Abstract
Background and Objectives:
Due to the important role of Streptococcus agalactiae, Group B streptococci (GBS), in production of invasive disease in neonates, investigation regarding the pathogenicity and antibiotic resistance factors is necessary in selecting the appropriate therapeutic agents. Beside capsule, the pilus has been currently recognized as an important factor in enhancing the pathogenicity of GBS. Resistance of GBS to selected antibiotics is noticeably increasing which is mainly due to the anomalous use of these drugs for treatment. The aim of this study was to determine the prevalence of pili genes followed by antibiotic susceptibility of GBS, previously serotyped, isolated from pregnant women in the city of Yazd, Iran.
Materials and Methods:
Fifty seven GBS from pregnant women were subjected to multiplex PCR for determination of PI-1, PI-2a and PI-2b pilus-islands and simultaneously, the phenotype of antibiotic resistance to penicillin, tetracycline, erythromycin, clindamycin, gentamycin and levofloxacin was determined. Antibiotic resistance genes (ermA, ermB, mefA, tetM, int-Tn) were further diagnosed using PCR and multiplex PCR.
Results:
PI-1+PI-2a with 71.9%; followed by PI-2a (21.1%) and PI-2b (7%) were observed. PI-1+PI-2a in serotype III was (73.2%), serotype II, Ia, Ib and V were 12.2%, 9.8%, 2.4% and 2.4% respectively. GBS penicillin sensitive was 89.5% and 96.5% resistance to tetracycline. The frequency of resistance genes were as follows: tetM (93%), ermA (33.3%), ermB (8.8%), int-Tn (80.7%) and mefA (0).
Conclusion:
Majority of GBS contained PI-1+PI-2a. Hence presence of this pilus stabilizes the colonization, therefore designing a program for diagnosing and treatment of infected pregnant women seems to be necessary.
Keywords: Streptococcus agalactiae, Pili, Antibiogram, Women, Yazd
INTRODUCTION
Streptococcus agalactiae, or group B Streptococcus (GBS) may colonize both genitourinary and gastrointestinal tract of human population (1 –3). Different investigations revealed that 11–30% of pregnant women harbor this bacterium in their vagina and as a result their infant may get infected during the birth (2, 4). Investigation expressed that overall of 4% of infected infants develop sepsis, rarely pneumonia and meningitis before or after delivery (1, 5–7). The site for colonization of GBS is mucosal epithelial cells and for colonizing the bacterium needs an adherence factor (8). Three adherence factors have been recognized as capsule, surface protein and pili (9). Recently, ten serotypes of GBS have been recognized according to antigenic properties of their polysaccharide capsules as Ia, Ib, II ...VIII and IX (5, 10, 11). In addition to capsule, pili play an important role as adherence and pathogenicity factors for further successful colonization and invasion to the host tissue (1, 12). According to latest reports, three pilus island genes rendering the pilus–like structure on the surface of the GBS (PI-1, PI-2a, PI-2b). Pili are composed of three subunit; a back bone and two ancillary proteins (13, 14). More recently, investigations from European countries and USA expressed that GBS strain carry either PI-2a or PI-2b, but many have PI-1 (13, 15). Research performed in different societies represent that resistances of the GBS to selected antibiotics are noticeably increasing. This could be mainly due to uncontrolled of antibiotics utilization (16). The selected antibiotic for treatment of GBS is penicillin G (17) However, tetracycline, erythromycin and clindamycin are recommended for individuals who are allergic or infected with penicillin resistance GBS (18). Different investigation represent that some of GBS isolates are found resistance to one of the antibiotics ampicillin, vancomycine, penicillin and clindamycin (19, 20).
The aim of the present study was to determine the frequency of Pilus Island and antibiotic resistance profile of the perviously serotyped GBS isolated from pregnant women (4).
MATERIALS AND METHODS
Fifty seven GBS isolates which were previously collected (2015–2016), identified and serotyped were selected. These were subjected to pilus-island and antibiotic resistance determination using molecular technique. This study was considered and approved by Ethical Committee of Shahid Sadoughi University of Medical Sciences, Yazd, Iran [IR.SSU.MEDICINE.REC.1396.447]. All GBS isolates were re-inoculated on blood agar and incubated at 37°C for 24 hrs. We used purified DNA (GBS strain 389, 337) confirmed by Fanrong Kong from the centre for Microbiology Laboratory Services (New South Wales, Australia) as positive control and sterile water for negative control.
Genomic DNA extraction.
