Abstract
Background
The present study analyzed gene polymorphisms in the potassium voltage-gated channel KQT-like subfamily member 1 (KCNQ1) and the long noncoding RNA, KCNQ1OT1, and their impacts on genetic susceptibility and survival in a Chinese Han population with gastric cancer (GC).
Methods
We designed a case-control study that included 681 patients with GC and 756 healthy controls. Three single-nucleotide polymorphisms (SNPs) in the KCNQ1 gene region and eight SNPs in the KCNQ1OT1 gene region were selected for further research.
Results
Among the 11 SNPs, we found no significant differences in the genotype and allele frequencies between GC patients and the healthy population. Hierarchical analysis by the log-additive model indicated that the KCNQ1 rs231348 CT genotype was significantly associated with an increased GC risk in individuals aged ≥55 years, regardless of gender. The KCNQ1OT1 rs231352 CC and rs7128926 AA genotypes increased the risk of GC in individuals with stage III/IV tumors larger than 5 cm in diameter. On evaluating the genotype polymorphism and survival analysis, we detected that the AA genotypes of the KCNQ1OT1 rs7128926 and rs7939976 polymorphisms presented a significant survival advantage over the GA/GG genotypes, especially in patients with the following characteristics: age >55, Helicobacter pylori infection, BMI >24, tumor in the non-cardia region with a diameter greater than 5 cm, clinical stage II, and postoperative adjuvant chemotherapy.
Conclusions
Our results suggest that the KCNQ1 rs231348 and KCNQ1OT1 rs231352 polymorphisms might be independent predictors of the risk of GC susceptibility depending on certain factors, such as the age of the individual and the tumor stage and diameter. Simultaneously, genotype polymorphism of the rs7128926 and rs7939976 loci of the KCNQ1OT1 gene independently predicted the recurrence-free survival (RFS) and overall survival (OS) of GC patients.
Keywords: Polymorphisms, potassium voltage-gated channel KQT-like subfamily member 1 (KCNQ1), KCNQ1OT1, gastric cancer (GC), genetic susceptibility, survival, Chinese Han population
Introduction
Gastric cancer (GC), a common malignancy with a 5-year survival rate of <25%, is the second leading cause of cancer mortality worldwide (1). The highest GC incidence rate and mortality rate in both sexes are found in East Asia, with approximately 679,100 new cancer cases and 498,000 cancer deaths reported in 2018 in China (2). Carcinogenesis is a complex multistep process resulting from both genetic and environmental factors (3). The development of GC is caused by individual genetic susceptibility, potential environmental components, and/or dietary habits (4,5), such as tobacco smoking, alcohol use, water intake, and daily consumption of meat broth (6,7). Furthermore, genetic factors, among which single-nucleotide polymorphisms (SNPs) are the most common, also play an important role in the development and progression of tumors, including GC. With the continuous development of genetic research in recent years, especially the application of genome-wide association studies (GWAS), more GC susceptibility genes have been discovered (8). Thus far, all GWAS data are from East Asians, including Japanese, Korean, and Chinese populations. However, results from other populations are expected in the next few years (9).
The KCNQ1 gene is located on chromosome 11 and consists of 17 exons of different lengths, ranging from 47 bp (exon 14) to 1,122 bp (exon 16). KCNQ1 encodes the Q1 subfamily of the voltage-dependent potassium channel, a type of membrane protein ubiquitous in the body, that plays an important role in controlling gastric acid secretion and stabilizing resting potential (10). The KCNQ1 locus includes 8–10 maternal allele-encoded protein-coding genes and 1 long noncoding RNA (lncRNA), KCNQ1OT1 (11), which is an imprinted gene that is expressed from the paternal allele. The KCNQ1OT1 transcript regulates the silencing of other imprinted genes in the imprinted gene cluster at position 11p15.5. The KCNQ1 gene is highly expressed in peripheral blood leukocytes, the heart, prostate, inner ear blood vessels, stomach, small intestine, kidney, and pancreas, expressed in tissues that are critical for ion homeostasis (12,13). Humans carrying germline mutations in KCNQ1 develop a range of pathologies, most notably cardiac arrhythmia (long and short QT, Jervell and Lange-Nielsen syndrome), but also hearing loss, elevated gastrin levels, gastric hyperplasia, and in some cases gastric neoplasia (14-17). These phenotypes have been modeled in KCNQ1 knockout mice which develop inner ear defects, imbalance, chronic gastritis, gastric hyperplasia, and gastric metaplasia (18).
Because the transcription of the KCNQ1OT1 gene overlaps with most of the KCNQ1 transcription unit on the anti-strand, it is possible to affect the transcription of KCNQ1 and other non-overlapping genes through transcriptional interference (19). Meanwhile, KCNQ1OT1 can cause transcriptional silencing in chromosomal regions and can serve as an example of lncRNA-mediated gene transcription silencing (20); it has also been associated with poor patient survival in several gastrointestinal cancers, including colorectal cancer (CRC) (21), esophageal cancer (22), and hepatocellular carcinoma (HCC) (23). lncRNAs are closely related to the occurrence and development of tumors (24). Multiple polymorphic sites in the KCNQ1 and KCNQ1OT1 gene region are related to the occurrence and outcome of GC, and genetic variations are also associated with increased risk and overall survival (OS). KCNQ1OT1 dysregulation participates in carcinogenesis and progression of human cancers (25,26). However, the expression and potential functions of this gene region in GC are largely unknown. Therefore, this study selected SNP loci from the KCNQ1 and KCNQ1OT1 gene region and observed the relationship between SNP genotypes and the susceptibility and survival prognosis in Chinese GC patients.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-8052).
Methods
Study participants
From May 2010 to June 2016, 681 patients with GC were enrolled from the Second Affiliated Hospital of the Air Force Medical University and 756 healthy subjects were recruited as controls through strict physical examinations in the same hospital. All 681 patients were confirmed as having primary GC by endoscopic and histopathological analysis. Patients with other types of cancers, gastritis, or gastric ulcers, or those who underwent radiotherapy or chemotherapy were excluded. In addition, all participants were Chinese who were not directly related within the past three generations. Demographic and lifestyle habit data of all the participants were collected through a detailed questionnaire, including residence, age, ethnicity, sex, dietary habits, and previous disease history.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The current study was approved by the institutional ethics committees of the Second Affiliated Hospital of the Air Force Medical University (No. K201009-03). All the participants provided signed informed consent documents and a donation of approximately 5 mL of blood as part of this research.
Genomic DNA extraction and genotyping
Approximately 5 mL of peripheral blood were collected from each subject. Genomic DNA from whole blood samples was extracted using the TIANamp Blood DNA Purification Kit (Gold Mag Co. Ltd., Xi’an City, China) according to the manufacturer’s instructions. Extracted DNA was stored at −20 °C. The concentration and purity of the genomic DNA were determined using a NanoDrop Spectrophotometer (ND-1000, Thermo Fisher Scientific). The KCNQ1 and KCNQ1OT1 gene polymorphisms was genotyped on Agena Mass ARRAY RS1000 platform according to the standard protocol.
