SUMMARY
Organoids are powerful models for studying tissue development, physiology, and disease. However, current culture systems disrupt the inductive tissue-tissue interactions needed for the complex morphogenetic processes of native organogenesis. Here, we show that mouse embryonic stem cells (mESCs) can be coaxed to robustly undergo fundamental steps of early heart organogenesis with an in-vivo-like spatiotemporal fidelity. These axially patterned embryonic organoids (gastruloids) mimic embryonic development and support the generation of cardiovascular progenitors, including first and second heart fields. The cardiac progenitors self-organize into an anterior domain reminiscent of a cardiac crescent before forming a beating cardiac tissue near a putative primitive gut-like tube, from which it is separated by an endocardial-like layer. These findings unveil the surprising morphogenetic potential of mESCs to execute key aspects of organogenesis through the coordinated development of multiple tissues. This platform could be an excellent tool for studying heart development in unprecedented detail and throughput.
Graphical Abstract
In Brief
Rossi et al. describe an embryonic organoid model that mimics the early development of the heart, from the generation of cardiovascular precursor cells to the specification of the first and second heart fields. These axially patterned organoids support the formation of cardiac crescent and early cardiac tube-like structures while reproducing the cell diversity and tissue-tissue interactions typical of embryos.
INTRODUCTION
Stem-cell-derived organoids self-organize into complex structures that mimic aspects of the architecture, cellular composition, and function of tissues found in real organs (Clevers, 2016; Lancaster and Knoblich, 2014; Rossi et al., 2018; Sasai, 2013). Although most organoids simulate specific features of adult organs, embryonic organoids can capture key processes that occur during early embryonic development, from the pre-implantation blastocyst (Rivron et al., 2018) to early post-implantation development (Harrison et al., 2017; Shao et al., 2017a, 2017b; Sozen et al., 2018; Zheng et al., 2019) and gastrulation (Beccari et al., 2018; van den Brink et al., 2014). Although embryonic organoids have been shown to mimic many of the morphological and transcriptional hallmarks of the early embryo, their potential to undergo organogenesis has not yet been explored.
The heart is the first organ to form and function in the embryo. Short after gastrulation, heart progenitors are specified, progressively localize anteriorly, and organize in a crescent-shaped domain (by embryonic day 7.5 [E7.5]), the first cardiac compartment that is morphologically identifiable during development. Cardiogenesis is based on the interaction of two different types of progenitors, namely the first heart field (FHF) and the second heart field (SHF) progenitors, the latter originating from pharyngeal mesoderm (Kelly et al., 2014). The cardiac crescent, mainly formed by FHF progenitors, subsequently rearranges to form a linear heart tube (around E8.0-E8.5), which is the first beating structure. Successively, SHF progenitors contribute to heart tube elongation and the heart undergoes looping, ballooning, and septation, giving rise to the four-chambered structure typical of adulthood (Harvey, 2002). Heart organogenesis requires cardiac progenitors to interact with surrounding tissues through mechanical interactions and the secretion of cardiac-inducing factors (Miquerol and Kelly, 2013). These involved tissues especially include the endothelium (Brutsaert, 2003; Brutsaert et al., 1998; Narmoneva et al., 2004) and foregut (Hosseini et al., 2017; Kidokoro et al., 2018; Lough and Sugi, 2000; Nascone and Mercola, 1995; Schultheiss et al., 1995; Varner and Taber, 2012).
We hypothesized that, due to their embryo-like multi-axial organization and gene expression patterns, mouse gastruloids (Beccari et al., 2018; van den Brink et al., 2014, 2020) could offer a suitable template for modeling early heart development because they potentially preserve the crucial tissue-tissue interactions required for this organogenesis. Indeed, studying heart organogenesis in such a complex and spatially organized system could allow the modeling of developmental events in an embryo-like context, where cardiac cells are naturally exposed to the influence of other tissues.
Here, we show that self-organizing mouse embryonic stem cells (mESCs) can capture early heart organogenesis in vitro with a surprising temporal and spatial accuracy. Exposing small ESC aggregates to a cocktail of three cardiogenic factors in gastruloid culture conditions promotes cardiac development in vitro starting from Mesp1+ progenitors, which progressively become restricted to the anterior portion of the gastruloid. Through a combination of light-sheet and confocal microscopy, RNAscope imaging, fluorescence-activated cell sorting (FACS), and single-cell RNA sequencing, we demonstrate that these embryoids support the formation of Flk1+ cardiovascular progenitors, the generation of a vascular-like network, and the formation of progenitors with first and second heart field identities. Strikingly, we find evidence for the morphogenesis of an anterior cardiac crescent-like domain, which subsequently gives rise to a beating compartment exhibiting Ca2+-handling properties comparable to functional fetal cardiomyocytes. Morphogenesis is established in close spatial proximity to the most anterior portion of a co-developing gut-tube-like structure, which is separated from the cardiac domain by an endocardial-like layer. Therefore, this in vitro model of cardiac organogenesis uniquely captures interactions between embryonic tissues in the context of a spatially organized embryo-like entity.
RESULTS
Optimization of Culture Conditions to Promote Efficient Beating Portions in Gastruloids
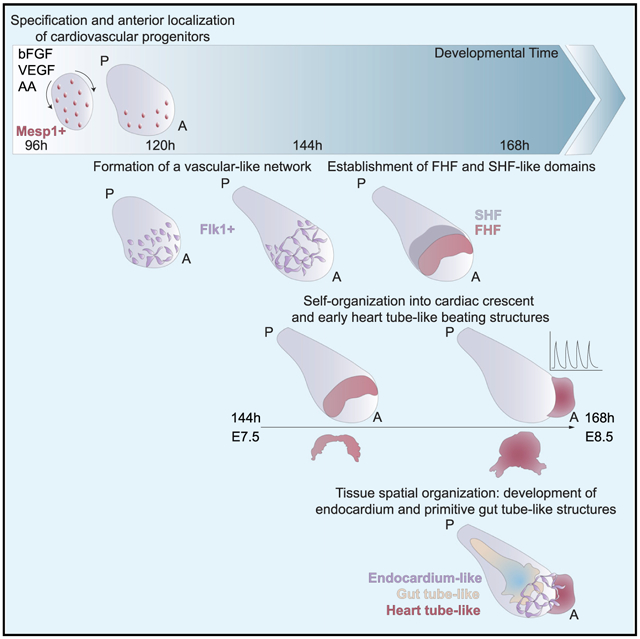
Gastruloids occasionally formed a beating domain that is exclusively located within their anterior region (38.5% ± 29.3% formed at 168 h) when cultured for 144 h or longer in N2B27 medium (Figure 1A). The location and activity of this beating structure suggested that it might correspond to a cardiac primordium. We tested whether well-known cardiogenic factors (Rajala et al., 2011) could increase the frequency of this event by adding basic fibroblast growth factor, ascorbic acid, and vascular endothelial growth factor 165 (VEGF) (Kattman et al., 2006), singly or in combination, and we increased nutrient and growth factor availability through volume optimization and shaking (Figures 1A and S1A). In these culture conditions (termed N2B27+++), the frequency of beating gastruloids increased by more than a factor of two (87.2% ± 15.6% formed at 168 h; Figures 1A and 1B; Video S1). We noticed that exposure to cardiogenic factors was most effective when applied in combination and between 96 and 144 h (Figures S1A–S1H), so we kept this protocol for the following experiments. Importantly, culturing in N2B27+++ did not alter the polarization of the gastruloids, the extent of their elongation (Figures S1I and S1J), or the timing of the emergence of the beating domains (Figures 1A and S1B–S1D) compared to standard conditions. Staining for Gata4 and cardiac troponin T (cTnT) confirmed that the beating structure was cardiac like (Figure 1C). FACS analysis of reporter-cell-derived gastruloids at different time points showed that gastruloids grown in N2B27 or N2B27+++ conditions were characterized by similar levels of cardiac progenitor markers (Gata6 and Flk1; Figures S1K and S1L). However, at 168 h in N2B27+++, there was an increase of expression of markers distinguishing the first and second heart fields (Hcn4 and Tbx1; Figures S1M and S1N). This is in keeping with previous literature showing that the three cardiogenic factors can induce cardiac differentiation from pluripotent stem cells (Kattman et al., 2006) and, more specifically, late-stage cardiac myogenesis in vitro (Sugi and Lough, 1995; Takahashi et al., 2003; Ye et al., 2013).
Figure 1. Embryonic Organoids Recapitulate Heart Organogenesis.
(A) Gastruloids form a beating portion on their anterior side at 168 h. White and red lines highlight the displacement of the beating domain over 10 ms.
(B) Frequency of beating structures at different time points in n = 6 independent experiments.
(C) Immunofluorescence for Gata4 and cTnT on gastruloids at 168 h.
(D) UMAP plot showing clusters obtained from 30,496 cells isolated from gastruloids at 96 h, 120 h, 144 h, and 168 h. n = 2 replicates per time point.
(E) Heatmap showing the expression of canonical cell type markers illustrating the cell type diversity in (D).
(F) Overlay of the time points analyzed (hours after aggregation) with the trajectories inferred from RNA velocity, projected on the uniform manifold approximation and projection (UMAP) in (D).
(G–K) Expression of typical cardiac genes.
Scale bars, 100 μm. A, anterior; P, posterior. See also Figures S1 and S2.
To characterize our embryonic organoid system at the single-cell level, we performed single-cell RNA sequencing (scRNA-seq) analysis of gastruloids every 24 h from 96 h to 168 h. Unsupervised clustering identified 59 clusters, which were assigned to 32 cell types based on canonical markers (Table S1). Our analysis revealed the presence of a multitude of cell types (Figures 1D and 1E), in line with the embryo-like organization of gastruloids (Beccari et al., 2018). Trajectories estimated from RNA velocity (La Manno et al., 2018; Bergen et al., 2020) were in line with previously known lineage relationships (Figure 1F). Cells differentiated from an epiblast-like stage into three main branches, corresponding to the three germ layer derivatives (Figures 1D and 1F). Interestingly, some cross-talk between germ layers was also identified, such as the contribution of neuromesodermal progenitors (NMPs) to both mesodermal and neural lineages. We also detected cells with presomitic and somitic identity (van den Brink et al., 2020; Veenvliet et al., 2020), further confirming that gastruloids grown in N2B27+++ maintained the main features of those grown in N2B27. Overall, we observed a broader variety of differentiating cell types compared to a recently published scRNA-seq analysis of mouse gastruloids (van den Brink et al., 2020), likely due to the broader time range that we analyzed. Comparison of our results with a recently published scRNA-seq dataset of mouse gastrulation and early organogenesis (Pijuan-Sala et al., 2019) showed a striking degree of similarity (Figures S2A–S2G).
