Summary
Group A Streptococcus (GAS) remains a top 10 deadliest human pathogen worldwide despite its sensitivity to penicillin. Although the most common GAS infection is pharyngitis (strep throat), it also causes life-threatening systemic infections. A series of complex networks between host and pathogen drive invasive infections, which have not been comprehensively mapped. Attempting to map these interactions, we examined organ-level protein dynamics using a mouse model of systemic GAS infection. We quantified over 11000 proteins, defining organ-specific markers for all analyzed tissues. From this analysis, an atlas of dynamically-regulated proteins and pathways was constructed. Through statistical methods, we narrowed organ-specific markers of infection to 34 from the defined atlas. We show these markers are trackable in blood of infected mice, and a subset has been observed in plasma samples from GAS-infected clinical patients. This proteomics-based strategy provides insight into host defense responses, establishes potentially useful targets for therapeutic intervention, and presents biomarkers for determining affected organs during bacterial infection.
Keywords: Multiplexed proteomics, systemic infection, Group A Streptococcus, S. pyogenes, Orbitrap, Tandem Mass Tags
eTOC blurb:
Group A Streptococcus (GAS) pathogenesis has largely been studied from a bacterial-centric perspective. Utilizing multiplexing proteomics, we characterize organ specific and systemic responses to infection from a host-centric perspective. Analysis of clinical blood samples suggest a potential application for the identified biomarkers to diagnose the site of GAS infection.
Graphical Abstract

Introduction
Group A Streptococcus (GAS), Streptococcus pyogenes, remains among the top 10 infection-associated mortality agents worldwide though it remains sensitive to penicillin (Walker et al., 2014). Estimates suggest that over 18 million cases of invasive disease and greater than 500000 deaths occur annually due to GAS infection (Carapetis et al., 2005). The most common manifestation of GAS is pharyngitis, or strep throat, though more severe disease states, i.e. necrotizing fasciitis, bacteremia, and toxic shock syndrome occur; high morbidity and mortality is associated with each of these conditions (Walker et al., 2014).
Reports (Hsieh and Huang, 2011, Lau et al., 2012, Yang et al., 2013, Davies et al., 2015) have identified GAS strains resistant to tetracycline and macrolides, which are responsible for outbreaks of scarlet fever in China. Numerous sequelae have been linked to repeated GAS infection including rheumatic heart disease (Walker et al., 2014), neurological issues (Kurlan et al., 2008) and other autoimmune disorders (Kirvan et al., 2003). Sequelae and emerging antibiotic resistance highlight the need to develop a vaccine to prevent GAS infection. Despite numerous attempts, a safe, effective and human-approved vaccine still does not exist (Dmitriev and Chaussee, 2010, Walker et al., 2014). This is in part due to molecular mimicry of GAS antigens with human tissues (Cunningham, 2000, Kirvan et al., 2003), contributing to the development of autoimmune disease.
Traditionally, virulence factors and host defense mechanisms have been defined through genetics approaches. While fruitful, there is still an unmet challenge in identifying what type of immune response or components provide clearance of GAS and resolve infection. Defining the host response by constructing an organ-specific (OS) proteomic atlas may shed light on the functional mechanisms through which hosts overcome infections and reveal efficacy markers for numerous therapeutics.
Greater understanding of the host response to pathogens can yield insight into molecular mechanisms of immune protection. Host-centric approaches have the potential to uncover pathways that, if targeted by antibiotic alternatives, would boost host immunity and preserve the beneficial microbiota of the host (Nizet, 2015). Remarkably, immune boosting compounds have yet to be approved (Nizet, 2015), despite examples of beneficial activity (Kyme et al., 2012). Lack of approval is likely due to the risk of autoimmune disorders or an overwhelming immune response.
GAS studies have been bacteria-centric, with a recent call for systems-based approaches to study the host response during infection (Bessen and Lizano, 2010, Kilsgard et al., 2016, Tsatsaronis et al., 2014). Systems-level studies have the advantage of identifying global host responses, allowing the possibility for development of host-centric therapies (Cunningham, 2000). However, a challenge in prioritizing the large number of results for further study remains.
GAS is a human-specific pathogen, so model organisms (Roberts et al., 2006, Piepmeier et al., 1995, Ashbaugh et al., 2000, Rivera-Hernandez et al., 2014) cannot easily be used to study mechanisms associated with bacterial pathogenesis. Although no model is a perfect surrogate for human GAS infection, they have led to meaningful biological discoveries (Watson ME Jr., 2016). Two strains of GAS, M1 serotype strains 5448 and 5448AP, display increased virulence in mouse models (Kilsgard et al., 2016).
We used multiplexed proteomics to map the host-specific responses during systemic GAS infection in mice on an organ-to-organ basis. The goal of this study is twofold: first, construct an atlas of OS dynamics associated with infection response and second, statistically prioritize the large number of results to define OS infection markers. Our results indicate a concerted response to infection across organs, and identify OS Markers of Systemic Infection (MSI). Several MSI were detected in the blood of infected mice and a subset was observed in GAS-infected human samples, and may be clinically relevant as diagnostics. This study represents a significant advance towards understanding the dynamic proteome during systemic GAS infection from a host-centric perspective.
Results
Construction of a Murine Organ Atlas
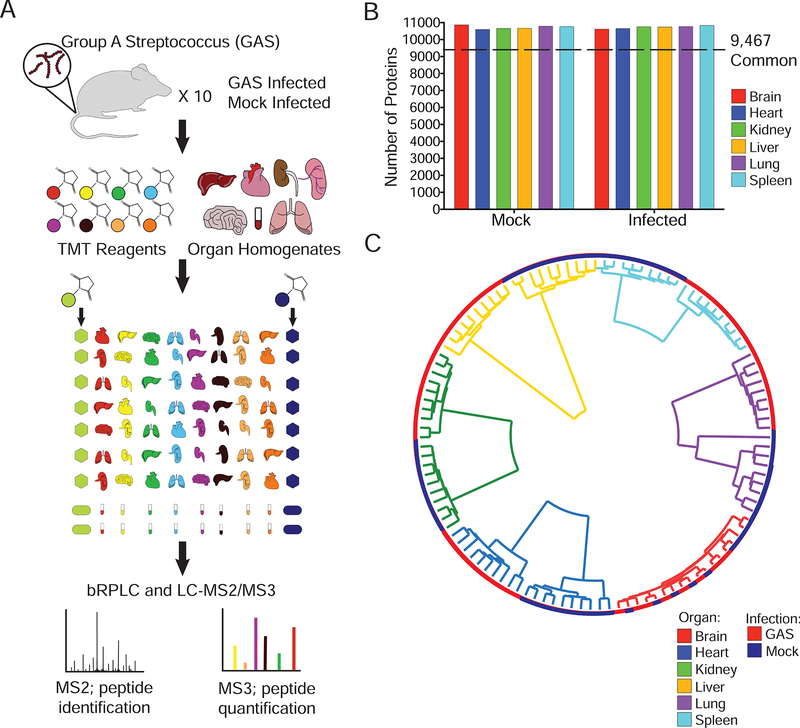
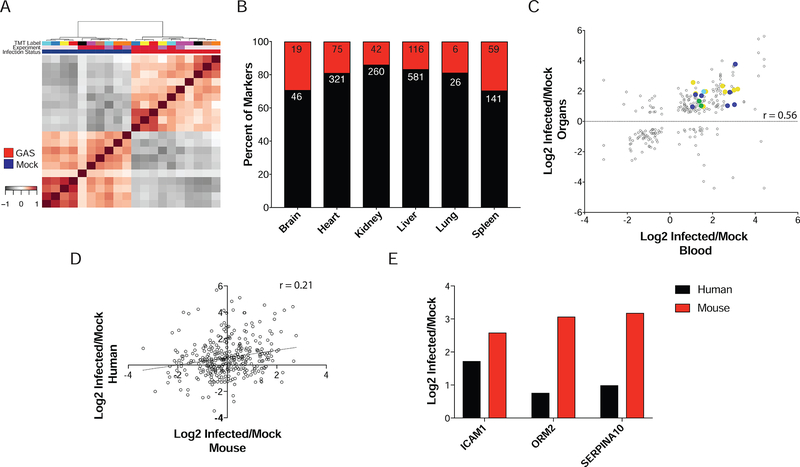
To determine OS responses during GAS infection, 10 biological replicates of brain, heart, lungs, kidney, spleen, liver and blood were analyzed from 8-week-old mice that were either GAS or mock-infected. Organs bacterial loads were quantified by assessing colony forming units (CFUs) (Figure S1). Homogenates were then lysed and trypsin-digested, with resulting peptides randomly labeled with Tandem Mass Tag (TMT) 10plex reagents (Table S11) and fractionated by basic pH reverse-phase chromatography. In total, 11394 proteins were quantified across all organs and treatment types (Figure 1A, Table S1, 1% protein and peptide false discovery). Blood proteomes were excluded, as they were analyzed independently of the organ tissues to prevent skewing of data from highly abundant serum proteins. Over 10700 proteins per organ treatment were quantified, with 9467 proteins common to all organs and treatments (Figure 1B, Table S2).
Figure 1– Quantitative Proteomics of a Murine GAS Infection Model.
(A) 10 mice per group were GAS or mock infected. At 48 hours post tail vein injection, mice were sacrificed and organs (brain, liver, kidney, spleen, lungs, heart and blood) harvested. Samples were randomized for TMT labeling; blood processed separately. Bridge channels containing equal portions of each organ homogenate were used for normalization purposes. All 10plexes were subjected to MS2 peptide identification and MS3 quantification. (B) A total of 11396 proteins were quantified. Plotted is the total number of proteins per organ in mock and infected organs. A total of 9467 proteins were common to all organs and infection statuses. (C) Spearman rank correlation coefficients were used in hierarchical clustering of samples. Organ origin drives primary clustering (Inner trees). Infection status is represented by the external ring.
No obvious batch effects were observed by correlational clustering of protein abundances (Figure 1C). Samples clustered by organ of origin, with GAS and mock-infected samples distinct from one another, except for the brain (Figure 1C). Notably, CFUs were present in the GAS infected brains, a result likely due to incomplete perfusion. We hypothesized that the lack of separation of GAS and mock-infected samples in the brain was a result of GAS’s inability to cross the blood-brain barrier (BBB) (Dileepan et al., 2016). To test this hypothesis, 10 biological replicates of Group B Streptococcus (GBS), a pathogen known to disrupt and cross the BBB (van Sorge and Doran, 2012), were analyzed. Clustering with these samples illustrated that brains clustered together, but GBS brains formed a distinct cluster separate from GAS and mock-infected brains (Figure S2). Comparing GBS-infected GAS and mock-infected brains, a total of 35 proteins were enriched in GBS-infected samples (>2 fold change, Benjamini-Hochberg (BH) FDR<0.01, Figure S2, Table S3).
