Abstract
Background and Objectives:
Serratia marcescens has emerged as a nosocomial pathogen responsible for human infections, where antibiotic resistance further complicates the treatments. In S. marcescens, biofilm formation and virulence factor production are controlled via quorum sensing (QS) system. QS is a signaling system that enables gene regulation to control diverse physiological functions in bacteria. Essential oils have shown to be potential in diminishing the pathogenicity and virulence of drug-resistant bacteria. This study was performed to determine whether eugenol would affect QS system, biofilm formation and virulence factor production of S. marcescens.
Materials and Methods:
Biofilm formation, extracellular virulence factor production (hemolysin and protease), swarming motility and pigment formation of S. marcescens ATCC 13880 and S. marcescens Sm2 were assessed after eugenol exposure at 1.25 and 2.5 µg/ml concentrations. The expression of genes involved in motility (flhD), attachment (fimC), biofilm formation (bsmB, bsmA), and QS regulatory (swrR) were also evaluated.
Results:
Eugenol treatment at 1.25 and 2.5 µg/ml concentrations caused a significant reduction in biofilm formation. The pigment, hemolysin and protease production of two studied S. marcescens strains, also reduced significantly by eugenol treatments (p<0.05). The bsmA, bsmB, flhD and fimC genes were down-regulated after eugenol treatment. The swrR gene expression was also reduced significantly by eugenol in both S. marcescens strains (p<0.05).
Conclusion:
Eugenol inhibited quorum sensing-regulated functions of two studied S. marcescens strains.
Keywords: Serratia marcescens, Virulence factors, Biofilm formation, Quorum sensing, Eugenol
INTRODUCTION
Serratia marcescens is a major nosocomial pathogen which is often isolated from human respiratory and urinary infections. Immunocompromised patients are more susceptible to infection by this opportunistic pathogen than healthy individuals (1). Infections with S. marcescens are hard to treat owing to its broad-spectrum resistance to antibiotics including β-lactam, aminoglycosides, quinolones and polypeptide antibiotics (2). Development of antibiotic resistance in S. marcescens compels the search of novel and effective treatment approaches.
Recent discoveries in the field of bacterial communication system known as quorum sensing (QS) has attracted a great attention in this alternative treatment to conventional chemotherapy. QS system in bacteria regulates the expression of specific genes and depends on the cell density. The QS system is mediated by autoinducers (AIs), secreted from each bacteria (3). Two acyl homoserine lactone (AHL)-dependent quorum sensing systems has been identified in Serratia spp. as swrI/swrR (4) and smaI/smaR (5). Serratia strains utilize AHLs like C4-homoserine lactone (C4-HSL), and C6-HSL to manage the expression of genes encoding virulence factors such as production of prodigiosin, protease, lipase, nuclease, hemolysin and biofilm formation (6, 7). Therefore, the quenching of AHL-mediated QS develops into a promising therapeutic target to attenuate the pathogenicity of S. marcescens (8). Recently, substantial efforts have been made for the identification of effective anti-QS and anti-biofilm agents (9). The anti-QS compounds, unlike antimicrobial agents, attenuate virulence of the pathogens rather than their growth, hence the risk of development of antibiotic resistance is almost nullified (10).
Most anti-QS agents have not been applied in clinic because of their toxicity and instability. So, studies have been focused on searching for new anti-QS agents from herbal extracts, which are rich source of bioactive and low toxic compounds (11). Essential oils (oily aromatic liquids) are extracted from natural plant materials and exhibit antimicrobial effects against bacteria, viruses, fungi and yeasts (12–14). Some of the antimicrobial properties of plant biologically active compounds may be attributed to QS inhibition, whereas the growth of the microorganisms may not be inhibited (15). Eugenol (4-allyl-2-methoxyphenol), the major clove extract constituent, exhibited significant bactericidal activity against different organisms like Escherichia coli, Pseudomonas aeruginosa and Staphylococcus aureus (16). Also, it has been reported that it possesses anti-QS properties (17, 18). Therefore, in the present study the anti-QS activity of eugenol against S. marcescens was evaluated.
MATERIALS AND METHODS
Bacterial strains.
S. marcescens ATCC 13880 and a clinical isolate (blood sample, Serratia sp. named Sm2) were studied in this research. S. marcescens ATCC 13880 was obtained from Iranian Biological Resource Center. Clinical isolate was identified by 16S rRNA gene sequencing (GenBank accession number MK371794). Luria-Bertani (LB) agar was used to culture the bacterial strains at 37°C. For all assays, bacteria from pure bacterial colonies (overnight culture) were inoculated in phosphate-buffered saline (PBS, pH 7.2). The density of inoculum was adjusted to 0.5 McFarland standard (108 CFU/ml). For Biofilm biomass quantification assay, the bacteria were cultured in tryptic soya broth (TSB). All assays were carried out two times in triplicate.
