Abstract
Background
Vascular dementia (VaD) is a degenerative cerebrovascular disease that leads to progressive decline of patients’ cognitive ability and memory. Yizhi Tongmai (YZTM) decoction is an empirical prescription first formulated by Professor Guomin Si. Our previous experiments proved the effectiveness of this prescription in the treatment of VaD. In this study, we aimed to use network pharmacology and molecular docking technology to systematically explain the potential anti-VaD mechanism of YZTM.
Methods
We identified the core compounds of YZTM and their potential targets through the TCMSP, BATMAN, and SwissTargetPrediction databases. Then, we identified the molecular targets of YZTM in VaD using the Online Mendelian Inheritance in Man and GeneCards databases. The common targets of YZTM and VaD were screened out, and then the pathways of these target genes were analyzed using the Database for Annotation, Visualization and Integrated Discovery v6.8. Molecular docking was used to verify the relationship between the core compounds and proteins.
Results
Through network pharmacology analysis, we discovered that the 5 core compounds in YZTM exert an anti-VaD effect. The potential mechanism of YZTM anti-VaD may be through inhibiting the NLRP3 inflammasome, TNF signaling pathway, and toll-like receptor signaling pathways. Subsequently, key compounds were docked with related proteins in the NLRP3 inflammasome (NLRP3, ASC, caspase-1, interleukin-18, and interleukin-1 β) using molecular docking technology. The compounds were found to spontaneously bind to the proteins.
Conclusions
YZTM may exert an anti-VaD effect through inhibition of the NLRP3 inflammasome. In addition, TNF signaling pathway and toll-like receptor signaling pathway may also be its underlying mechanism. The application of network pharmacology and molecular docking technology may provide a novel method for research of Chinese herbal medicine. YZTM may also provide a complementary treatment option for patients with VaD
Keywords: Yizhi Tongmai decoction (YZTM decoction), vascular dementia (VaD), network pharmacology, molecular docking
Introduction
Vascular dementia (VaD) is the second most common type of dementia after Alzheimer’s disease (AD), comprising more than 20% of global dementia cases (1). VaD is a clinical syndrome characterized by a severe decline in cognitive function resulting from the death of hypoxic brain tissue caused by decreased cerebral vascular perfusion (2). Essentially, VaD is caused by a decrease in the brain’s blood supply, which may be related to stroke or its risk factors (3). VaD has received less attention than Alzheimer’s, and despite the suffering, short life expectancy and heavy social burden associated with the disease, no drug therapy has been approved for its treatment (4). Studies have shown that old age, diabetes, hypertension, smoking, and atrial fibrillation are risk factors for VaD (5). VaD can coexist with various cerebrovascular diseases that can affect the cognition of older people, and is characterized by decreased cognitive ability and an increased risk of hippocampal atrophy (6). VaD can be exacerbated by the global or focal effects of vascular diseases. It is also characterized by neurocognitive disorder, as well as behavioral symptoms and motor abnormalities, such as Parkinsonian gait, dysarthria, and autonomic nervous dysfunction (7). The number of known drug targets is not yet large enough to develop a treatment for VaD; therefore, no definite treatment method for VaD exists (8). Although memantine and cholinesterase inhibitors have demonstrated excellent efficacy in the treatment of AD, neither the regulatory authorities nor a steering group have recommended their use in the treatment of VaD. The reasons for this are that their overall effect on VaD is not significant and their use may be accompanied by side effects (9). Therefore, the development of complementary and alternative therapies for VaD is crucial.
For thousands of years, traditional Chinese medicine (TCM) has been widely used to treat numerous diseases. Because many drugs used in TCM are available naturally, they have fewer side effects and cost less (10). Chinese herbal medicine has attracted the attention of dementia researchers because of its rich history as a treatment for the disease (11). According to the theory of TCM, deficiency, phlegm dampness, and stasis are important pathological factors of VaD. Yizhi Tongmai decoction (YZTM) is an effective prescription for the treatment of VaD that was created by Professor Guomin Si. This prescription contains Daemonorops draco Bl. (“XueJie” in Chinese, XJ), Pheretima (“DiLong” in Chinese, DL), Scolopendridae (“WuGong” in Chinese, WG), Panax Ginseng C. A. Mey (“RenShen” in Chinese, RS), Rehm,annia glutinosa Libosch (“DiHuang” in Chinese, DH), Alpinia oxyphylla Miq (“YiZhiRen” in Chinese, YZR), Gastrodia elata Bl. (“TianMa” in Chinese, TM), and Whitmania pigra Whitman (“ShuiZhi” in Chinese, SZ). The compatibility of the prescription accords with the thought of tonifying kidney and activating blood circulation in TCM, and our previous studies showed that YZTM could improve the vascular endothelial growth factor (VEGF) content in ischemic brain tissue, promote angiogenesis in VaD rats (12), and reduce the damage of neuronal nitric oxide (nNOS) neurons in the later stage of ischemia (13). Multidrug compatibility is regarded as the essence of TCM theory (14). However, due to the complex components and numerous targets involved, fully elucidating its mechanism using traditional methods is challenging. Therefore, it is necessary to reveal the potential mechanism of YZTM in the treatment of VaD at the systemic level.
