Abstract
Background
Serial profiling of circulating tumor DNA (ctDNA) could reflect dynamic molecular changes in response to treatment and potentially predict impending disease progression (PD). Herein, we investigated the molecular factors and dynamic changes in ctDNA that can serve as predictors of survival outcomes of patients with epidermal growth factor receptor (EGFR)-mutated advanced non-small cell lung cancer (NSCLC) who received osimertinib therapy after progression from prior EGFR inhibitor regimen.
Methods
Capture-based targeted sequencing was performed on the baseline and longitudinal plasma samples collected from 72 and 57 patients, respectively, using a 168-gene panel.
Results
Analysis revealed that inferior overall survival (OS) was correlated with various baseline molecular features including high allelic fraction (AF) of EGFR sensitizing mutations (P=0.045), high maximum AF (maxAF, P=0.060), or harboring concurrent genomic alterations such as copy number amplification (CNA) in EGFR (P=0.026) or in other genes (P=0.026), and genes involved in the cell cycle (P=0.004) or TP53 signaling pathway (P=0.032). Moreover, ctDNA clearance at first follow-up after 6 weeks of osimertinib therapy was correlated with significantly longer progression-free survival (PFS) (P=0.022) and OS (P=0.009). Molecular PD, reflected by the emergence of new mutation or increased AF of existing mutations, was detected at an average lead time of 2.5 months prior to radiological PD. Patients with molecular PD were more likely to harbor CNA (P=0.035) and TP53 mutations (P=0.023).
Conclusions
Molecular factors derived from serial ctDNA profiling can serve as predictive and prognostic markers, which could allow early detection of PD, preceding imaging modalities by 2.5 months.
Keywords: Circulating tumor DNA clearance (ctDNA clearance), molecular PD, osimertinib, serial assessment of ctDNA
Introduction
Tyrosine kinase inhibitors (TKI) targeting actionable mutations in the epidermal growth factor receptor (EGFR) have dramatically improved the clinical outcome of patients with EGFR-mutant advanced non-small cell lung cancer (NSCLC) (1,2). However, despite remarkable treatment responses, resistance inevitably occurs within 1 year of treatment and has become a growing challenge (3,4). With the approval of osimertinib, NSCLC patients are required to undergo rebiopsy for molecular testing of EGFR T790M status upon progression from EGFR-TKI therapy (5). Tissue rebiopsy procedures are invasive and less feasible in some patients (6). Alternatively, circulating tumor DNA (ctDNA) purified from the plasma has been increasingly used as a source of tumor-derived DNA for the detection of somatic mutations in cancer diagnosis, including T790M (7-12). Since ctDNA originates from the apoptotic and necrotic turnover of cancer cells (13), its genomic profile is consistent with the profile derived from the tumor DNA (7,14). The potential of ctDNA in dynamic monitoring of treatment response during the course of EGFR-TKI treatment has been actively explored and is growing more essential in the assessment of treatment response and the emergence of drug resistance (11,15-18). Numerous studies have demonstrated that a reduction in the allelic abundance of EGFR sensitizing mutation reflects the sensitivity to EGFR-TKI therapy (18,19). In addition to treatment monitoring, serial assessment of ctDNA levels has been explored as a potential noninvasive real-time prognostic and predictive biomarker (18,20-23). The prognostic value of assessing ctDNA levels has been demonstrated in detecting minimal residual disease in patients who underwent surgery (24-28). Detectable ctDNA levels at the time of diagnosis and presence of ctDNA at follow-up after initiating treatment were shown to be associated with poor prognosis (20,21). However, more effort is still needed to identify specific molecular markers from ctDNA that could predict clinical outcomes of NSCLC patients. In our study, we aimed to identify molecular factors at baseline and certain dynamic changes during osimertinib treatment derived from serial ctDNA profiling that could serve as predictive and prognostic markers in patients with EGFR-mutated advanced NSCLC who are undergoing osimertinib treatment. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-371).
Methods
Patients
Patients with EGFR T790M-positive advanced NSCLC who progressed on prior first-generation or second-generation EGFR-TKI therapy from 6 hospitals were prospectively enrolled in the study from November 1, 2016 until May 31, 2017. NSCLC patients who were diagnosed with non-resectable disease were included in the study based on the following criteria: (I) those who progressed from prior first-generation or second-generation EGFR-TKI, (II) confirmed EGFR T790M mutation evaluated in tissue samples by real-time polymerase chain reaction-based COBAS EGFR mutation test (Roche Diagnostics), and (III) have not received osimertinib therapy. Plasma samples were collected from the patients prior to initiating osimertinib therapy and longitudinally during the course of the treatment until radiologic confirmation of progressive disease (PD). NSCLC was diagnosed according to the 2015 World Health Organization histological classification of lung tumors (29). Clinical staging was according to the seventh edition of the American Joint Committee on Cancer (30). Treatment responses were assessed by the investigators based on the Response Evaluation Criteria in Solid Tumors version 1.1 (31). Medical records were retrieved to collect clinicopathologic data, treatment history, and survival outcomes. This study has been approved by the ethics committee of Jinling Hospital, Nanjing University School of Medicine (approval number: 2017NZKY-001-03) and performed in accordance with the Declaration of Helsinki as revised in 2013. Written informed consent was provided by all the patients included in the study.
