Abstract
Aims
The elevated expression of phospholamban (PLB) has been observed in heart failure and cardiac remodelling, inhibiting the affinity of Ca2+ pump to Ca2+ thereby impairing heart relaxation. However, the mechanisms underlying the regulation of PLB remains to be further studied. The present study aims to test the role of RNA-binding protein HuR in the regulation of PLB and the impact of this regulatory process in cardiac remodelling.
Methods and results
A mouse model specifically deleted HuR in cardiomyocytes were used for testing the role of HuR in regulating PLB during isoproterenol (ISO)-induced cardiac remodelling. HuR deficiency did not significantly influence the phenotype and function of mouse heart under static status. However, deletion of HuR in cardiomyocytes mitigated the effect of ISO in inducing PLB expression and reducing β1-AR expression, in turn aggravating ISO-induced myocardial hypertrophy and cardiac fibrosis. In H9C2 cells, association of HuR with PLB and β1-AR mRNAs stabilized PLB mRNA and destabilized β1-AR mRNA, respectively.
Conclusion
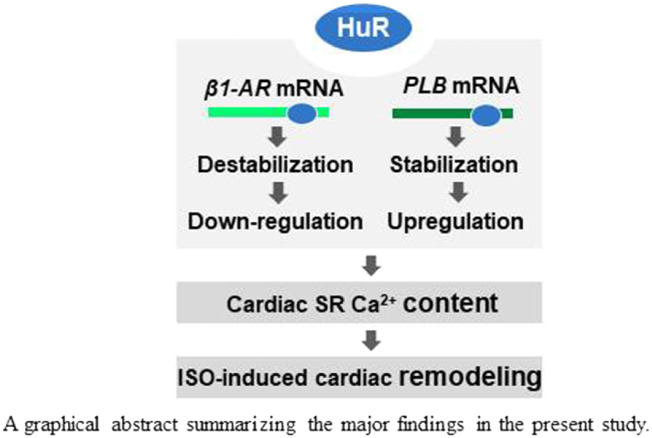
HuR stabilizes PLB mRNA and destabilizes β1-AR mRNA. The HuR-PLB and HuR-β1-AR regulatory processes impact on ISO-induced cardiac remodelling.
Keywords: HuR, PLB, β1-AR, mRNA turnover, Cardiac remodelling
Graphical Abstract
Graphical Abstract.
1. Introduction
The synchronized movements of Ca2+ into and out of the cell and that between the cytosol and sarcoplasmic reticulum (SR) are of critical importance for the physiological and pathological functions of cardiomyocytes.1–3 The alteration of myocardial Ca2+ uptake and release by SR has been observed in various animal heart disease models and human heart disorders.4 Phospholamban (PLB), the reversible inhibitor of the SR Ca2+-ATPase (SERCA2A), has been recognized as a key factor regulating the SR Ca2+ sequestration, in turn the excitation–contraction coupling of myocytes and myocardial remodelling.5,6 The activity of PLB could be influenced by protein phosphorylation and dephosphorylation. In response to adrenaline stimuli, β-adrenergic receptors (β-ARs) activate G protein-cAMP-PKA signal pathway.7 PKA phosphorylates PLB, repressing the role of PLB in inhibiting SERCA2A. On the other hand, PP1 and PP2A could dephosphorylate the phosphorylated form of PLB,8 recovering the inhibitory effect of SERCA2A by PLB.
In addition to protein phosphorylation, accumulating evidence indicates that PLB levels are also altered in failing hearts and in myocardial remodelling. Regulation of PLB at transcriptional level by transcriptional factors including glucocorticoid nuclear receptor,9 thyroid hormone receptor alpha1,10 as well as NF-YA and NF-YB11–13 has been documented. E3 ubiquitin protein ligase 1 could also mediate the ubiquitination-dependent protein degradation of PLB.14 However, whether PLB expression in myocardial remodelling could be regulated by other regulatory process remains to be studied.
HuR [‘human antigen R’, also known as ELAVL1 (embryonic lethal abnormal vision-like 1)], the ubiquitously expressed member of Hu/ELAV RNA-binding protein family, can specifically bind to the U-rich or AU-rich elements (AREs) locating in the 3′-UTRs or 5′-UTRs of mRNAs such as p21, SIRT1, cyclin A, cyclin B1, COX-2, p53, p16, p27, thereby regulating many cell activities (proliferation, survival, apoptosis, senescence, and differentiation) and affecting broader processes such as cancer and aging.15–17 Studies have described that HuR is involved in the regulation of β1-AR and may also partially inhibit β2-AR in cardiac remodelling or heart development.18,19 However, the in vivo significance of HuR-mediated post-transcriptional regulation in cardiac remodelling and the mechanisms underlying remain to be further studied.
By using a mouse model with deleted HuR in cardiomyocytes, we further investigated the role of HuR in cardiac remodelling and the mechanisms underlying. We found that deletion of HuR in cardiomyocytes greatly aggravated isoproterenol (ISO)-induced myocardial remodelling, which involves the regulation of PLB and β1-AR by HuR.
2. Methods
2.1 Ethics statement
The experimental animal facility has been accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). All mouse experiments conformed to the NIH guidelines (Guide for the care and use of laboratory animals), the Guide for the Care and Use of Laboratory Animals of the Health Science Center of Peking University, and the Institutional Animal Care and Use Committee (IACUC) of Model Animal Research Center of Nanjing University. Mice were housed in groups with 12-h dark-light cycles and had free access to food and water. Mice were sacrificed under the isoflurane inhalation (∼1.4%) and followed by cervical dislocation.
2.2 Mice and ISO treatment
HuR floxed mice were purchased from the Jackson Laboratories.20 Cardiomyocyte-specific HuR-deletion mice were generated by crossing HuR flox mice (HuRF/F) with α-MHC-Cre mice. All mouse lines were maintained in a C57BL/6J genetic background. Primer pairs ATGCTTCTGTCCGTTTGC and CAATATGGATTAACATTCTCCC were used for PCR analysis to confirm the success of HuR knockout.
To induce cardiac remodelling using ISO, 6- to 8-week-old HuR Flox/Flox male mice and their wild-type (WT) littermates were injected subcutaneously with ISO (7.5 mg/kg/day, Sigma-Aldrich, MO, USA) or with 0.9% saline for 2 weeks.
To induce MI model, animals were anaesthetized using mixture of ketamine (90 mg/kg) and pentobarbital sodium (30 mg/kg), fixed in the supine position and endotracheal intubation was performed. Then the heart was exposed and left coronary artery was tied. Then the chest cavity was closed. The heart tissues were dissected and detected 1 week after surgery.
To induce cardiac remodelling using angiotensin II (Ang II, 1.5 mg/kg/day, Sigma-Aldrich), mice described above were injected subcutaneously with Ang II at 1.5 mg/kg/day for 2 weeks or with phosphate buffer in pH = 7.2.
2.3 Cell culture and transfection
Rat cardiomyocyte H9C2 cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, CA, USA) with 1.5 g NaHCO3/L and supplemented with 10% foetal bovine serum at 37°C in 5% CO2, with 100 U/mL penicillin and 100 μg/mL streptomycin.
For plasmid transfections, Lipofectamine 2000 (Invitrogen) was used following the manufacturer’s manuscript. Cells were harvested 48 h after transfection for further analysis. To silence HuR, a HuR siRNA (GAGGCAAUUACCAGUUUCAUU) or a control siRNA (AAGAGGCAAUUACCAGUUUCA) was transfected by using RNAimax (Invitrogen) following the manufacturer’s manuscript. Cells were harvested 72 h after transfection for further analysis.
