Abstract
Modification of eukaryotic RNA by methylation of adenosine residues to generate N6-methyladenosine (m6A) is a highly prevalent process. m6A is dynamically regulated during cell metabolism and embryo development, and it is mainly involved in various aspects of RNA metabolism, including RNA splicing, processing, transport from the nucleus, translation, and degradation. Accumulating evidence shows that dynamic changes to m6A are closely related to the occurrence and development of cancer and that methyltransferases, as key elements in the dynamic regulation of m6A, play a crucial role in these processes. Therefore, in this review, we describe the role of methyltransferases as m6A writers in cancer and summarize their potential molecular mechanisms of action.
Keywords: RNA, m6A, methyltransferase, METTL3, METTL14, WTAP, cancer
Graphical Abstract
Wang and colleagues demonstrated that m6A is an important gene regulation mode involved in cancer progression. This review focuses on the key roles of methyltransferases, m6A “writers,” in cancer and future medical treatment.
Main Text
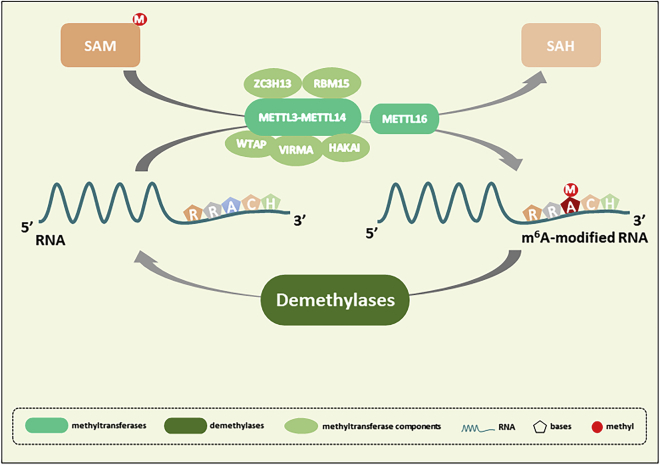
According to central genetic dogma, genetic information is transferred from DNA to RNA and then from RNA to protein by the transcription and translation processes, respectively. Epigenetic modification mainly involves DNA, RNA, and proteins. During the past few decades, investigation of human epigenetic modification has focused on DNA and proteins, with analysis of chromatin remodeling, gene silencing, DNA methylation, and histone modification. In comparison, RNA-related modification has received less attention, although in recent years, with the continuous development of detection technology, hundreds of different RNA modifications have been discovered, among which N6-methyladenosine (m6A) RNA methylation is the most abundant.1 m6A in RNA is an epigenetic modification in which a hydrogen atom (–H) connected to the sixth nitrogen atom (N6) on adenine is replaced by a methyl group (–CH3) (Figure S1). With the advent of high-throughput sequencing technology, many m6A RNA methylation sites have been discovered, which are mainly concentrated in exons and 3′ untranslated regions (UTRs), with the highest concentration near the termination codon. Each transcript contains three to five or more m6A modification sites, accounting for 0.1%–0.4% of total adenine (m6A/A).2 In eukaryotic RNAs, m6A mostly occurs in the critically conserved motif RRACH (R = G, A, and U; H = U, A, and C) (Figure 1),3,4 and it is most common in mRNA but is also widely found in long non-coding RNA (lncRNA), tRNA, rRNA, and microRNA (miRNA).
Figure 1.
m6A methylation is a dynamic and reversible process, installed by methyltransferases, and removed by demethylases
m6A methyltransferases include METTL3, METTL14, WTAP, VIRMA, HAKAI, ZC3H13, RBM15, METTL16.
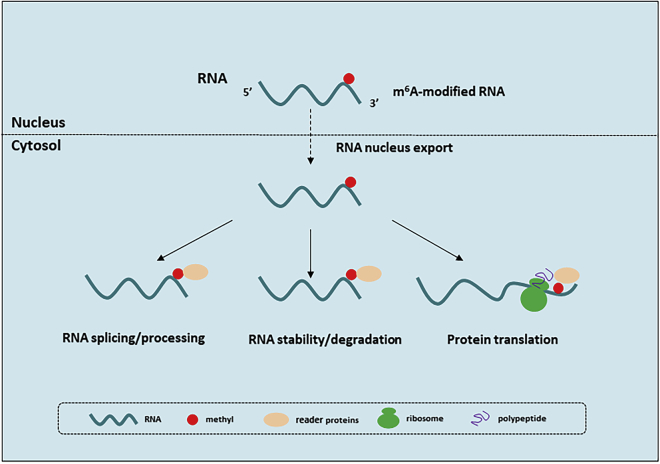
A large number of studies have confirmed that m6A RNA mainly interacts with three types of proteins, namely methyltransferases, demethylases, and reader proteins. Similar to DNA methylation, RNA methylation is also dynamic and reversible, and the related reactions are mainly catalyzed by methyltransferases and demethylases; methyltransferases catalyze the installation of m6A on RNA, while the demethylases remove m6A modifications (Figure 1). The biological function of dynamic RNA methylation is mainly mediated by reader proteins, which act by recognizing and binding to m6A and reading the biological information, mainly for splicing processing, nucleus to cytoplasm transport, and stabilization, translation, and degradation of RNA (Figure 2). Members of the YT521-B homology (YTH) domain family have been identified as m6A readers, including YTH domain family protein 1 (YTHDF1), YTH domain family protein 2 (YTHDF2), YTH domain family protein 3 (YTHDF3), YTH domain containing 1 (YTHDC1), and YTH domain containing 2 (YTHDC2), and they have conserved m6A-binding domains that can bind to the RNA m6A site. In addition, the heterogeneous nuclear ribonucleoprotein (HnRNP) family, insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs), and eIF3 can be combined with the m6A site and function as readers. According to relevant reports, YTHDF1,5 YTHDC2,6 and eIF37 have been shown to promote target mRNA translation, and YTHDF2 participates in RNA degradation,8 while YTHDF3 plays a synergistic role, interacting with YTHDF1 to promote RNA translation, and binding with YTHDF2 to promote RNA degradation.9 YTHDC1 facilitates RNA splicing and output,10 and HnRNP is involved in the splicing of RNA.11 IGF2BPs enhances the expression of target mRNA by promoting its stability.12
Figure 2.
m6A is recognized by reader proteins and affects RNA metabolism, including RNA export, splicing, processing, degradation, and protein translation
As the writers of m6A RNA methylation, methyltransferases and their components can regulate a variety of cellular physiological processes, including cell cycle, cell growth, cell differentiation, and apoptosis. In addition, methyltransferase components are involved in phenotypic transformation of Drosophila13 and embryonic development of mice.14 Furthermore, the relationship between methyltransferases and cancers has become an intense area of investigation. Herein, we review the key roles of methyltransferases in human cancers and their mechanisms of tumorigenesis.
RNA m6A methyltransferase
In the 1990s, Bokar et al.15 isolated 200-kDa (MT-A) and 800-kDa (MT-B) nuclear subcomplexes from HeLa cells and identified an m6A methyltransferase multi-component complex. This complex utilizes S-adenosylmethionine (SAM) as a methyl donor to provide a methyl group that replaces the hydrogen atom of the sixth nitrogen center of adenine.15,16 Subsequently, a 70-kDa protein, MT-A70, was extracted from the MT-A complex and was named methyltransferase-like 3 (METTL3).17 Further investigation led to the discovery of important methyltransferase components, such as methyltransferase-like 14 (METTLl4), Wilms tumor 1-associated protein (WTAP), protein virilizer homolog (VIRMA), E3 ubiquitin-protein ligase Hakai (HAKAI), zinc finger CCCH domain-containing protein 13 (ZC3H13), RNA-binding protein 15 (RBM15), and methyltransferase-like 16 (METTL16) (Table 1; Table S1). These discoveries have greatly advanced our understanding of m6A RNA methylation.