Cell-free DNA isolation and capture-based targeted DNA sequencing
Circulating cell-free DNA was recovered from 4–5 mL of plasma using the QIAamp Circulating Nucleic Acid kit (Qiagen, Hilden, Germany). A minimum of 50 ng of DNA is required for next-generation sequencing (NGS) library construction. Samples that did not meet the minimum requirement were excluded and not processed further. A 168-gene panel, consisting of 68 lung cancer-related genes and 100 other genes related to cancer development spanning 273 kilobases of the human genome, was used for targeted sequencing (OncoScreen Target, Burning Rock Biotech, Guangzhou, China). Indexed samples were sequenced on Nextseq500 (Illumina, Inc., USA) with paired-end reads and average sequencing depth of 10,000×. Sequencing analysis was performed using an optimized bioinformatics pipeline that enabled accurate detection of somatic variants by discriminating sequencing artifacts from real mutations (14).
Statistical analysis
Differences in the groups were calculated and presented using either Fisher’s exact test or two-tailed Student’s t-test, as appropriate. Associations of mutation status with survival outcomes were analyzed using multivariate Cox proportional hazards regression analysis. Progression-free survival (PFS) was defined from the date osimertinib was administered until the evaluation of PD. Overall survival (OS) was defined from the date osimertinib was administered until the day of death or the last day of follow-up, whichever is applicable. PFS and OS curves were estimated using the Kaplan-Meier method and the differences among the groups were evaluated using the log-rank test. P value <0.05 was considered as statistically significant. All the data were analyzed using R statistics package (R version 3.4.0; R: The R-Project for Statistical Computing, Vienna, Austria).
Results
Patient characteristics
A total of 72 patients with EGFR T790M-positive advanced NSCLC were included in the study. All the patients had stage IIIB to IV lung adenocarcinoma, progressed from prior therapy with either first- or second-generation EGFR-TKI, and were EGFR T790M-positive based on Cobas EGFR mutation test of tissue biopsy samples prior to inclusion in the study. The cohort had a median age of 60.5 years, ranging from 35 to 81 years. Among them, 43.1% (31/72) were males and 56.9% (41/72) were females. A majority of the patients (73.6%, 53/72) were never smokers, while 9.7% (7/72) of the patients had a smoking history. The remaining 16.7% (12/72) of the patients did not provide their smoking information.
After the collection of baseline plasma samples, the patients received osimertinib at 80 mg orally once daily, with the exception of 1 patient who received a dose reduction to 40 mg once daily due to adverse events. During the course of the treatment, plasma samples were collected longitudinally at follow-up every 6±1 weeks until the evaluation of PD by radiological imaging. The clinical features of the patients from this cohort were summarized in Table 1.
Table 1. Clinicopathologic characteristics of the cohort.
| Clinicopathologic features | n=72 |
|---|---|
| Age (years) (median, range) | 60.5 [35–81] |
| Gender | |
| Male | 31 (43.1%) |
| Female | 41 (56.9%) |
| Smoking history | |
| Without history of smoking | 53 (73.6%) |
| With history of smoking | 7 (9.7%) |
| Data unavailable | 12 (16.7%) |
| Histology | |
| Adenocarcinoma | 72 (100%) |
| Clinical stage | |
| IV | 69 (95.8%) |
| Others (IIIB, IIIC) | 3 (4.2%) |
| Metastasis | |
| Brain | 27 (37.5%) |
| Lymph node | 49 (68.1%) |
| History of EGFR-TKI prior to osimertinib therapy | |
| 1 | 70 (97.2%) |
| >1 | 2 (2.8%) |
| Prior history of chemotherapy | |
| None | 32 (44.4%) |
| With prior chemotherapy | 39 (54.2%) |
| Unknown | 1 (1.4%) |
| Baseline tumor tissue EGFR T790M positive | 72 (100%) |
Genomic profile at baseline
Capture-based targeted sequencing was performed on ctDNA samples collected from the 72 patients at baseline using a panel covering critical exons and introns of 168 cancer-related genes, achieving an average sequencing depth of 22,400× and median depth of 23,820× considering all samples. This panel was clinically validated to achieve a by-variant sensitivity of 87.2% for identifying non-copy number variation (CNV) mutations in plasma samples of patients with advanced lung cancer (14).
A total of 261 mutations in 43 genes were identified from 65 patients, including 159 single-nucleotide variants, 40 insertions, and deletions, 38 copy number amplifications (CNAs), 10 frameshifts, 9 splice variants, 4 copy number deletions, and 1 fusion. Somatic mutations in genes included in the targeted panel, including EGFR sensitizing mutations and T790M, were not detected in the ctDNA of 7 (9.7%, 7/72) patients. EGFR L858R and exon 19 deletion (19del) were detected in 40% (29/72) and 50% (36/72) of the patients, respectively. EGFR T790M was identified in 54 (75.0%, 54/72) patients, with 24 and 30 patients having concurrent EGFR L858R or 19del, respectively. Other oncogenic drivers such as ERBB2 amplifications (n=2), MET amplifications (n=1), BRAF V600E (n=1), and concurrent ERBB2 and MET amplifications (n=1) were detected concurrently with the EGFR sensitizing mutations. In addition, concurrent TP53 and PIK3CA were detected at mutation rates of 55.6% (40/72) and 6.9% (5/72), respectively (Figure S1).