2.4 Western blotting analysis
Western blot analysis was performed using standard procedures. Monoclonal anti-β1-AR and polyclonal anti-PLB antibodies were bought from Cell Signaling Technology (MA, USA). Monoclonal anti-β2-AR, monoclonal anti-CACNA1s, polyclonal anti-ADCY5, and monoclonal anti-SERCA2A antibodies were from Abcam (Cambridgeshire, UK). Monoclonal anti-HuR antibody was from Santa Cruz (TX, USA). Monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and monoclonal anti-α-tubulin antibodies were from EarthOx (CA, USA).
2.5 RNA isolation, reverse transcription (RT) and quantitative real-time qPCR analysis
Total cellular RNA was extracted by using TRIzol (Invitrogen) and treated with reverse transcriptase (Vazyme, Nanjing, China) and the mixture of oligo-dT and random hexamers (Vazyme). To quantify the levels of different mRNAs, RT-qPCR analysis was carried out by using the following primer pairs: primer pairs GCCGATCTGGTCATGGGACT and GGATGGGCAGGAAGGACACC for rat β1-AR, primer pairs CTCAGGAACGGGACGAAG and CTTGAAGGGCGATGTGATAG for rat β2-AR, primer pairs GTGACGATCACAGAAGCCAAG and TGATAGCCGAGCGAGTAAGGT for rat PLB, primer pairs GGCACAGTCAAGGCTGAGAATG and ATGGTGGTGAAGACGCCAGTA for GAPDH, primer pairs CTCATCGTGGTGGGTAACGTG and ACACACAGCACATCTACCGAA for mouse β1-AR, primer pairs ATGTCGGTTATCGTCCTGGC and GGTTTGTAGTCGCTCGAACTTG for mouse β2-AR, primer pairs AAAGTGCAATACCTCACTCGC and GGCATTTCAATAGTGGAGGCTC for mouse PLB, primer pairs GCTCCTCTTAGGGGCCACT and CCACGTCTCACCATTGGGG for mouse collagen I, and primer pairs ACGTAGATGAATTGGGATGCAG and GGGTTGGGGCAGTCTAGTG for mouse collagen III.
2.6 Preparation of transcripts
cDNA was used as a template for PCR amplification of RNA fragments. All 5′ primers contained the T7 promoter sequence (CCAAGCTTCTAATACGACTCACTATAGGGAGA). To prepare templates for the 5′-UTR, CR, and 3′-UTR fragments of β1-AR, PLB, and β2-AR mRNAs, we used following primer pairs:
(T7)TCCGAACCCTGCAACCT and (T7)CTACACCTTGGACTCCGAGGA for β1-AR CR (1–1401), (T7)GAGAGCCAGGCTCTCTGGG and AAAGATCTGCCCACCTCCC for β1-AR 3′UTR (1042–2492), (T7)GGCGCACTGCAAGGC and GGCTGGCAGGTTAGCAGG for β2-AR 5′UTR (1–228), (T7)ATGGAGCCACACGGGAAT and CTACAGTGGCGAGTCGTTTG for β2-AR CR (229–1485), (T7)TACAGGCTTTCTACTCTCTAAGACC and AAGCCACTCATCGGTCA for β2-AR 3′UTR (1486–2269), (T7)ACCTACTGCAGCTGCTCCCA and TATGCCAGGAAGGCAAAAGT for PLB 5′UTR (1-344), (T7)ATGGAAAAAGTGCAATACCTC and TCACAGAAGCATCACAATGATG for PLB CR (345–503), and (T7)AGAGCTGCCGCCACTCCA and CAAAACTCCATTTGAATATAGGTC for PLB 3′UTR (504–2480).
2.7 Constructs
To construct pGL3-derived reporter vectors, the 5′-UTR, CR, and 3′-UTR fragments of β1-AR, PLB, and β2-AR mRNAs were amplified by PCR. Primer pairs used are as follows:
GCTCTAGATCCGAACCCTGCAACCT and GCTCTAGACTACACCTTGGACTCCGAGGA for β1-AR CR, GCTCTAGAGAGAGCCAGGCTCTCTGGG and GCTCTAGAAAAGATCTGCCCACCTCCC for β1-AR 3′UTR, GGGGTACCGGCGCACTGCAAGGC and CCGCTCGAGGGCTGGCAGGTTAGCAGG for β2-AR 5′UTR, GCTCTAGAATGGAGCCACACGGGAAT and GCTCTAGACTACAGTGGCGAGTCGTTTG for β2-AR CR, GCTCTAGATACAGGCTTTCTACTCTCTAAGACC and GCTCTAGAAAGCCACTCATCGGTCA for β2-AR 3′UTR, GGGGTACCACCTACTGCA GCTGCTCCCA and CCGCTCGAGTATGCCAGGAAGGCAAAAGT for PLB 5′UTR, GCTCTAGAATGGAAAAAGTGCAATACCTC and GCTCTAGATCACAGAAGCATCACAATGATG for PLB CR, and GCTCTAGAAGAGCTGCCG CCACTCCA and GCTCTAGACAAAACTCCATTTGAATATAGGTC for PLB 3′UTR. The 5′UTR fragments of β2-AR and PLB were inserted between the NheI and XhoI sites of pGL3-promoter vector (Promega, WI, USA). The CR and 3′UTR fragments were inserted into the BamH I sites of pGL3-promoter vector (Promega).
2.8 RNA pulldown assays
PCR-amplified DNA fragments were used as templates to transcribe biotinylated RNA by using T7 RNA polymerase in the presence of biotin-UTP. One microgram of purified biotinylated transcripts was incubated with 30 µg of cell lysates for 30 min at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway), and the pulldown material was analysed by western blotting.
2.9 RNA half-life analysis
To analyse the half-lives of β1-AR and PLB mRNAs, actinomycin D (ActD, final concentration 2 μg/mL, Amresco, ID, USA) was added to H9C2 cell cultures. Forty-eight hours after siRNA transfection, the total RNAs were extracted at 0, 2, 4, 6, and 8 h after ActD treatment. The mRNA levels at different times were analysed by using RT-qPCR.
2.10 Luciferase reporter assay
H9C2 were seeded in 24-well plates (4 × 104 cells per well). Cells were transfected with siRNA targeting HuR (GAGGCAAUUACCAGUUUCAUU). Twenty-four hours later, cells were transfected with each of the pGL3-derived reporters together with a pRL-CMV (Cytomegalovirus) vector and cultured for additional 48 h. Firefly and renilla luciferase activities were measured with a double luciferase assay system (Promega) following the manufacturers’ instructions. All firefly luciferase measurements were normalized to renilla luciferase measurements from the same sample.
2.11 Haematoxylin and eosin (H&E), Masson, and WGA staining
Fresh heart tissues were fixed in 20% formalin overnight at 4°C. The fixed tissues were dehydrated by 70, 80, 95, and 100% ethanol (each for 1 h) at room temperature, following with 20 min incubation in xylene at room temperature. After pre-treatment by paraffin/xylene (1:1) for 1 h at 65°C, the tissues were embedded in fresh paraffin at room temperature and sectioned (7–10 μm). The slices then were stained with haematoxylin and eosin (H & E) or Masson. Images were taken using the Leica Q550 IW imaging workstation (Leica Microsystems, Hessen, Germany). Masson staining sections were analysed by an image processing system (NIS-Element-D3.2, Nikon, Chiba, Japan) by using the Image J software.
For wheat germ agglutinin staining, heart slices were fixed by using 4% paraformaldehyde (PFA) for 10 min, washed three times with 1× phosphate buffer saline (PBS) for 5 min, incubated with WGA (5.0 µg/mL, Thermo Fisher, MA, USA) for 15 min at room temperature in dark. Slices were attained with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg/mL) for 15 min to visualize the nucleus. The slices then were washed three times with 1× PBS for 5 min and images were obtained by using confocal microscope (Leica SP5, Leica Microsystems).