The associations between the functional SNPs of the KCNQ1 and KCNQ1OT1 genes and GC development were evaluated. The tag SNPs that represent the polymorphisms of a block were included in our study. Finally, three SNPs (rs6578283, rs231348, and rs760419) in KCNQ1, and eight SNPs (rs10832514, rs231361, rs231359, rs7128926, rs231356, rs231354, rs231352, and rs7939976) in KCNQ1OT1 were selected for further research. The characteristics of the sequences in this study are summarized in Table S1. Then, Agena Typer 4.0 software (Agena Bioscience) was used to analyze and manage our data.
Statistical analysis
Continuous variables, such as demographic variables, risk factors, and KCNQ1 and KCNQ1OT1 genotype distribution between the case and control groups, are shown as the mean ± standard deviation (SD) and were compared by Student’s t-test. Categorical variables are presented as frequencies with percentages (%) and were compared with the χ2 test or Fisher’s exact test when appropriate. The χ2 test was also used to assess whether the distribution of genotypes was consistent with the Hardy-Weinberg equilibrium (HWE). Univariate and multivariate logistic regression analyses were used to test the association between the SNP genotypes and the progression of GC based on the generated odds ratios (ORs) and 95% confidence intervals (18). Adjusted ORs were calculated using multivariate analysis adjusting for age and gender. The Kaplan-Meier method was used to estimate the OS and recurrence-free survival (RFS), the log-rank test was used to compare survival distributions, and the Cox proportional hazards model was used to determine the hazards ratio (HR). All statistical analyses were performed using SAS 9.4 software (IBM Corp.). Statistical significance was defined as a P value <0.05 with a two-tailed test.
Results
Characteristics of the participants
This study contained 681 GC patients and 756 healthy controls. Overall, the variable distribution of the selected demographic data did not differ between GC patients and controls. Table 1 shows the distributions of age, gender, and clinical stages of the study subjects. There were no significant statistical differences between the groups in terms of gender and age (P<0.001). The mean age was 57.57±10.826 years for GC patients and 52.58±8.709 years for the controls. The proportion of male subjects was significantly higher in both groups (77.4% vs. 64.7%, respectively). A follow-up of all patients was carried out according to our standard protocol, the median follow-up period for GC patients was 6.04 years (range, 0.12–10.68 years) and 6.15 years (range, 0.92–10.68 years) for the control cases. The latest follow-up data in this analysis was obtained in October 2019.
Table 1. Characteristics and clinical features of the GC group and the control group.
| Variables | GC (%) N=681 | Controls (%) N=756 | Pa |
|---|---|---|---|
| Age (years) | <0.001 | ||
|---|---|---|---|
| M ± SD | 57.57±10.826 | 52.58±8.709 | |
| ≥55 | 251 (36.86) | 429 (56.75) | |
| <55 | 430 (63.14) | 327 (43.25) | |
| Gender | <0.001 | ||
| Male | 527 (77.4) | 489 (64.7) | |
| Female | 154 (22.6) | 267 (35.3) | |
| BMI | <0.001 | ||
| <24 | 460 (67.5) | 432 (57.2) | |
| ≥24 | 221 (32.5) | 324 (42.8) | |
| Diameterb | |||
| <5 cm | 380 (55.8) | ||
| ≥5 cm | 273 (40.8) | ||
| H. pylori b | |||
| Yes | 392 | ||
| No | 187 |
| Stageb | |||
|---|---|---|---|
| ,0/I/II | 458 (67.25) | ||
| III/ IV | 204 (29.96) | ||
| R/Mb | |||
| No | 287 | ||
| Yes | 377 | ||
| Family historyb | |||
| No | 282 | ||
| Yes | 45 | ||
| ACTb | |||
| Yes | 430 | ||
| No | 233 | ||
| Positionb | |||
| Cardia | 125 (18.34) | ||
| No-cardia | 425(62.41) |
a, based on a two-sided χ2 test for distributions between gastric cancer and cancer-free controls; b, patient numbers may not add up to 100% of available subjects because of missing clinical data. P<0.05 indicates statistical significance. GC, gastric cancer; ACT, adjuvant chemotherapy; R/M, recurrence/metastasis.
Associations between KCNQ1 and KCNQ1OT1 SNPs and GC risk
The basic data on the three KCNQ1 SNP genotype frequencies (rs6578283, rs231348, and rs760419) and eight KCNQ1OT1 SNP genotype frequencies (rs10832514, rs231361, rs231359, rs7128926, rs231356, rs231354, rs231352, and rs7939976) are shown in Table S1, which contains the position, alleles, minor allele frequency (MAF) distribution, HWE P value, ORs, and 95% CIs of all the candidate SNPs. The MAF values for each SNP in the controls were similar to the values for the Chinese population in the database (0.208 vs. 0.195, 0.115 vs. 0.106, 0.335 vs. 0.332, 0.118 vs. 0.111, 0.214 vs. 0.217, 0.213 vs. 0.219, 0.090 vs. 0.092, 0.217 vs. 0.225, 0.123 vs. 0.133, 0.122 vs. 0.132, and 0.087 vs. 0.089, respectively). The HWE P values of the 11 SNPs were more than 0.05, and none showed a significant departure from the HWE. Using the Pearson χ2 test, we compared the SNP genotype and allele frequencies of KCNQ1 and KCNQ1OT1 between the casegroup and the control group and calculated ORs to evaluate associations with the GC risk. However, there were no statistically significant differences in genotype and allele frequencies between GC cases and controls in the Chinese population (P>0.05, Table S1). To further assess the possible association between the KCNQ1 and KCNQ1OT1 polymorphisms and the risk of GC under different genetic models, we applied logistic regression analysis and assumed four genetic models (codominant, dominant, recessive, and log-additive) with adjustments for age and gender (shown in Table S2). Again, we observed no statistically significant differences between patients and controls (P>0.05, Table S2).
We next performed a stratified analysis according to age, gender, clinical stage and tumor diameter to evaluate the effect of these 11 SNPs on GC risk. Stratified analysis by age revealed significant associations between the rs231348 genotype and the risk of GC, as displayed in Table S3. The rs231348 CT genotype in KCNQ1 was identified to increase the risk of GC in individuals aged ≥55 years using the log-additive model (OR =1.48, CI: 1.01–2.16, P=0.042). No significant associations with GC risk were observed in either females or males (Table S3). Stratified analysis by clinical stage revealed significant associations between the KCNQ1OT1 rs231352 genotype and the risk of GC, as displayed in https://cdn.amegroups.cn/static/public/10.21037atm-20-8052-1.pdf. The rs231352 CC genotype in KCNQ1OT1 was identified to decrease the risk of GC in individuals at stages I–II as opposed to stages III–IV according to the recessive model (OR =0.31, CI: 0.08–1.12, P=0.048). In addition, stratified analysis by tumor diameter revealed a significant association between the KCNQ1OT1 rs7128926 genotype and the risk of GC, as displayed in https://cdn.amegroups.cn/static/public/10.21037atm-20-8052-1.pdf. The rs7128926 AA genotype in KCNQ1OT1 was identified to decrease the risk of GC in individuals with a tumor diameter <5 cm as opposed to ≥5 cm according to the recessive model (P=0.026) (https://cdn.amegroups.cn/static/public/10.21037atm-20-8052-1.pdf).