Correlation of baseline genomic features and survival outcomes
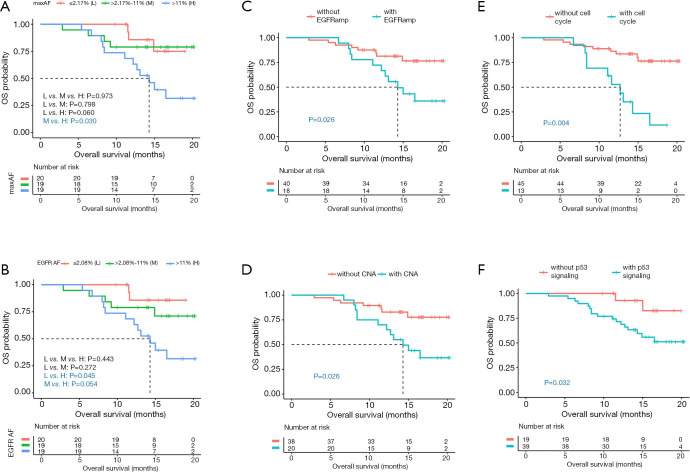
To identify the baseline molecular features potentially associated with survival outcomes, the correlation between allelic fraction (AF) and survival outcomes were analyzed by stratifying the maximum AF (maxAF) or AF of EGFR sensitizing mutations into tertiles as “high”, “median”, and “low” groups. AF is defined as the abundance of mutant allele detected in a particular mutation site, such as EGFR L858R, in a particular sample; whereas, maxAF is defined as the highest proportion of mutant allele, or highest AF, collectively observed in a particular sample, regardless of gene or mutation site. The details of the AF and maxAF of all the patient samples are shown in file (https://cdn.amegroups.cn/static/application/9b741657aae5b8109d04eca32228c9c8/tlcr-20-371-1.pdf). Log-rank analysis revealed that the patients with maxAF or AF of EGFR sensitizing mutation of >11% at baseline, considered as high level, had a trend of shorter OS than those with median or low maxAF [high vs. median, 14.3 months vs. not reached (NR), P=0.030; high vs. low, 14.3 months vs. NR, P=0.060; Figure 1A] or AF of EGFR sensitizing mutation (high vs. median, 14.3 months vs. NR, P=0.054; high vs. low, 14.3 months vs. NR, P=0.045; Figure 1B).
Figure 1.
Certain molecular features at baseline including (A) maxAF; (B) AF of EGFR sensitizing mutations; and concurrent alterations such as (C) EGFR amplification, (D) copy number amplifications, and genes involved in the (E) cell cycle and (F) TP53 signaling pathway are associated with significantly shorter OS. (A,B) Red line represents patients with low (L)-level maxAF or AF (n=20). Green line represents patients with medium (M)-level maxAF or AF (n=19). Blue line represents patients with high (H)-level maxAF or AF (n=19). (C,D,E,F) Red line represents patients without the indicated concurrent mutation. Blue line represents patients with indicated concurrent mutation. Dotted line indicates the median OS. The risk table below illustrates the number of patients included per time point.
Additional multivariate analysis revealed that patients with concurrent EGFR amplification (n=18) or CNA in other genes (n=20) at baseline had significantly inferior OS than patients without EGFR amplification (14.3 months vs. NR, P=0.026, Figure 1C) or CNA in other genes (14.3 months vs. NR, P=0.026, Figure 1D). In addition, patients with concurrent mutations in genes involved in the cell cycle or TP53 signaling pathway, detected in 14 and 43 patients, respectively, had significantly shorter OS than those without the mutation (cell cycle 12.7 months vs. NR, P=0.004, Figure 1E; TP53 signaling pathway NR vs. NR, P=0.032, Figure 1F). Mutations in genes involved in the cell cycle signaling detected in the cohort included CCND1, CDK4, CDK6, CDKN2A, MYC, and RB1; while mutations in genes involved in the TP53 signaling pathway detected in the cohort included TP53, CCND1, CDK4, CDK6, CDKN2A, and PTEN. No significant correlation between genomic features and PFS was found in our cohort.
Taken together, these data suggest that certain molecular factors at baseline, including AF of EGFR sensitizing mutations, maxAF, and concurrent gene amplifications and alterations in genes involved in the cell cycle and TP53 pathways, were significantly correlated with the OS outcome of the patients in our cohort.
Dynamic molecular changes during osimertinib treatment
Among the 72 patients included in the cohort, 15 patients had samples collected only at baseline and were not included in further analysis. Meanwhile, in addition to baseline samples, 57 patients had samples collected between 1 to 5 time points throughout the treatment course. Of them, 41 patients had samples collected at first follow-up, 6 weeks from initiating osimertinib treatment. Their best responses after 6 weeks of osimertinib therapy were partial response in 7 patients (17%, 7/41) and stable disease in 34 patients (83%, 34/41). Patients with and without ctDNA clearance at first follow-up had comparable best response to osimertinib therapy (P=0.438). The median PFS and OS of these 41 patients were 9.8 months (range, 2.8–18.7 months) and 14.1 months (range, 6.7–20.1 months), respectively. To identify molecular factors that could potentially predict response to osimertinib treatment, sequencing data of the samples from the first follow-up of 41 patients was compared with their corresponding baseline genomic profile.
Among the 41 patients, 78.0% (32/41) of the patients had EGFR T790M confirmed by NGS concurrent with either EGFR L858R or 19del, the remaining 4 and 5 patients only had either EGFR L858R or 19del, respectively. All the 32 T790M-positive patients lost the T790M at the first follow-up at 6 weeks post-osimertinib treatment (Figure S2A,B).
Expectedly, a significant decline in the AF of EGFR sensitizing mutation (P<0.001, Figure S3A) and maxAF (P<0.001, Figure S3B) were observed in a majority of the patients (92.7%, 38/41) at first follow-up. The remaining 3 patients have no detectable change. Of the 38 patients with dynamic AF, EGFR sensitizing mutations were lost in 24 patients; while 14 patients had significant AF reduction at first follow-up. Moreover, 20 patients had undetectable ctDNA levels at first follow-up, while 18 patients had detectable ctDNA, represented by maxAF ranging from 0.02% to 12.57%. Undetectable ctDNA level or ctDNA clearance is defined as having a maxAF of 0.01% or lower or the lack of detection of any mutations from the particular sample. Among the 18 patients with detectable ctDNA at first follow-up, 17 retained their baseline EGFR sensitizing mutations, while the remaining 1 patient retained TP53 mutation (P29). Only two patients were observed to acquire new mutations at first follow-up including PIK3CA E545K (P24) and KRAS G12V (P42).