Specific Markers Define Organ of Origin in Uninfected Samples
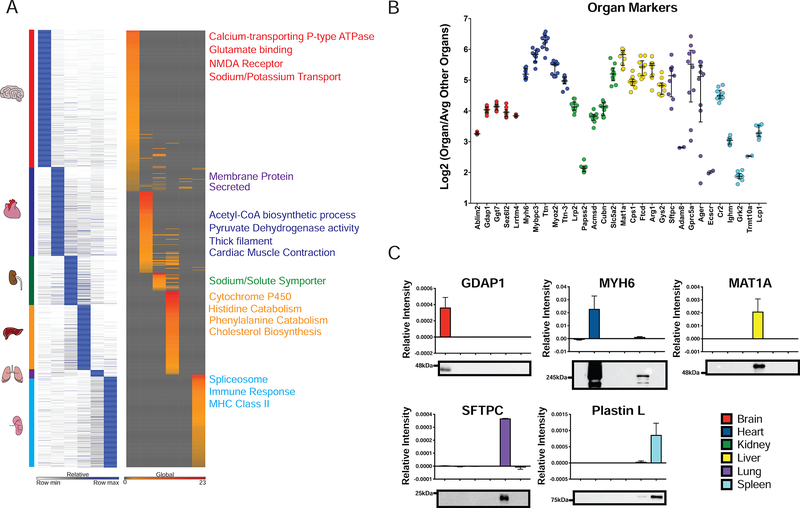
We hypothesized that each organ would have a unique signature of highly enriched proteins. To evaluate this hypothesis, we compared protein levels in each organ to all other organs. OS markers were then ranked on significance and fold change using π values (α≤1×10−5) (Xiao et al., 2014). OS markers ranged from 40 for lungs to 835 for the brain (Figure 2A). OS markers were analyzed in DAVID (Huang da et al., 2009b, Huang da et al., 2009a) for Gene Ontology (GO) and functional categories (Figure 2A, Table S4, S5). Minimal overlap in GO functional analysis was apparent by the lack of cross-organ representation of terms. Additionally, stratification of GO terms was appropriate on an organ-by-organ basis. For example, brain enriched proteins were associated with various neurological receptors (Figure 2A). The least distinct organ from a GO perspective was the lung, which had strong overlap with other organs despite several proteins defining its signature. GO enrichment in the lung was predominantly for membrane and secreted proteins.
Figure 2– Signatures of Organs based on Protein Markers.
(A) Heatmap of OS markers. Markers were identified by calculating Log2 ratio of the specified organ/average of all other organs and ranked by π-score (α≤1×10−5) – Left panel. Heat map of GO terms associated with organ markers is plotted in the right panel. Representative GO terms are listed to the right of the GO plot. (B) Top five OS markers ranked by π-score are plotted. With log2(Organ/Average other organs) on the y-axis and whiskers represent median and interquartile range of the data. (C) Western Blot validation of indicated OS markers. Relative intensity was normalized to total protein loaded on each gel. Mean relative intensity and standard deviation are plotted (n=2), and a representative blot is shown below each plot.
The expression levels of top organ markers for the brain (Gdap1, Ggt7, Sez6l2, Lrrtm4, Ablim2) are all known to be involved in synaptic transmission or other neurological processes (Figure 2B). Similar trends for OS involvement are known for top markers for the heart (Mybpc3, Ttn, Myoz2, Ttn-3, Myh6), kidney (Lrp2, Papss2, Acmsd, Cubn, Slc5a2), liver (Mat1a, Cps1, Ftcd, Arg1, Gys2), lung (Sftpc, Adam8, Ager, Ecscr, Gprc5a) and spleen (Cr2, Ighm, Grk2, Trmt10a, Lcp1). Among markers, a range of enrichment from 4 to 64-fold was observed and validated via Western Blot (Figure 2C). The reproducibility and enrichment attest to the fidelity of our proteome-based organ atlas.
Identification of Systemic and OS Infection Markers
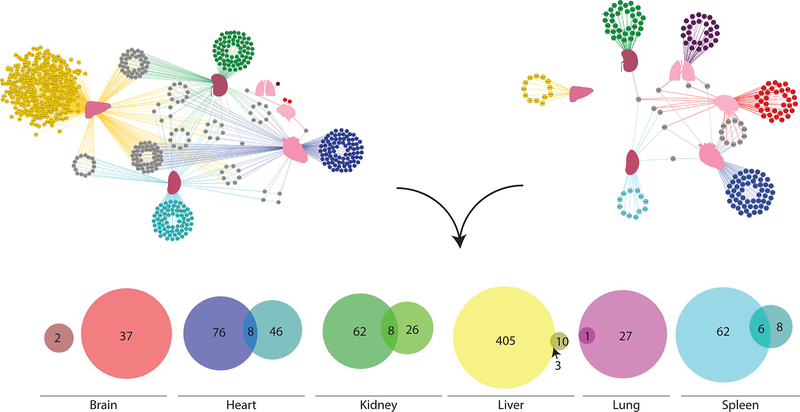
Quantitative proteomics data were analyzed on an organ-by-organ basis comparing infected to uninfected states (Figure S3). Proteins that were considered significant, BH-FDR<1% and at least twofold up or downregulated, were considered markers of infection (Table S6). Next, proteins were deemed OS if they were significant by the BH procedure and twofold or greater regulation in response to infection within that organ only. Separate analyses were performed for up and downregulated proteins. Proteins significantly regulated by these criteria in multiple organs were defined as systemic markers of infection. The specificity and cross-organ nature of the marker proteins was visualized via a protein network (Figure 3). In the protein network, proteins (nodes) specific to an organ were colored accordingly, and systemic proteins were colored gray. All edges were colored according to their organ of origin, simplifying visualization of the systemic markers.
Figure 3– Determination of OS Infection Markers.
Binary comparisons of infected vs mock were performed with BH FDR control at 1% and a twofold change were cut-offs for OS markers irrespective of directionality of fold-change. Markers were ranked by fold-change per organ. Proteins having a twofold change in multiple organs were considered systemic markers (protein network). Edges were colored according to organ of origin. In parallel, outlier analysis was performed for each organ, with log2 of fold-change of the organ relative to the mean of other organs plotted against the log2 of fold-change of the infected organ relative to the mean of other organs. Outliers were determined by the Tukey depth method. Proteins considered significant after BH correction were included in the analysis. Proteins having a twofold change in multiple organs were again considered systemic markers (protein network). Edges are colored according to organ of origin. Final markers were chosen based on the overlap of each statistical analysis (Venn diagrams). For simplicity, only upregulated proteins are depicted here, see Figure S5 for downregulated proteins.
In parallel, the proteomics data were subjected to outlier analysis via the Tukey depth method (Rousseeuw et al., 1999) (Figures 3 and S4). Input for outlier analysis was calculated in two parts. The first data set consisted of Log2 ratios of each protein for each infected organ relative to the average of other infected organs. The second data set consisted of Log2 ratios of each protein for each mock-infected organ relative to the average of other mock-infected organs. Only proteins that met BH-FDR controlled (<1%) significance in at least one of the scenarios were included in outlier analysis. We hypothesized that correlation of these ratios would demonstrate that most proteins within an organ do not change in the context of infection, while providing evidence that those that are most significantly changed in a specific organ would serve as markers for infection. This assumes that the most dynamic proteins with respect to the average response across the other organs will be identified as outliers. Concurrently, we postulated that outlier analysis would reduce the number of markers while having a higher specificity, identifying fewer systemic markers, due to the added context of the background response in the system.
After identification of outliers, proteins were directionally ranked based on the magnitude of their fold change as a ratio of ratios (Table S7). Markers were determined to be OS if there was an observed twofold or more change in only one organ; systemic markers exhibited twofold change or greater in multiple organs. The specificity of the marker proteins was plotted as a protein network (Figure 3). Outlier analysis identified fewer than 10% of proteins common to multiple organs. In contrast, the binary comparison approach identified 19.5%. The total number of specific markers was also reduced, as noted in the ratios, for the outlier analysis relative to the binary comparisons. The increased specificity and reduction in number of markers is directly evident in the protein network plots (Figure 3).
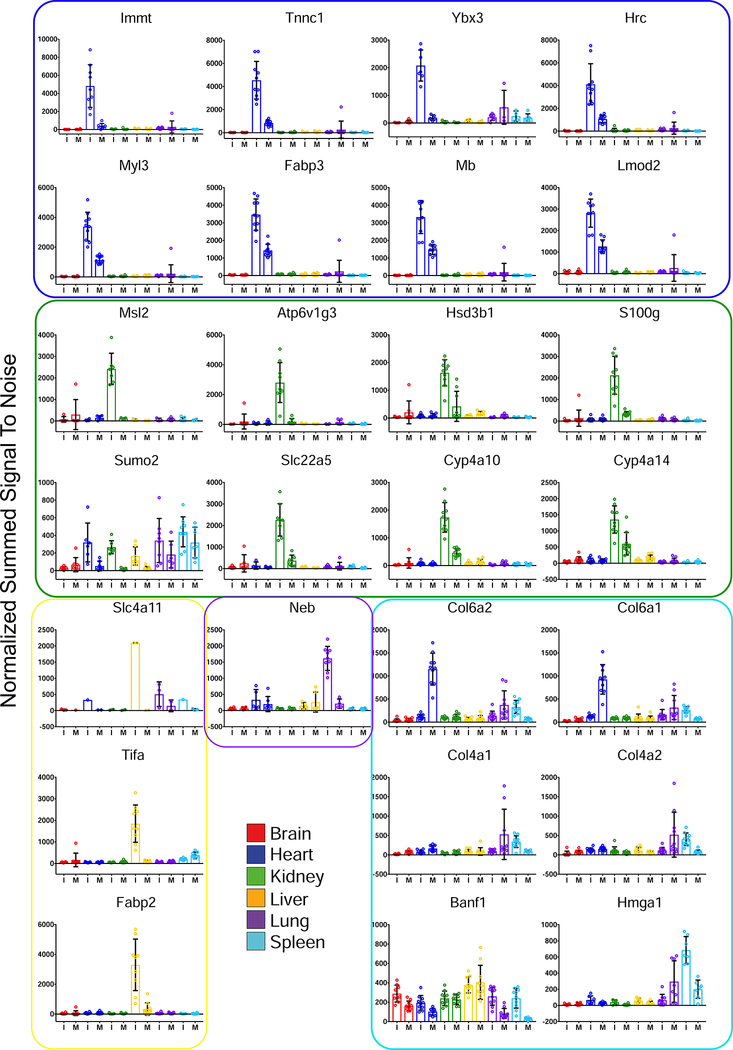
To prioritize OS markers, the intersection of these approaches was used (Figure 3). No overlap between brain analyses was observed, likely due to the inability of GAS to cross the BBB (Dileepan et al., 2016). Up and downregulated proteins taken from the intersection of these analyses were defined as MSI (Figures 4, S5). The union of markers is summarized in Figure 5.
Figure 4– Group A Streptococcus MSI.
Upregulated markers for infection are shown. Bars and data points are colored and grouped according to the organ of origin. Error bars represent standard deviation. Values plotted are normalized, summed signal-to-noise ratios. Proteins were defined from the overlap of the statistical methods (Figure 3). I – Infected and M – Mock.