Minimum inhibitory concentration (MIC) determination and growth analysis.
Eugenol was purchased from Merck (Germany). Stock solutions were prepared by adding 2 µl dimethyl sulfoxide to 1 ml of eugenol. The MIC and minimum bactericidal concentration (MBC) of eugenol was determined against S. marcescens strains using the Mueller–Hinton broth microdilution method (19). The effect of ½ MIC (2.5 µg/ml) concentration of eugenol on bacterial proliferation was determined by monitoring the growth curve of S. marcescens strains. Briefly, 1% of overnight cultures of microorganisms (OD600 nm ~ 0.4) was inoculated into LB broth (25 ml) supplemented with ½ MIC of eugenol. The flasks were incubated at 37°C with shaking at 150 rpm. The cell density of samples were assessed by spectrophotometer at 600 nm every 1-h interval (20).
Biofilm biomass quantification assay.
Strains were cultured in TSB at 37°C for 24 h. Bacterial suspensions were diluted (1:50) in TSB plus 0.2% glucose and incubated for 24 h at 37°C with different eugenol concentrations (final concentrations: 1.25 and 2.5 µg/ml). The wells were rinsed with PBS until the planktonic cells were removed. Biofilm formation was quantified by staining with Crystal Violet (0.4%) and a spectrophotometric assay (21).
Scanning electron microscopy (SEM).
Biofilms formed by eugenol-treated (1.25 µg/ml) and untreated cells on glass slides were treated and fixed with glutaraldehyde (2.5%, 2 h). After washing with distilled water, dehydration carried out by adding ethanol (35, 50, 70, 90 and 100%). After critical point drying and gold sputtering, the biofilm samples were tested under a scanning electron microscope (Philips, Japan) (22).
QS inhibition assays in S. marcescens.
QS inhibition assays (including prodigiosin, protease and hemolysin production) were performed by inoculating S. marcescens cells in LB medium and treatment with eugenol (1.25 and 2.5 µg/ml) at 37°C for 24 h. The pigment production was examined at 30°C.
Prodigiosin production assay.
Prodigiosin was extracted from treated and untreated cells. Two ml of cells were centrifuged (10,000 rpm, 10 min). To the pellet, 1 ml of acidified ethanol (4% of 1 M HCl in absolute ethanol) was added and vortexed. The mixture was centrifuged (10,000 rpm, 10 min) and the extracted pigment absorbance was measured at 534 nm (23).
Proteolytic assay.
Treated and untreated cells were centrifuged (11,000 rpm, 20 min, 4°C) and the proteolytic activity of supernatants were examined using azocasein as the substrate. 500 μl of substrate [0.3% azocasein (Sigma, USA) in 1 M PBS] was added to 500 μl of each supernatant. The mixture was incubated at 37°C for 1 h. After incubation, 500 μl of trichloro acetic acid (10%) was added and the reaction was terminated by incubation at −20°C (20 min) and then centrifuged (12,000 rpm −15 min). 100 μl of 1 M NaOH was added to supernatant (200 µl), and the absorbance was read at OD 440 nm (24).
Hemolysin assay.
Each bacterial group (treated and untreated cells) were centrifuged (11,000 rpm, 20 min, 4°C). 900 μl of sheep blood suspension (2%) in PBS was added to the supernatant (100 µl) and incubated at 37°C for 1 h. After centrifugation (3,000 rpm, 10 min), the supernatant absorbance was measured at 530 nm (25).
Swarming assay.
For swarming assay, each bacterial suspension (5 µl) was inoculated at the center of the swarming agar medium. The medium contains 1% peptone, 0.5% NaCl and 0.3% agar and eugenol at different concentrations. The plates were incubated in upright position at 30°C. The bacterial swarm zones were measured after 24 h (26).
Extracellular polysaccharide (EPS) assay.
Bacterial cells were inoculated with eugenol in 96-well sterile polystyrene microtitre plate and incubated for 24 h to form biofilm as described previously. To remove planktonic cells, wells were washed with PBS. A mixture composed of 0.9% NaCl (40 μl), 5% phenol (40 μl) and concentrated sulfuric acid (200 μl) were added to each well. After incubation of the mixture at dark for 1 h, absorbance was measured at 490 nm (27).
Quantitative real time PCR (qPCR).
