaging and quantification
Both 96-well and 384-well plates were imaged using the Operetta High Content imaging and analysis system (PerkinElmer). For 96-well and 384-well plates, 8 fields and 5-fields were captured from each well at 20x magnification respectively. Images were analyzed with PerkinElmer Harmony and Columbus software as described previously (Hubler et al., 2018; Najm et al., 2015). In brief, nuclei of live cells were identified using a threshold for area of DAPI staining to exclude pyknotic nuclei or debris. To identify oligodendrocytes, each DAPI positive nucleus was expanded by 50% to determine potential intersection with staining of an oligodendrocyte marker (O4/O1/MBP) in a separate channel. Expanded nuclei that intersected O4/O1/MBP staining were scored as oligodendrocytes. Percentage of oligodendrocytes was then calculated by dividing the number of oligodendrocytes by total number of DAPI positive cells per image.
Generation of CRISPR and CRISPRA OPCs
Guide sequences were curated from the Brie library (Doench et al., 2016) and cloned into the CRISPRv2 backbone (Addgene 52961) (Sanjana et al., 2014) for generating CRISPR-mediated knockout OPCs. Guide sequences were curated from the CRISPRav2 library (Horlbeck et al., 2016) and cloned into the lenti-SAMv2 backbone (Addgene 75112) (Joung et al., 2017) for CRISPR activation. The activation helper plasmid (Addgene 89308) (Joung et al., 2017) was co-transduced for all CRISPR activation OPCs. Plasmids containing cloning sites for the sgRNA were digested and annealed oligomers containing the sgRNA were then ligated to their respective backbones. sgRNAs used in this study include: Cas9 control (empty-no sgRNA), sgVhl (GCCCGGTGGTAAGATCGGGT), sgVhl.2 (TGTCCATCGACATTGAGGGA), CRISPRA-NTC (GATCGGTTATGTTTAGGGTT), CRISPRA-Ascl2 (CCAAGTTTACCAGCTTTACG), CRISPRA-Dlx3 (GTTAGGGGTAACAACAAAGA), CRISPRA-Vegfa (GAGGCCGTGGACCCTGGTAA), CRISPRA-Slx16a3 (GCCTGGCCGCTGTTCCCCGA), and CRISPRA-Bnip3 (GGGGGAGCGTGTGGCACGTG).
Positive stbl3 bacterial clones (Thermo Fisher, C737303) were confirmed to have the correct inserted oligomer using Sanger sequencing. HEK293T cells were then transfected using lenti-X shots following the manufacturer’s protocol (Clonetech, 631276). The next day, transfection media was switched to OPC growth media without growth factors for virus collection. After 2 days, the media from transfected HEK293T cells was collected, filtered, and supplemented with OPC growth factors PDGF and FGF. This lentivirus-containing media was added to OPCs at a ratio of 1:2 (v/v) with fresh OPC growth media. After 24 hours of incubation with virus, transduced cells were switched to fresh OPC growth media and allowed to recover for 48 hours. OPCs were then selected for 96 hours in OPC growth media supplemented with a lethal dose of puromycin (500ng/mL, Thermo Fisher, A1113802) for CRISPR knockout cells or a lethal dose of blasticidin (10μg/mL, Thermo Fisher, A1113903) and hygromycin (100μg/mL, Thermo Fisher, 10687010) for CRISPR activation cells. OPCs were then allowed to recover for at least 24 hours following removal of selection and frozen down in aliquots for future use. For all experiments, the lentivirally transduced CRISPR/CRISPRA targeting and non-targeting control OPCs were derived from the same batch of OPCs and infected and selected simultaneously. qPCR was performed to validate a reduction or overexpression of gene targets of interest for each batch of CRISPR/CRISPRA OPCs generated.
Generation of HA-Ascl2 OPCs
The lentiviral HA-ASCL2 overexpression construct was obtained from Genecopoeia (EX-Mm03774-Lv117) and transformed into stbl3 bacteria (Thermo Fisher, C737303). Positive clones were selected on ampicillin agar plates and amplified for maxi-prep (Qiagen, 12163) to obtain sufficient quantities of plasmid. This construct was used to generate lentivirus and transduce OPCs following the same protocol used to generate CRISPR knockout OPCs using puromycin (500ng/mL, Thermo Fisher, A1113802) to select for infected OPCs.
Validating CRISPR knockout in OPCs
Primers were identified surrounding the target cut site for the two sgVhl constructs (see previous section for sgRNAs) that generate 200–250 base pair amplicons.
For the sgVhl cut site the primers were:
F 5’ TCCCTACACGACGCTCTTCCGATCTCTCTCAGGTCATCTTCTGCAACC 3’
R 5’ AGTTCAGACGTGTGCTCTTCCGATCTGACAAGATGCTCGGGGTCGG 3’
For the sgVhl.2 cut site the primers were:
F 5’ TCCCTACACGACGCTCTTCCGATCTAATAAACAGGTGCCATGCCC 3’
R 5’ AGTTCAGACGTGTGCTCTTCCGATCTAGATTGACTATTAACCTGGCAATG 3’
PCR products were run on an agarose gel, excised by gel extraction (28115, Qiagen), and submitted to the Case Western Reserve University Genomics core for library preparation and sequencing. Libraries were prepared by adding unique indices by PCR using KAPA HiFi HotStart ReadyMix. Samples were then pooled evenly, quantified using NEBNext® Library Quant Kit for Illumina® (New England 641 Biolabs, E7630), and denatured and diluted per Illumina’s MiSeq instructions. These finished libraries were then sequenced using an Illumina MiSeq (250bp paired-end). Results were analyzed using Outknocker software (Schmid-Burgk et al., 2014) (http://www.outknocker.org/outknocker2.htm) to calculate the percentage of reads with insertions or deletions at the sgRNA target site.
Kinetics of OPC differentiation to oligodendrocytes
This protocol was adapted from a previous study from our lab (Elitt et al., 2018). Control and CRISPR/CRISRPA OPCs were seeded at 40,000 cells per well in 96-well poly-ornithine and laminin coated plates into oligodendrocyte differentiation medium. At day 1, 2, and 3 post-plating, cells were live stained with mouse anti-O4 (CCF Hybridoma Core Facility) or mouse anti-O1 (CCF Hybridoma Core Facility) followed by fixation with 4% PFA. Cells were then stained using rat anti-MBP (1:100, Abcam, ab7349), followed by counterstaining with DAPI (1 μg/ml, Sigma, D8417). 6–8 fields were captured per well and the percentage of MBP+ cells were quantified relative to total cell number (DAPI) to control for any possible variations in plating density.
Compound screening and assessment
Compound screening was carried out as described in (Lager et al., 2018). Poly-D-lysine-coated 384-well CellCarrier Ultra plates (PerkinElmer) were coated with laminin diluted in N2B27 base media using an EL406 Microplate Washer Dispenser (BioTek) equipped with a 5μl dispense cassette (BioTek) and were incubated at 37°C for at least 1 hour. A 3mM stock of the Selleck bioactive library in dimethylsulfoxide (DMSO) was then added to the plates using a 50nL solid pin tool attached to a Janus automated workstation (Perkin Elmer) at a 1:1000 dilution such that each well received a single compound at a final concentration of 3μM. Compounds for dose response validation were sourced from the Selleck library, except for ERK1/2 inhibitors SCH772984 (Selleck, S7101), AZD0364 (Selleck, S8708), and VX-11e (Selleck, S7709) and MEK5 inhibitors BIX02188 (Selleck, S1530) and BIX02189 (Selleck, S1531) which were purchased separately. OPCs were dispensed in oligodendrocyte differentiation media at 12,500 cells per well into the laminin-coated 384 well plates using the BioTek EL406 Microplate Washer Dispenser and differentiated at 37°C for 3 days. At this point, cells were fixed, washed and stained using the BioTek EL406 Microplate Washer Dispenser. Cells were stained with anti-MBP (1:100, Abcam, ab7349) along with DAPI (1 μg/ml, Sigma, D8417) and imaged using the Operetta High Content Imaging and Analysis system (PerkinElmer).