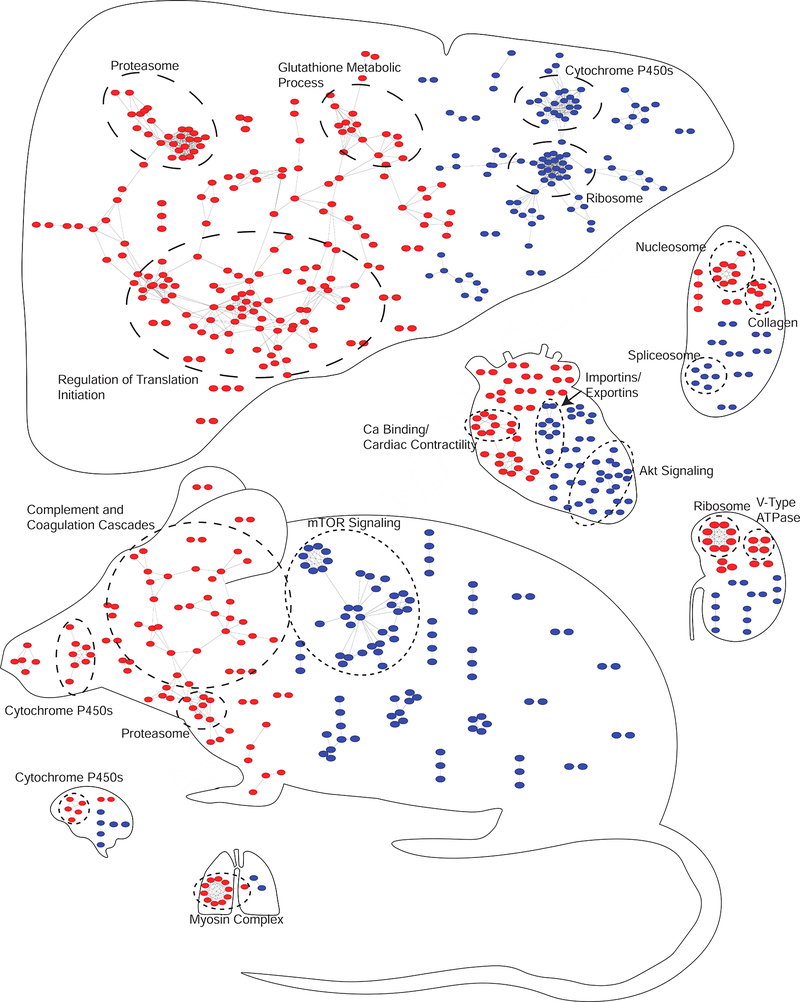
Figure 5– Assembly of a Marker Atlas for Systemic GAS Infection.
Organs are scaled according to the number of nodes contained within each organ. Nodes contained within the mouse are systemic markers, or those common to multiple organs. Placement of the nodes in the mouse is arbitrary. Nodes present in the individual organs are specific to that organ. Nodes represent proteins from the union of the outlier and binary comparison analyses that were significant and exhibited at least a twofold up (red) or down (blue) change. String analysis was performed with up and downregulated proteins independently. Only proteins with multiple connections are shown (significance > 0.7, no missing partners). GO functionally enriched clusters are circled and labeled accordingly.
Traceability of OS Markers of Infection in Blood
In total, 1640 proteins were quantified from whole blood (Table S8), with 617 unique to the blood analysis. The high degree of overlap between the blood and organ datasets could be due to the degree of vascularization of the organs analyzed. As with the organ analyses, clustering stratified replicates exactly into mock or GAS infected (Figure 6A).
Figure 6– Proteome Analysis of Blood Reveals Traceability of Organ Markers.
(A) Spearman’s Correlation clustering stratifies samples into mock and GAS infected subtypes. No obvious batch effects were observed. (B) SignalP analysis of infection markers derived from organs shows approximately equal percentages of proteins containing secretion signals. Red are those containing signal peptides and black those without. The numbers in each section of the bar indicate the raw counts of proteins in each category. (C) Correlation between blood levels and organ levels for infection markers. Overall Pearson correlation coefficient was 0.56, with a total of 19 infection markers traceable in blood. Spots are colored by tissue of specificity. (D) General discordance between GAS infected human and mouse blood proteomes. Pearson correlation coefficient=0.21, (E) Highlighted markers observed in blood from patients with GAS infection.
Recently, it was demonstrated that a protein’s tissue of origin could be inferred in plasma of GAS-infected mice (Malmstrom et al., 2016). We sought to determine if a non-targeted approach could provide similar inference of origin for infection markers defined herein, hypothesizing that proteins could be elevated in blood through secretion or necrotic processes. Therefore, the infection markers derived from the union of the statistical analyses were analyzed in SignalP (version 4.1) (Petersen et al., 2011) to determine if they contained a secretory signal sequence (Figure 6B, Table S9). Most protein markers did not, suggesting that if these proteins were found in blood they originated from necrotic processes. The liver had the highest number of proteins containing a signal sequence, which is expected as the liver is the primary source for plasma proteins (Miller et al., 1951).
Using the infection markers, OS protein levels were correlated with measured blood abundances (Figure 6C). Across all organs, 19 proteins were determined to be traceable in blood from an OS origin. Eight of these contained signal peptides, with five originating from liver. To confirm these makers, blood samples were obtained from GAS-infected patients and compared to healthy controls. An overall discordance between human and mouse blood levels was observed (Figure 6D). However, three markers showed a similar trend to that of GAS-infected mice (Figure 6E). Of these, Icam1 is known to be a general marker for sepsis (Sessler et al., 1995, Standage and Wong, 2011).
Specificity of MSI
To assess the specificity of MSI, spleens from mice infected systemically with the Gram-negative pathogen Salmonella enterica serovar Typhimurium (STM) were analyzed. Five mock-infected and five STM-infected spleens from two strains of mice were compared: C57BL/6 mice, a well-established model for STM, and CD1 mice, a genetic background control. Both CD1 and C57BL/6 mice had similar CFUs (Figure S7) and elicited a similar response with a high degree of correlation between shared proteins (R2=0.34, Figure S6A). These results include upregulation of Iigp1 (Table S12, S13), a protein associated with resistance to intracellular infection. Only two proteins were significant at BH-FDR<1%, Cd55 and Tspan15.
Next we assessed the specificity of the spleen MSI observed for GAS infection. A low degree of shared regulation between GAS infection and Salmonella infection was observed (R2=0.06, Figure S6B). Of the MSI markers observed in the GAS-infected spleens, four were related to collagen. None of the collagen-related proteins were significantly increased during STM infection. Other spleen MSI, Banf1 and HMGA1, were not significantly increased in the STM-infected spleens.
Discussion
Quantitative Proteomics Analysis of Organs
Comparison of organ protein profiles in the context of infection and mock infection led to several observable patterns. First, our data show OS enrichment for proteins, as seen in previous studies (Huttlin et al., 2010, Ponten et al., 2009). GO analysis of OS proteins shows correct stratification based on functional classifications. However, in some cases, such as the lung, GO analysis of OS markers did not achieve a distinct functional classification. Further examination of the respective classifications membrane proteins and secreted proteins showed that these are specific to the anatomy and function of the lung. These findings demonstrate the utility of GO functional classifications to define OS functions and highlight the potential to use these methods in the context of infection.
Murine Organ Atlas Defines Systemic and OS Infection Markers
We sought to define global immune responses and OS infection processes. The identification of the well-defined innate immune response served as a positive control (Figure 5). Given the numerous sequelae and complications associated with GAS infection, a key goal of this study was to gain insight into disease processes that occur specifically in each organ (Walker et al., 2014). An outlier analysis, similar to one predicting protein dysregulations (Lapek et al., 2017a), was utilized to identify proteins changing in each organ in response to infection while maintaining the context of changes occurring in other organs. This approach provided a greatly reduced number of OS infection markers (Figures 3 and 4), establishing a statistics-based means for marker prioritization.
Although rheumatic heart disease, a common sequela associated with GAS infection, does not occur in mouse models, numerous effects have been observed in the hearts of infected mice (Rush et al., 2014), making heart-specific markers worthy of further consideration. Among heart-specific MSI, Troponin C (Tnnc1) was noted. There are three troponin subunits, I, T and C, that associate with actin filaments and play an essential role in calcium-mediated regulation of muscle contraction (Takeda et al., 2003, Babuin and Jaffe, 2005). Traditionally, troponins are used as markers for acute cardiac events, including myocardial infarction. Since the cardiac isoform of troponin C is shared with slow-twitch skeletal muscles (Schreier et al., 1990), it is not used as part of the diagnostic panel for cardiac injury. Upon further examination of the proteomics data, troponin T was identified as a marker in the outlier analysis, but not the binary comparisons. Additionally, troponin I was identified as significantly upregulated in the heart in the context of heart infection vs other organs, but did not achieve statistical significance when comparing infected hearts to mock-infected hearts or by outlier analysis. Other heart MSI, including Myl3, Fabp3 and Mb, are also associated with cardiac injury (Hershberger et al., 2009, Cavus et al., 2006, Sallach et al., 2004). Mb and troponins have been examined in the context of other streptococcal infections (Watkin et al., 2004, Ammann et al., 2001). Intriguingly, Myl3 is thought to have some cross-reactivity with streptococcal antigens (Khavandi et al., 2008). These results indicate that markers of acute cardiac injury are upregulated in response to systemic infection, supporting previous findings (Rush et al., 2014).
Post-streptococcal glomerulonephritis is an inflammatory condition associated with GAS infections (Rodriguez-Iturbe and Haas, 2016). We postulated that kidney-specific MSI would be related to inflammatory biochemical processes. Upregulated kidney-specific MSI (i.e. Cyp4a10, Cyp4a14 and Slc22a5) have been linked to peroxisome proliferator-activated receptor signaling (PPARs) and phagocytic response. PPARs are known to increase levels of each of these proteins (Bartosiewicz et al., 2001, Kersten, 2014, Maeda et al., 2008). Cyp4a10 and Cyp4a14 both have roles in inducing an inflammatory response during infection (Nyagode et al., 2014). The observed increases in products of PPAR signaling are likely the result of cytokine storms, part of the innate immune response to GAS. Upregulation of these Cyps may also mediate clearance of apoptotic cells by macrophages (Majai et al., 2007). Additional MSI, Msl2 and Atp6v1g3, have been implicated in infection and the killing of phagocytosed bacteria (Franco et al., 2017, Morgan et al., 2009, Zodro et al., 2014).
Due to the role of the spleen in immune function, the markers derived from the spleen have the potential to provide valuable insight towards understanding host-pathogen (HP) dynamics. Among spleen-upregulated MSI were several collagen proteins. Type IV (T4) and type VI (T6) collagens are both extracellular matrix proteins that bind to GAS during infection (Dinkla et al., 2003b, Abdillahi et al., 2012, Dinkla et al., 2003a). T6 collagens have been shown to have antimicrobial effects at high concentrations (Abdillahi et al., 2012). The upregulation of these collagens in the spleen is interesting, as neither is associated with fibrotic scarring, a mechanism linked to type I collagens (Trojanowska et al., 1998). Increased titers of T4 in blood have been associated with other GAS serotypes in relation to rheumatic diseases (Dinkla et al., 2003b), suggesting that the increase of T4 and T6 collagens present in the spleen may be due to autoantigenic processing. In support of this hypothesis, a decrease in heart and lung collagen T4 and T6 was noted (Figure 4, Figure S5). Our observations regarding collagen may be specific to Gram-positive infections, as the findings were not observed within STM-infected mice.
To gain a broader perspective of the dynamic changes in response to infection, the union of markers derived from the binary comparisons and the outlier analysis were summarized (Figure 5). Irrespective of organ-of-origin, most markers were highly connected in both up and downregulated sets, suggesting proteins are functionally regulated at the complex or pathway level, and that fluxes in rogue proteins are not driving-markers of infection. Previous studies have suggested that proteins tend to be degraded when not part of their cognate complex (Goldberg and Dice, 1974, Hwang et al., 2010), supporting our observations. To assign mechanistic regulation to pathways, further analyses of the temporal dynamics of post-translational modifications at various states is needed.