To evaluate the effect of eugenol (2.5 µg/ml) on the expression level of QS-regulated genes, the total RNA of cells was extracted by RNX-PLUS (Sinaclone). Total RNA was dissolved in 0.1% diethylpyrocarbonate (DEPC)-treated water and treated by DNase I (1 U/μl, Thermo Scientific). 1 μl of 50 mM EDTA was added and the DNase was denatured. Treated RNA was reverse transcribed into cDNA by using cDNA reverse transcription kit (BIOFACT). The qPCR reactions were carried out by PCR Master Mix (Power SYBR Green, BIOFACT) and the real-time PCR system (MIC, Bio Molecular Systems). Targeted genes expression ( fimC, SwrR, flhD, bsmB and bsmA) was normalized by an internal control (rplU gene, 50S Ribosomal gene) (28). The sequences of the gene-specific primers are given in Table 1.
Table 1.
Primer sequences used in this study
| Gene | Forward primer (5′→ 3′) | Reverse primer (5′→ 3′) | Annealing temperature (°C) |
|---|---|---|---|
| flhD | TTGCCACTTCCGCTTTAACG | CTCTTTTCTTCGTCGGGCTAG | 60 |
| bsmA | TAGTCCGCACACTCATCGC | GATCTCCTGCGCCTGTGC | 60 |
| bsmB | GCGGATGTGTATGCCTTCG | GCCACGCATTTCTTCACTCA | 60 |
| fimC | ACCAGCCGTTTCAACAACAA | GGTTTGTACGGTGCGATCTT | 60 |
| swrR | GCCGAAATTCAAATGCTGCT | CTTACCTAAGCTCGCCCAGT | 48 |
| rplU | AAATCGGCGTTCCTTTCGTC | TGCTTACGGTGGTGTTTACG | 60 |
Statistical analysis.
All of the obtained values were displayed as mean ± standard error and were statistically analyzed by post hoc Tukey tests. P < 0.05 using GraphPad Prism 6. The statistical significance of real time PCR data was evaluated by the Relative Expression Software Tool (REST 2009).
RESULTS
Determination of MIC and growth curve analysis.
The MIC against two S. marcescens strains was 5 µg/ml (0.05% v/v). Since complete killing was observed at 10 µg/ml (0.1% v/v), this concentration was determined as MBC.
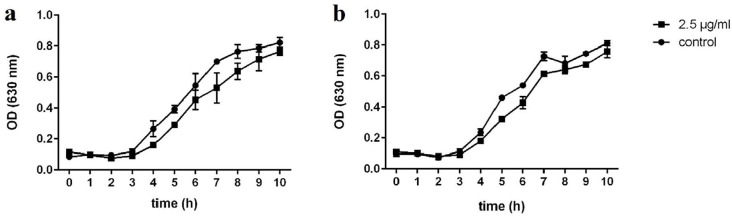
As shown in Fig. 1, there was no difference in cell density of S. marcescens strains in the presence of ½ MIC (2.5 μg/ml) of eugenol as compared to the untreated controls, indicating that eugenol at this concentration had no bactericidal effect on both studied S. marcescens strains. Hence, in the present study, 1.25 and 2.5 μg/ml of eugenol were used for further experiments.
Fig. 1.
Effect of eugenol (1/2 MIC:2.5 µg/ml) on growth of (a) S. marcescens ATCC 13880 and (b) S. marcescens Sm2. (Control: untreated group)
Biofilm formation inhibition.
As shown in Fig. 2, after eugenol treatments, the biofilm biomass of both strains decreased significantly (p<0.05). Eugenol concentrations of 1.25 and 2.5 µg/ml caused a dose-dependent decrease in biofilm formation (60 to 75%), when compared with the untreated controls.
Fig. 2.
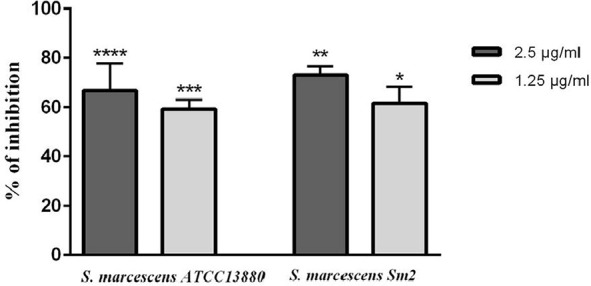
Biofilm formation inhibition of S. marcescens ATCC 13880 and S. marcescens Sm2 by eugenol treatments. The biofilm formation was estimated by crystal violet assay. Absorbance was measured at 492 nm. *significant at p < 0.05, **significant at p < 0.005, ***significant at p < 0.0005 and ****significant at p < 0.0001.
Scanning electron micrographs.
The SEM images of untreated controls clearly showed the coherent and integrated biofilm architecture, whereas the treated samples with 1.25 µg/ml eugenol exhibited a complete destruction of biofilm structure (Fig. 3). Furthermore, the eugenol-treated samples demonstrated considerable disruption of bacterial aggregation, in contrast to the elaborate aggregation of bacterial colonies in the untreated controls.