2.12 Echocardiography
A Vevo 770 UBM system (VisualSonics Inc., YTO, CA, USA) equipped with a 30 MHz sectorial probe was used for transthoracic echocardiography analysis. Mice were anaesthetized with a gas-mixture of 3% isoflurane. Parasternal long-axis was imaged by B-mode, while the levels of papillary muscles were acquired when the scanhead was rotated clockwise 90°. Wall thickness in the end-diastole and systole of left ventricular were obtained by the M-mode cursor. M-mode images for diastolic left ventricular posterior wall thickness (LVPW; d) and diastolic left ventricular anterior wall (LVAW; d) from three continuous cardiac cycles were recorded. Averaged ejection fraction (EF%) and fractional shortening (FS%) from three continuous cardiac cycles were analysed.
2.13 Bioinformatics analysis
CLIPdb2 database (https://omictools.com/clipdb-tool)21 was used to analyse validated HuR-binding RNAs by using Piranha and CIMS tools. The overlapping RNAs were used for KEGG Pathway enrichment analysis (https://www.genome.jp/kegg/pathway.html). A total of top 12 functional pathways which were of the most significant were chosen to show.
2.14 Detection of SR and cytosolic Ca2+
To detect the SR Ca2+ and cytosolic Ca2+, Mag-Fluo-4 Acetoxymethyl (Mag-Fluo-4 AM, 20401, AAT Bioquest, CA, USA) and Fluo-4 AM (20552, AAT Bioquest) were diluted in HANKS at final concentration of 5 μM and used for incubating H9C2 cells or primary cardiomyocytes at 37°C for 30 min following the instruction from the manufacturer. After washing with HANKS for three times, cells were incubated for 30 min to allow complete de-esterification of acetoxymethyl esters. To detect SR Ca2+ load, caffeine (10 μM, 205548, CALBIOCHEM, CA, USA) was used to generate immediate Ca2+ release from SR. Fluorescence signals then were monitored at 528 nm (excitation: 488 nm by argon-ion laser sources). Ca2+ images (x, y) and line-scan recordings (x, t) were acquired at interval of 50 ms for at least 3 min using laser-scanning confocal microscope (LSM880, Zeiss, BW, Germany).
2.15 Statistics
All data are presented as means ± SD. The statistical significance was analysed by using unpaired two tailed Student’s t-test when n ≥ 5. The statistical significance was analysed by using Mann–Whitney when n < 5. A value of P < 0.05(*) or P < 0.01(**) were considered as statistically significance.
3. Results
3.1 Deletion of HuR does not influence cardiac function under steady-state conditions
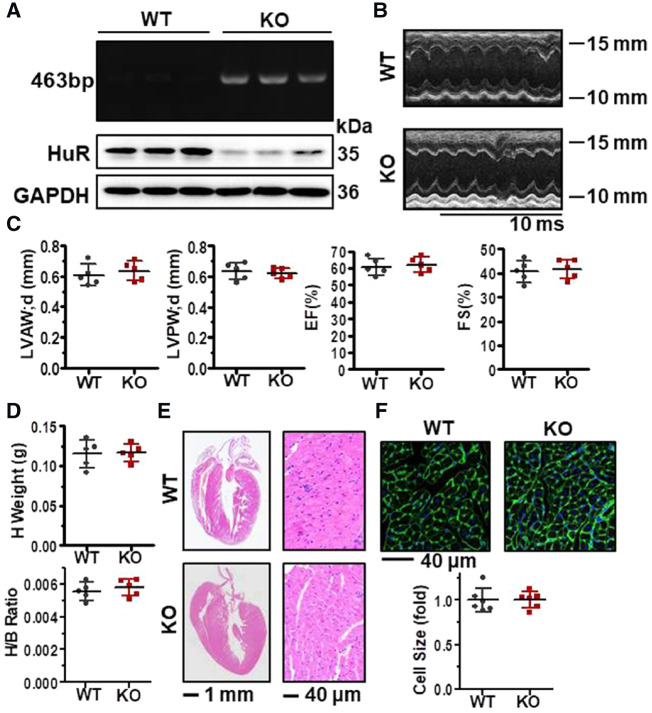
To investigate into the role of HuR in cardiac function and structure, we built a HuR-deletion mouse models. As α-MHC-Cre line is widely utilized to inhibit gene expression specifically in post-natal cardiomyocytes, we crossed the HuR flox mice (HuRF/F) with an α-MHC-Cre line to obtain cardiomyocyte-specific HuR knockout (α-MHC-Cre; HuRF/F) mice. DNA prepared from mice tails was used to test the existence of loxP and Cre genes by PCR analysis; protein lysates prepared from mouse heart tissues were used for western blot analysis to determine the protein levels of HuR. As shown in Figure 1A, in α-MHC-Cre; HuRF/F mice, loxP, and Cre genes could be detected (upper) and the protein levels of HuR in heart tissue were also significantly reduced (bottom), verifying the success of HuR deletion.
Figure 1.
Deletion of HuR does not influence cardiac function under steady-state conditions. (A) PCR and western blot analysis to test the presence of loxP and Cre genes as well as the protein of HuR in the heart tissues from cardiomyocyte-specific HuR-knockout (HuR KO) and wild-type (WT) mice (6–8 weeks old). (B) Representative images of the Doppler imaging of echocardiography. (C) Analysis of diastolic left ventricular anterior wall (LVAW; d), diastolic left ventricular posterior wall (LVPW; d), as well as the ejection fraction (EF) and fractional shortening (FS). Data are the means ± SD from five independent experiments. (D) Heart weight and the ratio between heart and body weight. Data are the means ± SD from five mice. (E) Representative images of H & E staining of the heart and heart slices. (F) WGA staining to evaluate the size of the cardiomyocytes from the heart slices. The relative cells size is shown as the means ± SD from three mice.
We next analysed the phenotype of HuR-deficient mice (6–8 weeks) under steady-state conditions. As shown in Figure 1B and C, deletion of HuR did not influence the Doppler imaging of echocardiography, LVAW; d, diastolic LVPW; d, as well as the EF% and FS%. The heart weight and the ratio between heart weight and body weight (Figure 1D) did not show significant difference after HuR-knockdown. In addition, the heart and cardiomyocyte morphology (H & E staining) (Figure 1E) and the size of cardiomyocytes (WGA staining) (Figure 1F) were comparable between WT and HuR-knockout mice. Similar results were also obtained in 8–9-month-old HuR-deficient mice (Supplementary material online, Figure S1). These results suggest that HuR ablation has no effect in the phenotype and function of mouse heart under steady-state conditions.
