Abstract
Post-SELEX modification of DNA aptamers is an established strategy to improve their affinity or inhibitory characteristics. In this study, we examined the possibility of increasing the recognition interface between the thrombin-binding aptamer HD1 (TBA) and thrombin by adding a chemically modified side chain to selected nucleotide residues. A panel of 22 TBA variants with N3-modified residues T3 and T12 was prepared by a two-step modification procedure. Aptamers were characterized by a combination of biophysical and biochemical methods. We identified mutants with enhanced affinity and improved anticoagulant activity. The crystal structures of thrombin complexes with three selected modified variants revealed that the modified pyrimidine base invariably allocates in proximity to thrombin residues Tyr76 and Ile82 due to the directing role of the unmodified TT loop. The modifications induced an increase in the contact areas between thrombin and the modified TBAs. Comparative analysis of the structural, biochemical, and biophysical data suggests that the non-equivalent binding modes of the mutants with thrombin in the T3- and T12-modified series account for the observed systematic differences in their affinity characteristics. In this study, we show that extending the recognition surface between the protein and modified aptamers is a promising approach that may improve characteristics of aptamer ligands.
Keywords: post-SELEX modifications, thrombin-binding aptamer, anticoagulants, crystal structure, G-quadruplex, binding affinity
Graphical Abstract
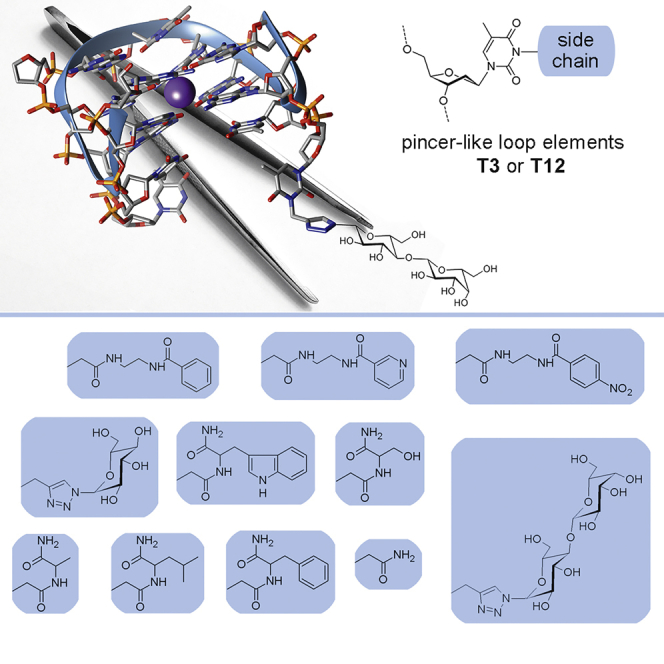
Smirnov et al. describe a post-SELEX approach for the thrombin aptamer HD1 that consists in expanding the aptamer-protein interface through attachment of non-natural side chains at specific aptamer residues. The authors report synthesis and characterization of 22 aptamer mutants and present crystal structures of thrombin complexes with three selected variants.
Introduction
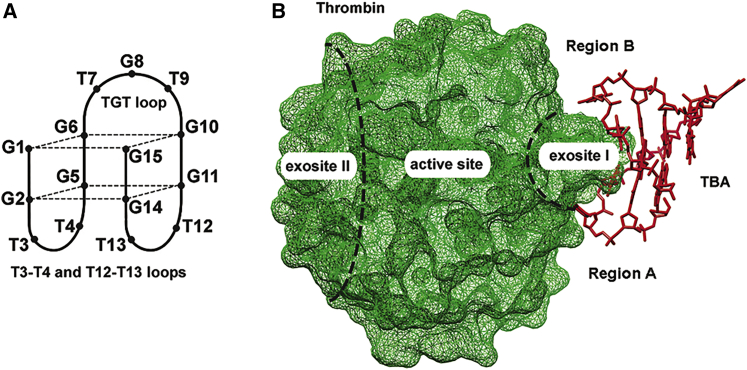
Nucleic acid aptamers are regarded as a promising class of high-specificity affinity ligands to a variety of biologically relevant targets with great potential in diagnostics and therapy. Aptamer selection through systematic evolution of ligands by exponential enrichment (SELEX) usually identifies a few high-affinity candidates that may be further optimized by post-SELEX modifications.1 The wide range of post-SELEX studies are firmly associated with thrombin-binding aptamer HD1 (TBA), a 15-nt G-rich oligonucleotide (5′-GGTTGGTGTGGTTGG-3′) that interferes with the coagulation cascade through specific binding with thrombin.2 Previous structural studies have revealed that TBA adopts a G-quadruplex architecture both in the free3,4 and ligand-bound state.5,6 According to the crystallographic structures of the thrombin-TBA complex,5,6 the aptamer folds as a chair-like anti-parallel G-quadruplex with a core formed by two stacked G-quartets that are covered on one side by a TGT loop that protrudes into the solvent and on the opposite side by two TT loops. The latter are formed by thymine nucleobases Thy3/4 and Thy12/13, which play a primary role in the recognition of the target protein (Figure 1). Modified TBA analogs have demonstrated high potential as promising aptamer anticoagulant drugs.7 Modification of the quadruplex core has been examined for a large number of non-natural nucleoside analogs.7, 8, 9, 10, 11 The impact of the core modifications on binding affinity or inhibitory characteristics is diverse and generally does not correlate with quadruplex stability.7, 8, 9, 10 The central TGT loop, although distal from the aptamer-protein interface, has been successfully modified by selected synthetic analogs to boost binding affinity or increase thermal stability.8,12, 13, 14, 15, 16 In regard to TT loops, Thy4 and Thy13 turned out to be the most intolerant to modifications.14,17, 18, 19 We found only two reports describing the highly beneficial substitution of Thy4/13 by synthetic analogs.15,20 In contrast, positions T3 or T12 in TBA accept a variety of unnatural thymidine analogs without compromising the ability of the aptamer to bind and inhibit thrombin.12,14,15,17,18,21,22 The tolerance of positions 3 and 12 to modifications is well illustrated by the fact that mutants with adenine19,23 or even those missing a base24 are able to support specific binding. Certain elasticity at T3/12 with respect to modifications opens the opportunity to examine a strategy other than a partial structural change of respective nucleotide units. Residues T3/12 may be used as a bridge for an extended segment of the mutant aptamer that could potentially contribute to affinity through specific interactions with remote protein residues. Implementation of this approach requires the screening of a variety of functional groups on the side chain of the respective residue to explore the protein surface in proximity to the established TBA-thrombin interface.
