Abstract
Glioma is the most common primary tumor of the central nervous system. We previously confirmed that zinc finger E-box binding homeobox (ZEB) 2 promotes the malignant progression of glioma, while microRNA-637 (miR-637) is associated with favorable prognosis in glioma. This study aimed to investigate the potential interaction between ZEB2 and miR-637 and its downstream signaling pathway in glioma. The results revealed that ZEB2 could directly bind to the E-box elements in the miR-637 promoter and promote cell proliferation, migration, and invasion via miR-637 downregulation. Subsequent screening confirmed that HMGA1 was a direct target of miR-637, while miR-637 could drive the malignant phenotype of glioma by suppressing HMGA1 both in vitro and in vivo. Furthermore, interaction between cytoplasmic HMGA1 and vimentin was observed, and vimentin inhibition could abolish increased migration and invasion induced by HMGA1 overexpression. Both HMGA1 and vimentin were associated with an unfavorable prognosis in glioma. Additionally, upregulated HMGA1 and vimentin were found in isocitrate dehydrogenase (IDH) wild-type and 1p/19q non-codeletion diffusely infiltrating glioma. In conclusion, we identified an oncogenic ZEB2/miR-637/HMGA1 signaling axis targeting vimentin that promotes both migration and invasion in glioma.
Keywords: glioma, ZEB2, miR-637, HMGA1, vimentin, migration, invasion
Graphical Abstract
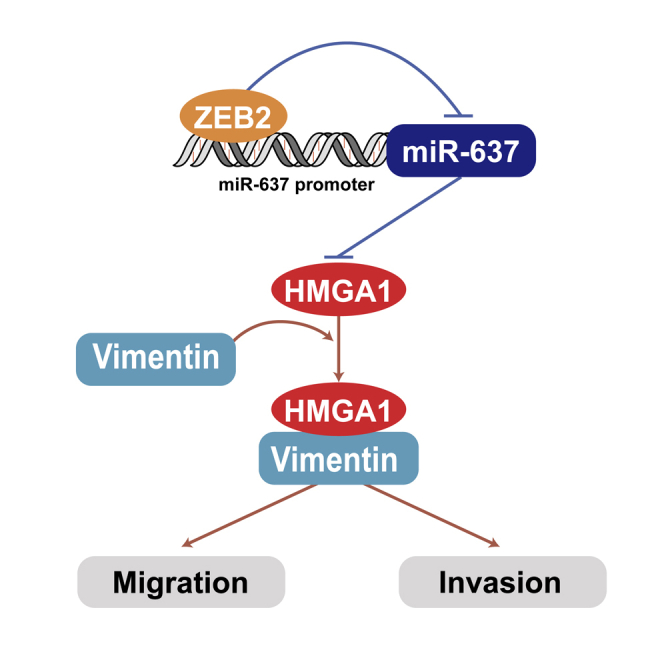
Transcription factor ZEB2 could directly bind to the E-box element in the miR-637 promoter region, leading to upregulation of HMGA1, one of the miR-637 target genes. Moreover, promoted HMGA1 could interact with vimentin and increase both migration and invasion capacity of GBM cells.
Introduction
Glioma is the most common primary brain tumor in adults. Glioblastoma (GBM) is the most malignant type of glioma, characterized by a high level of infiltration, therapeutic resistance, and poor prognosis, with a modest median survival of 14.2 months, even under radiation plus temozolomide therapy.1 Genetic changes, including isocitrate dehydrogenase (IDH) 1/2 mutation, 1p/19q codeletion, and O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation have recently been accepted as crucial molecular characteristics for prognosis prediction and subgroup stratification.2, 3, 4 Besides these genetic modifications, overexpression of oncogenic protein-coding genes and non-coding RNAs (ncRNAs)5 and dysregulated post-translational modification6 can also modulate the malignant phenotype of glioma. Thus, insights into their potential prognostic value are crucial.
Emerging evidence has indicated that ncRNAs, constituting a vast majority of the human transcriptome and previously considered as non-functional,7 could also regulate gene expression and are involved in the progression of a variety of cancers.8, 9, 10 As an important component of ncRNAs, microRNA could regulate oncogenesis and tumor progression by either blocking mRNA translation or promoting mRNA degradation.11,12 We previously confirmed that microRNA-637 (miR-637) is a favorable prognosis marker in glioma that targets the 3ʹ-untranslated region (UTR) of Akt1.13 Further targets of miR-637, such as signal transducer and activator of transcription 3 (STAT3) and SOX10,14,15 have been validated so far, but there remains a lack of knowledge regarding upstream regulation.
The zinc finger E-box binding homeobox (ZEB) family comprises two transcription factors, ZEB1 and ZEB2 (also known as SMAD-interacting protein-1 or SIP1), which both bind to E-box element CACCT(G) via zinc finger clusters and are critical regulators of cellular plasticity.16, 17, 18 In our previous study, we identified ZEB2 as a pivotal biomarker in glioma that promotes proliferation, migration, and invasion by inducing the epithelial–mesenchymal transition (EMT) process and cell cycle progression.19 A recent study showed that ZEB2 was inhibited by several miRNAs, such as miR-153,20 miR-155,21 and miR-30a,22 and these interactions significantly suppressed the EMT process. On the other hand, ZEB2 was also found to regulate miRNA expression as a repressive transcription factor. For example, there is a reciprocal interaction between ZEB2 and miR-203; while miR-203 inhibited ZEB2 through binding to its 3ʹ-UTR region, ZEB2 could directly abolish miR-203 expression by binding to an E-box element within the promoter of miR-203.23
Though miR-637 has been identified as a negative regulator of the EMT process,24 its potential association with ZEB2 remains unraveled. Herein, this study aimed to investigate the potential regulation of miR-637 by ZEB2 and extend current knowledge on the downstream signaling pathway of miR-637 in glioma.
Results
miR-637 is directly targeted by the transcriptional factor ZEB2 in glioma
Two putative ZEB2 binding sites (CACCT) were found within the promoter region of miR-637 (Figure 1A). To validate this result, we performed chromatin immunoprecipitation (ChIP) assays along with electrophoresis analysis using primers that specifically amplified these two regions. ZEB2 protein could recruit both predicted binding sites in the promoter region of miR-637 (Figures 1B and 1C). To further support these results, luciferase assays were then performed using constructs containing ZEB2 binding site 1 or 2 downstream of a constitutively active luciferase reporter gene. In accordance with the ChIP assay results, transfection with ZEB2-expressing plasmid significantly decreased luciferase expression when it was coupled to the binding site 1 or site 2 construct, while knockdown of ZEB2 with siZEB2 led to increased luciferase activity (Figure 1D). We further confirmed the ZEB2 binding site 1 via an electrophoretic mobility shift assay (EMSA) in which no band shift was detected when the nuclear extract was incubated with the wild-type (WT) ZEB2 probes, while a band shift was observed when mutant (Mut) ZEB site 1 probes were added (Figure S1).
Figure 1.
miR-637 is directly targeted by the transcriptional factor ZEB2 in glioma
(A) Schematic illustration of the two putative ZEB2-binding sites in the promoter region of miR-637. (B and C) ChIP assays along with electrophoresis analysis demonstrated amplification of both binding sites 1 and 2. (D) Relative luciferase activity of the indicated constructs was examined after the co-transfection of the ZEB2 plasmid or siZEB2 in 293T cells. (E) IHC analysis of ZEB2 and ISH analysis of miR-637 in glioma tissues were performed. (F) Expression of miR-637 was negatively correlated with that of ZEB2 in glioma tissues. Data are presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
Having confirmed that miR-637 was a direct target of ZEB2, we then examined their expression levels and correlation in clinical samples. Consistent with our previous results, a higher level of ZEB2 expression was detected in high-grade glioma (grade III and IV) compared with low-grade glioma (grade I and II), while a higher level of miR-637 expression was detected in low-grade glioma (Figure 1E). A negative correlation between the expression level of miR-637 and ZEB2 was evident (Figure 1F). Collectively, these results suggested that ZEB2 directly targeted an E-box element in the miR-637 promoter and might exert an inhibitory effect on miR-637 expression, although further evidence is needed.