The Formation of a Cardiac Domain Mimics In Vivo Heart Development
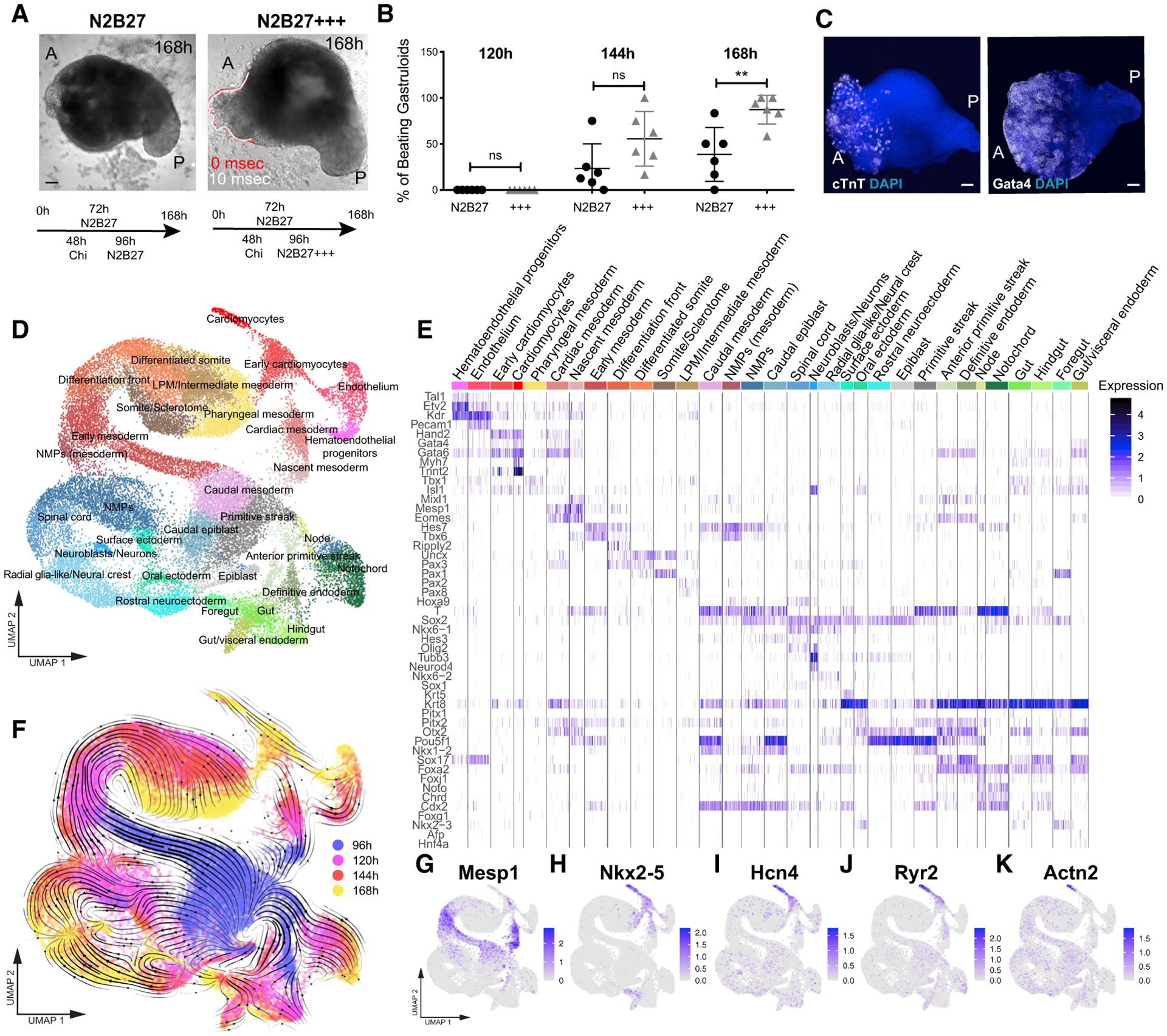
To understand whether the formation of the cardiac portion mimics developmentally relevant processes, we first analyzed the temporal expression of key genes involved in cardiovascular specification (Figures 1G–1K and S1O–S1S). Similar to what happens during embryonic development (Lescroart et al., 2014; Saga et al., 1996), the first upregulated cardiac gene was Mesp1, which was expressed around 96 h and then rapidly downregulated (Figures 1G, 2A, S1O, S2H, and S2I). Using a Mesp1-GFP reporter ESC line (Bondue et al., 2011) and live light-sheet imaging, we observed that Mesp1 expression started in a mosaic-like manner at 96 h and then Mesp1-positive cells were first confined to the anterior side before slowly disappearing after 120 h (Figure 2B; Video S2). Concurrently with the elongation and formation of an anterior-posterior axis, Gata6-expressing cells, marking early cardiac differentiation, were localized to the anterior side in gastruloids generated from Gata6-Venus ESCs (Freyer et al., 2015), opposite to the pole that was positive for Brachyury (Figures 2C, S2J, and S2K). Gata6 expression was maintained over time (Figures S2L and S2M).
Figure 2. Early Cardiac Development in Embryonic Organoids Mimics Embryonic Development.
(A) Spatial localization of Mesp1+ cells (stained for GFP) at 96 h as compared to Brachyury expression.
(B) Tracking of Mesp1+ cells with live light-sheet imaging from 96 to 110 h. Red, tracked cells; light blue, cell tracks forward.
(C) Spatial localization of Gata6+ cells at 96 and 120 h compared to Brachyury expression.
(D) Calcium imaging of gastruloids at 168 h using a Cal-520 AM calcium dye and light-sheet microscopy.
(E) Representative spiking profile.
(F) Spiking frequency of the gastruloid cardiac portion at 168 h for n = 21 gastruloids.
Data are represented as mean ± whiskers from min to max. Scale bars, 100 μm. See also Figure S2.
In the embryo, Wnt activity plays a key role in determining the fate and positioning of the developing cardiac cells (Gessert and Kühl, 2010; Naito et al., 2006). To test the involvement of a similar regulatory axis in vitro, we generated gastruloids from a Wnt/β-catenin signaling activity reporter ESC line (TCF/LEF-mCherry; Faunes et al., 2013; Ferrer-Vaquer et al., 2010). In line with published data (Turner et al., 2017), Wnt activity was found to be higher in the posterior region, coincident with the Brachyury-positive pole, and progressively decreased toward the anterior portion (Figures S2N and S2O), supporting a role for Wnt signaling in determining the positioning of the cardiac compartment along the anterior/posterior (A/P) axis.
From this stage onward, the early differentiation genes Nkx2–5 and Hcn4 were increasingly expressed (Figures 1H, 1I, S1P, and S1Q), followed by genes marking mature cardiomyocytes (α-actinin/Actn2 and Ryr2; Figures 1J, 1K, S1R, and S1S). This sequence of gene expression shows that gastruloids, stimulated with cardiogenic factors, recapitulate the temporal and spatial gene expression dynamics of cardiac development from the specification of cardiac progenitors to the formation of a beating cardiac structure.
In cardiomyocytes, physical contraction is coupled to electrical excitation through intracellular changes of Ca2+ (Tyser et al., 2016). To evaluate the functionality of the gastruloid cardiac domain, we thus assessed calcium transients, momentary spikes in voltage, by live gastruloid imaging via light-sheet microscopy (at 168 h). Image analysis revealed rhythmic calcium spiking in beating areas with a frequency comparable to beating rates observed in embryos from the crescent stage to the linear heart tube (Tyser et al., 2016; Figures 2D–2F; Video S3). Drugs interfering with calcium transport, including the l-type calcium channel blocker nifedipine and the β-adrenergic agonist isoproterenol, either completely inhibited these transients (Figure S2P) or caused an increase in spiking frequency (Figures S2Q and S2R), respectively. These data demonstrate that the cardiac compartments found in late gastruloids (168 h) exhibit Ca2+-handling properties compatible with functional cardiomyocytes.
Co-development of a Vascular Compartment within the Cardiac Domain
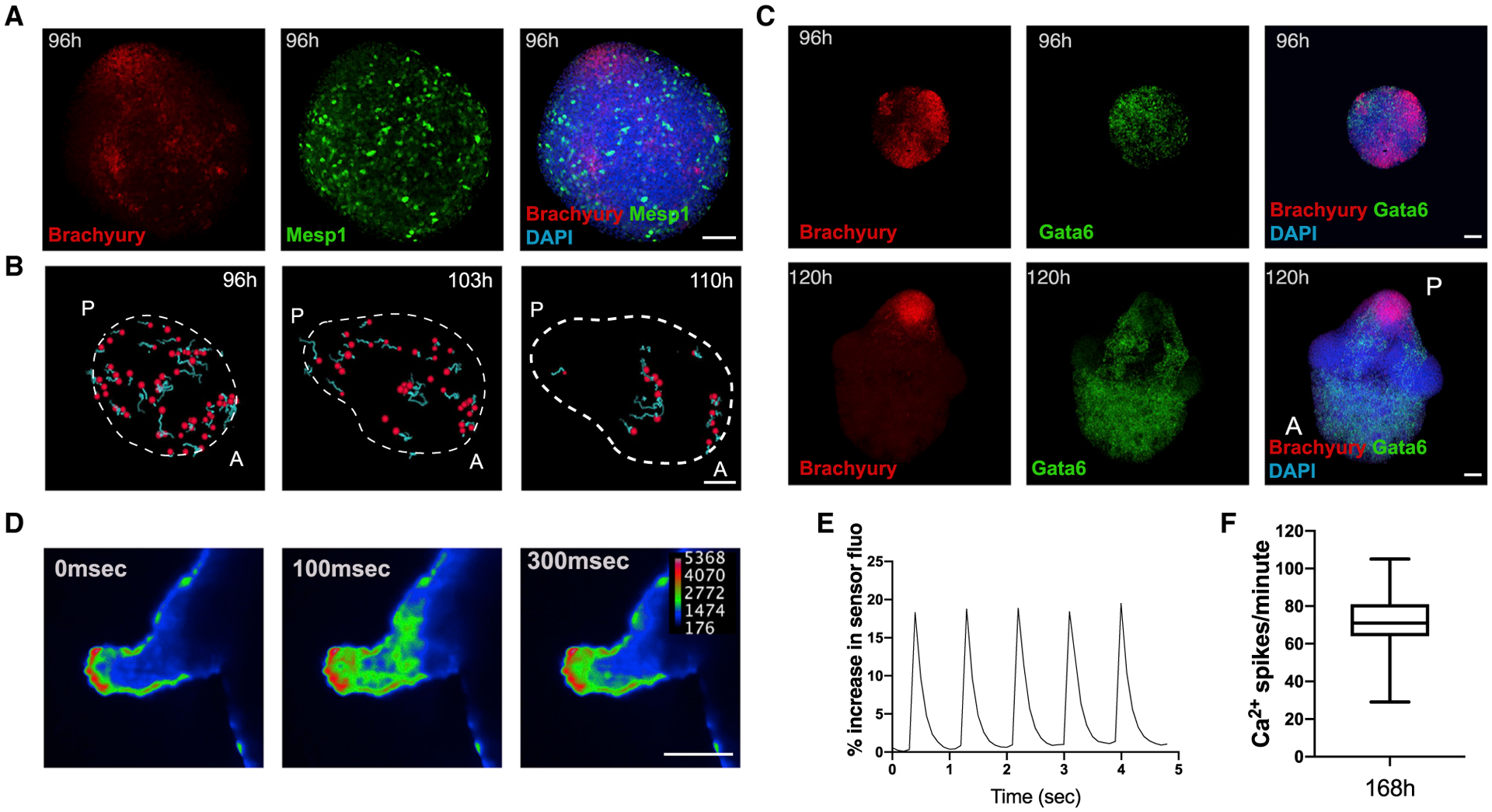
During embryonic development, the continuous cross-talk of endothelial cells with the developing heart is a prerequisite for cardiomyocyte maturation, function, and survival (Brutsaert, 2003). For this reason, we tested whether such tissue-tissue interactions could potentially take place in developing gastruloids, focusing on cardiovascular progenitors expressing the well-known marker Flk1 (also known as Kdr or Vegfr2; Kattman et al., 2006). In 96-h gastruloids derived from a Flk1-GFP reporter ESC line (Jakobsson et al., 2010), Flk1 was expressed at the anterior pole opposite to Brachyury (Figure 3A). Over time, Flk1 expression persisted in the anterior portion of the gastruloids (Figures 3B, S3A, and S3B), and Flk1-positive cells started to form a vascular-like network of spindle-shaped cells (Figure 3C; Video S4) that stained positive for the endothelial marker CD31 (Figures 3D and S3C). In an in vitro angiogenesis assay, Flk1-positive cells that were isolated by FACS from 168-h gastruloids and plated on Matrigel formed vascular-like networks similar to human umbilical vein endothelial cells (HUVECs) (Figures 3E and S3D). Interestingly, scRNA-seq analysis revealed the expression of typical endothelial markers (Figures 1E and 3F–3H) and suggested maturation of endothelial cells over time. Indeed, the upper portion of the endothelial cluster, corresponding to later stage (168 h) gastruloids (Figure 1F), expressed higher levels of genes associated with the Notch pathway (Figures 3I and 3J), which in vivo is involved in vascular morphogenesis, sprouting, and remodeling (Fischer et al., 2004; Krebs et al., 2000; Pitulescu et al., 2017). Moreover, endothelial cells at 168 h specifically expressed Cav1 (Figure 3K), a gene responsible for caveolae formation and involved in endothelial cell maturation, migration, and tubule formation (Lamalice et al., 2007; Liu et al., 2002). Collectively, these results suggest that gastruloids comprise regions that develop into a vascular-like compartment, which is associated with undergoing cardiovascular development.