The DNA was extracted using boiling method as described by Madzivhandila et al. (14). One ml of the microbial suspension was transferred into a sterile microtube and centrifuged at 3000 rpm for 10 min. The supernatant was removed and 1 ml of PBS was added to the pellet and centrifuged. This process was repeated three times. The pellet was then mixed with 100 μl of nuclease free water and incubated at 95°C in water bath. Following 10 min incubation, the supernatant was removed gently into another microtube and stored at −20°C for further molecular examination.
Detection of Pilus Island genes.
The presence of GBS pilus island PI-1, PI-2a and PI2b was detected by multiplex PCR as described by Martins et al. (15).
In order to confirm the PI-1 negative isolates that did not carry the pilus pathogenicity island or parts specific of its, using primers PI-1- All. All isolates that were negative for PI-1 gene were subjected to PCR with primers PI-1- All.
Antibacterial susceptibility testing.
Sensitivity to penicillin (10 µg), tetracycline (30 µg), erythromycin (15 µg), clindamycin (2 µg), gentamycin (120 µg) and levofloxacin (5 µg) (MAST, England) was determined using Kirby Bauer test as described by CLSI (2018) (21). Briefly, McFarland 0.5 suspension of GBS was swabbed over the surface of Muller Hinton agar plate (Merck, Germany) contained 5% sheep blood. Then the disk containing each antibiotic was placed onto the inoculated surface. In addition inducible clindamycin resistance was detected using the D-Zone test according to CLSI 2018 guidelines (21). Following overnight incubation, the inhibition zone of each disk was measured and the isolate was interpreted as either susceptible, intermediate or resistance. Consistently Streptococcus pneumoniae (ATCC 49619) was used as a control sample.
Determination of resistance genes.
All GBS strains were subjected to PCR technique for determination of resistance genes ermB, mefA as previously described (22). Also multiplex PCR was employed for detection of ermA, tetM, int-Tn (22).
Statistical analysis.
Chi-square test was assessed using SPSS version 20.
RESULTS
Amplification of pilus gene.
As Table 1 and Fig. 1 shows, all isolates of GBS represent at least one pilus gene. Majority of the isolates (71.9%) contained the gene PI1+PI2a followed by PI2a (21.1%) and PI2b (7%). The gene PI1+ PI2b was not detected in any of the isolate. As Table 1 indicates, 33 (57.9%) of GBS were serotype III, among which, 30 (90.9%) were found to contain PI1+ PI2a and 3 isolates (9.1%) with PI2a gene.
Table 1.
Distribution of genes encoding Pilus-Island across serotypes
| Pilus Island | Capsular Serotypes. n (%) | |||||
|---|---|---|---|---|---|---|
| Ia | Ib | II | III | V | Total | |
| PI1+PI2a | 4 (50) | 1 (100) | 5 (35.7) | 30 (90.9) | 1 (100) | 41 (71.9) |
| PI2a | 1 (12.5) | 0 | 8 (57.1) | 3 (9.1) | 0 | 12 (21.1) |
| PI2b | 3 (37.5) | 0 | 1 (7.1) | 0 | 0 | 4 (7) |
| Total | 8 (100) | 1 (100) | 14 (100) | 33 (100) | 1 (100) | 57 (100) |
P value= 0.000
Fig. 1.
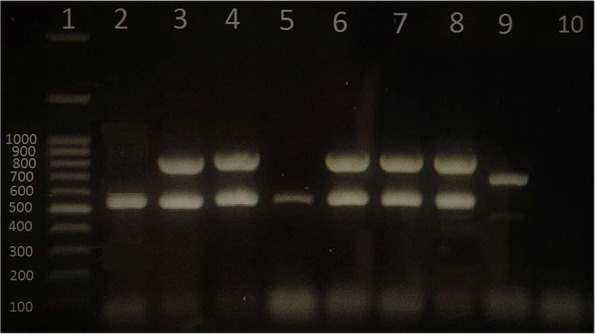
Gel electrophoresis of multiplex PCR amplification products for determination of Pilus Island genes of GBS isolates. Lane 1 is DNA ladder (100 bp, Fermentas USA). PI-1+PI-2a (lane 3, 4, 6, 7 and 8). PI-2a (Lane 2 and 5). PI-2b is lane 9. Lane 10 is negative control.
Antimicrobial susceptibility.
