Abstract
Cycads have developed a complex root system categorized either as normal or coralloid roots. Past literatures revealed that a great diversity of key microbes is associated with these roots. This recent study aims to comprehensively determine the diversity and community structure of bacteria and fungi associated with the roots of two Cycas spp. endemic to China, Cycas debaoensis Zhong & Chen and Cycas fairylakea D.Y. Wang using high-throughput amplicon sequencing of the full-length 16S rRNA (V1-V9 hypervariable) and short fragment ITS region. The total DNA from 12 root samples were extracted, amplified, sequenced, and analyzed. Resulting sequences were clustered into 61 bacteria and 2128 fungal OTUs. Analysis of community structure revealed that the coralloid roots were dominated mostly by the nitrogen-fixer Nostocaceae but also contain other non-diazotrophic bacteria. The sequencing of entire 16S rRNA gene identified four different strains of cyanobacteria under the heterocystous genera Nostoc and Desmonostoc. Meanwhile, the top bacterial families in normal roots were Xanthobacteraceae, Burkholderiaceae, and Bacillaceae. Moreover, a diverse fungal community was also found in the roots of cycads and the predominating families were Ophiocordycipitaceae, Nectriaceae, Bionectriaceae, and Trichocomaceae. Our results demonstrated that bacterial diversity in normal roots of C. fairylakea is higher in richness and abundance than C. debaoensis. On the other hand, a slight difference, albeit insignificant, was noted for the diversity of fungi among root types and host species as the number of shared taxa is relatively high (67%). Our results suggested that diverse microbes are present in roots of cycads which potentially interact together to support cycads survival. Our study provided additional knowledge on the microbial diversity and composition in cycads and thus expanding our current knowledge on cycad-microbe association. Our study also considered the possible impact of ex situ conservation on cyanobiont community of cycads.
Keywords: Cycas, coralloid root, diversity, metagenomics, root microbiome
Introduction
Plant roots are considered as repertoire of diverse community of microorganisms as it offers promising environment by providing organic compounds and stability in soil.1 They selectively invite and allows the assembly of microbial communities not only in the soil and root surface (rhizoplane and rhizosphere) but also within their compartments (endosphere).2 The assemblage of microbes in the root zone in turn contribute beneficial effects to their host plants by enhancing growth, elevating immunity toward microbial enemies, and stimulating tolerance to abiotic stress.3,4 Apparently, some plants display intricate root systems like the extraordinary event in the members of known ancient gymnosperm, Cycadales (Cycads).
Cycads are the most primitive living seed plants which is believed to exist around 300 million years ago but has encountered minimal or very slow evolution.5 They have faced extreme competitions and overcome environmental stressors, for example, severe drought and very low nutrient soil, a condition where most plants would have difficulty to survive.6 Past studies suggested that the secret to this success lies in the diverse microbial flora associated with their roots.7,8 Cycads can form two root types which differ morphologically and anatomically and categorize as coralloid and regular (normal) roots.8 The regular or normal roots are composed of tuberous primary roots arising from the main trunk of the plant with lateral secondary roots originating from the primary root. Normal roots can also develop aerially particularly arising at the base of their trunks.9 Of interest, like a case in the coniferous Pinus sp., the gymnosperm Cycadales also forms highly specialized coralloid roots as extension and irreversible modifications of their secondary roots. It grows laterally, forming dichotomous branching, small clusters of coral-like structures which spreads on the surface of the ground or above the soil up to 10 cm.6 Coralloid roots have been known to exist in all extant species of cycads.10 Their importance in Cycadales have been clearly demonstrated as it permits the invasion of symbiotic cyanobacteria where it forms a visible green ring around the cortex.11
Studies on cyanobacterial association with coralloid roots started in the early 1900s (Grilli Caiola, 1980)12 and developed significantly in recent years as high throughput sequencing technologies and bioinformatics pipelines becomes available. Other heterotrophic bacterial complexes, e.g. Rhizobiales (Proteobacteria) in addition to the dominating Nostocales (Cyanobacteria), were found in the coralloid roots of cycad species, that is, Mexican Dioon spp.,13,14 Ceratozamia mexicana, South African Encephalartos villosus,7 and epiphytic Panamian Zamia pseudoparasitica.15 Although receiving very little attention, eukaryotic microbes such as fungi were also detected in cycad roots. The first evidence of fungal colonization came from visual observation of Fisher and Vovides16 in the normal roots of some Zamiaceae species. Recently, metagenomics using amplicon sequencing uncovered high microbial population in Asian Cycas bifida8 and native Chinese Cycas panzhihuaensis.17 In these studies, a relatively high diversity of endophytic bacteria and fungi was recovered not only in roots but also in aerial compartments of cycads. It is believed that these microbes are providing beneficial effects on cycads to help these plants thrive in harsh environments. Cyanobacteria under the genus Nostoc were mostly the predominating symbionts of the coralloid roots across cycad species while members of Proteobacteria populate the normal roots.8,18 In view of the fungal communities, the communal families were under Ascomycota such as Trichocomaceae and Nectriaceae as reported in the recent articles.8,17 Despite these research efforts, root associated microbes of other cycad species especially those that are endemic in one area have not yet been conducted. Moreover, the similarity and differences of microbial community between coralloid and normal roots of different Cycas spp. have been rarely investigated.
Cycas, as the oldest and only genus in Cycadaceae, currently has 114 species which are present worldwide wherein 20% are native to China including Cycas debaoensis Zhong & Chen19 and Cycas fairylakea D.Y. Wang.20 The former species is native to Guangxi which grows on various soil types inhabiting sandy areas and karst limestone hills.21 On the other hand, the natural habitat of Cycas fairylakea in Guangdong province is characterized by humid condition, with fertile soil and moderate sunlight.22 Both species were included as critically endangered in the list of IUCN and hence subjected to ex situ conservation in botanical gardens in China. In fact, several adult individuals of C. debaoensis and C. fairylakea, also surveyed in this study have been uprooted from their natural population and transferred to botanical garden. The primary aim of the present study was to provide baseline information on root microbiomes of two Cycas spp. using amplicon-based sequencing. In particular, we hypothesized that there will be differences on the associated microbial communities between the two root types, and between the host species, Cycas debaoensis and Cycas fairylakea. Moreover, our study investigated and provided insights on the possible impact of ex situ conservation practice on the composition and diversity of symbiotic cyanobacteria in the coralloid roots of the two model Cycas spp. Knowledge on microbial diversity, abundance, and composition in the model plant hosts could further expand our understanding of host-microbe associations and the ecological importance of symbiosis. In addition, it serves as a primary step in discovering potential applications of microbes as practical solutions to common environmental and plant-related problems.
Materials and Methods
Sample collection, time, and location
Coralloid roots and tuberous non-coralloid roots, here termed as normal roots, were collected during April to June 2018 and April to May 2019, respectively. The weather condition at the collection sites during the time of sampling was mildly warm (22-28°C). The average rainfall ranges from 150 mm in April up to 350 mm in June with a relative humidity of 80%. These conditions favored growth of coralloid roots, and hence, sampling was conducted during this period. Different individuals of each host plant – Cycas debaoensis and Cycas fairylakea – with almost the same age range, that is, 15 to 20 years, originating from different environments: natural habitat and botanical garden (ex situ conservation area) were chosen for the collection of coralloid and normal roots. This resulted to a total of 12 root samples examined. Due to difficulty of finding good quality tuberous normal root at the time of sampling, normal root samples were not collected from the same plant individual where the coralloid roots were sampled. Description of the collected root samples was listed in Table 1.
Table 1.
List of 12 root samples used in the study. The host localities, approximate age of plant hosts in their present habitat at the time of sampling and other important remarks were indicated.
| Host species | Coralloid root | Locality | Age of the host plant | Years planted in the botanical garden | Additional remarks |
|---|---|---|---|---|---|
| Cycas debaoensis | CD1 | BGa | 15-20 years | 15 years | Located distantly (–300 m) from C. fairylakea, in slightly higher elevation, shady and hidden to visitors, planted near each other |
| CD2 | BG | 15-20 years | 15 years | ||
| CD3 | BG | 15-20 years | 15 years | ||
| Cycas fairylakea | CF1 | NH-BGb | 15-20 years | 1 year | Located at a lower elevation, expose to sunlight, near the entrance of Cycad Germplasm Conservation Center |
| CF2 | NHc | 15-20 years | – | ||
| CF3 | NH-BG | 15-20 years | 1 year | Located near CF1 | |
| Host species | Normal root | Locality | Additional remarks | ||
| Cycas debaoensis | ND1 | BG | 15-20 years | 15 years | On-trunk roots |
| ND2 | BG | 15-20 years | 15 years | Below-ground roots | |
| ND3 | BG | 15-20 years | 15 years | Below-ground roots | |
| Cycas fairylakea | NF1 | NH-BG | 15-20 years | 2 years | On-trunk roots |
| NF2 | NH-BG | 15-20 years | 2 years | Below-ground roots | |
| NF3 | NH | 15-20 years | – | Below-ground roots |
BG: Botanical Garden in Fairy Lake (FLBG), Shenzhen.
NH-BG: Transplanted adult individuals from natural habitat to botanical garden, the age range of the hosts and the years of existence in the botanical garden after transplantation were given.
NH: Natural habitat (wild population) in Shenzhen/North Guangdong.
Clusters of healthy and fresh coralloid root samples were collected from each plant. Note that coralloid root samples were visually examined for the presence of a cyanobacterial zone, or green ring as this differentiates them from precoralloid stage and indicates a mature symbiotic root. After collection, root samples were placed in ice and immediately transferred to the laboratory for further processing. Soil debris were gently washed off with iced milliQ water repeatedly until the washed-off water became clear. Due to the delicate structure and small size of coralloid roots, tissue sterilization using sodium hypochlorite and ethanol was not conducted to keep the roots intact and the cyanobacterial zone inside the roots protected. Therefore, all coralloid roots were further rinsed with cold sterile distilled water until clear water was finally observed. On the contrary, the normal roots were surface-sterilized with 1-minute washes of 2.5% sodium hypochlorite and 80% ethanol to rinse-off adhering soil and other debris. Clean coralloid and normal roots were then cut into small pieces, around 2 cm in length, using sterilized shears and immediately frozen in liquid nitrogen and stored at −80°C until further processing.
