Abstract
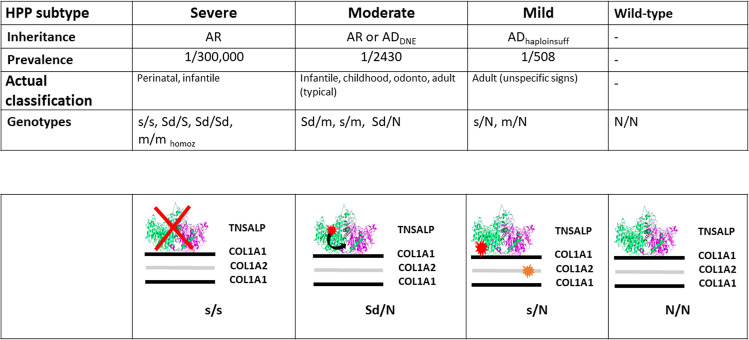
Hypophosphatasia (HPP) is caused by pathogenic variants in the ALPL gene. There is a large continuum in the severity, ranging from a lethal perinatal form to dental issues. We analyzed a cohort of 424 HPP patients from European geographic origin or ancestry. Using 3D modeling and results of functional tests we classified ALPL pathogenic variants according to their dominant negative effect (DNE) and their severity. The cohort was described by the genotypes resulting from alleles s (severe recessive), Sd (severe dominant), and m (moderate). Many recurrent variants showed a regional anchor pointing out founder effects rather than multiple mutational events. Homozygosity was an aggravating factor of the severity and moderate alleles were rare both in number and frequency. Pathogenic variants with DNE were found in both recessive and dominant HPP. Sixty percent of the adults tested were heterozygous for a variant showing no DNE, suggesting another mechanism of dominance like haploinsufficiency. Adults with dominant HPP without DNE were found statistically less severely affected than adults with DNE variants. Adults with dominant HPP without DNE represent a new clinical entity mostly diagnosed from 2010s, characterized by nonspecific signs of HPP and low alkaline phosphatase, and for which a high prevalence is expected. In conclusion, the genetic composition of our cohort suggests a nosology with 3 clinical forms: severe HPP is recessive and rare, moderate HPP is recessive or dominant and more common, and mild HPP, characterized by low alkaline phosphatase and unspecific clinical signs, is dominantly inherited and very common.
Subject terms: Calcium and phosphate metabolic disorders, Genetic testing, Disease genetics, Medical genetics
Introduction
Hypophosphatasia (HPP) (MIM 241510, 241500, and 146300) is a rare inherited disorder that mainly affects bone and teeth mineralization [1–3]. The prevalence of severe forms has been estimated to 1/300,000 in Europe [4] and 1/100,000 in North America [5]. HPP is caused by loss-of-function variants in the ALPL gene on 1p36.12 encoding the Tissue Nonspecific Alkaline Phosphatase (TNSALP or TNAP) [6, 7]. The clinical spectrum of HPP is extremely variable, ranging from the very severe, mostly lethal, perinatal form to the mild form of late adult onset presenting with nonpathognomonic symptoms such as arthropathy and musculoskeletal pain. A classification primarily based on the age of diagnosis and the presence or absence of bone symptoms is currently used: perinatal, prenatal benign, infantile, childhood, adult and odontohypophosphatasia (odontoHPP). The latter form presents as only dental manifestations. These classes of severity overlap, for instance a patient with HPP manifestation before 6 months may have milder symptoms than another patient with first symptoms appearing at 10 months [8, 9]. Features leading clinicians to suspect HPP differ depending on the age of the patient, and in the age group, some symptoms may be present or absent. The most common symptoms shared at any age are defective mineralization of bone and/or teeth, premature loss of teeth with intact roots, and, above all, reduced serum alkaline phosphatase (ALP) activity, which is a hallmark of ALPL variants [10].
Severe forms of HPP, mostly perinatal and infantile, are recessively inherited whereas moderate HPP may be dominantly or recessively inherited. Dominant inheritance is assumed to derive from the homodimeric conformation of TNSALP needed to its catalytic activity. In heterozygotes the mutant and the wild-type monomers can dimerize and negatively interact so that the activity of the wild-type monomer is disturbed by the mutant monomer (DNE). However, there is no reason to believe that other mechanisms of dominance are not involved. We recently showed that adults with mild HPP characterized by unspecific signs of the disease may carry dominant variants that do not have demonstrable DNE [11].
A treatment of HPP by enzyme replacement is now available [12]. Because biphosphonates used to treat OI are counter-indicated in HPP [13–15] the diagnosis of HPP versus its main differential diagnosis, osteogenesis imperfecta, is of importance. In pediatric forms, HPP is often diagnosed on the basis of clinical and biological data. Genetic testing will confirm the diagnosis and allow appropriate genetic counseling. Therefore, most of the patients diagnosed with HPP have their diagnosis confirmed by genetic testing [9, 16–18]. In prenatal context and in adults displaying the mildest symptoms, the identification of ALPL pathogenic variants is often decisive to provide the diagnosis. Indeed, imaging, clinical, and biological investigations are limited in prenatal context so that differential diagnoses may be difficult to exclude, especially OI. In adult HPP, when symptoms are nonspecific, the disease is difficult to prove even when AP is low, which has been reported in an increasing number of common pathologies including osteoporosis, arthropathies, chondrocalcinosis [19], and more recently ovarian and uterine disorders [20]. Genetic confirmation of the diagnosis is therefore very useful. However, we experienced that many patients with mild unspecific symptoms of HPP and low ALP are negative for ALPL variants. It was not clear whether it is because low AP may be found outside of any context of ALPL variant, or because we missed variants in deep introns or in regulatory sequences.
The study of ALPL variants is also crucial to understand genotype-phenotype relationships. A particular effort was done by various groups to test the effect of ALPL variants by in vitro assays [21–33]. More recently a series of 155 ALPL variants including variants responsible for HPP and variants from the general population listed in gnomAD database were tested [34]. These functional tests allow to classify the ALPL alleles according to their severity and their dominant effect, and therefore to refine genotype-phenotype correlation.
We review here genetic testing of patients addressed to our laboratory because of suspicion of HPP or for confirmation of the clinical diagnosis between 1997 and 2019. Our data provide a comprehensive picture of severe, moderate, and dominant ALPL variants in various European regions, allowing to decipher puzzling mild HPP, to estimate its prevalence, and to progress in understanding of the genotype-phenotype correlation.