Fig. 3.
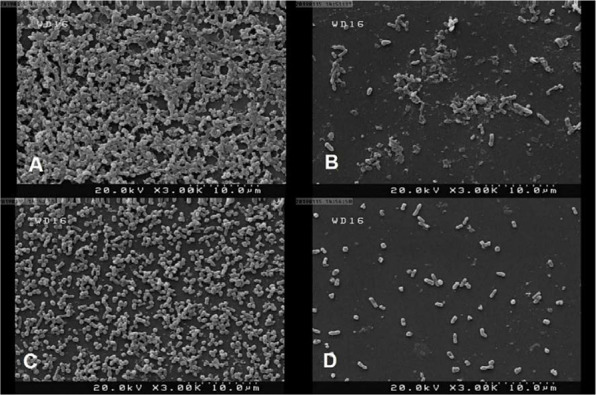
Scanning electron images of S. marcescens biofilms in the presence and absence of eugenol (1.25 µg/ml). A) S. marcescens ATCC 13880 untreated cells, B) S. marcescens ATCC 13880 treated control cells, C) S. marcescens Sm2 untreated control cells, D) S. marcescens Sm2 treated cells.
Prodigiosin inhibition.
The production of prodigiosin pigment decreased 30–50% by using 1.25 and 2.5 µg/ml concentrations of eugenol in S. marcescens ATCC 13880. Pigment production in S. marcescens Sm2 also decreased 45–70% after using the same concentrations of eugenol (Fig. 4).
Fig. 4.
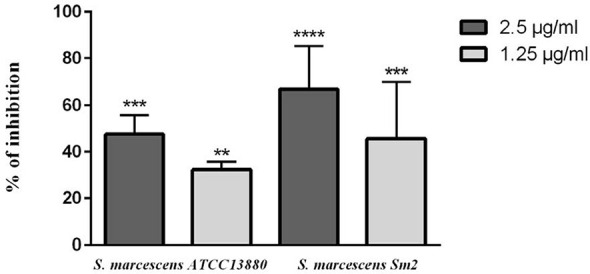
Prodigiosin inhibition in S. marcescens ATCC 13880 and S. marcescens Sm2 by eugenol. **significant at p < 0.005 and ***significant at p < 0.0005 and ****significant at p < 0.0001.
Protease inhibition.
As shown in Fig. 5, eugenol causeda significant decrease in protease production to about 20–35% and 25–35% by using 1.25 and 2.5 µg/ml eugenol in S. marcescens ATCC 13880 and S. marcescens Sm2, respectively.
Fig. 5.
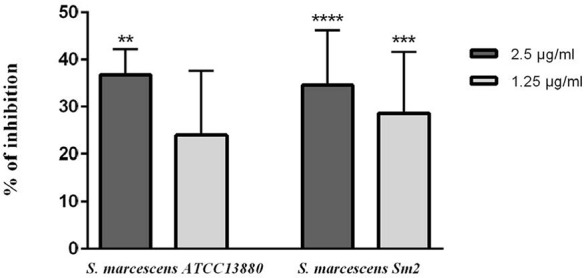
Inhibition of protease in S. marcescens ATCC 13880 and S. marcescens Sm2 by eugenol. **significant at p < 0.005 and ***significant at p < 0.0005 and ****significant at p < 0.0001.
Hemolysin inhibition.
Hemolysin production of two studied strains were decreased in the presence of eugenol (p<0.05). At 1.25 and 2.5 µg/ml eugenol, about 25% and 35% of hemolysin inhibition was observed in S. marcescens Sm2 and S. marcescens ATCC 13880, respectively (Fig. 6).
Fig. 6.
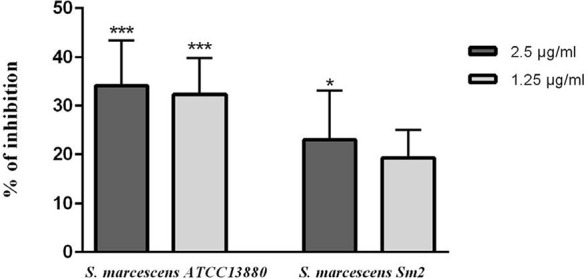
Hemolysin inhibition in S. marcescens ATCC 13880 and S. marcescens Sm2 by eugenol. *significant at p < 0.05 and ***significant at p < 0.0005.
Swarming inhibition.
As shown in Fig. 7, untreated control groups of S. marcescens ATCC 13880 and S. marcescens Sm2 exhibited swarming on the soft agar plates. The addition of eugenol (1.25 and 2.5 µg/ml) decreased swarming motility of the two studied strains.
Fig. 7.