DNA extraction
Frozen coralloid and normal root samples were pulverized to a fine powder using a sterile mortar and pestle. Total genomic DNA was isolated using a modified CTAB method. CTAB extraction buffer (2X CTAB, PVP and 0.2% mercaptoethanol) was mixed with 3 g pulverized coralloid or normal roots followed by 30 minutes incubation at 65°C. DNA was then extracted using phenol-chloroform-isoamylol (25:24:1) mixture and precipitated using isopropyl alcohol and 5 M NaCl. A series of washing steps using 76% and 100% ethanol was then conducted. DNA quality and concentrations were evaluated using NanoDrop 2000 at A260/280. Only the DNA extracts with ratios between 1.8 and 2.0 were used for further analyses.
Full-length 16S rRNA and ITS gene sequencing, processing and taxonomic assignment
Meta-barcoding has been proven useful in assessing associated microbiome in various plant niches as demonstrated in previous studies.8,17 However, utilization of short fragments (–500 bp) are sometimes insufficient to resolve complete bacterial identification.23 Thus, we integrated the third generation long read sequencing technology to amplify the full-length 16S rRNA comprising the nine (V1-V9) hypervariable regions (–1500 bp). The use of long-read sequencing over short-read sequencing provides better taxonomic accuracy with the ability to distinguish species and strain level.24 16S rRNA genes were amplified using the specific primers for V1-V9 hypervariable region with the pair combination of 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTAYGACTT-3′).25 For meta-barcoding of fungi, Internal Transcribe Spacer (ITS) region was amplified with primers ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) targeting the conserved regions of 5.8S and 28S rDNAs. PCR reactions contained 25 μl 2× Premix Taq (Takara Biotechnology, Dalian Co. Ltd., China), 1 μl of each primer (10 M) and 3 μl DNA template (20 ng/μl) in a final volume of 50 μl. PCR amplification was performed on a BioRad S1000 (Bio-Rad Laboratory, CA, USA) with the following protocol: initialization (5 minutes at 94°C), 30 cycles of denaturation (30 seconds at 94°C), annealing (30 s at 52°C), and extension (30 s at 72°C). A final elongation step was then conducted for 10 minutes at 72°C. The length and concentration of the PCR products were determined using 1% agarose gel electrophoresis. Only samples with bright bands between 16S rRNA V1-V9 (1400-1500 bp) were used for further experiments. PCR products were mixed in equal ratios according to the GeneTools Analysis Software (Version 4.03.05.0, SynGene). Then, the mixed PCR products were purified with EZNA Gel Extraction Kit (Omega, USA). Sequencing libraries were generated using NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (New England Biolabs, MA, USA) following manufacturer’s recommendations. Index codes were added afterwards. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Fisher Scientific, MA, USA) and Agilent Bioanalyzer 2100 system (Agilent Technologies, Waldbron, Germany). Finally, the library was sequenced on an IlluminaHiseq2500 platform and 250 bp paired-end reads were generated (Guangdong Magigene Biotechnology Co., Ltd. Guangzhou, China). Paired-end raw reads were quality-filtered using Trimmomatic (V0.33, http://www.usadellab.org/cms/?page=trimmomatic) and assembled using FLASH (V1.2.11, https://ccb.jhu.edu/software/FLASH/). Sequences were then assigned to each sample based on their unique barcode and primer using Mothur software (V1.35.1, http://www.mothur.org). Analysis of sequences were performed by using usearch software (V10, http://www.drive5.com/usearch/) where sequences with ⩾97% similarity were assigned to the matching OTU (Operational Taxonomic Unit). This software also removes chimeric and singleton OTU sequences. An OTU is hypothetically representing a species. The most frequently occurring sequence was extracted as representative sequence for each OTU and was screened for further annotation. For each representative sequence, the SILVA (for 16S rRNA, https://www.arb-silva.de/) and UNITE (for ITS, http://unite.ut.ee/index.php) databases were used to annotate taxonomic information wherein the confidence threshold was set to default (⩾0.5). OTUs tagged as chloroplast and mitochondria were then filtered prior to data normalization.
Diversity analysis
The relative abundance of species at each classification level was determined and used to draw histograms. Alpha and beta diversity indices were also used to analyze microbial community among root samples. For Alpha diversity indices, for example, Chao1, Shannon, and Simpson (1-D) indices were computed to analyze complexity of species diversity for a sample. The first index was selected to identify community richness while the latter two indices were used to determine community diversity. All these indices in our samples were calculated using software PAST (Paleontological Statistics; V4.03) and graphs were plotted in Microsoft Excel. For the analysis of Beta diversity, Principal Coordinate Analysis (PCoA) was performed to get principal coordinates and visualize complex, multidimensional data. A distance matrix of unweighted unifrac among samples were obtained prior to transforming the matrix to a new set of orthogonal axes, by which the maximum variation factor is demonstrated by first principal coordinate, and the second maximum one by the second principal coordinate, and so forth. PCoA analysis was presented by QIIME2 (V1.9.1) and ggplot2 package in R software (V2.15.3). Sample cluster analysis was performed as a UPGMA (Unweighted Pair-group Method with Arithmetic Means) method to interpret the distance matrix using average linkage and was conducted by upgma_cluster.py script (http://qiime.org/scripts/upgma_cluster.html) in QIIME software based on the unweighted unifrac distance matrix. Principal Coordinates Analysis (PCoA) using Bray-Curtis as distance matrix was also performed and visualized in the R environment.26
Phylogeny of cyanobacteria associated to two Cycas spp.
The 16S rRNA full-length sequences of the OTUs of cyanobacteria identified from two Cycas spp. were extracted from the datasets and aligned using the program MAFFT v.45027 with default parameters in Geneious Prime 2020.2.28 The 16S rRNA sequences of other cyanobacterial strains with symbiotic and free-living origin were retrieved from GenBank and included in the alignment. A total of 74 sequences with −1400 bp nucleotide sites was generated. Best fit model was determined under the Bayesian information criterion (BIC) using IQ-TREE v6.10.29 The resulting nucleotide substitution model was TIM3+I+G and used to perform maximum likelihood (ML) analysis with a total of 1000 replicates on IQ-TREE web server. Furthermore, Bayesian inference was also constructed in Geneious Prime-MrBayes3.2.630 with the general time reversible plus gamma distributed with invariant site (GTR+I+G) as the model. In the BI analysis, the setting of 2 MCMC running for 1 million generations, with 25% of the sampled trees discarded as burn-in was done. The phylogenetic trees were then viewed in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree) and Gloeobacter violaceus was selected to root the tree. Sequences of cyanobacteria were deposited in the NCBI GenBank under the accession numbers MW281057, MW281058, MW281059, MW281060.
Results
Sequence metrices and diversity analyses between root types and host species
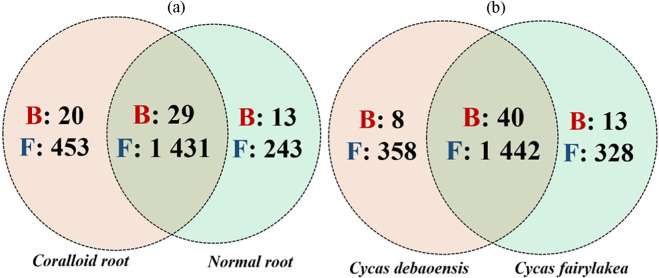
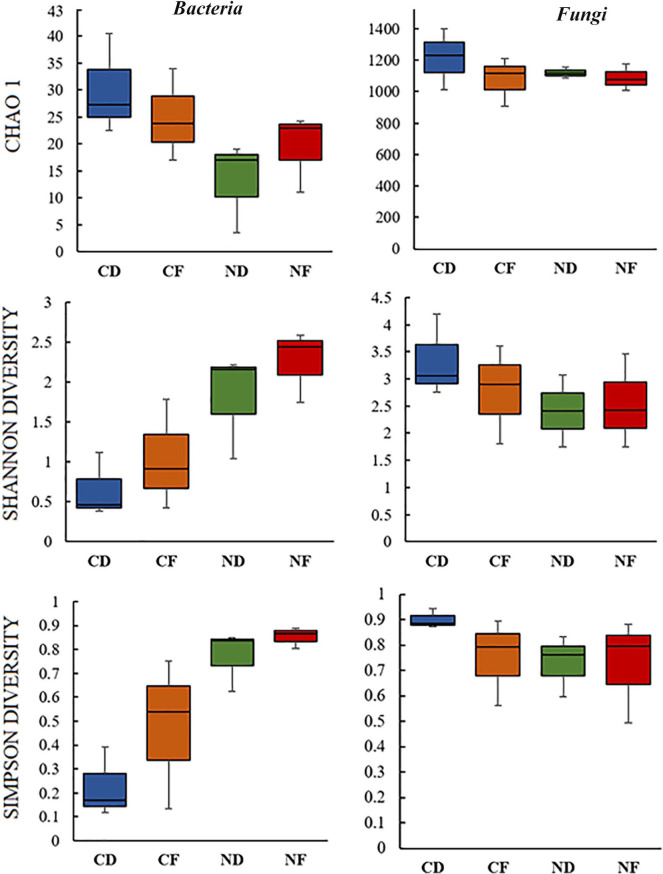
The present study focused on bacterial and fungal sequences, hence, the co-amplified archaeal, mitochondrial, and chloroplast sequences of 16S rRNA, and the sequence variants of other eukaryotic organisms such as plant, chromista, and other protists were removed prior to data analysis. A total of 2 691 656 high quality raw reads were generated, of which 2 680 923 were from ITS fragment (1 398 923 for coralloid roots; 1 282 000 for normal roots) and 10 733 (10 131 for coralloid roots; 602 for normal roots) were from full-length 16S rRNA. Clustering of these sequence reads retrieved a total of 61 bacterial OTUs from full length 16S rRNA and 2128 fungal OTUs from ITS. Overall, 49 bacterial OTUs were detected in coralloid roots and 42 in normal roots, of which 29 OTUs (47%) were shared in two root types regardless of host species (Figure 1a). For recovered fungal taxa, 1884 OTUs were noted in coralloid roots with 453 (21%) unique OTUs, while 1674 OTUs were noted in normal roots with 243 (11%) unique OTUs. A total of 1431 (67%) fungal OTUs were shared among coralloid and normal roots regardless of host species (Figure 1a). Between the two host Cycas spp., the highest number of OTU counts for bacteria was noted in Cycas fairylakea while Cycas debaoensis had the most number of recorded fungal OTUs (Figure 1b). A relatively high percentage in bacterial (65%) and fungal OTUs (67%) were common between the two host taxa. Looking at the equitability of individuals present in each sample, though the coralloid root is more species rich as shown by Chao 1 estimator, bacterial diversity (Shannon and Simpson diversity indices) (Figure 2) appears to be lower in coralloid roots than in normal roots regardless of the host trees, implying that the latter is significantly more diverse than the former as confirmed by a two-sample t-test (P < 0.05). Meanwhile, the diversity indices for fungi did not differ significantly between root and host types (P ⩾ 0.05) but the coralloid roots of two Cycas spp. showed the highest diversity values (Chao1, Shannon and Simpson diversity; Figure 2).