Recurrence risk evaluation and survival analysis
Recurrent and mortality events were recorded, and the RFS was calculated for the prognosis assessment. Of the 681 patients diagnosed with GC, more than half were classified as the most common stages 0 [26], I [126], and II [316]. Most patients showed a tumor diameter less than 5 cm [380], and during the entire observation period, 377 patients experienced recurrence. To further estimate the correlation between the KCNQ1 and KCNQ1OT1 polymorphisms and the recurrence or metastasis risk and survival analysis of GC, univariate analysis, multivariate analysis, and multiple inheritance models (dominant, recessive, and additive models) were applied to analyze potential associations by a logistic regression analysis adjusted for age and gender. As shown in https://cdn.amegroups.cn/static/public/10.21037atm-20-8052-2.pdf, after adjusting for age and gender, rs7128926 in KCNQ1OT1 decreased the GC recurrence and metastasis risk in both the codominant model (P=0.047) and the recessive model (P=0.026). Moreover, according to the recessive model, the KCNQ1OT1 rs7939976 GG genotype was associated with a decreased recurrence and metastasis risk of GC (P=0.026) compared with the rs7939976 AA/AG genotypes (https://cdn.amegroups.cn/static/public/10.21037atm-20-8052-2.pdf).
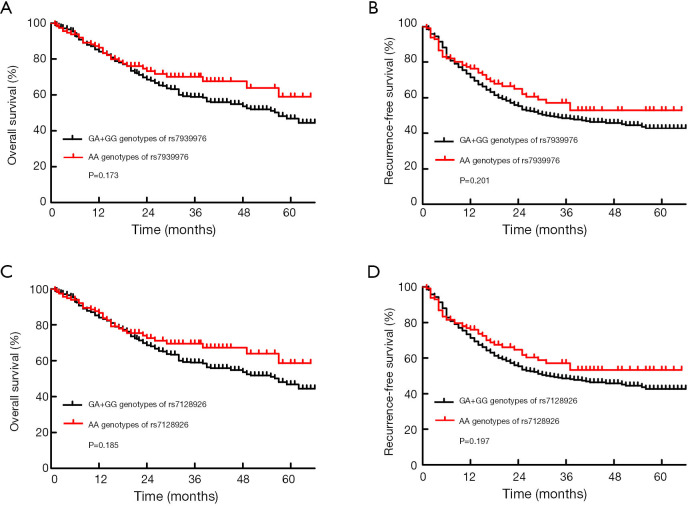
The results of the survival analysis in relation to the SNPs are listed in Table 2. Among the 11 SNPs, 2 KCNQ1OT1 SNPs, rs7939976 and rs7128926, were significantly associated with survival. Three genotypes, AA, GA and GG, were detected at the rs7939976 and rs7128926 loci of the KCNQ1OT1 gene. Compared with patients with the GG and GA genotype of the KCNQ1OT1 SNP rs7939976, we found that patients with the AA genotypes had longer OS and RFS after 1 year (Figure 1A,B), and patients carrying the A alleles had a substantially higher median OS compared with patients with no A allele (OR =20.749, CI: 2.68–160.616, P=0.004) (Table 2). Among all the cases, the MST was 57 months for those with the rs7939976 AA genotype (two A alleles), 47.8 months for those with the GA genotype (one A allele), and 4 months for those with the GG genotype (no A allele). This trend was also similar for the MST with RFS, with 32 months for AA, 11.2 months for GA, and 4 months for GG (P<0.01) (Table 2).
Table 2. Genotypes of KCNQ1 and KCNQ1OT1 polymorphisms with clinical outcome of gastric cancer patients.
| SNP ID | Genotype | Total/event | OS | Total/event | RFS | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Log-rank P | MST | HR (95% CI) | P | Log-rank P | MST | HR (95% CI) | P | ||||
| rs6578283 | AA | 421/140 | 62 | 0.804 | 421/179 | 41 | 0.503 | ||||
| GA | 211/77 | 0.773 | 57 | 0.958 (0.696–1.32) | 0.795 | 211/94 | 0.986 | 31 | 0.880 (0.660–1.174) | 0.386 | |
| GG | 32/9 | 47.3 | 1.234 (0.594–2.565) | 0.573 | 32/14 | 29 | 1.223 (0.670–2.231) | 0.512 | |||
| Dominant | 243/86 | 0.781 | 57 | 0.985 (0.725–1.339) | 0.925 | 243/108 | 0.87 | 30 | 0.918 (0.699–1.207) | 0.542 | |
| rs231348 | CC | 520/173 | 57 | 0.781 | 520/221 | 37 | 0.621 | ||||
| TC | 131/48 | 0.766 | 57 | 1.001 (0.704–1.451) | 0.953 | 131/59 | 0.343 | 32 | 0.996 (0.722–1.374) | 0.981 | |
| TT | 12/5 | 41.039 | 1.436 (0.524–3.937) | 0.481 | 12/7 | 17 | 1.561 (0.635–3.836) | 0.331 | |||
| Dominant | 143/53 | 0.754 | 57 | 1.040 (0.735–1.473) | 0.824 | 143/66 | 0.675 | 29 | 1.032 (0.757–1.406) | 0.842 | |
| rs760419 | AA | 304/105 | 62 | 0.174 | 304/131 | 31 | 0.302 | ||||
| GA | 281/90 | 0.517 | 59 | 0.787 (0.565–1.096) | 0.157 | 281/117 | 0.609 | 41 | 0.845 (0.632–1.128) | 0.253 | |
| GG | 79/31 | 39 | 1.165 (0.743–1.825) | 0.507 | 79/39 | 22 | 1.127 (0.752–1.687) | 0.563 | |||
| Dominant | 360/121 | 0.88 | 57 | 0.866 (0.638–1.176) | 0.358 | 360/156 | 0.953 | 37 | 0.904 (0.690–1.183) | 0.462 | |
| r231356 | TT | 406/138 | 57 | 0.827 | 406/172 | 40 | 0.358 | ||||
| TA | 228/79 | 0.961 | 57 | 1.038 (0.758–1.421) | 0.818 | 228/102 | 0.864 | 31 | 1.063 (0.806–1.403) | 0.664 | |
| AA | 30/9 | 46.776 | 1.289 (0.561–2.959) | 0.55 | 30/12 | 26 | 1.637 (0.827–3.241) | 0.157 | |||
| Dominant | 258/88 | 0.986 | 57 | 1.056 (0.778–1.433) | 0.726 | 258/115 | 0.613 | 31 | 1.102 (0.843–1.441) | 0.478 | |
| rs231354 | CC | 507/172 | 57 | 0.917 | 507/213 | 40 | 0.556 | ||||
| TC | 150/52 | 0.961 | 57 | 1.043 (0.740–1.470) | 0.812 | 150/71 | 0.636 | 29 | 1.150 (0.854–1.550) | 0.356 | |
| TT | 7/2 | 47.7 | 0.714 (0.099–5.167) | 0.739 | 7/3 | 17 | 0.582 (0.081–4.193) | 0.591 | |||
| Dominant | 157/54 | 0.925 | 57 | 1.032 (0.734–1.450) | 0.858 | 157/74 | 0.342 | 29 | 1.132 (0.842–1.521) | 0.413 | |
| rs231352 | CC | 509/172 | 57 | 0.914 | 509/214 | 40 | 0.545 | ||||
| TC | 148/52 | 0.936 | 57 | 1.045 (0.741–1.473) | 0.803 | 148/70 | 0.634 | 26 | 1.154 (0.857–1.555) | 0.346 | |
| TT | 7/2 | 47.7 | 0.714 (0.099–5.169) | 0.739 | 7/3 | 17 | 0.582 (0.081–4.195) | 0.591 | |||
| Dominant | 155/54 | 0.826 | 57 | 1.034 (0.736–1.453) | 0.848 | 155/73 | 0.34 | 26 | 1.135 (0.844–1.526) | 0.401 | |