Figure 3. Development of a Vascular-like Network.
(A and B) Spatial localization of Flk1+ cells at (A) 96 and (B) 120 h compared to Brachyury expression. Image in (B) is a composite of two separate images.
(C) Light-sheet live imaging of the anterior portion of Flk1-GFP gastruloids from 120 to 144 h, highlighting the formation of a vascular-like network.
(D) The Flk1+ vascular-like network is positive for CD31.
(E) Angiogenesis assay showing tube formation in isolated Flk1+ cells compared to HUVECs.
(F–K) UMAP plots showing the expression of endothelial marker genes (F–H) and genes related to endothelial cell maturation and morphogenesis (I–K) in the scRNA-seq dataset.
Scale bars, 100 μm. See also Figure S3.
In Vitro Cardiac Development Entails First and Second Heart Field Development
A key feature of cardiogenesis is a coordinated interaction between two mesodermal progenitor populations: the FHF, which contributes to the left ventricle and part of the atria, and the SHF, which gives rise to the outflow tract, right ventricle, and part of the atria (Harvey, 2002; Miquerol and Kelly, 2013). After migration from the posterior primitive streak, FHF progenitors form the cardiac crescent and early heart tube, located anteriorly. SHF progenitors, originating from cardiopharyngeal mesoderm (Cortes et al., 2018), are characterized by delayed differentiation and are located medially to the crescent, from where they contribute to heart-tube elongation. Both of these populations are characterized by a specific pattern of gene expression that was reproduced in embryonic organoids; focusing on FHF and SHF trajectories in our scRNA-seq dataset (Figure 4A), we observed that expression of typical FHF (Figures 4B and S3E) and SHF (Figures 4B and S3F) markers (Bruneau et al., 1999; Buckingham et al., 2005; Guo et al., 2011; Liang et al., 2013; McFadden et al., 2005; de Soysa et al., 2019; Wang et al., 2019; Zhou et al., 2017) increased over time and labeled two different populations (Figures 4B and S3E–S3I).
Figure 4. Embryonic Organoids Form a First and Second Heart Field.
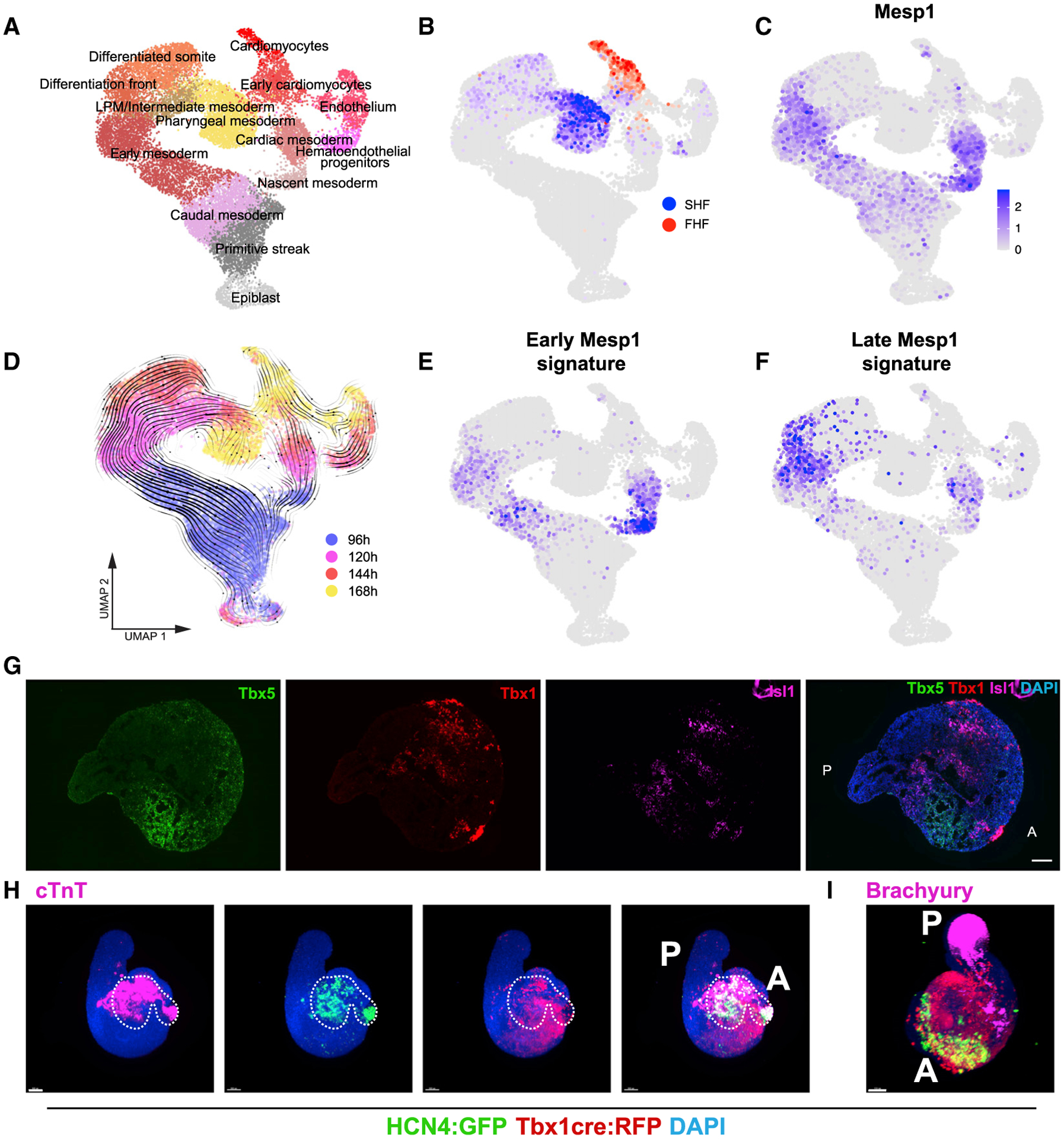
(A) UMAP plot showing clusters lying on the trajectories from epiblast to heart fields. n = 2 replicates per time point.
(B) Localization of FHF on the UMAP in (A) based on expression of Tbx5, Hcn4, Nkx2–5, Hand1, and SHF based on the expression of Tbx1, Six1, and Six2.
(C) Expression of Mesp1.
(D) Overlay of time points and RNA velocity projected on the UMAP in (A).
(E and F) Gene signature scores for early (E) and late (F) Mesp1-expressing cells in vivo based on in vivo differentially expressed genes from Lescroart et al. (2014). The scores are only shown for Mesp1-positive cells.
(G) RNAscope showing spatial localization of FHF and SHF domains in gastruloids at 168 h. Images are stitched tile scans.
(H) Representative immunofluorescence pictures of Hcn4-GFP::Tbx1Cre-RFP gastruloids at 168 h, showing the presence of FHF and SHF progenitors in the anterior domain of gastruloids. The white dashed line highlights the position of the Hcn4-positive domain in all pictures. n = 48 gastruloids from 4 independent experiments.
(I) Representative immunofluorescence picture of Hcn4-GFP::Tbx1Cre-RFP gastruloids at 168 h, showing Brachyury expression in the posterior region. n = 32 gastruloids from 2 independent experiments.
Scale bars, 100 μm. See also Figures S3 and S4.
FHF and SHF progenitors have previously been shown to derive from two successive peaks of Mesp1 expression during development, which are associated with different transcriptional signatures (Lescroart et al., 2014, 2018). Strikingly, in gastruloids, we also observed an earlier (~96 h) and later (~120 h) window of Mesp1 expression (Figures 4C and 4D) in two distinct trajectories that separate at early stages during gastrulation. Genes expressed in vivo preferentially in the earlier Mesp1-positive precursors (Lescroart et al., 2014) were enriched by 96 h in gastruloid Mesp1-positive cells, although genes associated in vivo preferentially with the later Mesp1-positive precursors were enriched by 120 h in gastruloid Mesp1-positive cells (Figures 4D–4F). RNA velocity analysis (Figure 4D) suggested that progenitors expressing FHF markers (Figure 4B) might be derived from Mesp1-expressing cells enriched for markers of the earlier population (Figures 4B and 4E), whereas progenitors expressing SHF markers (Figure 4B) might be derived from Mesp1-expressing cells enriched for markers of the later population (Figures 4B and 4F). Overall, these data show that gastruloids co-develop FHF- and SHF-like progenitor cells, which are created by a temporal sequence that mimics embryonic development.
To corroborate the presence of progenitor cell populations from both heart fields in our cardiac organoids, we used three experimental paradigms. First, using a recently published protocol for FACS-isolating SHF progenitors based on the expression of the C-X-C chemokine receptor type 4 (CXCR4) (Figure S3J; Andersen et al., 2018), we observed that Gata6+/CXCR4+ cells express higher levels of SHF markers (Tbx1, Isl1, and Fgf10) but low or unchanged levels of FHF markers (Nkx2–5, Tbx5, and Hcn4) compared to Gata6+/CXCR4− cells (Figures S3K–S3P). Second, RNAscope imaging showed the expression of FHF (Tbx5) and SHF (Tbx1) markers in mutually exclusive cell populations and Isl1 mostly overlapping with Tbx1 expression (Figure 4G). Third, gastruloids generated from a dual FHF/SHF reporter/tracer mESC line (Hcn4:GFP;Tbx1-Cre:RFP; Andersen et al., 2018) showed the development of a cardiac portion in their anterior domain (Figure 4H), with an in vivo like spatial arrangement: FHF progenitor cells localized more anteriorly with respect to SHF progenitors (Figure S3Q). All cardiac markers were expressed at the opposite pole to the posterior marker Brachyury (Figures 4I and S3R), and Hcn4 signal was typically located in the beating portion of gastruloids at 168 h, where cTnT is expressed (Figures 4H and S3R). Together, these data show that late gastruloids contain key cell types that have the typical signature and spatial organization of first and second heart field progenitors.