Figure 1.
Venn diagram displayed the unique and overlapping OTUs of bacteria (B; red color, bold type) and fungi (F; blue color, bold type) between root types and host species. The total number of OTUs were accounted and compared: (a) between normal and coralloid roots regardless of the host species and (b) between Cycas debaoensis and Cycas fairylakea regardless of root types.
Figure 2.
Alpha diversity analysis of microbiomes in the roots of Cycas debaoensis and Cycas fairylakea. The datasets from three individuals for each root type and host species were pooled. Hence, coralloid and normal roots are coded as CD and ND for Cycas debaoensis, and CF and NF for Cycas fairylakea, respectively. The species richness and diversity of bacteria and fungi among these samples were analyzed with three different diversity indices. T-test revealed a statistically significant difference on the diversity of bacteria between root types (P < 0.05) but not with host species. The difference on the diversity of fungi between root types and host species was not significant (P ⩾ 0.05).
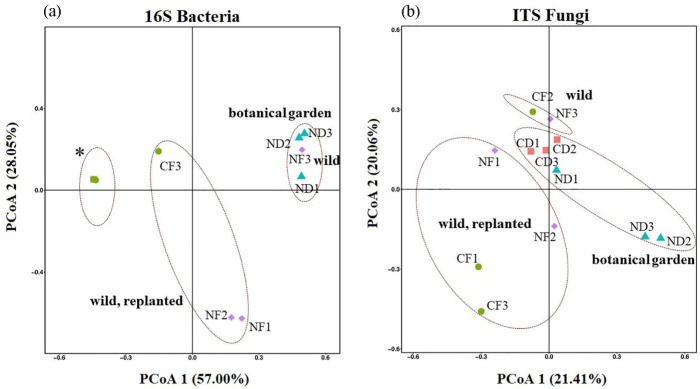
The beta-diversity was also compared between different individuals of C.debaoensis and C. fairylakea, all tree samples belonging to the same age group, in relation to the root types. Principal coordinates (PCo) plot revealed the distances between the microbiomes from each sample (Figure 3). PCoA 1 (57.00%) and PCoA 2 (28.05%) revealed a significant distinction between normal and coralloid root bacteriome, but not with the host species. Moreover, clustering of our samples based on localities of the host plants can be observed. For 16S rRNA, all coralloid root samples clustered together whereas CF3, initially grown from natural habitat and then transplanted to botanical garden for 1 year already, appeared to be slightly distant across all 6 replicates. For normal roots, no significant difference was observed with the clustering of the botanic garden samples with the one sample from a natural habitat (NF3) and with those transplanted individuals (NF1 and NF2) and grown in the botanic garden for 2 years (Figure 3a). In contrast to the PCoA of bacterial communities, the PCoA 1 of 21.41% and PCoA 2 of 20.06% for fungal communities based on the relative abundance of OTUs showed a slight difference among samples. The samples obtained from natural habitat and botanical garden clustered together while the points of the transplanted samples such as CF1, CF3, NF1, NF2 positioned closed to each other (Figure 3b). The significant difference observed in bacterial communities was found to be associated with the dominance of cyanobacteria recorded among the coralloid root samples which was separated and discussed further in the lower section of this paper.
Figure 3.
Principal coordinate analysis (PCoA) of bacteria (a) and fungal (b) communities using unweighted UniFrac metrics (β-diversity). In PCoA of bacteria (a), note the slightly distant point of one coralloid root sample of Cycas fairylakea (CF3) and the overlapping points (marked by asterisk *) of the remaining five coralloid root samples (CD1, CD2, CD3, CF1 and CF2). Normal root samples (NF1 and NF2) from replanted adult individuals of Cycas fairylakea were closed to each other while normal root sample (NF3) from host obtained in natural habitat is closer to normal roots of C. debaoensis from botanic garden. (b) A slight variation based on root type and host species was observed in the PCoA of fungal community.
Microbial composition and identified taxa associated in the root types of two Cycas spp
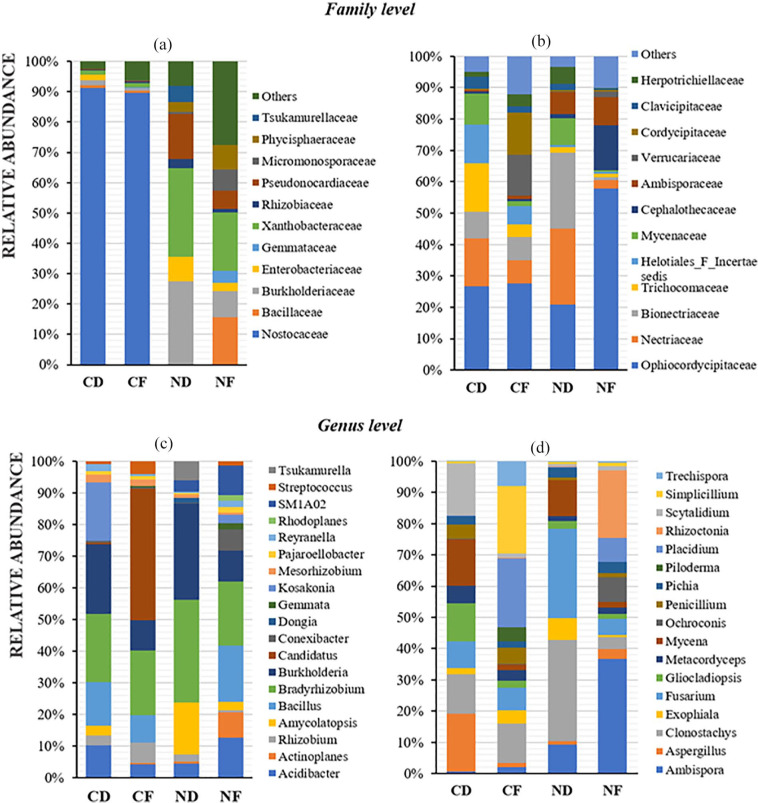
Based on the analysis of the 16S rRNA and ITS sequence reads, the OTUs obtained were mapped from higher to lower level of bacterial and fungal taxa. After sequence analysis, 98% to 99% of the total sequence reads were classified into nine bacterial phyla. In terms of lower taxonomic ranking, only about 30% and 87% of the total reads have been further classified to the genus level in coralloid root and normal roots, respectively (Figure 4a). Among the phyla identified, cyanobacteria dominated the coralloid roots with 91.28% relative abundance in C. debaoensis and 90.43% in C. fairylakea. Other bacterial phyla were also observed in the coralloid roots but in relatively low abundance, that is, <10% of the total microbial population in C. debaoensis and C. fairylakea are Proteobacteria (6.82% and 4.49%), Bacteroidetes (0.32% and 2.37%), Firmicutes (1.07% and 1.71%), Actinobacteria (0.38% and 0.87%), Patescibacteria (0.11% and 0.02%) and the candidate phylum WPS-2 (0.02% and 0.08%). The phylum Planctomycetes was only detected in C. fairylakea (0.02%). In the normal roots of C. debaoensis and C. fairylakea, Proteobacteria had the highest count with 76.00% and 54.76% relative abundance, respectively. This was followed by Actinobacteria (19.20% and 21.61%) and Planctomycetes (4.80% and 9.51%). Acidobacteria (1.44%) and WPS-2 (0.58%) were only detected in C. fairylakea as well as a single OTU count of Cyanobacteria (0.29%). At class and order levels, Nostocales under Oxyphotobacteria is the single most abundant in coralloid root, while Rhizobiales (Alphaproteobacteria), Betaproteobacteria and Gammaproteobacteria were dominant in normal roots. Among the bacterial families identified, we selected only the top 12 in coralloid and normal roots of two Cycas spp. for further comparison. Among the two root types, Nostocaceae (90%) was the most abundant bacteria in coralloid roots, while members of Xanthobacteraceae (24%) and Burkholderiaceae (18%) were the common population in normal roots (Figure 5a). The specific identified cyanobionts are further illustrated in the lower section of this paper. Hence, upon removing the cyanobacterial taxa in the datasets, the common bacterial taxon in the roots of two host species was also the symbiotic nitrogen-fixer Bradyrhizobium. Comparing the bacterial composition between two host species, high abundance of the genus Burkholderia was recorded in C. debaoensis than in C. fairylakea regardless of the root types while the genus Bacillus was notably high in C. fairylakea than C. debaoensis (Figure 5c). In addition, the genus Amycolatopsis along with the filamentous bacterium Actinoplanes were observed to be more prevalent in normal roots of C. debaoensis and C. fairylakea, respectively. Other bacterial genera were also detected but in low abundance like Acidibacter, Mesorhizobium, Kosakonia, and Rhizobium (Figure 5c).