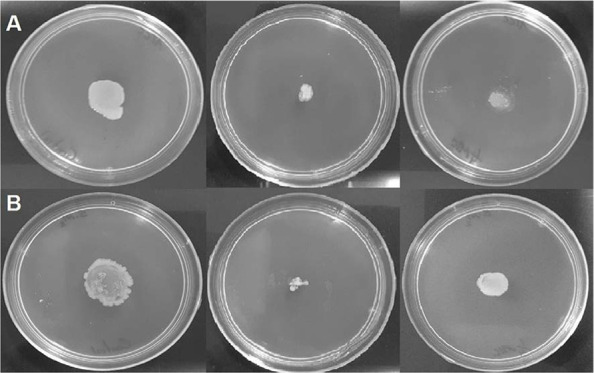
Swarming inhibition in (A) S. marcescens ATCC 13880 and (B) S. marcescens Sm2 by eugenol. Left to right: untreated cells, 2.5 µg/ml-treated cells, and 1.25 µg/ml-treated cells.
EPS inhibition.
At 1.25 and 2.5 µg/ml concentrations, eugenol showed significant inhibition in EPS production of about 50–60% in both studied strains (Fig. 8).
Fig. 8.
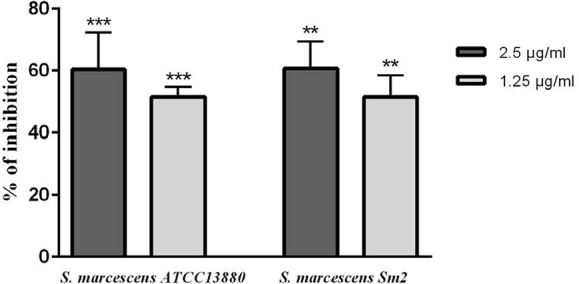
EPS inhibition in S. marcescens ATCC 13880 and S. marcescens Sm2 by eugenol. **significant at p < 0.005, ***significant at p < 0.0005.
qPCR analysis.
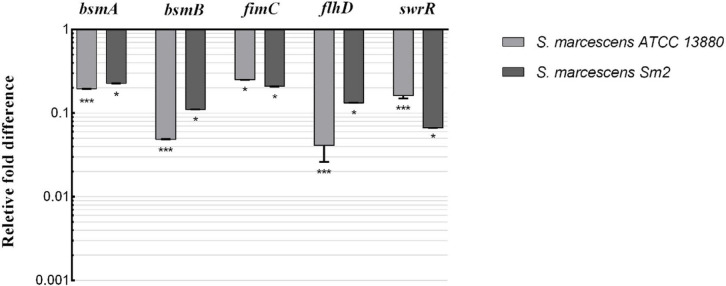
The qPCR data was presented as fold changes in transcriptional level of targeted genes of treated samples compared to their untreated controls (Fig. 9). Eugenol treatment resulted in down-regulation of the expression of flagellar gene (flhD), biofilm formation genes (bsmA and bsmB), fimbrial gene (fimC), and QS regulatory gene (swrR) in both S. marcescens strains (p<0.05).
Fig. 9.
Fold changes in S. marcescens ATCC 13880 and S. marcescens Sm2 gene expression after eugenol treatment (2.5 µg/ml). *significant at p < 0.05 and ***significant at p < 0.0005.
DISCUSSION
Traditional medicines such as essential oils have been found to have anti-QS function (29, 30). In the present study, the anti-QS potential of eugenol to inhibit QS-mediated virulence factor production and biofilm formation in S. marcescens was assessed. The results clearly demonstrate that eugenol hinders the biofilm formation and virulence factors production in S. marcescens, in a concentration-dependent manner. Since concentrations greater than 5 µg/ml resulted in bactericidal effects, further studies were performed at a concentration of 1.25 and 2.5 µg/ml eugenol.
Bacterial biofilms have the ability to resist the action of conventional antibiotics as well as the invading host immune responses (31). Therefore, new approaches are required to prevent the infections which are based on the biofilm formation. Our results showed that biofilm formation ability of both studied strains was decreased significantly by using eugenol. The SEM images of biofilms treated with eugenol (1.25 µg/ml) also exhibited very little adherence of cells on the surface. However, untreated cells showed a dense biofilm. Since the QS mechanism controls two stages of bacterial biofilm formation including the initiation and the maturation (32), eugenol acts as an anti-QS agent, which affects the bacterial adherence on the surface and also causes the subsequent dispersion of the biofilm. Thus, eugenol can make sessile cells susceptible to antibiotics. At the same time, eugenol helps the host immune system to clear the bacteria successfully. Overproduction of EPS leads to changes in biofilm structure, which will eventually be correlated with increased cell resistance to osmotic and oxidative stresses and killing by biocides (33). Herein, we observed that eugenol significantly reduced the production of EPS more than 50%.