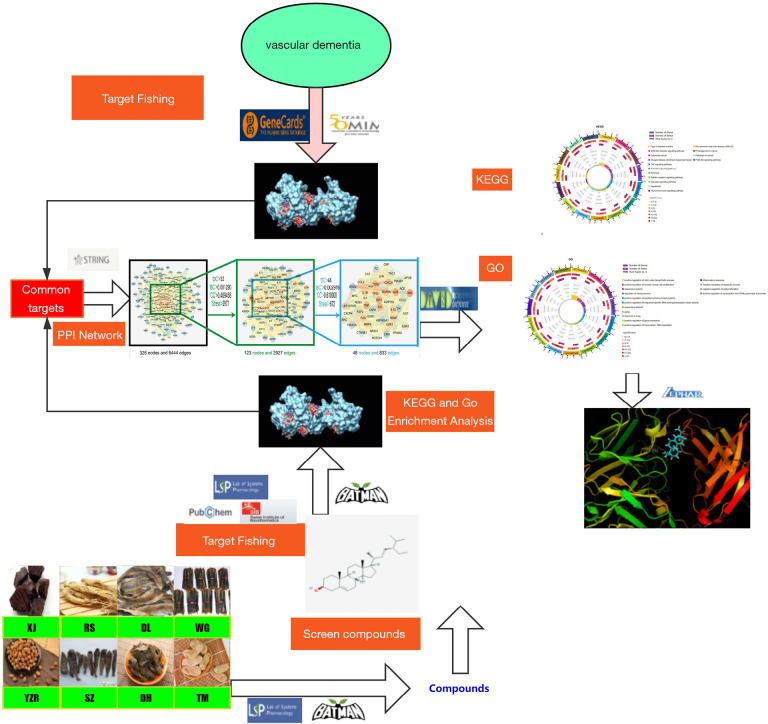
With the continuous innovation and development of systems biology and computer technology, network pharmacology has broad application value in drug target identification, active ingredient discovery, mechanism of action research, preclinical study of efficacy, safety evaluation, and other fields (15). Network pharmacology approaches can be used to study natural drugs with multiple compounds, multiple target genes, and multiple signal pathways, which embodies the holistic view of TCM. In this study, we used network pharmacology and molecular docking to predict the potential mechanism of YZTM in the treatment of VaD (Figure 1).
Figure 1.
The whole framework based on an integration strategy of network pharmacology.
Methods
Extraction of natural compounds in YZTM
The compounds of XJ, DL, WG, RS, DH, YZR, TM, and SZ in YZTM were extracted from the TCM Systems Pharmacology (TCMSP) database and BATMAN-TCM platform, which is one of the largest Chinese herbal medicine databases in the world. The database provides information on the relationships among herbs, natural compounds, target proteins, and diseases (16).
Screening of the active ingredients in YZTM
YZTM contains many natural compounds, but there are few components with pharmacological properties. In the TCMSP database, based on a previously reported model, we screened the various compounds in YZTM according to their pharmacokinetic absorption (A), distribution (D), metabolism (M), and excretion (E) parameters (17). Oral bioavailability (OB) refers to the percentage of an orally administered active substance that reaches the systemic circulation, and is one of most commonly assessed pharmacokinetic properties in drug screening. The OB value was calculated using a powerful in-house system (OBioavail1.1) (18). Drug-likeness (DL) is used to estimate which compounds have similar properties. A database-dependent method based on the Tanimoto coefficient was used to calculate the DL values of the components of YZTM using the formula shown below. In Eq. [1], A represents the molecular characteristic value of a certain Chinese medicine component, and B represents the average value of the molecular characteristic values of all Chinese medicine components in DrugBank (19). Based on previous studies and information obtained from the relevant Chinese herbal medicine database, we selected natural compounds with OB ≥30% and DL ≥0.18 for further analysis (20).
| [1] |
In the BATMAN-TCM platform, in order to collect candidate compounds, we set the potential TAR score threshold ≥20 and the P value threshold to <0.05.
Prediction of the target proteins of the selected compounds
All the active ingredients were input into the TCMSP database to obtain their known targets, and the 2-dimensional (2D) structure of the compounds was obtained from PubChem and imported into the SwissTargetPrediction database. For more accurate prediction of the target gene of each natural compound, relevant parameters were set (probability ≥0.6) (21).
Search for targets of YZTM in VaD
Two resources were used to obtain the target genes of YZTM in VaD: (I) online Mendelian Inheritance in Man (OMIM) database; (II) the GeneCards human gene database. We used as many databases as possible to ensure the credibility of the obtained targets. The official names of the genes about YZTM and VaD were obtained using UniProt, and the species was selected as “Homo sapiens”. Then, the names of the target genes were uniformly converted into gene and UniProt IDs (22). Finally, we mapped the target genes of the active constituents of YZTM and the VaD-related target genes, and then screened the common target genes to determine the potential targets of YZTM in VaD. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Construction of the protein–protein interaction network
Using the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database, and the STRING database is a database of known and predicted PPIs. We constructed a protein-protein interaction (PPI) network of the potential target genes of YZTM in VaD. Then put it into Cytoscape software. We used the “Analysis Network” tool in Cytoscape to get protein interaction network to obtain relevant parameters. Based on the four parameters of “degree (DC)”, “betweenness centrality (BC)”, “closeness centrality (CC)”, and “stress”, we do topology analysis on the PPI network to get HUB genes.
Enrichment analysis
To identify the biological processes and signaling pathways in which the main hub target genes are involved, Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8, a high-throughput functional labeling bioinformatics network, was used for pathway enrichment analysis. The target genes of YZTM in VaD were input into DAVID v6.8 for Gene Ontology (GO) biological process analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. GO biological processes with P≤0.01 and KEGG pathways with P≤0.01 were considered to be significantly enriched.
Construction of a network of herbs, natural compounds, and targets
On the basis of the natural compounds, target genes, and pathways of YZTM identified in the treatment of VaD, a network model of herbs, ingredients, targets, and diseases was constructed using Cytoscape 3.7.1 software, and the relationships among them were analyzed. We constructed the following: (I) a herbal-compound-target network diagram; (II) a PPI network diagram of the potential targets of YZTM in VaD; and (III)PPI topology analysis diagram. Herbs, compounds, and targets were represented in the network by nodes, and interactions between them were represented by edges linking the nodes.
Molecular docking
Using the LeDock software, we simulated the receptor ligand docking of the core targets in KEGG pathways to the core compounds in the YZTM. A binding energy of less than 0 indicates that the conformation of ligand and receptor molecules can be stably bound; the larger the absolute value, the stronger the stability. A binding energy of >0 indicates that the conformation of ligand and receptor molecules cannot be stably bound; the higher the absolute value, the more unstable the conformation (23).
Statistical analysis
The pathway information about the selected targets was retrieved from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The topological data were analyzed by Cytoscape version 3.7.2. for Windows, WPS Office version 11.1.0.9739-Release for Windows. Pathway enrichment analysis was run by Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8. All the relevant data analysis results are presented in “Results” Section below.