Western blot
For cell culture derived protein samples, at least 1 million OPCs were collected and lysed in RIPA buffer (Sigma, R0278) supplemented with protease and phosphatase inhibitor (Thermo Fisher, 78441) for at least 15 minutes and cleared by centrifugation at 13,000g at 4°C for 15 minutes. Protein concentrations were determined using the Bradford assay (Bio-Rad Laboratories). Protein was then diluted and added to Laemmli loading buffer, boiled at 95°C for 5 minutes, run using NuPAGE Bis-Tris gels (Thermo Fisher, NP0335BOX), and then transferred to PVDF membranes (Thermo Fisher, LC2002). Blocking and primary/secondary antibody solutions were performed for at least 30 min with 5% nonfat drug milk (Nestle Carnation) in TBS plus 0.1% Tween 20 (TBST). Primary antibodies used included anti-HIF1a (1:500, Abcam, ab2185), anti-phospho-p44/42 (ERK1/2) (100ng/mL, CST, 9101), anti-p44/42 (total ERK1/2) (100ng/mL, CST, 4696), anti-DLX3 (2μg/mL, abcam, ab178428), anti-SOX10 (1:100, R&D, AF2864), anti-B-Actin peroxidase (1:50,000, Sigma, A3854), anti-VHL (5μg/mL, BD Biosciences, 556347), anti-ASCL2 (1:10, EMD Millipore, MAB4417), anti-MBP (1μg/mL, Biolegend, 808401), anti-OLIG2 (100ng/mL, Millipore, MABN50), anti-HA (200ng/mL, Cell Signaling Technologies, 3724S), and anti-MAG (1μg/mL, Thermo Fisher, 346200). Membranes were then imaged using the Odyssey Fc Imaging System (LI-COR) and analyzed using Image Studio™ software that is integrated into the Odyssey imaging suite. Westerns were normalized to loading control Beta-Actin unless otherwise noted.
Co-immunoprecipitation
For immunoprecipitation, 15 million OPCs were lysed in co-IP buffer (50mM Tris-HCL, pH 7.4, 120mM NaCl, 1mM EDTA, 1% Nonidet P-40 with Complete EDTA-free protease inhibitor tablet) for 15 minutes with intermittent vortexing. Lysate was cleared by centrifugation at 16,000g at 4°C for 15 minutes. The supernatant was divided into three groups for input, immunoprecipitation, and IgG control. Samples were rotated at 4°C for 2 hours with 10uL of Protein G magnetic Dynabeads (Thermo Fisher, 10004D) to remove off target binding. Beads were removed by magnet. The supernatant of the input sample was stored at −80°C while the supernatant of the immunoprecipitation and IgG samples were combined with 15μg of HIF1a antibody (abcam, 2185), 10μg of OLIG2 antibody (Millipore, ab9610), or 2μg of normal rabbit IgG antibody (Cell signaling technologies, 2729S) and rotated overnight at 4°C. The following day, 20μL of protein G beads were added to the mixture and rotated overnight at 4°C. Samples were placed on a magnetic bar to remove supernatant, and beads were washed with 1mL of co-IP buffer 6 times. On the final wash, all supernatant was removed and 50uL of Laemmeli buffer was added. Sample was heated at 95°C for 5 minutes and collected to run for Western blot analysis.
qRT-PCR
At least 500,000 OPCs were lysed in TRIzol (Ambion) followed by phenol-chloroform extraction and processing with the RNeasy Mini Kit (Qiagen, 74104). RNA quality and quantity was determined using a NanoDrop spectrophotometer. cDNA was generated using the iSCRIPT kit following the manufacturer’s instructions (Biorad, 1708891). qRT-PCR was performed using pre-designed TaqMan gene expression assays (Thermo Fisher, 4369016). qPCR was performed using the Applied Biosystems 7300 real-time PCR system and probes were normalized to Rpl13a endogenous control. Taqman probe IDs and cat numbers used in the study include: Sox10 (Mm00569909_m1), Rpl13a (Mm05910660_g1), Vhl (Mm00494137_m1), Ascl2 (Mm01268891_g1), Tpi1 (Mm00833691_g1), Ciart (Mm01255905_g1), P4ha2 (Mm00477940_m1), Pdk1 (Mm00554300_m1), Pfkl (Mm00435587_m1), Slc16a3 (Mm00446102_m1), Dlx3 (Mm00438428_m1), Vegfa (Mm00437306_m1), Bnip3 (Mm01275600_g1), Pdgfra (Mm00440701_m1), Plp1 (Mm01297210_m1), and Myrf (Mm01194959_m1). As the taqman probe for Ascl2 bind regions outside of the Ascl2 cDNA sequence, SYBR qPCR (Kappa Biosystems, KK4605) was performed to verify the increase in Ascl2 expression in HA-ASCL2 OPCs using Rpl13a as a loading control with the following primers: Ascl2 (F: ATGGAAGCACACCTTGACTG, R: AGTGGACGTTTGCACCTTCA) Rpl13a (F: TTCTCCTCCAGAGTGGCTGT, R: GGCTGAAGCCTACCAGAAAG).
Cell viability assay
OPCs were seeded into 96 well plates at 25,000 cells/well (96-well plate, Fisher, 167008) and recovered for 24 hours. Cell viability was assessed using an MTS assay kit (abcam, ab197010) following the manufacturer’s protocol. Colorimetric measurements were recorded 9 hours after the addition of the MTS dye solution to the OPCs.
siRNA knockdown in OPCs
siRNAs were purchased from Horizon/Dharmacon (Ascl2: L-047262-01-0005, Dlx3: L-041957-01-0005, Non-Targeting Control: D-001206-14-05 5 nmol) and nucleofected into control and sgVhl OPCs using the Basic Nucleofector Kit for Primary Mammalian Glial Cells (Lonza, VPI-1006) following manufacturer’s instructions. In brief, 5 million OPCs were resuspended in nucleofection solution with 0.6μl of the desired 50μM siRNA solution, nucleofected using the Amaxa Nucleofector 2b (Lonza, AAB-1001) using the A033 setting, and then plated in differentiation media supplemented with penicillin-streptomycin (Thermo Fisher, 15070063) for 72 hours. Leftover OPCs were plated in growth media supplemented with penicillin-streptomycin (Thermo Fisher, 15070063) for 48 hours and then lysed with TRIzol (Ambion, 15596026) for RNA extraction and qPCR to assess knockdown efficiency.
RNA-seq sample preparation and analysis
At least 1 million OPCs were lysed in TRIzol and RNA was isolated as described for qPCR. Libraries were prepared following protocols from NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, E7490L) and NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, E7530L). In brief, samples were enriched for mRNA using oligo(dT) beads, which were then fragmented randomly and used for cDNA generation and subsequent second-strand synthesis using a custom second-strand synthesis buffer (Illumina), dNTPs, RNase H and DNA polymerase I. cDNA libraries then went through terminal repair, A-base ligation, adapter ligation, size selection and PCR enrichment. Final libraries were pooled evenly and sequenced on the Illumina NovaSeq with paired-end 150bp reads with a read-depth of at least 20 million reads per sample.
For gene expression analysis, reads were aligned to the mm10 genome build using salmon 0.14.1 (https://github.com/COMBINE-lab/salmon) (Patro et al., 2017) to quantify transcript abundance in transcripts per million (TPM) values. Transcripts were summarized as gene-level TPM abundances with tximport. A gene with TPM>1 was considered expressed. Differential expression analysis was then performed using DESEQ2 (https://bioconductor.org/packages/release/bioc/html/DESeq2.html) (Love et al., 2014). Significant genes were called based on p-adj and fold change values as described in the results section.
Violin plots
For violin plots in Figures 2, 6 and S2, RNAseq replicates were divided by the average of the respective control (TPM) on a per gene basis in each category and then all individual replicates were plotted together. For Figure S2, the average expression for each gene in each category in sgVhl OPCs was divided by the average expression in control OPCs. In all cases the genes in each category were limited to those that were expressed in sgVhl OPCs (TPM>1) and all TPM values were floored to 1.
Gene ontology analysis and heatmaps
Metascape (http://metascape.org/) (Zhou et al., 2019) was used to identify significant pathways from RNAseq data. Glycolysis genes used to construct the heatmap in Figure 2F were taken from Panther (“Glycolysis: P00024”). In Figure 6F, RNA-seq data was fit to the genes within “Oligodendrocyte Development” (GO: 0014003) that were downregulated in sgVhl OPCs compared to Cas9 control OPCs (fold change less than 0.75).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) scores were generated for gene sets in C5.bp datasets using classic scoring, 1000 gene-set permutations, and signal-to-noise metrics. Normalized enrichment scores, false discovery rate and FWER o-values were all calculated by GSEA software (https://www.gsea-msigdb.org/gsea/index.jsp) (Subramanian et al., 2005).