Table 1.
Multiple functions exerted by m6A RNA methyltransferases in various cancers
| Molecule | Role | Cancer | Target | Mechanism | References |
|---|---|---|---|---|---|
| METTL3 | oncogene | lung cancer | mRNA | METTL3-eIF3 cycle promotes translation of oncogenes | 18 |
| oncogene | AML | mRNA | enhances protein translation | 19 | |
| oncogene | breast cancer | mRNA | HBXIP-let-7g-METTL3-HBXIP positive feedback pathway | 20 | |
| oncogene | CRC, GC, HCC | mRNA | activates the glycolysis of tumor cells | 21, 22, 23 | |
| oncogene | HCC | mRNA | regulates sorafenib resistance by mediating autophagy | 24 | |
| oncogene | pancreatic cancer, HCC, CRC | miRNA | accelerates the maturation of oncogenic miRNA | 25, 26, 27, 28 | |
| oncogene | HCC | lncRNA | upregulates the expression of oncogenic LNC00958 | 29 | |
| suppressor | endometrial cancer | mRNA | inhibits the activation of AKT signaling pathway | 30 | |
| METTL14 | oncogene | AML | mRNA | inhibits myeloid differentiation of AMLs | 31 |
| oncogene | pancreatic cancer | mRNA | activates the signaling pathway | 32 | |
| suppressor | RCC | mRNA | inhibits the expression of the oncogene P2RX6 | 33 | |
| oncogene | breast cancer | miRNA | promotes the maturation of oncogenic miRNA | 34 | |
| suppressor | HCC | miRNA | promotes the maturation of cancer suppressor miRNA | 35 | |
| suppressor | CRC | lncRNA | downregulates the expression of carcinogenic lncRNA XIST | 36 | |
| WTAP | oncogene | GC | – | involved in the immune regulation of tumor cells | 37 |
| oncogene | DLBCL | – | stabilizes expression of transcriptional repressor BCL6 | 38 | |
| oncogene | AML | – | inhibits myeloid differentiation of AMLs | 39 | |
| VIRMA | oncogene | HCC | mRNA | inhibits the expression of tumor suppressor gene GATA3 | 40 |
| oncogene | PCa | lncRNA | upregulates the expression of oncogenic lncRNA CCAT1 and CCAT2 | 41 | |
| oncogene | breast cancer | mRNA | regulates the expression of CDK1 and regulates the tumor cell cycle | 42 | |
| HAKAI | oncogene | NSCLC | – | promotes EMT and mediates gefitinib resistance | 43 |
| oncogene | tumor cell line | mRNA | interacts with PSF to stabilize oncogene expression | 44,45 | |
| ZC3H13 | oncogene | CRC | – | MLL1-ZC3H13 gene fusion, affecting mitosis | 46 |
| suppressor | CRC | mRNA | inhibits activation of the Ras signaling pathway | 47 | |
| RBM15 | oncogene | AMKL | – | RBM15-MKL1 gene fusion | 48 |
| oncogene | breast cancer | – | RBM15 gene mutation | 49 | |
| METTL16 | oncogene | CRC | – | METTL16 gene frameshift mutation | 50 |
CRC, colorectal carcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; RCC, renal cell carcinoma; DLBCL, diffuse large B cell lymphoma; AML, acute myeloid leukemia; PCa, prostate carcinoma; NSCLC, non-small cell lung cancer; AMKL, acute megakaryocytic leukemia.
Methyltransferases and cancer
METTL3
Accumulating evidence shows that METTL3 can participate in the development of cancer through various mechanisms. For example, METTL3 regulates the level and stability of target gene mRNA by regulating the homeostasis of m6A, thereby promoting transcript translation and protein synthesis. METTL3 and METTL14 form stable complexes at a 1:1 ratio.51 In vitro, the combination of the two can significantly improve methylation efficiency. In vivo, METTL14 recognizes RNA substrates and serves as an RNA-binding platform, and METTL3 has the main methyltransferase catalytic domain. The METTL3-METTL14 complex is also the only known m6A methyltransferase heterodimer complex (Figure 1).52 Furthermore, METTL3 can recruit the translation initiation factor, eIF3, to the translation initiation complex when combined with certain mRNAs to enhance translation,7 for example, with epidermal growth factor receptor (EGFR) and bromodomain-containing protein 4 (BRD4).18 In addition, METTL3 is also a chromatin-based regulatory factor necessary for leukemia status.19 It locates to the transcription initiation point of SP1 and SP2, induces m6A modification of the relevant mRNA coding region, and enhances its translation by alleviating ribosomal stasis. In breast cancer,20 METTL3 promotes the expression of an oncoprotein, HBXIP, in an m6A-dependent manner. HBXIP inhibits the tumor suppressor miRNA let-7g, and because let-7g inhibits METTL3, the positive feedback pathway, HBXIP-let-7g-METTL3-HBXIP, promotes the progression of breast cancer. Similarly, METTL3 upregulates RAB2B expression to promote the growth of cervical cancer cells.53 In bladder cancer, METTL3 upregulates the expression of the adhesion molecule, ITGA6, to enhance the adhesion, migration, and invasion of tumor cells and to mediate signal output.54 METTL3 also promotes bladder cancer progression by regulating the AFF4-nuclear factor κB (NF-κB)-MYC multi-level signal network,55 and the METTL3-m6A-CDCP1 axis promotes the growth of chemically transformed cells and bladder cancer cells.56 In prostate carcinoma, researchers revealed a METTL3-m6A-MYC axis.57 METTL3 enhances MYC expression and plays a carcinogenic role by increasing the level of m6A in the MYC transcript.
In colorectal carcinoma (CRC),58,59 SOX2 and CCNE1 are downstream target genes of METTL3. As an oncogene, METTL3 maintains the expression of SOX2 and CCNE1 in CRCs through an m6A-dependent mechanism, maintains the stemness of CRCs, and promotes proliferation and metastasis. Furthermore, in CRC with high glucose metabolism,21 METTL3 induces CRC in a glycolysis-dependent manner. Mechanistically, METTL3 directly interacts with the mRNA of the glycolysis-related genes HK2 and GLUT1, thereby activating glycolysis in tumor cells. In gastric cancer22 and hepatocellular carcinoma (HCC),23 METTL3 promotes tumorigenesis by activating glycolysis. METTL3 upregulates the expression of HDGF by increasing the m6A modification of HDGF mRNA, and HDGF promotes tumor angiogenesis and glycolysis of cancer cells22 and can also activate the mTOCR1 pathway to promote glycolysis in cancer cells.23 It is noteworthy that Liu et al.60 and Yue et al.61 confirmed that METTL3 is closely related to the epithelial-mesenchymal transition (EMT) process of gastric cancer. Previous studies have found that METTL3 regulates the EMT process.62,63 METTL3 first modifies SNAIL mRNA, a key transcription factor of EMT, triggering the multi-ribosome-mediated translation of SNAIL mRNA. Upregulation of SNAIL then promotes the transforming growth factor β1 (TGFβ1)-induced EMT process. It is noteworthy that METTL3 also mediates sorafenib resistance in HCC by mediating autophagy.24 Additionally, there is a certain relationship between m6A modification and miRNA processing. METTL3 can promote the progression of cancer by accelerating the maturation of primary (pri-)miRNAs, including miR-25-2p in pancreatic cancer,25 pri-miR221/222 in HCC,26 and pri-miR-1246 and miR-6079 in CRC.27,28 Furthermore, METTL3 also upregulates the expression of oncogenic LINC00958 in HCC.29
METTL3 not only promotes cancer progression as an oncogene, but it also plays an anticancer role in some tumors. In glioblastoma, METTL3 inhibits the growth and self-renewal of tumor stem cells.64 In endometrial cancer,30 a low level of m6A promotes the proliferation of tumor cells, and low METTL3 expression activates the AKT pathway, leading to increased proliferation and tumorigenicity of endometrial cancer cells. In CRC,65 METTL3 inhibits the proliferation, migration, and invasion of tumor cells by inhibiting the phosphorylation of p38 and extracellular signal-regulated kinase (ERK).