Methods
Patients
The study was designed in accordance with the tenets of the Declaration of Helsinki. All patients were referred to our laboratory for diagnosis purpose and signed an informed consent for genetic testing. They were suspected to have HPP on the basis of low ALP and HPP symptoms depending on the age of diagnosis. Apart very few exceptions the patients were not seen by our hospital. The clinical form was assigned according to the description of the physician requesting the test, with much differences from a site to another one for clinical and laboratory data provided. The period of study ranges from 1997 to 2019 (22 years).
The patients were classified according to their age at the time of diagnosis: prenatal and neonatal period (perinatal form), before 6 months (infantile form), between 6 months and 18 years (childhood form), and >18 years (adult form). When it was known the age of first symptoms was taking into account for phenotypic classification.
Between 1997 and 2010 patients originated from 34 distinct countries, mostly Europe, North America and Australia, but also few patients originated from Japan and Middle East. North and West Europe were more represented than South and East Europe. After 2010 the ascertainment progressively became French as each country developed on-site genetic tests.
Genetic testing
Genetic testing was performed by sequencing exons and exon/intron borders of ALPL gene by Sanger methodology and large indels detected by using Taqman© probes of exons and qPCR. From 2014 sequencing was performed by NGS, together with other genes involved in PPi metabolism and in differential diagnosis of HPP [35], and large indels by using CovCop software [36] and/or ALPL-targeted arrayCGH.
Submission of variants and associated data
All the variants and associated data displayed here were submitted to the locus-specific database dedicated to ALPL variants (The TNSALP Gene Mutation Database at www.sesep.uvsq.fr/03_hypo_mutations.php).
Moderate and severe alleles
Functional tests were performed by various groups (see the ALPL mutation database http://www.sesep.uvsq.fr/03_hypo_mutations.php) but with similar and comparable methodologies. Briefly, eukaryotic cells were transiently transfected with either 100% of mutant plasmid or a mix corresponding to mutant plasmid/wild-type plasmid (ratio 50/50). Most of the labs used for the transfections MDCK or COS cells. After transient transfection the cells were lysed and ALP activities were measured. We empirically chose a threshold severe/moderate of 20% of wild-type activity and a threshold recessive/dominant of [0.9(100 + x)/2] of wild-type activity where x is the residual activity of the mutant.
Nomenclature of variants
The Human Genome Variation Society (HGVS) recommendations were used to standardize nomenclature of all analyzed variants [37]. The reference sequences used to specify ALPL variants were RefSeq sequence NM_000478.6 (identical to Ensembl sequence ENST000000374840.8) for the coding region canonical transcript. Protein variants are specified relative to the amino acid sequence resulting from translation of this transcript, specified in NP_000469.3 (identical to Ensembl sequence ENSP00000363973.3). For the noncoding regions (splice sites variants and large insdel) the reference sequence NG_008940.1 was used.
3D localization
3D localization of variants was obtained by using the 3D model of TNSALP from Mornet et al. (2001) [38].
Results
ALPL variants
The cohort consists in 424 unrelated HPP patients tested for ALPL variants between 1997 and 2019, 166 heterozygotes and 258 homozygotes or compound heterozygotes (Table 1 supplemental material). One hundred and sixty-six patients originated from France, 258 from 33 other countries, among whose 24 European, 4 with people from European ancestry (Australia, New Zeland, Canada, USA), 5 from Middle East (Soudi Arabia, Israel, Jordan, Tunisia, Turkey) and India and Japan. We identified 682 mutant alleles corresponding to 249 distinct variants. This confirms the very high genetic heterogeneity of HPP and the very low recurrence rate of most of the variants (in mean each allele is represented 2.7 times).
Table 1.
Distribution of the most common dominant variants in dominant and recessive HPP.
| Dominant allele | Number of dominant genotypes [moderate HPP] | Number of recessive genotypes [severe or moderate HPP] |
|---|---|---|
| c.1250A>G p.(N417S) | 11 | 9 |
| c.346G>A p.(A116T) | 7 | 5 |
| c.1001G>A p.(G334D) | 1 | 8 |
| c.668G>A p.(R223Q) | 1 | 7 |
| c.299C>T p.(T100M) | 1 | 7 |
| c.212G>A p.(R71H) | 6 | 1 |
| c.1172G>A p.(R391H) | 3 | 4 |
| c.550C>T p.(R184W) | 4 | 2 |
| c.1426G>A p.(E476K) | 4 | 2 |
| c.551G>A p.(R184Q) | 4 | 1 |
| c.1133A>T p.(D378V) | 3 | 1 |
| c.1015G>A p.(G339R) | 3 | 2 |
| c.512A>G p.(H171R) | 0 | 7 |
| Total | 48 | 56 |
Reference sequences used: NM_000478.6 (ENST000000374840.8).
Recurrent variants are due to founder effects
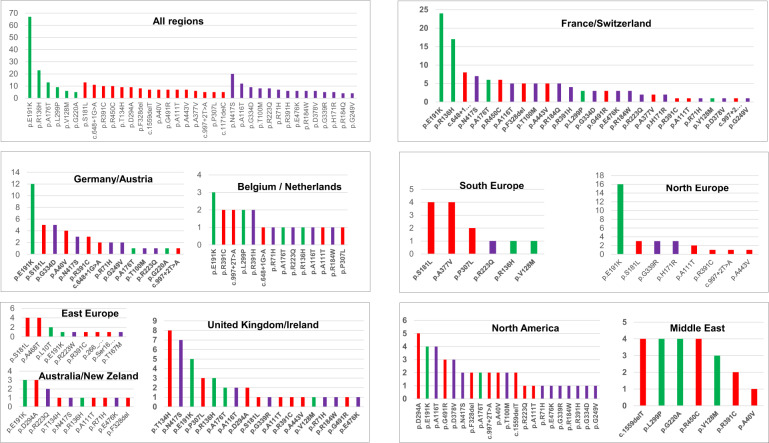
We noticed a regional anchor of many recurrent variants, c.648+1 G>A, c.407G>A p.(R136H), c.551G>A p.(R184Q) in France, c.400_401delinsCA p.(T134H) in UK, c.1001G>A p.(G334D) and c.119C>T p.(A40V) in Germany, c.1015G>A p.(G339R) in North Europe, c.542C>T p.(S181L) in South Europe, (c.1133A>T p.(D378V) in USA, c.1250A>G p.(N417S) in UK and France, c.881A>C p.(D294A) in Anglo-Saxon countries UK, USA and Australia (Fig. 1). Together with the extreme rarity of de novo ALPL variants [24, 39] this result indicates that the recurrence of these variants is due to founder effects and not to multiple mutational events. By contrast few variants such as c.1171C>T p.(R391C) and c.526G>A p.(A176T) were found almost everywhere, which may indicate multiple mutational events and/or more ancient mutations.