ZEB2 promotes the malignant phenotype of glioma by downregulating miR-637 in vitro
ZEB2 is a critical transcription factor involved in the EMT process, and we previously confirmed that ZEB2 was associated with the malignant phenotype of glioma.19 In contrast, miR-637 was identified as a marker of favorable prognosis in glioma.13 Building on these results led to an assumption that ZEB2 could facilitate the malignant phenotype of glioma by directly suppressing miR-637. To prove this, we transfected U251 and U87 cells with short hairpin RNA (shRNA) targeting ZEB2 and found increased miR-637 levels under ZEB2 knockdown (Figure 2A).
Figure 2.
ZEB2 promotes the malignant phenotype of glioma by downregulating miR-637 in vitro
(A) Quantitative real-time PCR assays demonstrated increased miR-637 levels under siZEB2 transfection. (B) The cell viability of U251 or U87 cells transfected with a ZEB2 expression plasmid and/or miR-637 lentiviral expression particles was evaluated by the MTT assay. (C–H) The proliferative rate, migration, and invasion of U251 or U87 cells transfected with a ZEB2 expression plasmid and/or miR-637 mimics were evaluated by the EdU incorporation assay (C and D), Transwell assay (E and F), and Boyden assay (G and H), respectively. Scale bar indicates 100 μm. Data are presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
Having observed that ZEB2 suppressed miR-637 expression, we then investigated the potential role of ZEB2/miR-637 interaction in the malignant phenotype of glioma. Both the 3-(4,5-dimethylthiazol-2-yl)-2,5- diphenyltetrazolium bromide (MTT) assay (Figure 2B) and 5-ethynyl-2ʹ-deoxyuridine (EdU) incorporation assay (Figures 2C and 2D) revealed that ectopic miR-637 expression by lentiviral particles or miR-637 mimics could abolish the increased cell viability and proliferation mediated by ZEB2 overexpression in U251 and U87 cells. Similar results were observed in the case of migration and invasion. In Transwell (Figures 2E and 2F) and Boyden assays (Figures 2G and 2H), while ectopic ZEB2 expression not only changed the shape of tumor cells into a small and round shape but also promoted the migratory and invasive activity of U251 and U87 cells, concomitant miR-637 overexpression significantly attenuated migration and invasion in glioma cells compared with ZEB2 overexpression alone (Figures 2E–2H). Collectively, ZEB2 promoted the malignant phenotype of glioma through transcriptional inhibition of miR-637.
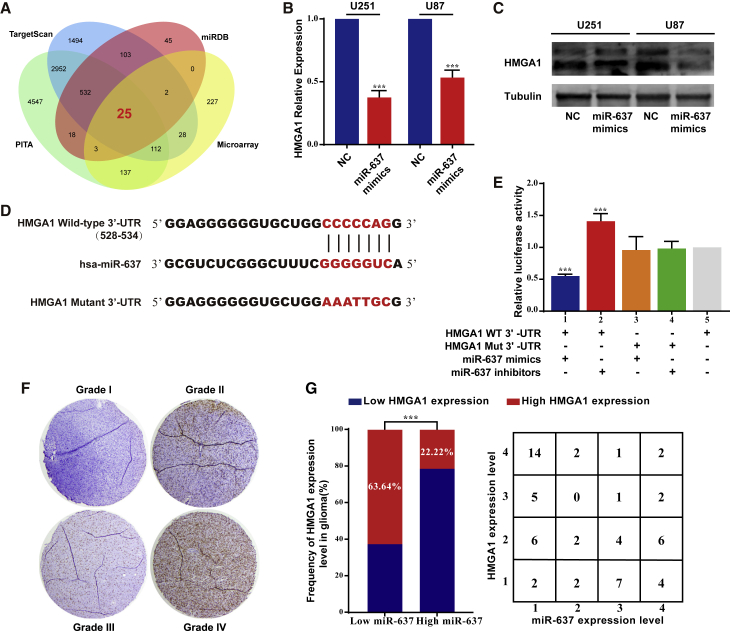
miR-637 directly targets HMGA1 in glioma
We then searched for potential target genes of miR-637 using the TargetScan, miRDB, and PITA databases to elucidate the molecular mechanism downstream of miR-637. Furthermore, we determined microarray-based gene expression signatures under ZEB2 knockdown compared with a negative control (NC), and genes with an absolute fold-change >1.5 and p <0.05 were considered as differentially expressed genes (DEGs). Since these genes might be indirect targets of ZEB2 via miR-637, the intersection between the downregulated genes under ZEB2 knockdown and the potential miR-637 target genes was examined. We found 25 genes with overlapping microarray and bioinformatic results (Figure 3A). In our previous study, we had already confirmed that ZEB2 was involved in the EMT process as well as in the cell cycle and apoptosis.19 A literature search led us to postulate that the PI3K/Akt signaling pathway could be a candidate downstream of ZEB2 due to its pleiotropic effects in glioma25, 26, 27 and because Akt1 was previously confirmed as a direct target of miR-637.13
Figure 3.
miR-637 directly targeted HMGA1 in glioma
(A) Venn diagram indicating the intersection of miR-637 target genes predicted in public databases (TargetScan, miRDB, and PICA) overlapping with microarray results. (B) Quantitative real-time PCR assay demonstrated decreased HMGA1 levels under miR-637 mimics transfection. (C) Western blot demonstrated decreased HMGA1 levels under miR-637 mimics transfection. (D) The putative miR-637-binding sites in the 3′-UTR region of HMGA1 are marked in red. (E) Relative luciferase activity was examined after the transfection of reporter vector with HMGA1 wild-type 3ʹ-UTR or mutant 3ʹ-UTR under co-transfection with miR-637 mimics or inhibitor in 293T cells. (F) IHC analysis of HMGA1 in glioma tissues was performed. (G) Expression of HMGA1 was negatively correlated with that of miR-637 in glioma tissues. Data are presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001). WT, wild type; Mut, mutant.
Therefore, we expanded our findings, focusing on HMGA1 protein, which is a crucial activator of the PI3K/Akt pathway.28 Both quantitative real-time PCR analysis (Figure 3B) and western blot (Figure 3C) revealed that ectopic miR-637 expression with miR-637 mimics significantly decreased the HMGA1 expression level. Furthermore, putative binding sites for miR-637 were predicted in the 3ʹ-UTR region of HMGA1, and a luciferase reporter vector was then constructed with the target region sequence (HMGA1 WT 3ʹ-UTR) or a Mut target region sequence (HMGA1 Mut 3ʹ-UTR) (Figure 3D). The luciferase reporter assay indicated that the co-transfection of miR-637 mimics significantly decreased the luciferase activity of the HMGA1 WT 3ʹ-UTR construct, while miR-637 inhibitor led to increased luciferase activity (Figure 3E, columns 1 and 2). A mutation of the putative binding sequence could successfully abrogate these effects (Figure 3E, columns 3 and 4).
Consistently, while a higher level of HMGA1 expression was identified in high-grade glioma (Figure 3F), a negative correlation between HMGA1 expression and miR-637 expression was found (Figure 3G). Collectively, our results indicated that miR-637 directly targeted HMGA1 and inhibited its expression.