Figure 4.
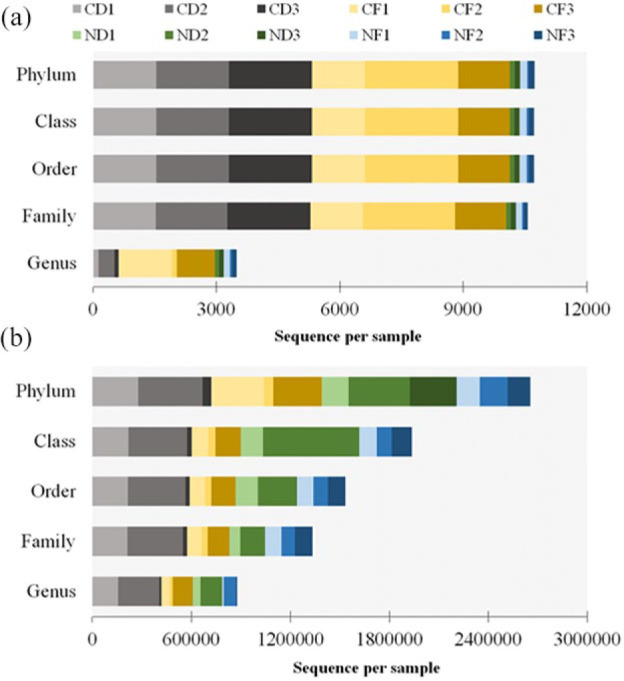
Relative number of total reads of bacteria (a) and fungi (b) assigned to taxonomic ranks. Samples in this study were coralloid roots (CD1, CD2, CD3, CF1, CF2, CF3) and normal roots (ND1, ND2, ND3, NF1, NF2, NF3) from Cycas debaoensis and Cycas fairylakea.
Figure 5.
Taxonomic composition and relative abundance of microbes detected in the roots of two Cycas spp. using full-length 16S rRNA for bacteria (a, c) and ITS for fungi (b, d). The datasets from three individuals for each root type and host species were averaged and presented per stacked bar. Abbreviations are as in Figure 2. To give a better view of other prevalent bacterial genera present in coralloid roots, members of Nostocaceae were excluded in the dataset as illustrated in C.
Meanwhile for the ITS data, 99% of the total sequences were ranked at phylum level with Ascomycota being the highest. However, the high variability of the ITS sequence used in our study may be the reason why 45% to 50% members of this phylum failed to classify to the lower taxonomic ranks (Figure 4b). Therefore, among the classified fungi at the class level, Sordariomycetes (40%-45%) was the highest. Furthermore, approximately 51% to 58% of the sequences could be specified into orders and families. At the lower taxonomic ranks, on average, only about 28% of the total reads have been successfully identified to genera (–373). However, these were unsuccessfully ranked at the species level.
The fungal Phylum Ascomycota inhabit the roots of cycads in high abundance, specifically 89.09% and 87.73% in coralloid roots of Cycas fairylakea and Cycas debaoensis, respectively, and 86.17% and 72.21% in their normal roots. Basidiomycota was also observed in all samples and high record was noted in the normal root of C. debaoensis (24.56%) while <10% was noted in other samples. Interestingly, Phylum Glomeromycota with abundance of 5.52% and 1.97% were noted in the normal roots of C. fairylakea and C. debaoensis, respectively, while only <1% was noted in the coralloid roots. Other fungal phyla such as Zygomycota, Chytridiomycota, and Rozellomycota were also observed but in significantly low count <0.2%. Unclassified fungi were also detected but with very low number (<1%) in each of the sample. Under Sordariomycetes, Order Hypocreales (30%) was the most abundant. At family level composition, Ophiocordycipitaceae, followed by Nectriaceae, Bionectriaceae, and Trichocomaceae were found to be the most frequently occurring fungal communities across samples. Furthermore, it is interesting to note the high prevalence of Ambisporaceae, a known group of arbuscular mycorrhizal fungi (AMF) in normal roots (Figure 5b). Members of this family are also present in coralloid roots but in very significant low frequency.
Among the identified fungal genera, the following were detected in relatively high abundance with their respective total number of OTUs mapped to their corresponding genus: Ambispora (66), Aspergillus (25), Penicillium (21), Clonostachys (7), Fusarium (5), Gliocladiopsis (1), Mycena (5), Scytalidium (4), Simplicillium (3), Placidium (1), and Rhizoctonia (1). Variations in fungal composition between root types of two Cycas spp. are more pronounced at increasingly specific taxonomic levels. In fact, some of these fungal taxa displayed specific association with host plant and root type. For example, Aspergillus, Scytalidium, and Gliocladiopsis were enriched in the coralloid roots of C. debaoensis, whereas Exophiala and Simplicillium were more noticeable in C. fairylakea. Rhizoctonia was notably higher in normal roots of C. fairylakea (Figure 5d). Furthermore, Clonostachys, Fusarium, and Mycena were more common in C. debaoensis than in C. fairylakea regardless of root types. On the contrary, the genus Placidium was common in C. fairylakea while low abundance was noted in C. debaoensis regardless of root types. Among the root types examined regardless of host species, the genus Ambispora was highly recorded in normal roots while Penicillium was slightly higher in coralloid roots than normal roots (Figure 5d).
Composition and phylogenetic placement of cyanobionts associated to two Cycas spp
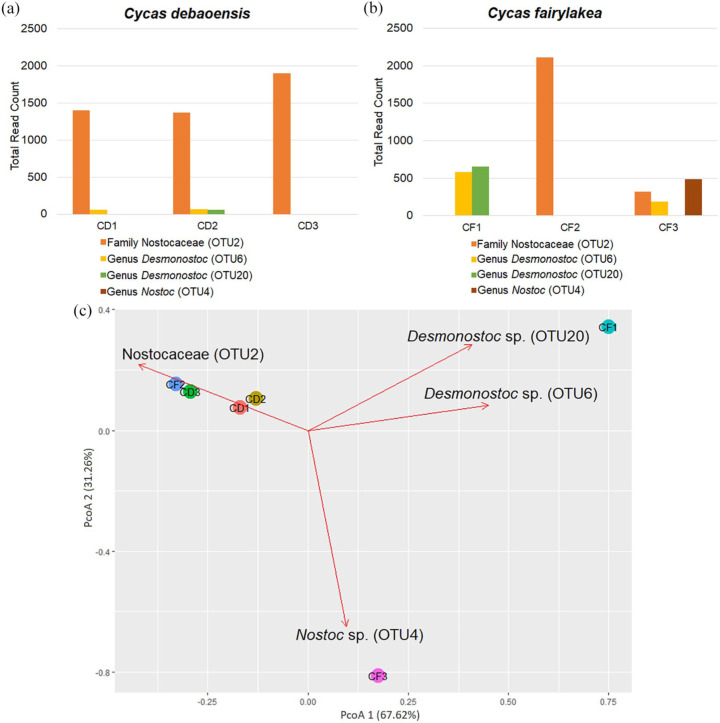
In the metagenomic dataset, high abundance of cyanobacteria was evident across all six coralloid samples which were identified as Nostocaceae (70.15%), Desmonostoc spp. (15.91%) and Nostoc sp. (4.81%; Figure 6a and b). Further analysis of cyanobacterial population in the two cycad species surveyed in this study revealed low specificity between symbionts and their respective cycad hosts. All three biological replicates of C. debaoensis were collected in the same habitat and although the full-length 16S rRNA sequence failed to assign OTU2 to a genus level classification, all three samples harbor a seemingly closely-related, if not a single species of Nostoc or related taxa. In addition, the two samples (CD1 and CD2) were also found to house two different strains of Desmonostoc but in low abundance. On the contrary, cyanobacterial population observed in C. fairylakea samples showed more individuality (Figure 6b). All four identified cyanobacterial OTUs were present in CF3 while high abundance of Desmonostoc spp. (OTU6 and OTU20) were found in CF1. It is worth noting that the abundance of unidentified Nostocaceae (OTU2) was high in sample from natural habitat (CF2) but recorded low in transplanted individuals (CF1 and CF3). Furthermore, OTU4 which was identified as Nostoc sp. strain PCC73102 was only found in CF3 (Figure 6b). Principal coordinates analysis (PCoA) plot shows the distances between the cyanobacteria composition from each sample (Figure 6c). PCoA 1 (67.62% variation) and PCoA 2 (31.26% variation) were consistent in revealing lower variation between C. debaoensis samples as compared to C. fairylakea where a clear separation among all three samples was evident. In addition, the PCoA plot elucidated significant groupings of our samples based on their localities. The coralloid root of C. fairylakea (CF2) growing in its natural habitat for about 15 years clustered together with samples taken from C. debaoensis hosts which were planted in the botanical garden at about the same time (15-20 years) whereas the cyanobacterial population of the transplanted individuals (CF1 and CF3), that is, host plants were initially growing in their natural habitat for 15 to 20 years and then were transplated in the botanic garden for 1 year now (Table 1), were slightly distinct from the rest of the samples.
Figure 6.
Cyanobacterial composition between coralloid roots of C. debaoensis (CD1, CD2, CD3) and C. fairylakea (CF1, CF2, CF3) using unweighted unifrac. (a) Histogram showed the total read count of the four cyanobacterial OTUs detected in the coralloid roots of the two host species. OTU2 showed the highest read among the two host species. (b) Transplanted samples of Cycas fairylakea (CF1 and CF3) separated in the principal coordinate analysis (PCoA) while the rest of the coralloid root samples from botanic garden and natural habitat (CD1, CD2, CD3, CF2) clustered together.