Results
The natural active ingredients in YZTM
Using the databases and criteria (OB, ≥30%; DL, ≥0.18 and TAR score, ≥20; P value to <0.05) described above, we screened out effective natural compounds of YZTM. We obtained 6, 2, 5, 19, 2, 18, 13, and 13 natural compounds of XJ, DL, WG, RS, DH, YZR, TM, and SZ, respectively.
Targets of the effective compounds of YZTM
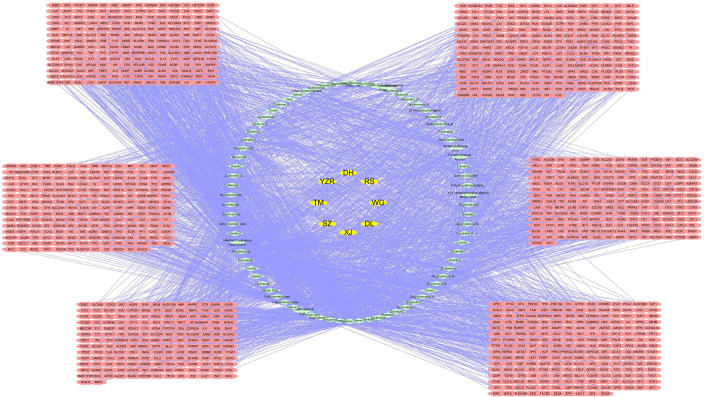
To predict the target genes of compounds in YZTM, we used the TCMSP, BATMAN and SwissTargetPrediction databases. In the TCM prescription in this study 303, 2, 215, 132, 45, 768, 216, and 342 target proteins were predicted for XJ, DL, WG, RS, DH, YZR, TM, and SZ, respectively. According to the above data, we established a herbal-compound-target network, comprising 1,312 nodes and 3,322 edges (Figure 2). The analysis network tool in Cytoscape was used to obtain the “Degree” parameter of the network. The results revealed that the top 5 compounds according to the degree value were Neopetasone, Abietic Acid, Pimaric Acid, Isopimaric Acid, and Sandaracopimaric Acid (Table 1). These compounds may be the core compounds of YZTM and thus, the material basis of YZTM’s anti-VaD effect. And, this diagram allowed the relationships among herbs, ingredients, and targets to be easily observed, and revealed the potential pharmacological effects of YZTM. The network graph results showed consistency with the multi-target effects of TCM and the synergistic effect of multidrug compatibility.
Figure 2.
The “herbal-compound-target” network diagram of YZTM in the treatment of VaD. Yellow represents herbals. Green represents the compounds. Pink represents the targets. XJ (Daemonorops draco Bl.), DL (Pheretima), WG (Scolopendridae), RS (Panax Ginseng C. A. Mey), DH (Rehm,annia glutinosa Libosch), YZR (Alpinia oxyphylla Miq), TM (Gastrodia elata Bl.), and SZ (Whitmania pigra Whitman). YZTM, Yizhi Tongmai.
Table 1. Compound information sheet.
| Molecule ID | Structure | Herb |
|---|---|---|
| Abietic Acid |
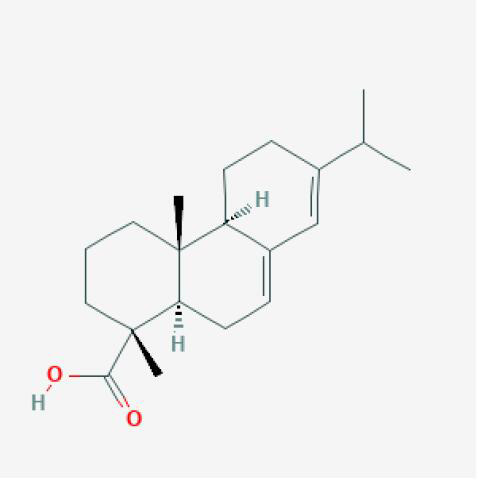
|
XJ |
| Isopimaric Acid |
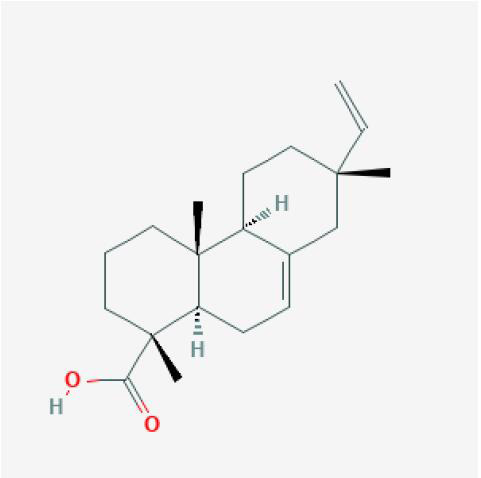
|
XJ |
| Neopetasone |
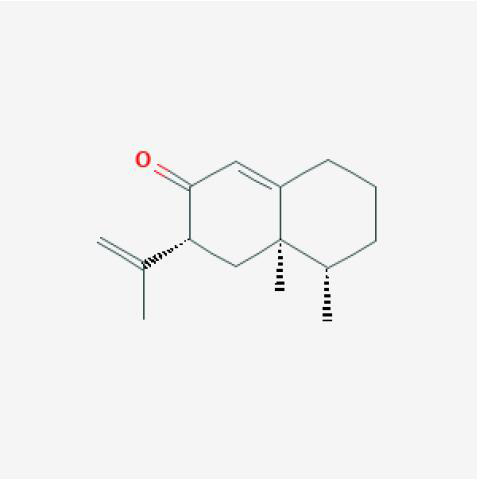
|
YZR |
| Pimaric Acid |
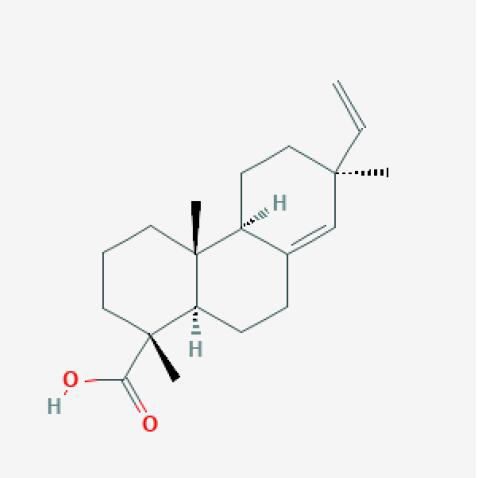
|
XJ |
| Sandaracopimaric Acid |
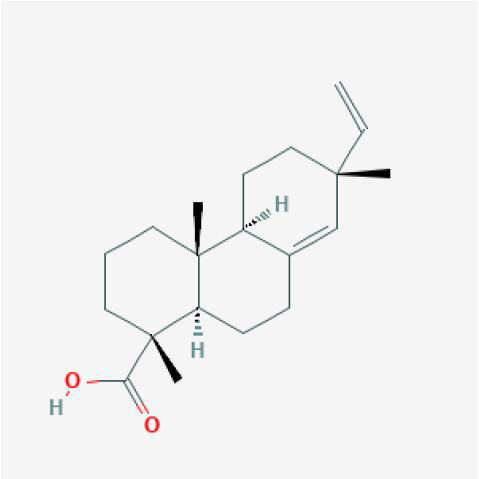
|
XJ |
XJ (Daemonorops draco Bl.); YZR (Alpinia oxyphylla Miq).
