Abstract
Patients with epidermal growth factor receptor (EGFR) mutations in lung adenocarcinoma can benefit from targeted therapy. However, noninvasively determination of EGFR mutation status before targeted therapy remains a challenge. This study constructed a nomogram based on a combination of radiomics features with the clinical and radiological features to predict the EGFR mutation status. The least absolute shrinkage and selection operator (LASSO) and Wilcoxon test were used for feature selection. Decision tree (DT), logistic regression (LR), and support vector machine (SVM) classifiers were used for radiomics model building. Used the clinical and radiological features establish clinical-radiology (C-R) model. The C-R model with the best radiomics model to establish clinical-radiological-radiomics (C-R-R) model. The predictive performance of the model was evaluated by ROC and calibration curves, and the clinical usefulness was assessed by a decision curve analysis. The current study showed that twelve radiomics features were significantly associated with EGFR mutations. The best radiomics signature model was obtained using the SVM classifier. The C-R-R model had the best distinguishing ability for predicting the EGFR mutation status, with an AUC of 0.849 (95% CI, 0.805-0.893) and 0.835 (95% CI, 0.761-0.909) in the development and validation cohorts, respectively. Our study provides a non-invasive C-R-R model that combines CT-based radiomics features with clinical and radiological features, which can provide useful image-based biological information for targeted therapy candidates.
Keywords: Computed tomography, EGFR, lung adenocarcinoma, radiomics
Introduction
An understanding of the pathological and molecular aspects of lung cancer has made great progress in recent years [1]. The discovery of lung cancer driver genes, especially the epidermal growth factor receptor (EGFR) gene, has increased the uptake of individualized targeted therapy [2,3]. Patients with EGFR mutations in lung adenocarcinoma respond well to EGFR tyrosine kinase inhibitors (TKI) [4-7]. By contrast, patients without these mutations are not eligible for treatment with the TKI at any disease stage [8]. These findings suggest that it is essential to determine patient EGFR mutation status ahead of treatment.
Currently, an analysis of the EGFR mutation status involves invasive procedures such as biopsy or surgical removal to acquire a specimen. However, these procedures are associated with limitations such as a small number of biopsy samples and the risk of sampling errors, among others [9,10]. In addition, tumors tend to be heterogeneous; thus, tissue samples obtained from a particular site might not be representative of the whole tumor [11]. Recently, a “liquid” biopsy has been proposed as an alternative approach; it involves the analysis of nucleic acids present in the peripheral blood and has high specificity for the detection of EGFR mutations. However, the sensitivity of this method is low, and the false negative rate is estimated at 30%; further, the recognized molecular variation may be unrelated to the tumor [12,13]. Overall, these findings suggest a need for a non-invasive and easy-to-use method to determine the EGFR mutation status.
Computed tomography (CT) is the preferred imaging method for lung cancer screening and diagnosis. Previous studies have shown that female sex, a non-smoking status, ground-glass opacity, air bronchogram findings, bubble-like lucencies, pleural retraction, or other CT signs are indicative of EGFR mutation [10,14-16]. However, these studies had a few limitations. For example, CT scan evaluation was performed subjectively by the observer and could not be quantified. In addition, the findings were inconsistent and their accuracy was relatively low.
Radiomics is a novel, non-invasive, and promising method, which uses advanced image algorithms in artificial intelligence to extract high-throughput features from medical images, quantify higher-dimensional features that cannot be observed in the human visual system, and apply useful image features to clinical decision-making [17,18]. Some studies have tried to predict EGFR mutation status with a CT-based radiomics model [11,19-25]. However, these studies were limited by a relatively small sample size, low accuracy, less radiomics characteristics, or the lack of validation data sets. Therefore, heterogeneity of the entire tumor cannot be effectively evaluated. In addition, clinical risk factors and radiological features were not included in the previous models despite evidence that including these features may improve the diagnostic performance of a model [11,20]. Therefore, we established a radiomics model based on the preoperative CT radiomics signature to predict the EGFR mutation status in this study. Further, we describe a user-friendly radiomics nomogram that combines the clinical risk factors, radiological features, and the radiomics signature.
Materials and methods
Patient selection
The institutional review board of our institution approved this retrospective study, and the need for informed consent was waived. We obtained the medical records of 780 patients with lung adenocarcinoma diagnosed by histopathology from January 2016 to May 2020, and collected their preoperative non-enhanced CT images and clinical data. The inclusion criteria were as follows: (1) patients with adenocarcinoma as the histological subtype according to the 2015 World Health Organization (WHO) lung cancer classification; (2) patients with thin-slice CT (1.25 mm) images and complete clinical data that can be used in the picture archiving and communication system (PACS); (3) age over 18 years; (4) patients who underwent a chest CT scan within 2 weeks before biopsy or surgery; (5) patients with no previous history of other malignant tumors; (6) and patients did not receive lung cancer-related treatment (such as chemotherapy, radiotherapy, or immunotherapy) prior to CT scanning. The exclusion criteria were as follows: (1) patients whose EGFR mutation status has not been tested; (2) patients with a histological subtype of lung cancer other than adenocarcinoma; (3) patients in whom the tumor boundary could not be easily delineated owing to massive pleural effusion or inflammation; (4) and patients whose CT image quality was poor. The flowchart of the inclusion and exclusion criteria is listed in the supplementary materials (Figure S1).
Based on the above criteria, a total of 420 patients [mean age ± standard deviation (SD), 57.43 ± 9.36 years; median age, 56.5 years; range age, 21-82 years, including 201 women (mean age ± SD, 56.30 ± 8.51 years; median age, 55.0 years; range age, 31-82 years) and 219 men (mean age ± SD, 58.46 ± 9.99 years; median age, 59.0 years; range age, 21-79 years)] were enrolled in this study. Patients were randomly divided into development and validation cohorts at a ratio of 7:3. Clinical variables included age, sex, smoking history [including non-smokers (never smokers) and smokers (previous and current smokers)], carcinoembryonic antigen (CEA) level, and 14 CT image features. The relationship between the clinical variables and radiological features of the two cohorts of patients and the EGFR mutation status is listed in the supplementary materials (Table S1).