Fig. 1. Distribution of recurrent ALPL variants according to the geographic origin of patients.
Variants are shown according to their frequency and their genetic status in ten regions defined to represent 32 countries: France/Switzerland, Germany/Austria, UK/Ireland, Belgium/Netherlands, North Europe (Denmark, Finland, Sweden, Norway, Lithuania), East Europe (Hungary, Croatia, Georgia, Poland, Czech republic), South Europe (Spain, Portugal, Greece, Italy), North America (USA/Canada), Australia/New Zeland and Middle East (Saudi Arabia, Israel, Japan, Jordan, Tunisia, Turkey). Moderate alleles are shown in green, severe alleles with no DNE in red and severe alleles with DNE in purple. Variants not classified are not shown.
The most frequent HPP allele is c.571G>A p.(E191K) that represents 9.8% (67/682) of all alleles. In France it represents 39.0% of the moderate alleles and 9.7% (24/247) of all alleles. It is also very common in Germany (12/70 = 17.1% of all alleles) and in North Europe i.e., Sweden, Denmark, Norway and Finland (15/40 = 37.5% of all alleles). In these countries 66.7% of patients with recessive moderate HPP carry at least one c.571G>A (p.E191K) allele. We confirm here our previous report suggesting that this variant occurred on a single chromosome in North Europe and spread throughout the rest of Europe and into the New World as a result of human migration (not shown) [40]. C.571G>A p.(E191K) account for the largest part of recessive moderate HPP and the phenotypes of patients carrying this variant range from the mildest adult form to relatively severe forms, never lethal however.
In US patients for which only 53 chromosomes were tested, we recurrently found the variants reported as common in this population: c.1133A>T p.(D378V), c.881A>C p.(D294A), c.571G>A p.(E191K), c.346G>A p.(A116T) [10]. The distribution was similar in US and in Europe, the greatest difference concerned c.881A>C p.(D294A) and c.1133A>T p.(D378V) that are relatively common in USA and clearly rare in Europe. Overall the distribution of variants reflects the geographic and historic proximity of the countries.
Moderate alleles are rare
According to the functional tests the alleles were classified in five types: normal (N), moderate (m), severe with no DNE (s), severe with DNE (Sd) and not classifiable (NC). NC alleles were missense variants not subjected to functional tests. Non missense variants including frameshift, termination, and splicing variants were classified s by default. With a cut off moderate/severe set up at 20% of residual activity, we counted only 30 (17.4%) distinct moderate alleles for 142 (82.5%) severe ones, 107 recessive and 35 dominant, and 77 non classifiable alleles. In term of allele frequencies, moderate alleles represent 29.5% (168/568) of all alleles for which the classification was possible. Severe recessive and dominant alleles represent 46.4% (264/568) and 23.9% (136/568), respectively. So, recessive moderate HPP is rare not only because recessive genotypes are statistically less probable but also because moderate alleles are less frequent than severe recessive alleles. However, there are two remarkable exceptions that show a high recurrence rate, c.571G>A p.(E191K), and c.407G>A p.(R136H).
Dominant alleles are common in recessive HPP
The most common severe dominant allele in Europe is c.1250A>G p.(N417S) that represents 2.9% (20/682) of all HPP alleles and 14.8% of the dominant alleles. It is particularly prevalent in UK and in France where it is the main cause of dominant HPP. The variant is common in Anglo-Saxon countries (UK, USA, Australia) and rare elsewhere except in France where it accounts for 13.5% (7/52) of dominant alleles. The other recurrent dominant variants in France are c.1426G>A p.(E476K), c.1015G>A p.(G339R), c.299C>T p.(T100M), c.346G>A p.(A116T) and c.1001G>A p.(G334D). The latter is the most common dominant allele in Germany, reflecting the German origin of the Canadian Mennonite community where this variant is almost the unique reported HPP variant [41]. The variant c.1133A>T p.(D378V) was previously reported specific of North American patients from European ancestry [10]. In a previous report, we estimated the proportion of dominant alleles among severe ones in European patients to 13.4% [4]. Here, thanks to additional functional tests we were able to refine this estimate to 35.1% (106/302), which indirectly change our previous estimate of the prevalence of moderate HPP [4]. Applying a proportion of 35.1% of dominant alleles would result in a prevalence of moderate HPP of 1/2430.
Among 160 recessive genotypes with classifiable alleles, 57 (35.6%) carry at least one dominant allele. This is critical for genetic counseling because relatives of patients affected with recessive severe HPP may carry a dominant allele and therefore can be affected with moderate HPP. The respective parts are very variable from a variant to another one, for instance c.1001G>A p.(G334D) was almost found in recessive HPP only and c.212G>A p.(R71H) in dominant HPP only (Table 1). Globally these recessive genotypes result in a severe phenotype.
Genotype-phenotype correlation
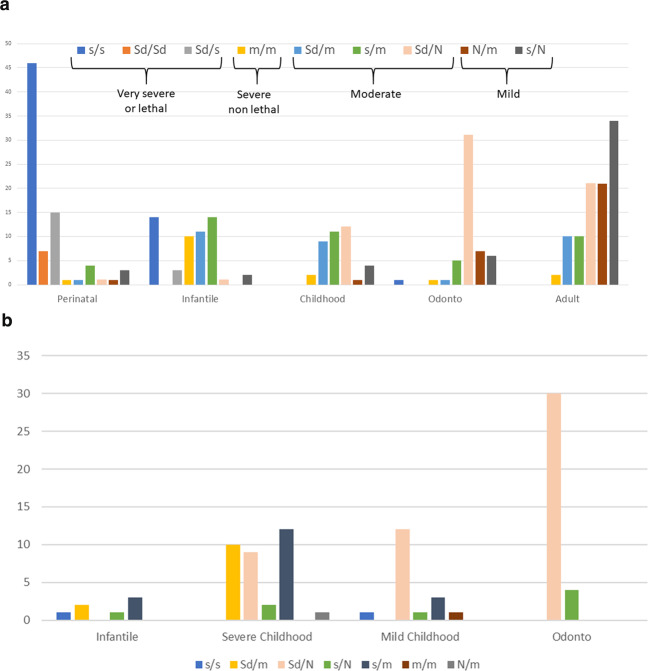
Allelic composition of the genotypes
The allelic composition of the genotypes is shown Fig. 2a. Excepted for the prenatal benign form, each phenotype shows genetic heterogeneity with 3–5 distinct genotypes. This reflects the complexity of the clinical spectrum. Assuming that the issue of prenatal benign HPP is in fact nonlethal HPP [42], i.e., all but one phenotype (perinatal lethal), we did not include this form in the figure. However, our results show that prenatal benign HPP was very similar to odontoHPP and is mostly due the genotype Sd/N.