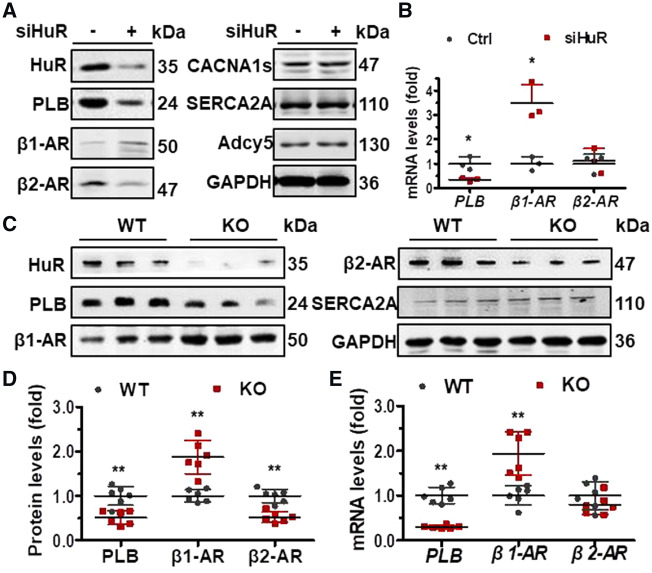
3.2 HuR regulates PLB, β1-AR, and β1-AR expression in vitro and in vivo
By bioinformatics analysis (KEGG Pathway analysis), HuR is involved in the cAMP, adrenergic, and calcium signalling pathways in cardiomyocytes (Supplementary material online, Figure S1). Although the mechanisms are not fully clear, studies have suggested that β1-AR and β2-AR mRNAs may be targets of HuR.18,19 To further confirm the regulation of β1-AR and β2-AR by HuR, the expression of β1-AR and β2-AR in cells silenced with HuR was evaluated. As shown, knockdown of HuR in H9C2 increased the levels of β1-AR protein but decreased that of β2-AR protein (Figure 2A). In cells silenced with HuR, the mRNA levels of β1-AR were increased, while the mRNA levels of β2-AR kept unchanged (Figure 2B). Importantly, the protein and mRNA levels of PLB reduced in cells with silenced HuR (Figure 2A and B). However, the observations that knockdown of HuR has no effect on the protein levels of CACNA1s, SERCA2A, and Adcy5 (Figure 2A); knockout of HuR did not significantly alter the protein levels of SERCA2A (Figure 2C). These results suggest that the regulation of PLB, β1-AR, and β2-AR by HuR is specific. Based on these findings, we tested the protein and mRNA levels of PLB, β1-AR, and β2-AR in HuR-knockout mice and their WT littermates. As shown in Figure 2C and D, knockout of HuR reduced the protein levels of PLB and β2-AR, but increased that of β1-AR. In keeping with the results shown in Figure 2B, knockout of HuR reduced the mRNA levels of PLB but increased that of β1-AR, while the mRNA levels of β2-AR kept unchanged (Figure 2E). The data are consistent with the hypothesis that HuR is able to regulate the expression of PLB, β1-AR, and β2-AR in vitro and in vivo.
Figure 2.
HuR regulates PLB, β1-AR, and β1-AR expression in vitro and in vivo. (A, B) H9C2 cells were transfected with a HuR siRNA (siHuR) for 48 h. Western blot (A) and RT-qPCR (B) were used to determine the protein and mRNA levels of HuR, PLB, β1-AR, β1-AR, CACNA1s, SERCA2A, Adcy5, and GAPDH as well as the mRNA levels of PLB, β1-AR, and β1-AR, respectively. Data in (B) are the means ± SD from three independent experiments; significance is analysed by Mann–Whitney U test (*P < 0.05). (C, D) Western blot analysis to test the protein levels of HuR, PLB, β1-AR, β1-AR, and TUBA in heart tissues from HuR-knockout (KO) and wild-type (WT) mice (C). The intensity of the blots in (C) is represented as means ± SD from six mice; significance is analysed by Student’s t-test (D) (**P < 0.01). (E) RT-qPCR analysis to test the mRNA levels of PLB, β1-AR, and β2-AR in tissues described in (C). Data are the means ± SD from six mice; significance is analysed by Student’s t-test (**P < 0.01).
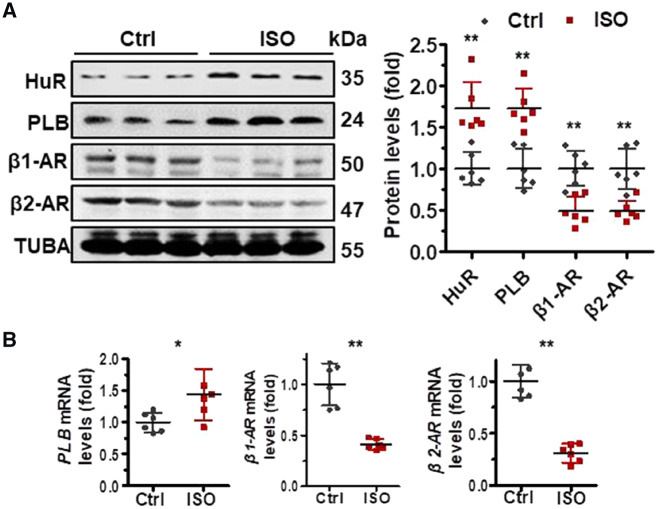
3.3 HuR ablation aggravates isoproterenol-induced cardiac remodelling
Next, we asked if HuR is involved in the process of ISO-induced myocardial remodelling. To this end, we treated HuR-deletion mice and their WT littermates with ISO, which is widely used as a non-selective β-ARs agonist to induce myocardial remodelling including ventricular hypertrophy and interstitial fibrosis.19,22 As shown in Supplementary material online, Figure S3A and B, treatment of mice with ISO reduced the contractility, increased the LVAW; d and the diastolic LVPW; d, and reduced the EF and FS. The weight of heart and the ratio between heart weight and the heart/body (H/B ratio) were also increased in response to ISO treatment (Supplementary material online, Figure S3C). The area of heart fibrosis and the size of cardiomyocytes were also increased markedly by ISO treatment (Supplementary material online, Figure S3D–F). These results confirm that ISO treatment could successfully induce myocardial remodelling. By western blot analysis, ISO treatment increased the protein levels of HuR and PLB, but decreased those of β1-AR and β2-AR (Figure 3A). In agreement with the findings in Figure 3A, ISO treatment increased the mRNA levels of PLB, but reduced that of β1-AR (Figure 3B). Knockdown of HuR reduced the protein levels of β2-AR (Figure 2A). However, in response to ISO treatment, increased HuR protein levels and reduced β2-AR protein levels were simultaneously observed (Figure 3A), suggesting that HuR is not responsible for the reduction of β2-AR in ISO-induced myocardial remodelling. Moreover, given that knockdown of HuR did not alter the β2-AR mRNA levels (Figure 2B), the observation that β2-AR mRNA levels decreased greatly in response to ISO treatment (Figure 3B) support a view that ISO may reduce β2-AR expression at the levels of transcription.
Figure 3.
The expression of HuR, PLB, β1-AR, and β1-AR alters in response to ISO. (A, Left) C57BL/6J mice were injected with isoproterenol (ISO, 7.5 mg/kg/day). Two weeks later, protein lysates prepared from heart tissues were subjected to western blot analysis to test the protein levels of HuR, PLB, β1-AR, β1-AR, and TUBA. (Right) the intensity of the blots in the left panels is represented as the means ± SD from six mice; significance is analysed by Student’s t-test (**P < 0.01). (B) RNA prepared from the tissues described in (A) was subjected to RT-qPCR to determine the mRNA levels of PLB, β1-AR, and β2-AR. Data are the means ± SD from six mice; significance is analysed by Student’s t-test (*P < 0.05; **P < 0.01).