ChIP-seq and analysis
Nuclei isolation and chromatin shearing were performed using the Covaris TruChIP protocol following manufacturer’s instructions for the “high-cell” format. In brief, 5–20 million cells (H3K27Ac) or 100 million cells (HIF1a) were crosslinked in “Fixing buffer A” supplemented with 1% fresh formaldehyde for 10 minutes at room temperature with oscillation and quenched for 5 minutes with “Quench buffer E.” These cells were then washed with PBS and either snap frozen and stored at −80°C, or immediately used for nuclei extraction and shearing per the manufacturer protocol. The samples were sonicated with the Covaris S2 using the following settings: 5% Duty factor 4 intensity for four 60-second cycles. Sheared chromatin was cleared and incubated overnight at 4 degrees with primary antibodies that were pre-incubated with protein G magnetic DynaBeads (Thermo Fisher, 10004D). Primary antibodies used included anti-H3K27Ac (9μg/sample, Abcam, ab4729), anti-HIF1a (25μg/sample, Abcam, ab2185), anti-Olig2 (15μg/samples, Millipore, ab9610), and anti-HA (15μg/sample, cell signaling technologies, 3724S). These beads were then washed, eluted, reverse cross-linked and treated with RNase A followed by proteinase K. ChIP DNA was purified using Ampure XP beads (Aline Biosciences, C-1003–5) and then used to prepare Illumina sequencing libraries as described previously (Schmidt et al., 2009). Libraries were sequenced on the Illumina HiSeq2500 with single-end 50bp reads with a read-depth of at least 20 million reads per sample.
For peak calling, reads were quality and adapter trimmed using Trim Galore! Version 0.4.1. Trimmed reads were aligned to mm10 with Bowtie2 version 2.3.2 (Langmead and Salzberg, 2012). Duplicate reads (potential artifacts of PCR in library preparation) were removed using Picard MarkDuplicates. Peaks were called with MACS version 2.1.1 (Feng et al., 2012) to define broad peaks for histone marks (H3K27Ac) and narrow peaks for transcription factors (HIF1a), OLIG2, and HA-ASCL2 and normalized to background genomic DNA with matched inputs. Thresholding was set at FDR<0.001 for calling both H3K27Ac, HIF1a, and OLIG2 peaks and was set at FDR<0.1 for calling HA-ASCL2 peaks. Peaks were visualized with the Integrative Genomics Viewer (IGV, Broad Institute). Peaks were assigned to the nearest expressed gene (TPM>1 in Cas9 control or sgVhl OPCs) using bedtools available in Galaxy (https://usegalaxy.org) (Afgan et al., 2018).
Omni ATAC-seq
Omni ATAC-Seq was performed on 50,000 OPCs in two independent biological replicates following the protocol outlined in (Corces et al., 2017). In brief, nuclei were extracted from cells and treated with transposition mixture containing Nextera Tn5 Transposase for (Illumina, FC-121–1030). Transposed fragments were then purified using Qiagen MinElute columns (Qiagen, 28004), PCR amplified, and libraries were purified with Agencourt AMPure XP magnetic beads (Aline Biosciences, C-1003–5) with a sample to bead ratio of 1:1.2. Final libraries were sequenced on the illumine HiSeq2500 with single-end 50bp reads with nearly 100 million reads per sample. Reads were aligned to the mm10 mouse genome following the same pipeline used for ChIP-seq data (see ChIP-seq and analysis) and peaks were called using the “narrowPeaks” function of MACS version 2.1.1 as outlined for HIF1a ChIP-seq (see ChIP-seq and analysis).
Diffbind analysis
H3K27Ac and Olig2 ChIP-seqs were performed in duplicate from two independent batches of Cas9 control and sgVhl OPCs. Differential H3K27Ac and Olig2 analysis between sgVhl and Cas9 control OPCs was performed using “Diffbind software” (https://bioconductor.org/packages/release/bioc/html/DiffBind.html) (Ross-Innes et al., 2012). A false discovery rate of 0.1 was used to call significantly enriched and depleted regions of H3K27Ac and 0.001 for Olig2.
HIF1a ChIP-seq overlap analysis
HIF1a ChIP-seq raw data were re-analyzed for: two HIF1a replicates ChIP-seqs and 1 input of E12.5 heart (GSM1500750, GSM150075, GSM1500749) (Guimaraes-Camboa et al., 2015), two HIF1a replicates ChIP-seqs and 1 input of melanocyte (GSM2305570, GSM2305571, GSM2305572) (Loftus et al., 2017), and two HIF1a ChIP-seqs and 2 inputs of Th17 cells (GSM1004819, GSM1004991, GSM1004820, GSM1004993) (Ciofani et al., 2012). For each of the HIF1a ChIP-seqs, the peaks were called as described above (MACS2, narrow peak, FDR<0.001). Peaks unique to OPCs were identified by finding peaks not found in the union of all the replicates of every other tissue sample using the bedtools (v 2.25.0) (Quinlan and Hall, 2010) “intersect” command. Common peaks across tissues were also identified using the bedtools “intersect”. RPCG-normed bigwigs were used to create aggregate plots for HIF1a, ATAC-seq, and H3K27ac using the deeptools (version 3.3.1) (Ramirez et al., 2016) “computeMatrix” and “plotHeatmap” commands, centering on the HIF1a peak locations in the sgVhl OPCs. Genes within 5Kb of these HIF1a peaks were called as HIF1a target genes. For melanocytes and T-cells, HIF1a target genes were called as only those at the intersection of both of the available replicates. For embryonic heart and OPCs, there were fewer peaks called compared to the other tissues, so HIF1a gene targets were called as the union of the two available replicates. These gene and peak lists were used to generate venn diagrams using the following webtool: (http://bioinformatics.psb.ugent.be/webtools/Venn/). Cell type specific gene targets of HIF1a were called if the gene was only a target of HIF1a in that specific cell type (see Table S2).
Motif enrichment analysis
Motifs were called under significant HIF1a peaks (FDR<0.001), OLIG2 peaks (FDR<0.001), and HA-ASCL2 peaks (FDR<0.1) or regions of significantly gained H3K27Ac (FDR<0.1) or OLIG2 (FDR<0.05) using HOMERv4.11.1 (Heinz et al., 2010). The FindMotifsGenome.pl tool was used with mm10 as the reference genome. Known motifs were compiled into supplementary Table S3 and Table S4 and ranked based on p-value. To calculate enrichment of motifs under tissue-specific HIF1a peaks, the percentile of the rank for each motif from HOMER was calculated in all four tissue types (OPCs, heart, melanocytes, and T cells). The average percentile for a motif in the three other tissues (heart, melanocyte, and T cells) was then subtracted from the percentile of the same motif in OPCs. The magnitude of the difference was then ranked and filtered for transcription factors expressed in OPCs to obtain the top motifs enriched under OPC-specific HIF1a peaks compared to other tissue types.
Mass spectrometry for lactate and pyruvate
OPCs were cultured in OPC growth media and after growth media was removed, cells were washed with ice-cold saline (3 times) and collected with 1 ml of 80% ethanol pre-chilled on dry ice. Cells were frozen at −80°C until analyses. This cell extract was then vortexed and sonicated for 30 seconds on, 30 seconds off, alternating for 10 minutes. Next, cells were pelleted by centrifugation at 4°C for 10 min at 14,000 rpm. Supernatant was transferred to GC/MS vials and evaporated to dryness under gentle stream of nitrogen. Keto- and aldehyde groups were reduced by addition of 10 μl of 1 N NaOH plus 15 μl NaB2H4 (prepared as 10 mg/ml in 50 mM NaOH). After mixing, samples were incubated at room temperature for 1 hour and then acidified by 55 μl of 1 N HCl by dropping the acid slowly. Next, samples were evaporated to dryness. Next, 50 μl of methanol were added to precipitate boric acid. Internal standard was added (10 μl of 17:0 FA, 0.1 mg/ml). Samples were evaporated to dryness and reacted with 40 μl of pyridine and 60 μl of tert-butylbis(dimethylsilyl) trifluoroacetamide with 10% trimethylchlorosilane (Regisil) TBDMS at 60°C for 1 hour. Resulting TBDMS derivatives were injected into GC/MS. Analyses were then carried out on an Agilent 5973 mass spectrometer equipped with 6890 Gas Chromatograph. A HP-5MS capillary column (60 m × 0.25 mm × 0.25 μm) was used in all assays with a helium flow of 1 ml/min. Samples were analyzed in Selected Ion Monitoring (SIM) mode using electron impact ionization (EI). Ion dwell time was set to 10 msec. Lactate and pyruvate were both measured for all samples.
