Abstract
Trafficking protein particle (TRAPP) complexes, which include the TRAPPC4 protein, regulate membrane trafficking between lipid organelles in a process termed vesicular tethering. TRAPPC4 was recently implicated in a recessive neurodevelopmental condition in four unrelated families due to a shared c.454+3A>G splice variant. Here, we report 23 patients from 17 independent families with an early-infantile-onset neurodegenerative presentation, where we also identified the homozygous variant hg38:11:119020256 A>G (NM_016146.5:c.454+3A>G) in TRAPPC4 through exome or genome sequencing. No other clinically relevant TRAPPC4 variants were identified among any of over 10,000 patients with neurodevelopmental conditions. We found the carrier frequency of TRAPPC4 c.454+3A>G was 2.4–5.4 per 10,000 healthy individuals. Affected individuals with the homozygous TRAPPC4 c.454+3A>G variant showed profound psychomotor delay, developmental regression, early-onset epilepsy, microcephaly and progressive spastic tetraplegia. Based upon RNA sequencing, the variant resulted in partial exon 3 skipping and generation of an aberrant transcript owing to use of a downstream cryptic splice donor site, predicting a premature stop codon and nonsense mediated decay. These data confirm the pathogenicity of the TRAPPC4 c.454+3A>G variant, and refine the clinical presentation of TRAPPC4-related encephalopathy.
Subject terms: Paediatric neurological disorders, Disease genetics
Introduction
Trafficking protein particle (TRAPP) complexes play a vital role in regulating membrane trafficking from the ER to the Golgi and plasma membrane. There are two TRAPP complexes identified in mammalian cells, TRAPPII and TRAPPIII, sharing core subunits TRAPPC1, TRAPPC2, TRAPPC3, TRAPPC4, TRAPPC5, TRAPPC6, and TRAPPC2L, with additional unique subunits specific to each of the complexes [1]. Biallelic variants in genes encoding human TRAPP subunits cause a range of disorders known as TRAPPopathies, including encephalopathy (TRAPPC2L, MIM:610970; TRAPPC6B, MIM:610397; TRAPPC12, MIM:614139), neurodevelopmental disorders (TRAPP6A, MIM:610396; TRAPPC9, MIM:611966), encephalomyopathy (TRAPPC11, MIM: 614138) and spondyloepiphyseal dysplasia tarda (TRAPPC2, MIM:300202) [2–6]. However, the phenotypic heterogeneity remains poorly understood.
Very recently, a homozygous, variant in a well-conserved splice donor site (hg38:11:119020256 A > G (NM_016146.5:c.454+3A>G) within TRAPPC4 (trafficking protein particle complex, subunit C4, (MIM:610971)) was identified in eight affected individuals from four independent families presenting a neurodevelopmental disorder characterized by severe-to-profound developmental delay/intellectual disability, microcephaly, early-onset seizures, and spastic quadriparesis [7, 8]. The TRAPPC4-related phenotype acronym is known as NEDESBA (MIM:618741), which stands for NEurodevelopmental Disorder with Epilepsy, Spasticity, and Brain Atrophy. Here, we report 23 additional cases from 17 independent families with an early-infantile-onset neurodegeneration harboring the same homozygous TRAPPC4 splice site variant, recurring in all the families from different ethnicities, and resulting in decreased TRAPPC4 expression due to aberrant splicing.
Materials and methods
Participant recruitment and sequencing
The institutional review boards of the University of California, San Diego, as well as the respective host institution approved this study. All study participants and/or their parents or guardians signed informed consent forms allowing for participation and to publishing of photographs and other identifying information. Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp DNA blood Midi kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. For families 1–6, 16, and 17, exome sequencing and data analysis were performed as previously described [9]. For families 7–10, exome sequencing and analysis were conducted at CENTOGENE AG and for families 12–15, exome sequencing and analysis were conducted at GeneDx [10, 11]. For family 15, genome sequencing was performed through the Undiagnosed Diseases Network. In families 11 and 12, trio genome sequencing and proband-only RNA sequencing were performed. Sequencing for the remaining families was performed at the Broad Institute (Cambridge, MA, USA). The candidate variant was confirmed, and segregation analysis was performed by Sanger sequencing. Further details are available in the Supplementary Material.