| rs7939976 | AA | 550/193 | 57 | 0.014 | 550/243 | 32 | 0.010 | ||||
| GA | 112/31 | 0.048 | 47.832 | 0.929 (0.595–1.449) | 0.745 | 112/42 | <0.001 | 11.258 | 1.030 (0.704–1.508) | 0.878 | |
| GG | 2/2 | 4 | 20.749 (2.680–160.616) | 0.004 | 2/2 | 2 | 23.238 (3.026–178.479) | 0.002 | |||
| Dominant | 114/33 | 0.263 | 47.06 | 0.972 (0.628–1.504) | 0.897 | 114/44 | 0.322 | 40.59 | 1.063 (0.730–1.548) | 0.750 | |
| rs10832514 | AA | 517/174 | 57 | 0.155 | 517/219 | 37 | 0.081 | ||||
| GA | 139/49 | 0.809 | 41.823 | 1.359 (0.955–1.933) | 0.088 | 139/64 | 0.812 | 29 | 1.346 (0.984–1.840) | 0.063 | |
| GG | 8/3 | 36.964 | 1.821 (0.574–5.783) | 0.309 | 8/4 | 20 | 2.041 (0.750–5.558) | 0.163 | |||
| Dominant | 147/52 | 0.517 | 41.778 | 1.383 (0.982–1.949) | 0.064 | 147/68 | 0.522 | 29 | 1.379 (1.017–1.870) | 0.039 | |
| rs231361 | AA | 409/142 | 57 | 0.975 | 409/179 | 35 | 0.608 | ||||
| GA | 225/76 | 0.832 | 57 | 1.031 (0.752–1.413) | 0.849 | 225/96 | 0.981 | 37 | 1.038 (0.787–1.371) | 0.791 | |
| GG | 29/8 | 48.286 | 1.066 (0.432–2.633) | 0.89 | 29/12 | 38.76 | 1.437 (0.700–2.952) | 0.323 | |||
| Dominant | 254/84 | 0.768 | 44.71 | 1.034 (0.761–1.405) | 0.832 | 254/108 | 0.856 | 37 | 1.066 (0.814–1.395) | 0.642 | |
| rs231359 | AA | 410/143 | 57 | 0.943 | 410/176 | 40 | 0.617 | ||||
| CA | 224/75 | 0.788 | 57 | 0.950 (0.692–1.303) | 0.749 | 224/99 | 0.986 | 31 | 1.029 (0.780–1.357) | 0.840 | |
| CC | 29/8 | 48.286 | 1.038 (0.420–2.562) | 0.936 | 29/12 | 38.76 | 1.433 (0.698–2.945) | 0.327 | |||
| Dominant | 253/83 | 0.611 | 45.069 | 0.956 (0.703–1.300) | 0.775 | 253/111 | 0.865 | 31 | 1.056 (0.808–1.381) | 0.690 | |
| rs7128926 | AA | 547/192 | 57 | 0.012 | 547/242 | 32 | 0.010 | ||||
| GA | 115/32 | 0.05 | 47.667 | 0.863 (0.553–1.346) | 0.515 | 115/43 | <0.001 | 11.293 | 0.946 (0.647–1.385) | 0.777 | |
| GG | 2/2 | 4 | 20.585 (2.659–159.37) | 0.004 | 2/2 | 2 | 22.981 (2.992–176.541) | 0.003 | |||
| Dominant | 117/34 | 0.277 | 46.924 | 0.902 (0.583–1.395) | 0.642 | 117/45 | 0.315 | 40.64 | 0.976 (0.670–1.421) | 0.899 | |
KCNQ1, potassium voltage-gated channel KQT-like subfamily member 1; OS, overall survival; RFS, recurrence-free survival; MST, median survival time;
Figure 1.
Kaplan-Meier curves of OS and RFS in patients with different KCNQ1OT1 rs7939976 and rs7128926 genotypes. (A,B) Median OS and RFS were longer in patients with KCNQ1OT1 rs7939976 AA genotypes than in those with GA + GG genotypes after 1 year (log-rank P=0.173 and 0.201, respectively). (C,D) Median OS and RFS were longer in patients with KCNQ1OT1 rs7128926 AA genotypes than in those with GG and GA genotypes after 1 year (log-rank P=0.185 and 0.197, respectively). OS, overall survival; RFS, recurrence-free survival. KCNQ1, potassium voltage-gated channel KQT-like subfamily member 1; OS, overall survival; RFS, recurrence-free survival.
Meanwhile, patients with the AA genotype of the KCNQ1OT1 rs7128926 SNP had longer OS and RFS after 1 year compared with patients with the GG and GA genotypes (Figure 1C,D), although there was no statistically significant difference (log-rank P=0.185 and 0.197, respectively). Meanwhile, as shown in Table 2, patients carrying the A allele of rs7128926 had a substantially higher OS (OR =0.863, CI: 0.553–1.346, P=0.012) and RFS (OR =0.946, CI: 0.647–1.385, P=0.01) compared with patients without the A allele. Among all the cases, the median survival time (MST) was 57 months for those with the rs7128926 AA genotype (two A alleles), 47 months for those with the GA genotype (one A allele), and only 4 months for those with the GG genotype (no A allele). This trend was similar for the MST with RFS, with 32 months for AA, 11.2 months for GA, and 2 months for GG (P<0.01). The other six SNP loci (rs10832514, rs231361, rs231359, rs231356, rs231354, and rs231352) of KCNQ1OT1 and the three SNP loci (rs6578283, rs231348 and rs760419) of KCNQ1 showed no significant correlation with OS and RFS in patients with GC (Table 2).
To further investigate the relationship between the KCNQ1OT1 rs7128926 and rs7939976 SNPs and survival time in GC patients, we carried out a stratified analysis of the correlation between the two SNP loci in patients with GC, and the effect of age, gender, BMI, Helicobacter pylori infection status, clinical stage, tumor diameter, and chemotherapy status on OS and RFS were evaluated using a codominant model (Tables 3,4). The analysis revealed that MST and OS were the lowest in patients with the GG genotype at the KCNQ1OT1 rs7128926 locus, especially in females over 60 years of age, followed by H. pylori infection, BMI >24, tumor in the non-cardia region with a diameter greater than 5 cm, clinical stage II, and postoperative adjuvant chemotherapy. This trend was similar for the MST and RFS among patients with the GG genotype at the KCNQ1OT1 rs7939976 locus (Table 3). We also found that patients carrying the GG genotype at the KCNQ1OT1 rs7939976 locus had a poorer MST with OS and RFS (Table 4).