PPI network amplification and analysis
We integrated data on VaD-related target genes from the OMIM and GeneCards databases. After screening using the relevant parameters and the removal of duplicate values, 1,337 VaD-related target genes were obtained.
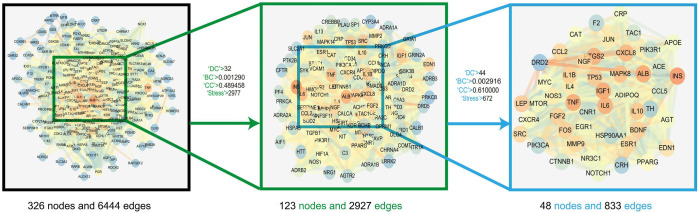
The target genes of YZTM were mapped to the VaD-related target genes, and 327 common target genes were obtained, representing the potential targets of YZTM in VaD (Figure 3). After inputting these common target genes into the STRING database and we obtained a PPI network comprising 326 nodes and 6,444 edges, depicting the biological processes of YZTM treatment of VaD in vivo. Then, we use the “Analysis network” tool in Cytoscape to get the protein interaction network to obtain the relevant parameters, and based on the four parameters of “DC”, “BC”, “CC” and “Stress”, the index above the median value is selected as the key index (24), and the HUB genes of YZTM acting on VaD is obtained (Figure 4). The threshold value of the first screening is “DC” ≥32, “CC” ≥0.489458, “BC” ≥0.001290, and “Stress” ≥2,977. Then through the topological analysis data screening again, 85 hub targets are obtained. The criteria for screening were “DC” ≥44, “CC” ≥0.002916, “BC” ≥0.610000, and “Stress” ≥672. We got 85 HUB nodes and established the related network diagram. The network diagram was composed of 48 nodes and 833 edges (Table 2).
Figure 3.
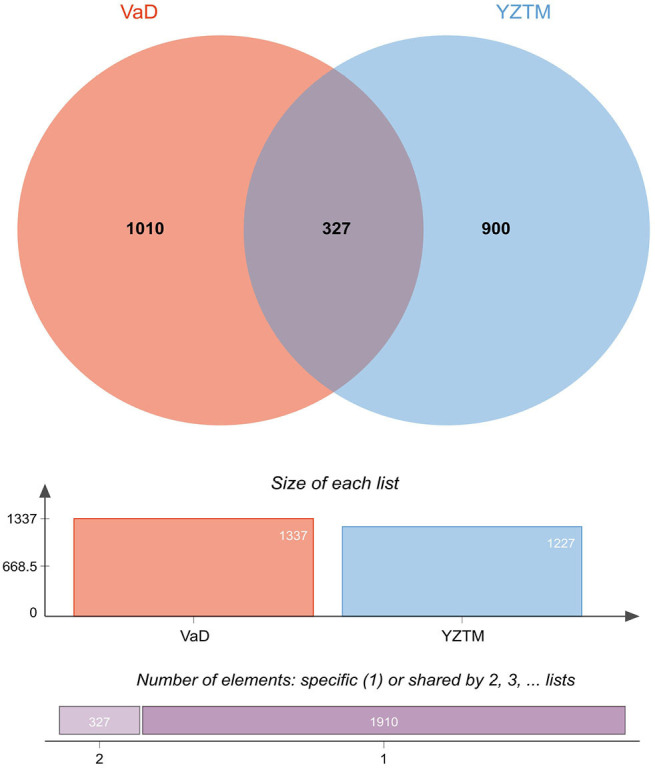
Venn diagram summarizing differentially targets of YZTM and VaD. YZTM (Yizhi Tongmai decoction); VaD (Vascular dementia). YZTM, Yizhi Tongmai.
Figure 4.
The process of topological screening for the protein-protein interaction network.
Table 2. Information on HUB targets.