Figure 1.
TBA schematic and location of the aptamer on the protein surface
(A) TBA schematic. (B) Aptamer location on the protein surface. (PDB: 4DII.6)
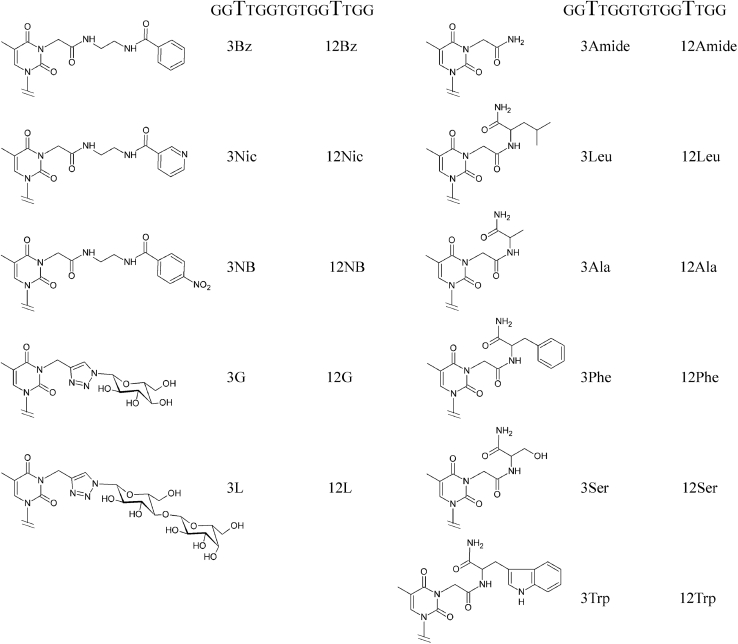
In the current study, we prepared a panel of TBA variants modified on the side chains of the pyrimidine bases at residues T3/T12. To expand the structural diversity of mutant variants, we used a two-step modification strategy. To incorporate a reactive thymidine residue into the respective position of TBA, we prepared three non-natural thymidine precursors containing a functional side chain at the N3 position of the pyrimidine base. Further transformation of partially modified thymidines with different chemical agents gave a library of 22 TBA mutants (Figure 2). Aptamer variants were characterized with respect to their biophysical properties, resistance to nuclease cleavage, affinity for thrombin, and anticoagulant activity. To gain insights into the structural properties of mutants with improved affinity, we present herein the crystal structures of three TBA variants in complex with thrombin. The crystallographic studies revealed specific features of the protein-aptamer interface in the modified variants that may account for their enhanced affinity with respect to unmodified TBA. The results reported herein provide clear evidence that the type of modification and position of the modified residue in the aptamer affect both the affinity and anticoagulant properties of mutant variants. It was found that modification of the T3 residue has higher potential to improve the characteristics of the natural prototype.
Figure 2.
Chemical structures of non-natural residues T3 and T12 in the modified TBA variants
Results
Synthesis of thymidine analogs and modified aptamers
Modification of thymidine residues was implemented through substitution at the N3 position of the pyrimidine nucleobase as described in Supplemental materials and methods. Briefly, we introduced amino, carboxyl, and alkynyl groups to the side chains of N3 thymidine derivatives to prepare modified predecessors of modified TBAs (Scheme S1). Final modification of a particular thymidine residue (Figure 2) was realized by using activated aromatic carboxylic acids, amino acid derivatives, or carbohydrate azides. The range of functionalities in the final aptamer library may be greatly increased by selecting a suitable array of synthetic agents containing the appropriate reactive groups.
Biophysical characterization of the mutant TBA variants
The solution structure of unmodified TBA3 suggests considerable conformational freedom for residues T3/12, which implies an insignificant contribution of T3/12 to the overall thermodynamic stability of the aptamer. Upon modification at N3, some substituents could potentially add to the stability of TBA through stacking or hydrophobic interactions with the neighboring Thy4/Thy13 base pair or exposed thymine of the TGT loop. A study of the thermal stability of TBA mutants by UV spectroscopy in 100 mM KCl (Table 1) showed that all types of modifications induced minor or moderate stabilization. In particular, a greater increase in the melting temperature (Tm) value was observed for variants 3/12Trp, 3/12Phe, 3Leu, and 12Bz. The presence of aromatic or hydrophobic groups in the more stable mutants is consistent with the presumable origin of the stabilization effect.
Table 1.