In Vitro Cardiac Development Occurs through Establishment of a Crescent-like Structure
After their specification in the mouse embryo, cardiac progenitors migrate antero-laterally and progressively fuse at the midline to define the first morphologically identifiable heart structure, the cardiac crescent, which appears around E7.5 (Miquerol and Kelly, 2013). Then, morphogenetic movements associated with foregut closure form a linear heart tube by E8.5 (Miquerol and Kelly, 2013). We explored whether gastruloids stimulated with cardiogenic factors could capture these morphological hallmarks of cardiogenesis. Remarkably, in gastruloids cultured for 144 h to 168 h, we observed a recapitulation of these events. Around 144 h, cTnT-positive cardiomyocytes were organized in crescent-like domains (Figures 5A and S4A). These further developed into denser crescent-like structures with beating areas, leading to beating epithelial protrusions on the anterior portion of the gastruloids at around 168 h (Figures 5A and S4B–S4E). Similar to mouse embryos (Ivanovitch et al., 2017; Le Garrec et al., 2017; Tyser et al., 2016), these phenotypes could be observed in gastruloids within 24 h and could be aligned to progressive morphological stages of embryonic cardiac development between E7.5 and E8.5. This is shown schematically in Figure 5B, which shows a comparison of the cardiac structures observed in gastruloids with a scheme of early stages of heart development in embryos. To provide a measure of the spectrum of shapes observed in gastruloids, we have generated artificial spherical, crescent-shaped, and concave shapes, which were used as a reference for a volumetric analysis, and we have measured geometric properties of both the artificial and observed shapes. The comparative volume analysis of the cTnT-positive gastruloid domains with the defined artificial shapes revealed a gradual transition from an almost spherical to a crescent-like shape (144–168 h) that then became concave (168 h; Figure 5C). These results highlight the remarkable capacity of embryonic organoids to reproduce some key features typical of morphogenetic processes in the early stages of heart development.
Figure 5. Embryonic Organoids Recapitulate Cardiac Morphogenesis.
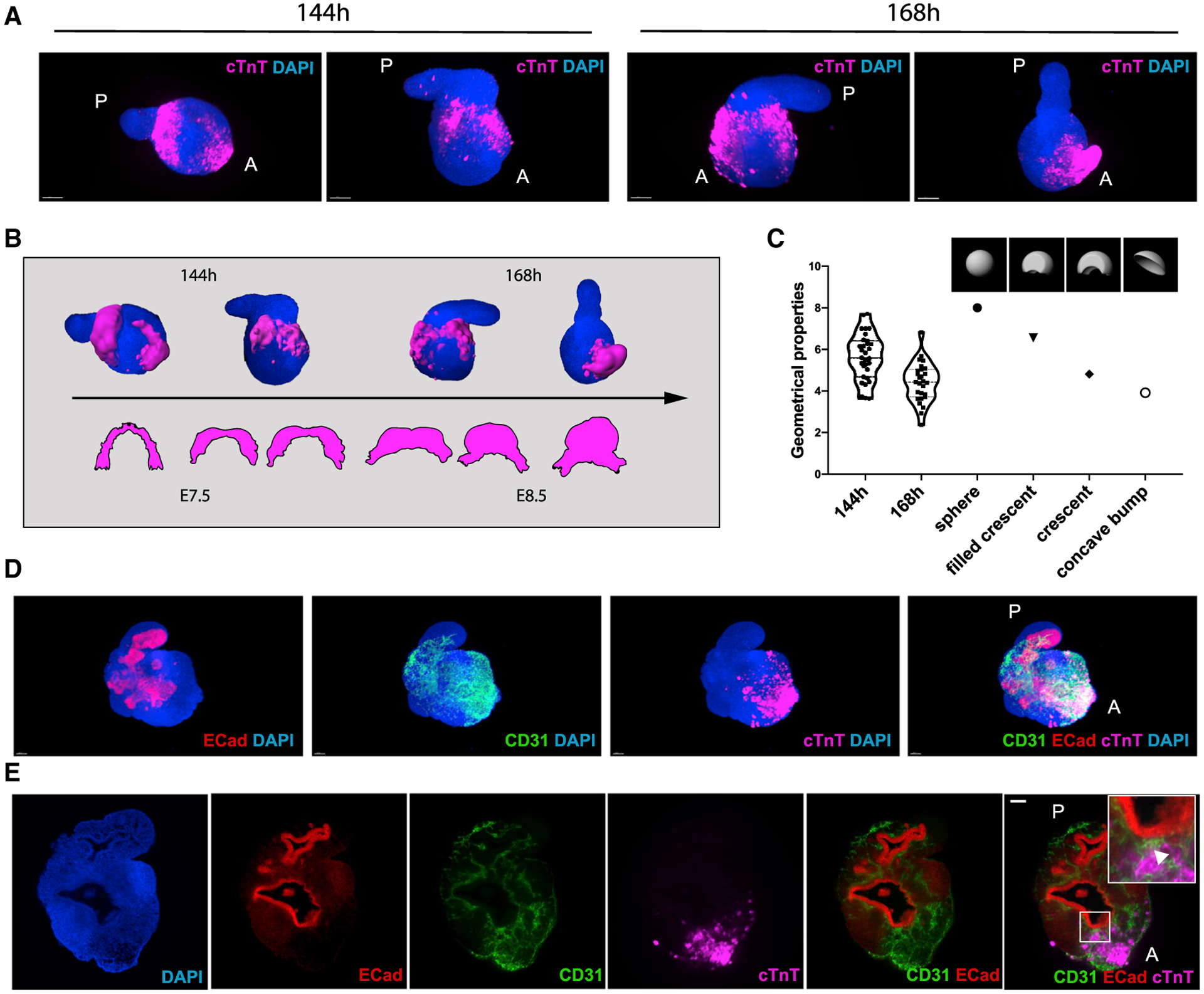
(A) Light-sheet imaging of cleared gastruloids from 144 to 168 h show an initial crescent-like domain that is condensed into a beating bud at 168 h.
(B) Schematic illustrating the comparison between gastruloid stages of cardiac development and embryonic stages from cardiac crescent to linear heart tube.
(C) Quantification of geometrical properties (spareness) of the gastruloid cardiac domains compared to those of defined artificial shapes, with n = 31 gastruloids at 144 h and n = 27 gastruloids at 168 h.
(D and E) The cardiac domain is localized near the anterior epithelial gut-tube-like structure, separated by a CD31+ endocardial layer.
Scale bars, 100 μm. See also Figure S4.
In Vitro Cardiogenesis Happens in the Context of Physiological Tissue-Tissue Interactions
The presence of a crescent-like structure and its evolution to a coherent beating group of cells led us to test whether this was accompanied by an association between the crescent and anterior endoderm, as is the case in the embryo (Ivanovitch et al., 2017; Lough and Sugi, 2000). Bulk transcriptomics analysis of late gastruloids revealed gene expression patterns (Sox17, Shh, and Cdx2) that suggested the presence of a primitive gut-tube-like tissue (Beccari et al., 2018) that was confirmed by our scRNA-seq analysis (Figures 1E and S2E). Strikingly, we found that the cTnT-positive cardiac domain in 168-h gastruloids was exclusively located next to a tube-shaped E-cadherin-positive epithelial tissue (n = 14/14), with a CD31-positive, putative endocardial-like layer in between (Figures 5D and 5E; Video S5). Moreover, we observed that, within the endothelial cluster in our scRNA-seq analysis (Figure 4A), the expression of genes previously associated with endocardium development (Tbx5, Tbx20, and Hand2; Figures S3E and S4F) and endocardium formation (Nfatc1 and Npr3; Figures S4G and S4H; Feng et al., 2019; Zhang et al., 2016) was polarized in a subset of cells more closely associated with the early cardiomyocyte cluster (arrowheads), suggesting an endocardial identity. This is reminiscent of the spatial arrangement of anterior structures in the developing embryo (Ivanovitch et al., 2017).
DISCUSSION
The heart develops through complex interactions between cardiac progenitors and surrounding tissues (Miquerol and Kelly, 2013). These interactions are crucial for sustaining cardiogenesis but have thus far not been reproduced in vitro. Indeed, even though numerous in vitro cardiac models, such as engineered heart muscles (Huebsch et al., 2016; Lind et al., 2017; Ma et al., 2015; Mills et al., 2019; Zhao et al., 2019) or cardiac spheroids (Giacomelli et al., 2017; Polonchuk et al., 2017), have been developed, the in vitro modeling of heart morphogenesis has been out of reach. Our results demonstrate that embryonic organoids can be stimulated to recapitulate in vitro the key steps of early cardiac development that in vivo require polarized interactions between different primordia. These interactions are critical for achieving the architectural and compositional complexity that are absent in conventional organoids (Rossi et al., 2018) but appear to be provided by the multi-axial properties and spatial organization of the different embryonic tissues that are characteristic of gastruloids (Beccari et al., 2018).
We show that embryonic organoids can be stimulated to form a cardiac portion that involves key developmental hallmarks, such as the generation and spatial arrangement of early cardiac progenitors and first and second heart field compartmentalization. Of note, precardiac spheroids were previously shown to generate progenitors with FHF and SHF identity; however, these spheroids did not develop an organized structure (Andersen et al., 2018). In contrast, in our model system, these populations emerged in an embryo-like, spatially organized structure. Moreover, the cardiac portion in our system co-developed with a vascular network and an endodermal component, both known to influence cardiac development (Brutsaert, 2003; Lough and Sugi, 2000). Due to the importance of these neighboring developing tissues, previous attempts were performed to derive spheroids composed of both endothelial and cardiac cells (Giacomelli et al., 2017; Polonchuk et al., 2017). However, these systems, based on an aggregation of cells pre-differentiated in 2D, lacked any visible structural organization. Future work will explore whether the morphogenetic processes seen in our system require the presence of spatial complexity (e.g., multi-axial embryonic patterning) and cross-talk with other tissues. In particular, the use of drugs or conditional knockout models to selectively inhibit the development of endoderm or endothelial compartments, as well as models based on diphtheria toxins to remove them in later stages, will be very useful to gain a mechanistic insight into the morphogenetic processes described here. These experiments would not only contribute to a better characterization of the embryonic organoid model presented here but would also be extremely useful to study in vitro how the influences of surrounding tissues are fine-tuned during heart development. Indeed, we believe that the unique cardiac morphogenetic behavior described in this system, which happens in the context of multi-axial 3D structures mimicking embryonic body plan development (Beccari et al., 2018), together with the concerted development of the other embryonic tissues, provides a useful tool for the study of long-standing developmental biology questions with unprecedented resolution and throughput.
