Abstract
Hypothesis-free Mendelian randomization studies provide a way to assess the causal relevance of a trait across the human phenome but can be limited by statistical power, sample overlap or complicated by horizontal pleiotropy. The recently described latent causal variable (LCV) approach provides an alternative method for causal inference which might be useful in hypothesis-free experiments across human phenome. We developed an automated pipeline for phenome-wide tests using the LCV approach including steps to estimate partial genetic causality, filter to a meaningful set of estimates, apply correction for multiple testing and then present the findings in a graphical summary termed causal architecture plot. We apply this pipeline to body mass index (BMI) and lipid traits as exemplars of traits where there is strong prior expectation for causal effects, and to dental caries and periodontitis as exemplars of traits where there is a need for causal inference. The results for lipids and BMI suggest that these traits are best viewed as contributing factors on a multitude of traits and conditions, thus providing additional evidence that supports viewing these traits as targets for interventions to improve health. On the other hand, caries and periodontitis are best viewed as a downstream consequence of other traits and diseases rather than a cause of ill health. The automated pipeline is implemented in the Complex-Traits Genetics Virtual Lab (https://vl.genoma.io) and results are available in https://view.genoma.io. We propose causal architecture plots based on phenome-wide partial genetic causality estimates as a new way visualizing the overall causal map of the human phenome.
Subject terms: Risk factors, Population genetics
Introduction
Associations between causal risk factors and disease can suggest new ways to improve health. Conventional epidemiological studies may uncover correlations but cannot easily disentangle noncausal or reverse-causal relationships where interventions on the putative risk factor will be ineffective. In this article, risk factors are described as “upstream” if they have effects on disease, or “downstream” if the putative risk factor is a marker of or a consequence of the disease.
Dental diseases are good examples of complex diseases, which are associated with a range of poor health outcomes and are hypothesized to be both a cause and consequence of ill health [1], but these associations could be confounded due to limitations of conventional epidemiological methods and may not reflect true causal relationships. In the context of recent calls to prioritize prevention and early interventions, address the global health problem of dental diseases and overcome isolation between dentistry and medicine [2, 3], there is a need to locate dental diseases in the context of causal flow through the human phenome. Conversely, lipid biomarkers such as low-density lipoprotein cholesterol (LDL-C) are good examples of complex traits, which are known to have effects on human health including cardiovascular disease [4, 5] and may act as a positive control for contemporary epidemiological methods, which aim to identify causal relationships.
In recent years, various techniques have been proposed which use genetic data to assess causality in observational studies [6] and these are particularly valuable in situations where large-scale interventional studies would be impractical or unethical. One example is Mendelian randomization (MR), an analytical paradigm that uses genetic variants as proxies for a putative risk factor to test for causal effects on an outcome [7]. In dental epidemiology, this method has been used to examine the effects of potentially modifiable risk factors like vitamin D and body mass index (BMI) on caries and periodontitis [8, 9], to assess the possible impact of periodontitis on hypertension [10] and undertake bidirectional analysis to test for causal relationships between dental diseases and cardio-metabolic traits in both directions [11]. To date, these studies have only explored on a small number of traits and the bespoke experimental design used for each study makes it difficult to compare estimates for different diseases. Dental diseases may therefore serve as a model for complex traits where it would be helpful to perform a causal inference analysis in a systematic manner across the whole phenome.
There are practical challenges meaning that MR may not be the preferred approach for a phenome-wide causal experiment in this context. At its heart, MR experiments rely of vertical pleiotropy, that is to say a genotype with effects on trait A is associated with trait B because trait A affects trait B. It can be difficult to distinguish this from horizontal pleiotropy, where a genetic variant has biological effects on both trait A and trait B. Many genetic variants have horizontally pleiotropic effects, leading to false positive findings or overestimation in effect sizes at true positive associations in classical MR experiments [11, 12]. Several estimation techniques have been developed that use the distribution of causal effect estimates across multiple variants in an attempt to detect and account for [13–15] or at least reduce the impact of horizontal pleiotropy [16]. These methods may, however, introduce additional assumptions about the distribution of effect estimates [17, 18] and run into problems when these assumptions are not met [19], suggesting each estimate produced using these methods may need interpretation on a case by case basis to assess whether the assumptions are reasonable. In addition, MR experiments can produce spurious findings due to sample overlap [20], which can be problematic in phenome-wide studies, as the same underlying population in a consortium or biobank may contribute to the available genome-wide association studies (GWAS) for many different traits. Finally, MR experiments use information from a small number of genetic variants and discard information from most of the genome, meaning that statistical power to detect causal relationship may be limited for a phenome-wide experiment for traits such as dental diseases, which have relatively few robust single variant association signals.