RNA preparation and short read RNA sequencing
RNA was isolated from cultured fibroblasts obtained from a skin biopsy from patient 15, in addition to 24 unrelated controls. RNA sequencing was performed at the Broad Institute (Cambridge, MA, USA). Libraries were prepared using a TruSeq Stranded mRNA Library Prep Kit (Cat. No. RS-122-2101, RS-122-2102, and RS-122-2103). Paired-end sequencing was performed using a read length of up to 100 bp on an Illumina HiSeq4000 instrument to achieve a minimum sequencing depth of 50 million paired-end reads.
RNA sequencing reads were processed using a Bpipe (Version 0.9.9.6, release 21/07/2018) pipeline for quality control checks, trimming, and alignment [12]. FastQC and Trimmomatic were used for sequencing quality checks and trimming of poor-quality reads [13]. Alignment was performed using STAR (version 2.7.3a, release 08 October 2019), in two-pass mode for read alignment to the Human Reference Genome Build 38 (excluding “ALT” contigs) [14]. Duplicate reads were marked with Picard MarkDuplicates and quantification was performed using FeatureCounts from the R Subread package Version 1.34.7, release 03/01/2019) [15]. Differential expression analysis was performed using the DESeq2 package (version 1.25.9, release 31 July 2019) comparing expression of the affected individual to 24 unrelated control samples [16]. Sashimi plots were prepared using ggsashimi [17].
Results
We report a cohort of 23 patients from 17 families with a TRAPPC4-related neurodevelopmental disorder (Fig. 1a). The families are of different ancestries (Iranian, Egyptian, Portuguese, English, mixed European-American) and were identified through collaboration among the involved study centers.
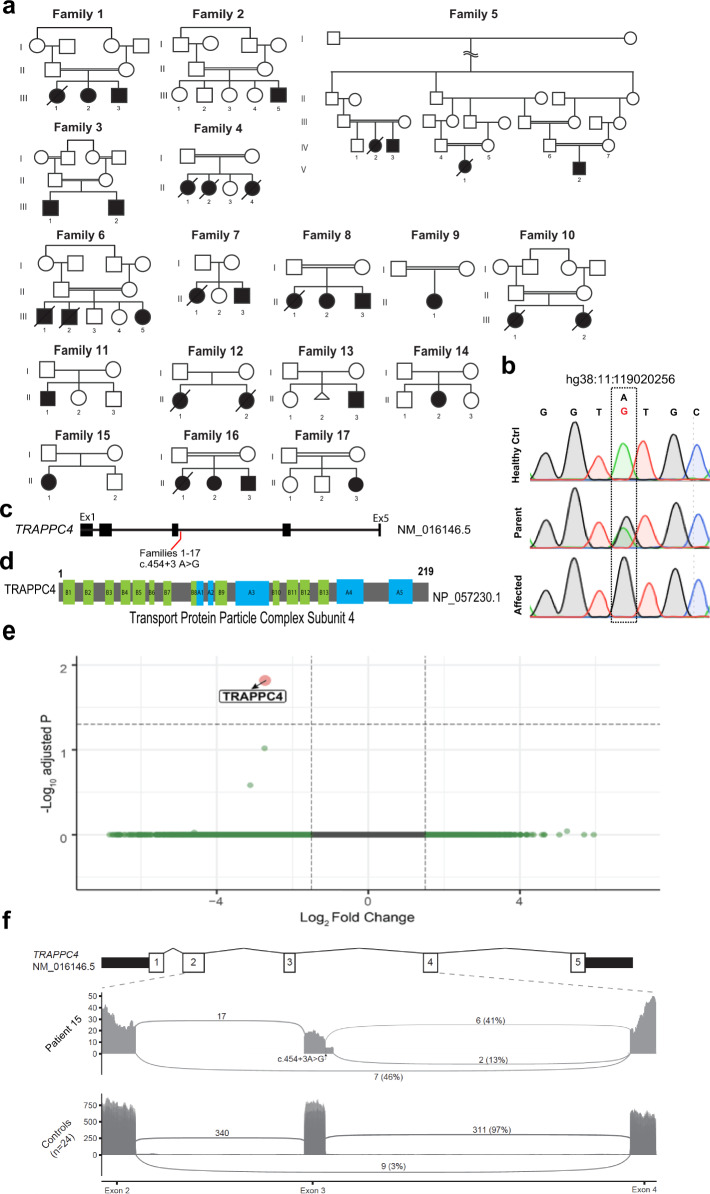
Fig. 1. Pedigrees, chromatogram, TRAPPC4 gene and protein, and mRNA analysis.