METTL14
As an important methyltransferase component, METTL14 may play multiple roles in the occurrence and progression of cancer. For example, in HCC,66 METTL14 not only promotes cancer progression, but it also acts as a tumor suppressor to inhibit tumor development. METTL14 is highly expressed in hematopoietic stem cells (HSPCs) and acute myeloid leukemia (AML) cells.31 METTL14 inhibits the myeloid differentiation of HSPCs and AML cells by regulating the m6A modification of MYB and MYC mRNA, thereby enhancing the self-renewal and proliferation of AML cells, ultimately leading to AML progression. METTL14 is also upregulated in pancreatic cancer tissues,32 and METTL14 knockdown increases the sensitivity of pancreatic cancer cells to cisplatin and strengthens mTOR pathway-dependent autophagy, thereby accelerating the apoptosis of tumor cells. Remarkably, m6A modification can not only stabilize mRNA, but it also downregulates the stability of mRNA. In renal cell carcinoma (RCC),33 METTL14 regulates the splicing of precursor (pre-)P2RX6 mRNA by regulating its level of m6A, thereby inhibiting expression of the P2RX6 mRNA and protein. This is different from the previously reported stabilization of target gene mRNAs and proteins by METTL14. Because P2RX6 increases ATP-dependent Ca2+ influx to promote the migration and invasion of RCCs, we suggest that METTL14 plays a tumor suppressor role in RCC. Gu et al.67 found a low level of m6A modification and METTL14 in bladder cancer. As mentioned above for RCC, METTL14-mediated m6A modification also suppressed the mRNA stability of the target gene, NOTCH1, which is involved in the occurrence of bladder cancer and the self-renewal of bladder cancer-initiating cells; therefore, METTL14 may inhibit the occurrence and development of bladder cancer by inhibiting NOTCH1 expression.
Yi et al.34 evaluated m6A levels in breast cancer and normal tissues, and they found that the abnormal expression of METTL14 led to a steady-state imbalance of m6A modification. Further investigation confirmed that METTL14 promotes the migration and invasion of breast cancer cells by upregulating miR-146a-5p. In HCC,35 METTL14 interacts with the microprocessor protein, DGCR8, and positively regulates the processing process of pri-miR126 to inhibit HCC metastasis. However, the absence of METTL14 reduces the binding of DOCR8 to pri-miR126, which inhibits the processing of pri-miR126 and decreases miR126 expression, which eventually accelerates HCC metastasis. Notably, METTL14 is also downregulated in CRC,68 which inhibits CRC progression through miR375-YAP1 and miR375-SP1 pathways. In addition, METTL14 is also involved in the regulation of lncRNA in CRC.36 Through RNA sequencing (RNA-seq), carcinogenic lncRNA XIST was identified as a downstream target of METTL14. METTL14 downregulated lncRNA XIST in an m6A-dependent manner and inhibited the proliferation and invasion of CRC.
WTAP
WTAP is a nuclear protein that is widely expressed in various tissues of the body. As a splicing factor of the WT1 gene, WTAP is involved in regulation of the cell cycle and embryonic development.69 As a key methyltransferase component, it is involved in the regulation of m6A RNA methylation.70 Although WTAP did not show any methyltransferase activity in vitro, knockdown of WTAP can significantly reduce the m6A peak of cellular RNA, even more significantly than knocking down METTL3 or METTL14.71 This shows that WTAP is an essential component for efficient RNA methylation. In addition, WTAP can interact with the METTL3-METTL14 complex to form a more stable METTL3-METTL14-WTAP complex and improve the catalysis of m6A RNA methylation.72 WTAP also promotes the development of HCC by participating in the regulation of the post-transcriptional inhibition of the tumor suppressor EST1 in an m6A-dependent manner.73 Furthermore, WTAP can also promote pancreatic cancer by combining with FAK mRNA and activating the FAK-phosphatidylinositol 3-kinase (PI3K)-AKT and FAK-SRC-GRB2-ERK axes.74
It is noteworthy that high expression of WTAP in gastric cancer is closely related to T lymphocyte infiltration.37 T regulatory cells and CD4 memory-activated T cells were significantly fewer in high WTAP expression gastric cancer patients than in low WTAP expression gastric cancer patients, indicating that tumor immunoregulation may be an important reason for the poor prognosis of high WTAP expression. In diffuse large B cell lymphoma (DLBCL),38 overexpression of WTAP can promote the proliferation of cancer cells and enhance their anti-apoptotic ability. In addition, WTAP can form a trimeric complex with heat shock protein 90 (Hsp90) and transcriptional repressor B cell lymphoma 6 in vivo and in vitro, which helps stabilize WTAP and maintain WTAP protein levels. WTAP also plays an important role in the differentiation of bone marrow cells, with increased expression in AML tissues.39 However, in in vitro experiments, the downregulation of WTAP promoted phorbol 12-myristate 13-acetate (PMA)-induced bone marrow differentiation and increased expression of the bone marrow differentiation markers CD14 and CD11b, indicating that WTAP is involved in the induction of abnormal proliferation and differentiation block in AML cells. Additionally, WTAP is overexpressed in glioblastoma and is involved in regulating the migration and invasion of glioblastoma.75 In xenograft models, overexpression of WTAP also increases tumorigenicity of tumor cells. Furthermore, WTAP can regulate the activity of EGFR.71 WTAP is therefore closely related to the progression and poor prognosis of various tumors, including cholangiocarcinoma,76 CRC,77 and ovarian cancer.78
VIRMA
Schwartz et al.71 identified a protein network that interacts with METTL3 and showed that VIRMA interacts with WTAP. Knockdown of VIRMA in human A549 cells decreased the peak value of m6A by approximately 4-fold compared with the control group. This was a significantly greater decrease than that observed upon knockdown of either METTL3 or METTL14, demonstrating that VIRMA is required for the full methylation program in mammals.71 VIRMA can be used as a bracket through which WTAP combines with METTL3-METTL14 to guide the enrichment of m6A in 3′ UTRs.79