An alternative analytical paradigm—the latent causal variable (LCV) method—has recently been proposed. LCV uses information aggregated across the whole genome to infer potential causal relationships between complex human traits and diseases [21]. In conjunction with large-scale genetic association studies made possible by resources such as UK Biobank [22] and automated pipelines for quality control and analysis such as the Complex-Traits Genetics Virtual Lab (CTG-VL) [23], this method now provides an opportunity to evaluate potentially causal relationships efficiently and at phenome-wide scale. Here we introduce a pipeline implemented in CTG-VL to perform a phenome-wide scan across hundreds of traits using the LCV method and the visualization of the results using causal architecture plots. We showcase this method using publicly available GWAS data for BMI, lipid levels, dental caries, and periodontitis [11].
Methods
Conceptual overview
The genetic correlation between two traits represents the correlation in genetic effect sizes at common genetic variants across traits [24]. The LCV approach initially estimates the genetic correlation between two traits using a modified linkage disequilibrium score regression technique, which can detect and account for sample overlap in genetic association studies [24]. Next, when there is evidence of genetic correlation, the model fits a single unobserved variable (termed L), which is causal for trait A and trait B and that mediates the observed genetic correlation (Fig. 1). To distinguish between horizontal and vertical pleiotropy, the LCV model compares the correlation between L and trait A with the correlation between L and trait B, and estimates a parameter termed genetic causality proportion (GCP). Positive values of GCP suggest the presence of vertical pleiotropy where trait A lies upstream of trait B and interventions on trait A are likely to affect trait B, while negative GCP values indicate that B lies upstream of trait A. GCP values near 0 imply that the genetic correlation between traits A and B is likely to be mediated by horizontal pleiotropy (where trait A and trait B are influenced by shared pathways but trait A and trait B do not lie in same pathway) and interventions on traits A or B are less likely to affect the other trait. A detailed description of the LCV method is provided in the original publication [21].
Fig. 1. Use of a latent variable for causal inference.
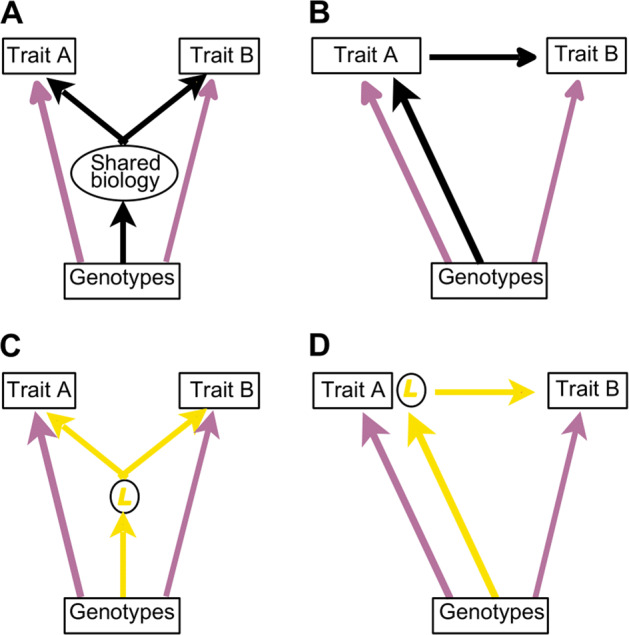
The genetic effects on traits A and B (purple arrows) are correlated in all scenarios. This could be due to horizontal pleiotropy ((a), true causal pathway drawn in black), vertical pleiotropy ((b), true causal pathway drawn in black), or a combination of both processes. In LCV analysis an inferred causal pathway is created which mediates the observed genetic correlation between traits A and B and must always pass through L. Where horizontal pleiotropy mediates the genetic correlation between traits A and B, the genetic correlation between L and traits A and B is similar in magnitude giving a GCP estimate near zero ((c), inferred causal pathway drawn in yellow, true causal pathway shown in (a)). In situations where L has a perfect genetic correlation with trait A, the only effects of genotypes on trait B must be through their effects on trait A ((d), inferred causal pathway drawn in yellow, true causal pathway shown in (b)), analogous to a positive finding in a classical MR experiment using a valid instrument and resulting in a GCP estimate of 1. (Color figure online).