Table 3. Stratified analysis of the KCNQ1OT1 rs7128926 polymorphism with gastric cancer OS and RFS.
| Variables | Genotype | OS | RFS | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total/event | MST | HR (95% CI) | P | Total/event | MST | HR (95% CI) | P | |||
| Age1 | ||||||||||
| ≥55 | AA | 244/89 | 56 | 1 | 0.033 | 244/109 | 26 | 1 | 0.012 | |
| GA | 43/13 | 46.124 | 1.182 (0.622–2.243) | 0.61 | 43/17 | 40.207 | 1.282 (0.731–2.248) | 0.386 | ||
| GG | 2/2 | 4 | 16.444 (1.976–136.829) | 0.01 | 2/2 | 2 | 23.626 (2.808–198.769) | 0.004 | ||
| <55 | AA | 303/103 | 57 | 1 | 303/133 | 38 | 1 | |||
| GA | 72/19 | 46.867 | 0.759 (0.407–1.416) | 0.386 | 72/26 | 40.437 | 0.798 (0.473–1.3484) | 0.399 | ||
| GG | NA | NA | ||||||||
| Gender3 | ||||||||||
| Male | AA | 423/145 | 57 | 1 | 423/182 | 34 | 1 | |||
| GA | 88/22 | 49.604 | 0.760 (0.448–1.290) | 0.309 | 88/30 | 43.837 | 0.937 (0.597–1.470) | 0.776 | ||
| GG | 1/1 | 32 | 1/1 | 5 | ||||||
| Female | AA | 124/47 | 49 | 1 | 0.029 | 124/60 | 28 | 1 | 0.025 | |
| GA | 27/10 | 57 | 1.111 (0.483–2.555) | 0.805 | 27/13 | 18 | 0.912 (0.429–1.941) | 0.812 | ||
| GG | 1/1 | 4 | 20.590 (2.226–190.458) | 0.008 | 1/1 | 2 | 20.027 (2.225–180.28) | 0.008 | ||
| BMIa | ||||||||||
| <24 | AA | 142/42 | 44.062 | 1 | 142/55 | 42 | 1 | 0.18 | ||
| GA | 21/4 | 53.505 | 0.664 (0.233–1.891) | 0.443 | 21/5 | 49.591 | 0.493 (0.175–1.387) | 0.18 | ||
| GG | NA | NA | ||||||||
| ≥24 | AA | 338/114 | 45.062 | 1 | 0.026 | 338/140 | 50 | 1 | 0.012 | |
| GA | 77/20 | 46.393 | 0.926 (0.566–1.515) | 0.761 | 77/29 | 37 | 1.082 (0.715–1.638) | 0.709 | ||
| GG | 1/1 | 4 | 16.775 (2.129–132.171) | 0.007 | 1/1 | 2 | 22.835 (2.891–180.37) | 0.003 | ||
| H. pylori infectiona | ||||||||||
| Yes | AA | 331/108 | 46.099 | 1 | 0.028 | 331/128 | 41.846 | 1 | 0.017 | |
| GA | 60/17 | 47.416 | 0.946 (0.556–1.608) | 0.837 | 60/20 | 44.019 | 0.963 (0.593–1.565) | 0.879 | ||
| GG | 1/1 | 4 | 16.661 (2.099–132.243) | 0.008 | 1/1 | 2 | 20.207 (2.547–160.309) | 0.004 | ||
| No | AA | 149/48 | 35.121 | 1 | 0.329 | 149/67 | 25 | 1 | 0.706 | |
| GA | 38/7 | 36.069 | 0.661 (0.288–1.517) | 0.329 | 38/14 | 37 | 0.886 (0.471–1.665) | 0.706 | ||
| GG | NA | NA | ||||||||
| GA | 70/18 | 47.349 | 0.796 (0.427–1.486) | 0.457 | 70/25 | 40.684 | 0.857 (0.507–1.448) | 0.564 | ||
| GG | NA | NA | ||||||||
| ACTa | ||||||||||
| No | AA | 190/51 | 62 | 1 | 0.145 | 190/55 | 45.538 | 1 | 0.092 | |
| GA | 43/7 | 50.692 | 0.520 (0.216–1.252) | 0.145 | 43/8 | 49.297 | 0.495 (0.218–1.122) | 0.092 | ||
| GG | NA | NA | ||||||||
| Yes | AA | 357/141 | 48 | 1 | <0.001 | 357/187 | 24 | 1 | 0.002 | |
| GA | 71/25 | 57 | 1.184 (0.702–1.997) | 0.527 | 71/35 | 26 | 1.179 (0.763–1.822) | 0.458 | ||
| GG | 1/1 | 4 | 21.275 (2.295–197.256) | 0.007 | 1/1 | 2 | 20.576 (2.282–185.538) | 0.007 | ||
| Tumor sitea | ||||||||||
| Cardia | AA | 106/44 | 39 | 1 | 0.696 | 106/54 | 18 | 1 | 0.56 | |
| GA | 19/6 | 44.485 | 0.836 (0.340–2.055) | 0.696 | 19/8 | 37 | 0.794 (0.366–1.722) | 0.56 | ||
| GG | NA | NA | ||||||||
| Non-cardia | AA | 348/103 | 47.599 | 1 | 0.005 | 348/130 | 42.709 | 1 | 0.005 | |
| GA | 76/16 | 50.944 | 0.879 (0.514–1.504) | 0.638 | 76/25 | 43.566 | 1.086 (0.695–1.697) | 0.716 | ||
| GG | 1/1 | 4 | 32.242 (3.866–268.872) | 0.001 | 1/1 | 2 | 30.802 (3.815–248.709) | 0.001 | ||
| Tumor diametera | ||||||||||
| <5 | AA | 309/82 | 48.422 | 1 | 0.307 | 309/107 | 43.251 | 1 | 0.617 | |
| GA | 68/11 | 54.953 | 0.701 (0.354–1.386) | 0.307 | 68/21 | 46.036 | 1.138 (0.686–1.889) | 0.617 | ||
| GG | NA | NA | ||||||||
| ≥5 | AA | 227/106 | 33 | 1 | 0.019 | 227/131 | 18 | 1 | 0.005 | |
| GA | 44/20 | 38 | 1.005 (0.551–1.833) | 0.987 | 44/20 | 31 | 0.709 (0.383–1.311) | 0.273 | ||
| GG | 2/2 | 4 | 20.254 (2.481–165.381) | 0.005 | 2/2 | 2 | 26.207 (3.200–214.648) | 0.002 | ||
| Clinical stagea | ||||||||||
| Early | AA | 110/11 | 1 | 0.98 | 110/14 | 57.535 | 1 | 0.416 | ||
| GA | 30/0 | 0 | 0.98 | 30/1 | 62.87 | 0.421 (0.052–3.383) | 0.416 | |||
| GG | NA | NA | ||||||||
| Middle | AA | 384/143 | 48 | 1 | 0.036 | 384/184 | 25 | 1 | 0.035 | |
| GA | 78/27 | 57 | 0.882 (0.538–1.446) | 0.618 | 78/38 | 26 | 0.920 (0.609–1.390) | 0.692 | ||
| GG | 2/2 | 4 | 14.270 (1.810–112.511) | 0.012 | 2/2 | 2 | 14.416 (1.859–111.822) | 0.011 | ||
| Late | AA | 49/36 | 13 | 1 | 0.269 | 49/41 | 7 | 1 | 0.336 | |
| GA | 6/5 | 15 | 0.530 (0.172–1.635) | 0.269 | 6/4 | 6 | 0.531 (0.146–1.930) | 0.336 | ||
| GG | NA | NA | ||||||||
1, adjusted by gender; 3, adjusted by age. a, patient numbers may not add up to 100% of available subjects because of missing clinical data. KCNQ1, potassium voltage-gated channel KQT-like subfamily member 1; OS, overall survival; RFS, recurrence-free survival; MST, median survival time; ACT, adjuvant chemotherapy; R/M, recurrence/metastasis.