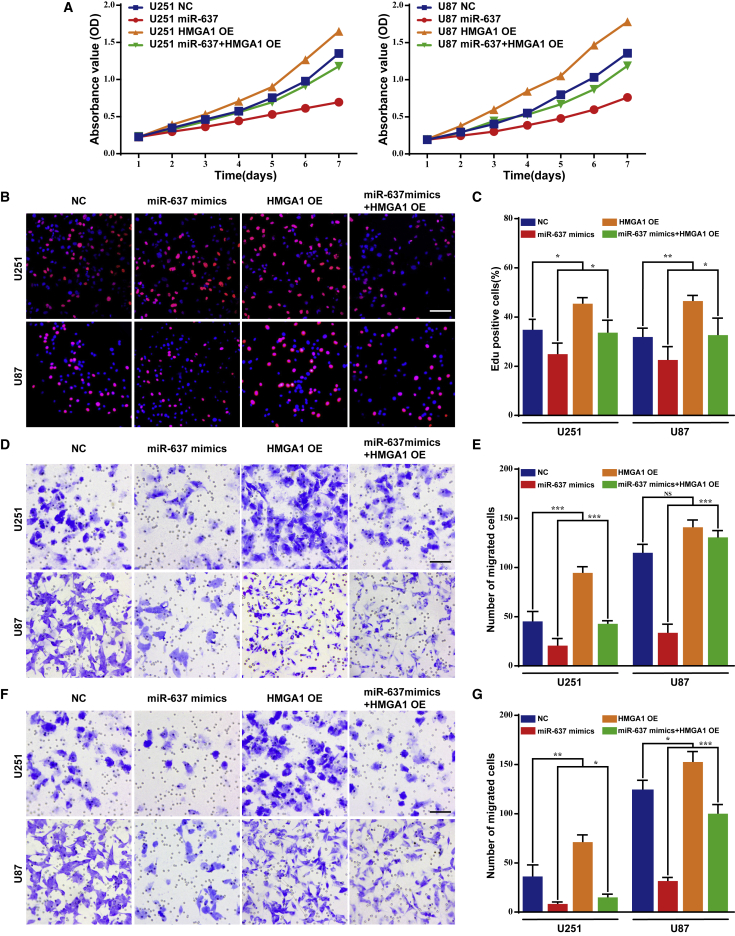
miR-637 suppresses the malignant phenotype of glioma by downregulating HMGA1 in vitro
Although HMGA1 was confirmed as a downstream target of miR-637, the effect of their interaction on the malignant phenotype of glioma remained unelucidated. Therefore, an MTT assay was first performed to evaluate their effects on cell viability, revealing that ectopic HMGA1 expression could successfully rescue the suppressed cell proliferation due to miR-637 overexpression in U251 and U87 cells (Figure 4A). Consistent with this, the EdU incorporation assay demonstrated an increased rate of cell proliferation in the rescued group compared with treatment with miR-637 mimics alone (Figures 4B and 4C). Regarding cell migration and invasion, in both U251 and U87 cells, the inhibited cell migration and invasion resulting from miR-637 overexpression were significantly rescued by ectopic HMGA1 expression (Figures 4D–4G). Briefly, these results confirmed that miR-637 suppressed the malignant phenotype of glioma, including cell proliferation, migration, and invasion, by downregulating HMGA1 in vitro.
Figure 4.
miR-637 suppresses the malignant phenotype of glioma by downregulating HMGA1 in vitro
(A) The cell viability of U251 or U87 cells transfected with miR-637 lentiviral expression particles and/or a HMGA1 expression plasmid was evaluated by the MTT assay. (B–G) The proliferative rate, migration, and invasion of U251 or U87 cells transfected with miR-637 mimics and/or a HMGA1 expression plasmid were evaluated by the EdU incorporation assay (B and C), Transwell assay (D and E), and Boyden assay (F and G), respectively. Scale bar indicates 100 μm. Data are presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
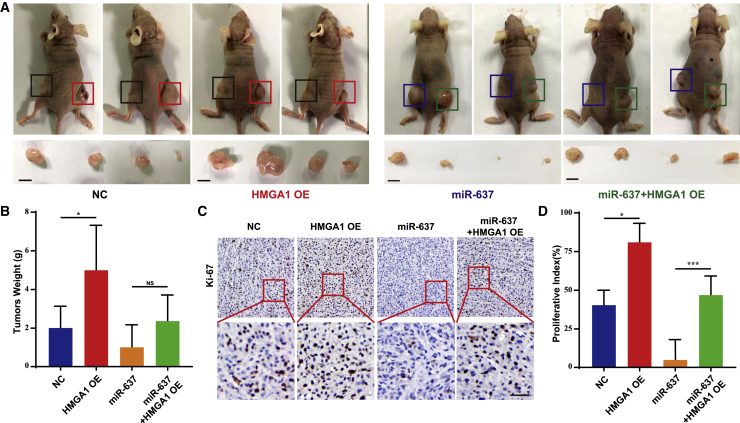
miR-637 modulates xenograft tumor growth by targeting HMGA1 in vivo
To further investigate whether HMGA1 could act as a direct target mediating the biological effects of miR-637, we established a subcutaneous xenograft model to examine the relationship between miR-637 and HGMA1 in a more biologically relevant setting. U87/NC, U87/HMGA1, U87/miR-637, and U87/miR-637/HMGA1 cells were subcutaneously implanted into nude mice, respectively. The mice were sacrificed at 28 days after tumor implantation, and the tumors were weighed. U87/HMGA1 cells formed larger tumors than U87/NC cells (Figure 5A, panels 1 and 2), while the U87/miR-637/HMGA1 group exhibited larger tumors than the U87/miR-637 group (Figure 5A, panels 3 and 4). Greater tumor weight was also evident in the U87/miR-637/HMGA1 group compared with that in the U87/miR-637 group, although this difference was not statistically significant due to small sample size (Figure 5B). We then evaluated the protein expression level of cell proliferation marker Ki-67 in these tumors. Consistently, a higher proliferative rate in the U87/HMGA1 group compared with that in the U87/NC group was confirmed, whereas a significantly higher proliferative rate was observed in the U87/miR-637/HMGA1 group compared with that in the in U87/miR-637 group (Figures 5C and 5D). Briefly, miR-637 suppressed xenograft tumor growth by targeting HMGA1 and then inhibiting the rate of cell proliferation in vivo.
Figure 5.
miR-637 modulates xenograft tumor growth by targeting HMGA1 in vivo
(A) Images of xenograft tumor models of mice injected with indicated U87 cells are shown. In the first group of four mice, U87/NC cells were injected in the left flank, and U87/HMGA1 cells were injected in the right flank; in the second group of four mice, U87/miR-637 cells were injected in the left flank, and U87/miR-637/HMGA1 cells were injected in the right flank. Scale bar indicates 1 cm. (B) Tumor weight was measured at the 28th day after inoculation in each group. (C and D) IHC analysis and quantification of Ki-67 expression in xenograft tumors from mice injected with indicated U87 cells were performed. Scale bar indicates 100 μm. Data are presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001). NC, negative control.