Collectively, our data show that embryonic organoids have the potential for modeling organogenesis. Herein, we have steered the self-organization of small aggregates of mESCs to form a cardiac primordium as a precursor of an embryonic heart. In the future, it will be interesting to study whether cardiac primordia can be further developed and matured and what type of tissue-tissue interactions are functionally involved in their development. Additionally, the principles presented here should be applicable to other organ systems once it is possible to overcome the significant challenge of the limited lifespan of embryonic organoids.
Limitations of Study
This study describes the emergence of multiple tissues in an embryonic organoid model. Although we observed a similar phenotype using different cell lines, it has been previously shown that distinct cell lines might show a different propensity to differentiate into specific cell types. Specifically, it has been suggested that the genetic background might skew the cell-type composition of gastruloids (van den Brink et al., 2020) and mESC differentiation in general (Ortmann et al., 2020). Such cell-line-associated heterogeneity has been observed also for human gastrulation in vitro models (Tewary et al., 2019). Moreover, regardless of the genetic background, mESC culture conditions can also impact their differentiation propensity (Ghimire et al., 2018). Similarly, we observed that different cell lines vary in their response to the Wnt agonist CHIR99021. All these factors should be taken into account when deriving gastruloids from a new cell line. Future studies could help to better decipher the mechanisms underlying these differences.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthias Lutolf (matthias.lutolf@epfl.ch).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The single cell RNA-seq data generated during this study are available at NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GEO: GSE158999. The complete code for reproduction or for exploration of the results is available on https://github.com/nbroguiere/Cardiac_Gastruloids. All scripts and settings used for volumetric crescent analysis are available at https://go.epfl.ch/Crescent_Analysis.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
mESCs were cultured at 37°C, 5%CO2 in DMEM supplemented with 10% Embryonic Stem Cell qualified FBS (GIBCO), NEAA, Sodium Pyruvate, β-mercaptoethanol, 3 μM CHIR99021 (Chi), 1 μM PD0325901 and 0.1 μg ml−1 LIF. For cell passaging, cells were detached using StemPro Accutase (GIBCO). Gata6-Venus (Freyer et al., 2015), Flk1-GFP (Jakobsson et al., 2010), Mesp1-GFP (Bondue et al., 2011) and Hcn4-GFP::Tbx1Cre-RFP (Andersen et al., 2018) cells were cultured on gelatin-coated tissue culture flasks; Sox1-GFP::Brachyury-mCherry (Deluz et al., 2016) and TCF/LEF-mCherry (Faunes et al., 2013; Ferrer-Vaquer et al., 2010) cells on tissue-culture flasks without coating. If not differently specified, Sox1-GFP::Brachyury-mCherry cells were used for our experiments. HUVECs were cultured in EGM-2 medium (Lonza). All cells were routinely tested for Mycoplasma with Mycoalert mycoplasma detection kit (Lonza) or by PCR.
METHOD DETAILS
Gastruloid culture
Gastruloids were generated as previously described (Baillie-Johnson et al., 2015). Briefly, 300–700 mESCs were plated in 40 μL N2B27 in 96-well Clear Round Bottom Ultra-Low Attachment Microplates (7007, Corning). After 48 h, 150 μL of N2B27 containing 3 μM Chi were added to each well. After 72 h, medium was changed with N2B27. Starting from 96 h, the protocol was optimized as described in Figure S1A. At 96 h, gastruloids were transferred in Ultra-Low Attachment 24-well Plates (3473, Corning) in 100 μL of medium, plus 700 μL of fresh N2B27 containing 30ng ml−1 bFGF (PMG0034, GIBCO), 5ng ml−1 VEGF 165 (PHC9394, GIBCO) and 0.5mM L-ascorbic acid phosphate (013–12061, Wako) (N2B27+++) and cultured on an orbital shaker placed at 37°C, 5%CO2 at 100rpm (VWR mini shaker). From 120 h onward, half medium was changed daily. Unless differently specified, N2B27+++ was applied from 96 to 144 h, while from 144 h to 168 h N2B27 was used for medium change. To generate Hcn4-GFP::Tbx1Cre-RFP gastruloids, 800–1200 cells were employed, due to initial difficulties in cell aggregation and extreme susceptibility to Chi treatment. For the same reason, Chi pulse for this line was done with 1 μM Chi, with the exact same modalities described for the other lines.
Live imaging and cell tracking
Bright-field live imaging of beating gastruloids was performed with a Nikon Ti inverted microscope equipped with an incubation chamber at 37°C, 5%CO2. Light-sheet live imaging of Flk1-GFP and Mesp1-GFP gastruloids was performed with a prototype of LS1 Live inverted light-sheet microscope (Viventis Microscopy Sarl, Switzerland), at 37°C, 5% CO2. A volume of 150–200 μm was acquired with a Z spacing of 2–3 μm between slices and pictures were captured every 20 min for Flk1-GFP gastruloids and every 10 min for tracking of Mesp1+ cells. Flk1-GFP light-sheet video montages were obtained with the Arivis Vision4D software. To track Mesp1+ cells in Gastruloids from 96 to 120 h, LS1 live light-sheet images were processed with the Fiji Mastodon plugin, using a semiautomatic tracking. Subsequently, Mastodon files were exported for Mamut, and the Fiji (Schindelin et al., 2012) Mamut plugin was used to display cell tracks as shown in Figure 1.
Immunofluorescence, confocal and light-sheet imaging on fixed samples
Immunofluorescence on whole mount gastruloids was performed as previously described (Baillie-Johnson et al., 2015). Briefly, gastruloids were washed in PBS and fixed in 4% PFA for 2 h at 4°C while shaking. Samples were washed 3 times in PBS and 3 times (10 min each) in blocking buffer (PBS, 10%FBS, 0,2%Triton X-100), then blocked for 1 h at 4°C in blocking buffer. Gastruloids were then incubated O/N with primary antibodies in blocking buffer, at 4°C while shaking. The day after, gastruloids were washed 4 times (20 min each) with blocking buffer, at 4°C while shaking, and incubated O/N with secondary antibodies and DAPI (2 μg ml−1, Sigma-Aldrich) in blocking buffer, at 4°C while shaking. The day after, gastruloids were washed for 1h with blocking buffer, at 4°C while shaking, then rinsed in PBS, 0,2%FBS, 0,2% Triton X-100 and mounted on Superfrost plus glass slides (ThermoFisher) with Floromount-G for confocal imaging (Southern Biotech). The following primary antibodies were used: mouse anti-Gata4 (1:500, Santa Cruz Biotechnology, G-4); chicken anti-GFP (1:750, Aves Labs); goat anti-Brachyury (1:300, Santa Cruz Biotechnology, C-19); rabbit anti-Brachyury (1:500, Abcam); rat anti-CD31 (1:100, BD, MEC 13.3), mouse anti-cardiac troponin T (1:100, ThermoFisher, 13–11), rabbit anti-E-Cadherin (1:500, Cell Signaling, 24E10). The following secondary antibodies were used: donkey anti-chicken 488 AlexaFluor (1:500, Jackson ImmunoResearch); donkey anti-goat AlexaFluor 568 (1:500, ThermoFisher); goat anti-rat AlexaFluor 568 (1:500, ThermoFisher); goat anti-mouse AlexaFluor 647 (1:500, ThermoFisher); donkey anti-rabbit 568 (1:500, ThermoFisher). Confocal pictures were acquired with a Zeiss LSM 700 inverted confocal microscope equipped with a Axiocam MRm black and white camera in the EPFL bioimaging and optics facility. Stitched images were obtained using the Fiji/ImageJ stitching plugin. For light-sheet imaging (Figures 4 and S4), samples were mounted in 1% low-melt agarose and cleared overnight with CUBIC mount solution (Lee et al., 2016). Light-sheet imaging was performed on a Zeiss Light-sheet Z1 microscope equipped with a Plan-Neofluar 20×/1.0 Corr nd = 1.45 objective. Light-sheet images were further processed with Imaris software, for 3D rendering and surface generation.
RNA extraction and qRT-PCR
RNA was extracted from gastruloids with the RNeasy Micro kit (QIAGEN), according to manufacturer’s instructions and quantified with a spectrophotometer (ND-1000, Nanodrop). 1 μg of RNA was reverse-transcribed with the iScript cDNA Supermix kit (Biorad). cDNA was diluted 1:10 and 1.5 μL of cDNA per reaction were used, in a total volume of 10 μl. 384 well plates were prepared using a robotized liquid handling platform (Hamilton Microlab Star). qPCR was run with a 7900HT Fast PCR machine (Applied Biosystems), using Power SYBR Green PCR Master Mix (Applied Biosystems), with an annealing temperature of 60°C. Gene expression was normalized on β-actin expression. Relative fold expression was calculated with the 2-ΔΔCT method. 500nM of the following primers were used: Mesp1 FOR GTCTGCAGCGGGGTGTCGTG; Mesp1 REV CGGCGGCGTCCAGGTTTCTA; Nkx2.5 FOR CACATTTTACCCGGGAGCCT; Nkx2.5 REV ACCAGATCTTGACCTGCGTG; Hcn4 FOR GTGGGGGCCACCTGCTAT; Hcn4 REV GTCGGGTGTCAGGCGGGA; α-actinin FOR GGGCTATGAGGAGTGGCTATT; α-actinin REV AGTCCTTCTGCAGCAAGATCT; RyR2 FOR TGCATGAGAGCATCAAACGC; RyR2 REV CGCGGAGAGAGGCATTACAT; Tbx5 FOR GGCATGGAAGGAATCAAGGTG; Tbx5 REV TTTGGGATTAAGGCCAGTCAC; Tbx1 FOR CTGTGGGACGAGTTCAATCAG; Tbx1 REV TTGTCATCTACGGGCACAAAG; Isl1 FOR ATGATGGTGGTTTACAGGCTAAC; Isl1 REV TCGATGCTACTTCACTGCCAG; FGF10 FOR TCAGCGGGACCAAGAATGAAG; FGF10 REV CGGCAACAACTCCGATTTCC; β-actin FOR CTGTCGAGTCGCGTCCACC; β-actin REV CGCAGCGATATCGTCATCCA.