Oligocortical spheroid immunohistochemistry
Oligocortical spheroids were treated every other day with DMSO or 300nM AZD8330 between days 70 and 74 and harvested on day 90. 2 hours prior to harvesting, spheroids were treated with 200uM Hypoxyprobe-1 (pimonidazole, Hypoxyprobe Inc, Burlington MA, HP1–100Kit). Spheroids were washed in PBS and fixed overnight in ice cold 4% Paraformaldehyde (Electron Microscopy Sciences, 15710) and then washed in PBS and cryoprotected in a 30% sucrose solution. Spheroids were frozen in OCT and sectioned at thickness of 15 μm. Slides were washed in PBS and incubated overnight with anti-MyRF (1:1000, gift from Michael Wegner) and anti-pimonidazole antibodies (1:250, Hypoxyprobe Inc, Burlington MA, HP1–100Kit) followed by labelling with appropriate Alexa-Fluor labeled secondary antibodies (2μg/mL, Thermo Fisher). Images were captured using a Hamamatsu Nanozoomer S60 Slide scanner with NDP 2.0 software. Images spanning the edge to the central region of each oligocortical spheroid were used for analysis. SOX10+ cells and MyRF+ oligodendrocytes in Hypoxyprobe positive and Hypoxyprobe negative areas within each image were quantified using ImageJ software.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism was used to perform statistical analyses unless otherwise noted. Statistical tests and replicate descriptions are detailed in each figure legend. In brief, black filled-in circles for bar graphs indicate biological replicates whereas open circles represent technical replicates.
Statistics were only performed on samples with biological replicates. Data is typically graphed as mean ± standard deviation (SD) or ± standard error of the mean (SEM) as detailed in the figure legend. A p-value less than 0.05 was considered significant unless otherwise noted.
Supplementary Material
Table S1. Top Functional Targets of HIF1a in OPCs, Related to Figure 2 and 4.
Overlap of genes with increased expression by RNA-seq, increased active chromatin by H3K27ac ChIP-seq and bound by HIF1a by HIF1a ChIP-seq reveals top functional targets of HIF1a in OPCs.
Table S2. Identification of Shared Canonical and Cell-Type-Specific HIF1a Peaks and Gene Targets, Related to Figure 2 and 3.
Overlap of OPC HIF1a gene targets with publicly available HIF1a ChIP-seq datasets in embryonic heart, T-cells, and melanocytes reveals canonical HIF1a target genes and peaks shared across all 4 cell types as well as target genes and peaks specific to each cell type.
Table S3. HOMER Motif Analysis Results, Related to Figure 2 and 3.
HOMER motif analysis was performed under HIF1a peaks (FDR<0.001) in sgVhl OPCs, peaks shared between cell types, and peaks that are cell-type specific. HOMER motif analysis was also performed under regions with significantly increased H3K27Ac (FDR<0.1) in sgVhl OPCs compared to Cas9 control OPCs.
Table S4. OLIG2 Binding Increases at Hypoxia Genes in sgVhl OPCs, Related to Figure 3.
HOMER motif analysis was performed under OLIG2 peaks (FDR<0.001) in Cas9 control and sgVhl OPCs and under OLIG2 peaks that were significantly increased (FDR<0.05, log2FC>1) in sgVhl OPCs compared to Cas9 control OPCs. Peak file and closest genes (within 50 Kb) for these increased OLIG2 peaks are listed.
Table S5. Primary Compound Screening and Filtering, Related to Figure 5.
Primary bioactive screening data showing the impact of 1753 compounds on total cell number (DAPI), number of oligodendrocytes (MBP+), and percentage of oligodendrocytes (MBP+/DAPI). To uncover top hits, compounds were first selected if they were considered nontoxic (DAPI > 0.7 fold change) and then selected if they increased oligodendrocyte formation (Oligodendrocyte percentage and number > 3 fold change and pass visual inspection).
Table S6. Validation Dose Curve of Top Compound Hits, Related to Figure 5.
Raw and normalized data for the percentage (MBP+/DAPI) of oligodendrocytes formed for all 3 replicates of the 8-point dose curve of the top 14 hits from the primary bioactive screen. This data was used to generate the heatmap presented in Figure 5D.
Highlights.
HIF1a activates both canonical hypoxia-response and unique OPC-specific genes
OLIG2 physically interacts with HIF1a and is enriched at OPC-specific HIF1a targets
OPC-specific HIF1a targets suppress Sox10 to impair oligodendrocyte formation
MEK inhibition increases Sox10 expression to rescue OPC function from hypoxia
ACKNOWLEDGEMENTS
This research was supported by grants from the National Institutes of Health F30HD096784 (K.C.A), T32NS077888 (K.C.A.), and T32GM007250 (K.C.A) and the New York Stem Cell Foundation (P.J.T. and M.A.S.) as well as institutional support from CWRU School of Medicine and philanthropic support from the Enrile, Peterson, Fakhouri, Long, Goodman, Geller, and Weidenthal families. Additional support was provided by the Small Molecule Drug Development and Genomics core facilities of the CWRU Comprehensive Cancer Center (P30CA043703), the CWRU Light Microscopy Imaging Center (S10-OD016164), and the University of Chicago Genomics Facility. The authors thank Brian Popko for tissue samples (obtained with funding from NIH grant R01NS034939). We are also grateful to C. Baecher-Allan, J. LaManna, K. Xu, D. Adams, Y. Federov, L. Barbar, K. McColl, D. Ramirez, E. Prendergast, E. Schwarz, P. Scacheri, D. Neu, W. Pontius, A. Saiakhova, Z. Nevin, M. Elitt, G. Beutler, J. Kerschner, and R. Sallari for technical assistance and/or discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
P.J.T. and M.M. are listed as inventors on pending patent claims filed by CWRU covering methods of generating oligocortical spheroids. P.J.T. is a co-founder and consultant for Convelo Therapeutics, which has licensed these claims and other patents unrelated to the current study. P.J.T. and CWRU retain equity in Convelo Therapeutics. All other authors have no competing interests.