Using the distribution of GCP estimates to infer the causal architecture of a trait
If traits A and trait B are swapped, the GCP estimate is unchanged in magnitude but the sign is reversed. In an experiment involving all pairwise comparisons between n traits this creates symmetry, which is to say for every positive signed GCP estimate observed in the experiment there must be an equal but negatively signed GCP estimate corresponding to the same pair of traits but with the order of traits reversed. If a randomly selected trait from group n has predominantly positive GCP estimates, this implies that the trait is an upstream factor of the majority of other traits in group n. Conversely, if the GCP estimates are predominantly negative, this implies that the trait is a downstream factor of most other traits in group n and interventions on this trait are less likely to change the other straits in group n.
We suggest that if two or more target traits are compared against the same panel of anchor traits, then differences in the distribution of GCP estimates between those traits provide an indication of which target traits may have a greater or lesser causal relevance (assuming GWAS of anchor traits are equally powered—see “Discussion”) for the human phenome, which traits represent upstream determinants of health and which are downstream consequences of other traits. We propose an automated pipeline for obtaining GCP estimates for trait A (hereafter the target trait) against a shared panel of traits B (hereafter the anchor traits) and visualizing the results in a causal architecture plot.
Pipeline stages and implementation
All traits conducted in studies of European ancestry that are available in CTG-VL catalog were selected. CTG-VL is a curated resource of genome-wide association (GWA) summary statistics and downstream analysis [23]. The complete list of GWA summary statistics and references is available in CTG-VL. Briefly, these data were derived from various international genetics consortia and UK Biobank, where the inclusion criteria was a nominally significant (P < 0.05) single nucleotide polymorphism-based heritability derived from LD-score regression [25]. In total, 1389 GWAS are currently available in CTG-VL. References for each of these GWAS are available in CTG-VL.
Traits selection
As a positive control, the analyses were first performed on GWAS summary statistics for high density lipoprotein cholesterol (HDL-C, n = 188,577), LDL-C (n = 188,578), total cholesterol (TC, n = 188,579), triglycerides (TG, n = 188,580) [26], and BMI (n = 339,224) [27] where we expected to observe effects of these traits on a multitude of traits and conditions. We then showcase this pipeline using GWAS summary statistics of dental caries and periodontitis due to a paucity of existing causal evidence. Genetic association data for dental disease traits were taken from GWAS, which combined clinical data from the GLIDE consortium with genetically validated proxy phenotypes from UK Biobank as previously described [11]. Data were combined using a z-score genome-wide meta-analysis weighted by effective sample size. The traits were (a) decayed, missing, and filled tooth surfaces (n = 26,792 from nine studies in GLIDE) and dentures (ncases = 77,714, ncontrols = 383,317 in UK Biobank) and (b) periodontitis (ncases = 17,353, ncontrols = 28,210 from seven studies) and loose teeth (ncases = 18,979, ncontrols = 442,052).
Analysis
The R version (https://github.com/lukejoconnor/LCV) for the LCV method made available by the original authors [21] was implemented in CTG-VL to carry out phenome-wide scans (https://vl.genoma.io). LCV models were fitted in a pairwise manner comparing each target trait against each of the up to 1389 anchor traits using the automated implementation of the LCV method in CTG-VL. For the analysis we used genetic variants present in the HapMap3 data set [28] and LD-scores obtained from European ancestry samples within the 1000 genomes project data (phase 3, 2018 release, provided by provided by Alkes Price’s group, https://data.broadinstitute.org/alkesgroup/LDSCORE/).
Post-processing
LCV estimates are only informative when there is evidence for genetic correlation between the target trait and the anchor trait. First, traits with evidence for a non-zero genetic correlation (Benjamini–Hochberg’s FDR < 5%) were carried forward. Next, we ran LCV analyses to estimate GCP in the remaining traits and again applied a Benjamini–Hochberg’s FDR < 5% to the GCP P value (H0: GCP = 0).