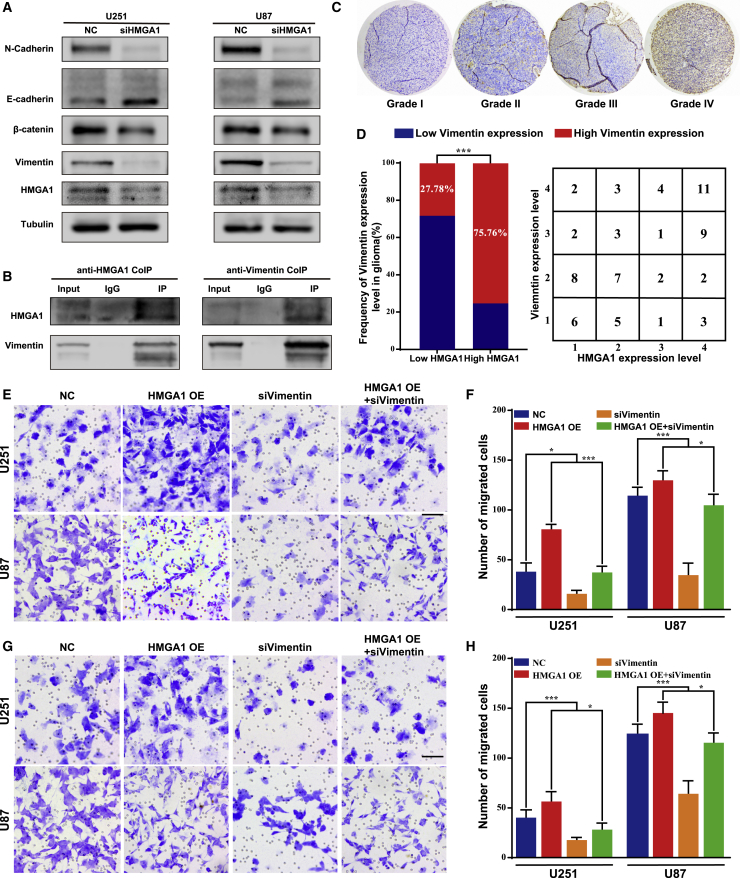
HMGA1 promotes glioma migration and invasion through vimentin in vitro
HMGA1 could regulate GBM cell stemness by modifying chromatin architecture at the promoter of SOX2, one of the major regulators of cell stemness.29 It was also reported that HMGA1 promoted tumor cell stemness and the EMT process in breast cancer.30 Consistently, we found significantly decreased levels of N-cadherin, vimentin, and β-catenin following HGMA1 knockdown in both U251 and U87 cells, while an increased level of E-cadherin expression was observed (Figure 6A). Interestingly, the anti-HGMA1 coimmunoprecipitation (coIP) assay demonstrated that vimentin, an intermediate filament expressed in immature astrocytes, could directly bind to HMGA1 (Figure 6B, panel 1). Reciprocal immunoprecipitation with anti-vimentin antibody similarly indicated this interaction (Figure 6B, panel 2). In glioma tissue samples, the level of vimentin expression was higher in high-grade glioma (Figure 6C), whereas a positive correlation was found between the expression levels of vimentin and HMGA1 (Figure 6D). A correlation analysis based on the CGGA as well as the TCGA glioma cohorts further supported our result (Figure S2). Vimentin is a critical member of the family of intermediate filaments and is involved in cell contraction and migration.31 Thus, we then focused on examining glioma cell migration and invasion. While HMGA1 significantly promoted both migration and invasion in vitro, concomitant inhibition of vimentin could successfully abolish this effect (Figures 6E–6H). Collectively, our results indicated that HMGA1 could interact with vimentin and facilitate both migration and invasion through vimentin in glioma.
Figure 6.
HMGA1 promotes glioma migration and invasion through vimentin in vitro
(A) The expression of crucial EMT-related proteins was examined by western blot assays. (B) The interaction between HMGA1 and vimentin was examined by coIP assay. (C) IHC analysis of vimentin in glioma tissues was performed. (D) Expression of vimentin was positively correlated with that of miR-637 in glioma tissues. (E–H) Migration and invasion of U251 or U87 cells transfected with an HMGA1 expression plasmid and/or siRNA targeting vimentin were evaluated by the Transwell assay (E and F) and Boyden assay (G and H), respectively. Scale bar indicates 100 μm. Data are presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001). NC, negative control.
HMGA1 and vimentin are associated with unfavorable prognosis in glioma
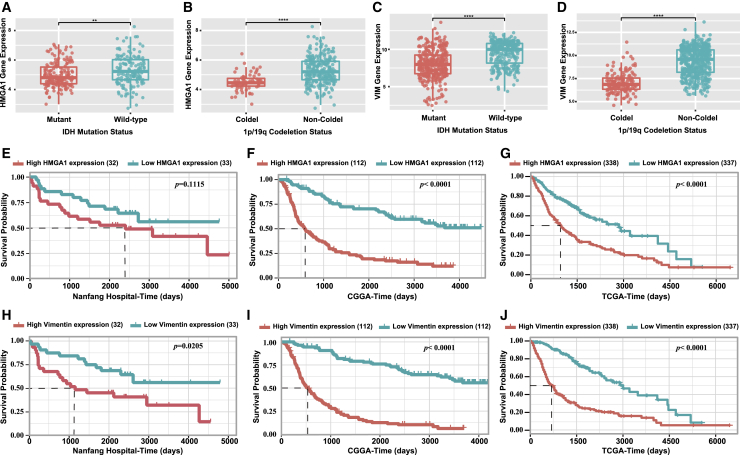
This oncogenic ZEB2/miR-637/HMGA1/vimentin signaling pathway was further confirmed with a cohort of 69 prospectively collected primary glioma tissues from our department. Since vimentin is an intermediate filament expressed in astrocytic progenitors, a high level of vimentin expression was observed in astrocytoma samples. There was no significant association between the expression level of vimentin or HMGA1 and the patient’s age or gender (Table 1). Notably, higher expression levels of ZEB2, HGMA1, and vimentin were observed in high-grade glioma (grade III and IV) compared with low-grade glioma (grade I and II), while a lower expression level of miR-637 was observed in high-grade glioma (Table 1). Interestingly, in the CGGA glioma cohort, we found that HMGA1 and vimentin were both markedly upregulated in IDH WT and 1p/19q non-codeletion glioma (Figures 7A–7D), which is considered a diffusely infiltrating glioma according to the 2016 World Health Organization (WHO) central nervous system (CNS) tumor classification.
Table 1.
Correlation between the clinicopathological factors and expression of ZEB2/miR-637/HMGA1/vimentin axis
| Characteristics | n | ZEB2 expression | miR-637 expression | HMGA1 expression | Vimentin expression | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | p | High | Low | p | High | Low | p | High | Low | p | ||
| Gender | |||||||||||||
| Male | 40 | 17 | 22 | 0.286 |
14 | 21 | 0.12 |
20 | 20 | 0.45 |
19 | 21 | 0.35 |
| Female | 29 | 14 | 14 | 13 | 12 | 13 | 16 | 16 | 13 | ||||
| Age | |||||||||||||
| >50 | 18 | 9 | 9 | 0.336 |
5 | 9 | 0.163 |
10 | 8 | 0.24 |
11 | 7 | 0.11 |
| <50 | 51 | 22 | 27 | 22 | 24 | 23 | 28 | 24 | 27 | ||||
| Histological type | |||||||||||||
| AT | 46 | 21 | 23 | 0.279 |
18 | 21 | 0.213 |
22 | 24 | 0.10 |
24 | 22 | 0.03 |
| OT | 15 | 6 | 9 | 5 | 8 | 5 | 10 | 4 | 11 | ||||
| Other | 8 | 4 | 4 | 4 | 4 | 6 | 2 | 7 | 1 | ||||
| WHO grade | |||||||||||||
| I + II | 34 | 4 | 29 | <0.001 | 24 | 8 | <0.001 | 8 | 26 | <0.001 | 8 | 26 | <0.001 |
| III + IV | 35 | 27 | 7 | 3 | 25 | 25 | 10 | 27 | 8 | ||||
Abbreviations: AT, astrocytic tumor; OT, oligodendroglial tumor; WHO, World Health Organization.
Figure 7.
HMGA1 and vimentin are associated with unfavorable prognosis in glioma
(A) Expression level of HMGA1 in glioma with different IDH status in CGGA cohort was examined. (B) Expression level of HMGA1 in glioma with different 1p/19q status in CGGA cohort was examined. (C) Expression level of vimentin in glioma with different IDH status in CGGA cohort was examined. (D) Expression level of vimentin in glioma with different 1p/19q status in CGGA cohort was examined. (E–G) HMGA1 was associated with an unfavorable prognosis in Nanfang Hospital, CGGA, and TCGA cohorts. (H–J) Vimentin was associated with an unfavorable prognosis in Nanfang Hospital, CGGA, and TCGA cohorts.
We have already confirmed that ZEB2 is a potential marker for poor prognosis in glioma,19 whereas miR-637 is correlated with favorable prognosis in glioma.13 Furthermore, a Kaplan-Meier analysis of overall survival in our cohort revealed that glioma patients with higher HMGA1 or vimentin expression exhibited poorer prognosis (Figures 7E and 7H), although the differences between the high HMGA1 expression group and low HMGA1 expression group were not statistically significant. We further validated these results on a larger scale with the CGGA and TCGA databases, and similar results were found (Figures 7F and 7I and 7G and 7J, respectively). Taken together, both HGMA1 and vimentin were associated with an aggressive growth pattern and poor prognosis in glioma, while the oncogenic ZEB2/miR-637/HMGA1/vimentin signaling pathway was evident in the glioma samples.