The thermal stabilities, binding affinities, and anticoagulant effects of TBA variants
| Mutant | Tm (°C) | KD (nM) | t (min) | Mutant | Tm (°C) | Kd (nM) | t (min) |
|---|---|---|---|---|---|---|---|
| TBA | 50.7 | 20.2 ± 1.3 | 4.81 | ||||
| 3Ala | 50.9 | 21.4 ± 2.8 | 3.67 | 12Ala | 51.7 | 51.0 ± 3.8 | 2.74 |
| 3Trp | 54.5 | >140 | 2.89 | 12Trp | 54.7 | 67.3 ± 12.1 | 3.12 |
| 3Leu | 54.3 | 10.9 ± 0.2 | 4.15 | 12Leu | 53.6 | 72.2 ± 0.9 | 4.14 |
| 3Ser | 51.7 | 14.6 ± 0.3 | 2.51 | 12Ser | 52.0 | 86.6 ± 4.5 | 2.34 |
| 3Phe | 54.3 | 22.6 ± 4.8 | 3.39 | 12Phe | 54.3 | 99.1 ± 6.3 | 4.07 |
| 3Amide | 52.6 | 29.2 ± 0.4 | 4.17 | 12Amide | 51.2 | 30.6 ± 6.1 | 3.97 |
| 3Bz | 52.3 | 30.0 ± 6.6 | 4.54 | 12Bz | 54.4 | >200 | 4.08 |
| 3NB | 51.7 | 163.5 ± 3.5 | 3.82 | 12NB | 53.1 | >200 | 3.60 |
| 3Nic | 52.1 | 27.1 ± 4.2 | 3.69 | 12Nic | 52.5 | >200 | 3.56 |
| 3G | 52.9 | 9.8 ± 0.6 | 3.34 | 12G | 53.4 | 20.7 ± 2.8 | 2.88 |
| 3L | 51.5 | 11.6 ± 0.5 | 5.28 | 12L | 51.0 | 27.2 ± 3.0 | 4.17 |
Tm was measured in 100 mM KCl; KD was measured in 100 mM potassium phosphate; t is the time required to reach 30% of the absorbance maximum at 360 nm; t = 0.87 min was observed for the blank sample (without aptamer). The errors in the Tm and t values were within 0.5°C and 0.1 min, respectively.
CD spectra of non-natural TBA analogs featured two positive bands at 247 and 295 nm and a negative band at 265 nm, confirming that all variants retained the TBA-like anti-parallel GQ structure (Figure S1). The intensities of the bands varied depending on the type of side chain at T3/12, thus providing evidence that the GQ core is responsive to changes at the periphery of the molecule.
Resistance to nuclease cleavage
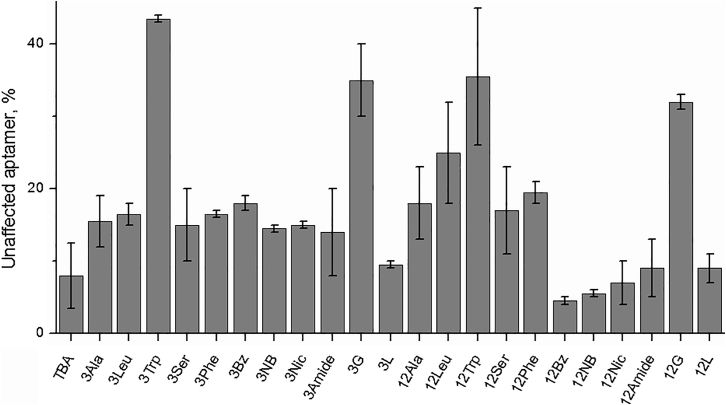
Increasing the lifetime of an aptamer in vivo is one of the major objectives of post-SELEX optimization. Formation of the GQ structure reduces the accessibility of core guanosines to rapid nuclease cleavage.25 In contrast, nucleotides in single-stranded loop regions are potential sites of nuclease cleavage. We compared the biochemical resistance of modified TBA variants with respect to S1 nuclease, which selectively cleaves single-stranded DNA. Although only one out of the three aptamer loops contained a modified residue, we observed notable variations in nuclease resistance (Figure 3; Figure S2). Under the selected conditions, mutants 12Bz, 12NB, and 12Nic showed the least stability, retaining approximately 5%–7% of the intact aptamer after 30 min of exposure. Nuclease resistance that was comparable to that of unmodified TBA or somewhat higher was observed for most amino acid variants and T3 aromatic series aptamers. Four samples associated with tryptophan or β-d-glycopyranoside side chains appeared to be the most stable. Particularly, variant 3Trp showed the presence of approximately 40% unaffected aptamer in the reaction mixture after 30 min of exposure.
Figure 3.
Resistance of the TBA variants to cleavage by S1 nuclease
Bar heights specify the proportion of unaffected aptamer after 30 min of exposure (SE, n = 2; for TBA: SE, n = 5).
Affinity to thrombin
Binding affinity to thrombin was estimated by microscale thermophoresis (MST) using the fluorescent Cy5 derivatives of TBA mutants in 100 mM K+ buffer (Table 1). Screening a panel of structurally diverse mutants revealed a clear relationship between the affinity and the location of the modified thymidine. Although dissociation constant (KD) values varied in a wide range for different modifications, non-natural 3T variants showed, with a single exception, consistently lower KD values than did their 12T counterparts. A systematically higher affinity for each modification in position 3 suggests a different arrangement of mutant variants relative to the protein in the T3 and T12 aptamer series. The only instance that broke this rule was the pair 3/12Trp.
In the 12T series, all mutants were less effective binders compared with TBA. Nevertheless, three mutants, 12Amide, 12G, and 12L, were characterized by relatively low KD values. Conversely, most aptamer variants in the 3T series retained high affinity to thrombin, showing KD values in the range of 10–30 nM, which is comparable to that of unmodified TBA. Approximately a 2-fold increase in affinity was observed for 3Leu, 3G, and 3L. High binding affinity was observed for all mutants with carbohydrate moieties. It is worth mentioning that a bulky disaccharide residue was able to accommodate in close proximity to the protein surface and contributed to the enhanced binding affinity of the aptamer 3L.
