Abstract
Summary
Large-scale data analysis in bioinformatics requires pipelined execution of multiple software. Generally each stage in a pipeline takes considerable computing resources and several workflow management systems (WMS), e.g. Snakemake, Nextflow, Common Workflow Language, Galaxy, etc. have been developed to ensure optimum execution of the stages across two invocations of the pipeline. However, when the pipeline needs to be executed with different settings of parameters, e.g. thresholds, underlying algorithms, etc. these WMS require significant scripting to ensure an optimal execution. We developed JUDI on top of DoIt, a Python based WMS, to systematically handle parameter settings based on the principles of database management systems. Using a novel modular approach that encapsulates a parameter database in each task and file associated with a pipeline stage, JUDI simplifies plug-and-play of the pipeline stages. For a typical pipeline with n parameters, JUDI reduces the number of lines of scripting required by a factor of O(n). With properly designed parameter databases, JUDI not only enables reproducing research under published values of parameters but also facilitates exploring newer results under novel parameter settings.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Large scale data analysis in bioinformatics requires many software to be executed in a pipeline where the output of an upstream stage is fed as an input to a downstream stage. Generally each stage takes a considerable amount of computing resources such as CPU time and memory. If the pipeline needs to be executed multiple times, may be due to input or parameter variations, it is desired that only the bare minimum stages that are affected by the variations are re-executed. To address such a requirement, many workflow management systems (WMS) have been developed. GNU Make (Stallman et al., 2004) is one of the earliest and widely used WMS where each pipeline stage is specified as a rule of commands that generate the output(s) from the input(s). Make automatically figures out the interdependency among the rules and subsequently, by using a directed acyclic graph (DAG) of the rules, executes the commands in a suitable order such that when a command is executed all inputs are already available. Moreover, if all inputs are unchanged, make skips re-execution of the command and saves computing resources.
Make has a steep learning curve, specifically for the biologists. Several newer WMS have been developed to specify Make rules in simpler languages, e.g. Snakemake (Köster and Rahmann, 2012) uses a simplified Python, Nextflow (Di Tommaso et al., 2017) uses Groovy, Common Workflow Language (Amstutz et al., 2016) uses JavaScript to hierarchically specify rules and files and so on. These WMS can also schedule the pipeline tasks efficiently on multiprocessor environments by exploiting the DAG of rules.
Several WMS to address the specific needs of scientific communities have also been developed, e.g. Deelman et al. (2005) developed one of the early NSF funded tool to build scientific software, Ramachandran (2018) developed for numerical computing, Wolstencroft et al. (2013) for web services, Stropp et al. (2012) for microarray data analysis, Shah et al. (2004) for processing biological sequences and so on. To ease building of workflows, further enhancements have also been developed, e.g. Pradal et al. (2008) provides a graphical interface for plant modelling, Freire et al. (2006) helps in managing rapidly evolving scientific workflows, Blankenberg et al. (2014) accelerates dissemination of workflows built with Galaxy and so on. Efforts have been made to address reproducibility of scientific results through containerized workflow builder (Di Tommaso et al., 2017; Freire et al., 2012; Köster and Rahmann, 2012). References to many such WMS available in the literature have been provided in the review articles (Cingolani et al., 2015; Cohen-Boulakia et al., 2017; Leipzig, 2017).
However, these WMS require significant effort in scripting to ensure optimal execution of the stages when the pipeline needs to be executed under different settings of parameters such as thresholds, algorithmic methods and so on. Generally these changes affect only a few parts of the pipeline leaving some scope for resource saving. Though Snakemake (Köster and Rahmann, 2012) and Nextflow (Di Tommaso et al., 2017) enable parameters based on ad hoc use of wildcard characters for easy scripting, a systematic handling of parameters is lacking in the literature. Moreover, these systems focus on each stage of the pipeline separately and the lack of information sharing across stages makes plug-and-play of stages difficult. For example, if a file generated by one stage is used as an input in multiple stages downstream then the details of the file need to be re-specified in each downstream stage making it tedious when there are several parameters.
2 JUDI
We develop JUDI on top of a Python-based build system, DoIt (http://pydoit.org), to systematically handle the issue of parameter settings using the principles of database management systems. The basic unit of execution in DoIt is a task roughly equivalent to a collection of rules in Make. Like other WMS, JUDI uses the dependency among tasks based on their input and output files. However, the novelty of JUDI is a consolidated way of capturing the variability under which the pipeline being build could possibly be executed. JUDI stores this variability in a simple parameter database table which contains one column for each parameter where each row indicates a value of the parameter (see details in Supplementary Section S2). Each file or task in JUDI is associated with a parameter database and hence does in fact represent a collection of physical files or DoIt tasks, respectively, each corresponding to one row of the parameter database (Supplementary Section S3–4). Which instances of a file are associated with a task instance is determined by applying few database operations on the parameter databases of the task and the file (Supplementary Section S5).