Discussion
To date, the engagement and regulation of ncRNAs remain inadequately characterized, including the potential interaction between protein-coding genes and ncRNAs as well as interactions among ncRNAs. A reciprocal inhibition between ZEB2 and miR-203 that regulates cancer stem cell growth was found in breast cancer,32 while ZEB2 was found to be downregulated under transforming growth factor-β-induced miR-192 overexpression, leading to p53 upregulation.33 Our previous study revealed that ZEB2 was involved in the malignant progression of glioma, while miR-637 was a favorable prognosis marker in glioma, but the relationship between ZEB2 and miR-637 was unelucidated. In this study, our findings built upon the current knowledge of miR-637 regulation by revealing that ZEB2 could directly bind to the promoter of miR-637 and repress its expression, which further led to the upregulation of HMGA1, a novel downstream target of miR-637. Targeting the EMT process, HMGA1 could further interact with vimentin and promote glioma migration and invasion.
miR-637 plays a pivotal role in a variety of cancers. It functions as a tumor suppressor via targeting and downregulating NUPR1 expression, which was shown to lead to the inhibition of proliferation, migration, and invasion in colorectal cancer cells.34 Moreover, supporting our previous results, miR-637 was found to target Akt1 and hamper tumorigenesis in both thyroid carcinoma and pancreatic ductal adenocarcinoma.35,36 miR-637 could also inhibit Akt phosphorylation and promote melanoma progression.37 Remarkably, although several circular RNAs or long non-coding RNAs (lncRNAs) were found to regulate miR-637 through the mechanism of competitive endogenous RNA, few studies have investigated the crosstalk between transcription factors and miR-637. Our results indicated that the critical EMT regulator ZEB2 could directly bind to the miR-637 promoter and suppress its expression, expanding the current understanding of miRNA regulation. Furthermore, HMGA1 was confirmed as another downstream target of miR-637, forming a complete regulatory network around miR-637.
HMGA1 is a member of the HMGA protein family, which does not possess intrinsic transcriptional activity but alters the chromatin structure by direct recognition of the A/T-rich sequences in the promoter and enhancer regions of multiple genes.38 HMGA1 overexpression was found in recurrent GBM patients,39 and knockdown of HMGA1 could suppress the stemness of GBM stem cells and sensitizes them to temozolomide.40 HMGA1 was also found to function as a critical regulator of GBM cell stemness by modifying chromatin architecture at the promoter of SOX2, and this effect was downstream of miR-296-5p.29 Moreover, the downregulation of lncRNA HIF1A-AS2 could impair the expression of HMGA1 and lead to inhibited growth of mesenchymal GBM stem-like cells.41 In accordance with a previous report,42 we found that HMGA1 was a key regulator of the EMT process and confirmed the interaction between HMGA1 and vimentin. Although HMGA1 is mostly reported as located in the nucleus, it was suggested that HMGA1 could be secreted through cytoplasmic translocation, and its cytoplasmic translocation could predict the aggressiveness of breast cancer.43 Supporting this result, our results further revealed that nuclear protein HMGA1 could be translocated into the cytoplasm and promoted the migration and invasion of glioma through interaction with vimentin, although further study on the precise mechanism of cytoplasmic translocation of HMGA1 is necessary.
Molecular features, such as IDH1/2 mutation and 1p/19q codeletion, were integrated with the histological appearance for a more accurate classification in the 2016 WHO CNS tumor classification.44 For example, diffuse astrocytoma and diffuse oligodendroglioma are grouped together based not only on their diffusely infiltrative growth pattern but also on their lack of IDH1/2 mutation.42 In IDH1 WT grade II glioma, intense expression of vimentin, an intermediate filament expressed in immature astrocytes, was observed, indicating the potential involvement of neural progenitors in IDH1 WT glioma.45 Oligodendroglioma characterized by whole-arm loss of 1p and 19q (1p/19q codeletion) was associated with better prognosis, although its association with HMGA1 and vimentin is unclear. Our subgroup analysis revealed that HMGA1 and vimentin were overexpressed in astrocytoma with intact 1p/19q loci, indicating that further insights into gliomagenesis considering different origins are needed.
In conclusion, we are the first to report that miR-637 was directly inhibited by transcription factor ZEB2, while miR-637 could further suppress HMGA1, which interacts with downstream vimentin. Based on these findings, an oncogenic ZEB2/miR-637/HMGA1 signaling axis targeting vimentin was identified, which drives both migration and invasion in glioma. Our study may shed new light on the integrated modulation of the malignant phenotype of glioma involving a transcription factor, microRNA, and a chromatin remodeling factor and facilitate a better understanding of glioma progression.
Materials and methods
Cell culture and clinical tissue collection
The human glioma cell lines U251 and U87 and human embryo kidney 293T cells were purchased from the Chinese Academy of Sciences (Shanghai, People’s Republic of China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, UT, USA) supplemented with 10% fetal calf serum (Hyclone) at 37°C in a humidified atmosphere of 5% CO2. We obtained 69 paraffin-embedded glioma samples from the Department of Neurosurgery, Nanfang Hospital, Southern Medical University, Guangzhou, People’s Republic of China. These glioma samples were from 40 male and 29 female patients aged 3–78 years (median age, 40 years). Of them, 65 were regularly and closely followed up, two lacked ZEB expression data, and nine lacked miR-637 expression data due to breaking of the sectioned sample during the immunohistochemistry (IHC) or in situ hybridization (ISH) assays. Prior consent was obtained from all of the patients for the use of their clinical materials for research purposes, and approval was obtained from the Ethics Committees of Nanfang Hospital. All specimens had a confirmed pathological diagnosis and were classified according to the WHO criteria.
ChIP assay
Protein-DNA complexes were immunoprecipitated from U251 and U87 cells using a Chromatin Immunoprecipitation Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s protocol with anti-ZEB2 (Bethyl Laboratories, Montgomery, TX, USA) and normal mouse IgG (Millipore) polyclonal antibodies, the latter serving as a control for nonspecific DNA binding. The precipitated DNA was then subjected to PCR with specific primers to amplify the putative ZEB2 binding region analyzed via electrophoresis (Table S1).
Quantitative real-time PCR, PCR, and gel electrophoresis analysis
RNA was extracted from glioma cells using Trizol reagent (Takara Bio, Shiga, Japan). For quantitative real-time PCR, cDNA was amplified using SYBR Green PCR Master Mix (Toyobo, Osaka, Japan) or miRNA PrimeScript RT Enzyme Mix (Takara Bio) and analyzed following gel electrophoresis. The relative expression level of mRNA or miRNA was normalized to β-actin or U6, respectively. The specific primers used for miR-637, HMGA1, and β-actin are shown in Table S1.
Luciferase reporter assay
To confirm that miR-637 was a direct target of ZEB2, fragments of miR-637 promoter containing two different ZEB2 binding sites were cloned into pGL3-Basic vectors. These vectors were co-transfected with ZEB2 overexpression plasmid or small interfering RNA (siRNA) targeting ZEB2 into 293T cells in 48-well plates and then harvested for luciferase assay 48 h after transfection. The luciferase activity was determined using a Dual-Luciferase Reporter Assay System (Promega Corp, Fitchburg, WI, USA) according to the manufacturer’s protocol.
To confirm that HMGA1 was a direct target of miR-637, a fragment of HMGA1 3ʹ-UTR containing the potential miR-637 binding sites was cloned into psiCHECK-2 vectors (named WT). Site-directed mutagenesis of the miR-637 binding site in the HMGA1 3ʹ-UTR region was then performed using the GeneTailor Site-Directed Mutagenesis System (Invitrogen, Waltham, MA, USA; named Mut). Subsequently, WT or Mut vectors were co-transfected with miR-637 mimics or inhibitors into 293T cells in 48-well plates and then harvested for luciferase assay 48 h after transfection. Luciferase activity was determined, as mentioned above.