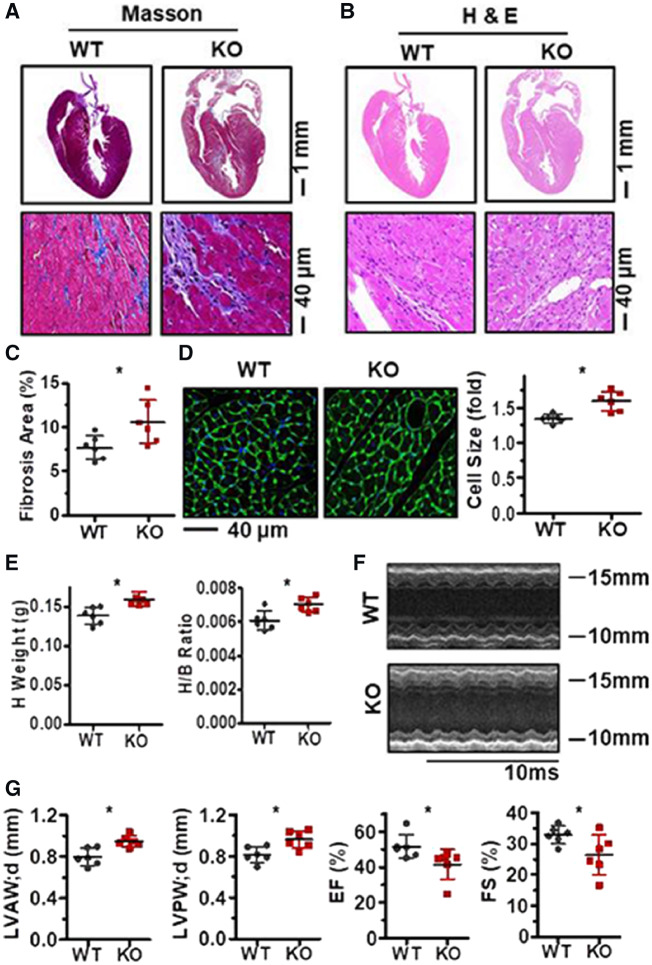
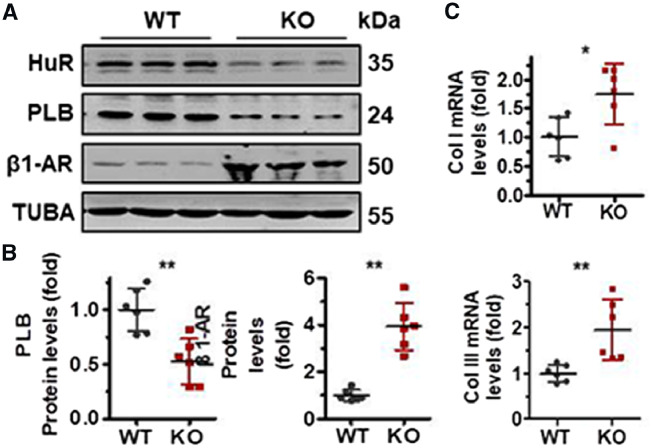
To further test the impact of HuR-PLB and HuR-β1-AR regulatory processes, HuR-knockout mice and their WT littermates were treated with ISO to induce myocardial remodelling. As shown, HuR ablation aggravated the phenotype of myocardial hypertrophy and fibrosis, as exhibited by Masson and H & E staining as well as the fibrosis area of heart and the size of cardiomyocytes (Figure 4A–D). As results depicted, deletion of HuR increased the heart weight and the ratio between heart and body weight, aggravated the effect of ISO treatment in impairing heart function, as reflected by the Doppler imaging of echocardiography, LVAW; d) and diastolic LVPW; d as well as the EF and FS (Figure 4E–G). In agreement with the results shown in Figure 2C and D, in response to ISO treatment, deletion of HuR reduced the protein levels of PLB but elevated that of β1-AR (Figure 5A and B). Of note, in the process of ISO-induced cardiac remodelling, HuR ablation also increased the mRNA levels of collagen I (Col I) and I collagen III (Col III) (Figure 5C). In sum, these data may link HuR-PLB and HuR-β1-AR regulatory processes to cardiomyocyte hypertrophy and fibrosis.
Figure 4.
HuR ablation aggravates ISO-induced cardiac remodelling. (A, B) HuR KO and wild-type mice (6–8 weeks old) were injected with ISO (7.5 mg/kg/day) for 2 weeks. Masson (A) and H & E (B) staining were performed to evaluate the cardiomyocyte fibrosis in heart and heart slices. (C) The percent of fibrosis area are shown as the means ± SD from six mice; significance is analysed by Student’s t-test (*P < 0.05). (D) WGA staining was performed to measure the size of cardiomyocytes in the heart slices. Data are means ± SD from three mice; significance is analysed by Mann–Whitney U test (*P < 0.05). (E) Statistical significance of heart weight and the ratio between heart and body weight from six mice were analysed by Student’s t-test (*P < 0.05). (F) Representative images of the Doppler imaging of echocardiography. (G) Echocardiography analysis to evaluate the diastolic left ventricular anterior wall (LVAW; d) and diastolic left ventricular posterior wall (LVPW; d) (left) as well as the ejection fraction (EF) and fractional shortening (FS) (right). Data are the means ± SD from six mice; significance is analysed by Student’s t-test (*P < 0.05).
Figure 5.
HuR ablation alters the expression of PLB and β1-AR in response to ISO treatment. (A, B) Protein lysates prepared from the heart tissues described in Figure 4A were subjected to western blot analysis to determine the protein levels of HuR, PLB, β1-AR, and TUBA (A). The intensity of the blots for PLB and β1-AR was shown as the means ± SD from 6 mice; significance is analysed by Student’s t-test (B) (*P < 0.05). (C) RT-qPCR analysis was used to test the mRNA levels of collagen I (Col I) and collagen III (Col III) in tissues described in (A). Data are the means ± SD from six mice; significance is analysed by Student’s t-test (**P < 0.01).
3.4 HuR regulates the mRNA turnover of PLB and β1-AR
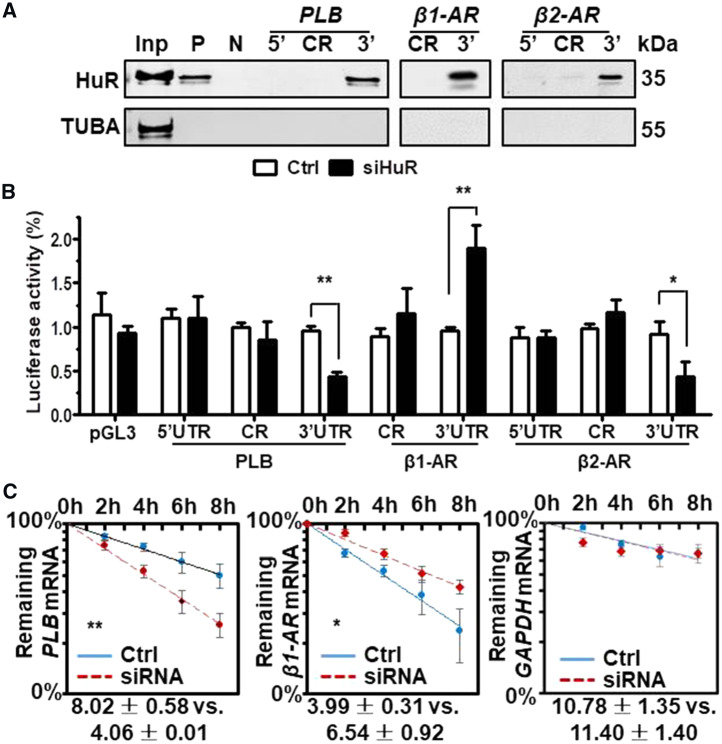
To further investigate the mechanisms underlying HuR-mediated PLB, β1-AR, and β2-AR regulation, in vitro-transcribed, biotinylated RNA fragments of the 5′UTR, CR (coding region), and 3′UTR of PLB, β1-AR, and β2-AR mRNAs (Supplementary material online, Figure S4A, Schematic) were used for RNA pulldown assays. As shown, HuR associated with the PLB, β1-AR, and β2-AR mRNA 3′UTRs, but not with other regions of these mRNAs (Figure 6A).