a Pedigrees of the 17 families, including an extended Iranian family composed of three smaller distantly related subfamilies (family 5). Double bar: consanguinity. Slash: deceased. b Sanger sequence chromatograms with the c.454+3A>G variant (NM_016146.5) in a healthy control (Healthy Ctrl), heterozygous carrier (Father) and patient from family 5 (Affected). c Schematic of NM_016146.5 TRAPPC4 transcript with five predicted exons and the c.454+3A>G variant (red) identified in the 17 families. Exons were numbered using the UC Santa Cruz (UCSC) Genome Browser as a reference. d TRAPPC4 219 amino-acid length protein (NP_057230.1) with predicted beta strands (B1–B13) and alpha helices (A1–A5). e Volcano plot illustrating total RNA transcript level of all transcripts mapped to the reference genome for fold change vs. adjusted p value of RNA sequencing from fibroblasts derived from Patient 15 compared with 24 control fibroblasts lines. TRAPPC4 (red dot) was the only gene significantly differentially expressed with a log2-fold change between 2 and 4 and a minus-log10 adjusted P value above genome-wide significance (gray dashed line) between 1 and 2. f Sashimi plot depicting individual reads from patient 15 (top) compared with sum of 24 controls (bottom) mapped to the reference genome from exon 2 to exon 4 of TRAPPC4. Site of location is shown with arrow. In control, splicing from exon 2 to 4 occurs in only 3% of reads in control but 46% of reads in patient 15. A novel splice donor site 40 bp downstream of the canonical site in exon 3 was seen in 13% of reads in patient 15, and not seen in control. Splicing from the canonical exon 3 donor site was seen in 97% of transcripts in controls but 41% in patient 15.
Identification of a homozygous TRAPPC4 c.454+3A>G variant
Among a collective cohort of over 10,000 patients with mostly recessive neurodevelopmental disease from the University of California, San Diego and University College London cohorts, ~50% remain unsolved after whole-exome sequencing. Among these, there were 23 patients from 17 families where both university groups independently identified the homozygous c.454+3A>G variant (hg38:11:119020256 A > G (NM_016146.5, rs375776811) in intron 3 of TRAPPC4, among a search for potential variants affecting splicing (Fig. 1b–d). TRAPPC4 encodes for Trafficking Protein Particle Complex Subunit 4 (TRAPPC4). No other clinically relevant variants in TRAPPC4 were identified in any of the remaining unsolved families. This variant had been initially filtered out from exome analysis because the allele frequency (AF) rose above our 0.02% AF-cutoff. It was encountered as homozygous only in affected members, and not in any unaffected members, and segregated with the phenotype consistent with a recessive mode of inheritance with full penetrance in each of the 17 families. Comparison of the exome calls among the probands of these families did not identify any shared haplotype, suggesting that either it is recurrently mutated or is an ancient genetic variant [7].
Search for the TRAPPC4 c.454+3A>G in clinical exome and genome databases
The gnomAD (v2.1.1) reports the minor allele frequency (MAF) of the TRAPPC4 c.454+3A>G variant to be 0.024% with 68 heterozygotes reported with representation in the South Asian, European, and African populations. In the GeneDx® database of patients referred for molecular diagnostics, there were four individuals that were homozygous for the TRAPPC4 c.454+3A>G variant (Table S1, families 12, 13, 14, and 15), all of whom consented to publication. In the Centogene® database, comprising clinically ascertained exomes from 47,811 individuals, there were eight individuals homozygous for the TRAPPC4 c.454+3A>G variant, and among these, consent to publish was obtained for four (Table S1, families 7, 8, 9, 10). All known individuals homozygous for the variant were found to be clinically affected, suggesting full penetrance, and bringing the total number of patients reported here to 27.