EMSA
We investigated ZEB2 binding activity on the promoter region of miR-637 using an EMSA Kit (Bioscience, Shanghai, People’s Republic of China) according to the manufacturer’s protocol. The samples with nuclear extracts from ZEB2-overexpressing glioma cells were used as positive controls, while samples without nuclear extracts were used as NCs. For competition experiments, a specific oligonucleotide competitor (unlabeled WT ZEB2 site 1 probe, site 1 WT; unlabeled Mut ZEB2 site 1 probe, site 1 Mut) was added to the binding mixture 10 min before the addition of the labeled probe. Following electrophoresis, the bands were visualized on a BioSens Gel Imaging System (BIOTOP, Shanghai, People’s Republic of China).
Microarray analysis
The total RNAs of U87 cells stably transfected with shRNA targeting ZEB2 and NC particles were isolated using Trizol reagent (Invitrogen) according to the manufacturer’s instructions, respectively. After quality-control checks, whole-genome expression microarray analysis (Affymetrix, Santa Clara, CA, USA) was performed by GeneChem (Shanghai, People’s Republic of China). Genes in the ZEB2-knockdown group with an absolute fold-change >1.5 and a p value <0.05 compared with those in the NC group were considered as DEGs.
IHC and ISH assays
Paraffin sections prepared from clinical samples and xenograft models were used for IHC or ISH assays to detect the expression levels of ZEB2, miR-637, HMGA1, vimentin, and Ki-67. For IHC staining, the indirect streptavidin-peroxidase method was used, as previously described.13 Following IHC staining, the tissue sections were examined and scored separately by two pathologists. The antibodies used were rabbit anti-ZEB2 (Cat. No. A302-473A, 1:500; Bethyl Laboratories), anti-HMGA1 (Cat. No. EPR7839, 1:200; Abcam, Cambridge, MA, USA), anti-vimentin (Cat. No. 10366-1-AP, 1:30; Proteintech, Rosemont, IL, USA), and anti-Ki67 (Cat. No. Ab16667, 1:100; Abcam). For the ISH assays, the tissue sections were hybridized with digoxigenin (DIG)-labeled miRCURY LNA probes (Bioscience) and were then incubated with alkaline-phosphatase-conjugated anti-DIG Fab fragments. Finally, the sections were stained with BM purple alkaline phosphatase substrate (Roche, Switzerland) according to the manufacturer’s instructions.
Lentivirus production and transfection
Lentiviral particles carrying hsa-miR-637 precursor vector, an expression construct for ZEB2 or HMGA1, or a counterpart NC sequence were constructed by GeneChem. U251 and U87 cells were then transfected with the indicated lentiviral vector, and polyclonal cells with green fluorescent protein signals were selected for further experiments using fluorescence-activated cell sorting.
Transfection with siRNAs, microRNA mimics/inhibitors, or plasmids
siRNA targeting ZEB2, HMGA1 or vimentin, miR-637 mimics/inhibitors, or NCs was designed and synthesized by RiboBio (Guangzhou, People’s Republic of China). An expression plasmid for ZEB2 and HMGA1 was designed and synthesized by Vigenebio (Jinan, People’s Republic of China). The efficiency was validated prior to the experiments by western blot or quantitative real-time PCR. Twelve hours prior to transfection, U87 or U251 cells were plated into 6-well or 96-well plates (Wuxi NEST Biotechnology, Jiangsu, People’s Republic of China) at 30%–50% confluence. During transfection, Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific, MA, USA) was used to facilitate the siRNA transfection into cells according to the manufacturer’s protocol. The cells were collected after 48–72 h for further experiments. The specific sequences are shown in Table S2.
Cell viability assay
Cell proliferation was analyzed using MTT assays. Briefly, cells were seeded into 96-well plates at a density of 1,000–1,500 cells/well and incubated for 1, 2, or 3 days. Approximately 20 μL of MTT (5 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) was added into each well and incubated at 37°C in a 5% CO2 atmosphere for 4 h. At the end of the incubation, the supernatants were removed, and 150 μL of dimethyl sulfoxide (Sigma-Aldrich) was added to each well. The absorbance value (optical density) of each well was measured at 490 nm at the same time for the following 7 days. For each experimental condition, five wells were used for replicates.
EdU incorporation assay
Proliferating U251 and U87 cells were examined using the Cell-Light EdU Apollo 488 In Vitro Imaging Kit (RiboBio) according to the manufacturer’s protocol. Briefly, the cells were incubated with 10 μM of EdU for 2 h before fixation with 4% paraformaldehyde, permeabilization by 0.3% Triton X-100, and EdU staining. The nuclei were stained with 5 μg/mL DAPI for 10 min. The number of EdU-positive cells was counted under a fluorescent microscope in five random fields (200× magnification). All assays were independently performed in triplicate.
In vitro migration and invasion assays
For the cell migration assay, 1 × 104 cells in 100 μL DMEM medium without fetal bovine serum (FBS) were seeded in the upper chamber of the Transwell apparatus (Corning Costar, Corning, NY, USA), and 500 μL of DMEM with 10% FBS was added to the lower chamber as a chemoattractant. Following incubation for 6 h at 37°C in a 5% CO2 atmosphere, the insert was washed with phosphate-buffered saline, and cells on the top surface of the insert were removed with a cotton swab. Cells adhering to the lower surface were fixed with methanol, stained with crystal violet solution, and counted under a microscope in five predetermined fields (200× magnification). All assays were independently repeated at least three times. The procedure for the Boyden assay was similar to the Transwell assay, except that the Transwell apparatus was first precoated with 24 μg/μL of Matrigel (R&D Systems, Minneapolis, MN, USA), and the cells were incubated for 12 h at 37°C in a 5% CO2 atmosphere. Cells adhering to the lower surface of the insert were counted in the same manner as for the cell migration assay.
Western blot analysis
Western blot was carried out as previously described19 with mouse polyclonal ZEB2 antibody (1:200; Santa Cruz Biotechnology, Dallas, TX, USA) and rabbit polyclonal HMGA1 (1:500; Abcam), E-cadherin, N-cadherin, and vimentin antibodies (1:1,000; Cell Signaling Technology, Danvers, MA, USA) as well as β-catenin antibodies (1:1,000; Abcam). Mouse monoclonal β-actin antibody (1:1,000; CoWin Biosciences, Beijing, People’s Republic of China) was used for normalization. A horseradish peroxidase (HRP)-conjugated anti-rabbit or anti-mouse IgG antibody was used as the secondary antibody (1:2,000; CoWin Biosciences). Signals were detected using enhanced chemiluminescence reagents (FdBio Science, Shenzhen, People’s Republic of China).
Subcutaneous xenograft model
We subcutaneously injected 1 × 106 U87 cells transfected with HMGA1 or/and miR-637 expressing lentiviral construct or the control lentiviral empty vector (mock; n = 4 per group) in 100 μL of DMEM without FBS into the dorsal flank areas of 6-week-old male BALB/c nude mice. The mice were sacrificed after 28 days, and the tumor tissues were excised and weighed. All mice were maintained in a barrier facility in racks of cages supplied with High efficiency particulate air (HEPA)-filtered air and fed an autoclaved laboratory rodent diet. All animal studies were conducted in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Animals under assurance number A3873-1.
Bioinformatic analysis
Sequencing data and clinical data from the Chinese Glioma Genome Atlas (CGGA) and The Cancer Genome Atlas (TCGA) cohorts were retrieved from the CGGA database (http://www.cgga.org.cn) and the GEPIA database (http://gepia.cancer-pku.cn/index.html), respectively. The gene expression levels of different samples were measured in terms of fragments per kilobase of transcript per million in the CGGA cohort or transcript per million fragments in the TCGA cohort. Expression and survival analyses were subsequently performed. Putative mi-637 targets were predicted with three public databases: TargetScan 7.2 (http://www.targetscan.org/vert_72), miRDB (http://mirdb.org), and PITA (http://genie.weizmann.ac.il/pubs/mir07/), which were then intersected with the microarray-based gene signatures to confirm potential targets of miR-637.