In testicular germ cell tumors,80 the abundance of m6A and the expression of VIRMA are different in different subtypes, with the level in seminoma higher than that in non-seminoma. Furthermore, the differences in expression levels of METTL14 and VIRMA can also distinguish seminoma from non-seminoma.81 Analysis of The Cancer Genome Atlas (TCGA) database shows differences in the expression of VIRMA between HCC and normal liver tissues,82 which are also observed in in vitro experiments. Through methylated RNA immunoprecipitation sequencing (MeRIP)-PCR analysis, VIRMA will upregulate ID2 mRNA m6A, thereby reducing ID2 expression and promoting cancer. VIRMA also regulates GATA3 pre-mRNA in an m6A-dependent manner, hindering the binding of HUR to GATA3 pre-mRNA, which in turn reduces GATA3 pre-mRNA expression, resulting in a decrease in GATA3 mRNA expression, eventually promoting HCC progression.40 In prostate cancer, VIRMA is a key factor for maintaining m6A levels,41 which upregulates the expression of carcinogenic lncRNA CCAT1 and CCAT2 through m6A modification, thereby increasing aggressiveness of the cancer. Remarkably, VIRMA can regulate cancer progression independently of m6A modified pathways. In breast cancer, VIRMA knockdown did not affect the m6A level of its target mRNA, cyclin-dependent kinase 1 (CDK1).42 In contrast, METTL3 knockdown significantly reduced the m6A modification level of CDK1 mRNA. Furthermore, knockdown of METTL3 did not change the amount of CDK1 mRNA interacting with VIRMA, indicating that the m6A modification did not interfere with the interaction of VIRMA and CDK1 mRNA and that VIRMA regulates CDK1 mRNA in an m6A-independent manner. In addition, in gastric cancer, VIRMA regulates c-JUN expression in an m6A-independent way, which ultimately promotes cancer.83
HAKAI
HAKAI is a key member of the methyltransferase complex.84 Knockdown of HAKAI in HeLa cells downregulated the level of m6A, which is consistent with the knockdown results of other methyltransferase members.79 Mammalian HAKAI, also known as Casitas B lineage lymphoma transformation sequence-like protein 1 (CBLL1), is an E3 ubiquitin-ligase that mediates the ubiquitination of E-cadherin to promote the endocytosis of E-cadherin at cell-cell contact. This ultimately leads to its lysosomal pathway degradation. E-cadherin, as a tumor suppressor, is downregulated during EMT, and its loss indicates a poor prognosis.85
In CRC,86 overexpression of HAKAI will drive the conversion of epithelial cells into mesenchymal cells, with the downregulation of E-cadherin and the upregulation of N-cadherin, making tumor cells more aggressive. Therefore, HAKAI is involved in two crucial processes of tumor progression, namely, reducing cell-matrix adhesion and enhancing cell invasion.85,86 In addition, overexpression of HAKAI can cause lung micrometastasis of tumor cells in vivo. Clinical tumor specimens of non-small cell lung cancer (NSCLC) with acquired gefitinib resistance also showed a decrease in E-cadherin and an increase in HAKAI expression.43 In vitro experiments confirmed that HAKAI mediates gefitinib resistance by promoting EMT. HAKAI knockout inhibited AKT activity in NSCLCs, making NSCLCs sensitive to cisplatin therapy.87 Of note, HAKAI can interact with PTB-associated splicing factor (PSF) to promote tumorigenesis.44,45 HAKAI enhances the binding of PSF to tumor-related protein mRNAs to post-transcriptionally regulate the target mRNA and reverse its tumor suppression effect. Interestingly, in breast cancer,88 HAKAI acts as a novel estrogen receptor-α core inhibitor to inhibit the proliferation and migration of estrogen-dependent tumor cells, but it does not affect the growth of estrogen-independent cells.
ZC3H13
ZC3H13 is a typical CCCH zinc finger protein involved in regulating RNA m6A. In Drosophila, the ZC3H13 homolog, Xio, was identified during female-male transformation studies.13 Xio colocalizes with the known methyltransferase complex and, importantly, the expression pattern of Xio is similar to that of other members of the m6A pathway. Xio also interacts with known methyltransferases, and its deletion causes a reduction in m6A levels. After knocking out Zc3h13 in mouse embryonic stem cells, m6A levels decreased by approximately 60%–70% compared with wild-type cells, suggesting that ZC3H13 significantly affects m6A modification in mice.89 In addition, knockout of ZC3H13 caused other methyltransferase components to be significantly transferred into the cytoplasm, and the expression levels of METTL3 and METTL14 in the nucleus were decreased. Similarly, knockdown of WTAP, VIRILIZER, or HAKAI also reduced the nuclear accumulation of METTL3 and METTL14, but the inhibition of WTAP, VIRILIZER, or HAKAI did not affect the nuclear localization of ZC3H13. These results indicate that the main role of ZC3H13 is to stabilize the ZC3H13-WTAP-VIRILIZER-HAKAI complex in the nucleus and regulate m6A. In addition, in the Zc3h13 mutant mouse, glioblastoma cells were more resistant to temozolomide.90
In humans, the fusion mutation MLL1-ZC3H13 disrupts the stability of chromosomes, affects mitosis, and promotes the formation of CRC tumors. It also upregulates the expression of the tumor stem cell marker CD44.46 In addition, Gewurz et al.91 found that ZC3H13 is a key upstream factor of NF-κB, which stabilizes NF-κB activation pathway-related mRNA and destabilizes NF-κB inhibitory pathway-related mRNA. Activation of NF-κB signaling accelerates tumor proliferation and invasion; therefore, ZC3H13 may play a role as an oncogene. Remarkably, ZC3H13 inactivates the Ras-ERK signaling pathway to inhibit the proliferation and invasion of CRC, and its reduction is also related to late tumor-node-metastasis (TNM) staging, indicating that ZC3H13 may be a tumor suppressor gene.47
RBM15
RBM15 and its analog RBM15B mediate the formation of m6A in mRNA and the lncRNA XIST.92 RBM15/RBM15B binds to the methyltransferase complex and recruits it to specific sites in RNA, mediating gene silencing in an m6A-dependent manner. In addition, RBM15 can also regulate a variety of signaling pathways that participate in the growth and apoptosis of many cells, including blood cells in particular. Some researchers have reported that RBM15 can promote the development of HSPCs and normal megakaryocytes by regulating the target, c-MYC,93 and can also affect myeloid differentiation by activation of NOTCH-stimulated transcription in HSPCs.94 In addition, RBM15 is also involved in the development of hematological tumors. The PRMT-RBM15 axis inhibits the terminal differentiation of megakaryocytes in acute megakaryocytic leukemia (AMKL), leading to defects in the formation of mature megakaryocytes.95 Mechanistically, RBM15 binds to the intron region of pre-mRNAs of important megakaryocytic genes, and it recruits splicing factors to the corresponding binding sites for selective splicing, while overexpression of PRMT1 inhibits the expression of RBM15, resulting in the failure of normal expression of megakaryocyte genes, thereby promoting the development of AMKL. Overexpression of PRMT1 or knockout of RBM15 inhibited the differentiation of CD34+ cells into mature megakaryocytes.96 In addition, the RBM15-MKL1 fusion gene is also associated with AMKL in adults and children.48