Analysis of the EGFR mutation status
EGFR mutations were detected using the EGFR detection kit (Beijing SinoMD Gene Detection Technology Co., Ltd., China). The polymerase chain reaction (PCR)-based amplified refractory mutation system (ARMS) method was used to confirm the mutations in EGFR exons 18, 19, 20, and 21. All methods were performed according to the manufacturers’ instructions.
CT scanning protocol
CT scanning was performed using two spiral CT systems (Discovery CT750 HD, GE Healthcare, Waukesha, WI, USA; Philips iCT 256, Koninklijke Philips N.V.). The scanning parameters of the CT scanners were as follows: tube voltage, 120 kVp; tube current, 150-200 mA; tube rotation time, 0.5-1.0 seconds; collimator width, 40 mm; matrix, 512 × 512; axial image layer thickness, 5 mm; layer spacing, 5 mm; reconstruction layer thickness, 1.25 mm; and reconstruction layer interval, 1.25 mm. The scan range was from the tip to the bottom of the lung.
CT image analysis
Two radiologists with 5 years and 16 years of experience in thoracic tumor diagnosis, who were blinded to the clinical and histological data, independently evaluated the radiological features (Table S1) of all patients on the PACS. In case of disagreement, a consensus was reached after discussion. All results were analyzed in the lung window (width, 1500 HU; level, -500 HU) and mediastinum settings (width, 300 HU; level, 40 HU).
Radiomics feature selection
Tumor segmentation and feature extraction
Tumor segmentation is a key step in radiomics feature extraction and model building. At present, manual segmentation is the most accurate and recognized segmentation method [19]. Preoperative thin-layer CT images were uploaded to ITK-SNAP 3.8 (http:/www.itksnap.org) in the medical digital imaging and communication (DICOM) format for three-dimensional (3D) manual segmentation of the regions of interest (ROI) [26]. Bin width was set to 25, that is, every 25 gray units corresponded to one gray level, for a total of 80 (2000/25) gray levels. To minimize between-observer differences [27], a radiologist (G.J.Z) with 5 years of experience in chest diagnostics manually delineated the ROI on the axial image of the CT lung window (width, 1500 HU; level, -500 HU), and then confirmed it in the coronal and sagittal positions. If the ROI was found to be inaccurate, further manual adjustment was required. Finally, the segmented region outlined on each slice was merged to generate the volume of interest (VOI) [28]. Each VOI was verified by another radiologist (J.Z) with 16 years of experience. Both readers were blinded to the clinical and histological data of all patients.
For each accurately segmented VOI, the open source Python software package PyRadiomics (https://pyradiomics.readthedocs.io/en/Latest/) was used to automatically extract the radiomics features in the VOI. A total of 1468 radiomics features were extracted from each VOI. The features were divided into three main categories: (1) first-order features; (2) shape features; (3) and texture features [including gray-level co-occurrence matrix features (GLCM); gray-level run-length matrix features (GLRLM); gray-level size zone matrix features (GLSZM); neighboring gray-tone difference matrix features (NGTDM); and gray-level dependence matrix features (GLDM)].
Feature selection
Radiomics feature extraction and analysis workflow is shown in Figure 1. Feature selection is important to improve the generalization ability and optimize the model [19]. A large number of radiomics features may lead to overfitting, reducing model classification ability. The radiomics features used to build the model account for only a small part of the total. Therefore, in order to reduce the redundancy between the features, we first standardized all radiomics features in the development cohort using the z-score method and applied the same way on the validation cohort. Subsequently, Wilcoxon rank sum test was used to retain features with a P-value of < 0.0000341 (0.05/1468, the significance level of the test level α = 0.05 divided by the number of radiomics features for Bonferroni correction) [29,30]. Least absolute shrinkage and selection operator (LASSO) regression was applied to select the most relevant predictive features. The radiomics features selection process is described in Figures S2 and S3.
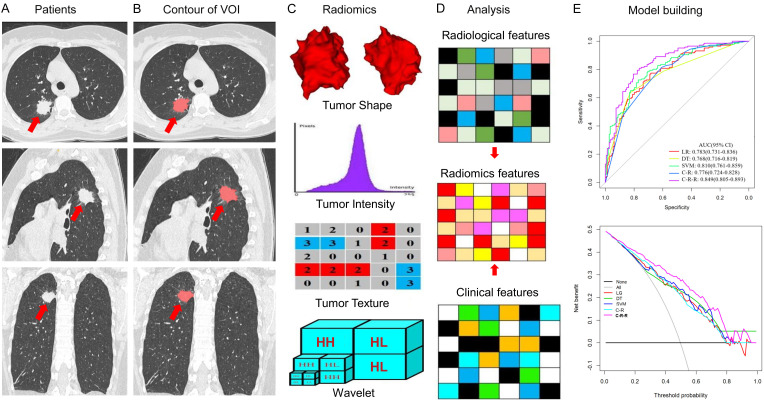
Figure 1.
Radiomics features extraction and analysis workflow. A. Original CT images of a patient with lung adenocarcinoma. B. Segmentation of the tumor volume of interest (VOI) on all CT slices by experienced radiologists. C. Feature extraction from the VOI, including tumor shape, intensity, texture, and wavelet features. D. Clinical, radiological, and radiomics feature analysis. E. Model building.
The selection of clinical and radiological features was based mainly on their correlation with the EGFR mutation status [14,15]. Firstly, univariate analysis was used to select the clinical and radiological features that were significantly different from the EGFR mutation status in the development cohort; next, multiple logistic regression analysis was further used to select the most relevant variables.
To evaluate the reproducibility and robustness of the feature extraction process, 3 months later, 40 patients were randomly selected from the development cohort, and the radiologist (G.Z.J) segmented the data again to construct a re-segmentation set. In addition, 40 patients were randomly selected from each CT scanner to establish different CT scanner sets to calculate intra-/interclass correlation coefficients (ICCs); values > 0.80 indicated good consistency.
Radiomics signature building
After eliminating the redundant features, we input the final selected radiomics features into the classifier to construct a radiomics signature for biological evaluation. In this study, three classifiers were evaluated, namely logistic regression (LR), decision tree (DT), and support vector machine (SVM). For DT, the size of the tree is controlled by complex parameters (CP). The CP value of the DT model was selected with the minimum ‘Bxerror’ (the mean value of 10-fold cross-validation error) [31]. SVM classifier uses radial basis function as the kernel function and employs 10-fold cross-validation to select the best-performing model during development.