REFERENCES
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, et al. (2018). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 46, W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas D, Bourbeau D, Sarlieve LL, Ittel ME, Dussault JH, and Puymirat J (1997). Oligodendrocyte maturation and progenitor cell proliferation are independently regulated by thyroid hormone. Glia 19, 324–332. [DOI] [PubMed] [Google Scholar]
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD, and Raff MC (1992). Cell death and control of cell survival in the oligodendrocyte lineage. Cell 70, 31–46. [DOI] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, and Raff MC (1994). A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development 120, 1097–1108. [DOI] [PubMed] [Google Scholar]
- Bartlett H, Veenstra GJ, and Weeks DL (2010). Examining the cardiac NK-2 genes in early heart development. Pediatr Cardiol 31, 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawner AT, Xu R, Liu D, and Jiang P (2017). Generating CNS organoids from human induced pluripotent stem cells for modeling neurological disorders. Int J Physiol Pathophysiol Pharmacol 9, 101–111. [PMC free article] [PubMed] [Google Scholar]
- Cassavaugh J, and Lounsbury KM (2011). Hypoxia-mediated biological control. J Cell Biochem 112, 735–744. [DOI] [PubMed] [Google Scholar]
- Chang KJ, Redmond SA, and Chan JR (2016). Remodeling myelination: implications for mechanisms of neural plasticity. Nat Neurosci 19, 190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry H, and Harris AL (2018). Advances in Hypoxia-Inducible Factor Biology. Cell Metab 27, 281–298. [DOI] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkhurst CN, Muratet M, et al. (2012). A validated regulatory network for Th17 cell specification. Cell 151, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton BL, Huang A, Dukala D, Soliven B, and Popko B (2017a). Neonatal Hypoxia Results in Peripheral Nerve Abnormalities. Am J Pathol 187, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton BL, Huang A, Kunjamma RB, Solanki A, and Popko B (2017b). The integrated stress response in hypoxia-induced diffuse white matter injury. J Neurosci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. (2017). An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nature methods 14, 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107, 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dengler VL, Galbraith M, and Espinosa JM (2014). Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol 49, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Jo J, Wang CY, Cristobal CD, Zuo Z, Ye Q, Wirianto M, Lindeke-Myers A, Choi JM, Mohila CA, et al. (2020). The Daam2-VHL-Nedd4 axis governs developmental and regenerative oligodendrocyte differentiation. Genes Dev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elitt MS, Shick HE, Madhavan M, Allan KC, Clayton BLL, Weng C, Miller TE, Factor DC, Barbar L, Nawash BS, et al. (2018). Chemical Screening Identifies Enhancers of Mutant Oligodendrocyte Survival and Unmasks a Distinct Pathological Phase in Pelizaeus-Merzbacher Disease. Stem Cell Reports 11, 711–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B (2010). Regulation of oligodendrocyte differentiation and myelination. Science 330, 779–782. [DOI] [PubMed] [Google Scholar]
- Fancy SP, Harrington EP, Yuen TJ, Silbereis JC, Zhao C, Baranzini SE, Bruce CC, Otero JJ, Huang EJ, Nusse R, et al. (2011). Axin2 as regulatory and therapeutic target in newborn brain injury and remyelination. Nat Neurosci 14, 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, and Liu XS (2012). Identifying ChIP-seq enrichment using MACS. Nat Protoc 7, 1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fufa TD, Baxter LL, Wedel JC, Gildea DE, Program NCS, Loftus SK, and Pavan WJ (2019). MEK inhibition remodels the active chromatin landscape and induces SOX10 genomic recruitment in BRAF(V600E) mutant melanoma cells. Epigenetics Chromatin 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao FB, Apperly J, and Raff M (1998). Cell-intrinsic timers and thyroid hormone regulate the probability of cell-cycle withdrawal and differentiation of oligodendrocyte precursor cells. Dev Biol 197, 54–66. [DOI] [PubMed] [Google Scholar]
- Giandomenico SL, and Lancaster MA (2017). Probing human brain evolution and development in organoids. Curr Opin Cell Biol 44, 36–43. [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Hofer MP, Syed YA, Amaral AI, Rundle J, Rahman S, Zhao C, and Kotter MRN (2016). Tamoxifen accelerates the repair of demyelinated lesions in the central nervous system. Sci Rep 6, 31599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann U, Reynolds R, Steck AJ, and Schaeren-Wiemers N (2003). Molecular changes in normal appearing white matter in multiple sclerosis are characteristic of neuroprotective mechanisms against hypoxic insult. Brain Pathol 13, 554–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinspan JB, Edell E, Carpio DF, Beesley JS, Lavy L, Pleasure D, and Golden JA (2000). Stage-specific effects of bone morphogenetic proteins on the oligodendrocyte lineage. J Neurobiol 43, 1–17. [PubMed] [Google Scholar]
- Guimaraes-Camboa N, Stowe J, Aneas I, Sakabe N, Cattaneo P, Henderson L, Kilberg MS, Johnson RS, Chen J, McCulloch AD, et al. (2015). HIF1alpha Represses Cell Stress Pathways to Allow Proliferation of Hypoxic Fetal Cardiomyocytes. Dev Cell 33, 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase VH (2009). The VHL tumor suppressor: master regulator of HIF. Curr Pharm Des 15, 3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankey GJ (2017). Stroke. Lancet 389, 641–654. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, et al. (2016). Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LE (2008). Carrot and stick: HIF-alpha engages c-Myc in hypoxic adaptation. Cell Death Differ 15, 672–677. [DOI] [PubMed] [Google Scholar]
- Hubler Z, Allimuthu D, Bederman I, Elitt MS, Madhavan M, Allan KC, Shick HE, Garrison E, M TK, Factor DC, et al. (2018). Accumulation of 8,9-unsaturated sterols drives oligodendrocyte formation and remyelination. Nature 560, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Fyffe-Maricich SL, Furusho M, Miller RH, and Bansal R (2012). ERK1/ERK2 MAPK signaling is required to increase myelin thickness independent of oligodendrocyte differentiation and initiation of myelination. J Neurosci 32, 8855–8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, and Kaelin WG Jr. (2001). HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. (2001). Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472. [DOI] [PubMed] [Google Scholar]
- Jablonska B, Gierdalski M, Chew LJ, Hawley T, Catron M, Lichauco A, Cabrera-Luque J, Yuen T, Rowitch D, and Gallo V (2016). Sirt1 regulates glial progenitor proliferation and regeneration in white matter after neonatal brain injury. Nat Commun 7, 13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonska B, Scafidi J, Aguirre A, Vaccarino F, Nguyen V, Borok E, Horvath TL, Rowitch DH, and Gallo V (2012). Oligodendrocyte regeneration after neonatal hypoxia requires FoxO1-mediated p27Kip1 expression. J Neurosci 32, 14775–14793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, and Zhang F (2017). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat Protoc 12, 828–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr., and Ratcliffe PJ (2008). Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30, 393–402. [DOI] [PubMed] [Google Scholar]
- Kaluz S, Kaluzova M, and Stanbridge EJ (2003). Expression of the hypoxia marker carbonic anhydrase IX is critically dependent on SP1 activity. Identification of a novel type of hypoxia-responsive enhancer. Cancer Res 63, 917–922. [PubMed] [Google Scholar]
- Kullmann JA, Trivedi N, Howell D, Laumonnerie C, Nguyen V, Banerjee SS, Stabley DR, Shirinifard A, Rowitch DH, and Solecki DJ (2020). Oxygen Tension and the VHL-Hif1alpha Pathway Determine Onset of Neuronal Polarization and Cerebellar Germinal Zone Exit. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupferschmidt K (2019). Cellular oxygen sensor system earns Nobel for trio. Science 366, 167. [DOI] [PubMed] [Google Scholar]
- Lager AM, Corradin OG, Cregg JM, Elitt MS, Shick HE, Clayton BLL, Allan KC, Olsen HE, Madhavan M, and Tesar PJ (2018). Rapid functional genetics of the oligodendrocyte lineage using pluripotent stem cells. Nat Commun 9, 3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Ko J, Ju C, and Eltzschig HK (2019). Hypoxia signaling in human diseases and therapeutic targets. Exp Mol Med 51, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Kim JW, Osborne O, Oh DY, Sasik R, Schenk S, Chen A, Chung H, Murphy A, Watkins SM, et al. (2014). Increased adipocyte O2 consumption triggers HIF-1alpha, causing inflammation and insulin resistance in obesity. Cell 157, 1339–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, and Fisher DE (2006). MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 12, 406–414. [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Munch AE, Chung WS, Peterson TC, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus SK, Baxter LL, Cronin JC, Fufa TD, Program NCS, and Pavan WJ (2017). Hypoxia-induced HIF1alpha targets in melanocytes reveal a molecular profile associated with poor melanoma prognosis. Pigment Cell Melanoma Res 30, 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan M, Nevin ZS, Shick HE, Garrison E, Clarkson-Paredes C, Karl M, Clayton BLL, Factor DC, Allan KC, Barbar L, et al. (2018). Induction of myelinating oligodendrocytes in human cortical spheroids. Nature methods 15, 700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, and Simon MC (2010). Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell 40, 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin MA, and Carmichael ST (2018). Mechanisms of demyelination and remyelination in the young and aged brain following white matter stroke. Neurobiol Dis. [DOI] [PubMed] [Google Scholar]