JUDI is available as a Python library and any JUDI pipeline first populates a global parameter database using a function add_param to avoid repeated local definition in each task. Each task is a derived class of Task with four class variables: (i) mask, (ii) inputs, (iii) targets and (iv) actions. Each of inputs and targets is a Python dictionary that maps each input or target file name to an object of class File. If the class variable mask is set in a task then it represents the list of parameters from the global parameters database that are not applicable to the current task. Each element of the Python list actions is a tuple (fun, args) where fun could be a Python string denoting the command line specification of the action and have placeholders {} which are replaced by the list args of values with the following exception: if a value is ‘$x’ then the placeholder is replaced by the physical path of file x instance associated with the task instance. A fun could also be a Python function and args could additionally have a value ‘#x’ which is replaced by a slice of the parameter database of file x applicable to the task instance (details of argument substitution in Supplementary Section S6).
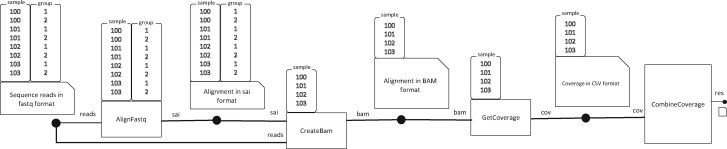
Figure 1 shows the tasks and files with their parameter databases for a slightly modified (Supplementary Fig. S2) four-stage pipeline used in (Köster and Rahmann, 2012, Fig. 1). In the first stage, each of eight FASTQ files of reads, one for each combination of four samples and two groups of pair-end reads are aligned to a reference genome producing an intermediate file. For each sample, the second stage converts the intermediate files for the two groups of pair-end reads to a BAM file, and the third stage generates a table of genome coverage information. The fourth stage consolidates genome coverage of all samples into a single table, unlike the Snakemake example where a coverage plot is generated separately for each sample. Listing 1 shows the Python script dodo.py for the pipeline of Figure 1 which can be executed from command line by doit -f dodo.py (details in Supplementary Section S7).
Fig. 1.
A slightly modified pipeline of Snakemake paper (Köster and Rahmann, 2012) visualized using JUDI concepts where each rectangle represents a task and each rectangle with a missing corner adjacent to a black dot represents a file. The parameter database table associated with a task (or a file) is also shown on the top of it
Listing 1.
dodo.py: JUDI script for the pipeline in Figure 1
1 from judi import File, Task, add_param, combine_csvs
2 add_param(’100 101 102 103’.split(), ’sample’)
3 add_param(’1 2’.split(), ’group’)
4 REF = ’hg_refs/hg19.fa’
5 path_gen = lambda x: ’{}_{}.fq’.format(x[’sample’], x[’group’])
6 class AlignFastq(Task):
7 inputs = {’reads’: File(’orig_fastq’, path = path_gen)}
8 targets = {’sai’: File(’aln.sai’)}
9 actions = [(’bwa aln {} {} > {}’, [REF,’$reads’,’$sai’])]
10 class CreateBam(Task):
11 mask = [’group’]
12 inputs = {’reads’: AlignFastq.inputs[’reads’],
13 ’sai’: AlignFastq.targets[’sai’]}
14 targets = {’bam’: File(’aln.bam’, mask = mask)}
15 actions = [(’bwa sampe {} {} {} | samtools view -Sbh --|
samtools sort --> {}’, [REF,’$sai’,’$reads’,’$bam’])]
16 class GetCoverage(Task):
17 mask = [’group’]
18 inputs = {’bam’: CreateBam.targets[’bam’]}
19 targets = {’cov’: File(’cov.csv’, mask = mask)}
20 actions = [(’(echo val; samtools rmdup {} --| samtools
mpileup --| cut -f4) > {}’, [’$bam’,’$cov’])]
21 class CombineCoverage(Task):
22 mask = [’group’, ’sample’]
23 inputs = {’cov’: GetCoverage.targets[’cov’]}
24 targets = {’res’: File(’combined.csv’, mask=mask, root=’.’)}
25 actions = [(combine_csvs, [’#cov’, ’#res’])]
To speed up the execution of a pipeline built using JUDI by utilizing multiple CPUs available in the modern processors, DoIt can be invoked by doit -n N -f dodo.py where N denotes the maximum number of independent tasks that DoIt should execute simultaneously. Moreover, to execute the pipeline in a high performance computing (HPC) cluster environment, the command string specified in the actions list of a task is prefixed with a cluster specific blocking command string. For example, each of the AlignFastq task instances in Listing 1 can be executed in a Slurm (Yoo et al., 2003) managed HPC node with four CPUs by prefixing srun -N 1 -n 1 -c 4 to the command string starting with bwa aln (details in Supplementary Section S9). Though srun is a blocking call, the desired parallel execution of multiple tasks is achieved by invoking doit with option -n.