| Gene | Description | UniProt | Degree |
|---|---|---|---|
| INS | Insulin | P01308 | 47 |
| ALB | Serum albumin | P02768 | 45 |
| TNF | Tumor necrosis factor | P01375 | 45 |
| IGF1 | Insulin-like growth factor I | P05019 | 45 |
| IL6 | Interleukin-6 | P05231 | 44 |
| PTGS2 | Prostaglandin G/H synthase 2 | P35354 | 44 |
| CXCL8 | Interleukin-8 | P10145 | 43 |
| MAPK8 | Mitogen-activated protein kinase 8 | P45983 | 42 |
| TP53 | Cellular tumor antigen p53 | P04637 | 41 |
| LEP | Leptin | P41159 | 41 |
| IL10 | Interleukin-10 | P22301 | 41 |
| BDNF | Brain-derived neurotrophic factor | P23560 | 40 |
| FOS | Proto-oncogene c-Fos | P01100 | 40 |
| FGF2 | Fibroblast growth factor 2 | P09038 | 40 |
| IL1B | Interleukin-1 beta | P01584 | 40 |
| NGF | Beta-nerve growth factor | P01138 | 39 |
| SRC | Proto-oncogene tyrosine-protein kinase Src | P12931 | 39 |
| MMP9 | Matrix metalloproteinase-9 | P14780 | 39 |
| ESR1 | Estrogen receptor | P03372 | 39 |
| CCL2 | C-C motif chemokine 2 | P13500 | 39 |
| EDN1 | Endothelin-1 | P05305 | 38 |
| JUN | Transcription factor AP-1 | P05412 | 37 |
| IL4 | Interleukin-4 | P05112 | 37 |
| NOS3 | Nitric oxide synthase, endothelial | P29474 | 37 |
| CXCR4 | C-X-C chemokine receptor type 4 | P61073 | 35 |
| PPARG | Peroxisome proliferator-activated receptor gamma | P37231 | 35 |
| MYC | Myc proto-oncogene protein | P01106 | 34 |
| CAT | Catalas | P04040 | 34 |
| EGR1 | Early growth response protein 1 | P18146 | 33 |
| MTOR | Serine/threonine-protein kinase mTOR | P42345 | 33 |
| NR3C1 | Glucocorticoid receptor | P04150 | 33 |
| AGT | Angiotensinogen | P01019 | 33 |
| CRP | C-reactive protein | P02741 | 33 |
| APOE | Apolipoprotein E | P02649 | 33 |
| NOTCH1 | Neurogenic locus notch homolog protein 1 | P46531 | 32 |
| ADIPOQ | Adiponectin | Q15848 | 32 |
| CCL5 | C-C motif chemokine 5 | P13501 | 27 |
| CTNNB1 | Catenin beta-1 | P35222 | 27 |
| TAC1 | Protachykinin-1 | P20366 | 27 |
| ACE | Angiotensin-converting enzyme | P12821 | 27 |
| PIK3R1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | P27986 | 25 |
| CNR1 | Cannabinoid receptor 1 | P21554 | 25 |
| HSP90AA1 | Heat shock protein HSP 90-alpha | P07900 | 23 |
| PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | P42336 | 23 |
| F2 | Prothrombin | P00734 | 23 |
| TH | Tyrosine 3-monooxygenase | P07101 | 20 |
| CRH | Corticoliberin | P06850 | 20 |
| DRD2 | D(2) dopamine receptor | P14416 | 17 |
KEGG pathway and GO biological process enrichment analysis
After inputting HUB targets into DAVID v6.8, we identified 75 KEGG pathways and 206 GO biological processes that met the enrichment criteria of P<0.01.
KEGG pathway analysis
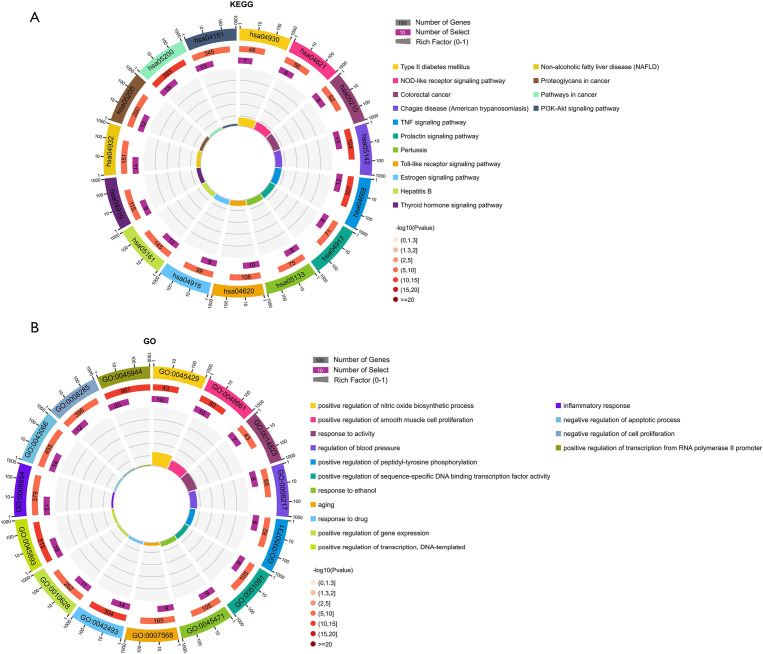
The top 15 most significantly enriched KEGG pathways were selected for analysis (Figure 5A). The potential target genes of YZTM in VaD were mainly enriched in the the nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway (hsa04621), tumor necrosis factor (TNF) signaling pathway (hsa04668), and the toll-like receptor signaling pathway (hsa04620). This finding suggests that YZTM may exert its effect against VaD by regulating the inflammatory response. Because the NOD-like receptor signaling pathway was the potential signal pathway, we speculated that in the treatment of VaD, YZTM may regulate the activity of the NLRP3 (NOD like receptor-3) inflammasome. The NLRP3 inflammasome, which consists of NLRP3, caspase-1, and apoptosis-related spot-like protein (ASC), helps to identify special microorganisms and dangerous substances. It is also the functional platform for caspase-1 activation, and is responsible for the maturation of interleukin (IL)-Iβ and IL-18 (25).
Figure 5.
KEGG and GO enrichment. (A) KEGG enrichment; (B) GO biological processes enrichment. KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Gene Ontology.
GO analysis
The top 15 most significantly enriched GO biological processes were selected for analysis (Figure 5B). The potential target genes of YZTM in VaD were mainly involved in inflammation, cell proliferation, and apoptosis.
Molecular docking
Molecular docking of NLRP3, ASC, caspase-1, IL-18, and IL-1 β to the core compounds in the first 5 positions was carried out using LeDock. The results showed that the binding energy of all molecules to proteins was less than 0, which indicated that ligands could spontaneously bind to receptors (Figure 6).
Figure 6.