Molecular changes during osimertinib treatment correlated with survival outcomes
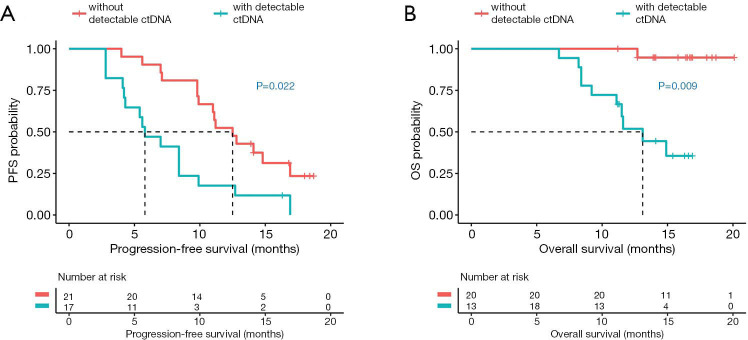
Next, correlation analysis was performed on the sequencing data and survival outcomes of the 38 patients to interrogate the potential of ctDNA clearance as a marker for predicting PFS and OS. Regression analysis revealed that ctDNA clearance at first follow-up was correlated with significantly longer PFS (12.5 vs. 5.8 months, P=0.022, Figure 2A) and OS (NR vs. 13.1 months, P=0.009, Figure 2B). The area under the curve (AUC) indicated that a maxAF cut-off of 0.01% as ctDNA clearance was an independent predictor of PFS of >9 months and OS of >12 months (AUC for PFS =0.821, sensitivity 81.0%, specificity 77.8%; Figure S4A; AUC for OS =0.850, sensitivity 74.1%, specificity 91.7%; Figure S4B).
Figure 2.
ctDNA clearance at 6 weeks after initiating osimertinib therapy (first follow-up) is associated with significantly longer PFS (A) and OS (B) as compared to patients with detectable ctDNA. Red line represents patients without detectable ctDNA at first follow-up (n=21). Blue line represents patients with detectable ctDNA at first follow-up (n=17). Dotted line indicates the median PFS or OS. The risk table below illustrates the number of patients included per time point.
Overall, these data demonstrate the correlation between favorable survival outcomes and the reduction of maxAF of up to 0.01%, termed as ctDNA clearance. These data suggest that ctDNA clearance can potentially serve as a predictive and prognostic marker in NSCLC patients undergoing osimertinib treatment.
Molecular changes correlated with disease progression
We further evaluated the potential of ctDNA mutation profiling in predicting PD at the molecular level. We analyzed the mutation profile of 57 patients with plasma samples for baseline and several time points during the course of osimertinib treatment prior to clinical PD.
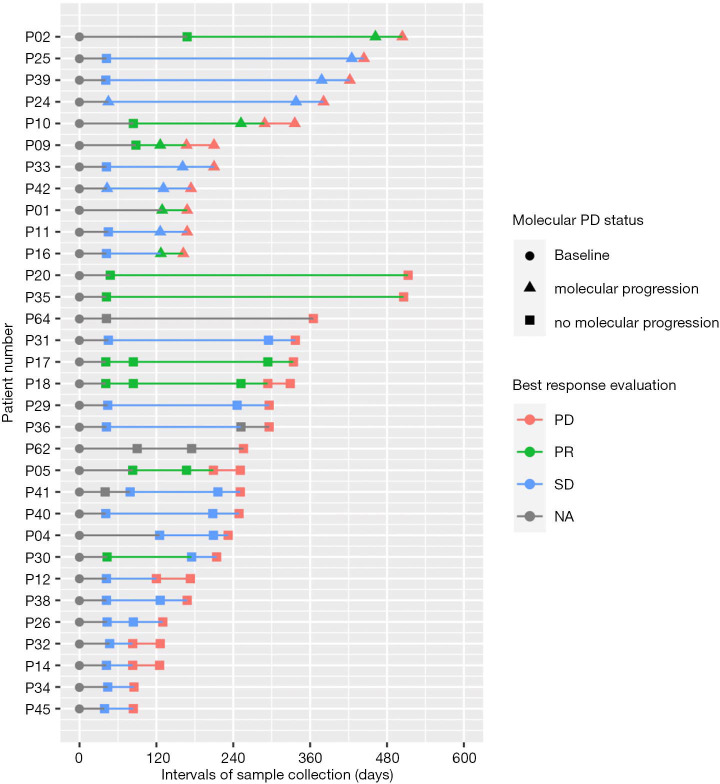
Of the 57 patients, 32 patients experienced radiologically-confirmed PD at a median follow-up of 5.6 months (168 days), ranging from 1.3 to 18.3 months (40 to 550 days). Among them, 34% (11/32) of the patients experienced molecular progression (molecular PD) at an average lead time of 2.5 months (74 days) prior to radiological PD (denoted with triangles in Figure 3). Molecular PD is defined as the emergence of acquired mutation or increased AF of existing mutations of more than 1.5 times from baseline levels. No particular molecular signatures reflecting molecular PD were detected in the other 21 patients (denoted with squares in Figure 3). Figure 3 summarizes the molecular PD status and the best response of the 32 patients with radiological PD.
Figure 3.
Summary of the molecular PD status and best response of the 32 patients with radiological PD. Swimmer’s plot illustrating the detection of molecular PD in 11 patients (P02–P16) and lack of molecular PD in 21 patients (P20–P45) and their best responses with osimertinib therapy. X-axis refers to the time point of sample collection. Y-axis refers to the patient number. Triangle denotes the time-point when molecular PD was detected. Colors represent best response evaluation.