Fig. 2. Composition of genotypes in HPP phenotypes.
Only genotypes with classifiable alleles are included. a In our cohort. For each phenotype the figure shows the number of genotypes according to our classification of alleles. The prenatal benign form is not shown because this particular form is assumed to finally result in all but one (perinatal lethal) phenotypes. The distribution allow to sort genotypes according to resulting phenotypes: severe (divisible into into subgroups: very severe and severe), moderate and mild. N normal (wild type); Sd severe dominant; s severe recessive (i.e., no DNE); m moderate; b In the pediatric US cohort [10] for comparison.
Perinatal lethal HPP
The extremities of the spectrum (perinatal lethal and odontoHPP) are much more homogenous when compared to other forms. Perinatal lethal HPP was due to genotypes s/s (57%), s/Sd (19%) and Sd/Sd (9%), the 2 latter correspond to the most severe genotypes and are specific for this clinical form. By contrast, s/s genotypes were also found in infantile HPP. Homozygotes represent 65% of the genotypes Sd/Sd and s/s, which is a very high rate of homozygosity when compared to recessive infantile HPP (28.6%) or in the whole cohort (28.3%). Thus homozygosity for s or Sd alleles is an aggravating factor of severity. Perinatal cases with other genotypes than s/s, Sd/s and Sd/Sd may actually have undetected pathogenic variants (genotypes m/s, m/Sd, m/m) or may have been misclassified, for instance when the pregnancy of a prenatal benign case was terminated on the basis of ultrasound signs.
Infantile HPP
Four genotypes were equally found in infantile HPP, s/s, Sd/m, s/m and m/m. This form shares genotypes with perinatal HPP (s/s) and with childhood HPP (Sd/m and s/m), supporting the idea that clinical heterogeneity of infantile HPP has a genetic basis. The presence of the m/m genotype is amazing since the expected phenotype is moderate. We noticed that all infantile patients with the genotype m/m were homozygous, suggesting again that homozygosity may be an aggravating factor of severity.
Childhood HPP
Childhood HPP shares genotypes with infantile HPP (Sd/m and s/m) and with adult and odontoHPP (Sd/N). Here again, the genetic heterogeneity reflects the clinical heterogeneity. In a North American cohort published in 2015 childhood HPP was divided in two subgroups, childhood moderate and childhood severe, mainly on the basis of radiographic findings [10]. We noticed that childhood HPP in our cohort and childhood severe HPP in the US cohort were perfectly superposable in term of type and proportion of genotypes (Fig. 2a, b) whereas the variants in the two groups were quite different. This result supports the basis of a genetic-based nosology of HPP.
Adult HPP
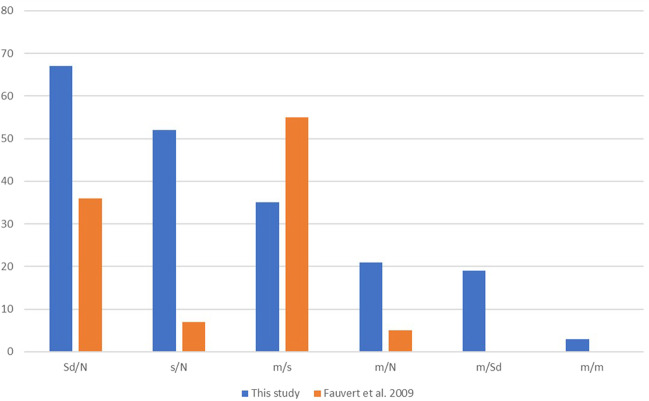
Adult patients were found to carry the genotypes Sd/N, Sd/m and s/m, but also the two unexpected and yet most frequent genotypes m/N and s/N. Interestingly these two genotypes seem specific for this clinical form whereas they were not observed in our previous study where we reported that moderate (and particularly adult) HPP mostly result from genotypes m/s and Sd/N [28] (Fig. 3).
Fig. 3. Comparison of the distributions of genotypes in moderate HPP with our previous study (Fauvert et al. 2009).
The actual distribution in blue is compared to our previous study in orange [28].
OdontoHPP and prenatal benign HPP
Patients with odonto HPP or prenatal benign HPP mostly carried the genotype Sd/N (60.7% and 66.7%, respectively).
Comparison whith the US cohort
In order to consolidate our hypothesis, we compared our results with those of the US cohort published in 2015 and composed of 173 pediatric patients (infantile, childhood, and odontoHPP) [10]. We found that the patterns of the two cohorts were very similar for pediatric cases and that the genotypes s/s and s/N, specific for perinatal and mild HPP, respectively, were quasi-inexistent in the US cohort (Fig. 2b) suggesting that the absence of these genotypes is linked to the pediatric nature of the cohort.
Evidence of a novel form of HPP: mild adult HPP with unspecific signs
Adults with m/N and s/N genotypes were previously underdiagnosed
The clinical picture of HPP dramatically changed between 1997 (a rare and mostly recessive bone disease) and 2019 (a multisystemic, frequent in its moderate form, and mostly dominant disease). This changes went together with genetic testing of more and more adult patients with low serum ALP and unspecific signs of the disease such as osteoporosis, chondrocalcinosis, musculo-squelettal pain and sometimes history of fractures with no or small trauma. In our cohort, the main reason for referral adult patients with m/N and s/N was low or borderline serum AP associated with unspecific signs of HPP. By dividing the cohort into 2 periods of ascertainment, 1997–2009 and 2010–2019, we found that 81% of adults s/N and 62% of adults m/N were studied between 2010 and 2019.