The receiver operating characteristic (ROC) curve was used to calculate the area under the curve (AUC), sensitivity, specificity, and accuracy to evaluate the performance of different radiomics models, and the radiomics model with the highest AUC was taken as the best model.
Used the clinical and radiological features independently related to EGFR mutation status after multivariate analysis to establish a clinical-radiology (C-R) model. Then, combined the C-R model with the best radiomics model to establish a clinical-radiological-radiomics (C-R-R) model and calculated the diagnostic efficiency of the C-R-R model.
The goodness of fit of the model was evaluated by the calibration curve and the Hosmer-Lemeshow test [32]. For the development and validation cohorts, a decision curve analysis (DCA) was used to calculate the net benefits of each model under different threshold probabilities to evaluate the clinical usefulness of the model.
Statistical analyses
All statistical analyses were performed with R 3.6.3 (http://www.rproject.org) and IBM SPSS Statistics for Windows 22.0 (IBM Corporation, USA). Radiomics feature extraction were performed on Python 3.6.3 (https://www.python.org) with PyRadiomics tool kit. The chi-square or Fisher’s exact tests were used to evaluate on category variables as appropriate. Independent sample t-test, and Mann-Whitney U test were used to evaluate on continue variables as appropriate. Receiver operating characteristic (ROC) curve analysis was performed to compare the results of models in both sets. Youden’s index was used to determine the optimal cutoff point in the ROC analysis and the corresponding accuracy, sensitivity and specificity were also calculated. The between-model differences in AUC values were compared by DeLong test. Two-sided P-values of less than 0.05 were considered of a statistically significant finding.
Results
Clinical and radiological features of patients
A total of 420 patients were enrolled in this study, of which 294 and 126 were in the development and validation cohorts, respectively. The rate of EGFR mutation in total, development and validation groups was 50.5% (212/420), 50.34% (148/294), and 50.8% (64/126), respectively. There was no significant difference in the EGFR mutation rate between the development and validation cohorts (P > 0.05). In addition, there was no significant difference between other clinical and radiological features between the two cohorts (all P > 0.05). The balance of data between the two cohorts demonstrates that the grouping of patients in this study was reasonable (Table S1).
The relationship between clinical and radiological features and the EGFR mutation status in the development cohort is shown in Table S2. Univariate analysis revealed that 12 features were significantly different between the EGFR mutant and wild-type groups. Multivariate analysis revealed that smoking history (odds ratio [OR], 0.373; 95% confidence interval [CI], 0.182-0.765; P = 0.007), bubble-like lucency (OR, 3.669; 95% CI, 1.975-6.816; P < 0.001), pleural attachment (OR, 0.296; 95% CI, 0.148-0.594; P = 0.001), and pleural retraction (OR, 2.207; 95% CI, 1.188-4.100; P = 0.012) were correlated independently with the EGFR mutation status.
Radiomics feature selection and signature building
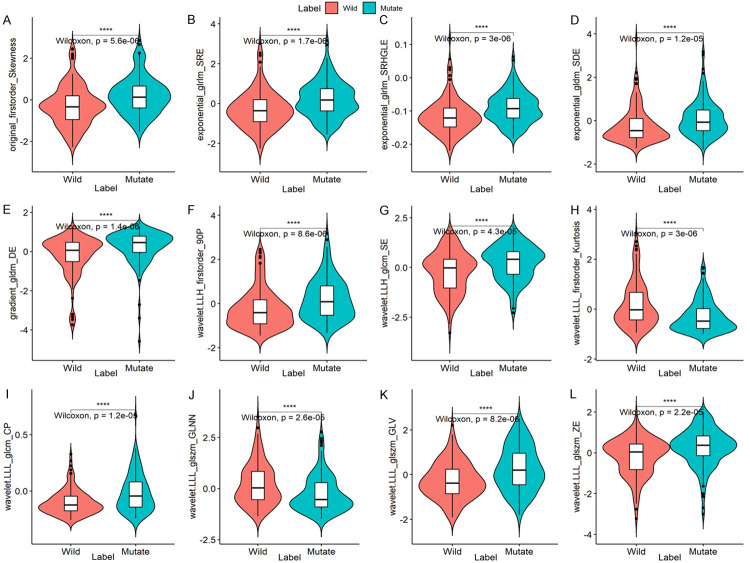
A total of 1468 radiomics features were successfully extracted from each patient’s VOI. In order to establish radiological markers, we first performed univariate analysis in the development cohort. According to Bonferroni correction, 57 radiomics features were selected, and then simplified to 12 potential predictors by LASSO regression (Table S3). In the development cohort, ‘wavelet.LLH_firstorder_Kurtosis’ and ‘wavelet.LLL_glszm_GLNN’ were significantly lower while the other 10 radiomics features were significantly higher in the EGFR mutant group than in the wild-type group (Figure 2). In the development cohort, the 12 radiomic features selected above were used to construct three different radiomics models for predicting EGFR mutations using LR, DT, and SVM classifiers. The correlation analysis of 12 radiomics features with one clinical variable and three radiological features is shown in Figure 3.
Figure 2.
Boxplot showing 12 radiomics features that are significantly different between the EGFR mutant and wild groups in the development cohort.
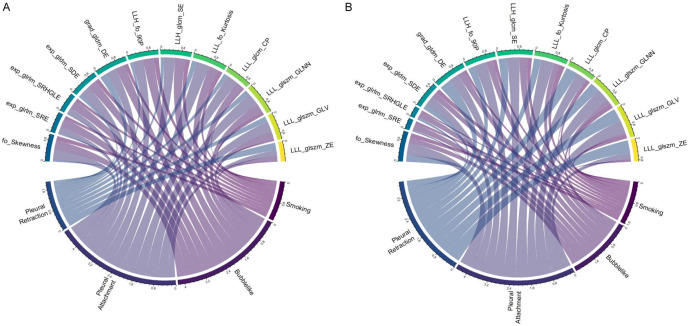
Figure 3.