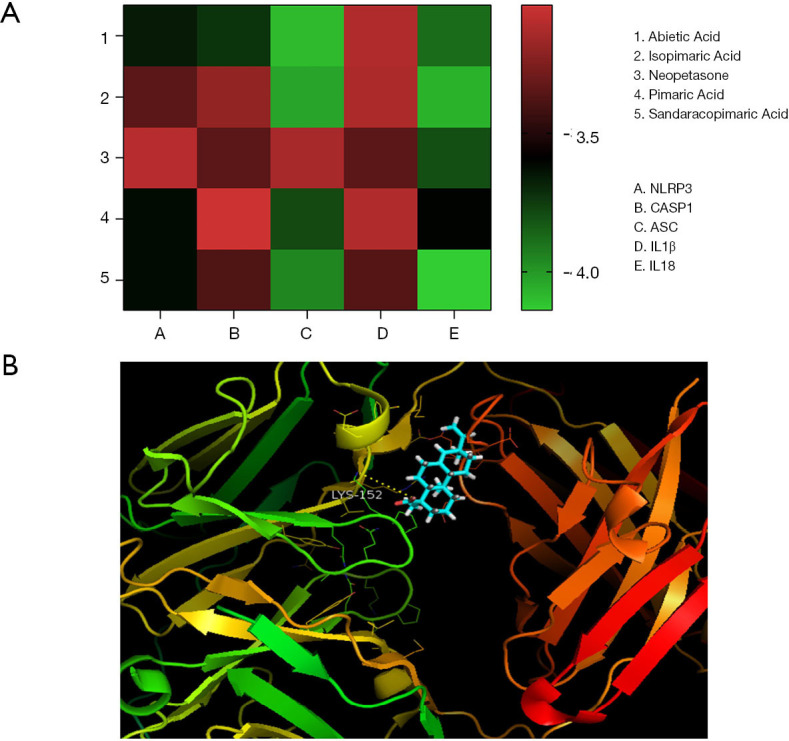
Molecular docking. (A) The five core ingredients and the NLRP3 inflammasome associated protein molecular mocking; (B) molecular docking model of Sandaracopimaric Acid and IL18.
Discussion
VaD is a complex brain disease caused by a decrease in blood supply to the brain and subsequent cognitive dysfunction (26). Studies have shown that VaD is more challenging to treat effectively than AD. At present, there is no specific drug for VaD, so the development of complementary and alternative therapies for this disease is extremely important. However, illuminating the complex mechanisms of YZTM in the treatment of VaD using traditional methods is challenging. Therefore, the integration of network pharmacology based on big data bioinformatics into the study of the molecular mechanisms of TCM in the treatment of diseases is of great significance. The results of the present study show that YZTM may inhibit the NLRP3 inflammasome to exert an effect against VaD.
From the network diagram of herbs, natural compounds, and targets, we identified 5 core compounds of YZTM: Neopetasone, Abietic Acid, Pimaric Acid, Isopimaric Acid, and Sandaracopimaric Acid. These compounds may have potential therapeutic effects on VaD. Abietic acid is an abietane diterpenoid compound mainly derived from Pimenta racemosa var. grissea (27), and it has strong antioxidant activity, anti-hypercholesterolemia, mucosal vascular protection and antitumor activity (28). Sandaracopimaric Acid has interesting biological activities, including cytotoxicity and anti-inflammatory effects (29). These compounds form the material basis of the mechanism of action of YZTM on VaD, and further research is necessary.
The mapped PPI network was amplified and subjected to enrichment analysis. We found that YZTM mainly exerted its anti-VaD effect via the NOD-like receptor signaling pathway (hsa04621), the TNF signaling pathway (hsa04668), and the toll-like receptor signaling pathway (hsa04620). Then, we performed GO biological process enrichment analysis, which showed that YZTM’s anti-VaD effect may be achieved via an inflammatory pathway. We molecularly docked the NLRP3 inflammasome-related proteins NLRP3, caspase-1, and IL-1β to the NOD-like receptor signaling pathway (hsa04621). We found that the inflammatory pathway of YZTM against VaD may be achieved through the NLRP3 inflammasome.
The role of inflammatory mechanism in the pathogenesis of VaD has been paid more and more attention. The current research tends to use drugs to inhibit the inflammatory reaction, so as to protect neurons, and then improve the symptoms of VaD and prevent the occurrence of VaD. NLRP3 is a member of the NOD-like receptor protein family, which is expressed in the cytoplasm. When PAMP or DAMP are activated, NLRP3 converts the adaptor molecule of ASC, which contains a caspase recruitment domain) into an oligomer, and forms a large inflammatory plaque in the cell (30,31). Pro-caspase-1 is also recruited into the activated complex. Active caspase-1 then processes pro-IL-1β and pro-IL-18 into their mature forms, which can be rapidly secreted from cells (32). Once formed, these can also enter into the extracellular space and further exacerbate inflammatory reactions (33,34). IL-1 is a major inflammatory mediator that is involved in a variety of vascular, metabolic and neurological diseases. IL-1 family cytokines are key mediators of inflammation. Proinflammatory IL-1a and IL-1β participate in both the central and systemic inflammatory mechanisms. IL-18 is another inflammatory cytokine, with involvement in acute and chronic inflammatory conditions and common brain diseases (35). Studies have shown that when the NLRP3 inflammasome is activated, pro-caspase-1 is converted into caspase-1, and pro-IL-1β is cleaved and IL-1β is secreted (36). The mechanism of IL-1β release after inflammasome activation is unclear, although it is known that cleavage of gasdermin D is a requirement. Gasdermin D is cleaved by caspase-1, which leads to plasma membrane instability and promotes the release of IL-1β (37). It is a receptor of the innate immune system response against pathogens. NLRP3 is a key receptor that can sense tissue injury and plays an important role in the activation of aseptic inflammation. Its maladjustment promotes the progression of many inflammatory, metabolic, degenerative and age-related diseases (38). Recent studies have shown that inflammatory corpuscles are involved in a broad range of pathophysiological processes in the brain as well as in chronic diseases, which are risk factors for neurodegenerative diseases or cerebrovascular diseases (37). In cerebral ischemia, the nlrp1 and NLRP3 inflammasome complexes can be formed in numerous ways, including through energy depletion, acidification, cathepsin release, decrease of intracellular K+ concentration, increase of reactive oxygen species production, mitochondrial DNA oxidation, increase of intracellular Ca2+ concentration, cell swelling, and activation of protein kinase R (PKR) (39). Studies have shown that necrotic cells in ischemia-reperfusion injury can activate NLRP3 and inhibiting the activity of the NLRP3 inflammasome can reduce acute inflammation, injury, and tissue injury (40). Moreover, NLRP3-deficient mice showed reduced neurovascular injury and prognosis after stroke, suggesting that inflammatory bodies may play an important role in poor prognosis in patients with stroke (41). The decrease of cognitive function and memory function in VaD model rats is related to the increase of Amyloid-beta (Aβ) and microtubule-associated protein tau (Tau protein) expression. Aβ and tau protein interact and participate in the damage of hippocampal neurons after ischemia, thus participating in the occurrence and development of VaD. Aβ can stimulate the excessive production of pro-inflammatory cytokines [including tumor necrosis factor (TNF)-α] in monocytes and microglia (42). In addition, Aβ can activate the inflammatory body of NLRP3 and promote the secretion of IL-1β. Inhibition of NLRP3 can reduce memory loss and Aβ deposition (43). NF-κB not only induces the production of a large number of inflammatory chemokines, cytokines, and cytokine precursors (including pro-il-1β), but it also activates NLRP3. Therefore, it is extremely important for the initiation and assembly of inflammatory bodies (44). Notably, NF-κB also has an anti-inflammatory function. It prevents premature and excessive activation of the NLRP3 inflammasome and inhibition of neutrophil protease activity by macrophages (38). The NF-κB pathway was shown to induce the release of proinflammatory cytokines and inflammatory response in VaD rats (45). Based on the above description, we also speculated that YZTM may play a role in the treatment of VaD via the NF-κB pathway. Toll-like receptors are involved in the progression of ischemic brain injury, and thus participate in the development of VaD (46). They can activate NF-κB signaling under the action of extracellular stimulators, thus increasing the transcription level of NLRP3 (47).