- Mei F, Fancy SPJ, Shen YA, Niu J, Zhao C, Presley B, Miao E, Lee S, Mayoral SR, Redmond SA, et al. (2014). Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med 20, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez-Montes I, Escobar B, Palacios B, Gomez MJ, Izquierdo-Garcia JL, Flores L, Jimenez-Borreguero LJ, Aragones J, Ruiz-Cabello J, Torres M, et al. (2016). Myocardial VHL-HIF Signaling Controls an Embryonic Metabolic Switch Essential for Cardiac Maturation. Dev Cell 39, 724–739. [DOI] [PubMed] [Google Scholar]
- Miska J, Lee-Chang C, Rashidi A, Muroski ME, Chang AL, Lopez-Rosas A, Zhang P, Panek WK, Cordero A, Han Y, et al. (2019). HIF-1alpha Is a Metabolic Switch between Glycolytic-Driven Migration and Oxidative Phosphorylation-Driven Immunosuppression of Tregs in Glioblastoma. Cell Rep 27, 226–237 e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, and Young RA (2011). Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147, 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata K, Jadhav U, Madha S, van Es J, Dean J, Cavazza A, Wucherpfennig K, Michor F, Clevers H, and Shivdasani RA (2020). Ascl2-Dependent Cell Dedifferentiation Drives Regeneration of Ablated Intestinal Stem Cells. Cell Stem Cell 26, 377–390 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Kobayashi M, Koyasu S, Chow CCT, and Harada H (2019). HIF-1-Dependent Reprogramming of Glucose Metabolic Pathway of Cancer Cells and Its Therapeutic Significance. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Madhavan M, Zaremba A, Shick E, Karl RT, Factor DC, Miller TE, Nevin ZS, Kantor C, Sargent A, et al. (2015). Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo. Nature 522, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Zaremba A, Caprariello AV, Nayak S, Freundt EC, Scacheri PC, Miller RH, and Tesar PJ (2011). Rapid and robust generation of functional oligodendrocyte progenitor cells from epiblast stem cells. Nature methods 8, 957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave KA (2010). Myelination and the trophic support of long axons. Nat Rev Neurosci 11, 275–283. [DOI] [PubMed] [Google Scholar]
- Newbern JM, Li X, Shoemaker SE, Zhou J, Zhong J, Wu Y, Bonder D, Hollenback S, Coppola G, Geschwind DH, et al. (2011). Specific functions for ERK/MAPK signaling during PNS development. Neuron 69, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue BW, Paulsen KD, O’Hara JA, Wilmot CM, and Swartz HM (2001). Estimation of oxygen distribution in RIF-1 tumors by diffusion model-based interpretation of pimonidazole hypoxia and eppendorf measurements. Radiat Res 155, 15–25. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner MP, von Teichman A, Nowicka A, Sulser T, Schraml P, and Moch H (2011). VHL gene mutations and their effects on hypoxia inducible factor HIFalpha: identification of potential driver and passenger mutations. Cancer Res 71, 5500–5511. [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmaso N, Jablonska B, Scafidi J, Vaccarino FM, and Gallo V (2014). Neurobiology of premature brain injury. Nat Neurosci 17, 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, and Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nature methods 11, 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafidi J, Hammond TR, Scafidi S, Ritter J, Jablonska B, Roncal M, Szigeti-Buck K, Coman D, Huang Y, McCarter RJ Jr., et al. (2014). Intranasal epidermal growth factor treatment rescues neonatal brain injury. Nature 506, 230–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, and Hornung V (2014). OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res 24, 1719–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, and Odom DT (2009). ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48, 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, and Mole DR (2011). High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117, e207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijers J, Junker JP, Mokry M, Hatzis P, Koo BK, Sasselli V, van der Flier LG, Cuppen E, van Oudenaarden A, and Clevers H (2015). Ascl2 acts as an R-spondin/Wnt-responsive switch to control stemness in intestinal crypts. Cell Stem Cell 16, 158–170. [DOI] [PubMed] [Google Scholar]
- Semenza GL (2007). Life with oxygen. Science 318, 62–64. [DOI] [PubMed] [Google Scholar]
- Semenza GL (2012). Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul YD, and Seger R (2007). The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta 1773, 1213–1226. [DOI] [PubMed] [Google Scholar]
- Shindo A, Liang AC, Maki T, Miyamoto N, Tomimoto H, Lo EH, and Arai K (2016). Subcortical ischemic vascular disease: Roles of oligodendrocyte function in experimental models of subcortical white-matter injury. J Cereb Blood Flow Metab 36, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythies JA, Sun M, Masson N, Salama R, Simpson PD, Murray E, Neumann V, Cockman ME, Choudhry H, Ratcliffe PJ, et al. (2019). Inherent DNA-binding specificities of the HIF-1alpha and HIF-2alpha transcription factors in chromatin. EMBO Rep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, and Wegner M (2004). Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development 131, 2349–2358. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, and Wegner M (2002). Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16, 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo N, Guo YE, He B, Gu H, and Xie X (2019). Inhibition of MAPK/ERK pathway promotes oligodendrocytes generation and recovery of demyelinating diseases. Glia 67, 1320–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabula Muris, C., Overall, c., Logistical, c., Organ, c., processing, Library, p., sequencing, Computational data, a., Cell type, a., Writing, g., et al. (2018). Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takubo K, Goda N, Yamada W, Iriuchishima H, Ikeda E, Kubota Y, Shima H, Johnson RS, Hirao A, Suematsu M, et al. (2010). Regulation of the HIF-1alpha level is essential for hematopoietic stem cells. Cell Stem Cell 7, 391–402. [DOI] [PubMed] [Google Scholar]
- Tee WW, Shen SS, Oksuz O, Narendra V, and Reinberg D (2014). Erk1/2 activity promotes chromatin features and RNAPII phosphorylation at developmental promoters in mouse ESCs. Cell 156, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. (2011). Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147, 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tilborg E, de Theije CGM, van Hal M, Wagenaar N, de Vries LS, Benders MJ, Rowitch DH, and Nijboer CH (2018). Origin and dynamics of oligodendrocytes in the developing brain: Implications for perinatal white matter injury. Glia 66, 221–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe JJ (2009). Brain injury in premature infants: a complex amalgam of destructive and developmental disturbances. Lancet Neurol 8, 110–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe JJ, Kinney HC, Jensen FE, and Rosenberg PA (2011). The developing oligodendrocyte: key cellular target in brain injury in the premature infant. Int J Dev Neurosci 29, 423–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang M, Arrington J, Shan T, Yue F, Nie Y, Tao WA, and Kuang S (2017). Ascl2 inhibits myogenesis by antagonizing the transcriptional activity of myogenic regulatory factors. Development 144, 235–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Tang D, Liu N, Xiong W, Huang H, Li Y, Ma Z, Zhao H, Chen P, Qi X, et al. (2017). Reciprocal Regulation between the Circadian Clock and Hypoxia Signaling at the Genome Level in Mammals. Cell Metab 25, 73–85. [DOI] [PubMed] [Google Scholar]
- Xia X, and Kung AL (2009). Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol 10, R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HJ, Vainshtein A, Maik-Rachline G, and Peles E (2016). G protein-coupled receptor 37 is a negative regulator of oligodendrocyte differentiation and myelination. Nat Commun 7, 10884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yohe ME, Gryder BE, Shern JF, Song YK, Chou HC, Sindiri S, Mendoza A, Patidar R, Zhang X, Guha R, et al. (2018). MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci Transl Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al. (2013). Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152, 248–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen TJ, Silbereis JC, Griveau A, Chang SM, Daneman R, Fancy SP, Zahed H, Maltepe E, and Rowitch DH (2014). Oligodendrocyte-encoded HIF function couples postnatal myelination and white matter angiogenesis. Cell 158, 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeis T, Graumann U, Reynolds R, and Schaeren-Wiemers N (2008). Normal-appearing white matter in multiple sclerosis is in a subtle balance between inflammation and neuroprotection. Brain 131, 288–303. [DOI] [PubMed] [Google Scholar]
- Zhang S, Kim B, Zhu X, Gui X, Wang Y, Lan Z, Prabhu P, Fond K, Wang A, and Guo F (2020). Glial type specific regulation of CNS angiogenesis by HIFalpha-activated different signaling pathways. Nat Commun 11, 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Top Functional Targets of HIF1a in OPCs, Related to Figure 2 and 4.
Overlap of genes with increased expression by RNA-seq, increased active chromatin by H3K27ac ChIP-seq and bound by HIF1a by HIF1a ChIP-seq reveals top functional targets of HIF1a in OPCs.
Table S2. Identification of Shared Canonical and Cell-Type-Specific HIF1a Peaks and Gene Targets, Related to Figure 2 and 3.
Overlap of OPC HIF1a gene targets with publicly available HIF1a ChIP-seq datasets in embryonic heart, T-cells, and melanocytes reveals canonical HIF1a target genes and peaks shared across all 4 cell types as well as target genes and peaks specific to each cell type.
Table S3. HOMER Motif Analysis Results, Related to Figure 2 and 3.
HOMER motif analysis was performed under HIF1a peaks (FDR<0.001) in sgVhl OPCs, peaks shared between cell types, and peaks that are cell-type specific. HOMER motif analysis was also performed under regions with significantly increased H3K27Ac (FDR<0.1) in sgVhl OPCs compared to Cas9 control OPCs.