As Fig. 2 indicates, 51 (89.5%) of isolates were sensitive to penicillin, while 55 (96.5%) were resistant to tetracycline. Twelve isolates (21.1%) with positive D-zone were inducible resistance to clindamycin (iMLSB). Table 2 represents phenotypical and genotypical resistance to both macrolide and tetracycline. As it shows, 8 isolates were found to be phenotypically resistant to macrolide but 5 expressed ermB and 19 were ermA gene positive. Further consideration revealed that 11 (91.7%) out of 12 isolates contained ermA gene with serotype III and the remainder 1 isolate was found to be serotype Ia. Six isolates were resistance to both clindamycin and erythromycin (cMLSB). Among which, 5 isolates contained ermB and one ermA gene. Two (5%) of serotype III were resistance to erythromycin but sensitive to clindamycin (M phenotype) and 2 serotypes Ia and II were resistance to clindamycin but sensitive to erythromycin (L phenotype). Correlation between the resistance gene of tetM (93%), ermA (33.3%), ermB (8.8%), mefA (0) and int-Tn (80.7%) with pili and different capsular serotype are all shown in Table 3.
Fig. 2.
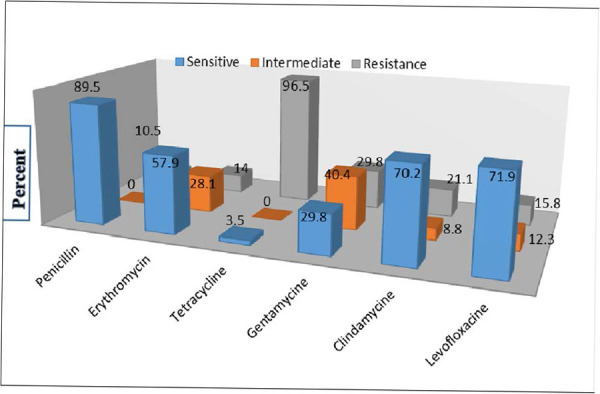
Antibiotic susceptibility of Group B Streptococcus isolates
Table 2.
Resistance phenotype and genotype (Macrolide – Tetracycline, n=57)
| Mcacrolide-resistance phenotype (n) | 8 |
|---|---|
| Macrolide-resistance genotype (n) | |
| ermB | 5 |
| ermA | 19 |
| mefA | 0 |
| ermA+ermB | 0 |
| Tetracycline resistance phenotype (n) | 55 |
| Tetracycline-resistance genotype (n) | |
| tetM | 53 |
| Int-Tn | 46 |
Table 3.
Distribution of serotypes and Pilus-Island among antimicrobial resistance genes of GBS isolates (N=57)
| Antibiotic resistance genotype (n/%) | Serotypes (n/%) | Pilus island (n/%) |
|---|---|---|
| ermA (19/33.3) | Ia (1/5.3), III (18/94.7) | PI2a (1/5.3), PI-1+PI2a (18/94.7) |
| ermB (5/8.8) | Ia (1/20), Ib (1/20), II (1/20), III (1/20), V (1/20) | PI2a (2/40), PI1+PI2a (3/60) |
| tetM (53/93) | Ia (6/11.3), Ib (1/1.9), II (13/24.5), III (32/60.4), V (1/1.9) | PI2a (10/18.9), PI2b (3/5.7), PI1+PI2a (40/75.7) |
| int-Tn (46/80.7) | Ia (6/13), II (8/17.4), III (31/67.4), V (1/2.2) | PI2a (6/13), PI2b (3/6.5), PI1+PI2a (37/80.4) |
| mefA (0) | 0 | 0 |
DISCUSSION
The present study was implicated one of the important virulence factor, Pilus-Island and tried to determine the correlation between the identified Pilus Island and the previous serotyped GBS in our area.
In addition, all isolates were further retested to find out the frequency of the resistance to selected antibiotics. All GBS were found to contain one of two genes encoded pili’s proteins (23). In addition, our study demonstrated that all isolates harbored at least one of PI gene alone or in combinations (Table 1).
As Table 1 shows, 41 (71.9%) of isolates contained PI-1+PI-2a. These results, however, concur with Margarit et al. (24) who reported that the most isolates they detected contained PI1+PI2a (46%). Although PI1+PI2b were not observed in our investigation, prevalence of PI2a and PI2b were found to be 21.1% and 7% respectively. Different studies represented that majority of pili gene detected from GBS strain are PI2a (25, 26). They claimed that this gene may contribute to biofilm synthesis, but PI1+PI2b does not play any function in biofilm synthesis. In a similar study, Rinaudo et al. showed that the most isolates of GBS (59.3%) harbor PI1+PI2a genes. The results they obtained are not compromised with our finding in which PI1+PI2a were found to be 71.9%. However, investigation concerning the role of PI1+PI2a in GBS revealed that the presence of this pilus-island can enhance the virulence of species in both colonization and formation of biofilm (26).