In addition to the patients carrying a homozygous TRAPPC4 c.454+3A>G variant, the same variant was found heterozygous in two individuals in our Middle Eastern cohort of over 5000 individuals (MAF = 0.040%) who underwent whole-exome sequencing, and in whom a causative gene other than TRAPPC4 was identified or families remain unsolved [18, 19]. In the GeneDx® unaffected adult cohort, TRAPPC4 c.454+3A>G was heterozygous in 64 out of 189,846 alleles (MAF = 0.033%). In the 100 K Genomes Project, the variant was seen in 64 of 59,464 participants (all heterozygous, MAF = 0.054%). In the NHLBI Exome Sequencing Project database the MAF = 0.046%, and in the UK10K database the MAF = 0.03%. The Centogene® database comprising clinically ascertained exomes of 47,811 individuals, we identified 14 carriers from unrelated families, with MAF = 0.026%. In the Baylor Genetics Laboratories database, of 14,775 sequenced individuals, 13 heterozygous individuals were identified (MAF = 0.088%). Six further heterozygous individuals were found in the data set of ~15,500 exomes of the Queen Square Genomic Center database (MAF = 0.039%). Accordingly, the carrier frequency of the TRAPPC4 c.454+3A>G variant ranges from 2.4–5.4 per 10,000 individuals in our collective cohort consisting mostly of individuals with neurological disease. There was no reported health consequence of carrying the heterozygous variant.
TRAPPC4 mRNA analysis reveals defective splicing
The c.454+3A>G variant occurred immediately following exon 3, suggesting defective splicing may result. RNA sequencing of dermal fibroblasts from patient 15 revealed TRAPPC4 as the only genome-wide significant differentially expressed gene, compared with 24 similarly sequenced controls (Adjusted p value = 0.016) (Fig. 1e), a finding that helped identify the TRAPPC4 c.454+3A>G variant as the cause in these families. Inspection of reads mapping to TRAPPC4 revealed exon 3 skipping in ~46% (7 of 15) of transcripts utilizing the acceptor splice site of exon 4 (Fig. 1f). This splice junction was also observed at low levels in the sequenced control samples where it was utilized by an average 3% of transcripts. A further small percentage (13%) of reads in the patient contain the NM_016146.5; c.454+3A>G variant and use a novel donor splice site at position c.454+41, resulting in a 40-nucleotide extension of exon 3. In summary, more than half (59%) of expressed TRAPPC4 transcripts from patient 15 fibroblasts display aberrant splicing while wild-type splicing of exon 3 was supported by 41% of reads. Both of the aberrant transcripts detected predict a frameshift and nonsense mediated decay, consistent with the observed decrease in TRAPPC4 expression. Real-time polymerase chain reaction (RT-PCR) and subsequent Sanger sequencing analysis of the RT-PCR fragments have been described by Van Bergen et al. [7].
Clinical evaluation of the affected individuals
Following a generally uncomplicated pregnancy and neonatal course, in the first few months of life all affected individuals developed a severe, progressive encephalopathy characterized by motor, language, and social developmental stagnation. Psychomotor regression was further observed in some cases (Table S1). With disease progression, patients experienced a critical cognitive decline and demonstrated acquired microcephaly. The degree of microcephaly was generally severe, with a mean occipito-frontal circumference of −5.77 standard deviations (SDs) at around 5 years of age as a mean. Subtle and non-specific facial dysmorphic features, including bitemporal narrowing, thick eyebrows, full cheeks, long philtrum, wide mouth, thin and tented upper lip, and pointed chin (Fig. 2a, b) were observed in most cases, in addition to all previously reported cases over 2 years of age [7, 8]. In all patients, seizure onset was in the first 6 months of life with highly variable seizure types (e.g., infantile spasms and focal, tonic-clonic, atonic, and tonic seizures) and frequencies. In most cases, there was a partial response to antiepileptic drugs (AEDs), including levetiracetam and clobazam, but seizures were refractory in patients 9 and 19. Electroencephalographic features were not specific, showing generalized disorganization and epileptiform discharges. Neurologic examination revealed spastic tetraplegia and hyperreflexia in all subjects (Video S1 and S2), similar to previously reported subjects where data were available [7, 8]. Abnormal movements mainly consisting of ataxia and dystonia were observed in patients 5, 6, 16, and 18. Patients 14, 15, and 18 showed bilateral cataracts. Despite the presence of visual fixation in most individuals, visual pursuits were almost invariably absent, suggesting impaired visual function probably independent of cataracts (Video S3). Hearing loss was observed in patients 11 and 12. Early death owing to infections occurred in five out of the 23 reported individuals at a mean of 8.8 years.