Anticoagulant activity of the modified aptamers
The anticoagulant properties of mutant aptamer variants were examined in a thrombin-induced fibrinogen polymerization assay. Fibrin gel formation in the presence of each aptamer was monitored over time by the increase in absorbance at 360 nm due to light scattering. For comparative analysis, we estimated the time required to reach 30% of the absorbance maximum (t). Data on the activity of TBA and modified aptamers are presented in Table 1 and Figure S3. In clotting experiments, we found a single sample (3L) that was a more potent inhibitor of thrombin than TBA. Apart from considering the highest inhibitory effect, it was interesting to compare the most successful inhibitors with respect to the chemical structure of the modified residue. Five chemical groups were present in aptamers that showed t values close to or above the threshold of 4 min: 3/12Leu, 3/12Amide, 3/12Bz, 3/12L, and 12Phe. Importantly, four out of five types of modifications were present in both the T3 and T12 aptamer series. Such a preference of particular chemical structures regardless of their location in the sequence indicates that the aptamer arrangement is consistent with the specific positioning of the modified residue.
Crystallographic structures of 3L, 3G, and 3Leu complexes with thrombin
For structural characterization of thrombin complexes with modified TBA, we selected three variants with the highest affinities. Trigonal P3221 crystals (Figure S4) with one 1:1 protein-DNA complex in the asymmetric unit were grown for the three complexes formed by thrombin and the mutants 3L, 3G, and 3Leu. The crystals were isomorphous and diffracted X-rays up to 1.58, 1.73, and 2.53 Å resolution, respectively. As expected, the binding of TBA variants does not substantially modify the thrombin structure. After superposition of the Cα heavy chain of the present structures with that of the PPACK inhibited enzyme (Protein Data Bank [PDB]: 1PPB),26 the root-mean-square deviation (RMSD) values were in the range of 0.44–0.57 Å.
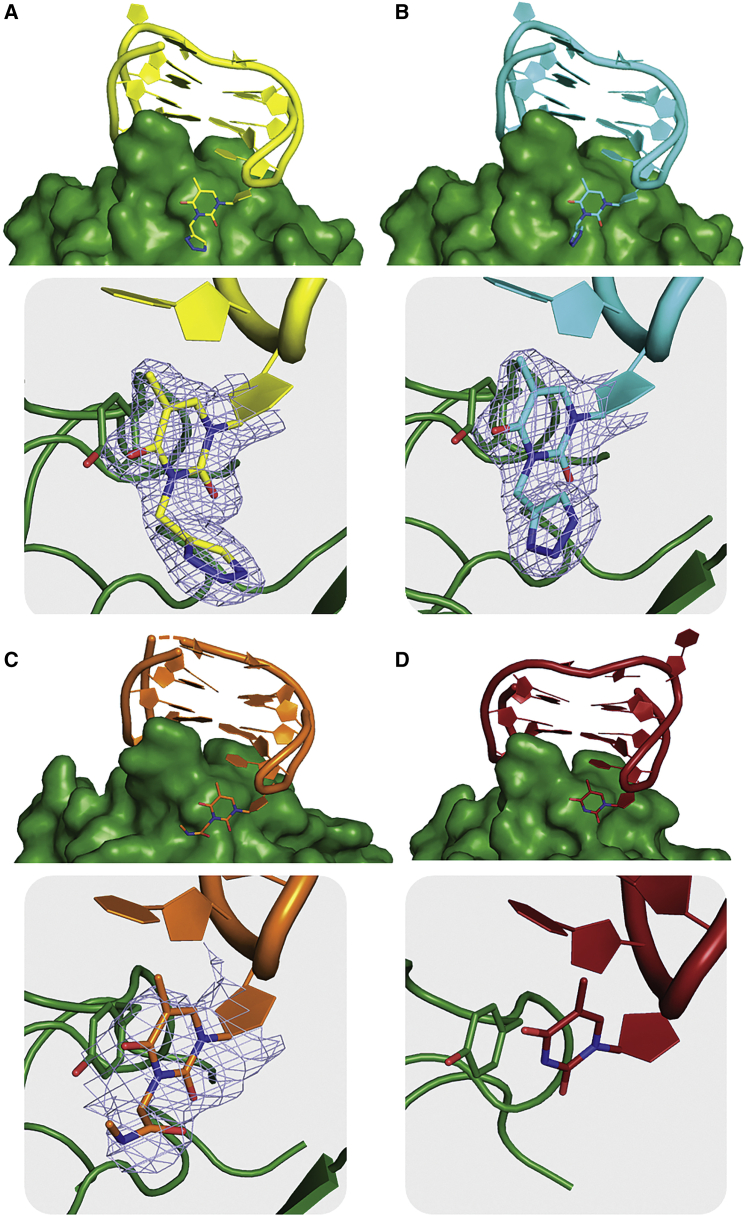
The refined models reveal the presence of the expected G-quadruplex domain, similar to that of the unmodified TBA.6 Resembling native TBA,6 the TT loops of the G-quadruplex domain act as a pincer-like system that captures the protruding region of exosite I (Figure 4). It is noteworthy to recall that TBA, due to its approximate 2-fold rotational symmetry, can bind the protein in two orientations related by a 180° rotation around the G-quadruplex axis.5,6 The two binding modes are basically equivalent, and the only significant difference is confined to the orientation of the TGT loop that is far away from the binding contacts with thrombin. In this context, exosite I can be described as formed by two subregions, referred to as A and B24 (Figure 1). In the former, a hydrophobic crevice on the protein surface forms a well-shaped joint that interlocks a thymine of a TT loop. Modifications that lower the symmetry of the two TT loops15,24,27 differentiate the energetics of the two binding modes. In the thrombin complexes with 3L, 3G, and 3Leu, the steric hindrance of the T3 modifications hampers the interactions with the exosite I A-region, removing the equivalence between the two binding modes of the unmodified aptamer, and strongly favoring the binding of the modified thymine at the B-region through π-π stacking with Tyr76 (Figure 4). The preference of a native TT loop to bind at the A-region strongly suggests that the A-region is already quite well modeled to host a native loop. Therefore, chemical modifications of one loop essentially act to modulate the binding affinity of the parent aptamer by increasing15,27 or decreasing24 the contacts of the oligonucleotide with the exosite I B-region. This, indeed, has been observed in the crystal structures of the three complexes, although only a part of the chemical substituents of T3 is detectable in the electron density and does not form specific interactions with the thrombin surface. Nevertheless, the visible part of the T3 substituents extends the covering of the protein surface by the aptamer and, in particular, shields a hydrophobic patch from water contact of this surface, including the side chain of Ile82.