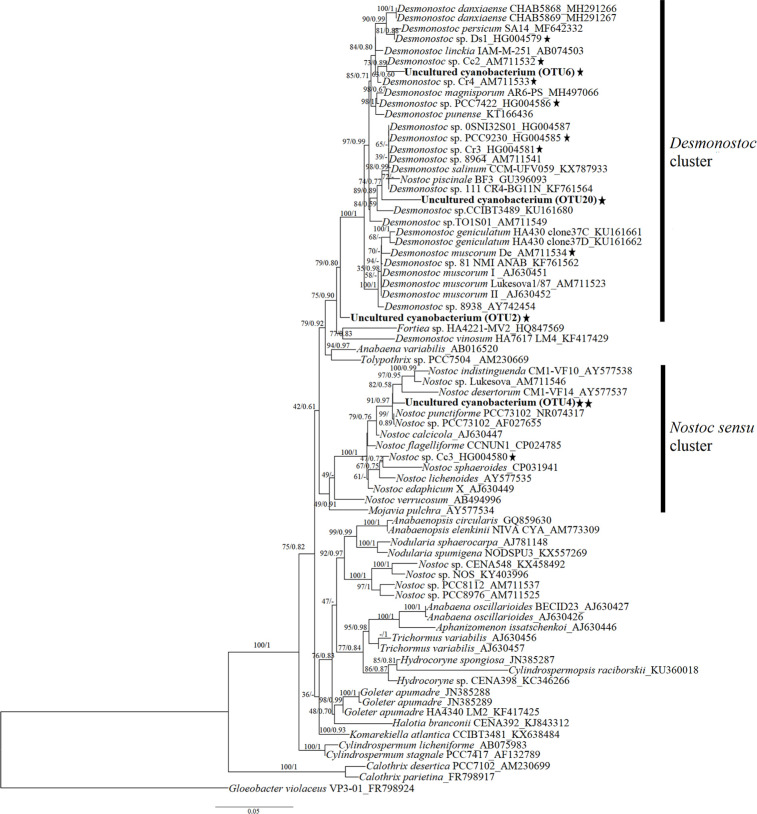
To specifically determine the relatedness of the cyanobionts associated to C. debaoensis and C. fairylakea within the Nostocacean group, a phylogenetic tree based on 16S rRNA gene was constructed (Figure 7). Consistent with the classification of cyanobacteria presented in the metagenome data, the phylogeny showed that the four sequences of cyanobionts fall within the cluster of Nostocaceae with a good bootstrap support. Specifically, OTU6 and OTU20 join the Desmonostoc cluster, while OTU4 falls within the Nostoc sensu stricto and phylogenetically closed to Nostoc punctiforme PCC73102. The OTU2 which was classified only at the family level fell outside the core cluster of Desmonostoc.
Figure 7.
Phylogenetic tree of cyanobacteria from the coralloid roots of Cycas debaoensis and Cycas fairylakea inferred by maximum likelihood (ML) and Bayesian inference (BI) based on 16S rRNA genes. The tree was constructed with 69 sequences of free-living and symbiotic strains of cyanobacteria under Order Nostocales downloaded from GenBank, and includes the sequences (OTU2, OTU6, OTU20, OTU4) from the present study. Bootstrap values and posterior probabilities (if available) are given at the nodes, respectively. Representative cyanobacterial strains reported on cycads were included and marked with black star beside the taxa. The sequences of four cyanobacteria analyzed in this study were shown in bold type. OTU4 was further highlighted with double black star as this was only recorded in Cycas fairylakea (CF3) individual originally planted in natural habitat and transferred to botanical garden.
Discussion
In this study, microbial communities associated with the coralloid and normal roots of two Cycas spp., C. debaoensis and C. fairylakea, were characterized by high throughput sequencing by targeting the full-length 16S rRNA for bacterial taxa and the short fragment of the ITS regions for fungal taxa. Our results showed that a wide variety of microbial taxa are associated with cycads uncovering diverse microbial composition across our samples. In fact, a relatively high number of fungi and bacteria has been retrieved in this study as shown by the total number of OTUs presented. We also particularly highlighted the composition and phylogenetic placement of the cyanobacteria recovered from coralloid root samples of two Cycas spp. as it lead to interesting results. With regards to fungi, almost 50% of the ITS sequence reads failed to specify the taxonomic rank below the level of phylum. As the fungal ITS sequences in the online database are relatively poor, amplifying other genes such as EF-1a, β-tubulin, and RNA polymerase would potentially aid the identification of fungi up to the lowest taxonomic ranking.31
Diversity of microbes between root types and host species
Studies in the past have demonstrated that Cycads house a diverse microbial community apart from the dominating cyanobacteria.13,17 The OTU richness for bacteria from this study was comparatively low as compared to the number reported in Cycas bifida,8 Cycas panzhihuaensis,17 and Dioon spp.14 using short fragment V3 to V5 of 16S rRNA gene. The sequencing of entire 16S rRNA gene may probably reveal low number of taxa as compared to the above studies but provides a more specific taxonomic resolution as demonstrated in this paper and previous work.24 In fact, 80% of the recorded bacterial taxa was assigned up to the genus level with full-length 16S rRNA data. Interestingly, regardless of the regions of 16S rRNA used, the reports in the paper of Zheng et al.8 and our study showed that the bacterial communities in normal roots were more diverse than in coralloid roots of Cycas spp. This supports the previous finding that overpopulation of cyanobacteria in the coralloid roots may restricts the colonization of other heterotrophic bacteria.8 Furthermore, the consistent abundance of cyanobacteria in the coralloid roots and its absence in the normal roots caused the data points to separate in the PCoA. It is worth noting that in both studies, the major composition of bacteria invading the normal roots are also present in the coralloid roots but in limited number. Therefore, the difference in bacterial diversity becomes insignificant when the cyanobacteria were disregarded in the dataset as was observed in Cycas bifida.8 Interestingly, the ability of the cyanobacteria to fix nitrogen explains why the invasion of facultative cyanobacteria occurs in the coralloid root of cycads. Though needs to be completely validated, the following grounds were emphasized in previous studies32-34: (1) the ancient coexistence of both cyanobacteria and cycads triggers the formation of coralloid roots for symbiosis, (2) the thin external layer and generation of lenticels in the coralloid root permits the easy invasion of cyanobacteria, (3) the production of chemical attractants by the host through the coralloid roots induces and prolongs the motile hormogonia phase of cyanobacteria for successful migration from environment to its root cortex, and (4) the cyanobacteria are probably creating a reciprocal relationship to the cycad host by regulating the type and amount of exudates to limit the growth of other microbes. The major ability of most filamentous cyanobacteria to fix atmospheric nitrogen into usable forms such as ammonia or nitrate has been exploited by cycads for long-term survival, and would explain its affinity to host these bacterial groups.
The insufficient quantity of roots available in the botanic garden led to the addition of samples from the natural populations of Cycas fairylakea to complete the three biological replicates. Interestingly, our analyses on accounting other bacterial taxa revealed that the alpha diversity from the pooled data in normal roots of C. fairylakea has higher richness and diversity than C. debaoensis. However, there was no significant difference in the beta diversity of bacterial communities among the samples but we observed a little evidence in the PCoA that points to normal roots from transplanted individuals (NF1 & NF2) of C. fairylakea were displayed differently. A similar observation was found in the study of Zheng and Gong17 that there was very little to almost no differences in the alpha and beta diversity of bacterial communities in Cycas panzhihuaensis between botanic garden and natural habitat. The differences on the diversity between the normal roots of the two Cycas spp. may be associated to the varying microenvironmental conditions in their habitats since diverse edaphic factors might have a significant effect on the richness and abundance of prokaryote in the roots of plants. For instance, bacterial composition on six Dioon spp. was taxonomically similar but the beta-diversity was differed between samples from botanic garden and natural habitat.14 Furthermore, a strong pattern based on biogeography and population types (wild vs cultivated) were observed with bacterial diversity in the nodules of woody legumes.35 As few samples were investigated in the present study, additional collections from natural populations of the two host species, C. debaoensis and C. fairylakea will help confirm the link on bacterial diversity with Cycads habitat and population type (wild, transplanted, and cultivated).
For fungal communities, a slightly higher diversity was observed in the coralloid roots than normal roots, albeit not significantly different. Less variation between the two species can also be observed in the PCoA. This may be attributed to the small spatial difference of the coralloid roots growing on the surface and the normal roots growing in a much deeper layer within the soil given that the topsoil surface is organic-rich supporting a relatively high microbial biomass available for host selection and eventually root colonization.36 In fact, Schlatter et al.37 reported that the fungal diversity and richness declined significantly with increasing soil depth. Moreover, garden management practices like regular soil tilling, continuous landscaping, and plant rearrangements could also impact the diversity of plant-associated fungi.38,39 The individuals of C. fairylakea are positioned near the entrance of the cycad center in the garden, are well-exposed to sunlight, and are also experiencing regular soil tilling as oppose to the C. debaoensis which are all located on slightly distant, shady, and elevated area, hidden to the garden visitors, and exposed to small or irregular tillage. Soil tilling probably favors bacterial community since it provides more oxygen in soil.40 However, over-tilling may not be good for fungi as it may disturb the entry and infection of fungal hypha or spores from soil to roots resulting to a slightly lower diversity as observed in roots of C. fairylakea. Furthermore, different environmental parameters such as humidity, solar radiation, and temperature influenced the abundance and species composition of fungi in their habitat41,42 as this may promotes spore germination and successful host infection.43 Thus, the shady and high elevation locality of all C. debaoensis plants in the botanical garden of Fairy Lake in Shenzhen provides a higher moisture which favors fungal growth resulting to a higher species richness.