Chord diagram of the correlation between a clinical variable and three radiological and 12 radiomics features. Correlation analysis between selected radiomics features and clinical and radiological features in the development (A) and validation (B) cohorts. Pearson’s correlation analysis showed that each link is significant correlation (P < 0.05). The width of the link indicates the relative strength. For example, pleural attachment was significantly correlated with LLL_glszm_GLNN and LLL_firstorder_Kurtosis in the development and validation cohorts.
Radiomics signature predictive performance and validation
The results of the radiomics signature and C-R model in the two cohorts are shown in Table 1 and Figure 4. The AUC values of SVM in the two cohorts [0.810 (95% CI, 0.761-0.859) and 0.796 (95% CI, 0.717-0.876), respectively] were higher than those of the other two classifiers (LR and DT). The AUC values of LR in the two cohorts were 0.783 (95% CI, 0.731-0.836) and 0.778 (95% CI, 0.695-0.862), and those of DT were 0.768 (95% CI, 0.716-0.819) and 0.761 (95% CI, 0.679-0.844). The AUC values of the C-R model in the two cohorts were 0.776 (95% CI, 0.724-0.828) and 0.739 (95% CI, 0.652-0.826), respectively.
Table 1.
Performance of the radiomics signature, C-R and C-R-R models
| AUC | Accuracy | Sensitivity | Specificity | |
|---|---|---|---|---|
| Development | ||||
| LR | 0.783 (0.731-0.836) | 0.728 (0.673-0.778) | 0.774 (0.664-0.849) | 0.682 (0.514-0.743) |
| DT | 0.768 (0.716-0.819) | 0.731 (0.677-0.781) | 0.637 (0.496-0.726) | 0.824 (0.695-0.877) |
| SVM | 0.810 (0.761-0.859) | 0.748 (0.695-0.797) | 0.726 (0.568-0.801) | 0.770 (0.648-0.831) |
| C-R | 0.776 (0.724-0.828) | 0.711 (0.655-0.762) | 0.720 (0.675-0.794) | 0.608 (0.455-0.693) |
| C-R-R | 0.849 (0.805-0.893) | 0.786 (0.734-0.831) | 0.808 (0.685-0.870) | 0.764 (0.642-0.845) |
| Validation | ||||
| LR | 0.778 (0.695-0.862) | 0.738 (0.652-0.812) | 0.773 (0.560-0.924) | 0.700 (0.533-0.834) |
| DT | 0.761 (0.679-0.844) | 0.762 (0.678-0.833) | 0.652 (0.368-0.731) | 0.883 (0.624-0.953) |
| SVM | 0.796 (0.717-0.876) | 0.746 (0.661-0.819) | 0.742 (0.499-0.909) | 0.750 (0.583-0.850) |
| C-R | 0.739 (0.652-0.826) | 0.714 (0.627-0.791) | 0.712 (0.448-0.803) | 0.717 (0.467-0.817) |
| C-R-R | 0.835 (0.761-0.909) | 0.802 (0.721-0.867) | 0.773 (0.485-0.879) | 0.833 (0.617-0.917) |
AUC, area under the curve; C-R, clinical-radiological; C-R-R, clinical-radiological-radiomics; DT, decision tree; LR, logistic regression; SVM, support vector machine.
Figure 4.

Receiver operating characteristic (ROC) curves from LG, DT classifier, SVM classifiers, C-R model, and C-R-R model used to predict EGFR mutations in development (A) and validation (B) cohorts. C-R, clinical-radiological; C-R-R, clinical-radiological-radiomics; DT, decision tree; LR, logistic regression; SVM, support vector machine.
The C-R-R model developed by combining the radiomics signature derived from the higher diagnostic efficiency of SVM and the C-R model produced higher AUC values in the development and validation cohorts: 0.849 (95% CI, 0.805-0.893) and 0.835 (95% CI, 0.761-0.909), respectively. In addition, the C-R-R model had the best discriminative ability in both cohorts, with sensitivities of 0.808 (95% CI, 0.685-0.870) and 0.773 (95% CI, 0.485-0.879) and specificities of 0.764 (95% CI, 0.642-0.845) and 0.833 (95% CI, 0.617-0.917). In the development cohort, the scores of the radiomics, C-R model, and C-R-R model were significantly higher in the EGFR mutant group than in the wild-type group. This result was confirmed in the validation cohort (Figure 5).
Figure 5.
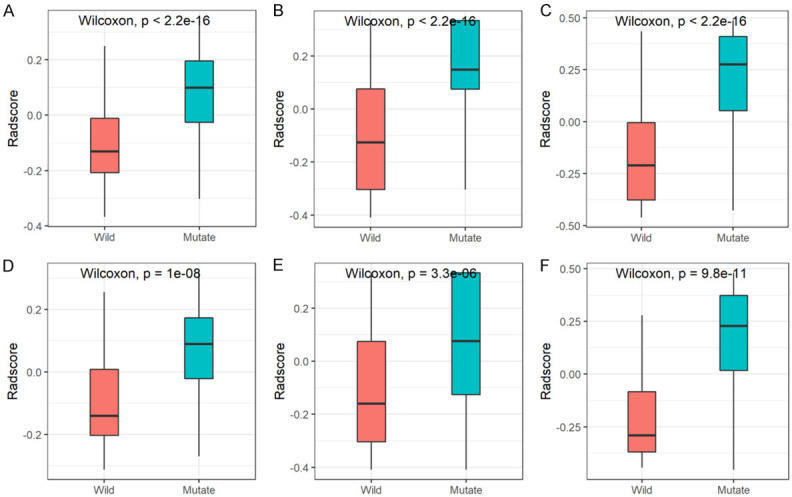
Boxplot showing the comparison of radiomics scores from the SVM classifier (A, D), C-R model (B, E), and C-R-R model (C, F) in the development (A-C) and validation (D-F) cohorts. C-R, clinical-radiological; C-R-R, clinical-radiological-radiomics; SVM, support vector machine.