Within the proteome atlas, the mTOR pathway was observed to be downregulated (Figure 5). mTOR is known to be inhibited by inflammatory cytokines (Frost and Lang, 2011). During pro-inflammatory states, the proteasome is known to activate numerous inflammatory proteins (Elliott et al., 2003). This function has led to the development of proteasomal inhibitors for attenuation of inflammation. In our mouse model, systemic upregulation of the proteasome was observed in response to GAS infection, suggesting a role in the pro-inflammatory state from which these samples were derived. Concurrent with mTOR downregulation, complement and coagulation cascades were upregulated. There are well defined roles for each of these pathways in the pathogenic processes of GAS, as GAS is known to be sequestered by the coagulation cascade (Loof et al., 2014) allowing the host to mount an effective response (Shannon et al., 2010). In response, GAS utilizes Streptokinase to escape entrapment and disseminate (Green, 1948).
Through our analyses, the brain and lung have the fewest markers of analyzed organs. The low number of markers associated with these organs was unsurprising due to the systemic nature of the infection. The brain and the lung have similar endothelial barrier function, though the lung barrier is more permeable than the BBB (Daneman et al., 2010). However, the markers that are present are still important. In the brain, upregulation of several Cyp2 and Cyp3 proteins related to the metabolism of arachidonic acid were noted. As arachidonic acid is metabolized into both pro and anti-inflammatory products (Serhan et al., 2008) and has ties to PPARγ regulation for protection against lipid oxidation (Wang et al., 2006), this regulation is likely a protective function in the brain against the innate immune response. Conversely, in the lung, upregulation of nebulin and a network of myosin proteins was observed. Combined with the observations related to T6 collagen in the lung (Figure 4, Figure S5), this suggests a compensatory response to the degradation of supporting extracellular matrix proteins. A potential explanation is reduced lung contractility due to decreased support proteins. To compensate, the lungs respond by acutely increasing expression of nebulin and myosins to maintain contractility, as each of these proteins have well-defined roles in contraction (Bang et al., 2009) and deficiencies associated with myopathies (Wallgren-Pettersson et al., 2002, Wallgren-Pettersson et al., 1999).
In the heart, a downregulated cluster was enriched for Akt pathway members. This is important because of the known connection of the Akt signaling axis to induction of autophagy processes (Li et al., 2015). Previously GAS was shown to be susceptible to autophagocytic killing (Nakagawa et al., 2004), and our data suggests a localized role for this process during systemic GAS infection.
Within the liver, cytochrome p450s are downregulated. Cytochrome p450s are known to be downregulated by infectious agents and inflammatory processes (Morgan, 1997). Conversely, proteins in the glutathione metabolic process are upregulated. As glutathione is an important antioxidant in protection from reactive oxygen species (ROS) (Aquilano et al., 2014), this result is expected. The glutathione metabolic processes present an interesting paradox in HP interactions: the balance in redox status that allows destruction of the pathogen while maintaining protection of the host. To a point, GAS appears to be well equipped to make this balance more complicated. GAS possesses a glutathione peroxidase that allows it to withstand high concentrations of hydrogen peroxide (Gibson et al., 2000, Brenot et al., 2004). These findings suggest that regulation of glutathione-related proteins in the host is likely in response to the pro-inflammatory, high ROS environment that GAS promotes for its own survival.
Through this analysis it became clear that signaling networks and protein complexes observed in the proteome atlas were being dynamically regulated in the context of systemic GAS infection. Much of this granularity would have been masked had the outlier analysis not been employed. While some of these pathways, such as mTOR (Frost and Lang, 2011), have known ties to inflammatory processes, this approach allows a more comprehensive exploration of the complexity of HP interactions. For example, our findings related to T4 and T6 collagens (Figures 4, S5) yield insight to the dynamic remodeling of tissues and host immune function that lead to sequelae associated with GAS.
It is important to note that the specificity of these observations to GAS or other Gram-positive bacteria has not been addressed. We have some evidence that these observations may be specific to Gram-positive infection as there was poor correlation (r=0.24) between the splenic response to GAS and STM (Figure S6B). Additionally, none of the defined splenic MSI were upregulated in STM infection. However, without analysis of other Gram-positive infections we cannot confidently discriminate whether these responses are specific to GAS or Gram-positive bacteria in general.
Blood Markers of Systemic Bacterial Infection
Results from our proteomic analysis of blood suggest that a combination of necrotic and secretory processes contribute to the presence of OS markers in the blood. In comparison with recent results, three markers that contain signal peptides followed similar patterns of increased abundance in blood following infection (Malmstrom et al., 2016). These were Alpha-1-acidglycoprotein 2, Plasma protease C1 inhibitor and Complement component C8 alpha chain, which are involved in acute phase response (Gabay and Kushner, 1999). The Malmstrom et al. study used a subcutaneous infection model with different bacterial strains and inoculums. The fact that some markers are common between these models suggests some core molecular mechanisms are shared, irrespective of route of infection, while others may be unique to the point of entry.
Analysis of human patient blood suggests a clinical application for a subset of MSI. An overall discordance between the human and mouse response to infection (Figure 6D) was observed, which may be the result of several factors. Patients had a variety of infection manifestations from necrotizing fasciitis to pneumonia (Table S14). It was not possible to obtain tissue biopsies, making it difficult to evaluate MSI. This discordance could also be attributed to the human-specific nature of GAS; the mouse model only represents certain aspects of the human infection (Watson ME Jr., 2016). Nevertheless, the trends observed in three markers, Icam1, Orm2, and Serpina10, can be useful diagnostic indicators of the localization of GAS infection in humans. Icam1 and Serpina10 may indicate infection in the liver, while Orm2 may indicate progression to the heart. Previous reports have shown that Icam1 expression is induced by GAS’s SlaA, increasing the adhesion of human monocytic leukemia cells (Oda et al., 2017). Icam1 deletion mutants have also displayed increased bacterial burden in mycobacterium infection, further evidence of a role for Icam1 during bacterial infection (de Paula et al., 2017). Less is known of Serpina10’s role in GAS infection, but studies suggest that it also increases immune cell penetration into the liver during sepsis (Butschkau et al., 2013). While Orm1 is known to predict patient outcomes in sepsis (Raju et al., 2016, Barroso-Sousa et al., 2013, Xiao et al., 2015), our studies implicate a similar potential for Orm2. Interestingly, EndoS secreted by GAS has the ability to hydrolyze Orm1 (Sjogren et al., 2013), and nitrosated forms of Orm1 can kill drug-resistant bacteria during sepsis (Watanabe et al., 2013). These proteins may have broad implications for improved OS diagnostics and may warrant further investigation.
Summary and Conclusions
While challenges remain in fully understanding the host response during bacterial infection, the construction of an organ proteome atlas is a powerful step forward. There have been calls to go beyond traditional bacteria-centric genetic approaches, and move towards host-centric systems biology approaches (Bessen and Lizano, 2010, Kilsgard et al., 2016, Tsatsaronis et al., 2014). We demonstrate the power of an unbiased systems level approach, through the use of quantitative proteomics, to define dynamic host responses to infection. This is advantageous over typical in vitro studies that lack the context of the entire system. Continued generation of datasets such as this, along with metabolomic and transcriptomic analyses, have the potential to provide even greater power. Integration of such datasets could more easily pinpoint pathways for targeted investigation. Through incorporation of additional layers, such as time, immune perturbations and post-translational modifications, finite mechanisms for host response can be defined.
The context of microenvironments, as assessed by dynamics in each organ, within hosts are important niches to consider during infection and treatment. Another important factor to consider is bioavailability of therapeutics at the site of infection. This can be affected by factors such as pH, cell type, and drug metabolism (Wagner et al., 2006). Each of these variables can lead to heterogeneous availability of drugs within and amongst tissues. The advent of techniques such as laser capture microdissection coupled to mass spectrometry (Banks et al., 1999) and new visualization techniques could eventually lead to more contextually relevant HP interaction studies. Applying this combination of techniques could lead to the reduction in host-specific background typical in HP interaction studies (Semanjski and Macek, 2016), and allow for examination of pathogen factors that are being expressed in vivo. This can also be achieved through targeted techniques (Erickson et al., 2017).
Over 200 different M-type strains have currently been identified (Bessen and Lizano, 2010). One of the most extensively utilized mediums for understanding infection, disease and defining biomarkers remains the blood (Malmstrom et al., 2016). While the plasma proteome is known to be variable among humans (Liu et al., 2015), blood biobanks are an invaluable resource for translational studies involving pathogens, including GAS. This study demonstrates the ability to detect protein dynamics resulting from bacterial infection, with tissue of origin inference, in blood samples.
The quantitative proteomics strategy outlined here shows potential to define an atlas of infection responses in mice and statistically prioritize protein biomarkers. Prioritization of potential markers in this manner directly addresses a common challenge in the field, where thousands of candidates are typically identified. Through a series of downstream analyses, we demonstrate the ability to further establish potential links to clinical relevance by tracking organ derived proteins in blood samples. Though this study was conducted in the context of a systemic bacterial infection, this approach is readily adaptable to numerous pathogens.
STAR METHODS:
CONTACT FOR REAGENT AND RESOURCE SHARING:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David J. Gonzalez (djgonzalez@ucsd.edu) +1 858 822–1218.
EXPERIMENTAL MODEL AND SUBJECT DETAILS:
Bacterial Strains.
M1T1 Group A Streptococcus (GAS) 5448 was originally isolated from a clinical necrotizing fasciitis patient sample (Chatellier et al., 2000). Serotype III Group B Streptococcus (GBS) COH1 strain was used for GBS infection experiments. For routine propagation, GAS and GBS were grown in liquid cultures of Todd-Hewitt broth (THB, Neogen) without aeration at 37 °C. IR715 is a fully virulent, nalidixic acid (Nal)-resistant derivative of Salmonella enterica serovar Typhimurium (STM) wild-type isolate ATCC 14028 (Stojiljkovic et al., 1995). STM was grown aerobically at 37°C in Luria-Bertani (LB) broth.
Mouse Infection Models.
All animal experiments were reviewed and approved by the University of California, San Diego Animal Care and Use Committee. For GAS and GBS studies, 8-week old female CD-1 mice (n=10) were used (Harlan Laboratories). To study the distribution of systemic GAS infection, mice were infected with 5×106 CFU/100 μL by tail vein injection and euthanized 48 hours post-infection for the collection of the blood, heart, spleen, brain, lungs, liver and kidneys, a modification of previous studies (Sakurai et al., 2003, Lin et al., 2015, Goldmann et al., 2004). Mice were perfused with PBS prior to organ collection. In parallel, an additional set of mice (n=10) were mock infected with PBS and organs were harvested similarly. All organs were subsequently homogenized using a Mini BeadBeater (Biospec) and samples were serially diluted in PBS and plated on Todd-Hewitt Agar (THA) to enumerate CFUs per gram of tissue. For GBS infection studies, mice were infected with 7.5×107 CFU/100 μL (van Sorge et al., 2009) by tail vein injection. Forty-eight hours post-infection, animals were sacrificed as with GAS infected animals and brains were collected. Brains were homogenized as before and CFUs enumerated on THA. For STM studies, C57BL/6 and CD1 mice were used. Mice were purchased from Envigo (Harlan). Groups of five, 8–10-week-old female mice were injected intraperitoneally with either 0.1 ml of DPBS (control) or 0.1 ml of a suspension of STM. C57BL/6 were inoculated with 1×103 CFUs while CD1 mice were inoculated with 1×105 CFUs. Mice were sacrificed at 4 days post-infection and the spleens were collected. The spleens were then weighed and homogenized. The organ suspensions were serially diluted, and plated on LB agar plates to determine bacterial counts.