Taxonomically and functionally diverse endophytic microbes in roots of two Cycas spp
Previous metagenomic studies revealed that the core microbiota in roots of healthy plants are members of the phyla Proteobacteria, Actinobacteria, Bacteroidetes, and sometimes, Firmicutes. These group of phyla seems to be active in the root as reported by studies in the model plant Arabidopsis thaliana,44 legumes,45 and other important plant crops.46,47 In the present study, a similar list of core bacteriome was observed in both root types when cyanobacteria was omitted. This finding is consistent with previous reports done on cycads microbiome.8,14,15 Although in lesser abundance, the uncharacterized candidate phylum WPS-2 was also identified in some of the coralloid root samples and normal root of C. fairylakea but not in any C. debaoensis normal root samples. Dry, bare soil environment and close association with boreal moss was the common niche of this phylum48 but rarely reported as associates of plant roots. Its first occurrence in cycad was seen in coralloid root of epiphytic Zamia pseudoparasitica.15 Therefore, this warrants the verification if these taxa were selected by cycads to be part of their root microbiome or simply a scavenger of their exudates. The non-photosynthetic genera comprising of the nitrogen-fixing soil bacteria Bradyrhizobium, Burkholderia, Rhizobium, and Mesorhizobium were also present in the roots of Cycas debaoensis and Cycas fairylakea and reportedly prevalent also in root-endosphere of Dioon spp.13,14 In addition, the genus Exiguobacterium were detected in all coralloid root samples only. Some of the enriched bacteria in the coralloid roots detected by full-length 16S rRNA are Bacillus and Rhizobium. Members of these taxa previously isolated from soil and endophytic plants were shown to stimulate plant growth, improve nitrogen fixation, and exhibit antagonistic activities against soil-borne oomycete and fungal pathogens.49,50
Although relatively unexplored, we believe that the fungi exhibit a significant place in cycads roots which interact and confer ecological importance to their hosts. Based on previous but limited reports, the primary components of fungal communities reported in roots of Cycas bifida8 and the endosphere compartments of Cycas panzhihuaensis17 belong to the families of Nectriaceae and Trichocomaceae. In this study, the two fungal families were also observed along with Bionectriaceae. However, we found that the family Ophiocordycipitaceae under Order Hypocreales was the most enriched fungal taxa in roots of the two Cycas spp. and have not been noted in the previous studies on cycads. Ophiocordycipitaceae are composed of species mostly known as parasitic fungi of insects.51 This family was previously noted as sister taxa of Clavicipitaceae,52 a major group of fungi consists of plant parasites and endophytes of grasses.53 To the best of our knowledge, our study is the first to significantly report high occurence of Ophiocordycipitaceae in roots implying that this group of fungal taxa has the potential to be plant pathogens or symbionts similar to Clavicipitaceae. This certainly merits additional studies, specifically on understanding the changes in host preferences by a specific group of fungi. Furthermore, the group of arbuscular mycorrhizal fungi (AMF) Ambisporaceae, specifically the genus Ambispora, were present in some of the coralloid root samples and highly prevalent in the normal roots of C. fairylakea. This group is a large component of the microbial biomass in soil where low host specificity54 but high host selectivity has been previously noted.55 Thus, the abundant occurrence of AMF, in general, and in our normal root samples maybe explained by soil condition, normal root size, and host selection of two Cycas spp. which supported the sporulation of these fungi similar to other group of plants like grasses which also reported high occurrence of AMF.56
The incidence of arbuscular mycorrhizal (AM) fungi has been previously investigated using hyphal and spore-based methods in the normal roots of some cycad species.16,57 However, contrasting with the results of earlier investigations where the absence of AM fungi was noted in the cortex of the coralloid roots, the present study recorded AM fungi in some of the coralloid root samples but in relatively low abundance. The use of the traditional microscopic examination in the earlier studies might have restricted the complete observation of AM fungi in coralloid roots or probably the colonizing AM fungi were not residing in the cortex since it has been already populated by cyanobacteria. Nevertheless, these group of fungal taxa are ecologically significant as they are also known as symbionts of many terrestrial plants and are believed to co-exist in legumes with nitrogen-fixing nodules.58 Since the process of nitrogen fixation demands phosphorus, it is likely that the function of AM fungi in Cycadales is similar with the Rhizobia nodulation by supplying the cyanobionts with adequate phosphorus to efficiently perform their task.16 This is not surprising since cycads are important relics which historically originated from the Triassic period, and naturally distributed in disturbed and nutrient-deficient environments where essential minerals like phosphorus are comparatively limited.59 Other fungal taxa, for example, Rhizoctonia and Gliocladiopsis, discovered in our study are mainly soil borne-fungi but has the potential to cause root rots and/or necrosis as reported in other plants,60 but their ecology on cycad roots is not yet known.
On the other hand, other genera prevalent in normal and coralloid roots in the present study are known to display high ecological and economical value for biocontrol and bioactive compounds. Among them, the genera Clonostachys and Simplicillium are generally known to potentially act as biological control agent and suppress nematodes and other plant parasites.61,62 For instance, root associated Clonostachys species have been reportedly effective against phytopathogens such as Botrytis cinerea, Sclerotinia sclerotiorum and Plasmodiophora brassicae.63,64 While some Simplicillium spp. can produce secondary metabolites with antimicrobial properties.65 These group of fungal taxa which has the potential to control pest were probably recruited by cycads to naturally develop control strategy as the population of C. fairylakea in Shenzhen were threatened by serious plant diseases and insect infestation.22 Finally, the most well-known and cosmopolitan members of Trichocomaceae and Nectriaceae such as Aspergillus and Fusarium were also found in our study together with Penicillium but in low frequency. These are composed of multifaceted fungi where some species are known to be saprotrophic competitors, opportunistic pathogens, and common endophytes providing various beneficial services to their hosts, both in terrestrial and aquatic habitats.66-68 For instance, strains under Aspergillus and Fusarium has been reported to produce secondary metabolites such as auxins and gibberellins to promote growth, induced systemic resistance and reduced stress in plants.69 The function of fungal partners in Cycadales have not yet been fully elucidated as most studies focused on the nitrogen-fixer cyanobionts. This study provides a strong evidence that not only taxonomically, but also functionally diverse group of fungi were associated with Cycas roots.
A phylogenetically related taxa of cyanobionts and the potential influence of ex situ conservation were observed in Cycas spp
Earlier reports indicated that the gymnospermous host Cycadales contains filamentous cyanobacteria predominantly belonging to the genus Nostoc and few strains under Calothrix, Scytonema, and Richelia.18,70 Inside the coralloid roots of two Cycas spp., 16S rRNA full-length sequencing uncovered strains belonging to Nostoc and Desmonostoc. The genus Desmonostoc is a recently established and separated distinct taxon as a result of the continous molecular taxonomic revision of Nostoc complex.71 In our study, the OTU6 displayed close relatedness to Desmonostoc strains previously isolated from cycad hosts Cycas circinalis (Cc2) and Cycas revoluta (Cr4) from a botanic garden in Italy.72 In another distant related clade, OTU20 join the free living strains isolated from soil and lake71,73 and the symbionts of Cycas and Gunnera which were also later assigned to Desmonostoc after 16S rRNA phylogenetic reconstruction was carried out.71,72 It is worth noting that OTU2 stood alone in a separate node and fell outside the main Desmonostoc cluster. Meanwhile, it is best to recall that this strain has been recorded abundantly in majority of our coralloid root samples but was taxonomically assigned at the family level only. In the revised classification system of cyanobacteria, it has been strongly recommended that strains occurring in a different place in a phylogenetic scheme should be given with new names.71,74 This is primarily to avoid confusion in the taxonomy of cyanobacteria as many genera have been reported with polyphyletic status.75 The placement of OTU2 in our 16S rRNA phylogeny clearly showed its close relationship to Desmonostoc but likely belonged to a different taxonomic unit (species or strain). This notable result suggest that further investigation involving isolation of cyanobacteria from the coralloid roots of Cycas spp. is required to completely address the potential novel taxonomic classification of the cyanobacteria reported in this study. The sequence of OTU4 which was found only in CF3 sample showed 99% similarity to the sequence of Nostoc punctiforme PCC73102, a strain of free living cyanobacterium recovered from aquatic environment. Interestingly, a similar strain has been previously reported in the roots of the cycad species Macrozamia from a natural habitat in Australia.70 Therefore, this cyanobiont strain was probably present only in the natural habitat, recruited and carried by the host Cycas fairylakea in its coralloid roots to its new local habitat inside the botanic garden.
The colonization of coralloid roots by cyanobacterial communities appears to be mainly depending on the community structure in the local soil surrounding the root.14 Interestingly, we found an indirect evidence on the possible influence of ex situ botanic garden conservation on the cyanobacterial communities among the coralloid root samples. As mentioned above, the associated cyanobiont communities between our coralloid root samples did not change much with an exemption of CF3 which harbors an additional strain of Nostoc sp. PCC73102. However, we observed that the recently transplanted individuals of Cycas spp. have lost their associated cyanobionts. For instance, Nostocaceae (OTU2) was found in high abundance on 15 to 20 year old Cycas fairylakea from a natural habitat (CF2) but was significantly decreased from the two individuals of similar-age C. fairylakea (CF1 and CF3), but at 12-month post-transplantation in the botanic garden. This may probably be attributed to the consequence of ex situ process which involves root-pruning during transfer, thereby resulted in the loss of the associated cyanobionts which were present in the coralloid roots prior to pruning and in differences in the “age” of the coralloid roots compared in this study. The roots of the natural habitat-derived sample of C. fairylakea were probably more mature than the roots of the two recently transplanted plants. Previous studies suggested that older roots could contain more cyanobacteria as the dermal breaks through lenticels were more proliferated resulting to an increase cyanobacterial invasion.14,32 Therefore, as the plants adjust to its new environment across time, the host also regenerates its coralloid roots and may restore the “lost” cyanobacterial communities as was observed with the individuals planted in the botanical garden over a long time (CD1, CD2, CD3) where high abundance of Nostocaceae has been also noted. Similar observation was noticed with pines where the associated bacterial community diverge gradually and was not completed after 12 months of transfer to a novel habitat.76 It seems that finding the right cyanobacterial partners is equally important for Cycas spp. to be successful in their new environment. Our result suggested that ex situ conservation may have an indirect but significant impact on Cycads microbial population, however, further work with increased number of samples from natural and ex situ populations is required to validate this observed pattern. In addition, collection and metagenome profiles of root samples of individuals prior to transplantation will also help to shed light on changes in microbial communities.