Causal architecture plots
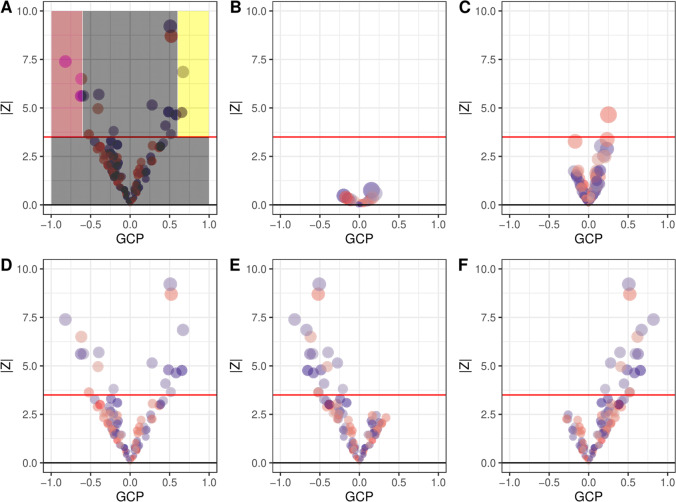
To visualize a target trait in the context of the human phenome, we propose a visual summary termed a causal architecture plot (Fig. 2). Each marker indicates an anchor trait where there is detectable genetic correlation with the target trait so plots with a complex target trait with few markers may indicate low heritability or an underpowered GWAS. The Y-axis represents the strength of evidence for causal relationship between the target trait and anchor trait with a red line indicating which relationships pass multiple test correction, allowing differences between traits with limited causal relevance or widespread causal relevance to be identified. A symmetrical funnel plot indicates equal numbers of upstream and downstream factors for the target trait (e.g., Fig. 2a, d), while an asymmetrical funnel indicates that the causal direction is predominantly from the anchor traits to the target trait (e.g., Fig. 2e) or from the target trait to the anchor traits (Fig. 2f). The markers are colored to show the direction of genetic correlation, which also indicates whether causal relationships are in a trait-increasing or trait-decreasing direction. Finally, the size of markers provides an indication about the precision of the LCV estimates.
Fig. 2. Interpretation of causal architecture plots.
Each panel represents phenome-wide LCV scan of a target trait (trait A), against each of hundreds anchor traits (trait B). Each dot represents a target trait tested against an anchor trait and the red line represents the statistical significance threshold (FDR < 5%). The size of each dot is proportional to GCP z-score, blue dots correspond to traits with positive genetic correlation (rG), while red dots correspond to traits with negative rG. a Schematic showing regions of the plot which represent noncausal relationships (gray), upstream (pink), and downstream (yellow) causal relationships, b Underpowered experiment, c well-powered experiment for a trait with limited causal relevance, d the trait has many causal relationships in both upstream and downstream directions, e a trait which is causally affected by other upstream traits, f a trait with downstream effects. (Color figure online).
Results
Lipid traits
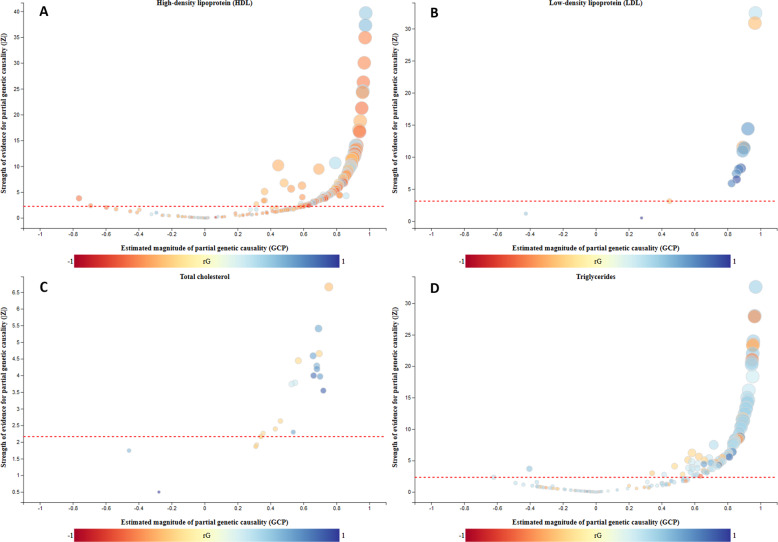
We observed that LDL, HDL, TG, and TC produced causal architecture plots showing only downstream effects on several traits (Fig. 3). Table 1 summarizes the number of causal relationships estimated by LCV per each trait and Supplementary Tables 1–4 show the complete list of results. HDL had trait-decreasing effects on many traits, while TG had predominantly trait-increasing effects. TC and LDL had relatively few genetic correlations, however, a large proportion of these were partially due to causal effects, again, predominantly in a downstream direction.
Fig. 3. Comparison of the causal architecture of lipid traits.