Osimertinib resistance mechanisms
At osimertinib progression, several resistance mechanisms emerged. Among the subgroup of patients who had radiological PD, a majority of the patients (78.1%, 25/32) were T790M-positive at baseline (Figure S5A). Of them, 80% (20/25) lost the T790M prior to radiological PD (Figure S5B) and at the time radiological PD was confirmed (Figure S5C). Two of the 5 patients who retained the T790M had an increase in AF, while the other 3 patients had a reduction in AF of T790M. Acquired mutations previously reported as osimertinib resistance mechanism (referred to as acquired mutations hereafter) were detected in 19 patients. Of them, resistance mutations were acquired during osimertinib treatment in 16 patients, while these mutations were detected at baseline in the remaining 3 patients. Tertiary EGFR mutations including C797S and L718Q were acquired by 4 and 1 patients, respectively. Acquired mutations in genes involved in bypass pathways were also detected from our cohort including BRAF V600E (n=1), MET amplification (n=2), MET splice variants resulting in exon 14 skipping (n=1), ERBB2 amplification (n=2), EML4-ALK fusion (n=1), and KRAS G12D (n=1). In addition, acquired RB1 mutations including c.1961-1G>C (AF: 6.35%) and p.D604G (AF: 1.02%) were detected from a patient each. Acquired TP53 mutations were detected from 6 patients, with 2 patients harboring mutations known to result in loss of heterozygosity. Moreover, acquired PIK3CA mutations were detected from 2 patients before PD (Figure S5B) and at PD (Figure S5C). Table 2 summarizes the acquired and primary resistance mechanisms detected from the cohort at PD from osimertinib treatment. The remaining 13 patients had mutations with unknown resistance mechanisms; with a majority of these patients (84.6%, 11/13) lost the EGFR T790M.
Table 2. Summary of osimertinib resistance mechanism detected in 32 patients.
| Resistance mechanism | EGFR T790M status | Mutation status | Patient count |
|---|---|---|---|
| EGFR L718Q | T790M loss | Acquired | 1 |
| EGFR C797S | T790M maintain (in cis) | Acquired | 2 |
| EGFR C797S + EML4-ALK fusion + ERBB2 amp | T790M maintain (in cis) | Acquired | 1 |
| EGFR C797S + MET amp | T790M maintain (in cis) | Primary + acquired | 1 |
| BRAF V600E | T790M loss | Primary | 1 |
| BRAF V600E | T790M loss | Acquired | 1 |
| ERBB2 amp | T790M loss | Acquired | 1 |
| ERBB2 amp + PIK3CA E545K | T790M loss | Primary + acquired | 1 |
| KRAS G12V | T790M loss | Acquired | 1 |
| MET exon 14 skipping c.2887+96G>A | T790M maintain | Acquired | 1 |
| MET amp | T790M loss | Acquired | 2 |
| MET amp + TP53 M237I + RB1 c.1961-1G>C | T790M loss | acquired (TP53 LOH) | 1 |
| PIK3CA E545K, Q546P + TP53 H193R | T790M loss | Acquired | 1 |
| TP53 L257V | T790M loss | Acquired | 1 |
| TP53 E286K | T790M loss | Acquired | 1 |
| TP53 E198X + RB1 D604G | T790M loss | Acquired | 1 |
| TP53 Y220C + RB1 V190M, p.V193fs + PTEN T319fs | T790M loss | Acquired (TP53 LOH) | 1 |
| Unknown | T790M loss | 11 | |
| Unknown | T790M loss | 2 |
Amp, amplifications; LOH, loss of heterozygosity.
Acquired mutations were detected at one time point before radiological PD in 62.5% (10/16) patients, while 6 patients had these mutations detected only at confirmation of PD. Interestingly, patients with acquired mutations detected prior to PD were more likely to have elevated maxAF (P=0.027, Figure S6A) and AF of EGFR sensitizing mutations (P=0.031, Figure S6B) prior to radiological PD as compared to patients who acquired these mutations later at PD. In patients who had acquired mutations detected early, both maxAF (P=0.04) and AF of EGFR sensitizing mutations (P=0.06) reached their maximum levels prior to confirmation of radiological PD. Regardless of whether acquired mutations were detected prior to or at PD, maxAF and AF of EGFR sensitizing mutations reached their maximum level at PD (maxAF, P=0.053; AF of EGFRm, P=0.067; Figure S6C,D). These data suggest that dynamic molecular changes during treatment, including the detection of acquired mutations and the elevation of either maxAF or AF of EGFR sensitizing mutations, can be detected as early as one time point prior to radiological PD and could help predict impending PD.
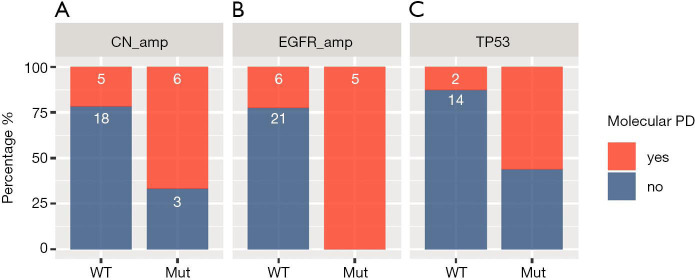
Moreover, molecular PD were more likely to be detected in patients who harbor CNA (P=0.035, Figure 4A), EGFR amplification (P=0.002, Figure 4B), or TP53 mutations (P=0.023, Figure 4C), whether at baseline or acquired, than patients who were wild-type for these alterations, suggesting that these alterations are associated with molecular PD.
Figure 4.