Adult with m/N and s/N genotypes are less severely affected
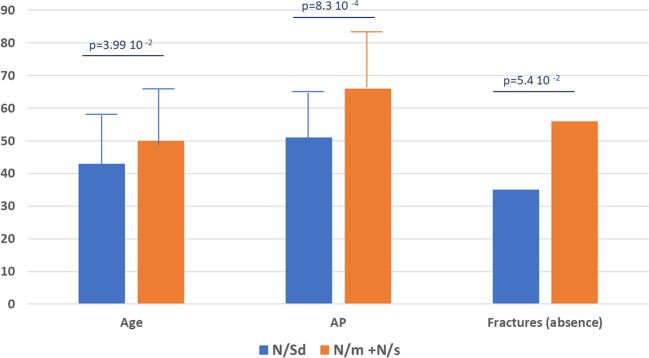
We hypothesized that patients with the genotypes m/N and s/N should be affected with the mildest form of HPP and possibly delineate a new phenotype distinct from adults previously tested, especially those with the genotype N/Sd. We previously discussed the fact that the alleles s in these genotypes have no DNE but still have a dominant effect, perhaps because of haploinsufficiency [11]. However, this mechanism remains to be experimentally established. By using three severity criteria, serum AP level, age at diagnosis and presence of fractures, we also showed that they were globally less severely affected. Here we confirm on a larger sample that adults with s/N and m/N genotypes are significantly less severely affected than adults with Sd/N (Fig. 4). These differences of severity did not affect dental issues that where not statistically different. However dental issues seemed less severe (caries, bad dental condition) in genotypes s/N and m/N than in other genotypes (characteristic loss of teeth).
Fig. 4. Comparison of moderate and mild genotypes for 3 markers of HPP severity.
The figure shows only adults with genotypes Sd/N in (blue) and genotypes m/N and s/N (orange). Age: age at diagnosis in years; AP: alkaline phosphatase in % of the lower limit for age and sex; Age is the age at diagnosis expressed in years. Fractures are given in % of patients reported with no fractures or history of nontraumatic or atypical fractures.
New insights in epidemiologic aspects
On the basis of our allele classification there are three forms of HPP: severe HPP that corresponds to perinatal and most of infantile severe forms, moderate HPP that includes infantile moderate, childhood, odontoHPP and adults with specific signs, and mild HPP corresponding to adults (and perhaps children) with less severe and often unspecific signs (Fig. 5).
Fig. 5. The 3 forms of HPP according to genetic data.
The figure summarizes a genetic-based nosology of HPP, with inheritance and prevalence, and a hypothesis about the mechanism of dominance in mild HPP.
Mild HPPs is expected to be very common in the European populations
Mild HPP may be due to s/N or m/N genotypes. If we consider that the proportion of DNE alleles among severe alleles is 35.1%, and that the frequency of heterozygotes for severe alleles in the population is 2 × 1 × √(1/300,000) = 1/274, the maximum frequency of s/N genotypes is (1–0.351) × 1/274 = 1/422. This corresponds to the case where all s/N are symptomatic and fully penetrant. The part due to m/N genotypes may be estimated by our finding that moderate alleles represent 29.5% of all classifiable alleles, so that it is expected that the frequency of m is ~1/3rd of the totality of alleles, Thus, the frequency of moderate alleles is 1/1278. So, mild HPP due to s/N or m/N genotypes would be [1/422) + [2 × 1/1278] = 1/254.
According to two studies reporting screening for adult patients with low AP and heterozygous for an ALPL variant, ~50% of heterozygotes for an ALPL variant [24/52 [43], and 53/105 [20]] express a phenotype. The patients reported carried m or s alleles only [20], or mostly m or s alleles [43]. On the basis of these published data we applied a penetrance of 50%, which lead to a maximal prevalence of mild HPP due to s/N or N/m genotypes estimated at 1/508.
A new mechanism of dominance in HPP
Beside the fact that the higher proportion of dominant alleles results in an increased estimate of the prevalence of moderate HPP (see above), our results suggest that the genetic model accounting for HPP is more complex than the one suggested until now (4 alleles, 3 phenotypes) [4, 7, 8, 44]. Heterozygotes for ALPL variants with no DNE express a mild form of HPP, and must be consequently assumed to have dominant HPP. Among them there are null alleles like frameshift and nonsense variants, and hypomorphic alleles like many missense variants. Expression of the disease in heterozygotes for null alleles argues in favor of haploinsufficiency while hypomorphic alleles are rather dominant by negative interaction between the mutant protein and another actor like the collagen matrix. Such interaction would be similar to DNE except that the interaction would not occur between the wild type and the mutated TNSALPs, but between the mutated TNSALP and the collagen matrix. Finally, we hypothesize that mild HPP is due in part to heterozygosity for variants with DNE, in part to heterozygosity for variants resulting in negative interaction with collagen and in part to heterozygosity for variants causing haploinsufficiency when they are found together with triggering factors. We previously suggested that the COL1A2 SNP p.(P549A) could be involved in such mechanism [11].
Discussion
We report here the genetic characteristics of the cohort of 424 HPP patients tested during 22 years.
The distribution of ALPL variants in Europe shows that most of recurrent variants result from a unique mutational event. The variant c.571G>A p.(E191K) was probably born in North Europe and seems to have spread through the whole Europe, but also North America and Australia. We did not find it in South Europe, which seems not due to the low number of chromosomes tested since the variant was detected only once among 40 patients originating from Spain or Portugal [43, 45]. According to the database gnomAD the allele frequency of c.571G>A p.(E191K) in the general European population ranges from 1/210 to 1/410 except in the Finish population where it can reach 1/60, i.e., 1 heterozygote for 30 persons and 1 homozygote for 3600 persons if we applied Hardy Weinberg rule. So, the allele frequency of c.571G>A p.(E191K) in the Finnish population is very high, similar to the frequency of the CFTR variant in cystic fibrosis NM_000592:c.1521_1523del (p.F508del) in North-West Europe (French Britany, UK). It has been suggested that the high frequency of p.F508del resulted from a positive selection of heterozygotes that could have been resistant to some infections. Because its prevalence the question of any possible advantage of c.571G > A p.(E191K) may be addressed. However, by contrast with p.F508del, c.571G > A p.(E191K) is moderate, which may explain its diffusion in European populations, even if there was no positive selection of heterozygotes.