Martins et al. in Spain examined 898 GBS strains and reported that majority of isolates contained PI1+PI2a with 49% but PI2b was the least (0.6%). Also they represented that 21% of the isolates were found to harbor PI1+PI2b. Their results did not correspond with our finding and seems probably due to small number of cases we investigated (27).
Through our investigation, it was found that the majority of isolates contained PI1+PI2a and belong to serotype III (30/57), followed in descending order by serotype II (5), Ia (4), Ib (1) and V (1). This figure of prevalence observed in our study, correspond with finding of Lue et al. who reported that 62/88 (70.5%) of PI1+PI2a were serotype III (23). It is important to note that serotype III is more concerned with neonate meningitis (2).
Our finding also concur with those of Khodaei et al. who revealed that 67% of the GBS isolates were PI1+PI2a and were mostly seen in serotype III (28).
Madzivhandila et al. in Africa demonstrated that the prevalence of pili type PI1+PI2b was as 45.1% followed by PI2a (29.8%), PI1+PI2a (24.8%) and PI2b with 0.2%. In addition, they expressed that the presence of PI1+PI2b can cause invasive disease and PI1+PI2a contribute the GBS for colonization. They showed that PI1+ PI2b were seen with serotype III and PI2a were found mostly in serotype Ia (29).
Margarit et al. found the most common serotypes of Ib, II and V contained two separate pili, whereas, majority of pilus-island PI2a was found in serotype Ia (24).
Fig. 2. indicates the result of susceptibility of detected GBS against selected antibiotics. Although the majority of isolates were resistance to tetracycline (96.5%), they showed the lowest resistance to penicillin (10.5%). Majority of the isolates harbored gene tetM (93%) followed by int-tn (80.7), mefA (0), ermA (33.3%) and ermB (8.8%). In a review study, Gizachew (30) reported 36.6% resistance rate for penicillin among 1974 positive GBS detected from pregnant women. In a another study from Iran, 88.5% of isolates GBS were found to be sensitive to penicillin (33).
As Table 2 indicates, 8 GBS isolates were found resistance to erythromycin but, however, 19 isolates were positive for ermA. This controversy may be explained that this gene is not expressed yet (32).
When our results were compared to Martins et al. it was found that 85.8% of their obtained isolates were resistance to tetracycline. They also reported that only 16.1% and 14.2% of the GBS were resistance to erythromycin and clindamycin respectively. Concerning the gene distribution, we found that 93% of the isolates contain tetM gene, whereas they reported that 97.3% of their detected GBS have had the above gene (33).
Another study conducted by Heelan et al. revealed that 85% of the isolates were tetracycline resistance which is almost similar with our finding. They also expressed that 22% of the isolates were resistant to erythromycin and 100% of isolates contained erythromycin resistance gene. Important point was that they showed cMLSB isolates carried the gene ermA and ermB which was similar to our result. Similarly, they found that the phenotype of iMLSB was directly related to gene ermA activity (34).
tetM is recognized as a most prevalent gene in tetracycline resistant Gram positive bacteria and is frequently associated with conjugate elements which belong to the family of Tn916. Today, it is well known that int-Tn encodes the integrase of Tn916 continuously and it is found in 88% of strains with tetM (22). Our investigation revealed that 80.7% of isolated GBS contained int-Tn. This finding indicates that tetracycline resistance GBS is probably due to their achievement to transposon of Tn916. As Table 3 shows, there was not any mefA gene among our tested GBS. It seems that resistance of isolates to erythromycin is accomplished by transposon (ermB, ermA) and or through efflux pomp like mefA gene.
CONCLUSION
Results obtained from this investigation allow us to conclude that majority of isolated GBS contain pili PI1+PI2a and resistance of these isolates to clindamycin, erythromycin and tetracycline are constantly increasing. Since presence of these pili is believed to enhance the bacterial colonization on host׳s target tissue, diagnostic program followed by treatment of infected pregnant women during pregnancy is strongly recommended.
ACKNOWLEDGEMENTS
We would like to thank Fanrong Kong for his kindly assessment and also who provided us the known references GBS serotypes for our previous published work.