Clinical patient selection and human sample collection.
Clinical blood samples were collected as part of the Molecular Diagnosis and Risk Stratification of Sepsis (MARS) initiative, a prospective cohort study in a tertiary mixed medical-surgical ICU in the Netherlands. Ethical approval for the study was provided by the Medical Ethics Committee of the University Medical Center Utrecht (UMCU), including an opt-out consent method (IRB No. 10–056C). Participants were notified of the study in writing by a brochure provided at ICU admission with an attached opt-out card that could be completed by the patient or by his or her legal representative in case of unwillingness to participate. All treated infections were registered daily by dedicated observers as described previously (Klein Klouwenberg et al., 2013). For this study we selected septic patients with culture-proven infection caused by GAS. Daily leftover citrate plasma was processed within 4 hours of sampling, and stored at −80 °C. Ten samples selected for this study were collected within ± 1 day from start antimicrobial treatment and matched to ten healthy controls. Additional patient information, including information relating to sex, age, and infection status, is contained in Table S14. Sample size was chosen based on plexing ability of TMT reagents. Ten healthy and ten controls were also chosen to correspond to the number of animals that were mock or infected from the mouse study.
METHOD DETAILS:
Western Blots.
Mouse organ lysates were prepared for immunoblot analysis as follows. Organs were added to 1X RIPA lysis buffer with 1X Roche Complete mini EDTA free protease inhibitors. Samples were bead beat three times for 1 min with a 1 min rest in between. Organ lysate was acquired and centrifuged for 10 min at 13,000 RPM 4 °C to pellet insoluble debris. Protein concentration was estimated using a Pierce™ BCA Protein Assay Kit (Thermo) and normalized across all samples. Total protein loaded on to the gel was confirmed by running a coomassie blue stained gel, and samples were normalized as previously described (Eaton et al., 2013). Organ lysates were then analyzed for corresponding marker proteins with antibodies purchased from Abcam according to manufacturer recommendations (GDAP1 - ab100905, MYH6 - ab50967, MAT1A - ab129176, SFTPC - ab211326 and LCP1 - ab109124). Samples were mixed with Laemmli SDS-PAGE sample buffer, boiled for 5 min then electrophoresed on a 12% Tris-HCl SDS-polyacrylamide gel. Electrophoresed proteins were transferred to a nitrocellulose membrane which was subsequently blocked with 5% milk in TBST and incubated with primary antibody overnight at 4 °C. Following incubation, membranes were washed with TBST and incubated with corresponding secondary antibodies for 1 hr according to manufacturer recommendations (anti-rabbit - ab671, anti-mouse - ab6789). After treatment, protein bands were detected using SignalFire™ ECL Reagent (Cell Signaling - #6883) and quantified using Bio-Rad ChemiDoc™ Touch Imaging System and Bio-Rad Image Lab 6.0.
Protein Digestion and TMT Labeling.
Organ homogenates were lysed in a buffer composed of 75 mM NaCl (Sigma), 3% sodium dodecyl sulfate (SDS, Fisher), 1 mM NaF (Sigma), 1 mM beta-glycerophosphate (Sigma), 1 mM sodium orthovanadate (Sigma), 10 mM sodium pyrophosphate (Sigma), 1 mM phenylmethylsulfonyl fluoride (PMSF, Sigma), and 1X Complete Mini EDTA free protease inhibitors (Roche) in 50 mM HEPES (Sigma), pH 8.5 (Villen and Gygi, 2008). To ensure full lysis, homogenates were passed through a 21-gauge syringe 20 times. Insoluble debris was then pelleted by centrifugation for 5 minutes at 14,000 rpm. Supernatants were transferred to new tubes and an equal volume of 8 M urea in 50 mM HEPES, pH 8.5 was added to each sample. Samples were then vortexed and sonicated for 5 minutes in a sonicating water bath to ensure protein denaturation.
Proteins were reduced with dithiothreitol (DTT, Sigma) and alkylated with iodoacetamide (Sigma) as previously described (Haas et al., 2006). Proteins were precipitated via methanol-chloroform precipitation (Wessel and Flugge, 1984). Precipitated proteins were re-solubilized in 300 μL of 1 M urea (Fisher) in 50 mM HEPES, pH 8.5. Vortexing, sonicating and manual grinding were used to aid solubility. Proteins were digested in a two-step digestion process. First, 3 μg of LysC (Wako) was added to each sample, and samples were digested overnight at room temperature. Next, 3 μg of trypsin was added, and samples were digested for six hours at 37 °C. Digests were acidified with trifluoroacetic acid (TFA, Pierce) to quench the digestion reaction. Peptides were desalted with C18 Sep-Paks (Waters) as previously described (Tolonen and Haas, 2014). Concentration of desalted peptides was determined with a BCA assay (Thermo Scientific), and peptides were aliquoted into 50 μg portions. Aliquots were dried under vacuum and stored at −80 °C until they were labeled with TMT reagents.
Peptides were labeled with 10-plex TMT reagents (Thermo Scientific) (McAlister et al., 2014, Thompson et al., 2003) as previously described (Wang et al., 2011). TMT reagents were reconstituted in dry acetonitrile (Sigma) at 20 μg/μL. Dried peptides were re-suspended in 30% dry acetonitrile in 200 mM HEPES, pH 8.5, and 7 μL of the appropriate TMT reagent was added to peptides. Reagents 126 and 131 (Thermo Scientific) were used to bridge between mass spec runs. Remaining reagents were used to label samples in random order. Labeling was carried out for 1 hour at room temperature, and was quenched by adding 8 μL of 5% hydroxylamine (Sigma). Labeled samples were acidified by adding 50 μL of 1% TFA and then pooled into appropriate 10-plex TMT samples, with pooled standard samples labeled with 126 and 131. Pooled samples were desalted with C18 Sep-Paks.
Basic pH Reverse-Phase Liquid Chromatography Sample Fractionation.
Sample fractionation was performed by basic pH reverse-phase liquid chromatography with concatenated fractions as previously described (Wang et al., 2011). Briefly, samples were re-suspended in 5% formic acid/5% acetonitrile and separated over a 4.6 mm x 250 mm C18 column (Thermo Scientific) on an Ultimate 3000 HPLC fitted with a fraction collector, degasser and variable wavelength detector. The separation was performed over a 22% to 35%, 60-minute linear gradient of acetonitrile in 10 mM ammonium bicarbonate (Fisher) at 0.5 mL/min. The resulting 96 fractions were combined as previously described (Wang et al., 2011). Fractions were dried under vacuum and re-suspended in 5% formic acid/5% acetonitrile and analyzed by liquid chromatography (LC)-MS2/MS3 for identification and quantitation.
LC-MS2/MS3 for Protein Identification and Quantitation.
All LC-MS2/MS3 experiments were carried out on an Orbitrap Fusion (Thermo Fisher Scientific) with an in-line Easy-nLC 1000 (Thermo Fisher Scientific) and chilled autosampler. Home-pulled, home-packed columns (100 μm ID x 30 cm, 360 μm OD) were used for analysis. Analytical columns were triple-packed with 5 μm C4 resin, 3 μm C18 resin, and 1.8 μm C18 resin (Sepax) to lengths of 0.5 cm, 0.5 cm, and 30 cm respectively. Peptides were loaded at 500 bar and eluted with a linear gradient of 11% to 30% acetonitrile in 0.125% formic acid over 165 minutes at a flow rate of 300 nL/minute, with the column heated to 60 °C. Nano-electrospray ionization was performed by applying 2000 V through a stainless-steel T-junction at the inlet of the microcapillary column.
The mass spectrometer was run in data-dependent mode, where a survey scan was performed over 500–1200 m/z at a resolution of 60,000 in the Orbitrap. Automatic gain control (AGC) was set to 2×105 for MS1 with a maximum ion injection time of 100 ms. The S-lens RF was set to 60 and centroided data was collected. Top-N mode was used to select the most abundant ions in the MS1 scan for MS2 and MS3 with N set to 10.
The decision tree option was used for MS2 analysis, using charge state and m/z range as qualifiers. Ions carrying 2 charges were analyzed from the m/z range of 600–1200, and ions carrying 3 and 4 charges were selected from the m/z range of 500–1200. An ion intensity threshold of 5×104 was used. MS2 spectra were obtained using quadrupole isolation at a 0.5 Th window and fragmented using Collision Induced Dissociation with a normalized collision energy of 30%. Fragment ions were detected and centroided data collected in the linear ion trap using rapid scan rate with an AGC target of 1×104 and maximum ion injection time of 35 ms.
MS3 analysis was performed using synchronous precursor selection (SPS) enabled to maximize TMT quantitation sensitivity (McAlister et al., 2014). A maximum of 10 MS2 precursors was specified for the SPS setting, which were simultaneously isolated and fragmented for MS3 analysis. Higher-Energy Collisional Dissociation fragmentation was used for MS3 analysis with a normalized collision energy of 55%. Resultant fragment ions (MS3) were detected in the Orbitrap at a resolution of 60,000 with a low mass cut-off of 110 m/z. AGC for MS3 spectra was set to 1×105 with a maximum ion injection time of 100 ms. Centroided data were collected, and MS2 ions between the range of 40 m/z below and 15 m/z above the precursor m/z were excluded by SPS.
QUANTIFICATION AND STATISTICAL ANALYSIS:
Data Processing and Analysis.
Data were processed using Proteome Discover 2.1 (Thermo Fisher Scientific). MS2 data were searched against concatenated Uniprot Human, Group A Streptococcus and Group B Streptococcus databases (downloaded 11–28-16) respectively using the Sequest algorithm (Eng et al., 1994). A decoy search was also conducted with sequences in reverse order (Elias and Gygi, 2007, Elias et al., 2005, Peng et al., 2003). A precursor mass tolerance of 50 ppm (Beausoleil et al., 2006, Huttlin et al., 2010) was specified and 0.6 Da tolerance for MS2 fragments. Static modification of TMT 10-plex tags on lysine and peptide n-termini (+229.162932 Da) and carbamidomethylation of cysteines (+57.02146 Da) were specified. Variable oxidation of methionine (+15.99492 Da) was also included in search parameters. Data were filtered to 1% peptide and protein level false discovery rates with the target-decoy strategy through Percolator (Kall et al., 2007, Spivak et al., 2009).
TMT reporter ion intensities were extracted from MS3 spectra for quantitative analysis, and signal-to-noise values were used for quantitation. Spectra were filtered and summed as previously described (Lapek et al., 2017b). Data were normalized in a multi-step process, whereby they were first normalized to the pooled standards (TMT-126 and −131) for each protein, and then to the median signal across the pooled standards from all experiments (Lapek et al., 2017a). An average of these normalizations was used for the next step. To account for slight differences in amounts of protein labeled, these values were then normalized to the median of the entire dataset and reported as final normalized summed signal-to-noise ratios per protein per sample.
Statistics and Bioinformatics Analyses.