Table 4. Stratified analysis of the KCNQ1OT1 rs7939976 polymorphism with gastric cancer OS and RFS.
| Variables | Genotype | Total/event | OS | Total/event | RFS | ||||
|---|---|---|---|---|---|---|---|---|---|
| MST | HR (95% CI) | P | MST | HR (95% CI) | P2 | ||||
| Age1 | |||||||||
| ≥55 | AA | 245/89 | 56 | 1 | 0.031 | 245/109 | 26 | 1 | 0.009 |
| GA | 42/13 | 45.762 | 1.221 (0.643–2.32) | 0.542 | 42/17 | 37 | 1.378 (0.784–2.424) | 0.265 | |
| GG | 2/2 | 4 | 16.54 (1.988–137.641) | 0.009 | 2/2 | 2 | 24.131 (2.866–203.164) | 0.003 | |
| <55 | AA | 305/104 | 57 | 1 | 305/134 | 38 | 1 | ||
| GA | 70/18 | 47.349 | 0.796 (0.427–1.486) | 0.457 | 70/25 | 40.684 | 0.857 (0.507–1.448) | 0.564 | |
| GG | NA | NA | |||||||
| Gender3 | |||||||||
| Male | AA | 424/145 | 57 | 424/182 | 34 | ||||
| GA | 87/22 | 49.503 | 0.800 (0.470–1.360) | 0.41 | 87/30 | 43.725 | 1.018 (0.646–1.604) | 0.938 | |
| GG | 1/1 | 32 | 1/1 | 5 | |||||
| Female | AA | 126/48 | 49 | 1 | 0.025 | 126/61 | 28 | 1 | 0.026 |
| GA | 25/9 | 57 | 1.272 (0.552–2.929) | 0.572 | 25/12 | 18 | 1.025 (0.482–2.179) | 0.949 | |
| GG | 1/1 | 4 | 21.275 (2.295–197.256) | 0.007 | 1/1 | 2 | 20.576 (2.282–185.538) | 0.007 | |
| BMIa | |||||||||
| <24 | AA | 140/42 | 43.675 | 1 | 140/55 | 42 | 1 | ||
| GA | 23/4 | 54.558 | 0.594 (0.209–1.687) | 0.328 | 23/5 | 51.132 | 0.439 (0.156–1.229) | 0.117 | |
| GG | NA | NA | 0.439 (0.156–1.229) | 0.117 | |||||
| ≥24 | AA | 343/115 | 45.225 | 1 | 0.027 | 343/141 | 50 | 1 | 0.008 |
| GA | 72/19 | 45.961 | 1.043 (0.638–1.706) | 0.866 | 72/28 | 37 | 1.239 (0.818–1.876) | 0.313 | |
| GG | 1/1 | 4 | 16.941 (2.151–133.427) | 0.007 | 1/1 | 2 | 23.274 (2.948–183.783) | 0.003 | |
| H. pylori infectiona | |||||||||
| Yes | AA | 333/109 | 46.116 | 1 | 0.028 | 333/129 | 41.865 | 1 | 0.017 |
| GA | 58/16 | 47.68 | 1.028 (0.605–1.747) | 0.918 | 58/19 | 43.953 | 1.026 (0.632–1.666) | 0.916 | |
| GG | 1/1 | 4 | 16.764 (2.113–133.028) | 0.008 | 1/1 | 2 | 20.401 (2.572) | 0.004 | |
| No | AA | 150/448 | 35.046 | 1 | 0.382 | 150/67 | 25 | 1 | |
| GA | 37/7 | 36.294 | 0.689 (0.299–1.589) | 0.382 | 37/14 | 37 | 0.974 (0.512–1.850) | 0.935 | |
| GG | NA | NA | |||||||
| ACTa | |||||||||
| No | AA | 192/51 | 62 | 1 | 0.233 | 192/55 | 45.801 | 1 | 0.144 |
| GA | 41/7 | 50.023 | 0.587 (0.244–1.410) | 0.233 | 41/8 | 48.662 | 0.544 (0.240–1.230) | 0.144 | |
| GG | NA | NA | |||||||
| Yes | AA | 358/142 | 48 | 1 | <0.001 | 358/188 | 23 | 1 | 0.001 |
| GA | 70/24 | 57 | 1.268 (0.750–2.143) | 0.375 | 70/34 | 29 | 1.296 (0.837–2.007) | 0.244 | |
| GG | 2/2 | 4 | 89.316 (9.807–813.41) | <0.001 | 2/2 | 2 | 42.558 (5.243–345.483) | <0.001 | |
| Tumor sitea | |||||||||
| Cardia | AA | 105/44 | 35 | 1 | 0.383 | 105/54 | 18 | 1 | 0.331 |
| GA | 20/6 | 45.681 | 0.656 (0.254–1.693) | 0.383 | 20/8 | 37 | 0.667 (0.295–1.510) | 0.331 | |
| GG | NA | NA | |||||||
| Non-cardia | AA | 351/103 | 47.847 | 1 | 0.006 | 351/130 | 42.963 | 1 | 0.004 |
| GA | 73/16 | 50 | 0.993 (0.581–1.697) | 0.98 | 73/25 | 42.314 | 1.233 (0.789–1.924) | 0.357 | |
| GG | 1/1 | 4 | 32.587 (3.909–271.665) | 0.001 | 1/1 | 2 | 31.240 (3.870–252.142) | 0.001 | |
| Tumor diametera | |||||||||
| <5 | AA | 308/82 | 48.354 | 1 | 0.261 | 308/107 | 43.174 | 1 | 0.692 |
| GA | 69/11 | 55.114 | 0.675 (0.341–1.338) | 0.261 | 69/21 | 46.241 | 1.108 (0.666–1.844) | 0.692 | |
| GG | NA | NA | |||||||
| ≥5 | AA | 230/106 | 33 | 1 | 0.017 | 230/131 | 18 | 1 | 0.008 |
| GA | 41/20 | 38 | 1.159 (0.634–2.119) | 0.631 | 41/20 | 23 | 0.852 (0.458–1.585) | 0.613 | |
| GG | 2/2 | 4 | 20.426 (2.503–166.657) | 0.005 | 2/2 | 2 | 26.846 (3.278–219.865) | 0.002 | |
| Clinical stagea | |||||||||
| Early | AA | 109/11 | 1 | 0.976 | 109/14 | 57.497 | 1 | 0.378 | |
| GA | 31/0 | 0 | 0.976 | 31/1 | 62.958 | 0.393 (0.049–3.146) | 0.378 | ||
| GG | NA | NA | |||||||
| Middle | AA | 387/143 | 49 | 1 | 0.04 | 387/184 | 25 | 1 | 0.038 |
| GA | 75/27 | 57 | 0.959 (0.584–1.576) | 0.87 | 75/38 | 25 | 0.999 (0.661–1.511) | 0.997 | |
| GG | 2/2 | 4 | 14.354 (1.821–113.143) | 0.011 | 2/2 | 2 | 14.494 (1.869–112.392) | 0.011 | |
| Late | AA | 50/37 | 13 | 1 | 0.269 | 50/42 | 7 | 1 | 0.336 |
| GA | 5/4 | 17 | 0.530 (0.172–1.635) | 0.269 | 5/3 | 4 | 0.531 (0.146–1.930) | 0.336 | |
| GG | NA | NA | |||||||
1, adjusted by gender; 3, adjusted by age. a, patient numbers may not add up to 100% of available subjects because of missing clinical data. KCNQ1, potassium voltage-gated channel KQT-like subfamily member 1; OS, overall survival; RFS, recurrence-free survival; MST, median survival time; ACT, adjuvant chemotherapy; R/M, recurrence/metastasis.