Based on the above results, the performance of the C-R-R model in the development cohort was significantly better than that of the SVM classifier (AUC, 0.81; P = 0.008), LR classifier (AUC, 0.783; P < 0.001), DT classifier (AUC, 0.768; P < 0.001), and C-R model (AUC, 0.776; P < 0.001). Similarly, the C-R-R model had the best performance in the validation cohort, which was significantly different from that of the C-R model (P = 0.004) but not from that of the three classifiers (P > 0.05). In short, the C-R-R model had a higher predictive value for EGFR mutations. In both cohorts, the AUC value, accuracy, and sensitivity of the C-R-R model in predicting the EGFR mutation status were higher than other classifiers and the C-R model, and the effect of the LR classifier was better than that of the DT classifier and the C-R model.
Clinical application of the C-R-R model
A nomogram was constructed based on the radiomics model with the highest AUC value in combination with one clinical variable and three radiological features. Among the five models, the radiomics nomogram had the best discriminating ability (Figures 4 and 6A).
Figure 6.
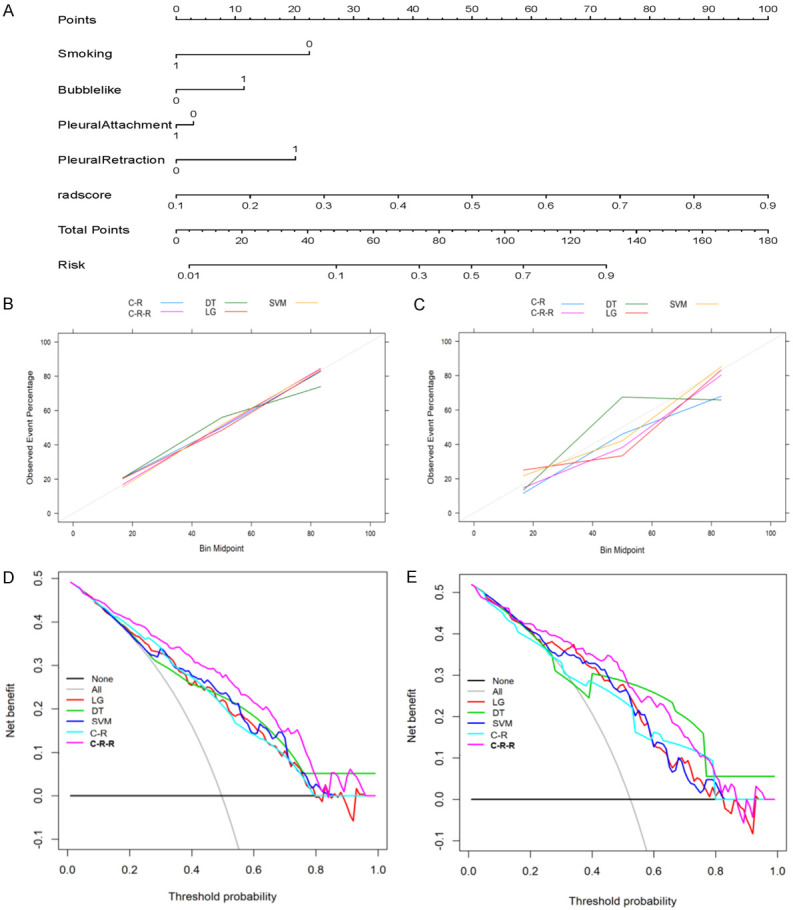
Building and performance of the C-R-R model. (A) The nomogram developed based on the C-R-R model. (B, C) Calibration curves of different classifiers and models generated from the development (B) and validation (C) cohorts. The goodness of fit of the predicted probability from different classifiers and models with the actual outcomes of EGFR mutations was evaluated. The x-axis represents the EGFR mutation probability calculated by different classifiers and models, and the y-axis represents the actual EGFR mutation probability. The diagonal line represents the ideal estimate of the ideal model. (D, E) Decision curves of different classifiers and models generated from the development (D) and validation (E) cohorts. The x-axis shows the threshold probability, and the y-axis measures the net income. C-R-R, clinical-radiological-radiomics.
The calibration curve revealed that the probability of predicting EGFR mutations by the nomogram was in good agreement with the actual probability (Figure 6B, 6C). The Hosmer-Lemeshow test was applied to the SVM classifier, C-R model, and C-R-R model with the best discrimination ability in the development and validation cohorts. The values of the SVM classifier in the two cohorts were 0.625 and 0.251, those of the C-R model were 0.646 and 0.111, and those of the C-R-R model were 0.426 and 0.313, respectively, indicating that the C-R-R model did not deviate from the perfect fit in both cohorts (Figure S4).
The decision curve was used to evaluate the clinical performance of different classifiers and models from the perspective of clinical application, thereby reflecting the clinical applicability of different classifiers and models (Figure 6D, 6E). The area under the decision curve of the C-R-R model was larger than that of other models, indicates that the model had the best clinical utility. When the threshold probability was in the range of 9%-83%, the C-R-R model provides net benefits.
Discussion
In this study, we constructed a C-R-R model based on the combination of a CT radiomics signature with clinical and radiological features to predict the EGFR mutation status of lung adenocarcinoma using a relatively large data set. We used three classifiers (LR, DT, and SVM) to calculate the diagnostic performance of radiomic signatures, and finally selected the SVM classifier with the highest AUC value. The C-R-R model was developed based on data from 294 patients. To further verify its diagnostic performance, we evaluated the model in a validation cohort of 129 patients. In the development and validation cohorts, the AUC values of the C-R-R model were high (0.849 and 0.835, respectively). Therefore, this model may help determine the EGFR mutation status of lung adenocarcinoma to guide personalized targeted therapy.
The EGFR mutation rate in this study was 50.5% (212/420), which was consistent with that in previous studies [10,14,15,33]. In the development and validation cohorts, the rate was 50.3% (148/294) and 50.8% (64/126), respectively. Our study found that sex and smoking history were significantly related to the EGFR mutation status: women and non-smokers were more likely to have EGFR mutations. In addition, our findings showed that 10 radiological features were significantly related to the EGFR mutation status (P < 0.05). For example, bubble-like lucency was more common in patients with EGFR mutations, while pleural attachment was more common in wild-type patients, which was consistent with previous studies [10,14]. In multivariate regression analysis, smoking history, bubble-like lucency, pleural attachment, and pleural retraction were independently correlated with EGFR mutation status (P < 0.05), while sex and seven other radiological features were not significantly different between the two groups (P > 0.05).
