Abstract
YEATS domain-containing protein 4 (YEATS4) is implicated in several oncogenic signaling pathways, and its expression is involved in various types of cancer; regardless, the pathophysiologic effects of YEATS4 on breast cancer remain unclear. This study finds that YEATS4 is increasingly expressed with breast cancer progression, and its expression is related to poor outcome and distant metastasis. YEATS4 overexpression in breast cancer cells strengthens their malignant characteristics in vitro and in vivo, particularly inducing epithelial-to-mesenchymal transition (EMT) and consequently, metastatic capability in breast cancer cells. By contrast, deleting YEATS4 in breast cancer cells with high-grade malignancy reduced these characteristics. With regard to the molecular mechanism, YEATS4 mediates histone H3K27ac at specific sites of the ZEB1 promoter to regulate its expression at the transcription level. Depleting ZEB1 blocks YEATS4-induced EMT, migration, invasion, and metastasis. YEATS4 expression is also positively correlated with ZEB1 expression in patients with breast cancer. Co-expression of YEATS4 and ZEB1 correlates with the shortest distant metastasis-free period. Taken together, our data reveal the critical role of YEATS4 in the progression and metastasis of breast cancer, as well as support YEATS4 as a potential therapeutic target and prognostic biomarker for breast cancer.
Keywords: YEATS4, ZEB1, breast cancer, EMT, metastasis
Introduction
Breast cancer is one of the most common malignancies with a rising incidence rate and the highest cancer mortality rate in females worldwide. An estimated 2.1 million new cases and 626,679 deaths were reported in 2018 [1,2]. Despite various methods for early diagnosis and multidisciplinary treatment to lower mortality rates [3], 25%-50% of breast cancer patients were predicted to ultimately develop distant metastases and then succumbed after decades of diagnosis and primary tumor resection [4,5].
Treatment modalities for breast cancer metastasis have thus far remained ineffective. Considerable efforts have been directed toward exploring the mechanisms of metastasis to identify a more effective treatment for breast cancer distant metastasis. The mechanism by which, metastasis occurs has yet to be determined. Accumulating evidence indicates that epithelial-to-mesenchymal transition (EMT), marked by loss of epithelial properties and acquisition of mesenchymal phenotypes, endows cancer cells with more aggressive behavior [6-8]. Thus, potential molecules in the adjustment of EMT need to be confirmed to identify new therapeutic targets for breast cancer progression and metastasis.
YEATS domain-containing protein 4 (YEATS4) located in the chromosomal region 12q13-15 has been initially identified in the nucleoli of glioma cells [9]. As a member of the protein family with a characteristic YEATS domain at the N-terminal [10], YEATS4 participates in chromatin modification and transcription regulation by involving the assembly of multisubunit complexes, particularly TIP60/TRRAP and SRCAP complexes [11,12], mediating histone proteins to participate in nucleosome remodeling, changing chromatin composition, and affecting gene transcription [13]. In multiple types of cancer, YEATS4 has been implicated in the progression of several types of cancer, including pancreatic cancer, gastric cancer, colorectal cancer, hepatocellular carcinoma, and non-small cell lung cancer [14-19]. Kim et al. also reported that YEATS4 is one of the key transcription factors inducing chemoresistance via intrinsic apoptosis-related pathways in ovarian cancer [20]. However, the pathophysiologic effects of YEATS4 on breast cancer remain unclear. Moreover, most of these studies have focused on the promoting effect of YEATS4 on cell proliferation. Whether and how YEATS4 contributes to cancer invasion and metastasis remains undetermined.
In the current study, we reveal that YEATS4 overexpression promotes proliferation and EMT in breast cancer cells, leading to aggressive phenotypes in vitro and in vivo. YEATS4 deletion restrains cell growth, results in mesenchymal-to-epithelial transition (MET), and inhibits metastases to distant sites. These effects on biological behavior are achieved by regulating ZEB1 transcription via H3K27 acetylation (H3K27ac). These results show the innovative role of YEATS4 in breast cancer progression and metastasis and provide a rational interpretation of the association of positive YEATS4 expression with progression and poor outcome. Thus, YEATS4 can be used as a potential therapeutic target for breast cancer.
Material and methods
Ethics statement
This experimental research relating to human beings and animals was authorized by the Ethical Review Committee of Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine. Written consent was obtained from each participant before the experiments.
Antibodies and reagents
Antibodies against histone H3, H3K4ac, H3K9ac, H3K14ac, H3K18ac, and H3K27ac were purchased from Abcam. E-cadherin, ZO-1, vimentin, N-cadherin, ZEB1, and GPADH were supplied by Cell Signaling Technology. YEATS4 were provided by Sigma-Aldrich. TRizol LS Reagent was purchased from Invitrogen. X-tremeGENE™ HP DNA Transfection Reagent was supplied by Roche.
Patients and specimens
The retrospective cohort consisted of 481 patients with stages I-III breast cancer who were diagnosed and enrolled in our research from 2005 to 2009 at the Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China. The total number included 42 cases of ductal carcinoma in situ (DCIS) and 439 cases of invasive carcinoma (IC), as well as 139 paired adjacent normal tissues (ANTs). Notably, 197 patients had lymph node metastasis in this retrospective cohort.
Overall survival (OS) is defined as the number of days after surgery to death from any cause. Distant metastasis-free survival (DMFS) is defined as the period after surgery to the date of diagnosis of distant metastasis derived from breast cancer. The follow-up deadline is the date of death, date of emigration, or September 30, 2019.
The following were the inclusion criteria: (1) Patients were diagnosed with unilateral breast cancer, which was pathologically confirmed. ANTs were pathologically sampled more than 5 cm from the margin of the tumor. (2) Complete clinicopathologic and follow-up data were available. The following were the exclusion criteria: (1) Patients with preoperative anticancer treatment such as radiotherapy, chemotherapy, and hormonal therapy were excluded. (2) Patients with severe complicating diseases such as heart and cerebral vascular diseases or other primary or familial malignancies were ruled out.
Immunohistochemistry
Sections of whole-tissue blocks from 481 breast cancer tissues and 139 ANTs were sliced into 4 µm sections and baked in a heat chamber for 1 h at 60°C. Deparaffination and antigen retrieval were conducted using the PT Link system (Agilent/Dako A/S). Slides were then immunoreacted for 1 h with the anti-YEATS4 primary antibody (diluted at 1:100; Sigma-Aldrich) or the anti-ZEB1 primary antibody (diluted at 1:100; Cell Signaling Technology) and then with a secondary antibody for 30 min. The OptiView DAB IHC Detection Kit (Ventana Medical Systems) was used for chromogen detection. Hematoxylin was used for counterstaining. The staining intensities and percentages of the positive cells were evaluated by 2 senior pathologists blinded to patient medical information. The staining intensity was graded as 0 (no stain), 1 (weak), 2 (moderate), or 3 (strong). The percentage of positive cancer cells was graded as 0 (< 5%), 1 (5%-25%), 2 (26%-50%), 3 (51%-75%), or 4 (> 75%). YEATS4 or ZEB1 expression was determined based on the semiquantitative immunoreactive score (IRS) [21], which was determined by multiplying the staining intensity with the percentage of positive cells. Slices with scores ranging from 0 to 5 were classified as negative for YEATS4 and ZEB1 expression, whereas those with scores from 6 to 12 were regarded as positive for YEATS4 and ZEB1 expression.
Cell lines and culture conditions
Human breast cancer cell lines MDA-MB-231, BT-474, MCF-7, ZR-75-1, T-47D, SK-BR-3, and MDA-MB-436 and human embryonic kidney cells 293T were obtained from the National Infrastructure of Cell Line Resource (Shanghai, China). The aforementioned cell lines were detected to ensure the absence of mycoplasma contamination. MDA-MB-231 and MDA-MB-436 cells were grown in Leibovitz’s L-15 Medium (ATCC), BT-474, ZR-75-1, T-47D, and SK-BR-3 cells in RPMI 1640 (Gibco), MCF-7 cells in Eagle’s Minimum Essential Medium (ATCC), and 293T cells in DMEM (Gibco). A base culture medium was added with fetal bovine serum (final concentration: 10%; Gibco), 1% penicillin-streptomycin, and 2 mM L-glutamine. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2.
Lentiviral construction and cell infection
Lentiviral construction and cell infection assays are described in detail in Supplementary materials and methods. The shRNA sequences are listed in Table S1.
RNA isolation and real-time quantitative polymerase chain reaction
Total RNA was extracted using TRIzolTM Reagent (Invitrogen) in accordance with the protocol. Real-time quantitative polymerase chain reaction (RT-qPCR) was conducted using SYBRTM Green PCR Master Mix (Applied BiosystemTM), following the instructions provided by the manufacturer. The primer sequences for RT-qPCR are listed in Table S1.
Cell proliferation assays
Cell proliferation assays were conducted as elaborately described in Supplementary materials and methods.
Colony formation and soft agar colony formation assays
Colony formation and soft agar colony formation assays are described in Supplementary materials and methods.
Wound healing assays
Wound healing assays are described in Supplementary materials and methods.
Transwell assays
Transwell-24 well plates (Corning) with a chamber pore size measuring 8 µm were used to perform cell migration and invasion assays. Procedures are elaborately described in Supplementary materials and methods.
Western blot analysis
Western blot analysis was conducted as described in Supplementary materials and methods.
Immunofluorescence
Immunofluorescence assays are described in detail in Supplementary materials and methods.
Dual luciferase reporter assays
Dual luciferase reporter assays are elaborately described in Supplementary materials and methods.