Single cell RNA-seq
Gastruloids at 96, 120, 144 and 168h were collected, washed twice in PBS, and dissociated with TrypLE Express (ThermoFisher) for 8 min at 37°C, with mechanical dissociation at half time. The enzymatic reaction was blocked with a medium containing 10% ES qualified serum. Samples were washed with PBS and centrifuged for 5 min at 200 g. Cells were resuspended in N2B27 medium and strained at 40 μm before counting. All samples showed a cell viability > 90% after processing, based on Trypan blue exclusion. Libraries for single cell gene expression profiling were constructed with 10x Genomics Chromium 3′ chemistry v3.1, and sequenced with Illumina Hiseq 4000 (read length: barcodes 28, index 8, transcript 91) at the EPFL Gene Expression Core Facility (GECF) according to standard protocols. Reads were aligned to the mouse genome mm10 with cellranger v3.1. The raw count matrices were imported in R v3.6 with Seurat v3.1 (Stuart et al., 2019). The complete code for reproduction or for exploration of the results is available on [https://github.com/nbroguiere/Cardiac_Gastruloids]. In short, live cells were selected based on gene counts (> 2000) and mitochondrial content (1.5%−15%). The in vivo gastrulation and early organogenesis dataset from Pijuan-Sala et al. (2019) was used as an atlas. An scmap classifier (Kiselev et al., 2018) trained on the in vivo data was used to do a first estimation of cell types present in the gastruloids, for the needs of doublet removal and alignment. Data was normalized to 10 000 counts per cell (raw counts of unique molecule identifiers were approx. 30–40 k per cell depending on the condition), and log transformed using natural logarithm and a pseudo-count of 1. Gene expression refers to such log-normalized count values throughout the manuscript. Doublets were removed with DoubletFinder 2.0.3 (McGinnis et al., 2019). For the alignment, extraembryonic tissues, which are absent in gastruloids, were excluded from the in vivo data. Alignment of in vitro and in vivo data was done based on the intersection of variable genes, centered to their mean and divided by their standard deviation (referred to as scaled gene expression throughout). Both datasets were projected in the principal component (PC) space of the atlas, and batch corrected individually with Harmony v1 (Korsunsky et al., 2019) in ‘atlas mode’ (keeping one of the replicates as a constant so that the batch corrected coordinates remain comparable with each other). Gastruloid and embryo data were then aligned with Harmony in atlas mode. During this alignment, the cell types almost entirely missing in gastruloids according to the scmap transfer (less than 2 cells per cell type, blood/erythroid cells and allantois) were temporarily excluded to limit over-alignment artifacts. The resulting Harmony coordinates were used to perform a refined identity transfer from the Pijuan-Sala et al. (2019) annotations, based on nearest centroid, by analogy with scmap, and to compute an integrated UMAP. To best preserve global structure despite of the large cell number, we first performed a UMAP on 200 cells per cell type, initialized on a diffusion map and with a number of neighbors of 300 in uwot v0.1.5. We then used this layout supplemented with noise to initialize the UMAP with all cells. Not all continuity relationships could simultaneously appear in 2D projections, so 3D and 4D UMAPs were also used for data exploration. The alignment was confirmed to be biologically meaningful by manually verifying that highly variable genes and canonical marker genes were overall well matched. In addition, the similarity of the gene expression by cell type when comparing embryo and gastruloids was used as a summary statistic for the alignment and identity transfer accuracy (based on the scaled expression of the union of variable genes, using cosine metric). Since biological details can be lost in aligned datasets, the gastruloids were then re-analyzed independently in a similar fashion: variable genes, scaled gene expression, principal components, and Louvain clustering were computed for gastruloids alone. Markers were defined as genes which are expressed in less than 50% of the cells in the dataset (we considered genes that are ubiquitously expressed not useful as markers), and are significantly upregulated in a given cluster (p < 1e-3, fold change > 150%). Clusters were assigned to cell types based on their markers, and since some Louvain clusters were fused by similarity, markers were re-computed by cell type (Table S1). The UMAP view of gastruloids alone was initialized on the aligned gastruloid-embryo UMAP view, to keep the overall orientation constant and thus aid visualization, but let to re-converge for 1000 epochs to ensure local structures are not influenced by the atlas. A close-up view of heart fields development was generated by subsetting the clusters which are on the trajectories between epiblasts and cardiac cells, and recomputing scaled gene expression, PCs, and UMAP (initialized on the previous view for the same reason). For signature visualization on UMAP plots, the sum of the log-normalized expression of genes in the signature was computed, and normalized between its average value, considered as zero enrichment, to its 99th quantile, considered most enriched. For RNA velocity, splicing information was recovered using velocyto v0.17 (La Manno et al., 2018) based on the cell ranger alignment. Trajectories were then inferred from the RNA-velocity using scVelo v0.2.1 (Bergen et al., 2020) in deterministic mode, importing the view and annotations from R to Python v3.7.2 using scanpy v1.5.1 (Wolf et al., 2018).
RNAscope
Gastruloids were washed in PBS and fixed O/N in 4% PFA, at 4°C while shaking. The day after, samples were washed 3 times in PBS and included in HistoGel (ThermoFisher) blocks. HistoGel blocks were then processed with a Tissue-Tek VIP 6 AI Vacuum Infiltration Processor (Sakura) and included in paraffin. Paraffin blocks were cut at 4 μm with a Hyrax M25 microtome (Zeiss). RNA-scope was performed with the ACDBio Manual assay kit using RNAscope Probe-Mm-Tbx1 (481911), RNAscope Probe-Mm-Isl1-C3 (451931-C3) and RNAscope Probe-Mm-Tbx5-C2 (519581-C2) probes, according to manufacturer’s instructions. Polr2a-C1, Ppib-C2 and Ubiquitin-C3 probes were used as positive and negative controls. Pictures were acquired with an upright Leica DM5500 scanning microscope equipped with a CCD DFC 3000 black and white camera. Tile scans were directly stitched by the Leica LAS X imaging software.
Flow cytometry analysis and FACS
Gastruloids were collected, washed in PBS, and digested in 4mg ml−1 dispase I (Roche), 3mg ml−1 collagenase IV (GIBCO) and 100 μg ml−1 DNase I (Roche) in PBS (2 digestion cycles at 37°C, 5 min each; gentle pipetting was applied between the two cycles to mechanically dissociate the gastruloids). Digestion was blocked with DMEM containing 10% FBS, then samples were centrifuged and the cell pellet was resuspended in sorting buffer (PBS, 5%FBS, 1mM EDTA, 1%P/S) for antibody staining. Samples were incubated for 1 h on ice with antibodies, and 30min on ice with Aqua live/dead fixable dead cell stain kit (405/525nm, Invitrogen) or 10 min on ice with DAPI. Unstained, FMO and single-color samples were used as controls. The following antibodies were used: anti CD31-PE 1:1200 (BD, MEC 13.3), anti VEGFR2/Flk1-APC 1:200 (Biolegend, Avas12); anti CXCR4-APC 1:100 (BD, 2B11). Samples were analyzed with a BD LSR II flow cytometer, data were processed using FlowJo. Cell sorting was performed using a BD FACSAria Fusion cell sorter.
In vitro angiogenesis assay
168-h Flk1-GFP gastruloids were collected and digested as described above for FACS analysis. Flk1+ and Flk1− cells were isolated through cell sorting using a BD FACSAria Fusion cell sorter. 5*104 cells per condition were plated in IBIDI μ-angiogenesis slides pre-coated with 10 μL reduced growth factor Matrigel (Corning) in the lower chamber. Undifferentiated mESCs and HUVEC were used as negative and positive controls, respectively. Live imaging of tube formation was performed with a Nikon Ti inverted microscope equipped with an incubation chamber at 37°C, 5% CO2, with acquisitions every 15 min.
Calcium imaging
To image calcium fluxes, gastruloids were incubated for 1 h with 8 μM Cal-520 (AAT Bioquest) at 37°C, 5% CO2. Gastruloids were then transferred to fresh medium before imaging. Imaging was performed with a Light-sheet Z1 microscope (Zeiss) equipped with an environmental chamber to maintain gastruloids at 37°C and 5% CO2. For imaging, gastruloids were embedded in 1% low melt agarose and the chamber was filled with culture medium. Nifidepine (Sigma Aldrich, 10 μM) and isoproterenol (isoprenaline hydrochloride, Sigma I5627, 1 μM) were added with a syringe directly to the imaging chamber during acquisition. The analysis of calcium spikes was performed with the Fiji Stacks-plot Z axis profile plugin. The baseline intensity was normalized to the minimum value over 10 s. The ratio of fluorescence intensity to baseline intensity was calculated and results are shown as the percentage of increase over the baseline, which shows the relative changes in intracellular Ca2+.
Crescent analysis
Analysis of crescent geometrical properties was performed on Sox1-GFP::Brachyury-mCherry gastruloids using a custom-made MATLAB script for Imaris files with a Imaris XT feature and EasyXT. Initially, we generated artificial shapes to be used as reference for defined geometrical metrics. Using a custom script (crescent_generator.m), 3D objects were generated to mimic crescent structures with different characteristics. To do so, the script creates two spot objects, makes channels from these spots and finally subtracts one channel to the other. To analyze surfaces, 3D images from non-beating organoids at 144 h and beating organoids at 168 h were acquired with a Light-sheet Z1 microscope (Zeiss) after clearing, as described above. For each individual stack, a surface was created using a dedicated user interface (GUI_DetectAndAnalyze.m), defining the object that should be created for each channel (settings_.m). Due to variability in background and signal intensity, the threshold (absolute intensity) was adjusted manually for each surface. Using a custom script (makeCroissantMeasure_final.m) geometrical measurements were computed and the results exported in csv table. In the graph, we plot the measure of Spareness, which is described as the ratio between the volume of the object and the volume of the best fitted ellipsoid. All scripts and settings used for analysis are available at https://go.epfl.ch/Crescent_Analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of immunofluorescence signals along the A/P axis
Quantification of immunofluorescence signals along the A/P axis of gastruloids was performed similarly to what previously described (Baillie-Johnson et al., 2015; Turner et al., 2017). Whole gastruloid light-sheet images were re-oriented along the A/P axis, to make sure both poles were visible in the resulting projected image, using a Fiji BigDataViewer (Pietzsch et al., 2015) plugin (https://github.com/bigdataviewer/bigdataviewer-playground). The first mipmap level of the re-oriented data was exported with a voxel size of 1 micron in XY and 2 microns in Z, and mean projected along the Z axis. Confocal images (for Gata6-Venus gastruloids at 96h) were directly processed. Average projections were used to draw a line with a thickness spanning the full or most of the gastruloid area from the posterior to the anterior pole of the gastruloid, to define directionality. Posterior pole was defined as the one expressing Brachyury. Due to frequent bending of the 168h gastruloid posterior portion on itself, the line was adjusted to avoid unwanted measuring of the same area twice, which would create aberrations. The “Straighten” tool in Fiji was used to double check for every image that the line was drawn to correctly capture gastruloid full length along the A/P axis, with no aberrations or multiple measures of the same area. A few 168h gastruloids with the posterior portion completely bended and touching the main gastruloid body could not be correctly straightened and were therefore excluded from analyses. To avoid background signal interference, a mask was created on the DAPI channel, and all background pixels outside this mask were converted to a “not a number” value. Finally, signal intensity was measured based on the Fiji “Plot Profile” tool, and values obtained were normalized to a 0–1 scale to be able to compare results from different gastruloids. The graphs display a smoothed curve, obtained with a LOWESS function in Graphpad Prism, and the relative standard deviation.
Statistics
All data shown in column graphs are expressed as mean ± SD, apart from the graph showing Calcium spikes frequency, which is expressed as mean ± whiskers from min to max. All other graphs show single data points. Statistical analysis between two columns was performed using two-tailed unpaired Student’s t test, whereas data containing more than two experimental groups were analyzed with one-way analysis of variance followed by Bonferroni’s test. To calculate the significance of the percentage of increase over baseline frequency after Isoproterenol administration, we applied a one sample t test. Statistical significance was calculated using the Graphpad Prism software, that was also used to generate all graphs. For each experiment, the exact value of n and what n represents can be found in the corresponding figure legends.