Harboring certain molecular alterations at PD, including copy number amplifications (CNA) (A), EGFR amplifications (B) and TP53 mutations (C), were associated with the detection of molecular PD. Red represents the patients with molecular PD. Blue represents patients who did not experience molecular PD. PD, progressive disease.
Overall, these data demonstrate that dynamic molecular changes and molecular features derived from serial ctDNA profiling can serve as a predictive marker for PD and highlights the importance of treatment monitoring in identifying molecular changes that enable early prediction of PD.
Correlation of acquired mutations with survival outcome
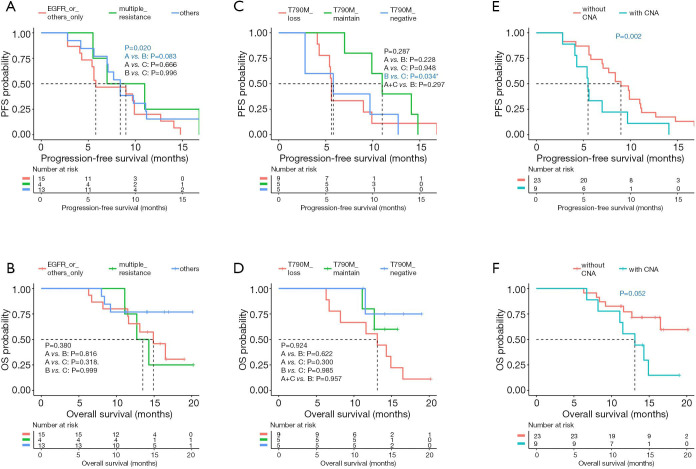
The correlation between the dynamic molecular changes observed during treatment progression and the survival of the patients in the cohort were further analyzed. Our analysis revealed that patients who acquired multiple mutations at PD had significantly longer PFS (P=0.020; median PFS of >1 vs. 1 vs. unknown, 9.0 vs. 5.8 vs. 8.4 months; Figure 5A) but a trend of shorter OS (P=0.380; median OS of >1 vs. 1 vs. unknown, 13.5 vs. 14.9 vs. NR months; Figure 5B) than those who only acquired one mutation in EGFR or other genes, or those with unknown mechanism of resistance. Furthermore, retention of T790M at PD was associated with a trend of longer PFS, with median PFS of 11.0 months (P=0.287; median PFS of loss vs. maintain vs. negative, 5.6 vs. 11.0 vs. 5.8 months; Figure 5C); however, statistical significance was not reached. Meanwhile, T790M status at PD was not associated with OS (P=0.924; median OS of loss vs. maintain vs. negative, 13.1 months vs. NR vs. NR; Figure 5D). Moreover, CNAs were acquired by 9 patients at PD. Patients who were detected with CNA at PD, whether retained from baseline or acquired at PD, commonly had significantly shorter PFS (5.5 vs. 9.0 months, P=0.002, Figure 5E) and had a trend of shorter OS (13.1 months vs. NR, P=0.052, Figure 5F) than those without CNA. The CNAs detected from our cohort included amplifications in EGFR, MET, ERBB2, MYC, and BRAF.
Figure 5.
Certain molecular alterations detected at PD, including the acquisition of more than 1 mutation (A,B), T790M status (C,D), and CNA (E,F), were associated with survival outcomes. Dotted line indicates the median PFS or OS. The risk table below illustrates the number of patients included per time point. PD, progressive disease; CNA, copy number amplification; PFS, progression-free survival; OS, overall survival.
No correlation was found between survival outcomes and early acquisition of mutations prior to radiological PD (PFS, P=0.1; OS, P=0.09) and detection of molecular PD (PFS, P=0.8; OS, P=0.9).
Taken together, these data demonstrate that serial mutation profiling of ctDNA could identify molecular factors that could potentially serve as markers to predict the prognosis of NSCLC patients undergoing osimertinib therapy.
Discussion
As a direct reflection of disease burden, ctDNA is now becoming a routine alternative source of tumor DNA for mutation detection in cancer diagnosis. When assessed longitudinally during the course of treatment, mutation profile derived from the longitudinal assessment of ctDNA could reflect the dynamic molecular changes occurring in response to the treatment and predict an impending PD. In our prospective multi-center study, we identified molecular factors through serial ctDNA profiling that are correlated with survival outcomes of osimertinib-treated advanced NSCLC patients. The serial assessment of the ctDNA mutation profile during osimertinib treatment not only identified baseline molecular features but also reflected dynamic molecular trends that correspond to treatment response and enable early prediction of PD as early as 2.5 months prior to radiologically-confirmed PD. The significant reduction in AF of EGFR sensitizing mutations and ctDNA clearance was associated with treatment response. Meanwhile, the emergence of new mutations and the increase in the AF of EGFR sensitizing mutations and overall maxAF are molecular changes that represent the early stages of developing drug resistance. Although the growth advantage of certain clones would ultimately translate into a clinically detectable PD, radiological imaging does not reflect the clonal evolution at the molecular level. Hence, the detection of dynamic molecular changes thru longitudinal assessment of ctDNA has the potential to predict impending disease relapse.
In general, mutations detected from plasma samples are the consequence of ctDNA shedding by tumors, which is positively correlated with tumor burden (12). T790M positivity evaluated using tissue rebiopsy samples was one of the inclusion criteria of our study; however, NGS-based molecular profiling of baseline blood samples only revealed 75% (54/72) concordance between tissue and plasma-based genotyping of T790M, indicating that 75% of our cohort were ctDNA shedders and 9.7% (7/72) who were T790M positive from tissue samples but had no mutation detected from ctDNA sample were non-shedders. Meanwhile, the remaining 15.3% (11/72) were positive for EGFR L858R or 19del but not T790M could also be considered shedders. Numerous reports have also observed the lower sensitivity in detecting T790M in plasma as compared to tissue samples (19,32-36). These observations might be due to the intrinsic intratumor heterogeneity, which is attributable to the presence of only a small subpopulation of cancer cells harboring T790M. Hence, patients who tested negative for T790M in plasma-based tests are recommended to have tissue-based genotyping for confirmation.