Causal architecture plots of HDL (a), LDL (b), total cholesterol (c) and Triglycerides (d). Each dot represents a target trait tested against an anchor trait and the red line represents the statistical significance threshold (GCP FDR < 5%). The size of each dot is proportional to GCP z-score, blue dots correspond to traits with positive genetic correlation (rG), while red dots correspond to traits with negative rG. Only traits with a genetic correlation at FDR < 5% are shown. (Color figure online).
Table 1.
Summary of the results of the LCV method for each trait.
| Trait | Number of genetic correlations with FDR < 5% | Number of potentially causal relationships (FDR < 5%) | Upstream/downstream traits |
|---|---|---|---|
| HDL | 253 | 140 | 2/140 |
| LDL | 14 | 12 | 0/12 |
| TC | 23 | 18 | 0/18 |
| TG | 186 | 96 | 2/94 |
| BMI | 647 | 133 | 23/110 |
| Caries | 527 | 71 | 71/0 |
| Periodontitis | 398 | 32 | 29/3 |
Each trait was tested against the same panel of 1389 partially heritable traits in CTG-VL catalog. All the results with FDR < 5% are provided in Supplementary Tables 1–7 and full results are browsable using CTG-View platform (https://view.genoma.io).
BMI, caries, and periodontitis
In part, the ability of LCV to resolve clear differences between the four lipid traits might be helped by the relatively simple genetic architecture of these traits. By contrast, complex traits such as BMI that are affected by many different biological processes may provide a more realistic control for comparison against caries and periodontitis.
For BMI, genetic correlations with 647 anchor traits were identified, of which 133 were partially due to causal relationships. The majority of GCP estimates were positively signed, suggesting that BMI may impact many other traits (Fig. 4a), however, there were also several negatively signed relationships, suggesting that BMI itself could potentially be amenable to several different interventions. The upstream trait with the greatest evidence on BMI “employment as a heavy goods vehicle driver,” while the downstream trait with greatest evidence was “vascular/heart problems diagnosed by doctor,” where a lower BMI was associated with greater odds of reporting no vascular or heart problems (Fig. 4a and Supplementary Table 5).
Fig. 4. Comparison of the causal architecture of BMI, DMFS/dentures (caries), and periodontitis.
Causal architecture plots of BMI (a), DMFS/dentures (caries) (b), and periodontitis (c). Each dot represents a target trait tested against an anchor trait and the red line represents the statistical significance threshold (GCP FDR < 5%). The size of each dot is proportional to GCP z-score, blue dots correspond to traits with positive genetic correlation (rG), while red dots correspond to traits with negative rG. All the statistically significance results (FDR < 5%) are shown in Supplementary Tables 5–7 and can be queried at https://view.genoma.io. (Color figure online).
For dental caries proxied by DMFS/dentures, there were detectable genetic correlations with 527 anchor traits, of which 71 supported partially causal relationships (Fig. 4b). All GCP estimates were negatively signed, suggesting that DMFS/dentures are more likely a downstream consequence of these traits rather than an upstream risk factor. Traits with evidence for partial genetic causality included harmful effects of variables capturing dietary habits, smoking, hypertensive diseases, and obesity, while a protective effect was observed for variables representing skilled employment and education (Fig. 4b and Supplementary Table 6).
For periodontitis proxied by the combination of periodontitis/loose teeth, 398 genetic correlations were detected at FDR < 5%, of which a relatively small faction (32 anchor traits) was modeled to be partially due to causal relationships. The directions of causal relationship were predominantly negatively signed (29 out of 32 traits), suggesting periodontitis was more likely the downstream consequence of these traits (Fig. 4c). The five traits with the strongest evidence for partial genetic causality were: (a) a harmful effect of drug or alcohol use for anxiety on periodontitis, (b) a protective effect of fairer skin color on periodontitis, (c) a harmful effect of peripheral artery disease on periodontitis, (d) an effect of periodontitis on dietary preference (proxied by preferred type of milk), and (e) a protective effect of a variable representing absence of problematic alcohol consumption. Periodontitis appeared to have a causal effect on other dental problems and increase in the use of dentures.