Statistical analysis
All quantified data represented an average of at least triplicate samples. SPSS 22.0 (IBM, Armonk, NY, USA) and GraphPad Prism 8.0 (GraphPad Software, San Diego, CA, USA) software were used for statistical analysis. Data are presented as mean ± standard deviation. Two-way analysis of variance or two-tailed Student’s t test was used for comparisons between groups. Chi-square or Fisher’s test was used to identify differences between categorical variables. Differences were considered statistically significant at p values <0.05. All statistical tests were two-sided. Single, double, and triple asterisks in the figures indicate statistical significance (∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001).
Acknowledgments
This study was supported by the National Nature Science Fund of China (nos. 81702473 and 81872064); the Natural Science Fund of Guangdong Province, China (no. 2020A1515010122); the President’s Fund of Nanfang Hospital (no. 2016C019); the Science and Technology Program of Guangzhou, China (no. 201607010350); and the Natural Science Fund of Tibet Autonomous Region (no. XZ2017ZR-ZYZ27). The funders had no role in study design, data collection, data analysis, decision to publish, or preparation of the manuscript.
Author contributions
Y.S. designed and supervised the study; Y.Z., T.Q., and J.L. performed most of experiments and statistical analysis; A.X., C.X., and X.Z. performed manuscript writing; J.L., S.D., and Z.Z. performed ChIP assay, luciferase reporter assay, and EMSA assay; H.L. and X.Z. performed language editing. All authors have read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.12.029.
Supplemental information
References
- 1.Johnson D.R., O’Neill B.P. Glioblastoma survival in the United States before and during the temozolomide era. J. Neurooncol. 2012;107:359–364. doi: 10.1007/s11060-011-0749-4. [DOI] [PubMed] [Google Scholar]
- 2.Brito C., Azevedo A., Esteves S., Marques A.R., Martins C., Costa I., Mafra M., Bravo Marques J.M., Roque L., Pojo M. Clinical insights gained by refining the 2016 WHO classification of diffuse gliomas with: EGFR amplification, TERT mutations, PTEN deletion and MGMT methylation. BMC Cancer. 2019;19:968. doi: 10.1186/s12885-019-6177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bund C., Guergova-Kuras M., Cicek A.E., Moussallieh F.M., Dali-Youcef N., Piotto M., Schneider P., Heller R., Entz-Werle N., Lhermitte B. An integrated genomic and metabolomic approach for defining survival time in adult oligodendrogliomas patients. Metabolomics. 2019;15:69. doi: 10.1007/s11306-019-1522-5. [DOI] [PubMed] [Google Scholar]
- 4.Harat M., Blok M., Harat A., Soszyńska K. The impact of adjuvant radiotherapy on molecular prognostic markers in gliomas. OncoTargets Ther. 2019;12:2215–2224. doi: 10.2147/OTT.S200818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reon B.J., Anaya J., Zhang Y., Mandell J., Purow B., Abounader R., Dutta A. Expression of lncRNAs in Low-Grade Gliomas and Glioblastoma Multiforme: An In Silico Analysis. PLoS Med. 2016;13:e1002192. doi: 10.1371/journal.pmed.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yin J., Park G., Lee J.E., Choi E.Y., Park J.Y., Kim T.H., Park N., Jin X., Jung J.E., Shin D. DEAD-box RNA helicase DDX23 modulates glioma malignancy via elevating miR-21 biogenesis. Brain. 2015;138:2553–2570. doi: 10.1093/brain/awv167. [DOI] [PubMed] [Google Scholar]
- 7.Kent O.A., Steenbergen C., Das S. In Vivo Nanovector Delivery of a Heart-specific MicroRNA-sponge. J. Vis. Exp. 2018;136:57845. doi: 10.3791/57845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma S., Pavlasova G., Seda V., Cerna K., Vojackova E., Filip D., Ondrisova L., Sandova V., Kostalova L., Zeni P.F. 2020. miR-29 Modulates CD40 Signaling in Chronic Lymphocytic Leukemia by Targeting TRAF4: an Axis Affected by BCR inhibitors. Blood Published online November 10, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tristán-Ramos P., Rubio-Roldan A., Peris G., Sánchez L., Amador-Cubero S., Viollet S., Cristofari G., Heras S.R. The tumor suppressor microRNA let-7 inhibits human LINE-1 retrotransposition. Nat. Commun. 2020;11:5712. doi: 10.1038/s41467-020-19430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wei Y., Lu C., Zhou P., Zhao L., Lyu X., Yin J., Shi Z., You Y.P. EIF4A3-induced circular RNA ASAP1(circASAP1) promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1/2 signaling. Neuro. Oncol. 2020 doi: 10.1093/neuonc/noaa214. Published online September 14, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding L., Lan Z., Xiong X., Ao H., Feng Y., Gu H., Yu M., Cui Q. The Dual Role of MicroRNAs in Colorectal Cancer Progression. Int. J. Mol. Sci. 2018;19:2791. doi: 10.3390/ijms19092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y. The novel regulatory role of lncRNA-miRNA-mRNA axis in cardiovascular diseases. J. Cell. Mol. Med. 2018;22:5768–5775. doi: 10.1111/jcmm.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Que T., Song Y., Liu Z., Zheng S., Long H., Li Z., Liu Y., Wang G., Liu Y., Zhou J. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34:4952–4963. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 14.Huang Z., Yuan C., Gu H., Cheng X., Zhou K., Xu J., Yin X., Xia J. Circular RNA circHIPK3 Promotes Cell Metastasis through miR-637/STAT3 Axis in Osteosarcoma. BioMed Res. Int. 2020;2020:2727060. doi: 10.1155/2020/2727060. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Jin C., Zhao J., Zhang Z.P., Wu M., Li J., Liu B., Liao X.B., Liao Y.X., Liu J.P. CircRNA EPHB4 modulates stem properties and proliferation of gliomas via sponging miR-637 and up-regulating SOX10. Mol. Oncol. 2020 doi: 10.1002/1878-0261.12830. Published online October 21, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Hill L., Browne G., Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int. J. Cancer. 2013;132:745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 17.Drápela S., Bouchal J., Jolly M.K., Culig Z., Souček K. ZEB1: A Critical Regulator of Cell Plasticity, DNA Damage Response, and Therapy Resistance. Front. Mol. Biosci. 2020;7:36. doi: 10.3389/fmolb.2020.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam E.H., Lee Y., Park Y.K., Lee J.W., Kim S. ZEB2 upregulates integrin α5 expression through cooperation with Sp1 to induce invasion during epithelial-mesenchymal transition of human cancer cells. Carcinogenesis. 2012;33:563–571. doi: 10.1093/carcin/bgs005. [DOI] [PubMed] [Google Scholar]
- 19.Qi S., Song Y., Peng Y., Wang H., Long H., Yu X., Li Z., Fang L., Wu A., Luo W. ZEB2 mediates multiple pathways regulating cell proliferation, migration, invasion, and apoptosis in glioma. PLoS ONE. 2012;7:e38842. doi: 10.1371/journal.pone.0038842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J., Xie M., Shi Y., Luo B., Gong G., Li J., Wang J., Zhao W., Zi Y., Wu X., Wen J. MicroRNA-153 functions as a tumor suppressor by targeting SET7 and ZEB2 in ovarian cancer cells. Oncol. Rep. 2015;34:111–120. doi: 10.3892/or.2015.3952. [DOI] [PubMed] [Google Scholar]