Chromatin immunoprecipitation assays
Cells were fixed and cross-linked with 1% formaldehyde solution for 20 min at room temperature, and cross-linking was terminated with 125 mM glycine. The nuclear material was sheared using a sonicator system (QSONICA). The chromatin preparation solution was incubated with rotation at 4°C for 1 h with 50 µL of Dynabeads Protein G magnetic beads (Invitrogen) that had been bound with 2-4 µg of the indicated antibodies. Chromatin immune complexes were washed sequentially with low-salt, high-salt, and LiCl buffers. Eluted DNA was purified using the PCR Purification Kit (Thermo Scientific) and analyzed by RT-qPCR using the SYBR Green Master Mix (Bio-Rad). The primer sequences used are presented in Table S1.
In vivo tumorigenicity and metastasis assays
In vivo tumorigenicity and metastasis assays are elaborately described in Supplementary materials and methods.
Statistical analysis
All experiments in this study were independently performed three times under similar conditions. The data were statistically analyzed using IBM SPSS 22.0 and GraphPad Prism 8.0. The relationship between YEATS4 and the clinicopathological characteristics were examined using the χ2 test. Different groups were compared, and the differences were analyzed using Student’s t test or one-way ANOVA. The difference in IRS between two groups was analyzed using the Mann-Whitney test. Survival data were analyzed using the Kaplan-Meier method and the log-rank test. The effect of YEATS4 and other clinicopathologic factors on OS and DMFS were evaluated by univariate and multivariate Cox proportional hazard regression analyses. The cBioPortal database was used to analyze the correlation between YEATS4 and core-EMT genes on the basis of the mRNA expression (http://cbioportal.org/) [22]. The P value and R-value were calculated by cBioPortal. P < 0.05 was considered statistically significant.
Results
YEATS4 is highly expressed in breast cancer tumors
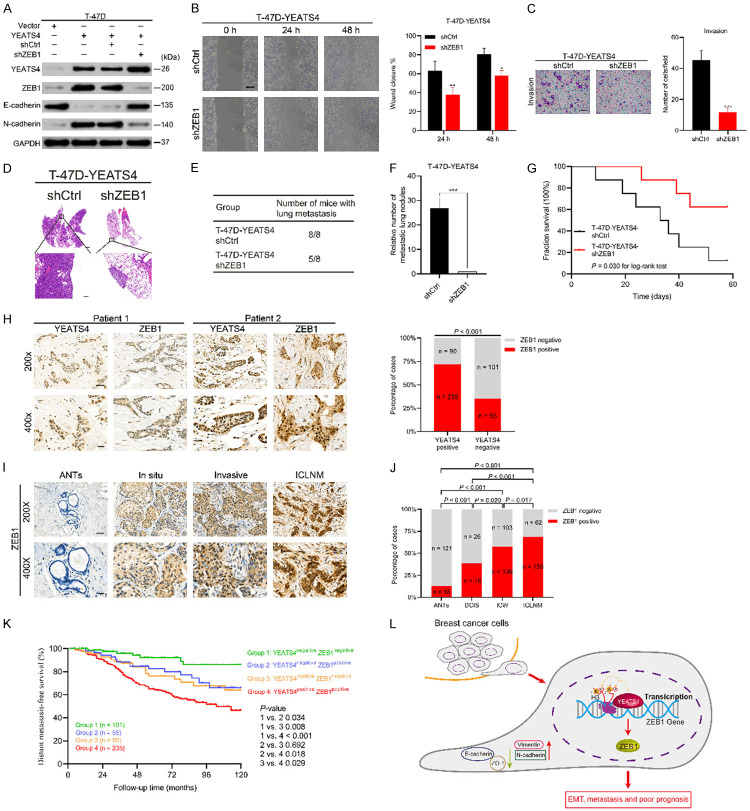
A total of 481 patients pathologically diagnosed with primary breast cancer from a retrospective cohort were enrolled in this study. All patients were female with a median age of 52 years (range 22-84 years). The median follow-up time was 123 months. Up to 41.0% of cases were diagnosed with lymph node metastasis during the postoperative pathological examination.
YEATS4 expression in all samples was determined using an antibody previously reported specifically for YEATS4 by immunohistochemistry (IHC) [16]. The reactivity was mainly recognized in the nucleus of breast cancer cells, which was basically in line with previous findings [15]. YEATS4 expression was positive in 325 (67.6%) of the 481 evaluable primary breast cancer tissues, whereas only 15.1% (21/139) of ANTs showed positive immunoreactivity for YEATS4. A significant difference in YEATS4 staining was found between ANTs and breast cancer tissues (Figure 1A). The median IRS of YEATS4 staining was markedly higher in breast cancer than in ANTs (Figure 1B). Representative images of YEATS4 staining in breast cancer tissues are presented in Figure 1C. Of the 481 patients, 68.6% (330) were estrogen receptor (ER)-positive, consisting of 6.1% (20) DCIS and 93.9% (310) IC. Notably, the median YEATS4 staining IRS was markedly higher in the ER-positive group than in the ER-negative group in breast cancer (Figure 1D), suggesting that YEATS4 was related to ER expression in breast cancer. The combined data indicate that YEATS4 expression was higher in breast cancer, particularly in the ER-positive group.
Figure 1.
YEATS4 is positively expressed in breast cancer. A. Proportion of positive YEATS4 expression in breast cancer tissues compared with that of adjacent normal tissues (ANTs) (P < 0.001). B. Median immunoreactive score (IRS) of YEATS4 staining in breast cancer tissues, which is significantly higher than that in ANTs (P < 0.001). C. Representative immunohistochemistry (IHC) imaging of different YEATS4 staining intensities in breast cancer tissues. Scale bar, 50 µm. D. Median IRS of YEATS4 staining being higher in ER-positive than ER-negative breast cancer tissues (P = 0.028).
YEATS4 expression is correlated with clinicopathologic features
Correlations between YEATS4 expression and clinicopathological parameters in breast cancer were analyzed (Table 1). YEATS4 positive expression correlated with pathologically large tumor size (χ2 = 8.107, P = 0.017), ER expression (χ2 = 5.356, P = 0.021), lymph node metastasis (χ2 = 6.520, P = 0.011), and advanced TNM stage (χ2 = 10.566, P = 0.005). For the molecular subtype, 73.6% (67/91) of breast cancer tissues showed positive YEATS4 expression in the Luminal A subtype, 69.9% (186/266) in the Luminal B subtype, 59.0% (46/78) in the HER2 type, and 56.5% (26/46) in the triple-negative breast cancer subtype. However, no significant difference in YEATS4 expression was found among several molecular subtypes of breast cancer (χ2 = 7.388, P = 0.061). YEATS4 expression showed no significant association with other clinicopathologic features, such as age at diagnosis, menopausal status, histologic grade, PR and HER2 expression, Ki-67 status, and P53 status. To summarize, positive YEATS4 expression was markedly related to pathologic tumor size, ER expression, lymph node involvement, and TNM staging.
Table 1.
Association between baseline characteristics and YEATS4 expression
| Clinicopathological criteria | Expression of YEATS4 | χ2 | P-value | ||
|---|---|---|---|---|---|
|
| |||||
| Positive (n = 325) | Negative (n = 156) | Total (n = 481) | |||
| Age at diagnosis (years) | |||||
| ≤ 35 | 59 (74.7%) | 20 (25.3%) | 79 | 2.184 | 0.139 |
| > 35 | 266 (66.2%) | 136 (33.8%) | 402 | ||
| Pathologic tumor size (cm) | |||||
| T1 | 93 (59.2%) | 64 (40.8%) | 157 | 8.107 | 0.017 |
| T2 | 201 (70.8%) | 83 (29.2%) | 284 | ||
| T3 | 31 (77.5%) | 9 (22.5%) | 40 | ||
| Menopausal status | |||||
| Premenopausal | 138 (65.7%) | 72 (34.3%) | 210 | 0.584 | 0.445 |
| Postmenopausal | 187 (69.0%) | 84 (31.0%) | 271 | ||
| Histological grade | |||||
| 1 | 50 (75.8%) | 16 (24.2%) | 66 | 4.426 | 0.219 |
| 2 | 135 (63.4%) | 78 (36.6%) | 213 | ||
| 3 | 114 (70.4%) | 48 (29.6%) | 162 | ||
| Unknown | 26 (65.0%) | 14 (35.0%) | 40 | ||
| ER | |||||
| Positive | 234 (70.9%) | 96 (29.1%) | 330 | 5.356 | 0.021 |
| Negative | 91 (60.3%) | 60 (39.7%) | 151 | ||
| PR | |||||
| Positive | 206 (64.2%) | 105 (35.8%) | 311 | 0.710 | 0.400 |
| Negative | 119 (70.0%) | 51 (30.0%) | 170 | ||
| HER2 | |||||
| Negative | 243 (66.4%) | 123 (33.6%) | 366 | 0.963 | 0.326 |
| Positive | 82 (71.3%) | 33 (28.7%) | 115 | ||
| Lymph node metastasis | |||||
| Negative | 179 (63.0%) | 105 (37.0%) | 284 | 6.520 | 0.011 |
| Positive | 146 (74.1%) | 51 (25.9%) | 197 | ||
| TNM | |||||
| I | 50 (56.2%) | 39 (43.8%) | 89 | 10.566 | 0.005 |
| II | 215 (67.8%) | 102 (32.2%) | 317 | ||
| III | 60 (80.0%) | 15 (20.0%) | 75 | ||
| Ki-67 status | |||||
| ≤ 20% | 102 (61.8%) | 63 (38.2%) | 165 | 3.789 | 0.052 |
| > 20% | 223 (70.6%) | 93 (29.4%) | 316 | ||
| P53 status | |||||
| Positive | 209 (67.9%) | 99 (32.1%) | 308 | 0.944 | 0.624 |
| Negative | 100 (65.8%) | 52 (34.2%) | 152 | ||
| Unknown | 16 (76.2%) | 5 (23.8%) | 21 | ||
| Molecular subtype | |||||
| Luminal A | 67 (73.6%) | 24 (26.4%) | 91 | 7.388 | 0.061 |
| Luminal B | 186 (69.9%) | 80 (30.1%) | 266 | ||
| HER2 type | 46 (59.0%) | 32 (41.0%) | 78 | ||
| TNBC | 26 (56.5%) | 20 (43.5%) | 46 | ||
YEATS4, YEATS domain-containing protein 4; TNM, tumor-node-metastasis; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNBC, triple-negative breast cancer. P-values that reach significance are in bold.