In addition to its key role in hematological disease, RBM15 is closely associated with other tumors. In pancreatic cancer, the expression of RBM15 has a significant correlation with overall survival.97 In breast cancer, RBM15 mutations are associated with the incidence of borderline and malignant phyllodes tumors.49
METTL16
METTL16 is a methyltransferase newly discovered in HeLa cells and the second catalytically active methyltransferase verified in humans.52 The METTL16 monomer consists of the N-terminal methyltransferase domain and the C-terminal vertebrate conserved region,98 which usually exists as a dimer.99 However, in in vitro experiments, METTL16 can assemble with target RNA in a monomeric form and regulate its m6A modification; therefore, the dimer is not necessary for its catalytic activity on RNA. METTL16 is associated with m6A of multiple ncRNAs, lncRNAs, and pre-mRNAs.100 METTL16 catalyzes the m6A modification of U6 snRNA at position 43,98 which post-transcriptionally regulates expression of the SAM synthase gene.101 This promotes interaction with cancer lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and promotes its expression.102 In addition, METTL16 is also necessary for the development of mouse embryos.14 METTL16-deficient mouse 16-cell embryos showed a decreased level of SAM synthase gene transcripts, causing a large number of transcriptome disorders in 64-cell blastocysts, which hindered mouse embryo development. Remarkably, a frameshift mutation of METTL16 in CRC can promote the progression of colon cancer.50
Conclusions
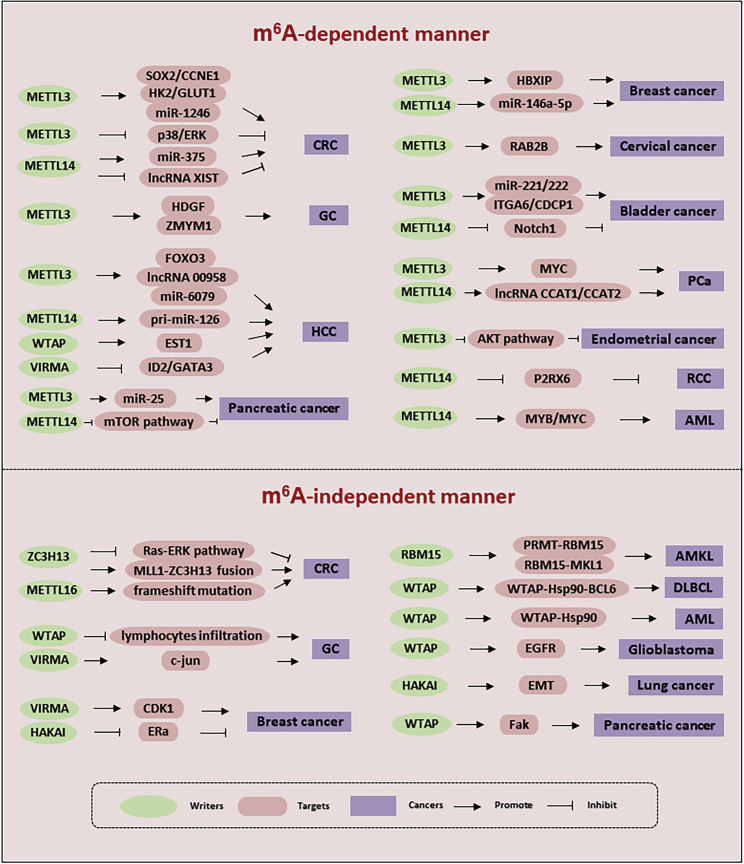
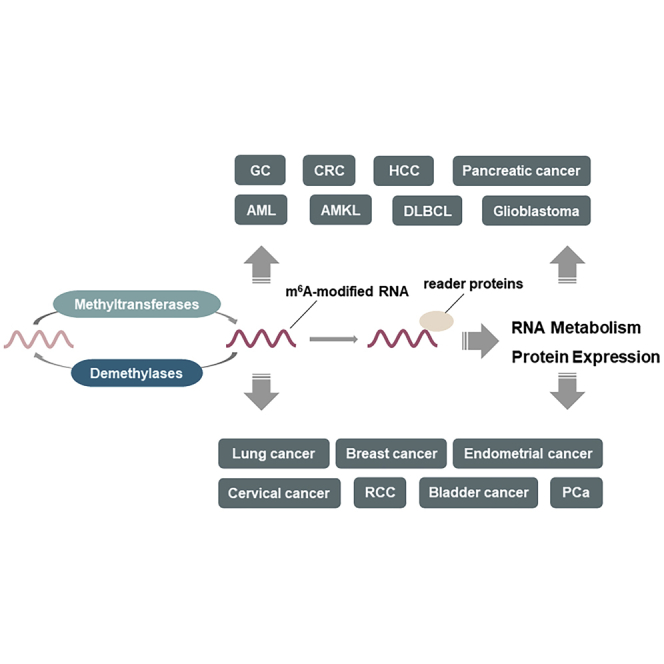
We have reviewed the role of methyltransferases and their constituent components in different cancers (Table 1; Figure 3). The regulation mechanisms of the various methyltransferases and their components in the m6A pathway have many similarities, and the stability and level of protein expression from target genes are regulated through the writing of m6A. The regulation can be roughly divided into three categories: first, the same components regulate a particular cancer through different target genes. For example, with regard to METTL3 in gastric cancer, its downstream target genes, HDGF22 and ZMYM1,61 are both involved in the progression of cancer. Second, different components regulate the same target gene in different cancers. For example, METTL3 regulates MYC in prostate carcinoma,57 and METTL14 regulates MYC in AML31 to promote cancer. Third, different components participate in the regulation of the same cancer through different target genes; for example, METTL3 and MTTL14 regulate the incidence and progression of CRC,34,58,59 bladder cancer,55,67 and pancreatic cancer25,32 through different targets, and METTL3,23 METTL14,35 WTAP,73 and VIRMA82 jointly regulate the progress of HCC. In addition, methyltransferases and their components can also accelerate the maturation of miRNA and regulate the expression of lncRNA in an m6A-dependent manner. Methyltransferases and their components also regulate cancer independently of m6A, mainly through two pathways: first, through the regulation of target genes, such as c-JUN,83 CDK1,42 and EGFR,75 and second, through mutations in methyltransferase genes; for example, the fusion mutation of RBM15 promotes the occurrence of AMKL,95 and the fusion mutation of ZC3H1346 and the frameshift mutation of METTL1650 promote the progression of CRC.
Figure 3.
Methyltransferases participate in cancer progression through an m6A-dependent manner and m6A-independent manner
Prospects
RNA m6A is involved in multiple cellular processes such as RNA maturation, protein translation, and molecular structure, and it is important in gene expression and regulation, especially in the occurrence and development of cancer. As a key regulator of m6A, the role that methyltransferases play in tumors cannot be ignored. With the advent of high-throughput sequencing, chemical immunity, and other precise techniques, as well as the emergence of highly specific antibodies, more and more methyltransferases and their components have been discovered, and it has become possible to determine the chemical basis and biological functions of various methyltransferases. Various studies have shown that due to the genetic heterogeneity and specificity of tumors, methyltransferases also exhibit significant tumor specificity, so they can play different roles in cancers. As far as methyltransferases themselves are concerned, different target genes regulated by methyltransferases and their mechanism of action are different in different tumors or the same tumor in different organisms, resulting in diverse results. In addition, in the pathway of m6A modification, the overall changes in the expression levels, including methyltransferases, demethylases, and reader proteins, regulate the m6A modification levels of target genes, while methyltransferases and demethylases regulate the m6A modification positions of the target genes, the interaction of the three through influencing m6A-modified levels and positions, and can also lead to their diverse functions in the course of the tumors. As an activator of cancer, methyltransferases regulate the level of target gene mRNA and protein through transcription and translation pathways, activate multi-level signaling pathways and metabolic pathways, regulate the cell cycle, promote the proliferation and metastasis of tumor cells, and enhance the aggressiveness of tumor cells through the EMT process. In the clinic, methyltransferases are also associated with overall survival rate, TNM stage, and drug resistance. As an inhibitor of cancer, methyltransferases can downregulate downstream oncogene mRNA and protein levels and inhibit the activation of signaling pathways.