Significance for proteomics data was assessed by Student’s T-test; variance was assessed by an F-test to ensure the correct statistical assumptions were used. Adjustment for multiple comparisons was performed through Benjamini-Hochberg procedure, with a false discovery rate of 1%. Outlier analysis, through Tukey’s depth method and visualized through bagplots (Rousseeuw et al., 1999), was utilized to determine organ specific markers of infection. Input included proteins which were significant (BH controlled FDR<0.01) in either infected organs or uninfected organs.
Gene ontology (GO) analysis through the DAVID server (Huang da et al., 2009b, Huang da et al., 2009a) was utilized to identify significant groups of proteins per organ, infected and mock-infected. Parameters were set as previously described (Nicolay et al., 2015). Gene heat maps of organ and infection specific markers and subsequent GO analyses were visualized in GENE-E (https://software.broadinstitute.org/GENE-E/index.html).
STRING-db (Szklarczyk et al., 2015) was utilized through the String App for Cytoscape (Version 3.5) to visualize connections between proteins. Connections were limited to high confidence (0.7) with interactions restricted to the query only.
Data and Software Availability.
The mass spectrometry proteomics data have been deposited into MassIVE (http://massive.ucsd.edu) and submitted to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) with the dataset identifiers MSV000081425; PXD007213 (organs) and MSV000081437; PXD007248 (blood); PXD008809 and MSV000081997 (STM/Clinical patient blood).
Supplementary Material
KEY RESOURCES TABLE:
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GDAP1 Rabbit polyclonal antibody | Abcam | ab100905 |
| Anti-MYH6 Mouse monoclonal antibody | Abcam | ab50967 |
| Anti-MAT1A Rabbit monoclonal antibody | Abcam | ab129176 |
| Anti-SFTPC Rabbit monoclonal antibody | Abcam | ab129176 |
| Anti-LCP1 Rabbit monoclonal antibody | Abcam | ab109124 |
| Bacterial and Virus Strains | ||
| M1T1 Group A Streptococcus (GAS) 5448 | (Chatellier et al., 2000) | N/A |
| Serotype III Group B Streptococcus (GBS) COH1 | (Wilson and Weaver, 1985) | N/A |
| Salmonella enterica serovar Typhimurium (STM) wild-type isolate ATCC 14028 | (Stojiljkovic et al., 1995) | N/A |
| Biological Samples | ||
| GAS infected human blood | University Medical Center Utrecht (UMCU) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Tandem Mass Tag Reagents | Thermo-Fisher | Cat#90110 |
| Deposited Data | ||
| Mouse organs proteomic data | This paper | MSV000081425; PXD007213 |
| Mouse blood proteomic data | This paper | MSV000081437; PXD007248 |
| Mouse STM spleen proteomic data and human clinical patient blood proteomic data | This paper | MSV000081997; PXD008809 |
| Experimental Models: Organisms/Strains | ||
| Mouse: female CD-1 | Harlan Laboratories | |
| Mouse: female C57BL/6 mice | Harlan Laboratories | |
| Software and Algorithms | ||
| GENE-E | https://software.broadinstitute.org/GENE-E/index.html | |
| DAVID | (Huang da et al., 2009a) | https://david.ncifcrf.gov/ |
| STRING-db | (Szklarczyk et al., 2015) | https://string-db.org/ |
| Proteome Discover | Thermo-Fisher | |
| R Studio (1.1.383) | https://www.rstudio.com/ | |
| SignalP | (Petersen, et al., 2011) | http://www.cbs.dtu.dk/services/SignalP/ |
Highlights:
Group A Streptococcus infected mouse organs have consistent proteomic responses.
Combining statistical methods reveals organ specific responses to infection.
Protein networks shift during infection in an organ specific and systemic fashion.
Selected blood biomarkers for site of infection are present in clinical patients.
Acknowledgements
This work is supported by the Ray Thomas Edwards Foundation and the University of California Office of the President (DJG), the National Institutes of Health (R01EY025947) (LZ), the Defense Threat Reduction Agency Joint Science and Technology Office for Chemical and Biological Defense (HDTRA1-14-1-0064) (LZ). JDL is an IRACDA fellow supported by NIGMS/NIH (K12GM068524). MR is supported by Public Health Service Grant AI126277, AI126465, AI114625, and AI121928 and by the Chiba University-UC San Diego Center for Mucosal Immunology, Allergy, and Vaccines. MR also holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Funding for collection of patient samples was provided by the Center for Translation Molecular Medicine (http://www.ctmm.nl), project MARS (grant 041-201). We acknowledge Dr. Kelly Doran for contributing Serotype III GBS COH1.
Footnotes
Declaration of Interests
All authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List:
- ABDILLAHI SM, BALVANOVIC S, BAUMGARTEN M & MORGELIN M (2012). Collagen VI encodes antimicrobial activity: novel innate host defense properties of the extracellular matrix. J Innate Immun, 4, 371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AMMANN P, FEHR T, MINDER EI, GUNTER C & BERTEL O (2001). Elevation of troponin I in sepsis and septic shock. Intensive Care Med, 27, 965–9. [DOI] [PubMed] [Google Scholar]
- AQUILANO K, BALDELLI S & CIRIOLO MR (2014). Glutathione: new roles in redox signaling for an old antioxidant. Front Pharmacol, 5, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHBAUGH CD, MOSER TJ, SHEARER MH, WHITE GL, KENNEDY RC & WESSELS MR (2000). Bacterial determinants of persistent throat colonization and the associated immune response in a primate model of human group A streptococcal pharyngeal infection. Cell Microbiol, 2, 283–92. [DOI] [PubMed] [Google Scholar]
- BABUIN L & JAFFE AS (2005). Troponin: the biomarker of choice for the detection of cardiac injury. CMAJ, 173, 1191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANG ML, CAREMANI M, BRUNELLO E, LITTLEFIELD R, LIEBER RL, CHEN J, LOMBARDI V & LINARI M (2009). Nebulin plays a direct role in promoting strong actin-myosin interactions. FASEB J, 23, 4117–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANKS RE, DUNN MJ, FORBES MA, STANLEY A, PAPPIN D, NAVEN T, GOUGH M, HARNDEN P & SELBY PJ (1999). The potential use of laser capture microdissection to selectively obtain distinct populations of cells for proteomic analysis--preliminary findings. Electrophoresis, 20, 689–700. [DOI] [PubMed] [Google Scholar]
- BARROSO-SOUSA R, LOBO RR, MENDONCA PR, MEMORIA RR, SPILLER F, CUNHA FQ & PAZIN-FILHO A (2013). Decreased levels of alpha-1-acid glycoprotein are related to the mortality of septic patients in the emergency department. Clinics (Sao Paulo), 68, 1134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTOSIEWICZ MJ, JENKINS D, PENN S, EMERY J & BUCKPITT A (2001). Unique gene expression patterns in liver and kidney associated with exposure to chemical toxicants. J Pharmacol Exp Ther, 297, 895–905. [PubMed] [Google Scholar]
- BEAUSOLEIL SA, VILLEN J, GERBER SA, RUSH J & GYGI SP (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol, 24, 1285–92. [DOI] [PubMed] [Google Scholar]
- BESSEN DE & LIZANO S (2010). Tissue tropisms in group A streptococcal infections. Future Microbiol, 5, 623–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENOT A, KING KY, JANOWIAK B, GRIFFITH O & CAPARON MG (2004). Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect Immun, 72, 408–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUTSCHKAU A, NAGEL P, GRAMBOW E, ZECHNER D, BROZE GJ JR. & VOLLMAR B (2013). Contribution of protein Z and protein Z-dependent protease inhibitor in generalized Shwartzman reaction. Crit Care Med, 41, e447–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARAPETIS JR, STEER AC, MULHOLLAND EK & WEBER M (2005). The global burden of group A streptococcal diseases. Lancet Infect Dis, 5, 685–94. [DOI] [PubMed] [Google Scholar]
- CAVUS U, COSKUN F, YAVUZ B, CIFTCI O, SAHINER L, AKSOY H, DENIZ A, OZAKIN E, AYTEMIR K, TOKGOZOGLU L & KABAKCI G (2006). Heart-type, fatty-acid binding protein can be a diagnostic marker in acute coronary syndromes. J Natl Med Assoc, 98, 1067–70. [PMC free article] [PubMed] [Google Scholar]
- CHATELLIER S, IHENDYANE N, KANSAL RG, KHAMBATY F, BASMA H, NORRBY-TEGLUND A, LOW DE, MCGEER A & KOTB M (2000). Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun, 68, 3523–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUNNINGHAM MW (2000). Pathogenesis of group A streptococcal infections. Clin Microbiol Rev, 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANEMAN R, ZHOU L, AGALLIU D, CAHOY JD, KAUSHAL A & BARRES BA (2010). The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS One, 5, e13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIES MR, HOLDEN MT, COUPLAND P, CHEN JH, VENTURINI C, BARNETT TC, ZAKOUR NL, TSE H, DOUGAN G, YUEN KY & WALKER MJ (2015). Emergence of scarlet fever Streptococcus pyogenes emm12 clones in Hong Kong is associated with toxin acquisition and multidrug resistance. Nat Genet, 47, 84–7. [DOI] [PubMed] [Google Scholar]
- DE PAULA RR, MARINHO FV, FAHEL JS & OLIVEIRA SC (2017). Contribution of intercellular adhesion molecule 1 (ICAM-1) to control Mycobacterium avium infection. Microbes Infect, 19, 527–535. [DOI] [PubMed] [Google Scholar]
- DILEEPAN T, SMITH ED, KNOWLAND D, HSU M, PLATT M, BITTNER-EDDY P, COHEN B, SOUTHERN P, LATIMER E, HARLEY E, AGALLIU D & CLEARY PP (2016). Group A Streptococcus intranasal infection promotes CNS infiltration by streptococcal-specific Th17 cells. J Clin Invest, 126, 303–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DINKLA K, ROHDE M, JANSEN WT, CARAPETIS JR, CHHATWAL GS & TALAY SR (2003a). Streptococcus pyogenes recruits collagen via surface-bound fibronectin: a novel colonization and immune evasion mechanism. Mol Microbiol, 47, 861–9. [DOI] [PubMed] [Google Scholar]
- DINKLA K, ROHDE M, JANSEN WT, KAPLAN EL, CHHATWAL GS & TALAY SR (2003b). Rheumatic fever-associated Streptococcus pyogenes isolates aggregate collagen. J Clin Invest, 111, 1905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DMITRIEV AV & CHAUSSEE MS (2010). The Streptococcus pyogenes proteome: maps, virulence factors and vaccine candidates. Future Microbiol, 5, 1539–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EATON SL, ROCHE SL, LLAVERO HURTADO M, OLDKNOW KJ, FARQUHARSON C, GILLINGWATER TH & WISHART TM (2013). Total protein analysis as a reliable loading control for quantitative fluorescent Western blotting. PLoS One, 8, e72457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELIAS JE & GYGI SP (2007). Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods, 4, 207–214. [DOI] [PubMed] [Google Scholar]