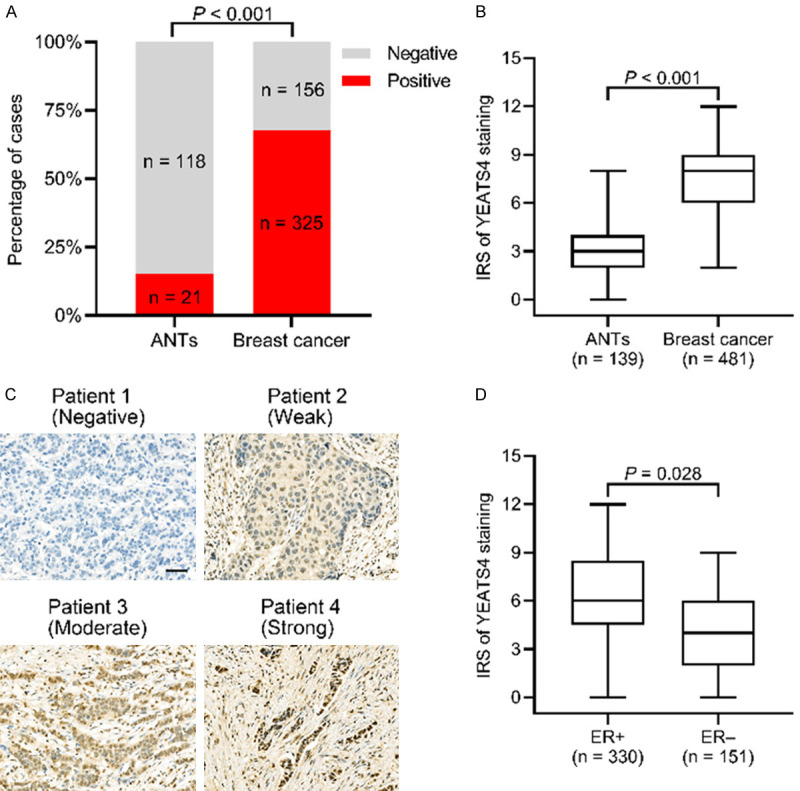
YEATS4 expression is related to breast cancer progression
We examined the YEATS4 expression levels in ANTs (n = 139), DCIS (n = 42), invasive cancer with no lymph node metastasis (ICW, n = 242), and invasive cancer with lymph node metastasis (ICLNM, n = 197). YEATS4 expression was found in 15.1% of ANTs, 47.6% of DCIS, 65.7% of ICW, and 74.1% of ICLNM specimens. Positive YEATS4 expression gradually increased with cancer progression; however, no significant difference was determined between ICW and ICLNM (P = 0.057; Figure 2A). All subtypes in cancer progression showed increased YEATS4 expression relative to that of ANTs (Figure 2A). The IRSs of the ANTs, DCIS, ICW, and ICLNM specimens were gradually increased, and significant differences among the groups were found (Figure 2B). Representative images of the YEATS4 staining of ANTs, DCIS, ICW, and ICLNM specimens are presented in Figure 2C. The YEATS4 expression of the IC subtype was higher than that of the DCIS subgroup independent of ER expression, whereas IRS was increased in both the IC subtype and the DCIS subgroup with ER-positive expression (Figure 2D). The finding further supports that YEATS4 expression was correlated with ER expression.
Figure 2.
YEATS4 promotes the progression of breast cancer. A. Percentage of positive YEATS4 expression in adjacent normal tissues (ANTs), ductal carcinoma in situ (DCIS), invasive carcinoma without lymph node metastasis (ICW), and invasive carcinoma with lymph node metastasis (ICLNM). B. Comparison of the median immunoreactive scores (IRSs) of YEATS4 staining among ANTs, DCIS, ICW, and ICLNM specimens. C. Representative images of YEATS4 staining of ANTs, DCIS, ICW, and ICLNM specimens. Scale bar, 50 µm for 200×; 20 µm for 400×. D. Median IRS being higher in invasive carcinoma than in DCIS under the same ER status. E. Percentage of positive YEATS4 expression at different TNM stages. F. Comparison of median IRS of YEATS4 staining among stages I, II, and III. G. Representative YEATS4 images at stages I, II, and III. Scale bar, 50 µm for 200×; 20 µm for 400×.
We then explored the relationship between YEATS4 expression and TNM stage. The positive rate of YEATS4 in stage I was 56.2%, which increased to 67.8% and 80.0% in stages II and III, respectively (Figure 2E). Significant differences in YEATS4 IRS were indicated among the subgroups based on the TNM stage (Figure 2F). Representative photomicrographs of the YEATS4 immunohistochemical staining of each TNM stage are presented in Figure 2G. Overall, YEATS4 expression is correlated with progression in breast cancer and can potentially play a critical role in cancer development.
YEATS4 expression is correlated with poor prognosis in breast cancer
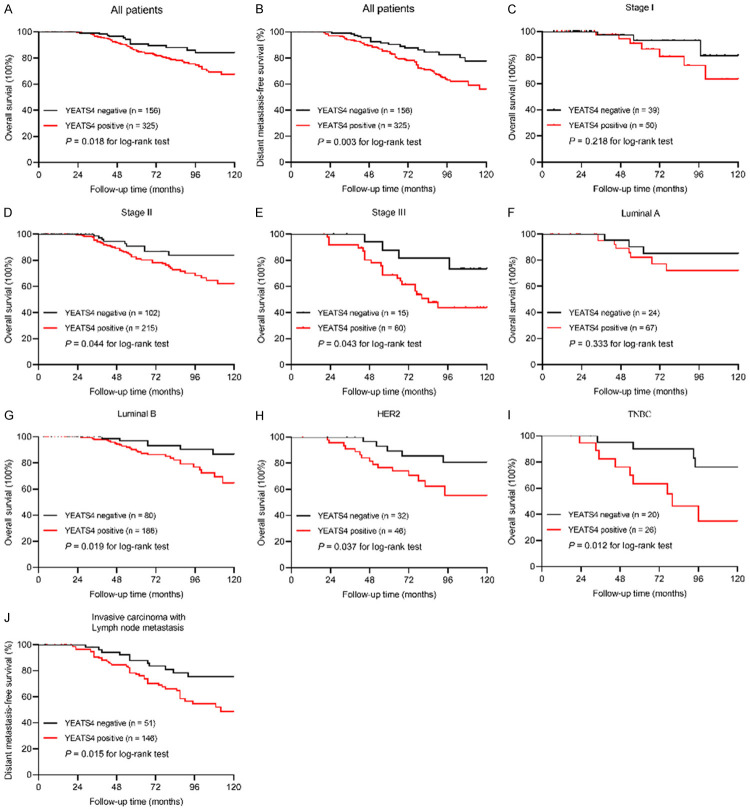
The effect of YEATS4 expression on clinical survival in patients with breast cancer was examined using the Kaplan-Meier method. Within the follow-up period, 62 (12.8%) patients died-49 (15.1%) from the YEATS4-positive group with 325 cases and 13 (8.3%) from the YEATS4-negative group with 156 cases. The 10-year OS was 84.0% in the YEATS4-negative group, and 67.4% in the YEATS4-positive group (univariate Cox regression HR 2.270, 95% CI 1.397-3.691, P = 0.001; Figure 3A; Table 2). Multivariate analysis indicated that positive YEATS4 expression was an independent poor prognostic indicator for OS (HR 1.969, 95% CI 1.146-3.382, P = 0.013; Table 2).
Figure 3.
YEATS4 has a prognostic value in breast cancer. A, B. Kaplan-Meier curves for the correlation between YEATS4 expression and overall survival (OS, P = 0.018) or distant metastasis-free survival (DMFS, P = 0.003). C-E. OS rate of patients with breast cancer according to different TNM stages. F-I. OS rate of patients with breast cancer, determined based on molecular subtype. J. Correlation of positive YEATS4 expression with a high risk of distant metastasis in invasive carcinoma with lymph node metastasis (P = 0.015).
Table 2.