To construct the radiomics signature, we screened 12 independent features highly correlated with EGFR mutations from 1468 candidate features, which were stable in the validation cohort. Since most of the selected radiomics features (11/12) were extracted from the filtered image, among which seven features were obtained through wavelet transform, so the texture and high-dimensional features were more strongly correlated with the EGFR mutation status. Wavelet transform is currently one of the commonly used methods for signal processing, such as noise elimination, and data smoothing and filtering. It can smooth the image and improve the ability to acquire features related to tumor heterogeneity [31]. Texture features cannot be recognized by the human visual system, nor can they be understood as specific meanings [34,35]. Our results showed that the radiomics features, including original_firstorder_Skewness, exponential_glrlm_SRE, and wavelet.LLL_glszm_ZE, were higher in the EGFR mutation group and were significantly related to EGFR mutations. Among them, exponential_glrlm_SRE was related to run length and fine textural texture. A larger value represented a shorter run length and a finer textural texture [36,37], while wavelet.LLL_glszm_ZE was related to texture heterogeneity; a higher value represented more extensive heterogeneity in texture patterns [38]. However, the features of wavelet.LLL_firstorder_Kurtosis and wavelet.LLL_glszm_GLNN were more extensive in the wild-type group, and were significantly correlated with wild-type EGFR. Among them, wavelet.LLL_firstorder_Kurtosis was related to the “peakedness” in the image ROI. A higher kurtosis value implies that the quality of the distribution was concentrated in the tail [39], and wavelet.LLL_glszm_GLNN was related to the gray value in the image. The lower the value, the greater the similarity of the intensity value [38]. This indicated that the finer the texture, the greater the heterogeneity within the tumor and the greater the variability of gray values, alongside the higher risk of EGFR mutations. These findings are consistent with those of a previous study [11]. Moreover, it also shows that these features are closely related to radiological characteristics. For example, in the development and validation cohorts, pleural attachment was significantly correlated with features such as LLL_glszm_GLNN and LLL_firstorder_Kurtosis, pleural retraction was significantly correlated with features such as LLL_glszm_ZE and LLL_glszm_GLV, bubbe-like was significantly correlated with features such as LLL_glcm_SE and grad_gldm_DE, and smoking was significantly correlated with features such as exp_gldm_SDE and fo_Skewness. The correlation between these features indicated that although texture-based features are not observable to the naked eye, specific combinations of several texture features can to some extent be explained by certain radiological features.
Medical images are routinely collected in the hospital during the diagnosis and treatment of patients with cancer, and the genotypes of tumors can be reflected to a certain extent by evaluating the characteristics of these images. For example, some studies have found that radiological features can predict the state of EGFR mutation status [10,14,15]; however, these studies have selected a large number of radiological features related to EGFR mutations, which will undoubtedly increase the clinical workload; and using a large number of features is associated with an increased risk of error. In addition, some studies have investigated the association between radiomics features and tumor genotypes [11,19-25]. However, these studies usually have a small sample size or low diagnostic efficiency, and more importantly, some studies have lack validation, which is an important part of radiomics analysis [18,40]. Our study selected the independent predictors most associated with EGFR mutation status through multivariate analysis of clinical and radiological features. The radiological features included in the model were mainly because certain features were related to the EGFR mutation status. However, when manually delineating the tumor ROI, a small part of these radiological features, such as pleural depression and fine spiculation, were too small to be included in the ROI. Therefore, the omission of these features may lead to the reduction of feature information, which cannot fully reflect the heterogeneity of the entire tumor. Our research showed that the AUC of the model containing only the radiomics signature was 0.81, while the AUC of the model containing clinical variables and radiological features (C-R model) was only 0.776, and the AUC of the fusion model (C-R-R model) was increased to 0.849. This demonstrated that the inclusion of clinical variables and radiological features in the model can improve the diagnostic performance of the model. In addition, the diagnostic performance of the radiomics model was better than that of the C-R model.
To our knowledge, this is the first report on prediction of the EGFR mutation status using a relatively large data set based on CT radiomics signature combined with clinical and radiological features. Three different classifiers were applied to evaluate the performance of the radiomic model, and finally the SVM classifier with the highest diagnostic performance was selected. Yang et al. [11] have reported that a model based on the CT radiomics signature can be used to predict the EGFR mutation status of patients with lung adenocarcinoma, with an AUC of 0.826 in the training cohort and only 0.779 in the validation cohort. Velazquez and colleagues [20] used the CT radiomics signature combined with clinical variables to predict the EGFR mutation status, and the AUC obtained only 0.75, and lack of verification, limiting its clinical applicability. In this study, the proposed C-R-R model shows good prediction performance in the development cohort (AUC, 0.849), and is well calibrated and stable (AUC, 0.835) in the verification cohort. Moreover, the DCA confirmed the clinical usefulness of the C-R-R model.
TKI therapy can provide significant clinical benefits to patients with EGFR mutations. Compared to treatment with platinum-based chemotherapy alone, approximately 70% of patients receiving TKI therapy experience symptom alleviation, improved quality of life, and prolonged progression-free survival [3,41,42]. Therefore, determining the EGFR mutation status is a prerequisite for receiving targeted therapy. Although image analysis cannot replace histological examination, it can provide additional information and help identify high-risk patients with EGFR mutations [3,10]. For example, when radiomics predicts a high possibility of EGFR mutation in patients with false-negative histological results, the tumors should be re-sampled for biopsy; otherwise, these patients will not benefit from targeted therapy [10]. Similarly, in patients with multiple tumors, radiomics can be used to select the most suspicious tumor for biopsy [43]. Therefore, when analysis based on histological examination is not feasible, imaging-based biomarkers can potentially be used in clinical practice.
There are several limitations to this study. First, like any other retrospective study, the results of this study may be affected by selection bias, and we encourage researchers from more centers to use different CT scanners and parameters to prospectively verify our findings. Second, this study was limited to the analysis of lung adenocarcinoma and did not involve other pathological subtypes because most EGFR mutations are present in adenocarcinoma. Finally, the 3D delineation of all tumors in this study was performed manually by a radiologist; in future studies, tumors may be effectively segmented automatically.