Numerous studies have explored the value of ctDNA levels as a prognostic marker and found the significant correlation between the level of ctDNA and survival outcomes in patients with non-metastatic or metastatic colorectal cancer (23,26,37), NSCLC (21,38,39), and various solid tumors (20). A study has observed disease recurrence in 77% (10/13) of the patients with detectable ctDNA levels, wherein ctDNA levels were detected preceding radiologic and clinical evidence of recurrence by a median of 3 months (23). Meanwhile, 45 patients with undetectable ctDNA levels during follow-up were recurrence-free (23). Moreover, high levels of cell-free DNA and the presence of ctDNA was demonstrated to be associated with significantly inferior OS (20). Consistently, in our cohort, detectable ctDNA (with maxAF of at least 0.2%) during follow-up after 6 weeks from initiating osimertinib therapy was associated with significantly shorter PFS (P=0.022) and OS (P=0.009). In addition to ctDNA levels as a potential prognostic biomarker, certain molecular factors at baseline could be putative prognostic biomarkers. Our analysis revealed that EGFR sensitizing mutations with AF greater than 11%, overall maxAF greater than 11%, and the detection of concurrent gene amplifications at baseline was significantly correlated with poor survival outcome in our cohort. We speculate that this is due to the genetic constitution of the tumor, wherein the abundance of the inhibitor sensitizing mutations could reflect the major oncogenic driver that is responsive to inhibition, which clinically results in improved response and survival outcome. Interestingly, a recent study having a similar study cohort as ours has reported that higher levels of EGFR activating mutations at baseline are also associated with poor outcomes in osimertinib-treated advanced NSCLC patients (36). Moreover, they also reported that the clearance of mutations in plasma ctDNA is correlated with better survival outcomes consistent with other reports (18,36,40,41).
In addition to providing prognostic information, the serial profiling of ctDNA could also provide a snapshot of the dynamic molecular changes that could inform treatment efficacy and enable prediction of an impending PD (41). Consistent with previous findings on the AF reduction of EGFR sensitizing mutations in response to EGFR-TKI treatment (19,39), the significant reduction in the AF of EGFR sensitizing mutation (P<0.001) and maxAF (P<0.001) were observed in our cohort at first follow-up within 6 weeks of osimertinib therapy. Molecular mechanisms associated with osimertinib resistance have already been well-elucidated (35,42,43). Consistent with these studies, various acquired mutations were detected in our cohort. Interestingly, the detection of these acquired mutations, even as early as 2.5 months prior to clinical PD, could enable the early prediction of PD. In addition to the emergence of acquired mutations, the elevation of AF of existing mutations such as EGFR sensitizing mutations, which contributes to the increase of overall maxAF to more than 1.5 times from baseline, are molecular changes that can be detected early and potentially allow the prediction of an impending PD. Interestingly, certain molecular alterations, including CNA (P=0.035), EGFR amplification (P=0.002), or TP53 mutations (P=0.023) were also significantly associated with the detection of molecular PD. The detection of these mutations in the ctDNA during treatment monitoring can guide subsequent treatment decisions.
Despite our interesting findings, our analysis is limited by the number of patients with longitudinal samples. A larger cohort is required to extend our findings. Moreover, since our present study focused only on the utility of plasma-based serial profiling in monitoring treatment responses, histological transformations observed in some patients in the cohort were not included in this study. It would be interesting to explore this mechanism of resistance particularly in patients who have unknown resistance mechanisms and in patients who were non-shedders.
In conclusion, our study identified various molecular features at baseline and dynamic molecular changes during treatment progression, which can serve as predictive and prognostic markers for NSCLC patients undergoing osimertinib therapy. Our study also highlights the clinical utility of serial ctDNA profiling to monitor dynamic molecular changes in ctDNA for the early detection of PD, which can precede imaging modalities with an average lead time of 2.5 months. Our study contributes to the growing evidence of the utility of longitudinal ctDNA profiling in the early detection of PD to improve the survival outcomes of patients with EGFR-mutant NSCLC.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors thank all the patients who participated in this study and their families. We also thank the investigators, study coordinators, operation staff, and the whole project team who worked on this study.
Funding: This work is supported by grants from the National Natural Science Foundation of China (NSFC) (grant numbers: 81773242 to SM, 81602015 and 81772500 to YS) and Major Project of Hangzhou Science and Technology Bureau (grant number: 20180417A01 to SM).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures involving human participants were performed in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Ethics Committee of Jinling Hospital, Nanjing University School of Medicine (approval number: 2017NZKY-001-03). Written informed consent was provided by all the patients included in the study.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-371
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-371
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-371). Dr. YS serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2020 to Aug 2022. Dr. AL, Ms. ML, Ms. HL, Dr. LZ, and Dr. XM report that they are employed by Burning Rock Biotech. The other authors have no conflicts of interest to declare.
References
- 1.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 2.Janne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 3.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol 2014;11:473-81. 10.1038/nrclinonc.2014.104 [DOI] [PubMed] [Google Scholar]
- 4.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.NCCN Clinical Practice Guidelines in Oncology. Non-Small Cell Lung Cancer Version 2.2020 [database on the Internet]. National Comprehensive Cancer Network (NCCN). 2020.