Conclusions
In this study, we combined network pharmacology with molecular docking computational biology methods to explore the mechanism underlying the effect of YZTM on VaD. The results indicated that YZTM may exert its anti-VaD effect via the NLRP3 inflammasome. In addition, TNF signaling pathway and toll-like receptor signaling pathway may also be its underlying mechanism. We expected that computer biology will provide a method for the modern research of TCM, and also hope to promote YZTM to become a complementary and alternative treatment method for VaD or vascular cognitive impairment.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This project is funded by the National Natural Science Foundation of China (no. 81473631).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-8165). The authors have no conflicts of interest to declare.
References
- 1.Wolters FJ, Ikram MA. Epidemiology of Vascular Dementia. Arterioscler Thromb Vasc Biol 2019;39:1542-9. 10.1161/ATVBAHA.119.311908 [DOI] [PubMed] [Google Scholar]
- 2.Chan ES, Bautista DT, Zhu Y, et al. Traditional Chinese herbal medicine for vascular dementia. Cochrane Database Syst Rev 2018;12:CD010284. 10.1002/14651858.CD010284.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venkat P, Chopp M, Chen J. Models and mechanisms of vascular dementia. Exp Neurol 2015;272:97-108. 10.1016/j.expneurol.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagunin AA, Ivanov SM, Gloriozova TA, et al. Combined network pharmacology and virtual reverse pharmacology approaches for identification of potential targets to treat vascular dementia. Sci Rep 2020;10:257. 10.1038/s41598-019-57199-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang ZY, Liu Z, Deng HH, et al. Effects of acupuncture on vascular dementia (VD) animal models: a systematic review and meta-analysis. BMC Complement Altern Med 2018;18:302. 10.1186/s12906-018-2345-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorelick PB, Scuteri A, Black SE, et al. Vascular contributions to cognitive impairment and dementia: a statement for healthcare professionals from the american heart association/american stroke association. Stroke 2011;42:2672-713. 10.1161/STR.0b013e3182299496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalaria RN. The pathology and pathophysiology of vascular dementia. Neuropharmacology 2018;134:226-39. 10.1016/j.neuropharm.2017.12.030 [DOI] [PubMed] [Google Scholar]
- 8.Kwon CY, Lee B, Suh HW, et al. Efficacy and Safety of Auricular Acupuncture for Cognitive Impairment and Dementia: A Systematic Review. Evid Based Complement Alternat Med 2018;2018:3426078. 10.1155/2018/3426078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Román GC. Vascular dementia may be the most common form of dementia in the elderly. J Neurol Sci 2002;203-204:7-10. 10.1016/S0022-510X(02)00252-6 [DOI] [PubMed] [Google Scholar]
- 10.Zhu N, Hou J, Ma G, et al. Network Pharmacology Identifies the Mechanisms of Action of Shaoyao Gancao Decoction in the Treatment of Osteoarthritis. Med Sci Monit 2019;25:6051-73. 10.12659/MSM.915821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Liu N, Wei Y, et al. Efficacy and safety of Shenmayizhi decoction as an adjuvant treatment for vascular dementia: Study protocol for a randomized controlled trial. Medicine (Baltimore) 2019;98:e18326. 10.1097/MD.0000000000018326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma H, Si G, Sun P. The effect of Yizhi Tongmai Granule on the expression of VEGF in brain tissue of vascular dementia rats. Chinese Medical Emergency 2009;18:80 + 98
- 13.Wang J, Si G, Ma H. The effect of Yizhi Tongmai Granule on the expression of nNOS in brain tissue of vascular dementia rats. Journal of Shandong University of traditional Chinese Medicine 2008;6:510-3 [Google Scholar]
- 14.Gill D, Georgakis MK, Koskeridis F, et al. Use of Genetic Variants Related to Antihypertensive Drugs to Inform on Efficacy and Side Effects. Circulation 2019;140:270-9. 10.1161/CIRCULATIONAHA.118.038814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Zou J, Jia Y, et al. A Study on the Mechanism of Lavender in the Treatment of Insomnia Based on Network Pharmacology. Comb Chem High Throughput Screen 2020;23:419-32. 10.2174/1386207323666200401095008 [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci Rep 2016;6:21146. 10.1038/srep21146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gan D, Xu X, Chen D, et al. Network Pharmacology-Based Pharmacological Mechanism of the Chinese Medicine Rhizoma drynariae Against Osteoporosis. Med Sci Monit 2019;25:5700-16. 10.12659/MSM.915170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012;13:6964-82. 10.3390/ijms13066964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:D1074-82. 10.1093/nar/gkx1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li K, Li H, Xu W, et al. Research on the Potential Mechanism of Gypenosides on Treating Thyroid-Associated Ophthalmopathy Based on Network Pharmacology. Med Sci Monit 2019;25:4923-32. 10.12659/MSM.917299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang SY, Zhao J, Lu Y, et al. Network pharmacology-based identification for therapeutic mechanism of Ling-Gui-Zhu-Gan decoction in the metabolic syndrome induced by antipsychotic drugs. Comput Biol Med 2019;110:1-7. 10.1016/j.compbiomed.2019.05.007 [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Liu T, Yang L, et al. Study on the multi-targets mechanism of triphala on cardio-cerebral vascular diseases based on network pharmacology. Biomed Pharmacother 2019;116:108994. 10.1016/j.biopha.2019.108994 [DOI] [PubMed] [Google Scholar]