In conclusion, preoperative prediction of the EGFR mutation status will help guide individualized targeted therapy. On radiomics analysis, 12 radiomic features were highly correlated with EGFR mutations, which was confirmed in the validation cohort. A C-R-R model was constructed based on CT radiomic features combined with the clinical variables and radiological features. The model showed excellent diagnostic performance and high sensitivity in predicting EGFR mutation status and could provide useful image-based biological information for patients eligible for targeted therapy.
Acknowledgements
This work was supported by the Talent Innovation and Entrepreneurship Project of Lanzhou (grant number 2016-RC-58); Open Fund project of Key Laboratory of Medical Imaging of Gansu Province (grant number GSYX202010).
Disclosure of conflict of interest
None.
Abbreviations
- AUC
area under the curve
- C-R
clinical-radiological
- C-R-R
clinical-radiological-radiomics
- CT
computed tomography
- DT
decision tree
- EGFR
epidermal growth factor receptor
- LR
logistic regression
- LASSO
least absolute shrinkage and selection operator
- PACS
picture archiving and communication systems
- ROI
region of interest
- TKI
tyrosine kinase inhibitors
- SVM
support vector machine
- VOI
volume of interest
Supporting Information
References
- 1.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, Li W, Hou M, Shi JH, Lee KY, Xu CR, Massey D, Kim M, Shi Y, Geater SL. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 3.Wu SG, Shih JY. Management of acquired resistance to EGFR TKI-targeted therapy in advanced non-small cell lung cancer. Mol Cancer. 2018;17:38. doi: 10.1186/s12943-018-0777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, de Castro J, Milella M, Reguart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Muñoz-Langa J, Valdivia J, Isla D, Domine M, Molinier O, Mazieres J, Baize N, Garcia-Campelo R, Robinet G, Rodriguez-Abreu D, Lopez-Vivanco G, Gebbia V, Ferrera-Delgado L, Bombaron P, Bernabe R, Bearz A, Artal A, Cortesi E, Rolfo C, Sanchez-Ronco M, Drozdowskyj A, Queralt C, de Aguirre I, Ramirez JL, Sanchez JJ, Molina MA, Taron M, Paz-Ares L. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 5.Yang JC, Hirsh V, Schuler M, Yamamoto N, O’Byrne KJ, Mok TS, Zazulina V, Shahidi M, Lungershausen J, Massey D, Palmer M, Sequist LV. Symptom control and quality of life in LUX-Lung 3: a phase III study of afatinib or cisplatin/pemetrexed in patients with advanced lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 2013;31:3342–3350. doi: 10.1200/JCO.2012.46.1764. [DOI] [PubMed] [Google Scholar]
- 6.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 7.Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, Su WC, Bennouna J, Kato T, Gorbunova V, Lee KH, Shah R, Massey D, Zazulina V, Shahidi M, Schuler M. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 8.Ettinger DS, Wood DE, Aggarwal C, Aisner DL, Akerley W, Bauman JR, Bharat A, Bruno DS, Chang JY, Chirieac LR, D’Amico TA, Dilling TJ, Dobelbower M, Gettinger S, Govindan R, Gubens MA, Hennon M, Horn L, Lackner RP, Lanuti M, Leal TA, Lin J, Loo BW Jr, Martins RG, Otterson GA, Patel SP, Reckamp KL, Riely GJ, Schild SE, Shapiro TA, Stevenson J, Swanson SJ, Tauer KW, Yang SC, Gregory K OCN. Hughes M. NCCN guidelines insights: non-small cell lung cancer, version 1.2020. J Natl Compr Canc Netw. 2019;17:1464–1472. doi: 10.6004/jnccn.2019.0059. [DOI] [PubMed] [Google Scholar]
- 9.Sacher AG, Dahlberg SE, Heng J, Mach S, Jänne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol. 2016;2:313–320. doi: 10.1001/jamaoncol.2015.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Kim J, Qu F, Liu S, Wang H, Balagurunathan Y, Ye Z, Gillies RJ. CT features associated with epidermal growth factor receptor mutation status in patients with lung adenocarcinoma. Radiology. 2016;280:271–280. doi: 10.1148/radiol.2016151455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Dong X, Wang J, Li W, Gu Z, Gao D, Zhong N, Guan Y. Computed tomography-based radiomics signature: a potential indicator of epidermal growth factor receptor mutation in pulmonary adenocarcinoma appearing as a subsolid nodule. Oncologist. 2019;24:e1156–e1164. doi: 10.1634/theoncologist.2018-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, Colasacco C, Dacic S, Hirsch FR, Kerr K, Kwiatkowski DJ, Ladanyi M, Nowak JA, Sholl L, Temple-Smolkin R, Solomon B, Souter LH, Thunnissen E, Tsao MS, Ventura CB, Wynes MW, Yatabe Y. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13:323–358. doi: 10.1016/j.jtho.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell RL, Kosche C, Burgess K, Wadhwa S, Buckingham L, Ghai R, Rotmensch J, Klapko O, Usha L. Misdiagnosis of Li-fraumeni syndrome in a patient with clonal hematopoiesis and a somatic TP53 mutation. J Natl Compr Canc Netw. 2018;16:461–466. doi: 10.6004/jnccn.2017.7058. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa M, Sakai F, Ishikawa R, Kimura F, Ishida H, Kobayashi K. CT features of epidermal growth factor receptor-mutated adenocarcinoma of the lung: comparison with nonmutated adenocarcinoma. J Thorac Oncol. 2016;11:819–826. doi: 10.1016/j.jtho.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Choi CM, Kim MY, Hwang HJ, Lee JB, Kim WS. Advanced adenocarcinoma of the lung: comparison of CT characteristics of patients with anaplastic lymphoma kinase gene rearrangement and those with epidermal growth factor receptor mutation. Radiology. 2015;275:272–279. doi: 10.1148/radiol.14140848. [DOI] [PubMed] [Google Scholar]