Univariate and multivariate analyses of overall survival in breast cancer patients
| Variables | Category | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | ||
| Age (Years) | > 35 vs. ≤ 35 | 0.631 | 0.388-1.027 | 0.063 | 1.080 | 0.245-4.749 | 0.918 |
| Tumor size | T2, T3 vs. T1 | 1.055 | 0.732-1.520 | 0.770 | |||
| Histological grade | Grade 3 vs. grade 1, 2 | 1.826 | 1.068-3.111 | 0.027 | 1.015 | 0.676-1.524 | 0.947 |
| ER | Negative vs. positive | 1.765 | 1.028-4.536 | 0.041 | 2.472 | 1.126-4.614 | 0.020 |
| PR | Negative vs. positive | 0.405 | 0.629-2.541 | 0.329 | |||
| HER2 | Positive vs. negative | 1.280 | 0.854-1.919 | 0.230 | |||
| Lymph node metastasis | Positive vs. negative | 8.157 | 3.542-17.034 | < 0.001 | 5.873 | 2.906-11.840 | < 0.001 |
| TNM stage | III vs. I + II | 3.392 | 1.677-6.880 | 0.001 | 3.325 | 1.336-8.250 | 0.011 |
| Ki-67 status | > 20% vs. ≤ 20% | 1.193 | 0.990-1.438 | 0.062 | 1.110 | 0.643-1.913 | 0.705 |
| P53 status | Positive vs. negative | 0.742 | 0.437-1.258 | 0.268 | |||
| YEATS4 | Positive vs. negative | 2.270 | 1.397-3.691 | 0.001 | 1.969 | 1.146-3.382 | 0.013 |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNM, tumor-node-metastasis; YEATS4, YEATS domain-containing protein 4. P-values that reach significance are in bold.
Distant metastasis occurred in 93 patients during the follow-up period-77 (23.4%) from the YEATS4-positive group with 325 cases and 16 (10.3%) from the YEATS4-negative group with 156 cases. The 10-year DMFS was 77.8% in the YEATS4-negative group and 56.1% in the YEATS4-positive group (univariate Cox regression HR 1.892, 95% CI 1.137-3.188, P = 0.015; Figure 3B; Table 3). When all eligible prognostic variables were subjected to multivariate Cox regression analysis, coupled with lymph node metastasis, positive YEATS4 expression was considered as an independent indicator for DMFS (HR 1.784, 95% CI 1.016-3.002, P = 0.029; Table 3).
Table 3.
Univariate and multivariate analyses of distant metastasis-free survival in breast cancer patients
| Variables | Category | Univariate | Multivariate | ||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| HR | 95% CI | P-value | HR | 95% CI | P-value | ||
| Age (Years) | > 35 vs. ≤ 35 | 0.972 | 0.389-2.434 | 0.952 | |||
| Tumor size | T2, T3 vs. T1 | 1.954 | 0.936-4.083 | 0.075 | 1.005 | 0.634-1.960 | 0.725 |
| Histological grade | Grade 3 vs. grade 1, 2 | 1.270 | 0.518-3.132 | 0.607 | |||
| ER | Negative vs. positive | 1.775 | 1.079-2.941 | 0.025 | 1.443 | 0.862-2.387 | 0.166 |
| PR | Negative vs. positive | 1.806 | 1.194-3.007 | 0.007 | 1.107 | 0.589-1.752 | 0.640 |
| HER2 | Positive vs. negative | 1.651 | 0.827-3.304 | 0.157 | |||
| Lymph node metastasis | Positive vs. negative | 4.017 | 2.209-7.303 | < 0.001 | 3.543 | 1.939-6.440 | < 0.001 |
| TNM stage | III vs. I + II | 2.521 | 1.146-5.541 | 0.021 | 2.244 | 1.018-4.944 | 0.450 |
| Ki-67 status | > 20% vs. ≤ 20% | 1.277 | 0.736-2.213 | 0.385 | |||
| P53 status | Positive vs. negative | 0.714 | 0.165-3.014 | 0.656 | |||
| YEATS4 | Positive vs. negative | 1.892 | 1.137-3.188 | 0.015 | 1.784 | 1.061-3.002 | 0.029 |
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; TNM, tumor-node-metastasis; YEATS4, YEATS domain-containing protein 4. P-values that reach significance are in bold.
The influence of YEATS4 expression on OS was further stratified by the TNM stage and the molecular subtypes to better identify patients who were at high risk. YEATS4 expression was negatively related to OS in stage II and III groups (P = 0.044, P = 0.043, respectively), instead of the stage I group (P = 0.218) (Figure 3C, 3E), indicating that positive YEATS4 expression could be a potential index for poor clinical outcome in patients with breast cancer. In addition, in the subgroup analyses based on molecular subtype (Figure 3F-I), YEATS4 expression was correlated with shortened OS in the subtype of Luminal B (P = 0.019), HER2 (P = 0.037), and TNBC (P = 0.012), but not Luminal A (P = 0.333).
We then explored the association between YEATS4 expression and DMFS in the ICLNM subgroup (Figure 3J). ICLNM with positive YEATS4 expression showed a high incidence of distant metastasis (P = 0.015). These results indicated that YEATS4 expression is an independent poor prognostic indicator for DMFS, particularly in ICLNM.
YEATS4 promotes the proliferation of breast cancer cells in vitro and in vivo
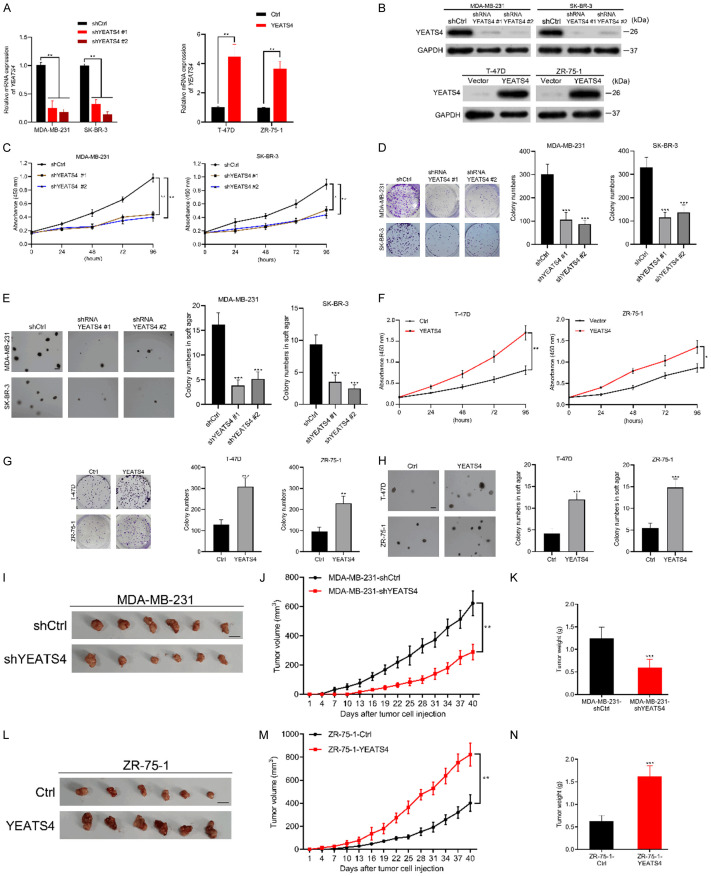
The aforementioned results indicated that positive YEATS4 expression was related to disease progression in patients with breast cancer. Therefore, we determined whether YEATS4 was involved in tumor growth. We established YEATS4-deleting MDA-MB-231 and SK-BR-3 cells (named as MDA-MB-231-shYEATS4 and SK-BR-3-shYEATS4) and YEATS4-overexpressing T-47D and ZR-75-1 cells (designated as T-47D-YEATS4 and ZR-75-1-YEATS4) by lentivirus infection. YEATS4 expression was verified by RT-qPCR and Western blot analysis (Figure 4A, 4B). The proliferation rate was significantly reduced in shYEATS4 breast cancer cells (Figure 4C). Plate colony formation assay and soft agar colony formation assay indicated that compared with control cells, MDA-MB-231-shYEATS4 and SK-BR-3-shYEATS4 cells formed fewer colonies (Figure 4D, 4E). Increased YEATS4 expression also markedly promoted the proliferation of T-47D and ZR-75-1 cells (Figure 4F). The colony-forming ability of T-47D -YEATS4 and ZR-75-1-YEATS4 cells in the regular plate or the soft agar was evidently improved (Figure 4G, 4H). We continued to evaluate the effect of YEATS4 on tumorigenicity in subcutaneous xenograft nude mice models. YEATS4 depletion evidently reduced the volume and weight of xenograft tumors (Figure 4I-K), whereas YEATS4 overexpression significantly promoted tumor growth (Figure 4L-N). The aforementioned findings suggested that YEATS4 played an important role to promote the growth of breast cancer cells in vitro and in vivo.
Figure 4.