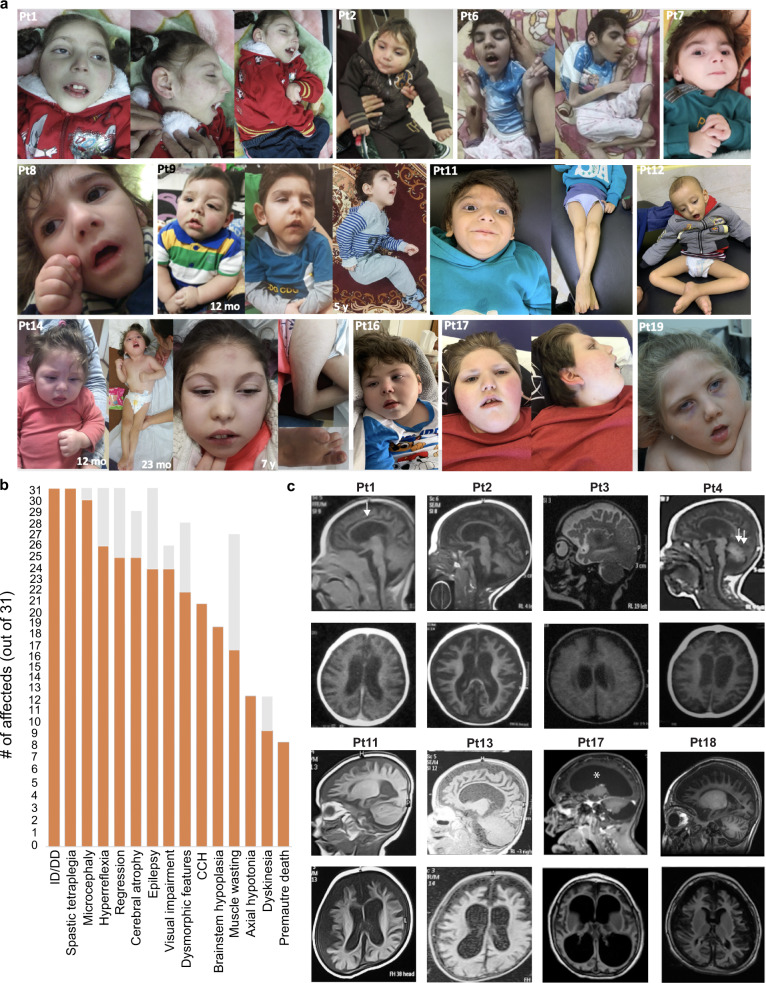
Fig. 2. Clinical and radiological characterization of TRAPPC4 patients.
a Clinical features of patients carrying the homozygous c.454+3A>G TRAPPC4 variant showing subtle and non-specific dysmorphic features (sloping forehead, thick eyebrows with medial sparing, depressed nasal bridge, long philtrum, wide mouth with tented upper lip, pointed chin, and uplifted earlobes), and motor deficits including associated contractures. Reduced muscle mass was evident in all patients, as evidenced by images of patients 6, 11, 12, and 14. Patient 14 also had foot anomalies mainly consisting of severe clinodactyly of the 5th toes and sandal gap toes. b Bar graph showing the distribution of the most relevant clinical features from the 17 ascertained families and the eight patients identified from clinical testing facilities. Orange: number of patients out of 27 showing each feature. Gray: data not available. c Brain MRI scans showing global cerebral atrophy in all patients, including cortex, corpus callosum (arrow in Patient 1), cerebellum (double arrows in patient 4), and ventricular enlargement (asterisk in patient 17).