3 Conclusion
We introduced a novel way of handling files and parameter settings in WMS with the motto: define once, reuse many times. We used DoIt for implementing our ideas quickly, however, JUDI can be implemented in other WMS too. Though Listing. 1 takes a similar number of lines as Snakemake, the usefulness of JUDI can be demonstrated by two tasks: CreateBam has only parameter ‘sample’ whereas its input files ‘reads’ and ‘sai’ have both parameters ‘sample’ and ‘group’. For each sample, the physical paths of sai and fastq files for both pair-end reads are passed to the aligner bwa sampe automatically by JUDI through the argument substitutions $sai and $reads. On the contrary, the same task needed hard-coding of file names due to the masked parameter ‘group’ in lines 11–12, Listing 1 of Köster and Rahmann (2012). The effort required could be significant if there were many masked parameters with a large number of values (Pal et al., 2019). In general JUDI takes O(n) times less number of lines for a pipeline with n parameters (Supplementary Section S8). Similarly, CombineCoverage shows how the coverage tables for the masked parameter ‘sample’ can be easily combined using JUDI utility function combine_csvs and argument substitutions.
Currently DoIt, and thus JUDI do not support docker/singularity containerization for reproducing task execution environments unlike other WMS (Di Tommaso et al., 2017; Köster and Rahmann, 2012). However, there is another aspect of reproducible research that JUDI facilitates far better than other WMS. For example, suppose the results using a pipeline built upon JUDI was published for a parameter P-value cutoff = {0.05, 0.01}. The same results for an unpublished cutoff 0.001 can be obtained just by adding the new value in the parameter definition. This usage can be further enhanced when JUDI is combined with graphical interfaces like iPython Notebooks.
Supplementary Material
Acknowledgement
The authors would like to thank the developers of DoIt. S.P. would like to thank Sunayna for the help on the manuscript.
Funding
This work was supported by the Intramural Research Program of the National Library of Medicine, National Institues of Health, USA.
Conflict of Interest: none declared.
Contributor Information
Soumitra Pal, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Teresa M Przytycka, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
References
- Amstutz P. et al. (2016) Common workflow language, v1.0. In: Common Workflow Language Working Group. Figshare, Cambridge, MA, London.
- Blankenberg D. et al. ; the Galaxy Team. (2014) Dissemination of scientific software with Galaxy ToolShed. Genome Biol., 15, 403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P. et al. (2015) BigDataScript: a scripting language for data pipelines. Bioinformatics, 31, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Boulakia S. et al. (2017) Scientific workflows for computational reproducibility in the life sciences: status, challenges and opportunities. Future Gener. Comp. Syst., 75, 284–298. [Google Scholar]
- Deelman E. et al. (2005) Pegasus: A Framework for Mapping Complex Scientific Workflows onto Distributed Systems. Scientific Programming, 13, 219–237. [Google Scholar]
- Di Tommaso P. et al. (2017) Nextflow enables reproducible computational workflows. Nat. Biotechnol., 35, 316–319. [DOI] [PubMed] [Google Scholar]
- Freire J. et al. (2006) Managing rapidly-evolving scientific workflows. In: Moreau L., Foster I. (eds.) Provenance and Annotation of Data, Lecture Notes in Computer Science. Springer, Berlin Heidelberg, pp. 10–18. [Google Scholar]
- Freire J. et al. (2012) Computational reproducibility: state-of-the-art, challenges, and database research opportunities. In: Proceedings of the 2012 ACM SIGMOD International Conference on Management of Data, SIGMOD ’12. Association for Computing Machinery, New York, NY, USA, pp. 593–596.
- Köster J., Rahmann S. (2012) Snakemake—a scalable bioinformatics workflow engine. Bioinformatics, 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Leipzig J. (2017) A review of bioinformatic pipeline frameworks. Brief. Bioinform., 18, 530–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. et al. (2019) Co-SELECT reveals sequence non-specific contribution of DNA shape to transcription factor binding in vitro. Nucleic Acids Res., 47, 6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradal C. et al. (2008) OpenAlea: a visual programming and component-based software platform for plant modelling. Funct. Plant Biol., 35, 751–760. [DOI] [PubMed] [Google Scholar]
- Ramachandran P. (2018) Automan: a python-based automation framework for numerical computing. Comput. Sci. Eng., 20, 81–97. [Google Scholar]
- Shah S.P. et al. (2004) Pegasys: software for executing and integrating analyses of biological sequences. BMC Bioinformatics, 5, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallman R.M. et al. (2004) GNU Make: A Program for Directed Recompilation: GNU Make Version 3.81. Free Software Foundation, Boston, MA. [Google Scholar]
- Stropp T. et al. (2012) Workflows for microarray data processing in the Kepler environment. BMC Bioinformatics, 13, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstencroft K. et al. (2013) The Taverna workflow suite: designing and executing workflows of Web Services on the desktop, web or in the cloud. Nucleic Acids Res., 41, W557–W561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A.B. et al. (2003) SLURM: simple Linux utility for resource management. In: Feitelson D. et al. (eds.) Job Scheduling Strategies for Parallel Processing, Lecture Notes in Computer Science. Springer, Berlin, Heidelberg, pp. 44–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.