Abstract
Coronavirus disease 2019 (COVID-19) is an emerging challenging area for the researchers to buckle up against the spread and control of the virus. Since earlier times, the diagnosis has been an important procedure in estimating the fate of epidemics by indicating the extent to which disease has been spread and to the extent, further disease prognosis would occur. The absence of anti-viral therapies and vaccines for COVID-19 at present suggests early diagnosis and isolation of the patients as the only smart approach available as of now. Presently, the increasing death rates, faster rates of transmission, non-availability of vaccines, and treatment have over-pressurized the researchers, health professionals, and government officials to develop effective clinical strategies in diagnosis and to come up with guidelines to be followed during conduction of each diagnostic procedure for maintaining healthcare systems. Since the incubation period of this virus is 2–14 days, a patient can transmit the infection without showing symptoms. Therefore, early diagnosis and isolation of susceptible individuals are the only way to limit the spread of the virus. Significance of diagnosis and triaging, information on specimen collection, safety considerations while handling, transport, and storage of samples have been highlighted in this paper to make people more aware and develop better clinical strategies in the future.
Keywords: COVID-19, diagnosis guidelines, specimen collection, safety consideration, SARS-COV-2
Introduction
Severe Acute Respiratory Syndrome-Coronavirus 2 (SARS-CoV-2) is a modified version of previous strain SARS-CoV whose outbreak had happened in 2003.1 Earlier, SARS-CoV had spread to 26 different countries and showed a fatality rate of over 10%, then a decade later Middle-east Respiratory Syndrome-Coronavirus (MERS-CoV) emerged in 2012 with a fatality rate of 35%.2 In a recent third outbreak, SARS-CoV-2 has spread to more than 218 countries and continues further.3 As of now, ~96 million people have been infected with SARS-CoV-2 and more than 2.0 million people have lost their lives (accessed on 18th January 2021, https://www.worldometers.info/coronavirus/). The virus has originated from Wuhan city of China in 2019 and named Coronavirus disease-2019 (COVID-19) by WHO officials.4 Despite sharing 80% genomic similarity with SARS-CoV, SARS-CoV-2 has proven to be more dangerous in terms of its transmission rate.5,6 This might have happened because of modification in the spike (S) protein of SARS-CoV-2 that increases its affinity towards angiotensin-converting enzyme 2 (ACE2) receptors in humans.7,8 This virus causes acute lung injuries, pneumonia, and sometimes may lead to the death of patients with severe symptoms. This pathological manifestation is due to the production of the cytokine storm during the active replication of the virus in human lung cells.9 To contain and prevent the further spread of COVID-19, it becomes important to gather all the information such as transmission history, mode of entry, replication, genetic structure, inflammatory response, and available diagnostic approaches to overcome the loss that has happened and/or may happen in the future to reduce its adverse effect.10 In Table 1, we have described social, clinical, and characteristic features including epidemiology of COVID-19 which would be helpful to have first insight into the pathophysiology of disease.11,12
Table 1.
An Overview of Recent COVID-19 Epidemiology and Its Characteristic Features
| Parameters of COVID-19 | Characteristic Features |
|---|---|
| Associated organism | SARS-CoV-2 (7th member of coronavirus family) |
| Origin | Wuhan, China in December 2019. |
| Clinical manifestation | Pneumonia and acute lung injuries |
| Symptoms | Fever, dry cough, shortness of breath, headache. |
| Risk group |
|
| Transmission | Reservoir- bats Identical host- pangolins Final host- human beings |
| Epidemiology | Spread to countries and territories- 218 |
| International conveyances- 02 | |
| Total confirmed cases- 56.7 million | |
| Total number of deaths- 1.3 million | |
| Mortality rate | 6% as of now but depends upon other factors such as sex (2.8% in males and 1.7% in females), comorbid state (5.6% for cancer, 6.0% for hypertension, 6.3% for a respiratory disorder, 7.3% for diabetes, and 10.5% for cardiovascular diseases), power of transmission (2.9% in Hubei province whereas 0.2% in other areas of China), age (0.2% for people in the age between 11–19 and 14.8% for people more than 80 years old) and conditions of the country (depends upon control, prevention strategies, mitigation policies, preparedness, and status of healthcare systems) |