Brain Magnetic resonance imagings (MRIs) showed a variable degree of cerebral atrophy in all cases (Fig. 2c). This was also evident in five out of the eight previously reported cases where MRIs were available [7, 8]. In many subjects, loss of white matter, ventriculomegaly, and cerebellar atrophy were also dramatic, and appeared more severe in older children, suggestive of progressive atrophy. The degree of ventriculomegaly varied considerably, but all subjects showed enlarged subarachnoid spaces and widely spaced cortical gyri. Hypoplasia of the corpus callosum and brainstem were also observed (Table S1).
Discussion
This study supports the pathogenicity of the recurrent c.454+3A>G variant in TRAPPC4 as the cause of a severe developmental encephalopathy characterized by a profound global developmental delay, and frequent psychomotor regression and lethality, and delineates the carrier frequencies throughout sampled populations. Affected individuals developed debilitating motor impairments, with spastic tetraplegia, cognitive decline, and movement disorders. Common facial dysmorphism includes bitemporal narrowing, thick eyebrows, full cheeks, long philtrum, wide mouth, thin and tented upper lip, and pointed chin. Our report of additional affected individuals helps clarify the expressivity in NEDESBA.
Early-onset epilepsy with variable seizure phenotypes and electroencephalographic findings is a feature of NEDESBA. In most subjects, there is a partial response to AEDs, however refractory seizures were observed in some patients. Interestingly, two affected siblings with gelastic seizures and episodes of temperature dysregulation have been previously reported, one of whom also had premature adrenarche and osteopenia [7]. These findings, suggestive of hypothalamic dysregulation, have not been observed in the other cases or in our cohort. Similarly, dystonia or ataxia were only observed in some affected individuals. Accordingly, NEDESBA might have a wide phenotypic spectrum, and variable expressivity. The report of our further cases helps to clarify the expressivity of these rare features.
An additional critical aspect of NEDESBA involves psychomotor regression. In previous reports, this condition was mainly described as a progressive encephalopathy. Most subjects from our cohort showed absent development of motor, verbal, and social skills. Although, some children attained developmental milestones in the first few months of life, these were invariably lost with disease progression. Psychomotor regression was observed in all phenotyped individuals, representing a cardinal feature of this disorder (Table S1). Together with the progressive course of the neuroimaging abnormalities, this observation suggests that TRAPPC4 encephalopathy displays early-onset neurodegenerative features.
An individual with TRAPPC4 variant displaying severe muscle involvement associated with biochemical alterations suggestive of episodic rhabdomyolysis (increased plasma lactate, creatine phosphokinase, and transaminases) has been very recently reported, resembling the progressive early-onset encephalopathy with episodic rhabdomyolysis (PEERB, MIM:618331) caused by recessive TRAPPC2L variants (MIM:618331) [8]. Muscle wasting was observed in our cohort, most likely secondary to the profound limitation of voluntary movements. In one case (patient 19), blood tests revealed elevated lactate on one occasion (level 424 U/L, reference range 20–200 U/L), but this was not observed on repeat testing. Although primary muscle involvement is still possible, many TRAPPC4 subjects also present clinical manifestations, which might lead to a transient increase in serum muscle enzymes (e.g., seizures, spasticity, and dystonia). Accordingly, the assessment of biochemical alterations for creatine kinase and other muscle enzymes on a case-by-case basis is essential.
Although not pathognomonic, the neuroimaging features of TRAPPC4 subjects demonstrate consistent cerebral atrophy with predominant involvement of subcortical white matter and ventricular enlargement, often in association with basal ganglia, cerebellar, and brainstem atrophy. The remarkable white matter involvement is also supported by observation of thin corpus callosum evident in most patients. However, in contrast to cases from a previous report [7], a significant sparing of the basal ganglia was not noted in our cohort. Accordingly, the exact progression of the neurodegenerative process and the temporal relationship between cerebral basal ganglia, brainstem, and cerebellar atrophy remain to be elucidated.
The c.454+3A>G variant is relatively common across populations worldwide, as shown by the relatively high carrier frequency in public and private databases. This is also supported by the absence of consanguinity in some of our families (9, 11, 12, 13, and 15) and in one previously reported family [19]. This variant might have derived from a possible shared common ancestor, however, neither our study nor previous studies were able to detect a shared haplotype. We showed that this variant leads to a failed splicing based on the presence of TRAPPC4 aberrant transcripts. These findings support the pathogenicity of the TRAPPC4 variant as the cause of the disorder in the studied subjects. Furthermore, the identification of the c.454+3A>G variant in RNAseq data in patient 15 stresses the power of RNAseq as an ancillary diagnostic tool in individuals with undiagnosed rare diseases, especially in cases where exome-sequencing yields negative results [20].