- ELIAS JE, HAAS W, FAHERTY BK & GYGI SP (2005). Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods, 2, 667–75. [DOI] [PubMed] [Google Scholar]
- ELLIOTT PJ, ZOLLNER TM & BOEHNCKE WH (2003). Proteasome inhibition: a new anti-inflammatory strategy. J Mol Med (Berl), 81, 235–45. [DOI] [PubMed] [Google Scholar]
- ENG JK, MCCORMACK AL & YATES JR (1994). An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom, 5, 976–89. [DOI] [PubMed] [Google Scholar]
- ERICKSON BK, ROSE CM, BRAUN CR, ERICKSON AR, KNOTT J, MCALISTER GC, WUHR M, PAULO JA, EVERLEY RA & GYGI SP (2017). A Strategy to Combine Sample Multiplexing with Targeted Proteomics Assays for High-Throughput Protein Signature Characterization. Mol Cell, 65, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANCO LH, NAIR VR, SCHARN CR, XAVIER RJ, TORREALBA JR, SHILOH MU & LEVINE B (2017). The Ubiquitin Ligase Smurf1 Functions in Selective Autophagy of Mycobacterium tuberculosis and Anti-tuberculous Host Defense. Cell Host Microbe, 21, 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FROST RA & LANG CH (2011). mTor signaling in skeletal muscle during sepsis and inflammation: where does it all go wrong? Physiology (Bethesda), 26, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GABAY C & KUSHNER I (1999). Acute-phase proteins and other systemic responses to inflammation. N Engl J Med, 340, 448–54. [DOI] [PubMed] [Google Scholar]
- GIBSON CM, MALLETT TC, CLAIBORNE A & CAPARON MG (2000). Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J Bacteriol, 182, 448–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLDBERG AL & DICE JF (1974). Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem, 43, 835–69. [DOI] [PubMed] [Google Scholar]
- GOLDMANN O, ROHDE M, CHHATWAL GS & MEDINA E (2004). Role of macrophages in host resistance to group A streptococci. Infect Immun, 72, 2956–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREEN J 1948. Production of streptokinase and haemolysin by haemolytic streptococci. Biochem J, 43, xxxii. [PubMed] [Google Scholar]
- HAAS W, FAHERTY BK, GERBER SA, ELIAS JE, BEAUSOLEIL SA, BAKALARSKI CE, LI X, VILLEN J & GYGI SP (2006). Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics, 5, 1326–37. [DOI] [PubMed] [Google Scholar]
- HERSHBERGER RE, LINDENFELD J, MESTRONI L, SEIDMAN CE, TAYLOR MR, TOWBIN JA & HEART FAILURE SOCIETY OF, A. (2009). Genetic evaluation of cardiomyopathy--a Heart Failure Society of America practice guideline. J Card Fail, 15, 83–97. [DOI] [PubMed] [Google Scholar]
- HSIEH YC & HUANG YC (2011). Scarlet fever outbreak in Hong Kong, 2011. J Microbiol Immunol Infect, 44, 409–11. [DOI] [PubMed] [Google Scholar]
- HUANG DA W, SHERMAN BT & LEMPICKI RA (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG DA W, SHERMAN BT & LEMPICKI RA (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc, 4, 44–57. [DOI] [PubMed] [Google Scholar]
- HUTTLIN EL, JEDRYCHOWSKI MP, ELIAS JE, GOSWAMI T, RAD R, BEAUSOLEIL SA, VILLEN J, HAAS W, SOWA ME & GYGI SP (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell, 143, 1174–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HWANG CS, SHEMORRY A & VARSHAVSKY A (2010). N-terminal acetylation of cellular proteins creates specific degradation signals. Science, 327, 973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KALL L, CANTERBURY JD, WESTON J, NOBLE WS & MACCOSS MJ (2007). Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods, 4, 923–5. [DOI] [PubMed] [Google Scholar]
- KERSTEN S (2014). Integrated physiology and systems biology of PPARalpha. Mol Metab, 3, 354–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHAVANDI A, WHITAKER J, ELKINGTON A, PROBERT J & WALKER PR (2008). Acute streptococcal myopericarditis mimicking myocardial infarction. Am J Emerg Med, 26, 638 e1–2. [DOI] [PubMed] [Google Scholar]
- KILSGARD O, KARLSSON C, MALMSTROM E & MALMSTROM J (2016). Differential compartmentalization of Streptococcus pyogenes virulence factors and host protein binding properties as a mechanism for host adaptation. Int J Med Microbiol, 306, 504–516. [DOI] [PubMed] [Google Scholar]
- KIRVAN CA, SWEDO SE, HEUSER JS & CUNNINGHAM MW (2003). Mimicry and autoantibody-mediated neuronal cell signaling in Sydenham chorea. Nat Med, 9, 914–20. [DOI] [PubMed] [Google Scholar]
- KLEIN KLOUWENBERG PM, ONG DS, BOS LD, DE BEER FM, VAN HOOIJDONK RT, HUSON MA, STRAAT M, VAN VUGHT LA, WIESKE L, HORN J, SCHULTZ MJ, VAN DER POLL T, BONTEN MJ & CREMER OL (2013). Interobserver agreement of Centers for Disease Control and Prevention criteria for classifying infections in critically ill patients. Crit Care Med, 41, 2373–8. [DOI] [PubMed] [Google Scholar]
- KURLAN R, JOHNSON D, KAPLAN EL & TOURETTE SYNDROME STUDY G (2008). Streptococcal infection and exacerbations of childhood tics and obsessive-compulsive symptoms: a prospective blinded cohort study. Pediatrics, 121, 1188–97. [DOI] [PubMed] [Google Scholar]
- KYME P, THOENNISSEN NH, TSENG CW, THOENNISSEN GB, WOLF AJ, SHIMADA K, KRUG UO, LEE K, MULLER-TIDOW C, BERDEL WE, HARDY WD, GOMBART AF, KOEFFLER HP & LIU GY (2012). C/EBPepsilon mediates nicotinamide-enhanced clearance of Staphylococcus aureus in mice. J Clin Invest, 122, 3316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAPEK JD, GRENINGER P, MORRIS R, AMZALLAG A, PRUTEANU-MALINICI I, BENES CH & HAAS W (2017a). Detection of Dysregulated Protein Assoiciation Networks by High-Throughput Proteomics Predicts Cancer Vulnerabilities. Nat Biotechnol, In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAPEK JD JR., LEWINSKI MK, WOZNIAK JM, GUATELLI J & GONZALEZ DJ (2017b). Quantitative Temporal Viromics of an Inducible HIV-1 Model Yields Insight to Global Host Targets and Phospho-Dynamics Associated with Vpr. Mol Cell Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAU EH, NISHIURA H, COWLING BJ, IP DK & WU JT (2012). Scarlet fever outbreak, Hong Kong, 2011. Emerg Infect Dis, 18, 1700–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI P, SHI J, HE Q, HU Q, WANG YY, ZHANG LJ, CHAN WT & CHEN WX (2015). Streptococcus pneumoniae induces autophagy through the inhibition of the PI3K-I/Akt/mTOR pathway and ROS hypergeneration in A549 cells. PLoS One, 10, e0122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN AE, BEASLEY FC, KELLER N, HOLLANDS A, URBANO R, TROEMEL ER, HOFFMAN HM & NIZET V (2015). A group A Streptococcus ADP-ribosyltransferase toxin stimulates a protective interleukin 1beta-dependent macrophage immune response. MBio, 6, e00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU Y, BUIL A, COLLINS BC, GILLET LC, BLUM LC, CHENG LY, VITEK O, MOURITSEN J, LACHANCE G, SPECTOR TD, DERMITZAKIS ET & AEBERSOLD R (2015). Quantitative variability of 342 plasma proteins in a human twin population. Mol Syst Biol, 11, 786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOOF TG, DEICKE C & MEDINA E (2014). The role of coagulation/fibrinolysis during Streptococcus pyogenes infection. Front Cell Infect Microbiol, 4, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAEDA T, WAKASAWA T, FUNABASHI M, FUKUSHI A, FUJITA M, MOTOJIMA K & TAMAI I (2008). Regulation of Octn2 transporter (SLC22A5) by peroxisome proliferator activated receptor alpha. Biol Pharm Bull, 31, 1230–6. [DOI] [PubMed] [Google Scholar]
- MAJAI G, SARANG Z, CSOMOS K, ZAHUCZKY G & FESUS L (2007). PPARgamma-dependent regulation of human macrophages in phagocytosis of apoptotic cells. Eur J Immunol, 37, 1343–54. [DOI] [PubMed] [Google Scholar]
- MALMSTROM E, KILSGARD O, HAURI S, SMEDS E, HERWALD H, MALMSTROM L & MALMSTROM J (2016). Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics. Nat Commun, 7, 10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MCALISTER GC, NUSINOW DP, JEDRYCHOWSKI MP, WUHR M, HUTTLIN EL, ERICKSON BK, RAD R, HAAS W & GYGI SP (2014). MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem, 86, 7150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER LL, BLY CG, WATSON ML & BALE WF (1951). The dominant role of the liver in plasma protein synthesis; a direct study of the isolated perfused rat liver with the aid of lysine-epsilon-C14. J Exp Med, 94, 431–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORGAN D, CAPASSO M, MUSSET B, CHERNY VV, RIOS E, DYER MJ & DECOURSEY TE (2009). Voltage-gated proton channels maintain pH in human neutrophils during phagocytosis. Proc Natl Acad Sci U S A, 106, 18022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORGAN ET (1997). Regulation of cytochromes P450 during inflammation and infection. Drug Metab Rev, 29, 1129–88. [DOI] [PubMed] [Google Scholar]
- NAKAGAWA I, AMANO A, MIZUSHIMA N, YAMAMOTO A, YAMAGUCHI H, KAMIMOTO T, NARA A, FUNAO J, NAKATA M, TSUDA K, HAMADA S & YOSHIMORI T (2004). Autophagy defends cells against invading group A Streptococcus. Science, 306, 1037–40. [DOI] [PubMed] [Google Scholar]
- NICOLAY BN, DANIELIAN PS, KOTTAKIS F, LAPEK JD JR., SANIDAS I, MILES WO, DEHNAD M, TSCHOP K, GIERUT JJ, MANNING AL, MORRIS R, HAIGIS K, BARDEESY N, LEES JA, HAAS W & DYSON NJ (2015). Proteomic analysis of pRb loss highlights a signature of decreased mitochondrial oxidative phosphorylation. Genes Dev, 29, 1875–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIZET V (2015). Stopping superbugs, maintaining the microbiota. Sci Transl Med, 7, 295ed8. [DOI] [PubMed] [Google Scholar]
- NYAGODE BA, WILLIAMS IR & MORGAN ET (2014). Altered inflammatory responses to Citrobacter rodentium infection, but not bacterial lipopolysaccharide, in mice lacking the Cyp4a10 or Cyp4a14 genes. Inflammation, 37, 893–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ODA M, DOMON H, KUROSAWA M, ISONO T, MAEKAWA T, YAMAGUCHI M, KAWABATA S & TERAO Y (2017). Streptococcus pyogenes Phospholipase A2 Induces the Expression of Adhesion Molecules on Human Umbilical Vein Endothelial Cells and Aorta of Mice. Front Cell Infect Microbiol, 7, 300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENG J, ELIAS JE, THOREEN CC, LICKLIDER LJ & GYGI SP (2003). Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res, 2, 43–50. [DOI] [PubMed] [Google Scholar]