| Genome structure | 5` UTR- replicase gene- structural genes- accessory genes- 3`UTR
|
| Incubation period | 2–14 days |
| Receptor | ACE2 |
| Replication pathway | Discontinuous mRNA production of structural genes leading to exocytosis of mature virus particles through the endomembrane system in the host. |
| Inflammatory response |
|
| Diagnostic approach |
|
Abbreviations: ORF, open reading frame; ACE2, angiotensin-converting enzyme 2; IL, interleukin; TNF α, tumor necrosis factor; TH 17, T helper cells; GM-CSF, granulocyte macrophage colony-stimulating factor; rRT-PCR, real-time reverse transcriptase-polymerase chain reaction; CXR, chest X-ray; CT, computed tomography. References.9,12,15,17
Since no effective vaccines and drugs are available for this disease, early detection, diagnosis, and isolation of the infected individuals is the best solution to address COVID-19 patients.13 Without the availability of suitable anti-viral drugs, all eyes are on diagnostic strategies that have the potential of determining the current scenario of the severity of the infection and to follow-up with the prognosis in the future.14
Timely diagnosis is the first step to prevent the spread of infection and providing care to the infected individuals; however, an alarming rate of its spread has generated concern in conducting diagnosis on a larger scale.15,16 Further, how well a country is responding to the current crises depends mainly upon its advancements in diagnostics, and therapeutic strategies, they may be adopting.17 Thus, in the present condition, we must look for a rapid, efficient, and error-free diagnosis on a larger scale.18
The first step in conducting a diagnostic test is the collection of a sample from the suspected person.19 The collection of the right specimen at right time is required to ensure an error-free diagnosis and accuracy of results. Thus, a set of prescribed protocols must be followed.20 The type of specimen collection, safety consideration upon their handling, transport, and storage are enlisted in this paper.21 Current diagnostic approaches and their limitations are also discussed systematically.
Significance and Need for Diagnosis
The SARS-CoV-2 infection was first identified in the Wuhan city of China where the outbreak happened.22 People reporting chest pain and signs of respiratory distress were immediately screened for common respiratory infections but none of the results were found to be positive; suggesting the presence of some other contaminant.23 Later, the analysis of bronchoalveolar lavage (BAL) fluid showed the sequence similarity to that of viruses belonging to the CoV family. However, it was not exactly similar to the previously known human pathogenic strains.23 For confirmation, immediate isolation of the virus on eukaryotic culture system and characterization was performed, indicated the emergence of seventh member and exposure to a third viral outbreak caused by members of the CoV family.23,24 This novel strain was named by the International Committee on Taxonomy of Viruses as SARS-CoV-2 because of its 80% homology to SARS-CoV.25 It also showed 59% sequence homology to MERS-CoV.26
The fatality rate exhibited by this virus was lesser than the earlier viral strains but its intensity of transmission has urged to declare COVID-19 pandemic as a health emergency.27 A higher incubation period of 2–14 days exhibited by SARS-CoV-2 indicates its possibility of transmission to healthy individuals from infected ones before the onset of symptoms. This means patients can transmit infection without even knowing they are already being infected when symptoms can take about 14 days to appear.28 This has generated a matter of concern among healthcare workers and health officials who have the sole responsibility of detecting an infection at an earlier stage.29 Not only the diagnostic techniques should be robust, reproducible, and accurate but it should be rapid as well.30 Any delay in diagnosing an infection can turn down the possibility of patient recovery and can risk the health of individuals close to the patients. This scenario has made the situation worse in various countries where diagnosis techniques are not very accurate and meticulously followed.17 By the time a person develops a symptom, it has already infected many other healthy people coming in its contact and therefore, creates difficulty in controlling the spread of the virus.31