Figure 6.
HuR regulates the mRNA turnover of PLB and β1-AR in cells. (A) RNA pulldown assays were performed by using H9C2 cell lysates and in vitro-transcribed RNAs depicted in Supplementary material online, Figure S3A to test the association of HuR with PLB, β1-AR, and β2-AR mRNAs. p27 5′UTR and CR (coding region) served as positive (P) and negative (N) controls, respectively. A 5-µg aliquot input and binding to TUBA were also assessed. (B) H9C2 cells were transfected with a siHuR for 24 h. Cells were further transfected with each of the pGL3-derived reporters together with a pRL-CMV vector and cultured for an additional 48 h, where upon the relative luciferase activities were determined. Data are the means ± SD from three independent experiments; significance is analysed by Mann–Whitney U test (*P < 0.05; **P < 0.01). (C) H9C2 cells were transfected with a siHuR for 48 h, the half-lives of PLB, β1-AR, and GAPDH mRNAs were evaluated as described in Methods section. The means ± SD from three independent experiments and the statistical significance (Mann–Whitney U test) are indicated (*P < 0.05; **P < 0.01).
To test if the association of HuR with PLB, β1-AR, and β2-AR mRNAs was functional, pGL3-derived reporters bearing fragments of PLB, β1-AR, and β2-AR mRNAs were constructed (Supplementary material online, Figure S4B, Schematic). H9C2 cells were transfected with each of these reporters and 24 h later, they were transfected with siRNAs (control or HuR-directed) and cultured for an additional 48 h. Knockdown of HuR reduced the luciferase activity of pGL3-derived vectors bearing the PLB and β2-AR 3′UTRs, but increased that bearing β1-AR 3′UTR (Figure 6B). However, knockdown of HuR has no effect on the activity of reporters bearing regions that did not interact with HuR (Figure 6B). Because knockdown of HuR reduced the mRNA levels of PLB but increased that of β1-AR, we further tested the half-lives of PLB and β1-AR mRNAs in cells with silenced HuR. As shown in Figure 6C, knockdown of HuR shortened the half-life of PLB mRNA but extended that of β1-AR mRNA. As a control, the half-life of GAPDH mRNA was not altered by HuR-knockdown. Together, HuR regulates the expression of PLB and β1-AR by stabilizing PLB mRNA and destabilizing β1-AR mRNA, respectively.
3.5 HuR-PLB and HuR-β1-AR regulatory processes impact on SR Ca2+ content
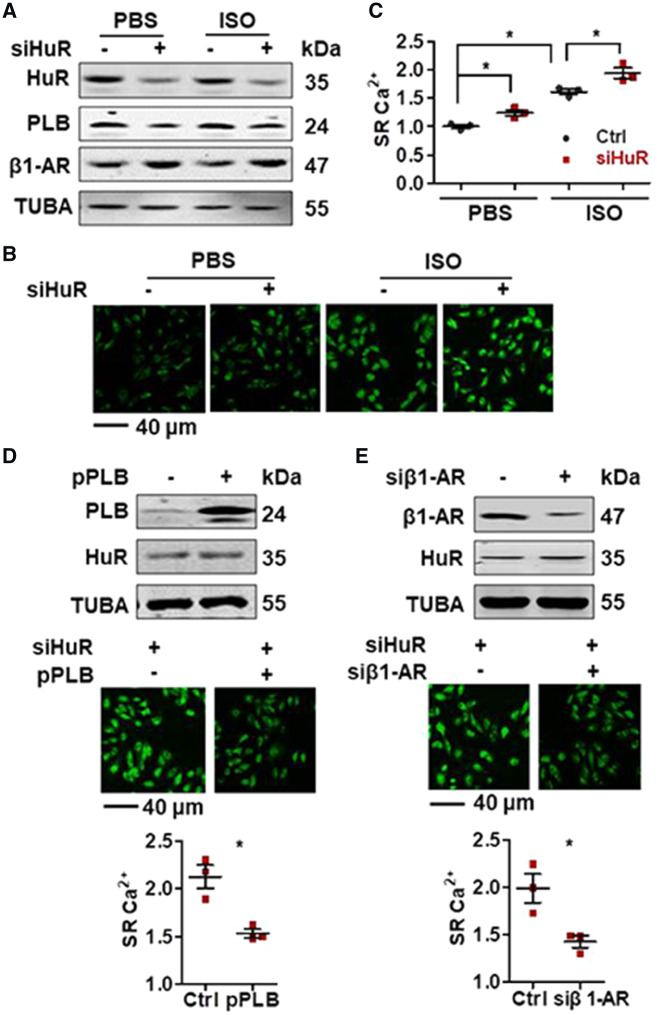
To further test the impact of HuR-PLB and HuR-β1-AR regulatory processes in SR Ca2+ content, H9C2 cells with silenced HuR were treated with ISO or saline (PBS) for 15 min, whereupon the SR Ca2+ content was observed. Because the treatment was too short, ISO treatment did not show effect in altering the protein levels of HuR, PLB, and β1-AR (Figure 7A). However, the SR Ca2+ concentration markedly increased in response to ISO treatment (Figure 7B and C). Knockdown of HuR reduced the protein levels of PLB but increased that of β1-AR (Figure 7A) and increased the SR Ca2+ concentration in the presence and absence of ISO as compared with control cells (Figure 7B and C). The increased SR Ca2+ by HuR-knockdown were nearly lost when cells were pre-treated with caffeine to clean the SR content, suggesting that fluorescence signal was mainly from SR (Supplementary material online, Figure S5A). Expression of PLB or knockdown of β1-AR could rescue the effect of HuR-knockdown in in enhancing ISO-induced SR Ca2+ release (Figure 7D and E). Furthermore, in primary isolated mouse cardiomyocytes, HuR-deficiency increased the cytosolic Ca2+ concentration in response to caffeine stimulation, suggesting that HuR-deficiency increased SR Ca2+ content (Supplementary material online, Figure S5B). Together, HuR-PLB and HuR-β1-AR regulatory processes may be linked to SR-related Ca2+ sequestration.
Figure 7.
Expression of PLB or knockdown of β1-AR rescues the effect of HuR-knockdown in aggravating SR Ca2+. (A–C) H9C2 cells were transfected with a siHuR for 48 h and further treated with ISO (20 μM) or saline for 15 min. The protein levels of HuR, PLB, β1-AR, and TUBA were analysed by western blot analysis (A). The levels of SR Ca2+ were tested (B); the intensity of the fluorescence was shown as the means ± SD from three independent experiments and used to analyse the significance by using Mann–Whitney U test (C) (*P < 0.05). (D, E) H9C2 cells were transfected with a siHuR for 24 h. Cells then were further transfected with a vector expressing PLB (pPLB) (D) or with a β1-AR siRNA (siβ1-AR) (E) and cultured for additional 24 h. After treatment with ISO (20 μM) for 15 min, the protein levels of HuR, PLB, and TUBA were analysed by western blot analysis (upper). The levels of SR Ca2+ were tested (middle); the intensity of the fluorescence in (D and C) was shown as the means ± SD from three independent experiments and analysed for significance by Mann–Whitney U test (bottom) (*P < 0.05).