We found that homozygosity is a factor of aggravating severity, at least in the perinatal form where the rate of homozygosity was very high and in infantile form where 100% of m/m genotypes with an unexpected severe phenotype were homozygous. This phenomenon has been observed in other inherited disorders such as hemochromatosis, phenylketonuria, Tay-Sachs disease, or congenital adrenal hyperplasia [46–49].
Another important point is that the three kind of alleles (m, s, Sd) were found in each region, and with a prevalence of similar scale. Besides the fact that this impacts genetic counseling (see above), this means that we expect a similar picture of HPP everywhere in Europe, with severe and moderate, recessive, and dominant HPP. The exception concerns extra-European countries (Middle East) where only recessive alleles s and m were detected, probably because consanguinity lead to a high number of homozygotes, but also perhaps because the number of tested chromosomes was too low to get a comprehensive view of ALPL alleles.
Our results show that dominant HPP is due to at least two distinct mechanisms resulting in two distinct degrees of severity, moderate HPP, and mild HPP. It particularly stands out that patients with mild HPP carry a great variety of alleles, missense variants but also splice site, frameshift, and nonsense variants. We did not test possible DNE of frameshift or splicing variants. However, these variants result in not functional proteins unable to dimerize, and consequently degraded in ER apparatus. It remains possible that the mechanism of dominance involves aggregation of the mutant protein (to be degraded) and the wild-type protein so that they could show a DNE. It has been shown that s alleles c.508A>G p.(N170D), c.602G>A) p.(C201Y), c.1559del p.(L520fs*) are retained in the ER or cis-Golgi network and subjected to degradation in the proteasome [50].
When we compare the allelic compositions of our cohort with the US cohort published in 2015 we found that the non ascertainment of patients with perinatal or adult HPP leads to the loss of s/s and s/N genotypes (Fig. 2b), which strongly suggests that these genotypes are specific for these phenotypes. A second strong argument in favor of specificity of genotypes is that most of the adult patients ascertained in the period 1998–2008 did not carry s/N.
Hypophosphatasia is extraordinarily heterogenous, multisystemic, with many endophenotypes: dental issues, bone fragility, chondrocalcinosis, pain, fatigue etc. We attempted to address the question of particular variants resulting in particular endophenotypes. However, the high allelic heterogeneity did not allow to identify correlations except in few recurrent heterozygous genotypes. This part of the genotype-phenotype correlation was previously studied and reported [11]. Here we just noticed that in heterozygotes some variants were associated with particular clinical forms: c.212G>A p.(R71H) (OdontoHPP), c.1172G>A p.(R391H) and c.648 + 1 G>A (Adult HPP), and c.571G>A p.(E191K) interestingly found associated with Adult HPP in France and with Odonto HPP in North Europe. If this double relationship is confirmed, this may corroborate the role of regional factors triggering haploinsufficiency.
Our results have some important consequences on genetic counseling. Parents of severe cases and heterozygous for s alleles were often reported with normal phenotype by comparison with the index case and his/her specific signs (fractures, low mineralization etc). However it is expected that the parents of patients with « recessive » severe HPP and carrying a s allele may express Mild HPP symptoms.
In conclusion, our data suggest that HPP is composed of three subtypes distinguishable by their genetic characteristics and their prevalence (Fig. 5). Severe HPP (1/300,000) is mostly due to homozygosity or compound heterozygosity for severe variants. Note that the severe phenotype may be easily subdivided into very severe or lethal, that includes genotypes s/s, s/Sd and Sd/Sd, and severe nonlethal (m/m homozygous). Moderate HPP (1/2430) is mainly due to the dominant negative effect of missense variants, and mild adult HPP, characterized by unspecific signs (1/508), is possibly due to a haploinsufficiency mechanism. The new mechanism of dominance is not clearly identified, especially whether other factors act to trigger haploinsufficiency or not. We hypothesize a negative interaction between TNSALP and another actor, possibly the collagen matrix (Fig. 5).
Supplementary information
Acknowledgements
The authors would like to thank the clinicians from 32 countries who gave us the opportunity to test their patients.
Data availability
The authors state the availability of data and materials for replication of findings.
Compliance with ethical standards
Conflict of interest
EM received honoraria from Alexion Pharmaceutical for expertize and presentations.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-020-00732-6) contains supplementary material, which is available to authorized users.
References
- 1.Mornet E. Hypophosphatasia. Orphanet J Rare Dis. 2007;2:40. doi: 10.1186/1750-1172-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mornet E, Nunes ME. Hypophosphatasia. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. GeneReviews((R)). Seattle (WA); University of Washington, 1993. [PubMed]
- 3.Whyte MP. Hypophosphatasia: an overview For 2017. Bone. 2017;102:15–25. doi: 10.1016/j.bone.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Mornet E, Yvard A, Taillandier A, Fauvert D, Simon-Bouy B. A molecular-based estimation of the prevalence of hypophosphatasia in the European population. Ann Hum Genet. 2011;75:439–45.. doi: 10.1111/j.1469-1809.2011.00642.x. [DOI] [PubMed] [Google Scholar]