A range of conditions are now described in the TRAPPopathies primarily involving the developing brain and muscle (Table S2). Our study highlights the involvement of this relatively common single allele as the cause for a severe recessive neurological disease characterized by progressive microcephaly, profound cognitive decline, infantile-onset epilepsy, and progressive spastic tetraplegia. Our observations expand the spectrum of TRAPPopathies, supporting the existence of phenotypic overlap in this complex group of disorders. To date, ~23 families of European, Mediterranean, Middle Eastern, and Indian ancestries are known to be homozygous for the c.454+3A>G variant in TRAPPC4. The highest frequency of this variant is found in individuals with European and Mediterranean ancestries, whereas it appears to be rare in African and East Asian populations. The commonality of the c.454+3A>G variant in the general human population warrants the search for TRAPPC4 variants in children with microcephaly, especially when associated with neurodegenerative features. The frequency of this variant in European and Mediterranean basin populations suggests that a genetic screening-based approach might prove useful in early carrier detection.
Supplementary information
Acknowledgements
The authors thank the patients and their families for participation in this study. We would like to thank Grazia Mancini for communicating unpublished results. The authors acknowledge Baylor Genetics Laboratories for supplying data. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. This research was conducted as part of the Queen Square Genomics group at University College London, supported by the National Institute for Health Research University College London Hospitals Biomedical Research Centre. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director under Award Number(s) U01HG007672 (PI- Shashi V, Duke University). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Drs. Jennifer Friedman and Dillon Chen for providing comments on the manuscript.
Funding
This work was supported by NIH grants R01NS098004, R01NS048453, and R01NS106387, SFARI research award to J.G.G. S.G. was sponsored by the Ruth L. Kirschstein Institutional National Research Service Award (T32 GM008666) from the National Institute on Deafness and Other Communication Disorders and by award F31HD095602 from the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development. Sequencing and analysis were provided by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG), and the Yale Center for Mendelian Genomics (Yale CMG, UM1HG006504) and supported by NHGRI, NEI, and NHLBI grant UM1 HG008900 and NHGRI R01 HG009141, the MRC (MR/S01165X/1, MR/S005021/1, G0601943), the NIH Research UCL Hospitals Biomedical Research Centre, Rosetree Trust, Ataxia UK, MSA Trust, Brain Research UK, Sparks GOSH Charity, Muscular Dystrophy UK (MDUK), Muscular Dystrophy Association (MDA USA), the Victorian Government’s Operational Infrastructure Support Program, and by the Australian Medical Research Future Fund project, “Massimo’s Mission”.
Data availability
The identified TRAPPC4 variant was submitted to the LOVD database at https://databases.lovd.nl (Individual ID #00306235, https://databases.lovd.nl/shared/individuals/00306235; genomic variant ID #0000673996, https://databases.lovd.nl/shared/variants/0000673996). The accession number for this variant in ClinVar is VCV000812649.1. The accession numbers in dbGAP are phs001272 and phs000744.
Compliance with ethical standards
Conflict of interest
A.B., A.C., J.J., and R.W. are employees of GeneDx, Inc. All other authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shereen G. Ghosh, Marcello Scala
These authors jointly supervised this work: Reza Maroofian, Joseph G. Gleeson
Deceased: S. Lane Rutledge.
Contributor Information
Reza Maroofian, Email: r.maroofian@ucl.ac.uk.
Joseph G. Gleeson, Email: jogleeson@ucsd.edu
Supplementary information
The online version of this article (10.1038/s41431-020-00717-5) contains supplementary material, which is available to authorized users.