Figure 4.
Cartoon/surface views of the thrombin-3L, thrombin-3G, and thrombin-3Leu interfaces
(A–C) Thrombin-3L (A), thrombin-3G (B), and thrombin-3Leu (C). In the panels, the 2Fo–Fc electron density maps of the modified nucleobases contoured at the 1.0 σ level. In all complexes, the modified thymine base stacks on Tyr76 in the B-region of thrombin exosite I. (D) As a comparison, the interface between thrombin and the parent TBA (PDB: 4DII) and the stacking interaction of its Thy12 and Tyr76 are also shown. Thrombin is colored in green, and 3L, 3G, 3Leu, and TBA are colored in yellow, cyan, orange, and dark red, respectively.
Moreover, these modifications cause slight variations in the conformation of the oligonucleotides with respect to TBA. In fact, albeit crystallized in the presence of potassium ions, the structures of the mutated TBAs are more similar to that of TBA observed in the presence of Na+ (TBA-Na, PDB: 4DIH) than that observed in the presence of K+ (TBA-K, PDB: 4DII)6 (Figure S5), as also indicated by the RMSD values reported in Table S3. It has been reported that the flexibility of unmodified aptamer increases in the presence of sodium.6 As a consequence, a better fit on the thrombin surface and optimization of the intermolecular contacts were observed when Na+ was bound to TBA. The number of protein-DNA interactions and the interface areas (Table S4) observed in the complexes of thrombin with the modified TBAs are higher with respect to those observed in the thrombin-TBA-K complex. Moreover, the contact areas between thrombin and the modified TBAs are even greater than those found in the thrombin-TBA-Na complex (Table S4). This finding suggests that even with a potassium ion bound in between the two tetrads, the improved adhesion to exosite I brought about by the modification introduced on T3 is capable of causing subtle adjustments of the oligonucleotide similar to those produced by the replacement of K+ by Na+ in the native aptamer. Altogether, these results could explain the observed increase in binding affinity toward thrombin for TBA variants 3L, 3G, and 3Leu with respect to the parent aptamer.
Discussion
Unlike many other DNA aptamers, directed chemical modification of TBA is facilitated by comprehensive structural characterization of the aptamer and its complex with thrombin. Aiming to expand the aptamer-protein interface, we took advantage of the specific arrangement of loop residues T3/12 in proximity to the thrombin surface. Twenty-two TBA variants with extended functionalities at Thy3 and Thy12 were prepared by a two-step modification approach to explore the protein landscape in proximity of the canonical thrombin-aptamer interface. We implemented a synthetic strategy based on the use of new thymidine derivatives with functional substituents at position N3 of the pyrimidine heterocycle. Commonly, to preserve hydrogen bonding capability, the pyrimidine base is modified solely at the C5 position. While this requirement is mandatory with respect to loop thymines 4 and 13, it does not apply to residues 3 and 12. The aptamer library was diversified with a variety of substituents from three different chemical groups: amino acids, aromatic carboxylic acids, and carbohydrates. Due to the peripheral arrangement of the modified fragment with respect to the GQ core, non-natural TBA mutants demonstrated insignificant variations in their biophysical characteristics. In contrast, we observed notable variations in the binding and biochemical properties of mutant aptamers. Structural studies of three non-natural aptamers revealed that the arrangement of N3 substituents of Thy3 in the B region of thrombin exosite I is governed by a strict requirement of an unmodified TT loop to be allocated in region A. As a result, the modified pyrimidine base is positioned in proximity to Tyr76 and the visible part of the modified side chain is engaged in a hydrophobic interaction with Ile82. Interestingly, although the binding modes of the two TT loops are equivalent in natural TBA,24 we found systematic differences in the binding affinity between T3 and T12 aptamer families, with the latter showing higher KD values. The orientation of the TGT loop, which is located far away from the protein, represents the only difference between the two equivalent modes of binding of the unmodified aptamer. It is reasonable to surmise that the structural organization of this loop, which is on the same G-quadruplex face as T12, influences, by through-bond propagation, the skeletal modes of the T12-modified oligonucleotides with a detrimental effect on protein binding.
Antithrombotic activity of TBA and its analogs is associated with competitive binding to exosite I. The binding pattern of fibrinogen, which is a substrate of thrombin and the natural TBA competitor, differs from that of the thrombin aptamer and affects residues Phe34, Ser36A, Leu65, Tyr76, Arg77A, Ile82, and Lys110.28, 29, 30 This area overlaps region B of thrombin in proximity to exosite I. In regard to anticoagulant activity, the alignment of a non-natural residue in this location is advantageous, as it may potentially interfere with anchoring fibrinogen. The improved anticoagulant activity of TBA variant 3L is consistent with the idea of a steric barrier that the bulky disaccharide unit builds between thrombin and fibrinogen. In this context, modification of the T3 residue at N3 with an extended side chain seems an attractive approach for improving the antithrombotic activity of TBA.