Given the prevalence of m6A modifications in different types of transcripts, further research into m6A methyltransferases is warranted to further clarify their influence on RNA metabolism and to fully understand their molecular mechanisms. Their biological relevance and diagnostic value in cancer requires further evaluation and may provide new prognostic markers for cancer and therapeutic targets. The development of m6A methyltransferase inhibitors and agonists, especially in combination with other therapeutic drugs, is expected to lead to the development of new cancer treatment methods.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (nos. 81871871 to Z.W. and 81902333 to X.C.); the Key Research and Development Plan (Social Development) of Science and Technology Department of Jiangsu Province (no. BE2019760 to Z.W.); the Medical Innovation Team Foundation of the Jiangsu Provincial Enhancement Health Project (no. CXTDA2017021 to Z.W.); the Postgraduate Research & Practice Innovation Program of Jiangsu Province (no. KYCX19_1170 to Z.C.); and the “123” advantageous disciplines, core technologies and “789” excellent talent training plan of the Second Affiliated Hospital of Nanjing Medical University (no. 789ZYRC202090146 to X.C.). We thank Jeremy Allen, PhD, from Liwen Bianji, Edanz Group China, for editing the English text of a draft of this manuscript.
Author contributions
J.H.and Z.C.wrote and drafted the manuscript and figures. X.C.and Z.C. revised the manuscript. J.C. retrieved literature. Z.W. designed this manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.12.021.
Contributor Information
Zhixiang Cheng, Email: zhixiangcheng@njmu.edu.cn.
Zhaoxia Wang, Email: wangzhaoxia@njmu.edu.cn.
Supplemental information
References
- 1.Yue Y., Liu J., He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015;29:1343–1355. doi: 10.1101/gad.262766.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rottman F., Shatkin A.J., Perry R.P. Sequences containing methylated nucleotides at the 5′ termini of messenger RNAs: possible implications for processing. Cell. 1974;3:197–199. doi: 10.1016/0092-8674(74)90131-7. [DOI] [PubMed] [Google Scholar]
- 3.Csepany T., Lin A., Baldick C.J., Jr., Beemon K. Sequence specificity of mRNA N6-adenosine methyltransferase. J. Biol. Chem. 1990;265:20117–20122. [PubMed] [Google Scholar]
- 4.Zhang C., Chen Y., Sun B., Wang L., Yang Y., Ma D., Lv J., Heng J., Ding Y., Xue Y. m6A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 5.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu P.J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., Qi M., Lu Z., Shi H., Wang J. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017;27:1115–1127. doi: 10.1038/cr.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., Liu C., He C. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roundtree I.A., Luo G.Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife. 2017;6:e31311. doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo J., Tang H.W., Li J., Perrimon N., Yan D. Xio is a component of the Drosophila sex determination pathway and RNA N6-methyladenosine methyltransferase complex. Proc. Natl. Acad. Sci. USA. 2018;115:3674–3679. doi: 10.1073/pnas.1720945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendel M., Chen K.M., Homolka D., Gos P., Pandey R.R., McCarthy A.A., Pillai R.S. Methylation of Structured RNA by the m6A writer METTL16 is essential for mouse embryonic development. Mol. Cell. 2018;71:986–1000.e11. doi: 10.1016/j.molcel.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 16.Narayan P., Rottman F.M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- 17.Tuck M.T. Partial purification of a 6-methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem. J. 1992;288:233–240. doi: 10.1042/bj2880233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., Du P., Kim W., Tang S., Sliz P. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millán-Zambrano G., Robson S.C., Aspris D., Migliori V., Bannister A.J., Han N. Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai X., Wang X., Cao C., Gao Y., Zhang S., Yang Z., Liu Y., Zhang X., Zhang W., Ye L. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19. doi: 10.1016/j.canlet.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 21.Shen C., Xuan B., Yan T., Ma Y., Xu P., Tian X., Zhang X., Cao Y., Ma D., Zhu X. m6A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer. 2020;19:72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q., Chen C., Ding Q., Zhao Y., Wang Z., Chen J., Jiang Z., Zhang Y., Xu G., Zhang J. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 23.Lin Y., Wei X., Jian Z., Zhang X. METTL3 expression is associated with glycolysis metabolism and sensitivity to glycolytic stress in hepatocellular carcinoma. Cancer Med. 2020;9:2859–2867. doi: 10.1002/cam4.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Z., Niu Y., Wan A., Chen D., Liang H., Chen X., Sun L., Zhan S., Chen L., Cheng C. RNA m6 A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020;39:e103181. doi: 10.15252/embj.2019103181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., Bai R., Li M., Ye H., Wu C., Wang C., Li S., Tan L., Mai D., Li G. Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 2019;10:1858. doi: 10.1038/s41467-019-09712-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han J., Wang J.Z., Yang X., Yu H., Zhou R., Lu H.C., Yuan W.B., Lu J.C., Zhou Z.J., Lu Q. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer. 2019;18:110. doi: 10.1186/s12943-019-1036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng W., Li J., Chen R., Gu Q., Yang P., Qian W., Ji D., Wang Q., Zhang Z., Tang J., Sun Y. Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:393. doi: 10.1186/s13046-019-1408-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y., Song S., Meng Q., Wang L., Li X., Xie S., Chen Y., Jiang X., Wang C., Lu Y. miR24-2 accelerates progression of liver cancer cells by activating Pim1 through tri-methylation of Histone H3 on the ninth lysine. J. Cell. Mol. Med. 2020;24:2772–2790. doi: 10.1111/jcmm.15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo X., Chen Z., Gao W., Zhang Y., Wang J., Wang J., Cao M., Cai J., Wu J., Wang X. M6A-mediated upregulation of LINC00958 increases lipogenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. J. Hematol. Oncol. 2020;13:5. doi: 10.1186/s13045-019-0839-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Eckert M.A., Harada B.T., Liu S.M., Lu Z., Yu K., Tienda S.M., Chryplewicz A., Zhu A.C., Yang Y. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng H., Huang H., Wu H., Qin X., Zhao B.S., Dong L., Shi H., Skibbe J., Shen C., Hu C. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191–205.e9. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong F., Liu X., Zhou Y., Hou X., He J., Li Q., Miao X., Yang L. Downregulation of METTL14 increases apoptosis and autophagy induced by cisplatin in pancreatic cancer cells. Int. J. Biochem. Cell Biol. 2020;122:105731. doi: 10.1016/j.biocel.2020.105731. [DOI] [PubMed] [Google Scholar]