Discussion
We collected data from 681 GC patients and 756 normal controls, detected 3 functional SNPs in the KCNQ1 gene and 8 functional SNPs in the KCNQ1OT1 gene, and assessed their associations with GC risk and survival in a Chinese population. There were no significant differences to the genotype and allele frequencies between the GC cases and risk in the healthy population; however, in the stratification analyses, the log-additive model indicated that the CT genotype at the KCNQ1 rs231348 locus was significantly associated with a decreased risk of GC in individuals aged ≥55 years. The CC genotype at the KCNQ1OT1 rs231352 locus also decreased the risk of GC in individuals at stages I–II as opposed to stages III–IV. In addition, a stratified analysis by tumor diameter revealed significant associations between the KCNQ1OT1 rs7128926 AA genotype and a decreased risk of GC for individuals with a tumor diameter <5 cm.
Consistent with our research, another recent study showed that KCNQ1 mutant mice are a powerful new tool for investigating the connection between acid balance, Helicobacter infection, and mucin disruption in the progression to GC, and that KCNQ1 mutations predispose mutant mice to metaplastic and pre-neoplastic changes in the stomach (27). In our study, KCNQ1 rs231348 and KCNQ1OT1 rs231352 independently predicted the risk of GC susceptibility under certain patient conditions, including the individual’s age, tumor stage, and tumor diameter. Simultaneously, polymorphism of the rs7128926 and rs7939976 loci of the KCNQ1OT1 gene independently predicted the OS and RFS of GC patients. One possible mechanism for this was that these genes for GC act as multiple DNA transposon-based forward genetic screens in patients (28). Than et al. (17) found that KCNQ1 was a potential tumor suppressor gene, and low expression of KCNQ1 was significantly associated with poor OS in patients with CRC. However, the relationship between KCNQ1 rs231348 and GC has not been reported previously, and this study provides the first report of an association between the KCNQ1 rs231348 polymorphism and the risk of GC susceptibility.
We also identified associations between the KCNQ1OT1 gene polymorphisms, rs7128926 and rs7939976, and OS and RFS, even after adjustment for other clinical factors. These represent novel associations that have not been identified previously. Because the transcription of the KCNQ1OT1 gene overlaps with most of the KCNQ1 transcription unit on the anti-strand, the transcription of KCNQ1 and other non-overlapping genes can be affected by transcriptional interference. In the evaluation of genotype polymorphism and survival analysis, we discovered that as the number of A alleles of the rs7128926 and rs7939976 loci of the KCNQ1OT1 gene increased, the MST with OS and RFS of GC cases showed an increasing trend, and that the AA plus GA genotypes presented a significant survival advantage over the GG genotypes. A further stratified investigation also showed that the AA/GA genotypes of the rs7128926 and rs7939976 polymorphisms improved survival in those with the following characteristics: age over 55, the presence of H. pylori infection, BMI >24, tumor in the non-cardia region with a diameter greater than 5 cm, clinical stage II, and postoperative adjuvant chemotherapy.
The KCNQ1 gene plays an important role in encoding the Q1 subfamily of the voltage-dependent potassium channel, and includes one paternally expressed lncRNA, KCNQ1OT1. The functions of ion channels influence a variety of cellular processes, many of which overlap heavily with these hallmarks of cancer (29). For these reasons, cancer has been described as a channelopathy (30). K+ channels play a major role in the maintenance of plasma membrane (PM) potential (29). With 77 genes coding for K+ channels, they are the largest and most diverse group of ion channels in the human genome. There is increasing evidence that K+ channels are implicated in a variety of cellular and tissue functions, including cell proliferation, differentiation, invasion, migration, and metastasis. KCNQ1 downregulation has also been observed in CRC, where KCNQ1 expression is associated with improved RFS at stages II–IV of the disease (29). Meanwhile, in agreement with our findings, previous studies found a significant association between a common KCNQ1OT1 promoter polymorphism (rs11023840) and the risk of symptomatic prolonged QT interval (31). The Previous study also examined the expression levels of KCNQ1OT1, which showed no significant alterations in early GC tissues compared with normal adjacent tissues by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (32).
The limitations of our study included its relatively small sample size and retrospective nature. Furthermore, the mechanisms whereby the identified SNPs influence the risk of GC susceptibility and GC survival were not elucidated. Further prospective studies with larger sample sizes are needed to clarify the correlations between the KCNQ1 and KCNQ1OT1 genes and GC. More basic studies using GC cells are also needed to identify the molecular mechanisms responsible for the effects of the KCNQ1 and KCNQ1OT1 genes on the susceptibility, risk, and survival of GC patients.
Conclusions
Our study showed that polymorphisms in the KCNQ1 and KCNQ1OT1 genes might have predictive or prognostic value in determining the susceptibility, risk, and survival of Chinese Han patients with GC. However, these results were only demonstrated to be good predictors in a specific population as a result of the heterogeneity of the tumor population and the relatively small sample size included in this study. Furthermore, the association between genetic factors and environmental factors was not fully investigated because of a lack of data regarding traits such as drinking and dietary habits. Further results are expected to clarify the influence and specific mechanisms by which KCNQ1 and KCNQ1OT1 gene polymorphisms affect the susceptibility, risk, and/or survival of GC patients.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors appreciate the contributions of the patients who participated in this study. We thank Catherine Perfect from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The authors state that they have obtained appropriate institutional review board approval (No. K201009-03) and have followed the principles outlined in the Declaration of Helsinki (as revised in 2013) for all human experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Footnotes
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-8052
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-8052
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-8052). The authors have no conflicts of interest to declare.