- PETERSEN TN, BRUNAK S, VON HEIJNE G & NIELSEN H (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods, 8, 785–6. [DOI] [PubMed] [Google Scholar]
- PIEPMEIER E JR., HAMMETT-STABLER C, PRICE M, PETERS J, KEMPER G & DAVIS M JR. (1995). Myositis and fasciitis associated with group A beta hemolytic streptococcal infections: development of a rabbit model. J Lab Clin Med, 126, 137–43. [PubMed] [Google Scholar]
- PONTEN F, GRY M, FAGERBERG L, LUNDBERG E, ASPLUND A, BERGLUND L, OKSVOLD P, BJORLING E, HOBER S, KAMPF C, NAVANI S, NILSSON P, OTTOSSON J, PERSSON A, WERNERUS H, WESTER K & UHLEN M (2009). A global view of protein expression in human cells, tissues, and organs. Mol Syst Biol, 5, 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAJU MS, V J, KAMARAJU RS, SRITHARAN V, RAJKUMAR K, NATARAJAN S, KUMAR AD & BURGULA S (2016). Continuous evaluation of changes in the serum proteome from early to late stages of sepsis caused by Klebsiella pneumoniae. Mol Med Rep, 13, 4835–44. [DOI] [PubMed] [Google Scholar]
- RIVERA-HERNANDEZ T, CARNATHAN DG, MOYLE PM, TOTH I, WEST NP, YOUNG PR, SILVESTRI G & WALKER MJ (2014). The contribution of nonhuman primate models to the development of human vaccines. Discov Med, 18, 313–22. [PMC free article] [PubMed] [Google Scholar]
- ROBERTS S, SCOTT JR, HUSMANN LK & ZURAWSKI CA (2006). Murine models of Streptococcus pyogenes infection. Curr Protoc Microbiol, Chapter 9, Unit 9D 5. [DOI] [PubMed] [Google Scholar]
- RODRIGUEZ-ITURBE B & HAAS M (2016). Post-Streptococcal Glomerulonephritis. In: FERRETTI JJ, STEVENS DL & FISCHETTI VA (eds.) Streptococcus pyogenes : Basic Biology to Clinical Manifestations; Oklahoma City (OK). [Google Scholar]
- ROUSSEEUW PJ, RUTS I & TUKEY JW (1999). The bagplot: A bivariate boxplot. American Statistician, 53, 382–387. [Google Scholar]
- RUSH CM, GOVAN BL, SIKDER S, WILLIAMS NL & KETHEESAN N (2014). Animal models to investigate the pathogenesis of rheumatic heart disease. Front Pediatr, 2, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAKURAI A, OKAHASHI N, NAKAGAWA I, KAWABATA S, AMANO A, OOSHIMA T & HAMADA S (2003). Streptococcus pyogenes infection induces septic arthritis with increased production of the receptor activator of the NF-kappaB ligand. Infect Immun, 71, 6019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALLACH SM, NOWAK R, HUDSON MP, TOKARSKI G, KHOURY N, TOMLANOVICH MC, JACOBSEN G, DE LEMOS JA & MCCORD J (2004). A change in serum myoglobin to detect acute myocardial infarction in patients with normal troponin I levels. Am J Cardiol, 94, 864–7. [DOI] [PubMed] [Google Scholar]
- SCHREIER T, KEDES L & GAHLMANN R (1990). Cloning, structural analysis, and expression of the human slow twitch skeletal muscle/cardiac troponin C gene. J Biol Chem, 265, 21247–53. [PubMed] [Google Scholar]
- SEMANJSKI M & MACEK B (2016). Shotgun proteomics of bacterial pathogens: advances, challenges and clinical implications. Expert Rev Proteomics, 13, 139–56. [DOI] [PubMed] [Google Scholar]
- SERHAN CN, CHIANG N & VAN DYKE TE (2008). Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol, 8, 349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SESSLER CN, WINDSOR AC, SCHWARTZ M, WATSON L, FISHER BJ, SUGERMAN HJ & FOWLER AA 3RD (1995). Circulating ICAM-1 is increased in septic shock. Am J Respir Crit Care Med, 151, 1420–7. [DOI] [PubMed] [Google Scholar]
- SHANNON O, RYDENGARD V, SCHMIDTCHEN A, MORGELIN M, ALM P, SORENSEN OE & BJORCK L( 2010). Histidine-rich glycoprotein promotes bacterial entrapment in clots and decreases mortality in a mouse model of sepsis. Blood, 116, 2365–72. [DOI] [PubMed] [Google Scholar]
- SJOGREN J, STRUWE WB, COSGRAVE EF, RUDD PM, STERVANDER M, ALLHORN M, HOLLANDS A, NIZET V & COLLIN M (2013). EndoS2 is a unique and conserved enzyme of serotype M49 group A Streptococcus that hydrolyses N-linked glycans on IgG and alpha1-acid glycoprotein. Biochem J, 455, 107–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPIVAK M, WESTON J, BOTTOU L, KALL L & NOBLE WS (2009). Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J Proteome Res, 8, 3737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANDAGE SW & WONG HR (2011). Biomarkers for pediatric sepsis and septic shock. Expert Rev Anti Infect Ther, 9, 71–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STOJILJKOVIC I, BAUMLER AJ & HEFFRON F (1995). Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol, 177, 1357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SZKLARCZYK D, FRANCESCHINI A, WYDER S, FORSLUND K, HELLER D, HUERTA-CEPAS J, SIMONOVIC M, ROTH A, SANTOS A, TSAFOU KP, KUHN M, BORK P, JENSEN LJ & VON MERING C (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res, 43, D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TAKEDA S, YAMASHITA A, MAEDA K & MAEDA Y (2003). Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature, 424, 35–41. [DOI] [PubMed] [Google Scholar]
- THOMPSON A, SCHAFER J, KUHN K, KIENLE S, SCHWARZ J, SCHMIDT G, NEUMANN T, JOHNSTONE R, MOHAMMED AK & HAMON C (2003). Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem, 75, 1895–904. [DOI] [PubMed] [Google Scholar]
- TOLONEN AC & HAAS W (2014). Quantitative proteomics using reductive dimethylation for stable isotope labeling. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TROJANOWSKA M, LEROY EC, ECKES B & KRIEG T (1998). Pathogenesis of fibrosis: type 1 collagen and the skin. J Mol Med (Berl), 76, 266–74. [DOI] [PubMed] [Google Scholar]
- TSATSARONIS JA, WALKER MJ & SANDERSON-SMITH ML (2014). Host responses to group a streptococcus: cell death and inflammation. PLoS Pathog, 10, e1004266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN SORGE NM & DORAN KS (2012). Defense at the border: the blood-brain barrier versus bacterial foreigners. Future Microbiol, 7, 383–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN SORGE NM, QUACH D, GURNEY MA, SULLAM PM, NIZET V & DORAN KS (2009). The group B streptococcal serine-rich repeat 1 glycoprotein mediates penetration of the blood-brain barrier. J Infect Dis, 199, 1479–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VILLEN J & GYGI SP (2008). The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc, 3, 1630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WAGNER C, SAUERMANN R & JOUKHADAR C (2006). Principles of antibiotic penetration into abscess fluid. Pharmacology, 78, 1–10. [DOI] [PubMed] [Google Scholar]
- WALKER MJ, BARNETT TC, MCARTHUR JD, COLE JN, GILLEN CM, HENNINGHAM A, SRIPRAKASH KS, SANDERSON-SMITH ML & NIZET V (2014). Disease manifestations and pathogenic mechanisms of Group A Streptococcus. Clin Microbiol Rev, 27, 264–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WALLGREN-PETTERSSON C, DONNER K, SEWRY C, BIJLSMA E, LAMMENS M, BUSHBY K, GIOVANNUCCI UZIELLI ML, LAPI E, ODENT S, AKCOREN Z, TOPALOGLU H & PELIN K (2002). Mutations in the nebulin gene can cause severe congenital nemaline myopathy. Neuromuscul Disord, 12, 674–9. [DOI] [PubMed] [Google Scholar]
- WALLGREN-PETTERSSON C, PELIN K, HILPELA P, DONNER K, PORFIRIO B, GRAZIANO C, SWOBODA KJ, FARDEAU M, URTIZBEREA JA, MUNTONI F, SEWRY C, DUBOWITZ V, IANNACCONE S, MINETTI C, PEDEMONTE M, SERI M, CUSANO R, LAMMENS M, CASTAGNA-SLOANE A, BEGGS AH, LAING NG & DE LA CHAPELLE A (1999). Clinical and genetic heterogeneity in autosomal recessive nemaline myopathy. Neuromuscul Disord, 9, 564–72. [DOI] [PubMed] [Google Scholar]
- WANG Y, YANG F, GRITSENKO MA, WANG Y, CLAUSS T, LIU T, SHEN Y, MONROE ME, LOPEZ-FERRER D, RENO T, MOORE RJ, KLEMKE RL, CAMP DG 2ND & SMITH RD (2011). Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics, 11, 2019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG ZJ, LIANG CL, LI GM, YU CY & YIN M (2006). Neuroprotective effects of arachidonic acid against oxidative stress on rat hippocampal slices. Chem Biol Interact, 163, 207–17. [DOI] [PubMed] [Google Scholar]
- WATANABE K, ISHIMA Y, AKAIKE T, SAWA T, KURODA T, OGAWA W, WATANABE H, SUENAGA A, KAI T, OTAGIRI M & MARUYAMA T (2013). S-nitrosated alpha-1-acid glycoprotein kills drug-resistant bacteria and aids survival in sepsis. FASEB J, 27, 391–8. [DOI] [PubMed] [Google Scholar]
- WATKIN RW, LANG S, SMITH JM, ELLIOTT TS & LITTLER WA (2004). Role of troponin I in active infective endocarditis. Am J Cardiol, 94, 1198–9. [DOI] [PubMed] [Google Scholar]
- WATSON ME NM JR., CAPARON MG. (2016). Animal Models of Streptococcus pyogenes Infection. In: FERRETTI JJ, S. D., FISCHETTI VA, EDITORS (ed.) Streptococcus pyogenes : Basic Biology to Clinical Manifestations 2016 Feb 10 ed. Internet: University of Oklahoma Health Sciences Center. [Google Scholar]
- WESSEL D & FLUGGE UI (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem, 138, 141–3. [DOI] [PubMed] [Google Scholar]
- WILSON CB & WEAVER WM (1985). Comparative susceptibility of group B streptococci and Staphylococcus aureus to killing by oxygen metabolites. J Infect Dis, 152, 323–9. [DOI] [PubMed] [Google Scholar]
- XIAO K, SU L, YAN P, HAN B, LI J, WANG H, JIA Y, LI X & XIE L (2015). alpha-1-Acid glycoprotein as a biomarker for the early diagnosis and monitoring the prognosis of sepsis. J Crit Care, 30, 744–51. [DOI] [PubMed] [Google Scholar]
- XIAO Y, HSIAO TH, SURESH U, CHEN HI, WU X, WOLF SE & CHEN Y (2014). A novel significance score for gene selection and ranking. Bioinformatics, 30, 801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG P, PENG X, ZHANG D, WU S, LIU Y, CUI S, LU G, DUAN W, SHI W, LIU S, LI J & WANG Q (2013). Characteristics of group A Streptococcus strains circulating during scarlet fever epidemic, Beijing, China, 2011. Emerg Infect Dis, 19, 909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZODRO E, JAROSZEWSKI M, IDA A, WRZESINSKI T, KWIAS Z, BLUYSSEN H & WESOLY J (2014). FUT11 as a potential biomarker of clear cell renal cell carcinoma progression based on meta-analysis of gene expression data. Tumour Biol, 35, 2607–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.