- 33.Gong D., Zhang J., Chen Y., Xu Y., Ma J., Hu G., Huang Y., Zheng J., Zhai W., Xue W. The m6A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca2+ influx modulating ERK1/2 phosphorylation and MMP9 signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:233. doi: 10.1186/s13046-019-1223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yi D., Wang R., Shi X., Xu L., Yilihamu Y., Sang J. METTL14 promotes the migration and invasion of breast cancer cells by modulating N6-methyladenosine and hsa-miR-146a-5p expression. Oncol. Rep. 2020;43:1375–1386. doi: 10.3892/or.2020.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J.Z., Yang F., Zhou C.C., Liu F., Yuan J.H., Wang F., Wang T.T., Xu Q.G., Zhou W.P., Sun S.H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 36.Yang X., Zhang S., He C., Xue P., Zhang L., He Z., Zang L., Feng B., Sun J., Zheng M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer. 2020;19:46. doi: 10.1186/s12943-020-1146-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Su Q., Li B., Lan L., Wang C., Li W., Wang G., Chen W., He Y., Zhang C. High expression of WTAP leads to poor prognosis of gastric cancer by influencing tumour-associated T lymphocyte infiltration. J. Cell. Mol. Med. 2020;24:4452–4465. doi: 10.1111/jcmm.15104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuai Y., Gong X., Ding L., Li F., Lei L., Gong Y., Liu Q., Tan H., Zhang X., Liu D. Wilms’ tumor 1-associating protein plays an aggressive role in diffuse large B-cell lymphoma and forms a complex with BCL6 via Hsp90. Cell Commun. Signal. 2018;16:50. doi: 10.1186/s12964-018-0258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bansal H., Yihua Q., Iyer S.P., Ganapathy S., Proia D.A., Penalva L.O., Uren P.J., Suresh U., Carew J.S., Karnad A.B. WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia. 2014;28:1171–1174. doi: 10.1038/leu.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lan T., Li H., Zhang D., Xu L., Liu H., Hao X., Yan X., Liao H., Chen X., Xie K. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer. 2019;18:186. doi: 10.1186/s12943-019-1106-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barros-Silva D., Lobo J., Guimarães-Teixeira C., Carneiro I., Oliveira J., Martens-Uzunova E.S., Henrique R., Jerónimo C. VIRMA-dependent N6-methyladenosine modifications regulate the expression of long non-coding RNAs CCAT1 and CCAT2 in prostate cancer. Cancers (Basel) 2020;12:771. doi: 10.3390/cancers12040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian J.Y., Gao J., Sun X., Cao M.D., Shi L., Xia T.S., Zhou W.B., Wang S., Ding Q., Wei J.F. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene. 2019;38:6123–6141. doi: 10.1038/s41388-019-0861-z. [DOI] [PubMed] [Google Scholar]
- 43.Weng C.H., Chen L.Y., Lin Y.C., Shih J.Y., Lin Y.C., Tseng R.Y., Chiu A.C., Yeh Y.H., Liu C., Lin Y.T. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38:455–468. doi: 10.1038/s41388-018-0454-2. [DOI] [PubMed] [Google Scholar]
- 44.Figueroa A., Kotani H., Toda Y., Mazan-Mamczarz K., Mueller E.C., Otto A., Disch L., Norman M., Ramdasi R.M., Keshtgar M. Novel roles of Hakai in cell proliferation and oncogenesis. Mol. Biol. Cell. 2009;20:3533–3542. doi: 10.1091/mbc.E08-08-0845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figueroa A., Fujita Y., Gorospe M. Hacking RNA: Hakai promotes tumorigenesis by enhancing the RNA-binding function of PSF. Cell Cycle. 2009;8:3648–3651. doi: 10.4161/cc.8.22.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parameswaran S., Vizeacoumar F.S., Kalyanasundaram Bhanumathy K., Qin F., Islam M.F., Toosi B.M., Cunningham C.E., Mousseau D.D., Uppalapati M.C., Stirling P.C. Molecular characterization of an MLL1 fusion and its role in chromosomal instability. Mol. Oncol. 2019;13:422–440. doi: 10.1002/1878-0261.12423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu D., Zhou J., Zhao J., Jiang G., Zhang X., Zhang Y., Dong M. ZC3H13 suppresses colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling. J. Cell. Physiol. 2019;234:8899–8907. doi: 10.1002/jcp.27551. [DOI] [PubMed] [Google Scholar]
- 48.Hsiao H.H., Yang M.Y., Liu Y.C., Hsiao H.P., Tseng S.B., Chao M.C., Liu T.C., Lin S.F. RBM15-MKL1 (OTT-MAL) fusion transcript in an adult acute myeloid leukemia patient. Am. J. Hematol. 2005;79:43–45. doi: 10.1002/ajh.20298. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Dios D.A., Levi D., Shah V., Gillett C., Simpson M.A., Hanby A., Tomlinson I., Sawyer E.J. MED12, TERT promoter and RBM15 mutations in primary and recurrent phyllodes tumours. Br. J. Cancer. 2018;118:277–284. doi: 10.1038/bjc.2017.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yeon S.Y., Jo Y.S., Choi E.J., Kim M.S., Yoo N.J., Lee S.H. Frameshift mutations in repeat sequences of ANK3, HACD4, TCP10L, TP53BP1, MFN1, LCMT2, RNMT, TRMT6, METTL8 and METTL16 genes in colon cancers. Pathol. Oncol. Res. 2018;24:617–622. doi: 10.1007/s12253-017-0287-2. [DOI] [PubMed] [Google Scholar]
- 51.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruszkowska A., Ruszkowski M., Dauter Z., Brown J.A. Structural insights into the RNA methyltransferase domain of METTL16. Sci. Rep. 2018;8:5311. doi: 10.1038/s41598-018-23608-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Y., Li Y., Huang Y., Jin Z., Wang C., Wang H., Xu J. METTL3 regulates the malignancy of cervical cancer via post-transcriptional regulation of RAB2B. Eur. J. Pharmacol. 2020;879:173134. doi: 10.1016/j.ejphar.2020.173134. [DOI] [PubMed] [Google Scholar]
- 54.Jin H., Ying X., Que B., Wang X., Chao Y., Zhang H., Yuan Z., Qi D., Lin S., Min W. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207. doi: 10.1016/j.ebiom.2019.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng M., Sheng L., Gao Q., Xiong Q., Zhang H., Wu M., Liang Y., Zhu F., Zhang Y., Zhang X. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene. 2019;38:3667–3680. doi: 10.1038/s41388-019-0683-z. [DOI] [PubMed] [Google Scholar]
- 56.Yang F., Jin H., Que B., Chao Y., Zhang H., Ying X., Zhou Z., Yuan Z., Su J., Wu B. Dynamic m6A mRNA methylation reveals the role of METTL3-m6A-CDCP1 signaling axis in chemical carcinogenesis. Oncogene. 2019;38:4755–4772. doi: 10.1038/s41388-019-0755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yuan Y., Du Y., Wang L., Liu X. The M6A methyltransferase METTL3 promotes the development and progression of prostate carcinoma via mediating MYC methylation. J. Cancer. 2020;11:3588–3595. doi: 10.7150/jca.42338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li T., Hu P.S., Zuo Z., Lin J.F., Li X., Wu Q.N., Chen Z.H., Zeng Z.L., Wang F., Zheng J. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu W., Si Y., Xu J., Lin Y., Wang J.Z., Cao M., Sun S., Ding Q., Zhu L., Wei J.F. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J. Cell. Mol. Med. 2020;24:3521–3533. doi: 10.1111/jcmm.15042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu T., Yang S., Sui J., Xu S.Y., Cheng Y.P., Shen B., Zhang Y., Zhang X.M., Yin L.H., Pu Y.P., Liang G.Y. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J. Cell. Physiol. 2020;235:548–562. doi: 10.1002/jcp.28994. [DOI] [PubMed] [Google Scholar]