- 23.Zhou F, He K, Guan Y, et al. Network pharmacology-based strategy to investigate pharmacological mechanisms of Tinospora sinensis for treatment of Alzheimer's disease. J Ethnopharmacol 2020;259:112940. 10.1016/j.jep.2020.112940 [DOI] [PubMed] [Google Scholar]
- 24.Wan Y, Xu L, Liu Z, et al. Utilising network pharmacology to explore the underlying mechanism of Wumei Pill in treating pancreatic neoplasms. BMC Complement Altern Med 2019;19:158. 10.1186/s12906-019-2580-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 2002;10:417-26. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- 26.Xia D, Sui R, Min L, et al. Fastigial nucleus stimulation ameliorates cognitive impairment via modulating autophagy and inflammasomes activation in a rat model of vascular dementia. J Cell Biochem 2019;120:5108-17. 10.1002/jcb.27787 [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Chen W, Liu Q, et al. Abietic acid suppresses non-small-cell lung cancer cell growth via blocking IKKβ/NF-κB signaling. Onco Targets Ther 2019;12:4825-37. 10.2147/OTT.S199161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang S, Zhang J, Yuan Y. Abietic acid attenuates IL-1β-induced inflammation in human osteoarthritis chondrocytes. Int Immunopharmacol 2018;64:110-5. 10.1016/j.intimp.2018.07.014 [DOI] [PubMed] [Google Scholar]
- 29.Takei M, Umeyama A, Shoji N, et al. Diterpenes drive Th1 polarization depending on IL-12. Int Immunopharmacol 2008;8:1602-8. 10.1016/j.intimp.2008.07.003 [DOI] [PubMed] [Google Scholar]
- 30.Cai X, Chen J, Xu H, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014;156:1207-22. 10.1016/j.cell.2014.01.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu A, Magupalli VG, Ruan J, et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 2014;156:1193-206. 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez-Castejon G, Brough D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev 2011;22:189-95. 10.1016/j.cytogfr.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baroja-Mazo A, Martín-Sánchez F, Gomez AI, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol 2014;15:738-48. 10.1038/ni.2919 [DOI] [PubMed] [Google Scholar]
- 34.Franklin BS, Bossaller L, De Nardo D, et al. The adaptor ASC has extracellular and 'prionoid' activities that propagate inflammation. Nat Immunol 2014;15:727-37. 10.1038/ni.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gracie JA, Robertson SE, McInnes IB. Interleukin-18. J Leukoc Biol 2003;73:213-24. 10.1189/jlb.0602313 [DOI] [PubMed] [Google Scholar]
- 36.Zhou R, Tardivel A, Thorens B, et al. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol 2010;11:136-40. 10.1038/ni.1831 [DOI] [PubMed] [Google Scholar]
- 37.Lénárt N, Brough D, Dénes Á. Inflammasomes link vascular disease with neuroinflammation and brain disorders. J Cereb Blood Flow Metab 2016;36:1668-85. 10.1177/0271678X16662043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong Z, Umemura A, Sanchez-Lopez E, et al. NF-κB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell 2016;164:896-910. 10.1016/j.cell.2015.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fann DY, Lee SY, Manzanero S, et al. Pathogenesis of acute stroke and the role of inflammasomes. Ageing Res Rev 2013;12:941-66. 10.1016/j.arr.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 40.Iyer SS, Pulskens WP, Sadler JJ, et al. Necrotic cells trigger a sterile inflammatory response through the Nlrp3 inflammasome. Proc Natl Acad Sci U S A 2009;106:20388-93. 10.1073/pnas.0908698106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward R, Li W, Abdul Y, et al. NLRP3 inflammasome inhibition with MCC950 improves diabetes-mediated cognitive impairment and vasoneuronal remodeling after ischemia. Pharmacol Res 2019;142:237-50. 10.1016/j.phrs.2019.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HJ, Yang SJ. Supplementation with Nicotinamide Riboside Reduces Brain Inflammation and Improves Cognitive Function in Diabetic Mice. Int J Mol Sci 2019;20:4196. 10.3390/ijms20174196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan MS, Yu JT, Jiang T, et al. The NLRP3 inflammasome in Alzheimer's disease. Mol Neurobiol 2013;48:875-82. 10.1007/s12035-013-8475-x [DOI] [PubMed] [Google Scholar]
- 44.Schroder K, Tschopp J. The inflammasomes. Cell 2010;140:821-32. 10.1016/j.cell.2010.01.040 [DOI] [PubMed] [Google Scholar]
- 45.Sun M, Shen X, Ma Y. Rehmannioside A attenuates cognitive deficits in rats with vascular dementia (VD) through suppressing oxidative stress, inflammation and apoptosis. Biomed Pharmacother 2019;120:109492. 10.1016/j.biopha.2019.109492 [DOI] [PubMed] [Google Scholar]
- 46.Wang R, Wang ST, Wang YD, et al. Stress-responsive heme oxygenase-1 isoenzyme participates in Toll-like receptor 4-induced inflammation during brain ischemia. Neuroreport 2016;27:445-54. 10.1097/WNR.0000000000000561 [DOI] [PubMed] [Google Scholar]
- 47.Sutterwala FS, Haasken S, Cassel SL. Mechanism of NLRP3 inflammasome activation. Ann N Y Acad Sci 2014;1319:82-95. 10.1111/nyas.12458 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as