- 21.Brown C.Y., Dayan S., Wong S.W., Kaczmarek A., Hope C.M., Pederson S.M., Arnet V., Goodall G.J., Russell D., Sadlon T.J., Barry S.C. FOXP3 and miR-155 cooperate to control the invasive potential of human breast cancer cells by down regulating ZEB2 independently of ZEB1. Oncotarget. 2018;9:27708–27727. doi: 10.18632/oncotarget.25523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.di Gennaro A., Damiano V., Brisotto G., Armellin M., Perin T., Zucchetto A., Guardascione M., Spaink H.P., Doglioni C., Snaar-Jagalska B.E. A p53/miR-30a/ZEB2 axis controls triple negative breast cancer aggressiveness. Cell Death Differ. 2018;25:2165–2180. doi: 10.1038/s41418-018-0103-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan X., Fu Z., Gao L., Zhou J., Deng X., Luo X., Fang W., Luo R. Direct interaction between miR-203 and ZEB2 suppresses epithelial-mesenchymal transition signaling and reduces lung adenocarcinoma chemoresistance. Acta Biochim. Biophys. Sin. (Shanghai) 2016;48:1042–1049. doi: 10.1093/abbs/gmw099. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q., Wang G., Xu L., Yao Z., Song L. Long non-coding RNA LINC00473 promotes glioma cells proliferation and invasion by impairing miR-637/CDK6 axis. Artif. Cells Nanomed. Biotechnol. 2019;47:3896–3903. doi: 10.1080/21691401.2019.1671431. [DOI] [PubMed] [Google Scholar]
- 25.Yu M., Xue Y., Zheng J., Liu X., Yu H., Liu L., Li Z., Liu Y. Linc00152 promotes malignant progression of glioma stem cells by regulating miR-103a-3p/FEZF1/CDC25A pathway. Mol. Cancer. 2017;16:110. doi: 10.1186/s12943-017-0677-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Koul D., Shen R., Kim Y.W., Kondo Y., Lu Y., Bankson J., Ronen S.M., Kirkpatrick D.L., Powis G., Yung W.K. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro-oncol. 2010;12:559–569. doi: 10.1093/neuonc/nop058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaushik N.K., Kaushik N., Yoo K.C., Uddin N., Kim J.S., Lee S.J., Choi E.H. Low doses of PEG-coated gold nanoparticles sensitize solid tumors to cold plasma by blocking the PI3K/AKT-driven signaling axis to suppress cellular transformation by inhibiting growth and EMT. Biomaterials. 2016;87:118–130. doi: 10.1016/j.biomaterials.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Wei F., Zhang T., Deng S.C., Wei J.C., Yang P., Wang Q., Chen Z.P., Li W.L., Chen H.C., Hu H., Cao J. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 2019;450:1–13. doi: 10.1016/j.canlet.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Bertoni H., Lal B., Michelson N., Guerrero-Cázares H., Quiñones-Hinojosa A., Li Y., Laterra J. Epigenetic modulation of a miR-296-5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells. Oncogene. 2016;35:4903–4913. doi: 10.1038/onc.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pegoraro S., Ros G., Piazza S., Sommaggio R., Ciani Y., Rosato A., Sgarra R., Del Sal G., Manfioletti G. HMGA1 promotes metastatic processes in basal-like breast cancer regulating EMT and stemness. Oncotarget. 2013;4:1293–1308. doi: 10.18632/oncotarget.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M., Zhang B., Sun B., Wang X., Ban X., Sun T., Liu Z., Zhao X. A novel function for vimentin: the potential biomarker for predicting melanoma hematogenous metastasis. J. Exp. Clin. Cancer Res. 2010;29:109. doi: 10.1186/1756-9966-29-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Polytarchou C., Iliopoulos D., Struhl K. An integrated transcriptional regulatory circuit that reinforces the breast cancer stem cell state. Proc. Natl. Acad. Sci. USA. 2012;109:14470–14475. doi: 10.1073/pnas.1212811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deshpande S.D., Putta S., Wang M., Lai J.Y., Bitzer M., Nelson R.G., Lanting L.L., Kato M., Natarajan R. Transforming growth factor-β-induced cross talk between p53 and a microRNA in the pathogenesis of diabetic nephropathy. Diabetes. 2013;62:3151–3162. doi: 10.2337/db13-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Jiang F., Xia X., Zhang B. LncRNA FAL1 promotes carcinogenesis by regulation of miR-637/NUPR1 pathway in colorectal cancer. Int. J. Biochem. Cell Biol. 2019;106:46–56. doi: 10.1016/j.biocel.2018.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Xu R.L., He W., Tang J., Guo W., Zhuang P., Wang C.Q., Fu W.M., Zhang J.F. Primate-specific miRNA-637 inhibited tumorigenesis in human pancreatic ductal adenocarcinoma cells by suppressing Akt1 expression. Exp. Cell Res. 2018;363:310–314. doi: 10.1016/j.yexcr.2018.01.026. [DOI] [PubMed] [Google Scholar]
- 36.Yuan Q., Liu Y., Fan Y., Liu Z., Wang X., Jia M., Geng Z., Zhang J., Lu X. LncRNA HOTTIP promotes papillary thyroid carcinoma cell proliferation, invasion and migration by regulating miR-637. Int. J. Biochem. Cell Biol. 2018;98:1–9. doi: 10.1016/j.biocel.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Liu W.L., Zhang L., Ge R., He F., Gao T.Y., Tian Q., Mu X., Chen L.H., Chen W., Li X. MiR-637 suppresses melanoma progression through directly targeting P-REX2a and inhibiting PTEN/AKT signaling pathway. Cell. Mol. Biol. 2018;64:50–57. [PubMed] [Google Scholar]
- 38.Pang B., Fan H., Zhang I.Y., Liu B., Feng B., Meng L., Zhang R., Sadeghi S., Guo H., Pang Q. HMGA1 expression in human gliomas and its correlation with tumor proliferation, invasion and angiogenesis. J. Neurooncol. 2012;106:543–549. doi: 10.1007/s11060-011-0710-6. [DOI] [PubMed] [Google Scholar]
- 39.Liu B., Pang B., Liu H., Arakawa Y., Zhang R., Feng B., Zhong P., Murata D., Fan H., Xin T. High mobility group A1 expression shows negative correlation with recurrence time in patients with glioblastoma multiforme. Pathol. Res. Pract. 2015;211:596–600. doi: 10.1016/j.prp.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Colamaio M., Tosti N., Puca F., Mari A., Gattordo R., Kuzay Y., Federico A., Pepe A., Sarnataro D., Ragozzino E. HMGA1 silencing reduces stemness and temozolomide resistance in glioblastoma stem cells. Expert Opin. Ther. Targets. 2016;20:1169–1179. doi: 10.1080/14728222.2016.1220543. [DOI] [PubMed] [Google Scholar]
- 41.Mineo M., Ricklefs F., Rooj A.K., Lyons S.M., Ivanov P., Ansari K.I., Nakano I., Chiocca E.A., Godlewski J., Bronisz A. The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J., Lv B., Su Y., Wang X., Bu J., Yao L. Exosome-Mediated Transfer of lncRNA HOTTIP Promotes Cisplatin Resistance in Gastric Cancer Cells by Regulating HMGA1/miR-218 Axis. Onco. Targets Ther. 2019;12:11325–11338. doi: 10.2147/OTT.S231846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Méndez O., Peg V., Salvans C., Pujals M., Fernández Y., Abasolo I., Pérez J., Matres A., Valeri M., Gregori J. Extracellular HMGA1 Promotes Tumor Invasion and Metastasis in Triple-Negative Breast Cancer. Clin. Cancer Res. 2018;24:6367–6382. doi: 10.1158/1078-0432.CCR-18-0517. [DOI] [PubMed] [Google Scholar]
- 44.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 45.Compes P., Tabouret E., Etcheverry A., Colin C., Appay R., Cordier N., Mosser J., Chinot O., Delingette H., Girard N. Neuro-radiological characteristics of adult diffuse grade II and III insular gliomas classified according to WHO 2016. J. Neurooncol. 2019;142:511–520. doi: 10.1007/s11060-019-03122-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.