- 6.Kawamura T, Kenmotsu H, Taira T, et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci 2016;107:1001-5. 10.1111/cas.12963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diehl F, Li M, Dressman D, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A 2005;102:16368-73. 10.1073/pnas.0507904102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. 10.1126/scitranslmed.3007094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spellman PT, Gray JW. Detecting cancer by monitoring circulating tumor DNA. Nat Med 2014;20:474-5. 10.1038/nm.3564 [DOI] [PubMed] [Google Scholar]
- 10.Yong E. Cancer biomarkers: Written in blood. Nature 2014;511:524-6. 10.1038/511524a [DOI] [PubMed] [Google Scholar]
- 11.Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. 10.1038/nrclinonc.2017.14 [DOI] [PubMed] [Google Scholar]
- 12.Ou SI, Nagasaka M, Zhu VW. Liquid Biopsy to Identify Actionable Genomic Alterations. Am Soc Clin Oncol Educ Book 2018;38:978-97. 10.1200/EDBK_199765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed] [Google Scholar]
- 14.Mao X, Zhang Z, Zheng X, et al. Capture-Based Targeted Ultradeep Sequencing in Paired Tissue and Plasma Samples Demonstrates Differential Subclonal ctDNA-Releasing Capability in Advanced Lung Cancer. J Thorac Oncol 2017;12:663-72. 10.1016/j.jtho.2016.11.2235 [DOI] [PubMed] [Google Scholar]
- 15.Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 16.Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. 10.1158/1078-0432.CCR-14-2594 [DOI] [PubMed] [Google Scholar]
- 17.Alix-Panabieres C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov 2016;6:479-91. 10.1158/2159-8290.CD-15-1483 [DOI] [PubMed] [Google Scholar]
- 18.Song Y, Hu C, Xie Z, et al. Circulating tumor DNA clearance predicts prognosis across treatment regimen in a large real-world longitudinally monitored advanced non-small cell lung cancer cohort. Transl Lung Cancer Res 2020;9:269-79. 10.21037/tlcr.2020.03.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. 10.1016/j.lungcan.2015.10.004 [DOI] [PubMed] [Google Scholar]
- 20.Ocana A, Diez-Gonzalez L, Garcia-Olmo DC, et al. Circulating DNA and Survival in Solid Tumors. Cancer Epidemiol Biomarkers Prev 2016;25:399-406. 10.1158/1055-9965.EPI-15-0893 [DOI] [PubMed] [Google Scholar]
- 21.Pecuchet N, Zonta E, Didelot A, et al. Base-Position Error Rate Analysis of Next-Generation Sequencing Applied to Circulating Tumor DNA in Non-Small Cell Lung Cancer: A Prospective Study. PLoS Med 2016;13:e1002199. 10.1371/journal.pmed.1002199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Li L, Cohen JD, et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol 2019;5:1118-23. 10.1001/jamaoncol.2019.0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beaver JA, Jelovac D, Balukrishna S, et al. Detection of cancer DNA in plasma of patients with early-stage breast cancer. Clin Cancer Res 2014;20:2643-50. 10.1158/1078-0432.CCR-13-2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klco JM, Miller CA, Griffith M, et al. Association Between Mutation Clearance After Induction Therapy and Outcomes in Acute Myeloid Leukemia. JAMA 2015;314:811-22. 10.1001/jama.2015.9643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625-34. 10.1136/gutjnl-2014-308859 [DOI] [PubMed] [Google Scholar]
- 28.Pantel K, Alix-Panabieres C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol 2019;16:409-24. 10.1038/s41571-019-0187-3 [DOI] [PubMed] [Google Scholar]
- 29.Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. 10.1097/JTO.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 30.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471-4. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz LH, Litiere S, de Vries E, et al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur J Cancer 2016;62:132-7. 10.1016/j.ejca.2016.03.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. 10.1158/1078-0432.CCR-15-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. 10.1016/j.jtho.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 35.Lin CC, Shih JY, Yu CJ, et al. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med 2018;6:107-16. 10.1016/S2213-2600(17)30480-0 [DOI] [PubMed] [Google Scholar]
- 36.Bordi P, Del Re M, Minari R, et al. From the beginning to resistance: Study of plasma monitoring and resistance mechanisms in a cohort of patients treated with osimertinib for advanced T790M-positive NSCLC. Lung Cancer 2019;131:78-85. 10.1016/j.lungcan.2019.03.017 [DOI] [PubMed] [Google Scholar]
- 37.Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015;26:1715-22. 10.1093/annonc/mdv177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Catarino R, Coelho A, Araujo A, et al. Circulating DNA: diagnostic tool and predictive marker for overall survival of NSCLC patients. PLoS One 2012;7:e38559. 10.1371/journal.pone.0038559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taus A, Camacho L, Rocha P, et al. Dynamics of EGFR Mutation Load in Plasma for Prediction of Treatment Response and Disease Progression in Patients With EGFR-Mutant Lung Adenocarcinoma. Clin Lung Cancer 2018;19:387-94.e2. 10.1016/j.cllc.2018.03.015 [DOI] [PubMed] [Google Scholar]
- 40.Thress KS, Markovets A, Barrett JC, et al. Complete clearance of plasma EGFR mutations as a predictor of outcome on osimertinib in the AURA trial. J Clin Oncol 2017;35:9018. 10.1200/JCO.2017.35.15_suppl.9018 [DOI] [Google Scholar]
- 41.Phallen J, Leal A, Woodward BD, et al. Early Noninvasive Detection of Response to Targeted Therapy in Non-Small Cell Lung Cancer. Cancer Res 2019;79:1204-13. 10.1158/0008-5472.CAN-18-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. 10.1001/jamaoncol.2018.2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib As First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:841-9. 10.1200/JCO.2017.74.7576 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as