Swarming motility has a crucial role in surface colonization and biofilm formation of S. marcescens (34). Eugenol showed a significant reduction in swarming motility in both S. marcescens strains.
In the present study, reduction of EPS production and swarming motility along with the inhibition of biofilm formation indicates that eugenol is effective in the early stages of biofilm formation as well as structural design of the biofilm.
QS system regulates the pigment production of S. marcescens through two signal molecules, N–butanoyl homoserine lactone and HHL and any interference with these QS systems will be followed by decrease in prodigiosin production (35). This pigment also linked to the hydrophobic surface attachment of bacteria and could be important in the colonization and dissemination of bacteria (36). In our study, the pigment production of both strains diminished significantly after eugenol treatment compared to their untreated controls.
Hemolytic activity and production of extracellular enzymes such as protease in S. marcescens, are also under QS control (7). It has been reported that the proteases of S. marcescens can induce inflammatory and immune responses and play a significant role as virulence factors (1). Almost all strains of S. marcescens secrete a hemolysin that causes hemolysis of animal and human erythrocytes (37) and the release of inflammatory mediators from leukocytes (38). It also acts as a cytotoxin on human epithelial cells and fibroblasts (39). According to our results, hemolysin and protease production decreased considerably after eugenol treatment in both strains. These results are consistent with the results of Srinivasan et al. who demonstrated that phytol, a diterpene alcohol compound majorly found in essential oils, decreased the level of extracellular enzymes and hemolysin production in S. marcescens (40).
According to Labbate et al. the major fimbrial proteins in S. marcescens are encoded by fimA and fimC genes (41). These QS-regulated genes have significant role in the attachment to biotic and abiotic surfaces by S. marcescens cells. In this study, the real-time PCR analysis showed that upon treatment with eugenol the expression of fimC was down-regulated almost 0.9-fold in both S. marcescens strains (Fig. 9). Recently, Salini and Pandian have also observed that methanolic extract of Anethum graveolens treatment decreased the expression of fimC gene (26).
The flagellar master operon flhD regulates swarming motility in S. marcescens (42). Eugenol treatment decreased the transcript level of flhD more than 0.9-fold in S. marcescens ATCC 13880 and S. marcescens Sm2 (Fig. 9). Reduction in expression of this gene is in accordance with the report of Srinivasan et al. which showed that the Piper betle ethyl acetate extract down-regulated the flhD gene expression (43). The C4-HSL mediated QS genes, bsmA and bsmB, modulate the biofilm formation in S. marcescens. Labbate et al. showed that bsmA and bsmB are involved in adhesion to the abiotic surfaces (41). In this study, the expressions of bsmA and bsmB in both S. marcescens strains were down-regulated by eugenol (Fig. 9). Salini and Pandian have also observed that the bsmA gene expression was down-regulated in samples treated with methanolic extract of A. graveolens (26). The SwrI/SwrR QS system regulates the biofilm formation, swarming motility, as well as production of protease, S-layer protein and serrawettin in S. marcescens (44). The expression of swrI gene leads to the production of an AHL synthase which synthesis the AHL autoinducers. An open reading frame, downstream of swrI, codes SwrR polypeptide. The SwrR has a major similarity to other AHL-dependent regulators such as members of the LuxR family (45). In the present study, after eugenol treatment, the swrR gene was down-regulated in S. marcescens ATCC 13880 and S. marcescens Sm2.
CONCLUSION
This study demonstrated that eugenol could prevent the production of QS-controlled virulence factors like production of prodigiosin pigment, protease and hemolysin. Moreover, the QS-mediated biofilm formation stages such as swarming motility, formation of microcolony, and extracellular polysaccharide production were inhibited by the eugenol treatment. Further, eugenol down-regulated the expression of genes responsible for QS system, adhesion, motility, and biofilm formation in S. marcescens. Thus, these results suggest that eugenol may have potential medicinal value including the anti-QS and anti-biofilm effects against S. marcescens strains.
ACKNOWLEDGEMENTS
This study was supported by the College of Science, University of Tehran, Tehran, Iran.