Table S4. OLIG2 Binding Increases at Hypoxia Genes in sgVhl OPCs, Related to Figure 3.
HOMER motif analysis was performed under OLIG2 peaks (FDR<0.001) in Cas9 control and sgVhl OPCs and under OLIG2 peaks that were significantly increased (FDR<0.05, log2FC>1) in sgVhl OPCs compared to Cas9 control OPCs. Peak file and closest genes (within 50 Kb) for these increased OLIG2 peaks are listed.
Table S5. Primary Compound Screening and Filtering, Related to Figure 5.
Primary bioactive screening data showing the impact of 1753 compounds on total cell number (DAPI), number of oligodendrocytes (MBP+), and percentage of oligodendrocytes (MBP+/DAPI). To uncover top hits, compounds were first selected if they were considered nontoxic (DAPI > 0.7 fold change) and then selected if they increased oligodendrocyte formation (Oligodendrocyte percentage and number > 3 fold change and pass visual inspection).
Table S6. Validation Dose Curve of Top Compound Hits, Related to Figure 5.
Raw and normalized data for the percentage (MBP+/DAPI) of oligodendrocytes formed for all 3 replicates of the 8-point dose curve of the top 14 hits from the primary bioactive screen. This data was used to generate the heatmap presented in Figure 5D.
Data Availability Statement
All datasets generated in this study have been deposited in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under SuperSeries accession codeGEO: GSE143474 with subseries for RNA-seq (GSE143473), ChIP-seq (GSE143472), and ATAC-seq (GSE155366).
Software used to analyze the datasets are freely or commercially available and detailed in the Key Resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD140a-APC | eBioscience | Cat#17-1401-81; RRID:AB_529482 |
| NG2-AF488 | Millipore | Cat#AB5320A4; RRID:AB_11203143 |
| OLIG2 | Proteintech | Cat#13999-1-AP; RRID:AB_2157541 |
| OLIG2 | Millipore | Cat#AB9610; RRID:AB_570666 |
| OLIG2 | Millipore | Cat#MABN50; RRID:AB_10807410 |
| MBP | Abcam | Cat#ab7349; RRID:AB_305869 |
| O1 | CCF Hybridoma Core Facility | N/A |
| O4 | CCF Hybridoma Core Facility | N/A |
| ASCL2 | Millipore | Cat#MAB4417; RRID:AB_10562654 |
| GFAP | Agilent/Dako | Cat#Z033401-2 |
| Donkey anti-rat AF488 | Thermo Fisher Scientific | Cat#A-21208; RRID:AB_2535794 |
| Donkey anti-mouse AF488 | Thermo Fisher Scientific | Cat# A-21202; RRID:AB_141607 |
| Donkey anti-mouse AF647 | Thermo Fisher Scientific | Cat#A31571; RRID:AB_162542 |
| Donkey anti-rabbit AF647 | Thermo Fisher Scientific | Cat#A31573; RRID:AB_2536183 |
| HIF1a | Abcam | Cat#Ab2185; RRID:AB_302883 |
| Phospho-p44/42. (phosphorylated-ERK1/2) | Cell Signaling Technologies | Cat#9101S; RRID:AB_331646 |
| P44/42 (total ERK1/2) | Cell Signaling Technologies | Cat#9102S; RRID:AB_330744 |
| Phospho-ERK5 (Thr218/Tyr220) | Cell Signaling Technologies | Cat#3371S; RRID:AB_2140424 |
| ERK5 (total) | Cell Signaling Technologies | Cat#3372S; RRID:AB_330491 |
| DLX3 | Abcam | Cat#Ab178428 |
| SOX10 | R&D Systems | Cat#AF2864; RRID:AB_442208 |
| B-Actin peroxidase | Sigma Aldrich | Cat#A3854; RRID:AB_262011 |
| VHL | BD Biosciences | Cat#556347; RRID:AB_396376 |
| MBP | Abcam | Cat#ab7349; RRID:AB_305869 |
| MBP | BioLegend | Cat#808401; RRID:AB_2564741 |
| MAG | Thermo Fisher Scientific | Cat#346200; RRID:AB_2533179 |
| H3K27Ac | Abcam | Cat#Ab4729; RRID:AB_2118291 |
| MyRF | Gift from Michael Wegner | |
| HA | Cell Signaling Technologies | Cat#3724S; RRID:AB_1549585 |
| Normal Rabbit IgG | Cell Signaling Technologies | Cat#2729S; RRID:AB_1031062 |
| SOX6 | abcam | Cat#30455; RRID:AB_1143033 |
| A2B5 | Millipore | Cat#MAB312; RRID:AB_94709 |
| NKX2-2 | DSHB | Cat#74.5A5; RRID:AB_531794 |
| Bacterial and Virus Strains | ||
| Stbl3 E.Coli | ThermoFisher | Cat#C737303 |
| Lenti-X shots for lenti-virus generation | Clonetech | Cat# 631276 |
| Biological Samples | ||
| MCH forebrain mouse protein lysate samples | Gift from Brian Popko | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| N2 Max | R&D Systems | Cat#AR009 |
| B27 | Thermo Fisher Scientific | Cat#12587010 |
| bFGF | R&D Systems | Cat#23-3FB-01M |
| PDGFA | Thermo Fisher Scientific | Cat#221-AA-050 |
| DMEM/F12 | Thermo Fisher | Cat#11320-033 |
| DMEM | Thermo Fisher | Cat# 11960-044 |
| Fetal Bovine Serum (FBS) | Fisher | Cat#A3160402 |
| MEM Non-essential amino acids | Thermo Fisher | Cat#11140-050 |
| Glutamax | Thermo Fisher | Cat#35050061 |
| 2-Mercaptoethanol | Sigma | Cat#M3148 |
| Poly-L-ornithine | Sigma Aldrich | Cat#P3655-50MG |
| Laminin | Sigma Aldrich | Cat#L2020-1MG |
| Noggin | R&D Systems | Cat#3344NG050 |
| IGF-1 | R&D Systems | Cat#291G1200 |
| cyclic AMP | Sigma Aldrich | Cat#D0260-100MG |
| NT3 | Thermo Fisher Scientific | Cat#267N3025 |
| T3 (Thyroid hormone) | Sigma Aldrich | Cat#T-6397 |
| Hb-EGF | R&D Systems | Cat#259-HE-050 |
| CNTF | R&D Systems | Cat#557-NT-010 |
| BMP4 | R&D Systems | Cat#314-BP-050 |
| CloneR | Stem Cell Technologies | Cat#5889 |
| LDN193189 | Sigma | Cat#SML0559 |
| SB-43152 | Sigma | Cat#S4317 |
| Normal donkey serum | Jackson ImmunoResearch | Cat#017-000-121 |
| Paraformaldehyde | Electron microscopy sciences | Cat#15710 |
| Triton X-100 | Sigma Aldrich | Cat#1086431000 |
| DAPI | Sigma Aldrich | Cat#D8417 |
| Puromycin | Thermo Fisher Scientific | Cat#A1113802 |
| Blasticidin | Thermo Fisher Scientific | Cat#A1113903 |
| Hygromycin | Thermo Fisher Scientific | Cat#10687010 |
| SCH772984 | Selleck Chemicals | Cat#S7101 |
| AZD0364 | Selleck Chemicals | Cat#S8708 |
| VX-11e | Selleck Chemicals | Cat#S7709 |
| BIX02188 | Selleck Chemicals | Cat#S1530 |
| BIX02189 | Selleck Chemicals | Cat#S1531 |
| RIPA Buffer | Sigma | Cat# R0278 |
| Protease and Phosphatase Inhibitor | Thermo Fisher Scientific | Cat#78441 |
| NuPAGE Bis-Tris gels | Thermo Fisher Scientific | Cat#NP0335BOX |
| PVDF membranes | Thermo Fisher Scientific | Cat#LC2002 |
| TRIzol | Ambion | Cat#15596-026 |
| Protein G magnetic DynaBeads | Thermo Fisher Scientific | Cat#10004D |
| Proteinase K | Thermo Fisher | Cat#AM2546 |
| Ampure XP beads | Aline Biosciences | Cat#C-1003-5 |
| Hypoxyprobe-1 | Hypoxyprobe Inc | Cat#HP1-100Kit |
| Penicillin-streptomycin | ThermoFisher | Cat#15070063 |