REFERENCES
- 1. Springman AC, Lacher DW, Waymire EA, Wengert SL, Singh P, Zadoks RN, et al. Pilus distribution among lineages of group b streptococcus: an evolutionary and clinical perspective. BMC Microbiol 2014; 14: 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sadeh M, Firouzi R, Derakhshandeh A, Khalili MB, Kong F, Kudinha T. Molecular characterization of Streptococcus agalactiae isolates from pregnant and non-pregnant women at Yazd University Hospital, Iran. Jundishapur J Microbiol 2016; 9 (2): e30412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Manshadi SAD, Alizadeh R, Salehi M, Seifi A, Seifi M. Bilateral septic arthritis of the knee caused by group B streptococci: a case report. Iran J Microbiol 2019; 11: 187– 190. [PMC free article] [PubMed] [Google Scholar]
- 4. Najarian N, Khalili MB, Astani A, Vakili M, Sadeh M. Serotype determination of Streptococcus agalactiae detected from vagina and urine of pregnant women in Yazd, Iran-2015. IJML 2018; 5: 49– 57. [Google Scholar]
- 5. Nazari A, Khalili MB, Astani A, Vakili M, Sadeh M, Mojibiyan M, et al. Determination of genotypes of Streptococcus agalactiae isolated from both urine and vagina of pregnant women referred to gynecology clinics of Yazd, Iran-2015. IJML 2017; 4: 180– 188. [Google Scholar]
- 6. Dehbashi S, Pourmand MR, Mashhadi R. Characterization of Afb, a novel bifunctional protein in Streptococcus agalactiae. Iran J Microbiol 2016; 8: 73– 79. [PMC free article] [PubMed] [Google Scholar]
- 7. Heath PT, Balfour GF, Tighe H, Verlander NQ, Lamagni TL, Efstratiou A, et al. Group B streptococcal disease in infants: a case control study. Arch Dis Child 2009; 94: 674– 680. [DOI] [PubMed] [Google Scholar]
- 8. Tamura G, Kuypers J, Smith S, Raff H, Rubens C. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect Immun 1994; 62: 2450– 2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wessels MR, Rubens CE, Benedi VJ, Kasper DL. Definition of a bacterial virulence factor: sialylation of the group B streptococcal capsule. Proc Natl Acad Sci U S A 1989; 86: 8983– 8987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, Creti R. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J Microbiol Methods 2010; 80: 212– 214. [DOI] [PubMed] [Google Scholar]
- 11. Hanh TQ, Van Du V, Hien PT, Chinh DD, Loi CB, Dung NM, et al. Prevalence and capsular type distribution of group B Streptococcus isolated from vagina of pregnant women in Nghe An province, Vietnam. Iran J Microbiol 2020; 12: 11– 17. [PMC free article] [PubMed] [Google Scholar]
- 12. Vengadesan K, Ma X, Dwivedi P, Ton-That H, Narayana SV. A model for group B Streptococcus pilus type 1: the structure of a 35-kDa C-terminal fragment of the major pilin GBS80. J Mol Biol 2011; 407: 731– 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sharma P, Lata H, Arya DK, Kashyap AK, Kumar H, Dua M, et al. Role of pilus proteins in adherence and invasion of Streptococcus agalactiae to the lung and cervical epithelial cells. J Biol Chem 2013; 288: 4023– 4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Madzivhandila M, Adrian PV, Cutland CL, Kuwanda L, Madhi SA, Team PT. Distribution of pilus islands of group B streptococcus associated with maternal colonization and invasive disease in South Africa. J Med Microbiol 2013; 62: 249– 253. [DOI] [PubMed] [Google Scholar]
- 15. Martins ER, Melo-Cristino J, Ramirez M. Evidence for rare capsular switching in Streptococcus agalactiae. J Bacteriol 2010; 192: 1361– 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Phares C, Lynfield R, Farley M, Mohle-Boetani J, Harrison L, Petit S, et al. Epidemiology of invasive group B streptococcal disease in the United States, 1999–2005. JAMA 2008; 299: 2056– 2065. [DOI] [PubMed] [Google Scholar]
- 17. Wang YH, Su LH, Hou JN, Yang TH, Lin TY, Chu C, et al. Group B streptococcal disease in nonpregnant patients: emergence of highly resistant strains of serotype Ib in Taiwan in 2006 to 2008. J Clin Microbiol 2010; 48: 2571– 2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gherardi G, Imperi M, Baldassarri L, Pataracchia M, Alfarone G, Recchia S, et al. Molecular epidemiology and distribution of serotypes, surface proteins, and antibiotic resistance among Group B streptococci in Italy. J Clin Microbiol 2007; 45: 2909– 2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sadowy E, Matynia B, Hryniewicz W. Population structure, virulence factors and resistance determinants of invasive, non-invasive and colonizing Streptococcus agalactiae in Poland. J Antimicrob Chemother 2010; 65: 1907– 1914. [DOI] [PubMed] [Google Scholar]