In summary, full-length 16S amplicon sequencing along with ITS regions identified rich and diverse microbial flora in the coralloid and normal roots of Cycas debaoensis and Cycas fairylakea. The main difference between the two root types is the high abundance of cyanobacterial population found in coralloid roots. Particularly, the full length was able to discern specific strains of Desmonostoc in addition to the well-observed Nostoc in the symbiotic coralloid roots of C. debaoensis and C. fairylakea which are potentially distinct from the previously described cyanobionts in Cycads. Based on our study, bacterial and fungal communities observed in cycad roots were perhaps highly influenced by the soil microbiome and habitat conditions. Thus, similar to cyanobionts, other microbes were also slightly or not at all selective or specific to their hosts and vice versa. Hence, for future studies to confirm this observation, it is best to consider biogeography of root samples in a larger scale and to obtain metagenomic profiles of both rhizosphere and bulk soil microbes. The application of culture-based approaches is equally important to further study the complete classification of cyanobacteria reported in this study, the mechanism of symbiosis between cyanobacteria and cycads, as well as to properly elucidate the functions of the other associated microbes. Furthermore, our study also highlighted the possible influence of ex situ conservation (transplantation) on microbial communities, specifically on the cyanobionts of Cycads. It would be an interesting future study to look at the specific impact of transplantation on specific microbial groups and at the ability of the host plant to recover or repopulate its roots with previously “lost” microbial communities. Finally, the results of this recent study also open another opportunity to further investigate if the microbes documented in the roots of two Cycas spp. are synergistically working with the established symbiotic cyanobacteria for cycads to continuously persist amidst unfavorable environment.
Acknowledgments
MHP and ACGC are thankful to the CAS-TWAS President’s Fellowship for their generosity and support for doctoral scholarship. Also, we would like to thank Mr. Christian Supsup of De La Salle University (Manila, Philippines) for his valuable assistance in data analysis using R Studio.
Footnotes
Author contributions: MHP, ACGC, TC, and NL conceptualized and designed the study. MHP and ACGC performed the experiment and analyzed the data. MHP, ACGC, TC, TEEDC, HR, and NL wrote the paper. All authors read, review, and approved the final manuscript.
Declaration of conflicting interests:The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding:The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the National Forestry and Grassland Administration for Rescues and Breeding of Wild Rare and Endangered Species (Li N. 2017, 2018, 2019), the Shenzhen Fairy Lake Botanical Garden for Metagenomics Studies on Microbial Diversity in the Coralloid Roots in Cycadales (Li N. FLSF-2020-02), the Shenzhen Urban Management Bureau (Li N. 201411), and the Shenzhen Key Laboratory of Subtropical Plant Diversity, Fairy Lake Botanical Garden, Chinese Academy of Sciences (Chen T. 2017-2020). There was no additional external funding received for this study.
ORCID iDs: Melissa H Pecundo  https://orcid.org/0000-0002-5697-5419
https://orcid.org/0000-0002-5697-5419
Data availability statement: All 16S rRNA and ITS sequences, including the datasets used and analyzed in this study are available upon request from primary authors.
References
- 1. Bergelson J, Mittelstrass J, Horton MW. Characterizing both bacteria and fungi improves understanding of the Arabidopsis root microbiome. Sci Rep. 2019;9:24. doi: 10.1038/s41598-018-37208-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196-1206. [DOI] [PubMed] [Google Scholar]
- 3. Bulgarelli D, Rott M, Schlaeppi K, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488:91-95. [DOI] [PubMed] [Google Scholar]
- 4. Hacquard S, Garrido-Oter R, Gonzales A, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17:603-616. [DOI] [PubMed] [Google Scholar]
- 5. Chang ACG, Lai Q, Chen T, et al. The complete chloroplast genome of Microcycas calocoma (Miq.) A. DC. (Zamiaceae, Cycadales) and evolution in Cycadales. PeerJ. 2020;8:e8305. doi: 10.7717/peerj.8305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rai AN, Bergman B, Rasmussen U. Cyanobacteria in symbiosis. Kluwer Academic Publishers; 2002. [Google Scholar]
- 7. Lobakova ES, Orazova MK, Dobrovol’skaya G. Microbial complexes occurring on the apogeotropic roots and in the rhizosphere of cycad plants. Microbiology. 2003;72:628-633. [PubMed] [Google Scholar]
- 8. Zheng Y, Chiang TY, Huang CL, Gong X. Highly diverse endophytes in roots of Cycas bifida (Cycadaceae), an ancient but endangered gymnosperm. J Microbiol. 2018;56:337-345. doi: 10.1007/s12275-018-7438-3 [DOI] [PubMed] [Google Scholar]
- 9. Bryan GS. Fascicled roots of Cycas revoluta. Am J Botany. 1936;23:334-335. [Google Scholar]
- 10. Lindblad P. Cyanobacteria in symbiosis with cycads. Microbiol Monogr. 2009;8:225-233. doi: 10.1007/7171_2008_118 [DOI] [Google Scholar]
- 11. Chang ACG, Chen T, Li N, Duan J. Perspectives on endosymbiosis in coralloid roots: association of cycads and cyanobacteria. Front Microbiol. 2019;10:1888. doi: 10.3389/fmicb.2019.01888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grilli Caiola M. On the phycobionts of the cycad coralloid roots. New Phytol. 1980;85:537-544. doi:10.1111/j.1469-8137.1980.tb00769.x [Google Scholar]
- 13. Gutiérrez-García K, Bustos-Díaz ED, Corona-Gómez JA, et al. Cycad coralloid roots contain bacterial communities including cyanobacteria and Caulobacter spp. that encode niche-specific biosynthetic gene clusters. Genome Biol Evol. 2019;11:319-334. doi: 10.1093/gbe/evy266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suárez-Moo PDJ, Vovides AP, Griffith MP, Barona-Gómez F, Cibrián-Jaramillo A. Unlocking a high bacterial diversity in the coralloid root microbiome from the cycad genus Dioon. PLoS ONE. 2019;14:e0211271. doi: 10.1371/journal.pone.0211271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bell-Doyon P, Laroche J, Saltonstall K, Aguilar JCV. Specialized bacteriome uncovered in the coralloid roots of the epiphytic gymnosperm, Zamia pseudoparasitica. Environ DNA. 2020;2:418-428. [Google Scholar]
- 16. Fisher JB, Vovides AP. Mycorrhizae are present in cycad roots. Botanical Rev. 2004;70:16-23. [Google Scholar]
- 17. Zheng Y, Gong X. Niche differentiation rather than biogeography shapes the diversity and composition of microbiome of Cycas panzhihuaensis. Microbiome. 2019;7:152. doi: 10.1186/s40168-019-0770-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thajuddin N, Muralitharan G, Sundaramoorthy M, et al. Morphological and genetic diversity of symbiotic cyanobacteria from cycads. J Basic Microbiol. 2010;50:254-265. doi: 10.1002/jobm.200900343 [DOI] [PubMed] [Google Scholar]
- 19. Zhong YC, Chen CJ. Cycas debaoensis Y.C. Zhong et C.J. Chen- a new cycad from China. Acta Phytota xon. 1997;35:571. [Google Scholar]
- 20. Wang DY. Taxonomy of Cycas in China. In: Wang FX, Liang HB, Chen TQ, Wang DY, eds. Cycads in China. Guangdong Science and Technology Press; 1996:13-19. [Google Scholar]
- 21. Zhan QQ, Wang JF, Gong X, Peng H. Patterns of chloroplast DNA variation in Cycas debaoensis (Cycadaceae): conservation implications. Conserv Genet. 2011;12:959-970. [Google Scholar]
- 22. Jian S, Liu N, Gao Z, et al. Biological characteristics of wild Cycas fairylakea population in Guangdong Province, China. Front Biol China. 2006;4:430-433. doi: 10.1007/s11515-006-0058-z [DOI] [Google Scholar]
- 23. Mizrahi-Man O, Davenport ER, Gilad Y. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PLoS ONE. 2013;8:e53608. doi: 10.1371/journal.pone.0053608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson JS, Spakowicz DJ, Hong B, et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun. 2019;10:5029. doi: 10.1038/s41467-019-13036-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heuer H, Krsek M, Baker P, Smalla K, Wellington EM. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. RStudio Team. RStudio: integrated development for R. RStudio, PBC; 2020. Accessed February 2020 http://www.rstudio.com/ [Google Scholar]
- 27. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucl Acids Res. 2016;44:W232-W235. Iqtree.cibic.univie.ac.at [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754-755. [DOI] [PubMed] [Google Scholar]
- 31. Tekpinar AD, Kalmer A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwigia. 2019;109:187-224. doi: 10.1127/nova_hedwigia/2019/0528 [DOI] [Google Scholar]
- 32. Ahern CP, Staff IA. Symbiosis in cycads: the origin and development of coralloid roots in Macrozamia communis (Cycadeceae). Am J Bot. 1994;81:1559-1570. doi: 10.1002/j.1537-2197.1994.tb11467.x [DOI] [Google Scholar]
- 33. Nilsson M, Rasmussen U, Bergman B. Cyanobacterial chemotaxis to extracts of host and nonhost plants. FEMS Microbiol Ecol. 2006;55:382-390. doi: 10.1111/j.1574-6941.2005.00043.x [DOI] [PubMed] [Google Scholar]