Discussion
Previous approaches to obtaining phenome-wide causal maps have been based around the MR paradigm [29]. The LCV method has attractive properties for phenome-wide analysis as it is robust to sample overlap, has greater statistical power than MR [17], and is unconfounded by horizontal pleiotropy [21]. We implemented a pipeline to automate LCV analysis and visualize results in causal architecture plots, and applied this to lipid traits and BMI as positive controls, and to caries and periodontitis as exemplars of complex traits where there is a need for additional causal evidence. The results suggest that, at a high level, dental diseases are embedded in the human phenome but best viewed as a downstream marker of biological events and a consequence of other diseases rather than as a driver of biological changes, which lead to large or widespread changes in other traits. The results therefore support the current drive to target upstream determinants of dental diseases [3] and potentially provide a framework for prioritizing subsets of traits, which have shown causal relevance for further validation or translational research. Specifically, the results for caries and periodontitis prioritize socio-economic status, cardiovascular health, diet, and mental health/alcohol use as traits, which could be targeted to improve dental health. Conversely, the results for HDL-C confirm that interventions on HDL-C are likely to have protective effects on many traits and diseases, and that BMI is a trait with many causal relationships in both upstream and downstream directions. We suggest that this pipeline may be helpful to researchers undertaking initial characterization of a phenotype, and have implemented it as part of CTG-VL, a freely available online resource (https://vl.genoma.io).
The LCV method requires GWAS summary statistic data and needs to identify a genetic correlation between the target trait and anchor trait for the results to be meaningful. It was therefore only possible to examine traits that have been studied using a large enough GWAS to yield a stable heritability estimate. While this captures many important diseases, risk factors and intermediate traits reflected by the large number of anchor traits, there are natural limitations to the results, which are available at this moment in time. For example, risk factors or outcomes such as the oral microbiota composition, oral health quality of life, dental anxiety, and satisfaction with dental appearance and function may be causally related to dental diseases but are not represented by current GWAS. For dental diseases specifically, this illustrates the need to ensure that oral and dental health is represented in epidemiological studies using current methods to avoid perpetrating the underrepresentation of dentistry in the next generation of epidemiological research. As the number of curated GWAS summary statistics in CTG-VL catalog increases over time, this limitation will become less important. It will become possible to construct more detailed causal architecture plots for any given target trait, and it may be possible to move from single-trait causal profiles toward multi-trait profiles, which present an overall causal map of the human phenome.
This pipeline is primarily intended to give an overview of a trait’s putative causal profile and to help identify novel and interesting relationships that can then be investigated further through additional epidemiology and statistical genetics methods including MR. In this study, one interesting pair of findings was that hair color appears to be an upstream determinant of dental caries and that skin color appears to be a risk factor for periodontitis. These findings may have a biological explanation (e.g., both ancestry and skin color are associated with periodontitis in observational studies [30, 31], skin color is associated with caries in children with a possible mechanism related to vitamin D [32], and hair keratins have a role in enamel formation, which might predispose to caries [33, 34]). Alternatively, the findings may also reflect complexity introduced by the scale and sampling frame of UK Biobank. Although the LCV model is more robust than MR to biases due to horizontal pleiotropy and sample overlap, the LCV model may become biased by correlation between genetic variation and environmental factors, which affect disease [17, 35]. This aggregation might be due to factors such as ancient ancestry [36], genetic nurture effects [37], or sampling phenomena [38] and is a concern in the UK Biobank sample [39] where much of the data used in this experiment were obtained. Interpreted in this light, it is possible that environmental factors that are more prevalent in groups of people with certain hair type or skin color are a cause of dental diseases. This example may therefore illustrate some of the challenges created by population stratification, but also the opportunities for genetic information to inform research about social and environmental factors, which may affect disease.
Previous studies using MR methods have found some evidence for causal effects of caries and periodontitis on cardiovascular health traits [10, 11], which was not recapitulated using the LCV method. In part, this may be because LCV aims to captures the overall or predominant direction of causality mediated by a single latent variable and may therefore be a poor fit to systems with complex features such as polytonicity, nonlinear effects, or bidirectional causality. We suggest that the causal architecture plots are used to provide an overall causal context to a trait as an adjunct to other methods to assert causality, which have different strengths and limitations. Despite this, profiles for lipid traits were obtained under the same analytical conditions but appear strikingly different, providing a clear indication that the method can resolve major differences in causal architecture between diseases.
It is important to recognize the limitations of this work. Here, we presented a pipeline to do a phenome-wide scan of potential causal associations. However, it is important to note that the current set of GWAS does not encompass the complete phenome and this is biased toward well-powered GWAS and thus restricted to common diseases and traits. As the range of GWAS studies increases with time, this limitation will become less prominent. It is also important to recognize that GCP estimates are also tied to the statistical power of the GWAS, thus impacting the ability to detect causal associations for specific traits. Low statistical power of GWAS does not, however, bias the model toward positive or negative values of GCP, so the distribution of positive or negative values of GCP estimates will still be informative even when there are relatively few causal associations identified. Another limitation is that in contrast to MR methods, the LCV approach does not infer the magnitude of effects of risk factors on a trait. Finally, the model assumes that the GWAS for both traits and reference LD data are drawn from the same underlying population, which at present limits this pipeline to analysis of studies of European ancestry participants. As the number of GWAS studies in diverse populations increases and additional reference data sets become available, it may be possible to extend this method to non-European populations.