*p < 0.05; **p < 0.01; ***p < 0.001; confidence intervals 95%; alpha level 0.05.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Gata4 (clone G-4) | Santa Cruz Biotechnology | Cat # sc-25310; RRID: AB_627667 |
| Chicken polyclonal anti-GFP | Aves labs | Cat # GFP-1020; RRID: AB_2307313 |
| Goat polyclonal anti-Brachyury (C-19) | Santa Cruz Biotechnology | Cat # sc-17745; RRID:AB_2200243 |
| Rabbit monoclonal anti-Brachyury (clone EPR18113) | Abcam | Cat # ab209665; RRID: AB_2750925 |
| Rat monoclonal anti-CD31 (clone MEC 13.3) | BD | Cat # 553370; RRID:AB_394816 |
| Mouse monoclonal anti-cardiac troponin (clone 13–11) | ThermoFisher | Cat # MA5–12960; RRID:AB_11000742 |
| Rabbit monoclonal anti-E-Cadherin (24E10) | Cell Signaling Technology | Cat # 3195; RRID:AB_2291471 |
| PE rat monoclonal anti-CD31 (clone MEC 13.3) | BD | Cat # 561073; RRID:AB_10563931 |
| APC rat monoclonal anti-VEGFR2/Flk1 (clone Avas12) | Biolegend | Cat # 136405; RRID:AB_2044066 |
| APC rat monoclonal anti-CXCR4 (clone 2B11) | BD | Cat # 558644; RRID:AB_1645219 |
| RNAscope Probe-Mm-Tbx1 | ACD bio | Cat #481911 |
| RNAscope Probe-Mm-Isl1-C3 | ACD bio | Cat # 451931-C3 |
| RNAscope Probe-Mm-Tbx5-C2 | ACD bio | Cat # 519581-C2 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CHIR99021 | Merck/Millipore | Cat # 361559; CAS: 252917-06-9 |
| PD0325901 | Selleck Chemicals | Cat # S1036; CAS: 391210-10-9 |
| Recombinant human VEGF | GIBCO | Cat # PHC9394 |
| Recombinant mouse FGFb | GIBCO | Cat # PMG0035 |
| L-ascorbic acid phosphate | Wako | Cat #013–12061; CAS: 1713265-25-8 |
| Cal-520 | AAT Bioquest | Cat# 21130 |
| Nifidepine | Sigma Aldrich | Cat # N7634; CAS: 21829-25-4 |
| DL-Isoproterenol hydrochloride | Sigma Aldrich | Cat # I5627; CAS: 51-30-9 |
| Deposited Data | ||
| Single cell RNA-seq data | This paper | GEO: GSE158999 |
| Experimental Models: Cell Lines | ||
| Gata6-H2BVenus (ColA1TetO-Gata4-mCherry/+; R26M2rtTA/+;Gata6H2B-Venus/+) | Laboratory of Alfonso Martinez Arias | KH2 (C57BL6 × 129Sv) |
| Flk1-GFP | Laboratory of Alexander Medvinsky | E14Tg2a |
| Mesp1-GFP | Laboratory of Cédric Blanpain | N/A |
| Hcn4-GFP::Tbx1Cre-RFP | Laboratory of Chulan Kwon | N/A |
| Sox1-GFP::Brachyury-mCherry | Laboratory of David Suter | CGR8, strain 129 |
| TCF/LEF-mCherry | Laboratory of Alfonso Martinez Arias | E14Tg2A |
| Oligonucleotides | ||
| Primers, see STAR Methods | This Paper | N/A |
| Software and Algorithms | ||
| Fiji/ImageJ | Schindelin et al., 2012 | https://imagej.net/Fiji |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Arivis Vision4D | Arivis | https://imaging.arivis.com/imaging-science/arivis-vision4d |
| Imaris | Oxford Instruments | https://imaris.oxinst.com |
| FlowJo | BD | https://www.flowjo.com |
| MATLAB | MathWorks | https://ch.mathworks.com/products/matlab.html |
| Seurat v3.1 | Stuart et al., 2019 | https://satijalab.org/seurat/ |
| Velocyto v0.17 | La Manno et al., 2018 | http://velocyto.org |
| scVelo v0.2.1 | Bergen et al., 2020 | https://pypi.org/project/scvelo/ |
| Scanpy v1.5.1 | Wolf et al., 2018 | https://scanpy.readthedocs.io/en/stable/ |
| Harmony v1 | Korsunsky et al., 2019 | https://github.com/immunogenomics/harmony |
| DoubletFinder 2.0.3 | McGinnis et al., 2019 | https://github.com/chris-mcginnis-ucsf/DoubletFinder |
| Cellranger v3.1 | 10x Genomics I nc | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Other | ||
| Scripts and settings used for crescent analysis | This paper | https://go.epfl.ch/Crescent_Analysis |
| scRNA-seq analysis scripts | This paper | https://github.com/nbroguiere/Cardiac_Gastruloids |
Highlights.
Gastruloids generate cardiovascular progenitors and form a vascular-like structure
Both first and second heart field-like progenitors are specified
Cardiac progenitors self-organize into crescent and heart tube-like beating domains
Cellular diversity and tissue-tissue interactions mimic embryonic development
ACKNOWLEDGMENTS
We thank Alfonso Martinez Arias for great support throughout the project. We thank Denis Duboule and Nadia Mercader for useful feedback on the manuscript. We thank Gioele La Manno for helpful discussion on single-cell RNA-seq. We thank members of the Lutolf laboratory for discussions and sharing materials, Cédric Blanpain (ULB) for providing Mesp1-GFP cells, Alexander Medvinsky (MRC Edinburgh) for providing Flk1-GFP cells, Andrew Oates (EPFL) and Peter Strnad (Viventis Microscopy Sàrl) for making light-sheet LS1 Live microscope available and for technical support, Olivier Burri for EasyXT development, Thierry Laroche for support with Z1 light-sheet imaging, Arne Seitz and other members of Bioimaging and Optics Facility (EPFL) for microscopy support, Jessica Sordet-Dessimoz and Gian-Filippo Mancini from Histology Core Facility for RNAscope, and all personnel of Histology Core Facility, Flow Cytometry Core Facility, and Gene Expression Core Facility (EPFL) for their technical support. This work was funded by EPFL.
Footnotes
DECLARATION OF INTERESTS
A.B. is part of Viventis Microscopy Sàrl that has commercialized the LS1 light-sheet microscope used in this study for time-lapse imaging of gastruloids. The EPFL (with Cambridge Enterprise Limited) has filed for patent protection (PCT/GB2019/052668) on the embryoid technology described herein, and M.P.L. and G.R. are named as inventors on the patent.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.stem.2020.10.013.
REFERENCES
- Andersen P, Tampakakis E, Jimenez DV, Kannan S, Miyamoto M, Shin HK, Saberi A, Murphy S, Sulistio E, Chelko SP, and Kwon C (2018). Precardiac organoids form two heart fields via Bmp/Wnt signaling. Nat. Commun 9, 3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie-Johnson P, van den Brink SC, Balayo T, Turner DA, and Martinez Arias A (2015). Generation of aggregates of mouse embryonic stem cells that show symmetry breaking, polarization and emergent collective behaviour in vitro. J. Vis. Exp 53252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari L, Moris N, Girgin M, Turner DA, Baillie-Johnson P, Cossy A-C, Lutolf MP, Duboule D, and Arias AM (2018). Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272–276. [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol Published online August 3, 2020. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- Bondue A, Tännler S, Chiapparo G, Chabab S, Ramialison M, Paulissen C, Beck B, Harvey R, and Blanpain C (2011). Defining the earliest step of cardiovascular progenitor specification during embryonic stem cell differentiation. J. Cell Biol 192, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, and Seidman CE (1999). Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev. Biol 211, 100–108. [DOI] [PubMed] [Google Scholar]
- Brutsaert DL (2003). Cardiac endothelial-myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol. Rev 83, 59–115. [DOI] [PubMed] [Google Scholar]
- Brutsaert DL, Fransen P, Andries LJ, De Keulenaer GW, and Sys SU (1998). Cardiac endothelium and myocardial function. Cardiovasc. Res 38, 281–290. [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, and Zaffran S (2005). Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet 6, 826–835. [DOI] [PubMed] [Google Scholar]
- Clevers H (2016). Modeling development and disease with organoids. Cell 165, 1586–1597. [DOI] [PubMed] [Google Scholar]
- Cortes C, Francou A, De Bono C, and Kelly RG (2018). Epithelial properties of the second heart field. Circ. Res 122, 142–154. [DOI] [PubMed] [Google Scholar]
- de Soysa TY, Ranade SS, Okawa S, Ravichandran S, Huang Y, Salunga HT, Schricker A, Del Sol A, Gifford CA, and Srivastava D (2019). Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 572, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluz C, Friman ET, Strebinger D, Benke A, Raccaud M, Callegari A, Leleu M, Manley S, and Suter DM (2016). A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 30, 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faunes F, Hayward P, Descalzo SM, Chatterjee SS, Balayo T, Trott J, Christoforou A, Ferrer-Vaquer A, Hadjantonakis A-K, Dasgupta R, and Arias AM (2013). A membrane-associated β-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 140, 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Chen L, Nguyen PK, Wu SM, and Li G (2019). Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front. Cardiovasc. Med 6, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Vaquer A, Piliszek A, Tian G, Aho RJ, Dufort D, and Hadjantonakis A-K (2010). A sensitive and bright single-cell resolution live imaging reporter of Wnt/ß-catenin signaling in the mouse. BMC Dev. Biol 10, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Schumacher N, Maier M, Sendtner M, and Gessler M (2004). The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18, 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer L, Schröter C, Saiz N, Schrode N, Nowotschin S, Martinez-Arias A, and Hadjantonakis A-K (2015). A loss-of-function and H2B-Venus transcriptional reporter allele for Gata6 in mice. BMC Dev. Biol 15, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert S, and Kühl M (2010). The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res 107, 186–199. [DOI] [PubMed] [Google Scholar]
- Ghimire S, Van der Jeught M, Neupane J, Roost MS, Anckaert J, Popovic M, Van Nieuwerburgh F, Mestdagh P, Vandesompele J, Deforce D, et al. (2018). Comparative analysis of naive, primed and ground state pluripotency in mouse embryonic stem cells originating from the same genetic background. Sci. Rep 8, 5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E, Bellin M, Sala L, van Meer BJ, Tertoolen LGJ, Orlova VV, and Mummery CL (2017). Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 144, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Sun Y, Zhou B, Adam RM, Li X, Pu WT, Morrow BE, Moon A, and Li X (2011). A Tbx1-Six1/Eya1-Fgf8 genetic pathway controls mammalian cardiovascular and craniofacial morphogenesis. J. Clin. Invest 121, 1585–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SE, Sozen B, Christodoulou N, Kyprianou C, and Zernicka-Goetz M (2017). Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810. [DOI] [PubMed] [Google Scholar]
- Harvey RP (2002). Patterning the vertebrate heart. Nat. Rev. Genet 3, 544–556. [DOI] [PubMed] [Google Scholar]
- Hosseini HS, Garcia KE, and Taber LA (2017). A new hypothesis for foregut and heart tube formation based on differential growth and actomyosin contraction. Development 144, 2381–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N, Loskill P, Deveshwar N, Spencer CI, Judge LM, Mandegar MA, Fox CB, Mohamed TMA, Ma Z, Mathur A, et al. (2016). Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci. Rep 6, 24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovitch K, Temiño S, and Torres M (2017). Live imaging of heart tube development in mouse reveals alternating phases of cardiac differentiation and morphogenesis. eLife 6, e30668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson L, Franco CA, Bentley K, Collins RT, Ponsioen B, Aspalter IM, Rosewell I, Busse M, Thurston G, Medvinsky A, et al. (2010). Endothelial cells dynamically compete for the tip cell position during angiogenic sprouting. Nat. Cell Biol 12, 943–953. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, and Keller GM (2006). Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell 11, 723–732. [DOI] [PubMed] [Google Scholar]