Conclusions
We examined a diverse panel of TBA mutants chemically modified at Thy3 and Thy12 with respect to their biophysical, biochemical, and structural characteristics. Aptamer variants with enhanced affinity or improved anticoagulant activity were identified within a family of T3 mutants. Inspection of the spatial organization of three non-natural aptamers in complex with human thrombin by crystallographic methods provided proof of a general rule that guides the arrangement of modified residues. Specifically, our results confirm the directing role of the unmodified TT loop of the aptamer, which preferably binds to region A of thrombin exosite I. Additionally, it was shown that modifications were able to induce an increase in the interface area between the modified aptamer and the protein. Another important finding of the current study suggests that the difference between the two possible binding modes of mutant aptamers with thrombin accounts for notably lower affinity in the T12 aptamer family. The arrangement of modified residues in the B region of thrombin exosite I opens the opportunity for efficient interference with fibrinogen binding and cleavage. In this respect, expanding the functionality at the Thy3 side chain may be acknowledged as a promising strategy in the search for efficient aptamer anticoagulants.
Materials and methods
Most reagents were purchased from Sigma-Aldrich and Acros. Research-grade human thrombin from plasma for microscale thermophoresis and clotting studies was purchased from Renam (Russia). Human d-Phe-Pro-Arg-chloromethylketone (PPACK)-inhibited thrombin for crystallography was purchased from Haematologic Technologies (USA). Fibrinogen from human plasma was obtained from Sigma-Aldrich. Synthetic details for the N3-modified phosphoramidites and oligonucleotides are given in Supplemental materials and methods.
Ultraviolet thermal denaturation
Absorbance versus temperature profiles were obtained with a Cary 50 spectrophotometer equipped with a Peltier cell holder. Melting experiments were performed at 295 nm in 10 mM sodium cacodylate (pH 7.2) and 100 mM KCl. The heating/cooling rate was 0.5°C/min. The melting points were determined from derivative plots of the melting curves. The oligonucleotide concentration was 5 μM.
Circular dichroism spectroscopy
Circular dichroism (CD) measurements were performed at 20°C with an aptamer concentration of 5 μM in 10 mM sodium cacodylate (pH 7.2) and 100 mM KCl by using a Chirascan CD spectrometer (Applied Photophysics).
Microscale thermophoresis
The KD values of the aptamer-thrombin complexes were measured with a Monolith NT.115 instrument (NanoTemper) with the Cy5 detection channel. The concentration of Cy5-labeled aptamers was 20 nM in 10 mM Tris HCl, 100 mM potassium phosphate (pH 7.4), and 0.5% Tween 20. A standard series of dilutions in the same buffer was used for thrombin with the highest final concentration in a capillary of 1 μM. The T-jump region was used for automatic KD calculations.
S1 nuclease cleavage
Cy5-labeled oligonucleotides were dissolved at a concentration of 1 μM in 50 μL of 1× reaction buffer. Samples were heated to 95°C and cooled slowly before digestion. Ten units of S1 nuclease (Thermo Scientific) were added to the aptamer samples. After incubation for 10 or 30 min at 25°C, the reaction mixtures were precipitated with 2% LiClO4 in acetone and analyzed by electrophoresis in a polyacrylamide gel. Electrophoresis was performed with a 20% polyacrylamide gel in 7 M urea and 1× TBE (pH 8.4).
Fibrinogen clotting in the presence of aptamers
Human thrombin (100 μL, 10 U/mL) was added to a solution of fibrinogen (2 mg/mL) and aptamer (30 nM) in 1 mL of PBS in a quartz cuvette in a temperature-controlled cuvette holder of a spectrophotometer at 25°C. Monitoring of the absorbance at 360 nm started immediately after the addition of thrombin and was stopped after the curve reached a plateau. The blank clotting curve was determined by measuring the absorbance in the absence of aptamer.
Crystallography
In order to induce folding, the three TBA mutant samples were annealed in 10 mM potassium phosphate buffer (pH 7.4) and 100 mM KCl by heating to 90°C for 5 min and then slowly cooling for 50–60 min and storage at 20°C overnight. A standard protocol was followed for the preparation of the complexes between thrombin and 3L, 3G, and 3Leu.6,24,27,31, 32, 33 Crystals suitable for X-ray diffraction data collection (Figure S4) were grown at 20°C by hanging drop vapor diffusion, where 0.5 μL of complex solution was mixed with 0.5 μL of reservoir solution containing 200 mM KCl, 35% (v/v) pentaerythritol propoxylate, and 50 mM HEPES (pH 7.5) for the thrombin-3L and thrombin-3G complexes, or 18% (v/v) 2-propanol, 18% (w/v) polyethylene glycol 4000, and 100 mM tri-sodium citrate (pH 5.6) for the thrombin-3Leu complex. No cryoprotection was required for freezing of the crystals prior to data collection.
Diffraction data were collected at the i04 beamline of the Diamond Light Source synchrotron (Harwell Science and Innovation Campus, Oxfordshire, UK) using λ = 0.9795 Å. Datasets were processed using autoPROC34, 35, 36, 37, 38 and STARANISO39 software. The phase problem was solved by molecular replacement using Phaser40 from the CCP4 package36 and, as a search model, the coordinates of the native protein (PDB: 1PPB).26 Refinement and manual model building were carried out using the REFMAC536,41 and WinCoot42 programs, respectively. Detailed statistics on data collection and refinement are reported in Table S2.