In summary, we present a pipeline to estimate and visualize GCP across traits with GWAS summary statistics implemented in CTG-VL. All the results are freely available for download in https://view.genoma.io.
Supplementary information
Acknowledgements
SH is funded by a National Institute for Health Research (NIHR) Academic Clinical Fellowship. MER is funded NHMRC and Australian Research Council (ARC), through a NHMRC-ARC Dementia Research Development Fellowship (GNT1102821). PFK is funded by an Australian Government Research Training Program Ph.D. Scholarship and QIMR Berghofer Postgraduate Top-Up Scholarship. GCP is funded by an Australia Research Council Discovery Early Career Researcher Award (DE180100976).
Data availability
No participant-level data were accessed to produce this article. The sources of GWAS summary statistics and reference used to perform analysis are described in full in the methods. Links and references for specific datasets are available at https://view.genoma.io.
Compliance with ethical standards
Conflict of interest
GC-P contributed to this study while employed by the University of Queensland, but he is now an employee of 23andMe Inc. He still maintains CTG-VL along PFK, MER and LDH. All other authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-020-00734-4) contains supplementary material, which is available to authorized users.
References
- 1.Chapple ILC, Bouchard P, Cagetti MG, Campus G, Carra M-C, Cocco F, et al. Interaction of lifestyle, behaviour or systemic diseases with dental caries and periodontal diseases: consensus report of group 2 of the joint EFP/ORCA workshop on the boundaries between caries and periodontal diseases. J Clin Periodontol. 2017;44:S39–51. doi: 10.1111/jcpe.12685. [DOI] [PubMed] [Google Scholar]
- 2.Peres MA, Macpherson LMD, Weyant RJ, Daly B, Venturelli R, Mathur MR, et al. Oral diseases: a global public health challenge. Lancet. 2019;394:249–60.. doi: 10.1016/S0140-6736(19)31146-8. [DOI] [PubMed] [Google Scholar]
- 3.Watt RG, Daly B, Allison P, Macpherson LMD, Venturelli R, Listl S, et al. Ending the neglect of global oral health: time for radical action. Lancet. 2019;394:261–72.. doi: 10.1016/S0140-6736(19)31133-X. [DOI] [PubMed] [Google Scholar]
- 4.Armitage J, Baigent C, Barnes E, Betteridge DJ, Blackwell L, Blazing M, et al. Efficacy and safety of statin therapy in older people: a meta-analysis of individual participant data from 28 randomised controlled trials. Lancet. 2019;393:407–15.. doi: 10.1016/S0140-6736(18)31942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baigent C, Keech A, Kearney PM, Blackwell L, Buck G, Pollicino C, et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–78. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 6.Pingault J-B, O’Reilly PF, Schoeler T, Ploubidis GB, Rijsdijk F, Dudbridge F. Using genetic data to strengthen causal inference in observational research. Nat Rev Genet. 2018;19:566–80.. doi: 10.1038/s41576-018-0020-3. [DOI] [PubMed] [Google Scholar]
- 7.Lawlor DA. Commentary: two-sample Mendelian randomization: opportunities and challenges. Int J Epidemiol. 2016;45:908–15.. doi: 10.1093/ije/dyw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudding T, Thomas SJ, Duncan K, Lawlor DA, Timpson NJ. Re-examining the association between vitamin D and childhood caries. Plos ONE. 2015;10:13. doi: 10.1371/journal.pone.0143769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shungin D, Cornelis MC, Divaris K, Holtfreter B, Shaffer JR, Yu Y-H, et al. Using genetics to test the causal relationship of total adiposity and periodontitis: Mendelian randomization analyses in the Gene-Lifestyle Interactions and Dental Endpoints (GLIDE) Consortium. Int J Epidemiol. 2015;44:638–50.. doi: 10.1093/ije/dyv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czesnikiewicz-Guzik M, Osmenda G, Siedlinski M, Nosalski R, Pelka P, Nowakowski D, et al. Causal association between periodontitis and hypertension: evidence from Mendelian randomization and a randomized controlled trial of non-surgical periodontal therapy. Eur Heart J. 2019;40:3459–70. 10.1093/eurheartj/ehz646. [DOI] [PMC free article] [PubMed]
- 11.Shungin D, Haworth S, Divaris K, Agler CS, Kamatani Y, Keun Lee M, et al. Genome-wide analysis of dental caries and periodontitis combining clinical and self-reported data. Nat Commun. 2019;10:2773. doi: 10.1038/s41467-019-10630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8. doi: 10.1038/s41588-018-0099-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25.. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhaom Q, Wang J, Hemani GH, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. arXiv. 2018. https://arxiv.org/abs/1801.09652.