- 5.Fraser D. Hypophosphatasia. Am J Med. 1957;22:730–46.. doi: 10.1016/0002-9343(57)90124-9. [DOI] [PubMed] [Google Scholar]
- 6.Mornet E. Hypophosphatasia: the mutations in the tissue-nonspecific alkaline phosphatase gene. Hum Mutat. 2000;15:309–15.. doi: 10.1002/(SICI)1098-1004(200004)15:4<309::AID-HUMU2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 7.Mornet E. Genetics of hypophosphatasia. Arch Pediatr. 2017;24:5S51–5S6. doi: 10.1016/S0929-693X(18)30014-9. [DOI] [PubMed] [Google Scholar]
- 8.Mornet E. Hypophosphatasia. Metabolism. 2018;82:142–55.. doi: 10.1016/j.metabol.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Linglart A, Biosse-Duplan M. Hypophosphatasia. Curr Osteoporos Rep. 2016;14:95–105. doi: 10.1007/s11914-016-0309-0. [DOI] [PubMed] [Google Scholar]
- 10.Whyte MP, Zhang F, Wenkert D, McAlister WH, Mack KE, Benigno MC, et al. Hypophosphatasia: validation and expansion of the clinical nosology for children from 25 years experience with 173 pediatric patients. Bone. 2015;75:229–39.. doi: 10.1016/j.bone.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 11.Taillandier A, Domingues C, Dufour A, Debiais F, Guggenbuhl P, Roux C, et al. Genetic analysis of adults heterozygous for ALPL mutations. J Bone Min Metab. 2018;36:723–33.. doi: 10.1007/s00774-017-0888-6. [DOI] [PubMed] [Google Scholar]
- 12.Whyte MP, Greenberg CR, Salman NJ, Bober MB, McAlister WH, Wenkert D, et al. Enzyme-replacement therapy in life-threatening hypophosphatasia. N Engl J Med. 2012;366:904–13.. doi: 10.1056/NEJMoa1106173. [DOI] [PubMed] [Google Scholar]
- 13.Whyte MP. Atypical femoral fractures, bisphosphonates, and adult hypophosphatasia. J Bone Min Res. 2009;24:1132–4. doi: 10.1359/jbmr.081253. [DOI] [PubMed] [Google Scholar]
- 14.Sutton RA, Mumm S, Coburn SP, Ericson KL, Whyte MP. “Atypical femoral fractures” during bisphosphonate exposure in adult hypophosphatasia. J Bone Min Res. 2012;27:987–94.. doi: 10.1002/jbmr.1565. [DOI] [PubMed] [Google Scholar]
- 15.Deeb AA, Bruce SN, Morris AA, Cheetham TD. Infantile hypophosphatasia: disappointing results of treatment. Acta Paediatr. 2000;89:730–3. doi: 10.1080/080352500750044106. [DOI] [PubMed] [Google Scholar]
- 16.Baujat G, Michot C, Le Quan Sang KH, Cormier-Daire V. Perinatal and infantile hypophosphatasia: clinical features and treatment. Arch Pediatr. 2017;24:5S61–5S5. doi: 10.1016/S0929-693X(18)30016-2. [DOI] [PubMed] [Google Scholar]
- 17.Linglart A, Salles JP. Hypophosphatasia: the contribution of imaging. Arch Pediatr. 2017;24:5S74–5S9. doi: 10.1016/S0929-693X(18)30019-8. [DOI] [PubMed] [Google Scholar]
- 18.Bloch-Zupan A. Hypophosphatasia: diagnosis and clinical signs - a dental surgeon perspective. Int J Paediatr Dent. 2016;26:426–38.. doi: 10.1111/ipd.12232. [DOI] [PubMed] [Google Scholar]
- 19.Briot K, Roux C. Adult hypophosphatasia. Curr Opin Rheumatol. 2016;28:448–51.. doi: 10.1097/BOR.0000000000000286. [DOI] [PubMed] [Google Scholar]
- 20.Dahir KM, Tilden DR, Warner JL, Bastarache L, Smith DK, Gifford A, et al. Rare variants in the gene ALPL that cause hypophosphatasia are strongly associated with ovarian and uterine disorders. J Clin Endocrinol Metab. 2018;103:2234–43.. doi: 10.1210/jc.2017-02676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zurutuza L, Muller F, Gibrat JF, Taillandier A, Simon-Bouy B, Serre JL, et al. Correlations of genotype and phenotype in hypophosphatasia. Hum Mol Genet. 1999;8:1039–46.. doi: 10.1093/hmg/8.6.1039. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Wang L, Geng J, Yu T, Yao RE, Shen Y, et al. Characterization of six missense mutations in the tissue-nonspecific alkaline phosphatase (TNSALP) gene in Chinese children with hypophosphatasia. Cell Physiol Biochem. 2013;32:635–44.. doi: 10.1159/000354467. [DOI] [PubMed] [Google Scholar]
- 23.Takinami H, Goseki-Sone M, Watanabe H, Orimo H, Hamatani R, Fukushi-Irie M, et al. The mutant (F310L and V365I) tissue-nonspecific alkaline phosphatase gene from hypophosphatasia. J Med Dent Sci. 2004;51:67–74. [PubMed] [Google Scholar]
- 24.Taillandier A, Sallinen SL, Brun-Heath I, De Mazancourt P, Serre JL, Mornet E. Childhood hypophosphatasia due to a de novo missense mutation in the tissue-nonspecific alkaline phosphatase gene. J Clin Endocrinol Metab. 2005;90:2436–9. doi: 10.1210/jc.2004-1456. [DOI] [PubMed] [Google Scholar]
- 25.Taillandier A, Lia-Baldini AS, Mouchard M, Robin B, Muller F, Simon-Bouy B, et al. Twelve novel mutations in the tissue-nonspecific alkaline phosphatase gene (ALPL) in patients with various forms of hypophosphatasia. Hum Mutat. 2001;18:83–4. doi: 10.1002/humu.1154. [DOI] [PubMed] [Google Scholar]
- 26.Lia-Baldini AS, Muller F, Taillandier A, Gibrat JF, Mouchard M, Robin B, et al. A molecular approach to dominance in hypophosphatasia. Hum Genet. 2001;109:99–108. doi: 10.1007/s004390100546. [DOI] [PubMed] [Google Scholar]
- 27.Fukushi-Irie M, Ito M, Amaya Y, Amizuka N, Ozawa H, Omura S, et al. Possible interference between tissue-non-specific alkaline phosphatase with an Arg54->Cys substitution and acounterpart with an Asp277->Ala substitution found in a compound heterozygote associated with severe hypophosphatasia. Biochem J. 2000;348:633–42. [PMC free article] [PubMed] [Google Scholar]
- 28.Fauvert D, Brun-Heath I, Lia-Baldini AS, Bellazi L, Taillandier A, Serre JL, et al. Mild forms of hypophosphatasia mostly result from dominant negative effect of severe alleles or from compound heterozygosity for severe and moderate alleles. BMC Med Genet. 2009;10:51. doi: 10.1186/1471-2350-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orimo H, Girschick HJ, Goseki-Sone M, Ito M, Oda K, Shimada T. Mutational analysis and functional correlation with phenotype in German patients with childhood-type hypophosphatasia. J Bone Min Res. 2001;16:2313–9. doi: 10.1359/jbmr.2001.16.12.2313. [DOI] [PubMed] [Google Scholar]