YEATS4 promotes breast cancer cell growth in vitro and in vivo. (A, B) Verification of the efficiency of YEATS4 deletion by two shRNAs in MDA-MB-231 and SK-BR-3 cells and YEATS4 overexpression in T-47D and ZR-75-1 cells by RT-qPCR (A) and Western blot analysis (B). (C) Influence of YEATS4 deletion on the cell viability of MDA-MB-231 and SK-BR-3 cells, as determined using the CCK-8 assays. (D, E) Effect of YEATS4 knockdown on plate (D) and soft agar (E) colony formation in MDA-MB-231 and SK-BR-3 cells. Scale bar, 200 µm for soft agar colony formation. (F) Effect of YEATS4 overexpression on the cell viability of T-47D and ZR-75-1 cells. (G, H) Influence of YEATS4 overexpression on plate (G) and soft agar (H) colony formation as determined in T-47D and ZR-75-1 cells. Scale bar, 200 µm for soft agar colony formation. (I-K) MDA-MB-231 cells stably transfected with shYEATS4 or shCtrl were subcutaneously injected into the nude mice at a predetermined number of cells (n = 6 for each group). Nude mice were euthanized after 40 d. Representative xenograft tumors in nude mice (I). Scale bar, 1 cm. YEATS4 deletion in MDA-MB-231 cells reduced the volume (J) and weight (K) of tumors. (L-N) ZR-75-1-YEATS4 or control cells were subcutaneously injected into the nude mice (n = 6 for each group). Representative xenograft tumors in the nude mice (L). Scale bar, 1 cm. As shown in (M, N), YEATS4 overexpression in ZR-75-1 cells significantly promoted the tumor growth. Data are presented as mean ± s.d. for 3 independent assays. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
YEATS4 promotes the migration and invasion of cancer cells in vitro and in vivo
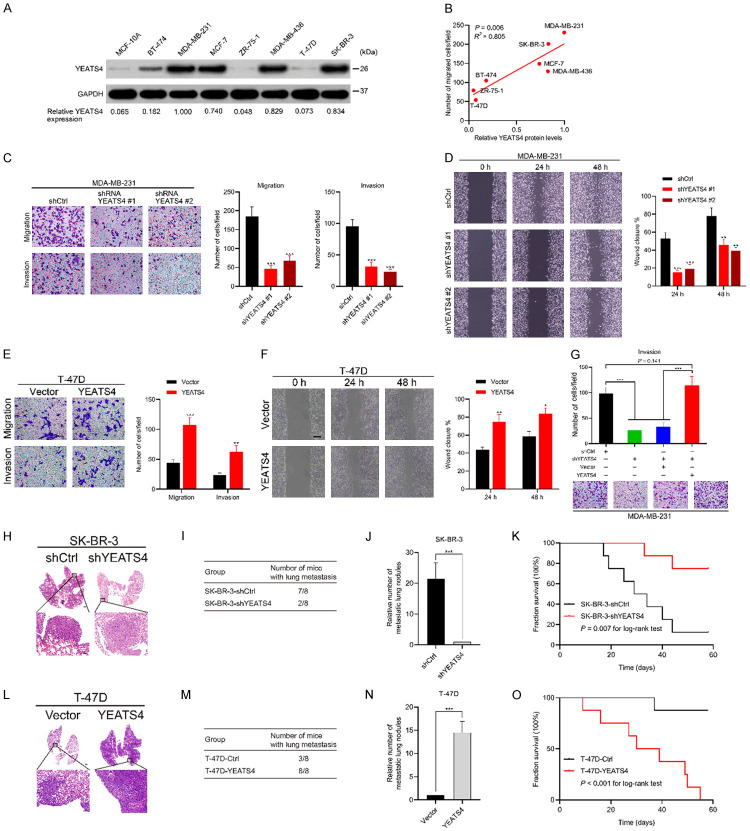
YEATS4 was closely related to distant metastasis in patients with breast cancer, prompting the investigation of the function of YEATS4 in cancer metastasis. YEATS4 expression in normal breast epithelial cells and several breast cancer cell lines was evaluated. YEATS4 was highly expressed in MDA-MB-231, MCF-7, MDA-MB-436, and SK-BR-3 cells but poorly expressed in ZR-75-1 and T-47D cells (Figure 5A). The relative expression of YEATS4 was significantly correlated with the migration ability of the cells (P = 0.006) (Figure 5B). Moreover, YEATS4 deletion in MDA-MB-231 cells evidently reduced their migration and invasion abilities in both the transwell assay and wound healing assay (Figure 5C, 5D). By contrast, YEATS4 overexpression in T-47D cells strengthened cell migration and invasion (Figure 5E, 5F). Notably, the recuperation of YEATS4 expression restored the cell invasion ability of YEATS4-deleting MDA-MB-231 cells (Figure 5G). To explore the role of YEATS4 in metastasis in vivo, SK-BR-3-shYEATS4 or T-47D-YEATS4 cells (with the corresponding control cells) were injected intravenously into the nude lung metastasis mouse models. YEATS4 deletion reduced the incidence of lung metastasis and decreased the number of metastatic lung nodules while prolonging the overall survival time in the SK-BR-3-shYEATS4 group (Figure 5H-K). However, YEATS4 overexpression increased the incidence of lung metastasis and the number of metastatic lung nodules while reducing the overall survival time in the T-47D-YEATS4 group (Figure 5L-O). Collectively, these results suggested that YEATS4 promoted the invasion and metastasis of breast cancer cells in vitro and in vivo.
Figure 5.
YEATS4 promotes cell migration and invasion in breast cancer cells in vitro and in vivo. (A) YEATS4 expression detected in several breast cancer cell lines by Western blot analysis. Relative expression of YEATS4 to GAPDH is shown below. (B) Correlation analysis between relative YEATS4 expression and migration ability of cancer cells by using the Pearson correlation method. (C) Migration and invasion capabilities assessed in MDA-MB-231 cells stably infected with lentivirus carrying YEATS4 shRNAs or negative control shRNA (shCtrl) by transwell assays. Scale bar, 50 µm. (D) Migration ability in MDA-MB-231 cells transfected with YEATS4 shRNAs or shCtrl by wound healing assays. Scale bar, 200 µm. (E) Migration and invasion abilities in stable T-47D-YEATS4 cells, determined using transwell assays. Scale bar, 50 µm. (F) Migration capability in stable T-47D-YEATS4 cells, detected by using wound healing assays. Scale bar, 200 µm. (G) YEATS4 was first downregulated by shYEATS4 expressing lentiviruses infection. These cells were then infected with YEATS4 overexpression lentiviruses or control vector overexpression lentiviruses. These different groups of MDA-MB-231 cells were subjected to transwell invasion assays. Scale bar, 50 µm. (H-K) Stable SK-BR-3-shYEATS4 or negative control cells intravenously injected into mice in lung metastasis assays (n = 8 for each group). Representative images of H&E-stained lung tissues from the different groups (H). Scale bar, 1 mm for upper panel and 100 µm for lower panel. Number of nude mice with lung metastasis in each group (I). Relative number of lung metastasis nodules in each group (J). Overall survival time in the different groups of nude mice (K). (L-O) T-47D-YEATS4 or control cells intravenously injected into mice in lung metastasis assays (n = 8 for each group). Representative images of H&E-stained lung tissues from the different groups (L). Scale bar similar to that in (H). Number of nude mice with lung metastasis in each group (M). Relative number of lung metastasis nodules in each group (N). Overall survival time in the different groups of nude mice (O). **, P < 0.01; ***, P < 0.001.
YEATS4 regulates epithelial-to-mesenchymal transition in breast cancer cells
Growing evidence suggests that EMT is activated during tumor progression and metastasis in response to a deregulated genome [7,23]. Transcriptomic analyses of YEATS4 in several types of cancer cells indicate that YEATS4 regulates various functional gene groups and pathways, including EMT [24-27]. We first investigated the expression of EMT markers mediated by the differential expression of YEATS4. YEATS4 depletion increased the expression of epithelial markers (E-cadherin and ZO-1) and reduced the expression of mesenchymal markers (vimentin and N-cadherin) in MDA-MB-231 and SK-BR-3 cells (Figure 6A-C). By contrast, YEATS4 overexpression decreased the expression of epithelial markers and increased the expression of mesenchymal markers in T-47D and ZR-75-1 cells (Figure 6D-F). The findings suggest that YEATS4 is a determinant to promote EMT in breast cancer cells.
Figure 6.