- 34. Khayatan B, Bains DK, Cheng MH, et al. A putative O-linked β-N-acetylglucosamine transferase is essential for hormogonium development and motility in the filamentous cyanobacterium Nostoc punctiforme. J Bacteriol. 2017;199:e00075-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ramoneda J, Le Roux JJ, Frossard E, Frey B, Gamper HA. Geographical patterns of root nodule bacterial diversity in cultivated and wild populations of a woody legume crop. FEMS Microbiol Ecol. 2020;96:fiaa145. doi: 10.1093/femsec/fiaa145 [DOI] [PubMed] [Google Scholar]
- 36. Bhattarai A, Bhattarai B, Pandey S. Variation of soil microbial population in different soil horizons. J Microbiol Exp. 2015;2:75-78. doi: 10.15406/jmen.2015.02.00044 [DOI] [Google Scholar]
- 37. Schlatter DC, Kahl K, Carlson B, Huggins DR, Paulitz T. Fungal community composition and diversity vary with soil depth and landscape position in a no-till wheat-based cropping system. FEMS Microbiol Ecol. 2018;94:fiy098. doi: 10.1093/femsec/fiy098 [DOI] [PubMed] [Google Scholar]
- 38. Kabir Z. Tillage or no-tillage: impact on mycorhizzae. Canadian J Plant Sci. 2005;85:23-29. [Google Scholar]
- 39. Solis MJ, Yurkov A, dela Cruz TE, Unterseher M. Leaf-inhabiting endophytic yeasts are abundant but unevenly distributed in three Ficus species from botanical garden greenhouses in Germany. Mycol Prog. 2015;14:1019. doi: 10.1007/s11557-014-1019-6 [DOI] [Google Scholar]
- 40. McNabb DH, Startsev AD. Effects of compaction on aeration and morphology of boreal forest soils in Alberta, Canada. Canadian J Soil Sci. 2009;89:45-56. [Google Scholar]
- 41. Garbeva P, van Veen JA, van Elsas JD. Microbial diversity in soil: selection of microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol. 2004;42:243-270. doi: 10.1146/annurev.phyto.42.012604.135455 [DOI] [PubMed] [Google Scholar]
- 42. Hazard C, Gosling P, van der Gast CJ, Mitchell DT, Doohan FM, Bending GD. The role of local environment and geographical distance in determining community composition of arbuscular mycorrhizal fungi at the landscape scale. ISME J. 2013;7:498-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cooke RC, Whipps JM. Ecophysiology of fungi. Blackwell Scientific Publications; 1993. [Google Scholar]
- 44. Lundberg DS, Lebeis SL, Dang JL. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiao X, Chen W, Zong L, et al. Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments. Mol Ecol. 2017;26:1641-1651. doi: 10.1111/mec.14027 [DOI] [PubMed] [Google Scholar]
- 46. Dong CJ, Wang LL, Li Q, Shang QM. Bacterial communities in the rhizosphere, phyllosphere and endopshere of tomato plants. PLoS ONE. 2019;14:e0223847. doi: 10.1371/journal.pone.0223847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cordero J, de Freitas R, Germida JJ. Bacterial microbiome associated with the rhizosphere and root interior crops in Saskatchewan, Canada. Can J Microbiol. 2020;66:71-85. [DOI] [PubMed] [Google Scholar]
- 48. Holland-Moritz H, Stuart J, Lewis LR, et al. Novel bacterial lineages associated with boreal moss species. Environ Microbiol. 2018;20:2625-2638. doi: 10.1111/1462-2920.14288 [DOI] [PubMed] [Google Scholar]
- 49. Coenye T, Vandamme P. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ Microbiol. 2003;5:719-729. doi: 10.1046/j.1462-2920.2003.00471.x [DOI] [PubMed] [Google Scholar]
- 50. Singh VK, Chaudhari AK, Notarte KIR, Kumar A, Singh R, Bhadouria R. Metal-oxidizing microbes and potential application in bioremediation. In: Kumar A, Singh VK, Singh P, Mishra VK, eds. Microbe mediated remediation of environmental contaminants. Elsevier; 2020:107-114. doi:10.1016/B978-0-12-821199-1.00011-0 [Google Scholar]
- 51. Schardl CL, Young CA, Moore N, et al. Genomes of plant-associated clavicipitaceae. Adv Bot Res. 2014;70:291-327. doi: 10.1016/B978-0-12-397940-7.00010-0 [DOI] [Google Scholar]
- 52. Kepler RM, Sung GH, Ban S, et al. New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia. 2012;104:182-197. [DOI] [PubMed] [Google Scholar]
- 53. Clay K, Schardl CL. Evolutionary origins and ecological consequences of endophyte symbiosis with grasses. Am Nat. 2002;160:S99-S127. [DOI] [PubMed] [Google Scholar]
- 54. Opik M, Vanatoa A, Vanatoa E, et al. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol. 2010;188:223-241. [DOI] [PubMed] [Google Scholar]
- 55. Zhang Q, Yang R, Tang J, Yang H, Hu S, Chen X. Positive feedback between mycorrhizal fungi and plants influences plant invasion success and resistance to invasion. PLoS ONE. 2010;5:e12380. doi: 10.1371/journal.pone.0012380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Muneer MA, Wang M, Jing Z, et al. Low host specificity of arbuscular mycorrhizal fungi associated with dominant steppe plants in inner Mongolia. Appl Ecol Environ Res. 2019;17. doi: 10.15666/aeer/1705_1207312089 [DOI] [Google Scholar]
- 57. Muthukumar T, Udaiyan K. Arbuscular mycorrhiza in cycads of Southern India. Mycorhizza. 2002;12:213-217. doi: 10.1007/s00572-002-0179-4 [DOI] [PubMed] [Google Scholar]
- 58. Smith SE, Read DJ. Mycorrhizal symbiosis. 2nd ed. Academic Press; 1997. [Google Scholar]
- 59. Halliday J, Pate J. Symbiotic nitrogen fixation by coralloid roots of the Cycad Macrozamia riedlei: physiological characteristics and ecological significance. Aust J Plant Physiol. 1976;3:349-358. [Google Scholar]
- 60. Parkinson LE, Shivas RG, Dann EK. Novel species of gliocladiopsis (nectriaceae, hypocreales, ascomycota) from avocado roots (Persea americana) in Australia. Mycoscience. 2017;58:95-102. doi: 10.1016/j.myc.2016.10.004 [DOI] [Google Scholar]
- 61. Meyer SL, Roberts DP. Combinations of biocontrol agents for management of plant parasitic nematodes and soilbone plant-pathogenic fungi. J Nematol. 2002;34:1-8. [PMC free article] [PubMed] [Google Scholar]
- 62. Toju H, Tanaka Y. Consortia of anti-nematode fungi and bacteria in the rhizosphere of soybean plants attacked by root-knot nematodes. R Soc Open Sci. 2019;6:181693. doi: 10.1098/rsos.181693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zou CG, Tu HH, Liu XY, Tao N, Zhang KQ. PacC in the nematophagous fungus Clonostachys rosea controls virulence to nematodes. Environ Microbiol. 2010;12:1868-1877. doi: 10.1111/j.1462-2920.2010.02191.x [DOI] [PubMed] [Google Scholar]
- 64. Baloyi MA, Laing MD, Yobo KS. Use of mixed cultures of biocontrol agents to control sheep nematodes. Vet Parasitol. 2012;184:367-370. doi: 10.1016/j.vetpar.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 65. Dong QL, Dong RZ, Xing XY, Li YK. A new antibiotic produced by the cyanobacterium-symbiotic fungus Simplicillium lanosoniveum. Nat Prod Res. 2018;32:1348-1352. doi: 10.1080/14786419.2017.1343320 [DOI] [PubMed] [Google Scholar]
- 66. Notarte KI, Yaguchi T, Suganuma K, dela Cruz TE. Antibacterial, cytotoxic and trypanocidal activities of marine-derived fungi isolated from Philippine macroalgae and seagrasses. Acta Bot Croat. 2018;77:141-151. doi: 10.2478/botcro-2018-0016 [DOI] [Google Scholar]
- 67. Notarte KIR, Devanadera MKP, Mayor ABR, Cada MCA, Pecundo MH, Macabeo APG. Toxicity, antibacterial, and antioxidant activities of fungal endophytes Colletotrichum and Nigrospora spp. isolated from Uvaria grandiflora. Philipp J Sci. 2019;148:503-510. [Google Scholar]
- 68. dela Cruz TEE, Notarte KIR, Apurillo CCS, Tarman K, Bungihan ME. Biomining fungal endophytes from tropical plants and seaweeds for drug discovery. In: Ozturk M, Egamberdieva D, Pešić M, eds. Biodiversity and biomedicine. Academic Press; 2020. doi: 10.1016/B978-0-12-819541-3.00004-9 [DOI] [Google Scholar]
- 69. Hung R, Lee Rutgers S. Applications of aspergillus in plant growth promotion. In: Gupta VK, ed. New and future developments in microbial biotechnology and bioengineering. Elsevier; 2016:223-227. [Google Scholar]
- 70. Gehringer MM, Pengelly JJL, Cuddy WS, Fieker C, Forster PI, Neilan BA. Host selection of symbiotic cyanobacteria in 31 species of the Australian cycad genus: Macrozamia (Zamiaceae). Mol Plant Microbe Interact. 2010;23:811-822. doi: 10.1094/MPMI-23-6-0811 [DOI] [PubMed] [Google Scholar]
- 71. Hrouzek P, Lukešová A, Mareš J, Ventura S. Description of the cyanobacterial genus Desmonostoc gen. nov. including D. muscorum comb. nov. as a distinct, phylogenetically coherent taxon related to the genus Nostoc. Fottea. 2013;13:201-213. [Google Scholar]
- 72. Papaefthimiou D, Hrouzek P, Mugnai MA, et al. Differential patterns of evolution and distribution of the symbiotic behaviour in nostocacean cyanobacteria. Int J Syst Evol Microbiol. 2008;58:553-564. [DOI] [PubMed] [Google Scholar]
- 73. de Alvarenga L, Vaz M, Genuario D, et al. Extending the ecological distribution of Desmonostoc genus: proposal of Desmonostoc salinum sp. nov., a novel Cyanobacteria from a saline-alkaline lake. Int J Syst Evol Microbiol. 2018;68. doi: 10.1099/ijsem.0.002878 [DOI] [PubMed] [Google Scholar]
- 74. Cai F, Li X, Yang Y, Jia N, Huo D, Li R. Compactonostoc shennongjiaensis gen. & sp. nov. (nostocales, cyanobacteria) from wet rocky wall in China. Phycologia. 2019;58:200-210. [Google Scholar]
- 75. Komárek J, Kastovsky J, Mares J, Johansen JR. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) using a polyphasic approach. Preslia. 2014;86:295-335. [Google Scholar]
- 76. Rigg JL, Offord CA, Zimmer H, Anderson IC, Singh BK, Powell JR. Conservation by translocation: establishment of Wollemi pine and associated microbial communities in novel environments. Plant Soil. 2016;411:209-225. doi: 10.1007/s11104-016-3010-2 [DOI] [Google Scholar]