- 30.Satou Y, Al-Shawafi HA, Sultana S, Makita S, Sohda M, Oda K. Disulfide bonds are critical for tissue-nonspecific alkaline phosphatase function revealed by analysis of mutant proteins bearing a C(201)-Y or C(489)-S substitution associated with severe hypophosphatasia. Biochim Biophys Acta. 2012;1822:581–8. doi: 10.1016/j.bbadis.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Brun-Heath I, Taillandier A, Serre JL, Mornet E. Characterization of 11 novel mutations in the tissue non-specific alkaline phosphatase gene responsible for hypophosphatasia and genotype-phenotype correlations. Mol Genet Metab. 2005;84:273–7. doi: 10.1016/j.ymgme.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Mentrup B, Girschick H, Jakob F, Hofmann C. A homozygous intronic branch-point deletion in the ALPL gene causes infantile hypophosphatasia. Bone. 2017;94:75–83. doi: 10.1016/j.bone.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 33.Spentchian M, Merrien Y, Herasse M, Dobbie Z, Glaser D, Holder SE, et al. Severe hypophosphatasia: characterization of fifteen novel mutations in the ALPL gene. Hum Mutat. 2003;22:105–6. doi: 10.1002/humu.9159. [DOI] [PubMed] [Google Scholar]
- 34.Del Angel G, Reynders J, Negron C, Steinbrecher T, Mornet E. Large-scale in vitro functional testing and novel variant scoring via protein modeling provide insights into alkaline phosphatase activity in hypophosphatasia. Hum Mutat. 2020;41:1250–62. [DOI] [PMC free article] [PubMed]
- 35.Taillandier A, Domingues C, De Cazanove C, Porquet-Bordes V, Monnot S, Kiffer-Moreira T, et al. Molecular diagnosis of hypophosphatasia and differential diagnosis by targeted Next Generation Sequencing. Mol Genet Metab. 2015;116:215–20.. doi: 10.1016/j.ymgme.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derouault P, Parfait B, Moulinas R, Barrot CC, Sturtz F, Merillou S, et al. ‘COV’COP’ allows to detect CNVs responsible for inherited diseases among amplicons sequencing data. Bioinformatics. 2017;33:1586–8. doi: 10.1093/bioinformatics/btx017. [DOI] [PubMed] [Google Scholar]
- 37.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat. 2016;37:564–9. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 38.Mornet E, Stura E, Lia-Baldini AS, Stigbrand T, Menez A, Le DuMH. Structural evidence for a functional role of human tissue nonspecific alkaline phosphatase in bone mineralization. J Biol Chem. 2001;276:31171–8. doi: 10.1074/jbc.M102788200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Ke YH, Wang C, Yue H, Hu WW, Gu JM, et al. Identification of the mutations in the tissue-nonspecific alkaline phosphatase gene in two Chinese families with hypophosphatasia. Arch Med Res. 2012;43:21–30. doi: 10.1016/j.arcmed.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Herasse M, Spentchian M, Taillandier A, Mornet E. Evidence of a founder effect for the tissue-nonspecific alkaline phosphatase (TNSALP) gene E174K mutation in hypophosphatasia patients. Eur J Hum Genet. 2002;10:666–8. doi: 10.1038/sj.ejhg.5200857. [DOI] [PubMed] [Google Scholar]
- 41.Orton NC, Innes AM, Chudley AE, Bech-Hansen NT. Unique disease heritage of the Dutch-German Mennonite population. Am J Med Genet A. 2008;146A:1072–87.. doi: 10.1002/ajmg.a.32061. [DOI] [PubMed] [Google Scholar]
- 42.Wenkert D, McAlister WH, Coburn SP, Zerega JA, Ryan LM, Ericson KL, et al. Hypophosphatasia: nonlethal disease despite skeletal presentation in utero (17 new cases and literature review) J Bone Min Res. 2011;26:2389–98.. doi: 10.1002/jbmr.454. [DOI] [PubMed] [Google Scholar]
- 43.Riancho-Zarrabeitia L, Garcia-Unzueta M, Tenorio JA, Gomez-Gerique JA, Ruiz Perez VL, Heath KE, et al. Clinical, biochemical and genetic spectrum of low alkaline phosphatase levels in adults. Eur J Intern Med. 2016;29:40–5. doi: 10.1016/j.ejim.2015.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Mornet E. Molecular Genetics of Hypophosphatasia and Phenotype-Genotype Correlations. Subcell Biochem. 2015;76:25–43. doi: 10.1007/978-94-017-7197-9_2. [DOI] [PubMed] [Google Scholar]
- 45.Tenorio J, Alvarez I, Riancho-Zarrabeitia L, Martos-Moreno GA, Mandrile G, de la Flor Crespo M, et al. Molecular and clinical analysis of ALPL in a cohort of patients with suspicion of Hypophosphatasia. Am J Med Genet A. 2017;173:601–10.. doi: 10.1002/ajmg.a.37991. [DOI] [PubMed] [Google Scholar]
- 46.Avigad S, Kleiman S, Weinstein M, Cohen BE, Schwartz G, Woo SL, et al. Compound heterozygosity in nonphenylketonuria hyperphenylalanemia: the contribution of mutations for classical phenylketonuria. Am J Hum Genet. 1991;49:393–9. [PMC free article] [PubMed] [Google Scholar]
- 47.Allen KJ, Gurrin LC, Constantine CC, Osborne NJ, Delatycki MB, Nicoll AJ, et al. Iron-overload-related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358:221–30.. doi: 10.1056/NEJMoa073286. [DOI] [PubMed] [Google Scholar]
- 48.Ohno K, Suzuki K. Multiple abnormal beta-hexosaminidase alpha chain mRNAs in a compound-heterozygous Ashkenazi Jewish patient with Tay-Sachs disease. J Biol Chem. 1988;263:18563–7. [PubMed] [Google Scholar]
- 49.Mornet E, Crete P, Kuttenn F, Raux-Demay MC, Boue J, White PC, et al. Distribution of deletions and seven point mutations on CYP21B genes in three clinical forms of steroid 21-hydroxylase deficiency. Am J Hum Genet. 1991;48:79–88. [PMC free article] [PubMed] [Google Scholar]
- 50.Komaru K, Ishida-Okumura Y, Numa-Kinjoh N, Hasegawa T, Oda K. Molecular and cellular basis of hypophosphatasia. J Oral Biosci. 2019;61:141–8. doi: 10.1016/j.job.2019.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors state the availability of data and materials for replication of findings.