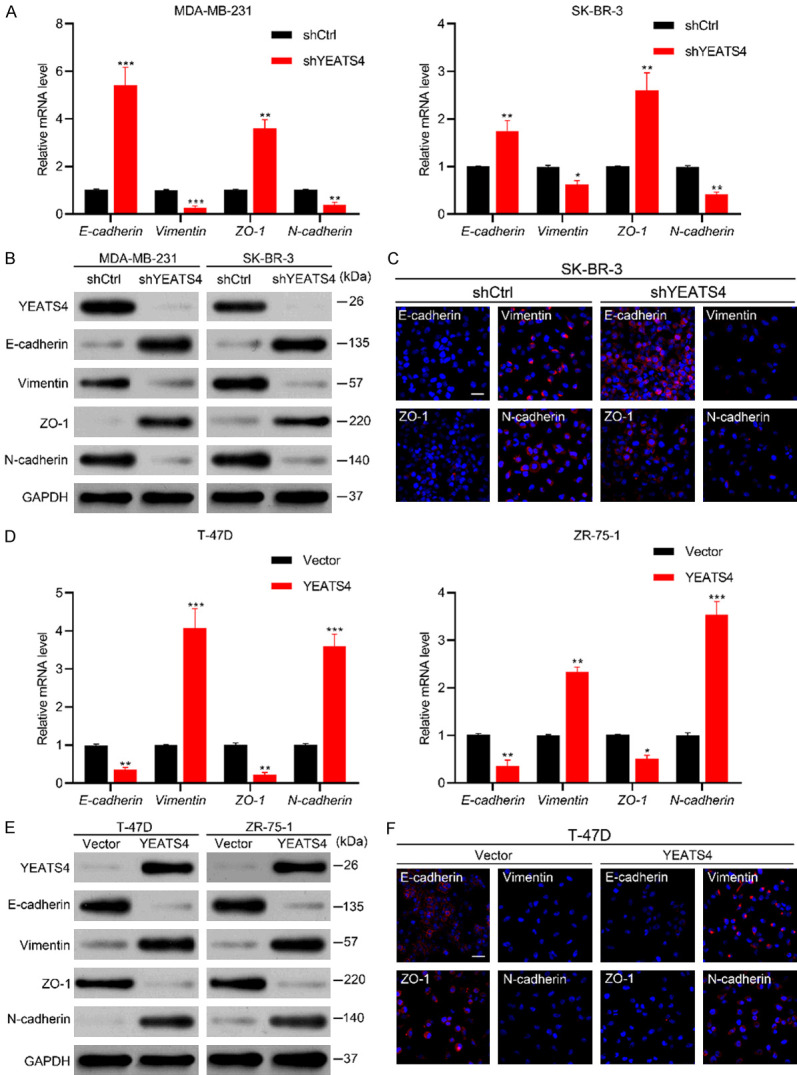
YEATS4 mediates EMT in breast cancer cells. (A) mRNA expression changes in EMT markers, including epithelial markers E-cadherin and ZO-1 and mesenchymal markers vimentin and N-cadherin in MDA-MB-231 and SK-BR-3 cells after stable YEATS4 deletion in RT-qPCR assays. (B) Expression changes in EMT markers were evaluated by Western blot analysis in MDA-MB-231 and SK-BR-3 cells after YEATS4 downregulation. (C) Immunofluorescence images in YEATS4 deleting SK-BR-3 and control cells. Scale bar, 20 µm. (D-F) EMT markers in stable T-47D-YEATS4 and ZR-75-1-YEATS4 cells compared with control cells, as determined by RT-qPCR (D), Western blot analysis (E), and immunofluorescence staining (F; scale bar, 20 µm). Data are presented as mean ± s.d. for 3 independent assays in RT-qPCR. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
YEATS4 transcriptionally regulates ZEB1 expression
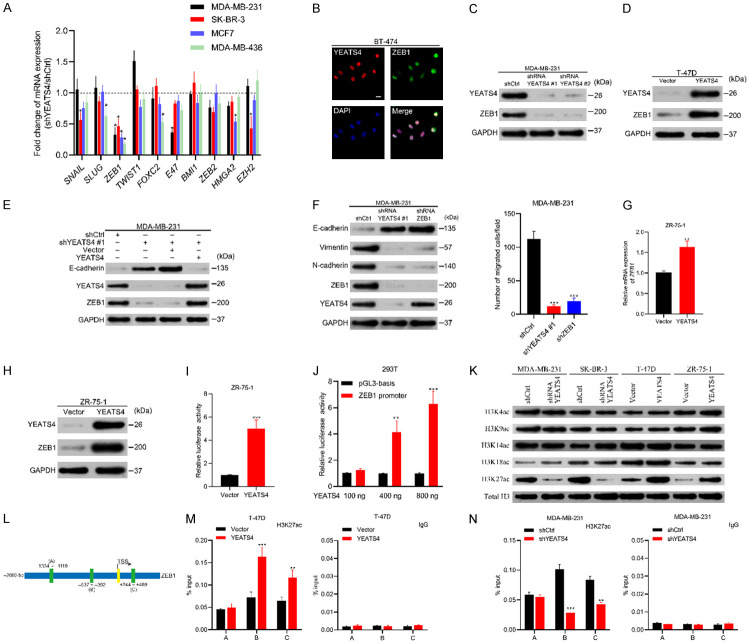
EMT is regulated by a complicated network, consisting of EMT transcriptional factors (EMT-TF), namely, TWIST1, SNAIL, SLUG, ZEB families, FOXC2, E47, HMGA2 and epigenetic regulators such as BMI1 and EZH2 [7,28]. We first integrated and analyzed the RNA sequencing (RNA-seq) data in breast cancer samples from the METABRIC dataset to determine the correlation between the mRNA expression levels of YEATS4 and EMT-TF by using the cBioPortal database [22]. YEATS4 was positively associated with most EMT-TF genes in varying degrees, but ZEB1 was the most relevant gene (Figure S1). Subsequently, we screened the mRNA expression changes of the above EMT regulatory factors in four breast cancer cell lines after stable YEATS4 deletion. Although several genes responded to YEATS4 knockdown, the ZEB1 mRNA expression levels markedly and steadily decreased in all four cell lines (Figure 7A). Immunofluorescence experiments indicated a high association between YEATS4 and ZEB1 expression in BT-474 cells (Figure 7B), which was further demonstrated by the results that ZEB1 was reduced or elevated with YEATS4 downregulation or overexpression (Figure 7C, 7D). Moreover, YEATS4 restoration in YEATS4-downregulated cells allowed the recovery of ZEB1 expression and maintained a low level of E-cadherin expression (Figure 7E). However, ZEB1 deletion did not influence YEATS4 expression (Figure 7F). Meanwhile, similar to YEATS4 depletion, ZEB1 deletion exhibited a similar effect on both cell phenotypes and migration capability in the presence of YEATS4, suggesting that ZEB1 is integral for YEATS4-mediated EMT (Figure 7F).
Figure 7.
YEATS4 regulates ZEB1 expression transcriptionally. (A) Fold changes in the mRNA expression levels of epithelial-to-mesenchymal transition (EMT) transcriptional factors in four breast cancer cell lines after YEATS4 depletion. Fold changes calculated by normalizing mRNA levels of YEATS4 deletion cells to that of the control cells. (B) Immunofluorescence images of YEATS4 and ZEB1 staining in BT-474 cells. YEATS4 and ZEB1 simultaneously expressed in cancer cells. Scale bar, 20 µm. (C) Expression of YEATS4 and ZEB1 in stable YEATS4-deleting MDA-MB-231 cells, determined by Western blot analysis. (D) YEATS4 and ZEB1 expression in YEATS4-overexpressing T-47D cells, detected by Western blot analysis. (E) Expression changes in ZEB1 and E-cadherin after restoring YEATS4 in YEATS4-deleting MDA-MB-231 cells. (F) Comparison of EMT markers and migration capabilities between YEATS4-deleting or ZEB1-deleting MDA-MB-231 cells and the control group. (G-I) ZEB1 mRNA (G), protein expression levels (H), and promoter activities (I) were investigated after 48 h transient transfection of YEATS4 overexpression or empty vector plasmids in ZR-75-1 cells. (J) Enhancement of the ZEB1 promoter activity by YEATS4 overexpression in a dose-dependent manner. (K) The abundance of H3 lysine acetylation was assessed by Western blot analysis; total H3 was used as a loading control. (L) Schematic of three regions relative to the ZEB1 transcription start site used as primers to test histone-occupied abundance. (M, N) ChIP was conducted to assess H3K27ac occupancy in T-47D-YEATS4 (M) and MDA-MB-231-shYEATS4 (N) cells. IgG was used as a negative control. Percentage of input indicates the ratio of the DNA fragment of each promoter region bound by H3K27ac to the total of the input DNA fragment without the H3K27ac antibody pull-down. Data are presented as mean ± s.d. for 3 independent assays. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We determined whether YEATS4 as a transcription factor mediates ZEB1 via transcription. YEATS4 overexpression plasmids were transiently transfected into ZR-75-1 cells. Consequently, the expression levels of ZEB1 mRNA and protein were elevated (Figure 7G, 7H). In addition, ZEB1 promoter activity was markedly enhanced in 48 h, indicating that the regulatory effect of YEATS4 on ZEB1 expression occurs at the level of transcription (Figure 7I). Corresponding to the aforementioned results, YEATS4 increased the ZEB1 promoter activity dose-dependently (Figure 7J).
We determined how this modulatory effect is demonstrated at the transcript level. The YEATS domain proteins were recently identified as a group of histone acetyl-lysine readers to influence gene transcription [25,29,30]. To explore whether YEATS4 mediates histone modification, acetylated histone modification markers were evaluated by altering the level of YEATS4 expression. Among H3K4, H3K9, H3K14, H4K18, and H3K27, only acetylated-H3K27 (H3K27ac) was influenced by changes in YEATS4 expression. YEATS4 overexpression increased H3K27ac, whereas YEATS4 deletion reduced this modification (Figure 7K).
Owing to the correlation of H3K27ac with active transcription [31,32], we determined whether YEATS4 regulates H3K27ac modification in the ZEB1 promoter. The JASPAR open-access database was used to analyze the occupancy site of the ZEB1 promoter by H3K27ac. The result indicated the presence of three putative-binding sites for histone-occupied abundance (Figure 7L). Chromatin immunoprecipitation assay was conducted on T-47D-YEATS4 and MDA-MB-231-shYEATS4 cells. The results showed that YEATS4 was related to the elevated expression of H3K27ac in the -637 to -392 bp and +244 to +469 bp regions of the ZEB1 promoter in T-47D-YEATS4 cells (Figure 7M). The ZEB1 promoter site occupancies by H3K27ac significantly decreased in MDA-MB-231-shYEATS4 cells (Figure 7N). However, the occupancies of other acetylated histones such as H3K4, H3K9, H3K14, and H3K18 at the corresponding ZEB1 promoter sites did not change with the variation in YEATS4 expression (Figure S2). The aforementioned findings were consistent with a previous report [25] and suggested that YEATS4 mediated the transcriptional regulation of ZEB1 by modulating H3K27ac and recruiting H3K27ac to the specific sites of the ZEB1 promoter.
ZEB1 is an effector for YEATS4-induced EMT, migration, invasion, and metastasis
We evaluated the role of ZEB1 in the YEATS4-mediated pro-metastatic effect. ZEB1 deletion in T-47D-YEATS4 cells caused the upregulation of epithelial markers and downregulation of mesenchymal markers at the transcriptional (Figure S3) and protein levels (Figure 8A). Moreover, ZEB1 knockdown reduced the migration and invasion capabilities of T-47D-YEATS4 cells (Figure 8B, 8C). In vivo metastasis assay showed that ZEB1 deletion apparently reduced the incidence of lung metastasis and the number of metastatic lung nodules while increasing the overall survival time in the T-47D-YEATS4 cells (Figure 8D-G). These findings indicate that ZEB1 is crucial for YEATS4-mediated breast cancer invasion and metastasis both in vitro and in vivo.
Figure 8.