REFERENCES
- 1. Kida Y, Inoue H, Shimizu T, Kuwano K. Serratia marcescens serralysin induces inflammatory responses through protease-activated receptor 2. Infect Immun 2007; 75: 164– 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hejazi A, Falkiner F. Serratia marcescens. J Med Microbiol 1997; 46: 903– 912. [DOI] [PubMed] [Google Scholar]
- 3. Hawver LA, Jung SA, Ng W-L. Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiol Rev 2016; 40: 738– 752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eberl L, Christiansen G, Molin S, Givskov M. Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J Bacteriol 1996; 178: 554– 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coulthurst SJ, Williamson NR, Harris AK, Spring DR, Salmond GP. Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology (Reading) 2006; 152: 1899– 1911. [DOI] [PubMed] [Google Scholar]
- 6. Rice S, Koh K, Queck S, Labbate M, Lam K, Kjelleberg S. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J Bacteriol 2005; 187: 3477– 3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Houdt R, Givskov M, Michiels CW. Quorum sensing in Serratia. FEMS Microbiol Rev 2007; 31: 407– 424. [DOI] [PubMed] [Google Scholar]
- 8. Morohoshi T, Shiono T, Takidouchi K, Kato M, Kato N, Kato J, et al. Inhibition of quorum sensing in Serratia marcescens AS-1 by synthetic analogs of N-acylhomoserine lactone. Appl Environ Microbiol 2007; 73: 6339– 6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brackman G, Coenye T. Quorum sensing inhibitors as anti-biofilm agents. Curr Pharm Des 2015; 21: 5– 11. [DOI] [PubMed] [Google Scholar]
- 10. Rasmussen TB, Givskov M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int J Med Microbiol 2006; 296 (2-3): 149– 161. [DOI] [PubMed] [Google Scholar]
- 11. Khan MSA, Zahin M, Hasan S, Husain FM, Ahmad I. Inhibition of quorum sensing regulated bacterial functions by plant essential oils with special reference to clove oil. Lett Appl Microbiol 2009; 49: 354– 360. [DOI] [PubMed] [Google Scholar]
- 12. Reichling J, Schnitzler P, Suschke U, Saller R. Essential oils of aromatic plants with antibacterial, antifungal, antiviral, and cytotoxic properties–an overview. Forsch Komplementmed 2009; 16: 79– 90. [DOI] [PubMed] [Google Scholar]
- 13. Kalemba D, Kunicka A. Antibacterial and antifungal properties of essential oils. Curr Med Chem 2003; 10: 813– 829. [DOI] [PubMed] [Google Scholar]
- 14. Bakkali F, Averbeck S, Averbeck D, Idaomar M. Biological effects of essential oils–a review. Food Chem Toxicol 2008; 46: 446– 475. [DOI] [PubMed] [Google Scholar]
- 15. Vattem DA, Mihalik K, Crixell SH, McLean RJ. Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia 2007; 78: 302– 310. [DOI] [PubMed] [Google Scholar]
- 16. Walsh SE, Maillard JY, Russell A, Catrenich C, Charbonneau D, Bartolo R. Activity and mechanisms of action of selected biocidal agents on Gram-positive and-negative bacteria. J Appl Microbiol 2003; 94: 240– 247. [DOI] [PubMed] [Google Scholar]
- 17. Zhou L, Zheng H, Tang Y, Yu W, Gong Q. Eugenol inhibits quorum sensing at sub-inhibitory concentrations. Biotechnol Lett 2013; 35: 631– 637. [DOI] [PubMed] [Google Scholar]
- 18. Rathinam P, Vijay Kumar H, Viswanathan P. Eugenol exhibits anti-virulence properties by competitively binding to quorum sensing receptors. Biofouling 2017; 33: 624– 639. [DOI] [PubMed] [Google Scholar]
- 19. Wikler MA. “Performance Standards for Antimicrobial Susceptibility Testing, Sixteenth Informational Supplement, M100-S16,” Clinical and Laborator y Standards Institute (CLSI), Vol. 26, No. 3, 2006, Pennsylvania: [Google Scholar]
- 20. Adonizio A, Kong K-F, Mathee K. Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrob Agents Chemother 2008; 52: 198– 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kashef N, Akbarizare M, Kamrava SK. Effect of sublethal photodynamic inactivation on the antibiotic susceptibility and biofilm formation of clinical Staphylococcus aureus isolates. Photodiagnosis Photodyn Ther 2013; 10: 368– 373. [DOI] [PubMed] [Google Scholar]
- 22. Gowrishankar S, Poornima B, Pandian SK. Inhibitory efficacy of cyclo (l-leucyl-l-prolyl) from mangrove rhizosphere bacterium–Bacillus amyloliquefaciens (MMS-50) toward cariogenic properties of Streptococcus mutans. Res Microbiol 2014; 165: 278– 289. [DOI] [PubMed] [Google Scholar]
- 23. Sethupathy S, Shanmuganathan B, Kasi PD, Pandian SK. Alpha-bisabolol from brown macroalga Padina gymnospora mitigates biofilm formation and quorum sensing controlled virulence factor production in Serratia marcescens. J Appl Phycol 2016; 28: 1987– 1996. [Google Scholar]