- 61.Yue B., Song C., Yang L., Cui R., Cheng X., Zhang Z., Zhao G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol. Cancer. 2019;18:142. doi: 10.1186/s12943-019-1065-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin X., Chai G., Wu Y., Li J., Chen F., Liu J., Luo G., Tauler J., Du J., Lin S. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019;10:2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Li J., Chen F., Peng Y., Lv Z., Lin X., Chen Z., Wang H. N6-methyladenosine regulates the expression and secretion of TGFβ1 to affect the epithelial-mesenchymal transition of cancer cells. Cells. 2020;9:296. doi: 10.3390/cells9020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.G. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deng R., Cheng Y., Ye S., Zhang J., Huang R., Li P., Liu H., Deng Q., Wu X., Lan P., Deng Y. m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. OncoTargets Ther. 2019;12:4391–4402. doi: 10.2147/OTT.S201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang B.H., Yan L.N., Yang J.Y. Pending role of METTL14 in liver cancer. Hepatobiliary Surg. Nutr. 2019;8:669–670. doi: 10.21037/hbsn.2019.10.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gu C., Wang Z., Zhou N., Li G., Kou Y., Luo Y., Wang Y., Yang J., Tian F. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N6-methyladenosine of Notch1. Mol. Cancer. 2019;18:168. doi: 10.1186/s12943-019-1084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X., Xu M., Xu X., Zeng K., Liu X., Sun L., Pan B., He B., Pan Y., Sun H. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 processing. Mol. Ther. 2020;28:599–612. doi: 10.1016/j.ymthe.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., Fray R.G. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y., Peng C., Chen J., Chen D., Yang B., He B., Hu W., Zhang Y., Liu H., Dai L. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer. 2019;18:127. doi: 10.1186/s12943-019-1053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B.Q., Liang Z.Y., Seery S., Liu Q.F., You L., Zhang T.P., Guo J.C., Zhao Y.P. WT1 associated protein promotes metastasis and chemo-resistance to gemcitabine by stabilizing Fak mRNA in pancreatic cancer. Cancer Lett. 2019;451:48–57. doi: 10.1016/j.canlet.2019.02.043. [DOI] [PubMed] [Google Scholar]
- 75.Jin D.I., Lee S.W., Han M.E., Kim H.J., Seo S.A., Hur G.Y., Jung S., Kim B.S., Oh S.O. Expression and roles of Wilms’ tumor 1-associating protein in glioblastoma. Cancer Sci. 2012;103:2102–2109. doi: 10.1111/cas.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jo H.J., Shim H.E., Han M.E., Kim H.J., Kim K.S., Baek S., Choi K.U., Hur G.Y., Oh S.O. WTAP regulates migration and invasion of cholangiocarcinoma cells. J. Gastroenterol. 2013;48:1271–1282. doi: 10.1007/s00535-013-0748-7. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J., Tsoi H., Li X., Wang H., Gao J., Wang K., Go M.Y., Ng S.C., Chan F.K., Sung J.J., Yu J. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut. 2016;65:1482–1493. doi: 10.1136/gutjnl-2014-308614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu H.L., Ma X.D., Tong J.F., Li J.Q., Guan X.J., Yang J.H. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. OncoTargets Ther. 2019;12:6191–6201. doi: 10.2147/OTT.S205730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lobo J., Costa A.L., Cantante M., Guimarães R., Lopes P., Antunes L., Braga I., Oliveira J., Pelizzola M., Henrique R., Jerónimo C. m6A RNA modification and its writer/reader VIRMA/YTHDF3 in testicular germ cell tumors: a role in seminoma phenotype maintenance. J. Transl. Med. 2019;17:79. doi: 10.1186/s12967-019-1837-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lobo J., Barros-Silva D., Henrique R., Jerónimo C. The emerging role of epitranscriptomics in cancer: focus on urological tumors. Genes (Basel) 2018;9:552. doi: 10.3390/genes9110552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng X., Li M., Rao X., Zhang W., Li X., Wang L., Huang G. KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. OncoTargets Ther. 2019;12:3421–3428. doi: 10.2147/OTT.S180954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Miao R., Dai C.C., Mei L., Xu J., Sun S.W., Xing Y.L., Wu L.S., Wang M.H., Wei J.F. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J. Cell Physiol. 2020;235:7420–7432. doi: 10.1002/jcp.29645. [DOI] [PubMed] [Google Scholar]
- 84.Růžička K., Zhang M., Campilho A., Bodi Z., Kashif M., Saleh M., Eeckhout D., El-Showk S., Li H., Zhong S. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215:157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodríguez-Rigueiro T., Valladares-Ayerbes M., Haz-Conde M., Aparicio L.A., Figueroa A. Hakai reduces cell-substratum adhesion and increases epithelial cell invasion. BMC Cancer. 2011;11:474. doi: 10.1186/1471-2407-11-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castosa R., Martinez-Iglesias O., Roca-Lema D., Casas-Pais A., Díaz-Díaz A., Iglesias P., Santamarina I., Graña B., Calvo L., Valladares-Ayerbes M. Hakai overexpression effectively induces tumour progression and metastasis in vivo. Sci. Rep. 2018;8:3466. doi: 10.1038/s41598-018-21808-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Z., Wu Y., Tao Z., Ma L. E3 ubiquitin ligase Hakai regulates cell growth and invasion, and increases the chemosensitivity to cisplatin in non-small-cell lung cancer cells. Int. J. Mol. Med. 2018;42:1145–1151. doi: 10.3892/ijmm.2018.3683. [DOI] [PubMed] [Google Scholar]
- 88.Gong E.Y., Park E., Lee K. Hakai acts as a coregulator of estrogen receptor alpha in breast cancer cells. Cancer Sci. 2010;101:2019–2025. doi: 10.1111/j.1349-7006.2010.01636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wen J., Lv R., Ma H., Shen H., He C., Wang J., Jiao F., Liu H., Yang P., Tan L. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chow R.D., Guzman C.D., Wang G., Schmidt F., Youngblood M.W., Ye L., Errami Y., Dong M.B., Martinez M.A., Zhang S. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat. Neurosci. 2017;20:1329–1341. doi: 10.1038/nn.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gewurz B.E., Towfic F., Mar J.C., Shinners N.P., Takasaki K., Zhao B., Cahir-McFarland E.D., Quackenbush J., Xavier R.J., Kieff E. Genome-wide siRNA screen for mediators of NF-κB activation. Proc. Natl. Acad. Sci. USA. 2012;109:2467–2472. doi: 10.1073/pnas.1120542109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Niu C., Zhang J., Breslin P., Onciu M., Ma Z., Morris S.W. c-Myc is a target of RNA-binding motif protein 15 in the regulation of adult hematopoietic stem cell and megakaryocyte development. Blood. 2009;114:2087–2096. doi: 10.1182/blood-2009-01-197921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ma X., Renda M.J., Wang L., Cheng E.C., Niu C., Morris S.W., Chi A.S., Krause D.S. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol. Cell. Biol. 2007;27:3056–3064. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang L., Tran N.T., Su H., Wang R., Lu Y., Tang H., Aoyagi S., Guo A., Khodadadi-Jamayran A., Zhou D. Cross-talk between PRMT1-mediated methylation and ubiquitylation on RBM15 controls RNA splicing. eLife. 2015;4:e07938. doi: 10.7554/eLife.07938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jin S., Mi Y., Song J., Zhang P., Liu Y. PRMT1-RBM15 axis regulates megakaryocytic differentiation of human umbilical cord blood CD34+ cells. Exp. Ther. Med. 2018;15:2563–2568. doi: 10.3892/etm.2018.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Geng Y., Guan R., Hong W., Huang B., Liu P., Guo X., Hu S., Yu M., Hou B. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival. Ann. Transl. Med. 2020;8:387. doi: 10.21037/atm.2020.03.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aoyama T., Yamashita S., Tomita K. Mechanistic insights into m6A modification of U6 snRNA by human METTL16. Nucleic Acids Res. 2020;48:5157–5168. doi: 10.1093/nar/gkaa227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lence T., Paolantoni C., Worpenberg L., Roignant J.Y. Mechanistic insights into m6A RNA enzymes. Biochim. Biophys. Acta. Gene Regul. Mech. 2019;1862:222–229. doi: 10.1016/j.bbagrm.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 100.Warda A.S., Kretschmer J., Hackert P., Lenz C., Urlaub H., Höbartner C., Sloan K.E., Bohnsack M.T. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004–2014. doi: 10.15252/embr.201744940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824–835.e14. doi: 10.1016/j.cell.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Brown J.A., Kinzig C.G., DeGregorio S.J., Steitz J.A. Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. USA. 2016;113:14013–14018. doi: 10.1073/pnas.1614759113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.