- 16.Bowden J, Davey, Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14.. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koellinger PD, de Vlaming R. Mendelian randomization: the challenge of unobserved environmental confounds. Int J Epidemiol. 2019;48:665–71.. doi: 10.1093/ije/dyz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rees JMB, Wood AM, Burgess S. Extending the MR-Egger method for multivariable Mendelian randomization to correct for both measured and unmeasured pleiotropy. Stat Med. 2017;36:4705–18.. doi: 10.1002/sim.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377–89.. doi: 10.1007/s10654-017-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40:597–608. doi: 10.1002/gepi.21998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor LJ, Price AL. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat Genet. 2018;50:1728–34.. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuéllar-Partida G, Lundberg M, Kho PF, D’Urso S, Gutiérrez-Mondragón LF, Ngo TT, et al. Complex-Traits Genetics Virtual Lab: a community-driven web platform for post-GWAS analyses. BioRxiv. 2019. 10.1101/518027.
- 24.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274. [DOI] [PMC free article] [PubMed]
- 27.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Felix R, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–U401.. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The International HapMap 3 Consortium. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52. [DOI] [PMC free article] [PubMed]
- 29.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naorungroj S, Slade GD, Divaris K, Heiss G, Offenbacher S, Beck JD. Racial differences in periodontal disease and 10-year self-reported tooth loss among late middle-aged and older adults: the dental ARIC study. J Public Health Dent. 2017;77:372–82.. doi: 10.1111/jphd.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peres MA, Antunes JLF, Boing AF, Peres KG, Bastos JLD. Skin colour is associated with periodontal disease in Brazilian adults: a population-based oral health survey. J Clin Periodontol. 2007;34:196–201. doi: 10.1111/j.1600-051X.2006.01043.x. [DOI] [PubMed] [Google Scholar]
- 32.Gyll J, Ridell K, Öhlund I, Karlsland Åkeson P, Johansson I, Lif, et al. Vitamin D status and dental caries in healthy Swedish children. Nutr J. 2018;17:11. doi: 10.1186/s12937-018-0318-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duverger O, Ohara T, Shaffer JR, Donahue D, Zerfas P, Dullnig A, et al. Hair keratin mutations in tooth enamel increase dental decay risk. J Clin Invest. 2014;124:5219–24.. doi: 10.1172/JCI78272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duverger O, Beniash E, Morasso MI. Keratins as components of the enamel organic matrix. Matrix Biol. 2016;52-54:260–5. doi: 10.1016/j.matbio.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawson DJ, Davies NM, Haworth S, Ashraf B, Howe L, Crawford A, et al. Is population stratification in the genetic biobank era irrelevant, a challenge, or an opportunity? Hum Genet. 2020;139:23–41. 10.1007/s00439-019-02014-8. [DOI] [PMC free article] [PubMed]
- 36.Leslie S, Winney B, Hellenthal G, Davison D, Boumertit A, Day T, et al. The fine-scale genetic structure of the British population. Nature. 2015;519:309. doi: 10.1038/nature14230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, et al. The nature of nurture: effects of parental genotypes. Science. 2018;359:424–8. doi: 10.1126/science.aan6877. [DOI] [PubMed] [Google Scholar]
- 38.Taylor AE, Jones HJ, Sallis H, Euesden J, Stergiakouli E, Davies NM, et al. Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2018;47:1207–16. 10.1093/ije/dyy060. [DOI] [PMC free article] [PubMed]
- 39.Haworth S, Mitchell R, Corbin L, Wade KH, Dudding T, Budu-Aggrey A, et al. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat Commun. 2019;10:333. doi: 10.1038/s41467-018-08219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No participant-level data were accessed to produce this article. The sources of GWAS summary statistics and reference used to perform analysis are described in full in the methods. Links and references for specific datasets are available at https://view.genoma.io.