- 24. Bakkiyaraj D, Sivasankar C, Pandian SK. Inhibition of quorum sensing regulated biofilm formation in Serratia marcescens causing nosocomial infections. Bioorg Med Chem Lett 2012; 22: 3089– 3094. [DOI] [PubMed] [Google Scholar]
- 25. Devi KR, Srinivasan S, Ravi AV. Inhibition of quorum sensing-mediated virulence in Serratia marcescens by Bacillus subtilis R-18. Microb Pathog 2018; 120: 166– 175. [DOI] [PubMed] [Google Scholar]
- 26. Salini R, Pandian SK. Interference of quorum sensing in urinary pathogen Serratia marcescens by Anethum graveolens. Pathog Dis 2015; 73: ftv038. [DOI] [PubMed] [Google Scholar]
- 27. Dubois M, Gilles KA, Hamilton JK, Rebers Pt, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem 1956; 28: 350– 356. [Google Scholar]
- 28. Fekrirad Z, Kashef N, Arefian E. Photodynamic inactivation diminishes quorum sensing-mediated virulence factor production and biofilm formation of Serratia marcescens. World J Microbiol Biotechnol 2019; 35: 191. [DOI] [PubMed] [Google Scholar]
- 29. Jakobsen TH, Bragason SK, Phipps RK, Christensen LD, van Gennip M, Alhede M, et al. Food as a source for quorum sensing inhibitors: iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl Environ Microbiol 2012; 78: 2410– 2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taganna JC, Quanico JP, Perono RMG, Amor EC, Rivera WL. Tannin-rich fraction from Terminalia catappa inhibits quorum sensing (QS) in Chromobacterium violaceum and the QS-controlled biofilm maturation and LasA staphylolytic activity in Pseudomonas aeruginosa. J Ethnopharmacol 2011; 134: 865– 871. [DOI] [PubMed] [Google Scholar]
- 31. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science 1999; 284: 1318– 1322. [DOI] [PubMed] [Google Scholar]
- 32. De Kievit TR, Gillis R, Marx S, Brown C, Iglewski BH. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl Environ Microbiol 2001; 67: 1865– 1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A 1999; 96: 4028– 4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shanks RM, Lahr RM, Stella NA, Arena KE, Brothers KM, Kwak DH, et al. A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One 2013; 8 (3): e57634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Packiavathy IASV, Priya S, Pandian SK, Ravi AV. Inhibition of biofilm development of uropathogens by curcumin–an anti-quorum sensing agent from Curcuma longa. Food Chem 2014; 148: 453– 460. [DOI] [PubMed] [Google Scholar]
- 36. Rosenberg M, Blumberger Y, Judes H, Bar-Ness R, Rubinstein E, Mazor Y. Cell surface hydrophobicity of pigmented and nonpigmented clinical Serratia marcescens strains. Infect Immun 1986; 51: 932– 935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schiebel E, Braun V. Integration of the Serratia marcescens haemolysin into human erythrocyte membranes. Mol Microbiol 1989; 3: 445– 453. [DOI] [PubMed] [Google Scholar]
- 38. König W, Faltin Y, Scheffer J, Schöffler H, Braun V. Role of cell-bound hemolysin as a pathogenicity factor for Serratia infections. Infect Immun 1987; 55: 2554– 2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hertle R, Hilger M, Weingardt-Kocher S, Walev I. Cytotoxic action of Serratia marcescens hemolysin on human epithelial cells. Infect Immun 1999; 67: 817– 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Srinivasan R, Mohankumar R, Kannappan A, Karthick Raja V, Archunan G, Karutha Pandian S, et al. Exploring the anti-quorum sensing and antibiofilm efficacy of phytol against Serratia marcescens associated acute pyelonephritis infection in Wistar rats. Front Cell Infect Microbiol 2017; 7: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Labbate M, Zhu H, Thung L, Bandara R, Larsen MR, Willcox MD, et al. Quorum-sensing regulation of adhesion in Serratia marcescens MG1 is surface dependent. J Bacteriol 2007; 189: 2702– 2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu J-H, Lai M-J, Ang S, Shu J-C, Soo P-C, Horng Y-T, et al. Role of flhDC in the expression of the nuclease gene nucA, cell division and flagellar synthesis in Serratia marcescens. J Biomed Sci 2000; 7: 475– 483. [DOI] [PubMed] [Google Scholar]
- 43. Srinivasan R, Devi KR, Kannappan A, Pandian SK, Ravi AV. Piper betle and its bioactive metabolite phytol mitigates quorum sensing mediated virulence factors and biofilm of nosocomial pathogen Serratia marcescens in vitro. J Ethnopharmacol 2016; 193: 592– 603. [DOI] [PubMed] [Google Scholar]
- 44. Mahlen SD. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 2011; 24 (4): 755– 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciens MG1. J Bacteriol 1999; 181: 1703– 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]