| Bioactive Compound Library-I | Selleck Chemicals | Cat#1700 |
| Critical Commercial Assays | ||
| QIAquick Gel Extraction Kit | Qiagen | Cat#28115 |
| Qiagen Plasmid Maxi Kit (25) | Qiagen | Cat#12163 |
| KAPA HiFi HotStart ReadyMix | KAPA Biosystems | Cat#kk2602 |
| KAPA Sybr Fast qPCR kit | KAPA Biosystems | Cat# KK4605 |
| Taqman gene expression master mix | Thermo Fisher | Cat#4369016 |
| NEBNext® Library Quant Kit for Illumina® | New England 641 Biolabs | Cat#E7630 |
| Protein Assay Kit | Bio-Rad | Cat#5000001 |
| RNeasy Mini Kit | Qiagen | Cat#74104 |
| iScript cDNA synthesis kit | Bio-Rad | Cat#1708891 |
| NEBNext Poly(A) mRNA Magnetic Isolation Module | NEB | Cat#E7490L |
| NEBNext Ultra RNA Library Prep Kit for Illumina | NEB | Cat#E7530L |
| truChIP Chromatin Shearing Kit with Formaldehyde | Covaris | Cat#520154 |
| Hypoxyprobe Kit | Hypoxyprobe Inc | Cat#HP1-100Kit |
| ON-TARGETplus Mouse siRNA smartpool: Ascl2 | Horizon/Dharmacon | Cat#L-047262-01-0005 |
| ON-TARGETplus Mouse siRNA smartpool: Dlx3 | Horizon/Dharmacon | Cat#L-041957-01-0005 |
| siGENOME Mouse siRNA pool: Non-Targeting Control #2 | Horizon/Dharmacon | Cat#D-001206-14-05 5 nmol |
| Basic NucleofectorTM Kit for Primary Mammalian Glial Cells | Lonza | Cat# VPI-1006 |
| Qiagen MinElute PCR purification Kit | Qiagen | Cat#28004 |
| MTS assay kit | abcam | Cat# ab197010 |
| Deposited Data | ||
| RNA-seq datasets | This paper | GSE143474; GSE143473 (subseries) |
| ChIP-seq datasets | This paper | GSE143474; GSE143472 (subseries) |
| ATAC-seq datasets | This paper | GSE143474; GSE155366 (subseries) |
| Experimental Models: Cell Lines | ||
| Lenti-X 293T Cell line | Takara Biosciences | Cat#632180 |
| Epiblast stem cell (EpiSC) derived OPCs | Najm et al. 2011 | N/A |
| Human iPSCs (H7) to derive Oligocortical spheroids | NIH hESC registry | NIHhESC-10-0061; RRID:CV-CL_9772 |
| Experimental Models: Organisms/Strains | ||
| Mouse, Male: 129S/SvEv E3.5 for EpiSC isolation | Jackson Laboratory | RRID:MGI:3050593 |
| Mouse, mixed sex, postnatal day 2 and 7: C57BL/6J | Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Oligonucleotides | ||
| Primer: sgVHL Forward: TCCCTACACGACGCTCTTCCGATCTCTCTCAGGTCATCTTCTGCAACC | This manuscript | IDT |
| Primer: sgVHL Reverse: AGTTCAGACGTGTGCTCTTCCGATCTGACAAGATGCTCGGGGTCGG | This manuscript | IDT |
| Primer: sgVHL.2 Forward: TCCCTACACGACGCTCTTCCGATCTAATAAACAGGTGCCATGCCC | This manuscript | IDT |
| Primer: sgVHL.2 Reverse: AGTTCAGACGTGTGCTCTTCCGATCTAGATTGACTATTAACCTGGCAATG | This manuscript | IDT |
| sgRNAs | See methods | IDT |
| Taqman probes | See methods | IDT |
| Recombinant DNA | ||
| CRISPRv2 constructs | Sanjana et al., 2014 | Addgene plasmid #52961; RRID:Addgene_52961 |
| lentiSAMv2 constructs | Joung et al., 2017 | Addgene plasmid #75112; RRID:Addgene_75112 |
| Activation helper | Joung et al., 2017 | Addgene plasmid #89308; RRID:Addgene_89308 |
| N-terminus tagged 3x HA Ascl2 construct | Genecopoeia | Cat# EX-Mm03774-Lv117 |
| Software and Algorithms | ||
| Columbus Image Data Storage and Analysis System | Perkin Elmer | www.perkinelmer.com/Product/columbus |
| Outknocker | Schmid-Burgk et al., 2014 | http://www.outknocker.org/outknocker2.htm |
| Graphpad Prism | Graphpad Software | https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
| Image Studio Acquisition software | LI-COR | LI-COR Image Studio Software; RRID:SCR_015795 |
| Salmon 0.14.1 | Patro et al. 2017 | https://github.com/COMBINE-lab/salmon |
| DESEQ2 | Love et al. 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html; RRID:SCR_015687 |
| Metascape | Zhou et al. 2019 | http://metascape.org/gp/index.html#/main/step1; RRID:SCR_016620 |
| GSEA | Subramanian et al. 2005 | https://www.gsea-msigdb.org/gsea/index.jsp; RRID:SCR_003199 |
| Trim Galore! Version 0.4.1 | Krueger, F. 2015 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/; RRID:SCR_011847 |
| Bowtie2 Version 2.3.2 | Langmead and Slazberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml; RRID:SCR_005476 |
| MACs Version 2.1.1 | Feng et al. 2012 | http://liulab.dfci.harvard.edu/MACS/; RRID:SCR_013291 |
| Integrative Genomics Viewer | Broad Institute | http://www.broadinstitute.org/igv/; RRID:SCR_011793 |
| Galaxy | Afgan et al. 2018 | http://galaxyproject.org/; RRID:SCR_006281 |
| Diffbind software | Ross-Innes et al. 2012 | https://bioconductor.org/packages/release/bioc/html/DiffBind.html; RRID:SCR_012918 |
| HOMERv4.11.1 | Heinz et al., 2010 | http://homer.ucsd.edu/; RRID:SCR_010881 |
| BEDTools Version 2.25.0 | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2; RRID:SCR_006646 |
| Deeptools Version 3.3.1 | Ramirez et al., 2016 | https://deeptools.readthedocs.io/en/develop/; RRID:SCR_016366 |
| Microsoft Excel | Microsoft | 2016 |
| Illustrator CC | Adobe | 2020 |
| Other | ||
| C-Chamber | BioSpherix | Cat#C-274 |
| 96-well plate | ThermoFisher | Cat#167008 |
| 384-well PDL-coated plate | PerkinElmer | Cat#6057500 |
| Operetta High Content Imaging and Analysis System | PerkinElmer | N/A |
| EL406 Microplate Washer Dispenser | BioTek | N/A |
| 5μl dispense cassette | BioTek | N/A |
| Janus automated workstation | PerkinElmer | N/A |
| Odyssey Fc Imaging System | LI-COR | N/A |
| 7300 real-time PCR system | Applied Biosystems | N/A |
| 5973 Mass Spectrometer | Agilent | N/A |
| Hamamatsu Nanozoomer S60 Slide Scanner | Hamamatsu | C13210-01 |
| SuperSignal West Pico PLUS Chemiluminescent Substrate | ThermoFisher | Cat#34580 |
| Halt Protease and Phosphatase Inhibitor Cocktail, EDTA-free (100x) | ThermoFisher | Cat#78441 |
| Nextera Tn5 Transposase | Illumina | Cat#FC-121-1030 |
| Covaris S2 Focused Ultrasonicator | Covaris | Cat#17015 |
| Amaxa Nucleofector 2b | Lonza | Cat#AAB-1001 |
| E12.5 heart HIF1a ChIP-Seq | Guimaraes-Camboa et al., 2015 | GSM1500750, GSM1500751, GSM1500749 |
| Melanocyte HIF1a ChIP-Seq | Loftus et al., 2017 | GSM2305570, GSM2305571, GSM2305572 |
| Th17 T-cell HIF1a ChIP-Seq | Ciofani et al., 2012 | GSM1004819, GSM1004991, GSM1004820, GSM1004993 |
| Mus musculus mm10 genome build | NCBI; Mouse Genome Sequencing Consotium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/ |