The SuperPose program36,43 was used to calculate RMSDs. Features of the thrombin-DNA complex interface and interactions between the two molecules were determined by the Contact (CCP4 package),36 CoCoMaps,44 and PISA36,45 programs. Molecular graphics figures were prepared with PyMOL (DeLano Scientific, Palo Alto, CA, USA) and the UCSF Chimera package.46 The coordinates of the structures were deposited in the PDB (PDB: 6Z8V, 6Z8W, and 6Z8X for thrombin-3L, thrombin-3G, and thrombin-3Leu, respectively).
MALDI mass spectrometry
Mass spectrometry analysis (Table S1, Data S1) was performed with Ultraflex II or Microflex (Bruker Daltonics) MALDI-TOF mass spectrometer using 0.25 M aqueous 3-hydroxypicolinic acid in deionized water as the matrix and 10 mM diammonium citrate as an additive. Before mixing with MALDI matrix, all samples were treated with cation exchange resin in ammonium form. Spectra were recorded in linear mode for positive ions.
Data availability
Atomic coordinates and structural factors for the thrombin-3L, thrombin-3G, and thrombin-3Leu complexes have been deposited with the PDB (PDB: 6Z8V, 6Z8W, and 6Z8X, respectively).
Acknowledgments
We thank the DLS-CCP4 Data Collection and Structure Solution Workshop 2019 at Diamond Light Source (Oxfordshire, UK), during which R. Troisi carried out the diffraction data collection. This work was partially supported by the Russian Fund for Basic Research (18-04-00614).
Author contributions
Conception and design, E.T., I.S., and F.S.; acquisition of data, E.T., N.K., I.S., and R.T.; analysis and interpretation of data, E.T., I.S., N.K., R.T., and F.S.; writing – review and/or revision of the manuscript, E.T., and F.S.; administrative, technical, or material support, E.T. and F.S.; study supervision, E.T. and F.S.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.01.004.
Supplemental information
References
- 1.Röthlisberger P., Hollenstein M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018;134:3–21. doi: 10.1016/j.addr.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 3.Schultze P., Macaya R.F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 4.Russo Krauss I., Napolitano V., Petraccone L., Troisi R., Spiridonova V., Mattia C.A., Sica F. Duplex/quadruplex oligonucleotides: role of the duplex domain in the stabilization of a new generation of highly effective anti-thrombin aptamers. Int. J. Biol. Macromol. 2018;107(Pt B):1697–1705. doi: 10.1016/j.ijbiomac.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 5.Padmanabhan K., Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta Crystallogr. D Biol. Crystallogr. 1996;52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 6.Russo Krauss I., Merlino A., Randazzo A., Novellino E., Mazzarella L., Sica F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012;40:8119–8128. doi: 10.1093/nar/gks512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avino A., Fabrega C., Tintore M., Eritja R. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012;18:2036–2047. doi: 10.2174/138161212799958387. [DOI] [PubMed] [Google Scholar]
- 8.Van Riesen A.J., Fadock K.L., Deore P.S., Desoky A., Manderville R.A., Sowlati-Hashjin S., Wetmore S.D. Manipulation of a DNA aptamer-protein binding site through arylation of internal guanine residues. Org. Biomol. Chem. 2018;16:3831–3840. doi: 10.1039/c8ob00704g. [DOI] [PubMed] [Google Scholar]
- 9.Ying G., Lu X., Mei J., Zhang Y., Chen J., Wang X., Ou Z., Yi Y. A structure-activity relationship of a thrombin-binding aptamer containing LNA in novel sites. Bioorg. Med. Chem. 2019;27:3201–3207. doi: 10.1016/j.bmc.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Kolganova N.A., Varizhuk A.M., Novikov R.A., Florentiev V.L., Pozmogova G.E., Borisova O.F., Shchyolkina A.K., Smirnov I.P., Kaluzhny D.N., Timofeev E.N. Anomeric DNA quadruplexes. Artif. DNA PNA XNA. 2014;5:e28422. doi: 10.4161/adna.28422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotkowiak W., Czapik T., Pasternak A. Novel isoguanine derivative of unlocked nucleic acid-Investigations of thermodynamics and biological potential of modified thrombin binding aptamer. PLoS ONE. 2018;13:e0197835. doi: 10.1371/journal.pone.0197835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scuotto M., Persico M., Bucci M., Vellecco V., Borbone N., Morelli E., Oliviero G., Novellino E., Piccialli G., Cirino G. Outstanding effects on antithrombin activity of modified TBA diastereomers containing an optically pure acyclic nucleotide analogue. Org. Biomol. Chem. 2014;12:5235–5242. doi: 10.1039/c4ob00149d. [DOI] [PubMed] [Google Scholar]
- 13.Varada M., Aher M., Erande N., Kumar V.A., Fernandes M. Methoxymethyl threofuranosyl thymidine (4′-MOM-TNA-T) at the T7 position of the thrombin-binding aptamer boosts anticoagulation activity, thermal stability, and nuclease resistance. ACS Omega. 2019;5:498–506. doi: 10.1021/acsomega.9b03042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai B., Yang X., Sun L., Fan X., Li L., Jin H., Wu Y., Guan Z., Zhang L., Zhang L., Yang Z. Stability and bioactivity of thrombin binding aptamers modified with d-/l-isothymidine in the loop regions. Org. Biomol. Chem. 2014;12:8866–8876. doi: 10.1039/c4ob01525h. [DOI] [PubMed] [Google Scholar]