ZEB1 mediates YEATS4-induced EMT and metastasis. (A) Recovery of epithelial marker expression and reduction of mesenchymal marker expression in T-47D-YEATS4 cells by ZEB1 deletion. (B, C) Reduction of YEATS4-induced migration and invasion abilities in T-47D-YEATS4 cells by ZEB1 depletion, as determined by wound healing (B) and transwell invasion (C) assays. Scale bar, 200 µm for (B); 50 µm for (C). (D) Representative images of H&E-stained lung tissues from the T-47D-YEATS4 cells with or without ZEB1 deletion. The scale bars represent 1 mm in upper panel and 100 µm in lower panel. (E) Total number of mice with lung metastasis in different groups. (F) Relative number of lung metastasis nodules in each group. (G) Overall survival of mice in each group. (H) Positive association of YEATS4 expression with ZEB1 expression in breast cancer tissues. Left, representative staining images of YEATS4 and ZEB1. Scale bar, 50 µm for 200×; 20 µm for 400×. (I) Representative images of ZEB1 staining of ANTs, DCIS, ICW, and ICLNM specimens. Scale bar, 50 µm for 200×; 20 µm for 400×. (J) Proportions of ANTs, DCIS, ICW, and ICLNM specimens with ZEB1-positive staining. (K) DMFS curves of patients with breast cancer, plotted based on the expression profiles of YEATS4 and ZEB1 as indicated. Co-expression of YEATS4 and ZEB1 (Group 4) indicating the shortest distant metastasis-free period vs. other groups. (L) Schematic demonstrating the role of YEATS4 in mediating ZEB1 expression, EMT, and metastasis in breast cancer. Data are presented as mean ± s.d. for 3 independent assays. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
ZEB1 expression was further examined in our retrospective cohort of human breast cancer patients. ZEB1 expression was detected in 290 (60.3%) of the 481 breast cancer tissues. YEATS4 expression showed a highly positive correlation with ZEB1, as determined by IHC (Figure 8H). Similar to that of YEATS4, the positive expression of ZEB1 was gradually increased as the breast cancer progressed (Figure 8I, 8J). To investigate the clinical relevance of the patterns of YEATS4 and ZEB1 expression in human breast cancer, we explored the association between the YEATS4 and ZEB1 expression and DMFS in our cohort. The patients were divided into four groups: YEATS4negative-ZEB1negative, YEATS4negative-ZEB1positive, YEATS4positive-ZEB1negative, and YEATS4positive-ZEB1positive. DMFS curves were plotted using the Kaplan-Meier method, and differences in survival between groups were compared using the log-rank test (Figure 8K). The YEATS4positive-ZEB1positive group exhibited a significantly low DMFS. These results are consistent with and verify the aforementioned in vitro and in vivo findings.
Discussion
The results of this study emphasize the correlation between YEATS4 expression and progression in breast cancer. YEATS4 expression facilitates cell growth, migration, invasion, and EMT in vitro and strengthens tumorigenesis and metastasis in vivo. By contrast, YEATS4 deletion reverses these malignant phenotypes in otherwise aggressive breast cancer cells. Mechanistically, the results indicate that YEATS4 transcriptionally regulates the expression of the EMT transcription factor ZEB1, which promotes EMT, cancer cell invasion, and metastasis in multiple human cancers [33,34]. This study reveals a regulatory mechanism by which YEATS4 mediates H3K27ac recruitment at the specific sites of the ZEB1 promoter to facilitate its transcription. ZEB1 deletion weakens the functions of YEATS4 and generates similar results induced by direct YEATS4 deletion. However, direct transactivation or epigenetic modification, which contributes more in YEATS4 transcriptionally regulating ZEB1, requires to be explored further.
Accumulating evidence indicates that YEATS4 is associated with cancer progression in various malignancies [15-17]. In the current study, the IRS of YEATS4 expression gradually increased from ANTs, DCIS, ICW to ICLNM as breast cancer progressed. A gradual increase in the IRS of YEATS4 expression was also observed in TNM stages I, II, and III. These findings suggest that YEATS4 is positively related to the progression of breast carcinoma, which is consistent with previous studies [15-17,20]. Subgroup analyses showed that the unfavorable effect of positive YEATS4 expression was more significant in stages II and III and luminal B, HER2, and TNBC patients. The reason could be the relatively poor prognosis of patients in these subgroups and the increased occurrence of distant metastasis. Although YEATS4 expression correlated with ER status, YEATS4 seemed to have exerted no effect on OS in luminal A patients. The reason could be that luminal A patients typically showed improved prognosis and reduced recurrence after hormone therapy [35,36], concealing the effect of YEATS4 expression. A powerful correlation between YEATS4 and shortened survival period was observed in the TNBC subgroup. Owing to the limited choice of treatment for this subgroup [37], YEATS4 concealment could be an effective treatment choice for these patients. In addition, positive YEATS4 expression was related to a significantly low DMFS in patients with ICLNM. These findings suggest that YEATS4 can potentially play an oncogenic role in promoting breast cancer progression and metastasis.
The negative regulatory effect of YEATS4 on multiple well-known tumor suppressors such as p53 and p21 [16,38], sufficiently proves the feasibility of YEATS4 as an oncogene. Pikor et al. identified YEATS4 as a candidate oncogene by integrative genetic analysis in lung cancer cells and showed that YEATS4 downregulation significantly attenuated cancer cell growth and tumor formation [16]. In the present study, YEATS4 facilitated cell proliferation and strengthened tumorigenicity in vivo. YEATS4 could induce EMT and consequently promote metastasis in breast cancer. Breast cancer cells with YEATS4 overexpression represent a mesenchymal phenotype, including relevant enhanced migratory and invasive capabilities in vitro. Our findings also indicate that YEATS4 deletion causes MET, suggesting that EMT-MET can be a transforming reciprocal process by manipulating YEATS4 expression. EMT is vital for cancer cells to leave the primary site and form neoplasia in other parts of the body [7]. Malignant phenotypes mediated by YEATS4 overexpression in breast cancer cells result in the aggravation of distant metastases in vivo. The results provide a molecular mechanism to explicate the clinical findings that breast cancer patients with positive YEATS4 expression are more likely to succumb to distant metastasis and that YEATS4 is an independent indicator for OS and DMFS.
Our results revealed that ZEB1 is potentially the primary mediator in YEATS4-induced EMT in breast cancer. However, YEATS4 is a multifunctional transcription factor with diverse target genes [10]. The transcriptomic analyses of YEATS4 in several types of cancer cells, including breast cancer cells, demonstrated that YEATS4 regulates various functional gene groups and pathways. In addition to widely-known genes relating to cell cycle, DNA replication, and DNA repair, YEATS4 also regulates some gene groups or pathways that are supposed to be involved in the EMT program, such as gene groups relevant to cell differentiation, cell adhesion, cell locomotory, and extracellular matrix receptor interactions [24-27]. However, transcriptomic data also showed that target genes identified by YEATS4 in different cell models tend to be different and enriched in various functional gene groups [24-26]. Thus, the transcriptional activities of YEATS4 are quite context-dependent. Our results revealed that YEATS4 is mainly involved in promoting ZEB1 transcription and EMT in breast cancer cells, but the underlying cellular context warrants further study.
The majority of the studies surrounding YEATS4 have focused primarily on cell growth and viability, and few have explored the role of YEATS4 in EMT, migration, and invasion of cancer cells; none of these studies have investigated breast cancer. YEATS4 promoted RNA polymerase II enrichment on the NOTCH2 promoter. Consequently, NOTCH2 expression increased, inducing EMT, migration, and invasion in gastric cancer and hepatocellular cancer [19,39]. Wild-type p53 forms a ternary complex with MDM2 and Slug to promote the degradation of this EMT inducer [40]. The YEATS4-PP2Cβ complex interacts with and dephosphorylates p53 at serine 366, consequently decreasing p53 stabilization [41]. Thus, YEATS4 can potentially enhance Slug expression. Another study also indicated that YEATS4 deletion by siRNA led to Snail downregulation in pancreatic cancer cells, suggesting the potential role of YEATS4 to mediate EMT [17]. Notably, as a histone acetylation reader, YEATS4 colocalizes with H3K27ac and H3K14ac to promote histone variant H2A.Z deposition onto chromatin to selectively regulate the expression of associated EMT proteins, such as upregulating fibronectin and downregulating E-cadherin in hepatocellular cell carcinoma [25,42].
The mechanistic link between YEATS4 and ZEB1 was previously unknown. Our results showed that altering the YEATS4 expression changes the acetylation status of H3K27 at the ZEB1 promoter, which subsequently controls ZEB1 expression at the transcription level. However, YEATS4 expression exerted no effect on the acetylation of H3K4, H3K9, H3K14, and H3K18; similarly, no recruitment of these acetylated histones on the ZEB1 promoter was observed. Therefore, YEATS4 transcriptionally regulates ZEB1 expression via the regulation of H3K27 acetylation and enrichment of H3K27ac at specific sites of the ZEB1 promoter. Consequently, YEATS4 promotes EMT, migration, invasion, and metastasis. H3K27ac is identified as an active mark for gene transcription [25,43]. The YEATS domain of YEATS4 adopts conserved aromatic residues within the “RK” motif to recognize H3K27ac [29]. A recent report suggests that YEATS4 recruits the Dot1l-RNA polymerase II complex onto the target gene promoter by recognizing H3K27ac to initiate transcription [44]. YEATS4 regulation of ZEB1 expression via H3K27ac requires further investigation.