References
- 1.Wei L, Niu F, Wu J, et al. Association study between genetic polymorphisms in folate metabolism and gastric cancer susceptibility in Chinese Han population: A case-control study. Mol Genet Genomic Med 2019;7:e633. 10.1002/mgg3.633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Ullah MF, Aatif M. The footprints of cancer development: Cancer biomarkers. Cancer Treat Rev 2009;35:193-200. 10.1016/j.ctrv.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 4.Hartgrink HH, Jansen EP, van Grieken NC, et al. Gastric cancer. Lancet 2009;374:477-90. 10.1016/S0140-6736(09)60617-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 2009;285:116-26. 10.1016/j.canlet.2009.04.031 [DOI] [PubMed] [Google Scholar]
- 6.Turdikulova S, Dalimova D, Abdurakhimov A, et al. Association of rs2294008 and rs9297976 Polymorphisms in PSCA Gene with Gastric Cancer Susceptibility in Uzbekistan. Cent Asian J Glob Health 2016;5:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo HD, Lee J, Choi IJ, et al. Dietary flavonoids and gastric cancer risk in a Korean population. Nutrients 2014;6:4961-73. 10.3390/nu6114961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu D, Zheng R, Xin J, et al. Evaluation of GWAS-Identified Genetic Variants for Gastric Cancer Survival. EBioMedicine 2018;33:82-7. 10.1016/j.ebiom.2018.06.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700-13. 10.1158/1055-9965.EPI-13-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Splawski I, Shen J, Timothy KW, et al. Genomic structure of three long QT syndrome genes: KVLQT1, HERG, and KCNE1. Genomics 1998;51:86-97. 10.1006/geno.1998.5361 [DOI] [PubMed] [Google Scholar]
- 11.Chiesa N, De Crescenzo A, Mishra K, et al. The KCNQ1OT1 imprinting control region and non-coding RNA: new properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum Mol Genet 2012;21:10-25. 10.1093/hmg/ddr419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Daghri NM, Alkharfy KM, Alokail MS, et al. Assessing the contribution of 38 genetic loci to the risk of type 2 diabetes in the Saudi Arabian Population. Clin Endocrinol (Oxf) 2014;80:532-7. 10.1111/cen.12187 [DOI] [PubMed] [Google Scholar]
- 13.Peroz D, Rodriguez N, Choveau F, et al. Kv7.1 (KCNQ1) properties and channelopathies. J Physiol 2008;586:1785-9. 10.1113/jphysiol.2007.148254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winbo A, Sandstrom O, Palmqvist R, et al. Iron-deficiency anaemia, gastric hyperplasia, and elevated gastrin levels due to potassium channel dysfunction in the Jervell and Lange-Nielsen Syndrome. Cardiol Young 2013;23:325-34. 10.1017/S1047951112001060 [DOI] [PubMed] [Google Scholar]
- 15.Rice KS, Dickson G, Lane M, et al. Elevated serum gastrin levels in Jervell and Lange-Nielsen syndrome: a marker of severe KCNQ1 dysfunction? Heart Rhythm 2011;8:551-4. 10.1016/j.hrthm.2010.11.039 [DOI] [PubMed] [Google Scholar]
- 16.Asad-Ur-Rahman FN, Hughes L, Talha Khan M, et al. Long QT Syndrome and Duodenal Ampullary Adenoma: A New Association. ACG Case Rep J 2016;3:e163. 10.14309/crj.2016.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Than BL, Goos JA, Sarver AL, et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene 2014;33:3861-8. 10.1038/onc.2013.350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MP, Ravenel JD, Hu RJ, et al. Targeted disruption of the Kvlqt1 gene causes deafness and gastric hyperplasia in mice. J Clin Invest 2000;106:1447-55. 10.1172/JCI10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandey RR, Mondal T, Mohammad F, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008;32:232-46. 10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- 20.Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol 2011;22:343-50. 10.1016/j.semcdb.2011.02.020 [DOI] [PubMed] [Google Scholar]
- 21.Bian Y, Gao G, Zhang Q, et al. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol Ther 2019;20:886-96. 10.1080/15384047.2019.1579959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Wen S, Zhang Y, et al. Identifying potential metastasis-related long non-coding RNAs, microRNAs, and message RNAs in the esophageal squamous cell carcinoma. J Cell Biochem 2019;120:13202-15. 10.1002/jcb.28594 [DOI] [PubMed] [Google Scholar]
- 23.Li C, Miao R, Zhang J, et al. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int J Oncol 2018;52:1603-12. 10.3892/ijo.2018.4313 [DOI] [PubMed] [Google Scholar]
- 24.Andresini O, Rossi MN, Matteini F, et al. The long non-coding RNA Kcnq1ot1 controls maternal p57 expression in muscle cells by promoting H3K27me3 accumulation to an intragenic MyoD-binding region. Epigenetics Chromatin 2019;12:8. 10.1186/s13072-019-0253-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng W, Wang C, Liang C, et al. The Dysregulated Expression of KCNQ1OT1 and Its Interaction with Downstream Factors miR-145/CCNE2 in Breast Cancer Cells. Cell Physiol Biochem 2018;49:432-46. 10.1159/000492978 [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis 2018;9:742. 10.1038/s41419-018-0793-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elso CM, Lu X, Culiat CT, et al. Heightened susceptibility to chronic gastritis, hyperplasia and metaplasia in Kcnq1 mutant mice. Hum Mol Genet 2004;13:2813-21. 10.1093/hmg/ddh307 [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Chen Z, Zhao X, et al. Effects of IGF2BP2, KCNQ1 and GCKR polymorphisms on clinical outcome in metastatic gastric cancer treated with EOF regimen. Pharmacogenomics 2015;16:959-70. 10.2217/pgs.15.49 [DOI] [PubMed] [Google Scholar]
- 29.Anderson KJ, Cormier RT, Scott PM. Role of ion channels in gastrointestinal cancer. World J Gastroenterol 2019;25:5732-72. 10.3748/wjg.v25.i38.5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litan A, Langhans SA. Cancer as a channelopathy: ion channels and pumps in tumor development and progression. Front Cell Neurosci 2015;9:86. 10.3389/fncel.2015.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coto E, Calvo D, Reguero JR, et al. Differential methylation of lncRNA KCNQ1OT1 promoter polymorphism was associated with symptomatic cardiac long QT. Epigenomics 2017;9:1049-57. 10.2217/epi-2017-0024 [DOI] [PubMed] [Google Scholar]
- 32.Lu Q, Yu T, Ou X, et al. Potential lncRNA diagnostic biomarkers for early gastric cancer. Mol Med Rep 2017;16:9545-52. 10.3892/mmr.2017.7770 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as