References
- 1.Brunet S, Sacher M. In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic. 2014;8:803–18. doi: 10.1111/tra.12183. [DOI] [PubMed] [Google Scholar]
- 2.Milev MP, Graziano C, Karall D, Kuper WFE, AL-Deri N, Cordelli DM, et al. Bi-allelic mutations in TRAPPC2L result in a neurodevelopmental disorder and have an impact on RAB11 in fibroblasts. J Med Genet. 2018;55:753–64. doi: 10.1136/jmedgenet-2018-105441. [DOI] [PubMed] [Google Scholar]
- 3.Gedeon AK, Colley A, Jamieson R, Thompson EM, Rogers J, Sillence D, et al. Identification of the gene (SEDL) causing X-linked spondyloepiphyseal dysplasia tarda. Nat Genet. 1999;22:400–4. doi: 10.1038/11976. [DOI] [PubMed] [Google Scholar]
- 4.Bogershausen N, Shahrzad N, Chong JX, von Kleist-Retzow JC, Stanga D, Li Y, et al. Recessive TRAPPC11 mutations cause a disease spectrum of limb girdle muscular dystrophy and myopathy with movement disorder and intellectual disability. Am J Hum Genet. 2013;93:181–90. doi: 10.1016/j.ajhg.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marin-Valencia I, Novarino G, Johansen A, Rosti B, Issa MY, Musaev D, et al. A homozygous founder mutation in TRAPPC6B associates with a neurodevelopmental disorder characterised by microcephaly, epilepsy and autistic features. J Med Genet. 2018;55:48–54. doi: 10.1136/jmedgenet-2017-104627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacher M, Shahrzad N, Kamel H. TRAPPopathies: an emerging set of disorders linked to variations in the genes encoding transport protein particle (TRAPP)-associated proteins. Traffic. 2019;1:5–26. doi: 10.1111/tra.12615. [DOI] [PubMed] [Google Scholar]
- 7.Van Bergen NJ, Guo Y, Al-Deri N, Lipatova Z, Stanga D, Zhao S, et al. Deficiencies in vesicular transport mediated by TRAPPC4 are associated with severe syndromic intellectual disability. Brain. 2019;143:112–30. doi: 10.1093/brain/awz374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaur P, Kadavigere R, Girisha KM, Shukla A. Recurrent bi-allelic splicing variant c.454+3A>G in TRAPPC4 is associated with progressive encephalopathy and muscle involvement. Brain. 2020;143:e29. doi: 10.1093/brain/awaa046. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med. 2013;16:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer P, Kandaswamy KK, Weiss MER, Paknia O, Werber M, Bertoli-Avella AM, et al. Development of an evidence-based algorithm that optimizes sensitivity and specificity in ES-based diagnostics of a clinically heterogeneous patient population. Genet Med. 2019;1:53–61. doi: 10.1038/s41436-018-0016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Retterer K, Juusola J, Cho MT, Vitazka P, Millan F, Gibellini F, et al. Clinical application of whole-exome sequencing across clinical indications. Genet Med. 2016;7:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 12.Sadedin SP, Pope B, Oshlack A. Bpipe: a tool for running and managing bioinformatics pipelines. Bioinformatics. 2012;11:1525–6. doi: 10.1093/bioinformatics/bts167. [DOI] [PubMed] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;15:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;1:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y, Smyth GK, Shi W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019;8:e47. doi: 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrido-Martin D, Palumbo E, Guigo R, Breschi A. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput Biol. 2018;8:e1006360. doi: 10.1371/journal.pcbi.1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon-Salazar TJ, Silhavy JL, Udpa N, Schroth J, Bielas S, Schaffer AE, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott EM, Halees A, Itan Y, Spencer EG, He Y, Azab MA, et al. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat Genet. 2016;48:1071–6. doi: 10.1038/ng.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonorazky HD, Naumenko S, Ramani AK, Nelakuditi V, Mashouri P, Wang P, et al. Expanding the boundaries of RNA sequencing as a diagnostic tool for rare Mendelian disease. Am J Hum Genet. 2019;104:466–83. doi: 10.1016/j.ajhg.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The identified TRAPPC4 variant was submitted to the LOVD database at https://databases.lovd.nl (Individual ID #00306235, https://databases.lovd.nl/shared/individuals/00306235; genomic variant ID #0000673996, https://databases.lovd.nl/shared/variants/0000673996). The accession number for this variant in ClinVar is VCV000812649.1. The accession numbers in dbGAP are phs001272 and phs000744.