- 15.Dolot R., Lam C.H., Sierant M., Zhao Q., Liu F.-W., Nawrot B., Egli M., Yang X. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018;46:4819–4830. doi: 10.1093/nar/gky268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsvetkov V.B., Varizhuk A.M., Pozmogova G.E., Smirnov I.P., Kolganova N.A., Timofeev E.N. A universal base in a specific role: tuning up a thrombin aptamer with 5-nitroindole. Sci. Rep. 2015;5:16337. [Google Scholar]
- 17.Pasternak A., Hernandez F.J., Rasmussen L.M., Vester B., Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011;39:1155–1164. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chai Z., Guo L., Jin H., Li Y., Du S., Shi Y., Wang C., Shi W., He J. TBA loop mapping with 3′-inverted-deoxythymidine for fine-tuning of the binding affinity for α-thrombin. Org. Biomol. Chem. 2019;17:2403–2412. doi: 10.1039/c9ob00053d. [DOI] [PubMed] [Google Scholar]
- 19.Nagatoishi S., Isono N., Tsumoto K., Sugimoto N. Loop residues of thrombin-binding DNA aptamer impact G-quadruplex stability and thrombin binding. Biochimie. 2011;93:1231–1238. doi: 10.1016/j.biochi.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Virgilio A., Petraccone L., Vellecco V., Bucci M., Varra M., Irace C., Santamaria R., Pepe A., Mayol L., Esposito V., Galeone A. Site-specific replacement of the thymine methyl group by fluorine in thrombin binding aptamer significantly improves structural stability and anticoagulant activity. Nucleic Acids Res. 2015;43:10602–10611. doi: 10.1093/nar/gkv1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng C.G., Damha M.J. G-quadruplex induced stabilization by 2′-deoxy-2′-fluoro-d-arabinonucleic acids (2′F-ANA) Nucleic Acids Res. 2007;35:4977–4988. doi: 10.1093/nar/gkm520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Virgilio A., Petraccone L., Scuotto M., Vellecco V., Bucci M., Mayol L., Varra M., Esposito V., Galeone A. 5-Hydroxymethyl-2′-deoxyuridine residues in the thrombin binding aptamer: investigating anticoagulant activity by making a tiny chemical modification. ChemBioChem. 2014;15:2427–2434. doi: 10.1002/cbic.201402355. [DOI] [PubMed] [Google Scholar]
- 23.Tsiang M., Gibbs C.S., Griffin L.C., Dunn K.E., Leung L.L.K. Selection of a suppressor mutation that restores affinity of an oligonucleotide inhibitor for thrombin using in vitro genetics. J. Biol. Chem. 1995;270:19370–19376. doi: 10.1074/jbc.270.33.19370. [DOI] [PubMed] [Google Scholar]
- 24.Pica A., Russo Krauss I., Merlino A., Nagatoishi S., Sugimoto N., Sica F. Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer. FEBS J. 2013;280:6581–6588. doi: 10.1111/febs.12561. [DOI] [PubMed] [Google Scholar]
- 25.Cao Z., Huang C.-C., Tan W. Nuclease resistance of telomere-like oligonucleotides monitored in live cells by fluorescence anisotropy imaging. Anal. Chem. 2006;78:1478–1484. doi: 10.1021/ac0517601. [DOI] [PubMed] [Google Scholar]
- 26.Bode W., Turk D., Karshikov A. The refined 1.9-A X-ray crystal structure of d-Phe-Pro-Arg chloromethylketone-inhibited human α-thrombin: structure analysis, overall structure, electrostatic properties, detailed active-site geometry, and structure-function relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo Krauss I., Merlino A., Giancola C., Randazzo A., Mazzarella L., Sica F. Thrombin-aptamer recognition: a revealed ambiguity. Nucleic Acids Res. 2011;39:7858–7867. doi: 10.1093/nar/gkr522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huntington J.A. Molecular recognition mechanisms of thrombin. J. Thromb. Haemost. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 29.Tsiang M., Jain A.K., Dunn K.E., Rojas M.E., Leung L.L.K., Gibbs C.S. Functional mapping of the surface residues of human thrombin. J. Biol. Chem. 1995;270:16854–16863. doi: 10.1074/jbc.270.28.16854. [DOI] [PubMed] [Google Scholar]
- 30.Pechik I., Madrazo J., Mosesson M.W., Hernandez I., Gilliland G.L., Medved L. Crystal structure of the complex between thrombin and the central “E” region of fibrin. Proc. Natl. Acad. Sci. USA. 2004;101:2718–2723. doi: 10.1073/pnas.0303440101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troisi R., Napolitano V., Spiridonova V., Russo Krauss I., Sica F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018;46:12177–12185. doi: 10.1093/nar/gky990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo Krauss I., Pica A., Merlino A., Mazzarella L., Sica F. Duplex-quadruplex motifs in a peculiar structural organization cooperatively contribute to thrombin binding of a DNA aptamer. Acta Crystallogr. D Biol. Crystallogr. 2013;69:2403–2411. doi: 10.1107/S0907444913022269. [DOI] [PubMed] [Google Scholar]
- 33.Russo Krauss I., Spiridonova V., Pica A., Napolitano V., Sica F. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Res. 2016;44:3969. doi: 10.1093/nar/gkw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G.W., McCoy A. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tickle I.J., Flensburg C., Keller P., Paciorek W., Sharff A., Vonrhein C., Bricogne G. Global Phasing; 2019. STARANISO. [Google Scholar]
- 40.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 44.Vangone A., Spinelli R., Scarano V., Cavallo L., Oliva R. COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics. 2011;27:2915–2916. doi: 10.1093/bioinformatics/btr484. [DOI] [PubMed] [Google Scholar]
- 45.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structural factors for the thrombin-3L, thrombin-3G, and thrombin-3Leu complexes have been deposited with the PDB (PDB: 6Z8V, 6Z8W, and 6Z8X, respectively).