- 20. Daramroodi AK, Keshavarzi F, Raissi F. The investigation of antibiotic resistance and rapid detection of group B Streptococcus (Bca) from vaginal specimens of pregnant women by colony PCR method. J Bas Res Med Sci 2018; 5: 27– 32. [Google Scholar]
- 21. Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing: Approved Twenty-: Document M100-S28. Wayne, PA, USA: CLSI; 2018. [Google Scholar]
- 22. Poyart C, Jardy L, Quesne G, Berche P, Trieu-Cuot P. Genetic basis of antibiotic resistance in Streptococcus agalactiae strains isolated in a French hospital. Antimicrob Agents Chemother 2003; 47: 794– 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu B, Chen X, Wang J, Wang D, Zeng J, Li Y, et al. Molecular characteristics and antimicrobial resistance in invasive and noninvasive Group B Streptococcus between 2008 and 2015 in China. Diagn Microbiol Infect Dis 2016; 86: 351– 357. [DOI] [PubMed] [Google Scholar]
- 24. Margarit I, Rinaudo CD, Galeotti CL, Maione D, Ghezzo C, Buttazzoni E, et al. Preventing bacterial infections with pilus-based vaccines: the group B streptococcus paradigm. J Infect Dis 2009; 199: 108– 115. [DOI] [PubMed] [Google Scholar]
- 25. Xia FD, Mallet A, Caliot E, Gao C, Trieu-Cuot P, Dramsi S. Capsular polysaccharide of Group B Streptococcus mediates biofilm formation in the presence of human plasma. Microbes Infect 2015; 17: 71– 76. [DOI] [PubMed] [Google Scholar]
- 26. DeLeo FR, Rinaudo CD, Rosini R, Galeotti CL, Berti F, Necchi F, et al. Specific involvement of Pilus type 2a in biofilm formation in Group B Streptococcus. PLoS One 2010; 5 (2): e9216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martins E, Andreu A, Melo-Cristino J, Ramirez M. Distribution of Pilus Islands in Streptococcus agalactiae that cause human infections: insights into evolution and implication for vaccine development. Clin Vaccine Immunol 2013; 20: 313– 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khodaei F, Najafi M, Hasani A, Kalantar E, Sharifi E, Amini A, et al. Pilus-encoding islets in S. agalactiae and its association with antibacterial resistance and serotype distribution. Microb Pathog 2018; 116: 189– 194. [DOI] [PubMed] [Google Scholar]
- 29. Madzivhandila M, Adrian PV, Cutland CL, Kuwanda L, Madhi SA, The Po PSTT. Distribution of pilus islands of group B streptococcus associated with maternal colonization and invasive disease in South Africa. J Med Microbiol 2013; 62: 249– 253. [DOI] [PubMed] [Google Scholar]
- 30. Gizachew M, Tiruneh M, Moges F, Tessema B. Streptococcus agalactiae maternal colonization, antibiotic resistance and serotype profiles in Africa: a meta-analysis. Ann Clin Microbiol Antimicrob 2019; 18: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghanbarzadeh N, Mehramiz M, Gannadkafi M, Namaei MH. The prevalence of group B streptococcus rectovaginal colonization and antimicrobial susceptibility pattern among pregnant women: a descriptive-analytical study. Mod Care J 2017; 14 (3): e66391. [Google Scholar]
- 32. Bolukaoto JY, Monyama CM, Chukwu MO, Lekala SM, Nchabeleng M, Maloba MR, et al. Antibiotic resistance of Streptococcus agalactiae isolated from pregnant women in Garankuwa, South Africa. BMC Res Notes 2015; 8: 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Martins ER, Pedroso-Roussado C, Melo-Cristino J, Ramirez M, The Portuguese Group for the Study of Streptococcal Infections. Streptococcus agalactiae causing neonatal infections in portugal (2005–2015): diversification and emergence of a CC17/PI-2b multi-drug resistant sublineage. Front Microbiol 2017; 8: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heelan JS, Hasenbein ME, McAdam AJ. Resistance of group B Streptococcus to selected antibiotics, including erythromycin and clindamycin. J Clin Microbiol 2004; 42: 1263– 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]