- Kelly RG, Buckingham ME, and Moorman AF (2014). Heart fields and cardiac morphogenesis. Cold Spring Harb. Perspect. Med 4, a015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro H, Yonei-Tamura S, Tamura K, Schoenwolf GC, and Saijoh Y (2018). The heart tube forms and elongates through dynamic cell rearrangement coordinated with foregut extension. Development 145, dev152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Yiu A, and Hemberg M (2018). scmap: projection of single-cell RNA-seq data across data sets. Nat. Methods 15, 359–362. [DOI] [PubMed] [Google Scholar]
- Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs LT, Xue Y, Norton CR, Shutter JR, Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. (2000). Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 14, 1343–1352. [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamalice L, Le Boeuf F, and Huot J (2007). Endothelial cell migration during angiogenesis. Circ. Res 100, 782–794. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, and Knoblich JA (2014). Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125. [DOI] [PubMed] [Google Scholar]
- Le Garrec J-F, Domínguez JN, Desgrange A, Ivanovitch KD, Raphaël E, Bangham JA, Torres M, Coen E, Mohun TJ, and Meilhac SM (2017). A predictive model of asymmetric morphogenesis from 3D reconstructions of mouse heart looping dynamics. eLife 6, e28951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Choi J, Jo Y, Kim JY, Jang YJ, Lee HM, Kim SY, Lee H-J, Cho K, Jung N, et al. (2016). ACT-PRESTO: Rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci. Rep 6, 18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescroart F, Chabab S, Lin X, Rulands S, Paulissen C, Rodolosse A, Auer H, Achouri Y, Dubois C, Bondue A, et al. (2014). Early lineage restriction in temporally distinct populations of Mesp1 progenitors during mammalian heart development. Nat. Cell Biol 16, 829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sànchez-Dànes A, Moignard V, Dubois C, Paulissen C, Kinston S, et al. (2018). Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 359, 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, and Evans SM (2013). HCN4 dynamically marks the first heart field and conduction system precursors. Circ. Res 113, 399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park S-J, Kotikian A, Nesmith AP, Campbell PH, et al. (2017). Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat. Mater 16, 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang XB, Park DS, and Lisanti MP (2002). Caveolin-1 expression enhances endothelial capillary tubule formation. J. Biol. Chem 277, 10661–10668. [DOI] [PubMed] [Google Scholar]
- Lough J, and Sugi Y (2000). Endoderm and heart development. Dev. Dyn 217, 327–342. [DOI] [PubMed] [Google Scholar]
- Ma Z, Wang J, Loskill P, Huebsch N, Koo S, Svedlund FL, Marks NC, Hua EW, Grigoropoulos CP, Conklin BR, and Healy KE (2015). Self-organizing human cardiac microchambers mediated by geometric confinement. Nat. Commun 6, 7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, and Olson EN (2005). The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132, 189–201. [DOI] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, and Gartner ZJ (2019). DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RJ, Parker BL, Quaife-Ryan GA, Voges HK, Needham EJ, Bornot A, Ding M, Andersson H, Polla M, Elliott DA, et al. (2019). Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell 24, 895–907.e6. [DOI] [PubMed] [Google Scholar]
- Miquerol L, and Kelly RG (2013). Organogenesis of the vertebrate heart. Wiley Interdiscip. Rev. Dev. Biol 2, 17–29. [DOI] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, and Komuro I (2006). Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc. Natl. Acad. Sci. USA 103, 19812–19817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narmoneva DA, Vukmirovic R, Davis ME, Kamm RD, and Lee RT (2004). Endothelial cells promote cardiac myocyte survival and spatial reorganization: implications for cardiac regeneration. Circulation 110, 962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascone N, and Mercola M (1995). An inductive role for the endoderm in Xenopus cardiogenesis. Development 121, 515–523. [DOI] [PubMed] [Google Scholar]
- Ortmann D, Brown S, Czechanski A, Aydin S, Muraro D, Huang Y, Tomaz RA, Osnato A, Canu G, Wesley BT, et al. (2020). Naive pluripotent stem cells exhibit phenotypic variability that is driven by genetic variation. Cell Stem Cell 27, 470–481.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzsch T, Saalfeld S, Preibisch S, and Tomancak P (2015). BigDataViewer: visualization and processing for large image data sets. Nat. Methods 12, 481–483. [DOI] [PubMed] [Google Scholar]
- Pijuan-Sala B, Griffiths JA, Guibentif C, Hiscock TW, Jawaid W, Calero-Nieto FJ, Mulas C, Ibarra-Soria X, Tyser RCV, Ho DLL, et al. (2019). A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulescu ME, Schmidt I, Giaimo BD, Antoine T, Berkenfeld F, Ferrante F, Park H, Ehling M, Biljes D, Rocha SF, et al. (2017). Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat. Cell Biol 19, 915–927. [DOI] [PubMed] [Google Scholar]
- Polonchuk L, Chabria M, Badi L, Hoflack J-C, Figtree G, Davies MJ, and Gentile C (2017). Cardiac spheroids as promising in vitro models to study the human heart microenvironment. Sci. Rep 7, 7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala K, Pekkanen-Mattila M, and Aalto-Setälä K (2011). Cardiac differentiation of pluripotent stem cells. Stem Cells Int. 2011, 383709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivron NC, Frias-Aldeguer J, Vrij EJ, Boisset J-C, Korving J, Vivié J, Truckenmüller RK, van Oudenaarden A, van Blitterswijk CA, and Geijsen N (2018). Blastocyst-like structures generated solely from stem cells. Nature 557, 106–111. [DOI] [PubMed] [Google Scholar]
- Rossi G, Manfrin A, and Lutolf MP (2018). Progress and potential in organoid research. Nat. Rev. Genet 19, 671–687. [DOI] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, and Taketo MM (1996). MesP1: a novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development 122, 2769–2778. [DOI] [PubMed] [Google Scholar]
- Sasai Y (2013). Cytosystems dynamics in self-organization of tissue architecture. Nature 493, 318–326. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss TM, Xydas S, and Lassar AB (1995). Induction of avian cardiac myogenesis by anterior endoderm. Development 121, 4203–4214. [DOI] [PubMed] [Google Scholar]
- Shao Y, Taniguchi K, Townshend RF, Miki T, Gumucio DL, and Fu J (2017a). A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun 8, 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Taniguchi K, Gurdziel K, Townshend RF, Xue X, Yong KMA, Sang J, Spence JR, Gumucio DL, and Fu J (2017b). Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater 16, 419–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozen B, Amadei G, Cox A, Wang R, Na E, Czukiewska S, Chappell L, Voet T, Michel G, Jing N, et al. (2018). Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol 20, 979–989. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugi Y, and Lough J (1995). Activin-A and FGF-2 mimic the inductive effects of anterior endoderm on terminal cardiac myogenesis in vitro. Dev. Biol 168, 567–574. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, and Lee RT (2003). Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 107, 1912–1916. [DOI] [PubMed] [Google Scholar]
- Tewary M, Dziedzicka D, Ostblom J, Prochazka L, Shakiba N, Heydari T, Aguilar-Hidalgo D, Woodford C, Piccinini E, Becerra-Alonso D, et al. (2019). High-throughput micropatterning platform reveals Nodal-dependent bisection of peri-gastrulation-associated versus preneurulation-associated fate patterning. PLoS Biol. 17, e3000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DA, Girgin M, Alonso-Crisostomo L, Trivedi V, Baillie-Johnson P, Glodowski CR, Hayward PC, Collignon J, Gustavsen C, Serup P, et al. (2017). Anteroposterior polarity and elongation in the absence of extra-embryonic tissues and of spatially localised signalling in gastruloids: mammalian embryonic organoids. Development 144, 3894–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyser RC, Miranda AM, Chen C-M, Davidson SM, Srinivas S, and Riley PR (2016). Calcium handling precedes cardiac differentiation to initiate the first heartbeat. eLife 5, e17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink SC, Baillie-Johnson P, Balayo T, Hadjantonakis A-K, Nowotschin S, Turner DA, and Martinez Arias A (2014). Symmetry breaking, germ layer specification and axial organisation in aggregates of mouse embryonic stem cells. Development 141, 4231–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink SC, Alemany A, van Batenburg V, Moris N, Blotenburg M, Vivié J, Baillie-Johnson P, Nichols J, Sonnen KF, Martinez Arias A, and van Oudenaarden A (2020). Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 582, 405–409. [DOI] [PubMed] [Google Scholar]
- Varner VD, and Taber LA (2012). Not just inductive: a crucial mechanical role for the endoderm during heart tube assembly. Development 139, 1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenvliet JV, Bolondi A, Kretzmer H, Haut L, Scholze-Wittler M, Schifferl D, Koch F, Pustet M, Heimann S, Buschow R, et al. (2020). Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. bioRxiv. 10.1101/2020.03.04.974949. [DOI] [PubMed] [Google Scholar]
- Wang W, Niu X, Stuart T, Jullian E, Mauck WM 3rd, Kelly RG, Satija R, and Christiaen L (2019). A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nat. Cell Biol 21, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Zhang S, Greder L, Dutton J, Keirstead SA, Lepley M, Zhang L, Kaufman D, and Zhang J (2013). Effective cardiac myocyte differentiation of human induced pluripotent stem cells requires VEGF. PLoS ONE 8, e53764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pu W, Li G, Huang X, He L, Tian X, Liu Q, Zhang L, Wu SM, Sucov HM, and Zhou B (2016). Endocardium minimally contributes to coronary endothelium in the embryonic ventricular free walls. Circ. Res 118, 1880–1893. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY, Aggarwal P, Zhang B, Conant G, Ronaldson-Bouchard K, et al. (2019). A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 176, 913–927.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Xue X, Shao Y, Wang S, Esfahani SN, Li Z, Muncie JM, Lakins JN, Weaver VM, Gumucio DL, and Fu J (2019). Controlled modelling of human epiblast and amnion development using stem cells. Nature 573, 421–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Wang J, Guo C, Chang W, Zhuang J, Zhu P, and Li X (2017). Temporally distinct Six2-positive second heart field progenitors regulate mammalian heart development and disease. Cell Rep. 18, 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The single cell RNA-seq data generated during this study are available at NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/) under the accession number GEO: GSE158999. The complete code for reproduction or for exploration of the results is available on https://github.com/nbroguiere/Cardiac_Gastruloids. All scripts and settings used for volumetric crescent analysis are available at https://go.epfl.ch/Crescent_Analysis.