- 16.Suh YJ, Lee HJ, Kim YJ, Kim KG, Kim H, Jeon YK, Kim YT. Computed tomography characteristics of lung adenocarcinomas with epidermal growth factor receptor mutation: a propensity score matching study. Lung Cancer. 2018;123:52–59. doi: 10.1016/j.lungcan.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 17.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 18.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Chen B, Liu X, Song J, Fang M, Hu C, Dong D, Li W, Tian J. Quantitative biomarkers for prediction of epidermal growth factor receptor mutation in non-small cell lung cancer. Transl Oncol. 2018;11:94–101. doi: 10.1016/j.tranon.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rios Velazquez E, Parmar C, Liu Y, Coroller TP, Cruz G, Stringfield O, Ye Z, Makrigiorgos M, Fennessy F, Mak RH, Gillies R, Quackenbush J, Aerts H. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77:3922–3930. doi: 10.1158/0008-5472.CAN-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Kim J, Balagurunathan Y, Li Q, Garcia AL, Stringfield O, Ye Z, Gillies RJ. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer. 2016;17:441–448. e446. doi: 10.1016/j.cllc.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tu W, Sun G, Fan L, Wang Y, Xia Y, Guan Y, Li Q, Zhang D, Liu S, Li Z. Radiomics signature: a potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer. 2019;132:28–35. doi: 10.1016/j.lungcan.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 23.Mei D, Luo Y, Wang Y, Gong J. CT texture analysis of lung adenocarcinoma: can radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging. 2018;18:52. doi: 10.1186/s40644-018-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Ding C, Zhang H, Song J, Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys. 2019;46:4545–4552. doi: 10.1002/mp.13747. [DOI] [PubMed] [Google Scholar]
- 25.Hong D, Xu K, Zhang L, Wan X, Guo Y. Radiomics signature as a predictive factor for EGFR mutations in advanced lung adenocarcinoma. Front Oncol. 2020;10:28. doi: 10.3389/fonc.2020.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 27.Meng X, Xia W, Xie P, Zhang R, Li W, Wang M, Xiong F, Liu Y, Fan X, Xie Y, Wan X, Zhu K, Shan H, Wang L, Gao X. Preoperative radiomic signature based on multiparametric magnetic resonance imaging for noninvasive evaluation of biological characteristics in rectal cancer. Eur Radiol. 2019;29:3200–3209. doi: 10.1007/s00330-018-5763-x. [DOI] [PubMed] [Google Scholar]
- 28.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolossváry M, Karády J, Szilveszter B, Kitslaar P, Hoffmann U, Merkely B, Maurovich-Horvat P. Radiomic features are superior to conventional quantitative computed tomographic metrics to identify coronary plaques with napkin-ring sign. Circ Cardiovasc Imaging. 2017;10:e006843. doi: 10.1161/CIRCIMAGING.117.006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oikonomou EK, Williams MC, Kotanidis CP, Desai MY, Marwan M, Antonopoulos AS, Thomas KE, Thomas S, Akoumianakis I, Fan LM, Kesavan S, Herdman L, Alashi A, Centeno EH, Lyasheva M, Griffin BP, Flamm SD, Shirodaria C, Sabharwal N, Kelion A, Dweck MR, Van Beek EJR, Deanfield J, Hopewell JC, Neubauer S, Channon KM, Achenbach S, Newby DE, Antoniades C. A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur Heart J. 2019;40:3529–3543. doi: 10.1093/eurheartj/ehz592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui Y, Liu H, Ren J, Du X, Xin L, Li D, Yang X, Wang D. Development and validation of a MRI-based radiomics signature for prediction of KRAS mutation in rectal cancer. Eur Radiol. 2020;30:1948–1958. doi: 10.1007/s00330-019-06572-3. [DOI] [PubMed] [Google Scholar]
- 32.Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: the Hosmer-Lemeshow test revisited. Crit Care Med. 2007;35:2052–2056. doi: 10.1097/01.CCM.0000275267.64078.B0. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G, Yang PC. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Q, Yao K, Liu Z, Li L, Zhao X, Wang S, Shang H, Lin Y, Wen Z, Zhang X, Tian J, Wang M. Radiomics analysis of placenta on T2WI facilitates prediction of postpartum haemorrhage: a multicentre study. EBioMedicine. 2019;50:355–365. doi: 10.1016/j.ebiom.2019.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Yao K, Liu P, Liu Z, Han T, Zhao Z, Cao Y, Zhang G, Zhang J, Tian J, Zhou J. A radiomics model for preoperative prediction of brain invasion in meningioma non-invasively based on MRI: a multicentre study. EBioMedicine. 2020;58:102933. doi: 10.1016/j.ebiom.2020.102933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu DH, Kurani AS, Furst JD, Raicu DS. Run-length encoding for volumetric texture. 2004 [Google Scholar]
- 37.Tustison N, Gee J. Run-length matrices for texture analysis. Or Insight. 2008 [Google Scholar]
- 38.Thibault G, Fertil B, Navarro C, Pereira S, Mari J. Texture indexes and gray level size zone matrix application to cell nuclei classification. 10th International Conference on Pattern Recognition and Information Processing. 2009 [Google Scholar]
- 39.Lorensen WE, Cline HE. Marching cubes: a high resolution 3D surface construction algorithm. 1987;21:163–169. [Google Scholar]
- 40.Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2:1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- 41.Robichaux JP, Elamin YY, Tan Z, Carter BW, Zhang S, Liu S, Li S, Chen T, Poteete A, Estrada-Bernal A, Le AT, Truini A, Nilsson MB, Sun H, Roarty E, Goldberg SB, Brahmer JR, Altan M, Lu C, Papadimitrakopoulou V, Politi K, Doebele RC, Wong KK, Heymach JV. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med. 2018;24:638–646. doi: 10.1038/s41591-018-0007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sequist LV, Han JY, Ahn MJ, Cho BC, Yu H, Kim SW, Yang JC, Lee JS, Su WC, Kowalski D, Orlov S, Cantarini M, Verheijen RB, Mellemgaard A, Ottesen L, Frewer P, Ou X, Oxnard G. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol. 2020;21:373–386. doi: 10.1016/S1470-2045(19)30785-5. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, Shi J, Ye Z, Dong D, Yu D, Zhou M, Liu Y, Gevaert O, Wang K, Zhu Y, Zhou H, Liu Z, Tian J. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53:1800986. doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.