4. Discussion
The present study reveals a role of HuR in ISO-induced cardiac remodelling by using a mouse model specifically deleted HuR in cardiac myocytes. Apart from β1-AR,18 which is reported as a target of HuR-mediated mRNA turnover, HuR could also stabilize the mRNA of PLB, thereby the expression of PLB in ISO-induced cardiac remodelling (Figures 2–6). The HuR-PLB and HuR-β1-AR regulatory processes may be linked to the alteration of the SR Ca2+ sequestration (Figure 7 and Supplementary material online, Figure S5). PLB ablation has been found to lose response to ISO and resulted in cardiac hypertrophy and decreased cardiac contractility.23,24 The elevated expression of PLB has been described in hypoplastic left heart syndrome, in patients with arrhythmogenic right ventricular cardiomyopathy/dysplasia, and in patients with preserved left ventricular EF and chronic isolated mitral regurgitation.25–27 However, reduced expression of PLB has been observed in failing heart.28,29 Therefore, the elevation of HuR in ISO-induced mouse model of cardiac remodelling might rather play a compensatory role in cardioprotection by up-regulation of PLB and down-regulation of β1-AR (Figure 3A). In addition, in mouse models of myocardial ischaemia (MI) and Ang II-induced cardiac remodelling, the protein levels of HuR and PLB were simultaneously induced (Supplementary material online, Figure S6), indicating that the impact of HuR-PLB regulatory process is not limited to ISO-induced cardiac remodelling.
An earlier study claimed that HuR might also inhibit the expression of β2-AR, since primary neonatal rat cardiac myocytes (NRCMs) silenced with HuR reversed mildly the reduction of β2-AR mRNA levels in response to ISO treatment.19 However, the present study indicates that HuR could promote the expression of β2-AR (Figures 2A, C, D and 6B). In addition, the findings that knockdown of HuR in H9C2 cells and knockout of HuR in mouse cardiomyocytes do not influence significantly the mRNA levels of β2-AR (Figures 2B and E) suggest that the regulation of β2-AR by HuR may occur in the level of translation. Furthermore, because the induction of HuR protein levels and the reduction of β2-AR protein levels are simultaneously observed in response to ISO treatment (Figure 3A), HuR-β2-AR regulatory process may not be responsible for the reduction of β2-AR expression in ISO-induced cardiac remodelling. This point is also confirmed by the observation that ISO treatment elevated β2-AR mRNA levels (Figure 3B).
The renin–angiotensin–aldosterone system is involved in cardiac fibrosis and remodelling. For examples, Ang II could increase the expression of transforming growth factor-β (TGF-β), interleukin-1 (IL-1), IL-6, and tumor Necrosis Factor-α (TNF-α), leading to enhanced generation of myofibroblasts.30,31 Studies have demonstrated that HuR could stabilize the mRNA encoding TGF-β1,32 in turn activating infarct myofibroblasts.33 Given that deletion of HuR aggravates the effect of ISO in inducing myocardial hypertrophy and cardiac fibrosis as well as in impairing heart function (Figures 4and5), HuR-TGF-β1 regulatory process may not impact on ISO-induced cardiac remodelling. In addition, loss of HuR activates NF-κB, thereby increasing cytokine induction in rodent fibroblasts.34 Interestingly, HuR-deficiency increased the mRNA levels of IL-1, IL-6, and TNF-α (Supplementary material online, Figure S7), suggesting that the effect of HuR-knockdown in ISO-induced cardiac remodelling may also correlated with the induction of these cytokines by HuR-NF-κB regulatory axis.
Because the mice we used in the present study are too young (6–8 weeks), we have not observed any heart phenotype and function changes in cardiomyocyte-specific HuR-knockdown mice under static status (Figure 1). HuR is well recognized as a regulator of aging and aging-related processes.35,36 Thus, whether aged cardiomyocyte-specific HuR-deficient mice are prone to exhibit much severe aging-related heart disorders than their WT littermates remains to be studied.
PLB is expressed in heart, aorta, and other tissues.37 In addition to β-AR mRNAs, HuR also associates with mRNAs that encode angiotensin receptors, iNOS, COX-2, and TNF-α, which are involved in heart failure, myocardial infarction, and hypertension.33,38,39 Accordingly, elevated HuR levels were also detected in several vascular pathologies, including intimal hyperplasia, genetic hypertension, atherosclerosis, sclerosis of venous graft, and fibromuscular dysplasia.40,41 Therefore, it is plausible to postulate that the HuR-PLB axis may also impacts in these diseases.
Authors’ contributions
W.W., D.Y., Z.L., M. G., and Y.Z. designed the study. H. H., J. M., Y.C., Z. Z., J. B., T. F., M. Z., and F. J. performed the experiments. M.G., W.W., and Y. Z. wrote the article.
Conflict of interest: none declared.
Funding
This work was supported by National Key Research and Development Program of China (2017YFA0504302); the National Natural Science Foundation of China (81420108016, 81741003, and 91749208). M.G. was supported by the NIA IRP and NIH.
Supplementary Material
Time for primary review: 34 days
Translational perspectives
PLB, the reversible inhibitor of SERCA2A, is of critical importance for the SR Ca2+ sequestration thereby the excitation–contraction coupling of myocytes and myocardial remodelling. The activity of PLB could be inhibited by β1-AR-cAMP-PKA pathway through PKA-mediated protein phosphorylation. The present study demonstrated that HuR is able to modulate cardiac remodelling by regulating the expression of PLB and β1-AR. These findings reveal novel mechanisms controlling myocardial remodelling and may provide new clues for strategies to interfere cardiac remodelling-related heart disorders in humans.
References
- 1. Ai X, Curran JW, Shannon TR, Bers DM, Pogwizd SM.. Ca2+/calmodulin-dependent protein kinase modulates cardiac ryanodine receptor phosphorylation and sarcoplasmic reticulum Ca2+ leak in heart failure. Circ Res 2005;97:1314–1322. [DOI] [PubMed] [Google Scholar]
- 2. Bers DM. Calcium cycling and signaling in cardiac myocytes. Annu Rev Physiol 2008;70:23–49. [DOI] [PubMed] [Google Scholar]
- 3. Sammels E, Parys JB, Missiaen L, De Smedt H, Bultynck G.. Intracellular Ca2+ storage in health and disease: a dynamic equilibrium. Cell Calcium 2010;47:297–314. [DOI] [PubMed] [Google Scholar]
- 4. Brini M, Carafoli E.. Calcium pumps in health and disease. Physiol Rev 2009;89:1341–1378. [DOI] [PubMed] [Google Scholar]
- 5. Simmerman HK, Jones LR.. Phospholamban: protein structure, mechanism of action, and role in cardiac function. Physiol Rev 1998;78:921–947. [DOI] [PubMed] [Google Scholar]
- 6. Kranias EG, Bers DM.. Calcium and cardiomyopathies. Subcell Biochem 2007;45:523–537. [DOI] [PubMed] [Google Scholar]
- 7. Ma YC, Huang XY.. Novel signaling pathway through the beta-adrenergic receptor. Trends Cardiovasc Med 2002;12:46–49. [DOI] [PubMed] [Google Scholar]
- 8. Heijman J, Dewenter M, El-Armouche A, Dobrev D.. Function and regulation of serine/threonine phosphatases in the healthy and diseased heart. J Mol Cell Cardiol 2013;64:90–98. [DOI] [PubMed] [Google Scholar]