In summary, we demonstrate that YEATS4 expression is increased and significantly associated with the progression of breast cancer. YEATS4 has an oncogenic function in mediating breast cancer EMT and metastasis by transcriptionally regulating ZEB1 expression (Figure 8L). The co-upregulation of YEATS4 and ZEB1 is correlated with poor DMFS in our cohort. Our results underscore the critical role of YEATS4 in cancerous signaling. We speculate that YEATS4 is a promising target for therapeutic intervention in breast cancer.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81670595, 81970568), Shanghai Natural Science Funds (16ZR1428200), and Excellent Youth Medical Talents Program of Shanghai General Hospital (06N1702011).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Jemal A, Wender RC, Gansler T, Ma J, Brawley OW. An assessment of progress in cancer control. CA Cancer J Clin. 2018;68:329–339. doi: 10.3322/caac.21460. [DOI] [PubMed] [Google Scholar]
- 4.Demicheli R, Dillekås H, Straume O, Biganzoli E. Distant metastasis dynamics following subsequent surgeries after primary breast cancer removal. Breast Cancer Res. 2019;21:57. doi: 10.1186/s13058-019-1139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weigelt B, Peterse JL, van ’t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 6.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 7.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 9.Fischer U, Heckel D, Michel A, Janka M, Hulsebos T, Meese E. Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum Mol Genet. 1997;6:1817–1822. doi: 10.1093/hmg/6.11.1817. [DOI] [PubMed] [Google Scholar]
- 10.Schulze JM, Wang AY, Kobor MS. YEATS domain proteins: a diverse family with many links to chromatin modification and transcription. Biochem Cell Biol. 2009;87:65–75. doi: 10.1139/O08-111. [DOI] [PubMed] [Google Scholar]
- 11.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J Biol Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 12.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Jin J, Chung MWH, Feng L, Sun H, Hao Q. Identification of the YEATS domain of GAS41 as a pH-dependent reader of histone succinylation. Proc Natl Acad Sci U S A. 2018;115:2365–2370. doi: 10.1073/pnas.1717664115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao K, Yang J, Hu Y, Deng A. Knockdown of YEATS4 inhibits colorectal cancer cell proliferation and induces apoptosis. Am J Transl Res. 2015;7:616–623. [PMC free article] [PubMed] [Google Scholar]
- 15.You S, Wang F, Hu Q, Li P, Zhang C, Yu Y, Zhang Y, Li Q, Bao Q, Liu P, Li J. Abnormal expression of YEATS4 associates with poor prognosis and promotes cell proliferation of hepatic carcinoma cell by regulation the TCEA1/DDX3 axis. Am J Cancer Res. 2018;8:2076–2087. [PMC free article] [PubMed] [Google Scholar]
- 16.Pikor LA, Lockwood WW, Thu KL, Vucic EA, Chari R, Gazdar AF, Lam S, Lam WL. YEATS4 is a novel oncogene amplified in non-small cell lung cancer that regulates the p53 pathway. Cancer Res. 2013;73:7301–7312. doi: 10.1158/0008-5472.CAN-13-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jixiang C, Shengchun D, Jianguo Q, Zhengfa M, Xin F, Xuqing W, Jianxin Z, Lei C. YEATS4 promotes the tumorigenesis of pancreatic cancer by activating beta-catenin/TCF signaling. Oncotarget. 2017;8:25200–25210. doi: 10.18632/oncotarget.15633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji S, Zhang Y, Yang B. YEATS domain containing 4 promotes gastric cancer cell proliferation and mediates tumor progression via activating the Wnt/β-catenin signaling pathway. Oncol Res. 2017;25:1633–1641. doi: 10.3727/096504017X14878528144150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Kiuchi J, Komatsu S, Imamura T, Nishibeppu K, Shoda K, Arita T, Kosuga T, Konishi H, Shiozaki A, Kubota T, Okamoto K, Fujiwara H, Ichikawa D, Tsuda H, Otsuji E. Overexpression of YEATS4 contributes to malignant outcomes in gastric carcinoma. Am J Cancer Res. 2018;8:2436–2452. [PMC free article] [PubMed] [Google Scholar]
- 20.Kim YR, Park MS, Eum KH, Kim J, Lee JW, Bae T, Lee DH, Choi JW. Transcriptome analysis indicates TFEB1 and YEATS4 as regulatory transcription factors for drug resistance of ovarian cancer. Oncotarget. 2015;6:31030–31038. doi: 10.18632/oncotarget.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D, Moers V, Lemaire S, De Clercq S, Minguijón E, Balsat C, Sokolow Y, Dubois C, De Cock F, Scozzaro S, Sopena F, Lanas A, D’Haene N, Salmon I, Marine JC, Voet T, Sotiropoulou PA, Blanpain C. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 24.Okabe H, Satoh S, Kato T, Kitahara O, Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y, Nakamura Y. Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res. 2001;61:2129–2137. [PubMed] [Google Scholar]
- 25.Hsu CC, Shi J, Yuan C, Zhao D, Jiang S, Lyu J, Wang X, Li H, Wen H, Li W, Shi X. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Genes Dev. 2018;32:58–69. doi: 10.1101/gad.303784.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Garcia F, Villagrasa P, Matsui J, Kotliar D, Castro V, Akavia UD, Chen BJ, Saucedo-Cuevas L, Rodriguez Barrueco R, Llobet-Navas D, Silva JM, Pe’er D. Integration of genomic data enables selective discovery of breast cancer drivers. Cell. 2014;159:1461–1475. doi: 10.1016/j.cell.2014.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, Chiang DY, Reva B, Mermel CH, Getz G, Antipin Y, Beroukhim R, Major JE, Hatton C, Nicoletti R, Hanna M, Sharpe T, Fennell TJ, Cibulskis K, Onofrio RC, Saito T, Shukla N, Lau C, Nelander S, Silver SJ, Sougnez C, Viale A, Winckler W, Maki RG, Garraway LA, Lash A, Greulich H, Root DE, Sellers WR, Schwartz GK, Antonescu CR, Lander ES, Varmus HE, Ladanyi M, Sander C, Meyerson M, Singer S. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Wen H, Xi Y, Tanaka K, Wang H, Peng D, Ren Y, Jin Q, Dent SY, Li W, Li H, Shi X. AF9 YEATS domain links histone acetylation to DOT1L-mediated H3K79 methylation. Cell. 2014;159:558–571. doi: 10.1016/j.cell.2014.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho HJ, Li H, Linhares BM, Kim E, Ndoj J, Miao H, Grembecka J, Cierpicki T. GAS41 recognizes diacetylated histone H3 through a bivalent binding mode. ACS Chem Biol. 2018;13:2739–2746. doi: 10.1021/acschembio.8b00674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439–445. doi: 10.1038/jhg.2013.66. [DOI] [PubMed] [Google Scholar]
- 32.Sato Y, Hilbert L, Oda H, Wan Y, Heddleston JM, Chew TL, Zaburdaev V, Keller P, Lionnet T, Vastenhouw N, Kimura H. Histone H3K27 acetylation precedes active transcription during zebrafish zygotic genome activation as revealed by live-cell analysis. Development. 2019;146:dev179127. doi: 10.1242/dev.179127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang P, He X, Wang Q, Huang Y, Jen KY, LaBarge MA, You L, Kogan SC, Gray JW, Mao JH, Wei G. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014;74:520–531. doi: 10.1158/0008-5472.CAN-13-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, Díez M, Viladot M, Arance A, Muñoz M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26–35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Gao JJ, Swain SM. Luminal A breast cancer and molecular assays: a review. Oncologist. 2018;23:556–565. doi: 10.1634/theoncologist.2017-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park JH, Ahn JH, Kim SB. How shall we treat early triple-negative breast cancer (TNBC): from the current standard to upcoming immuno-molecular strategies. ESMO Open. 2018;3:e000357. doi: 10.1136/esmoopen-2018-000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park JH, Roeder RG. GAS41 is required for repression of the p53 tumor suppressor pathway during normal cellular proliferation. Mol Cell Biol. 2006;26:4006–4016. doi: 10.1128/MCB.02185-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang G, Jiang H, Lin Y, Wu Y, Cai W, Shi B, Luo Y, Jian Z, Zhou X. lncAKHE enhances cell growth and migration in hepatocellular carcinoma via activation of NOTCH2 signaling. Cell Death Dis. 2018;9:487. doi: 10.1038/s41419-018-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, Li KC, Hong TM, Yang PC. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694–704. doi: 10.1038/ncb1875. [DOI] [PubMed] [Google Scholar]
- 41.Park JH, Smith RJ, Shieh SY, Roeder RG. The GAS41-PP2Cbeta complex dephosphorylates p53 at serine 366 and regulates its stability. J Biol Chem. 2011;286:10911–10917. doi: 10.1074/jbc.C110.210211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang HD, Kim PJ, Eun JW, Shen Q, Kim HS, Shin WC, Ahn YM, Park WS, Lee JY, Nam SW. Oncogenic potential of histone-variant H2A. Z.1 and its regulatory role in cell cycle and epithelial-mesenchymal transition in liver cancer. Oncotarget. 2016;7:11412–11423. doi: 10.18632/oncotarget.7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, Haverty PM, Bainer R, Cheung T, Arnott D, Flynn EM, Romero FA, Magnuson S, Gascoigne KE. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24:1722–1729. doi: 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 44.Liu B, Yang L, Zhu X, Li H, Zhu P, Wu J, Lu T, He L, Liu N, Meng S, Zhou L, Ye B, Tian Y, Fan Z. Yeats4 drives ILC lineage commitment via activation of Lmo4 transcription. J Exp Med. 2019;216:2653–2668. doi: 10.1084/jem.20182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.