- 9. Haghighi K, Chen G, Sato Y, Fan GC, He S, Kolokathis F, Pater L, Paraskevaidis I, Jones WK, Dorn GW 2nd, Kremastinos DT, Kranias EG.. A human phospholamban promoter polymorphism in dilated cardiomyopathy alters transcriptional regulation by glucocorticoids. Hum Mutat 2008;29:640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Belakavadi M, Saunders J, Weisleder N, Raghava PS, Fondell JD.. Repression of cardiac phospholamban gene expression is mediated by thyroid hormone receptor-{alpha}1 and involves targeted covalent histone modifications. Endocrinology 2010;151:2946–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McTiernan CF, Lemster BH, Frye CS, Johns DC, Feldman AM.. Characterization of proximal transcription regulatory elements in the rat phospholamban promoter. J Mol Cell Cardiol 1999;31:2137–2153. [DOI] [PubMed] [Google Scholar]
- 12. Tada M, Yabuki M, Toyofuku T.. Molecular regulation of phospholamban function and gene expression. Ann N Y Acad Sci 1998;853:116–129. [DOI] [PubMed] [Google Scholar]
- 13. Yabuki M, Toyofuku T, Otsu K, Nishida M, Kuzuya T, Hori M, Tada M.. Involvement of NF-Y in transcriptional regulation of the phospholamban gene. Eur J Biochem 1998;258:744–751. [DOI] [PubMed] [Google Scholar]
- 14. Teng AC, Miyake T, Yokoe S, Zhang L, Rezende LM Jr, Sharma P, MacLennan DH, Liu PP, Gramolini AO.. Metformin increases degradation of phospholamban via autophagy in cardiomyocytes. Proc Natl Acad Sci USA 2015;112:7165–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang W. Regulatory RNA-binding proteins in senescence. Ageing Res Rev 2012;11:485–490. [DOI] [PubMed] [Google Scholar]
- 16. Simone LE, Keene JD.. Mechanisms coordinating ELAV/Hu mRNA regulons. Curr Opin Genet Dev 2013;23:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grammatikakis I, Abdelmohsen K, Gorospe M, Posttranslational control of HuR function. Wiley Interdiscip Rev RNA2016;doi:10.1002/wrna. [DOI] [PMC free article] [PubMed]
- 18. Kirigiti P, Bai Y, Yang YF, Li X, Li B, Brewer G, Machida CA.. Agonist-mediated down-regulation of rat beta1-adrenergic receptor transcripts: role of potential post-transcriptional degradation factors. Mol Pharmacol 2001;60:1308–1324. [DOI] [PubMed] [Google Scholar]
- 19. Yin Q, Yang C, Wu J, Lu H, Zheng X, Zhang Y, Lv Z, Zheng X, Li Z.. Downregulation of β-Adrenoceptors in isoproterenol-induced cardiac remodeling through HuR. PLoS One 2016;11:e0152005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T.. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest 2009;119:3530–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang YC, Di C, Hu B, Zhou M, Liu Y, Song N, Li Y, Umetsu J, Lu ZJ.. CLIPdb: a CLIP-seq database for protein-RNA interactions. BMC Genomics 2015;16:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McClatchy DB, Ma Y, Liem DA, Ng DCM, Ping P, Yates JR 3rd. Quantitative temporal analysis of protein dynamics in cardiac remodeling. J Mol Cell Cardiol 2018;121:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo W, Grupp IL, Harrer J, Ponniah S, Grupp G, Duffy JJ, Doetschman T, Kranias EG.. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ Res 1994;75:401–409. [DOI] [PubMed] [Google Scholar]
- 24. Shanmugam M, Gao S, Hong C, Fefelova N, Nowycky MC, Xie LH, Periasamy M, Babu GJ.. Ablation of phospholamban and sarcolipin results in cardiac hypertrophy and decreased cardiac contractility. Cardiovasc Res 2011;89:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akdis D, Medeiros-Domingo A, Gaertner-Rommel A, Kast JI, Enseleit F, Bode P, Klingel K, Kandolf R, Renois F, Andreoletti L, Akdis CA, Milting H, Lüscher TF, Brunckhorst C, Saguner AM, Duru F.. Myocardial expression profiles of candidate molecules in patients with arrhythmogenic right ventricular cardiomyopathy/dysplasia compared to those with dilated cardiomyopathy and healthy controls. Heart Rhythm 2016;13:731–741. [DOI] [PubMed] [Google Scholar]
- 26. Miyamoto SD, Stauffer BL, Polk J, Medway A, Friedrich M, Haubold K, Peterson V, Nunley K, Nelson P, Sobus R, Stenmark KR, Sucharov CC.. Gene expression and β-adrenergic signaling are altered in hypoplastic left heart syndrome. J Heart Lung Transplant 2014;33:785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zheng J, Yancey DM, Ahmed MI, Wei C-C, Powell PC, Shanmugam M, Gupta H, Lloyd SG, McGiffin DC, Schiros CG, Denney TS, Babu GJ, Dell’Italia LJ.. Increased sarcolipin expression and adrenergic drive in humans with preserved left ventricular ejection fraction and chronic isolated mitral regurgitation. Circ Heart Fail 2014;7:194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yokoe S, Asahi M.. Phospholamban is downregulated by pVHL-Mediated degradation through oxidative stress in failing heart. Int J Mol Sci 2017;18:doi:10.3390/ijms18112232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vanderheyden M, Mullens W, Delrue L, Goethals M, de Bruyne B, Wijns W, Geelen P, Verstreken S, Wellens F, Bartunek J.. Myocardial gene expression in heart failure patients treated with cardiac resynchronization therapy responders versus nonresponders. J Am Coll Cardiol 2008;51:129–136. [DOI] [PubMed] [Google Scholar]
- 30. Lahera V, Cachofeiro V, de Las Heras N.. Interplay of hypertension, inflammation, and angiotensin II. Am J Hypertens 2011;24:1059.. [DOI] [PubMed] [Google Scholar]
- 31. Park S, Nguyen NB, Pezhouman A, Ardehali R.. Cardiac fibrosis: potential therapeutic targets. Transl Res 2019;209:121–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nabors LB, Gillespie GY, Harkins L, King PH.. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res 2001;61:2154–2161. [PubMed] [Google Scholar]
- 33. Prabhu SD, Frangogiannis NG.. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ Res 2016;119:91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hashimoto M, Tsugawa T, Kawagishi H, Asai A, Sugimoto M.. Loss of HuR leads to senescence-like cytokine induction in rodent fibroblasts by activating NF-κB. Biochim Biophys Acta 2014;1840:3079–3087. [DOI] [PubMed] [Google Scholar]
- 35. Wang W. HuR and post-transcriptional regulation in vascular aging. Sci China Life Sci 2014;57:863–866. [DOI] [PubMed] [Google Scholar]
- 36. Srikantan S, Gorospe M.. HuR function in disease. Front Biosci (Landmark Ed) 2012;17:189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fujii J, Zarain-Herzberg A, Willard HF, Tada M, MacLennan DH.. Structure of the rabbit phospholamban gene, cloning of the human cDNA, and assignment of the gene to human chromosome 6. J Biol Chem 1991;266:11669–11675. [PubMed] [Google Scholar]
- 38. Blaxall BC, Pellett AC, Wu SC, Pende A, Port JD.. Purification and characterization of beta-adrenergic receptor mRNA-binding proteins. J Biol Chem 2000;275:4290–4297. [DOI] [PubMed] [Google Scholar]
- 39. Misquitta CM, Iyer VR, Werstiuk ES, Grover AK.. The role of 3′-untranslated region (3′-UTR) mediated mRNA stability in cardiovascular pathophysiology. Mol Cell Biochem 2001;224:53–67. [DOI] [PubMed] [Google Scholar]
- 40. Pullmann R Jr, Juhaszova M, López de Silanes I, Kawai T, Mazan-Mamczarz K, Halushka MK, Gorospe M.. Enhanced proliferation of cultured human vascular smooth muscle cells linked to increased function of RNA-binding protein HuR. J Biol Chem 2005;280:22819–22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. KlöSs S, Rodenbach D, Bordel R, MüLsch A, Human-antigen R (HuR) expression in hypertension: downregulation of the mRNA stabilizing protein HuR in genetic hypertension. Hypertension 2005;45:1200–1206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.