Initially, identifying the travel history of the people coming from affected areas was the only way to identify susceptible cases.29 Such patients were kept under investigation in the isolation wards of hospitals but keeping in mind the risk associated with the lives of healthcare workers CDC has issued guidelines for conducting a diagnosis of patients only underclass II certified biosafety cabinets (BSL).32 This not only ensures the safety of healthcare providers but will further limit the spread of infection. Till now the infection is spreading continuously and the impact it will create on a global scale is unsaid. The current situation not only pressurizes government agencies, industries, public health sectors, and other clinical laboratories to join hands for advancing the diagnostic tools but also to be prepared for quality testing when the reemergence of such infections might occur shortly.29
Testing depends upon a process referred to as triaging where patients are divided into four categories –mild; moderate; severe and critical based on the symptoms they show.33 This process is to ensure that the critical and severe patients receive treatment on a priority basis and thus, helps in the reduction of the load caused to hospital authorities and the government for arranging the necessary infrastructure and resources for the patients.34 Approximately, 80% of the cases can be treated by supportive care which lies in mild and moderate categories.35 CDC has issued a statement indicating the need for decision making by the healthcare workers before the conduction of diagnostic tests in the people expected to be at risk.36 The decision should be based upon the course of illness and the reports coming from affected areas if any.37
Current Diagnostic Approaches
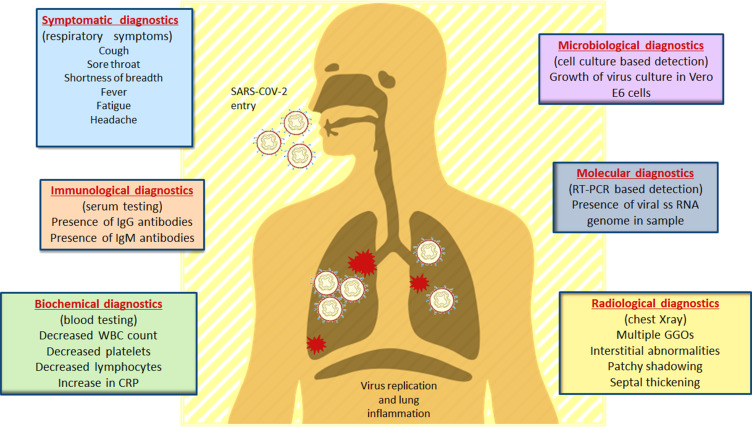
Diagnostic tools are prerequisites when it comes to initial identification and detection of an outbreak.38 Various multidisciplinary diagnostic tools are employed to detect any infection that has occurred on a regional, national, or global scale.39 For the detection and diagnosis of COVID-19, similar tools are used by clinicians to understand the pathophysiology and diagnosing the infection.40 Besides, Virologists in the early detection of SARS-CoV-2, immunology-based diagnostic approaches that rely upon the use of ELISA for detecting the IgM and IgG antibodies specific to the viral proteins are also employed.41 Microbiologists are contributing by culturing and isolating the virus on human cell lines.42 Biochemists are involved in understanding the changes in the blood constituents upon subsequent encounter with the pathogen43 and Radiologists are analyzing the clinical changes in the chest through X-rays of infected patients.44,45 Figure 1 describes the diagnostic approaches employed in SARS-CoV-2 diagnosis and prognosis. However, instead of focusing on the disciplinary-based detection methods, we have categorized them on their level of technology (Figure 2).
Figure 1.
Clinical and laboratory-based diagnostic approaches in COVID-19 diagnosis and prognosis. The clinical symptoms of COVID-19 along with their diagnostic techniques based on different research areas. As the virus gains entry into the host through nasal pathway, it travels up to the respiratory system where different clinical changes can be observed during the disease progression. Role of interdisciplinary biologists and their contribution in detection of COVID-19 is shown in this figure. Data from Tu et al.90
Abbreviations: CRP, C-reactive protein; WBC, white blood cells; RT-PCR, reverse transcriptase polymerase chain reaction; GGO, ground glass opacities.
Figure 2.
Method-based categorization of diagnostic approaches in SARS-CoV-2 detection. The current advances in the diagnostic tools for COVID-19 has been revolving around five major technology-based approaches. Serological methods are based upon serum testing that identifies either host antibodies or viral-specific proteins after a patient has been exposed to the virus. Molecular approaches focus upon detection of viral-specific sequence in the patient’s respiratory specimens. Various kits and devices (lab based and POC) have been approved based upon principles of molecular or serological detection. Radiology identifies the clinical changes within the respiratory tracts of the patients using medical imaging such as chest X ray and CT scan. Isolation of virus culture and growth on cell lines is a part of cell culture technique for detection of virus particles.
A detailed understanding of molecular approaches, serological techniques, radiological detection, and cell culturing methods would be discussed in Table 2. Currently, various lab and POC-based devices and diagnostic kits are also becoming popular in controlling COVID-19.46
Table 2.
Available Diagnostic Methods for the Detection of COVID-19
| Method | Basis | Mechanism | Advantage | Limitations |
|---|---|---|---|---|
| Molecular | Identification and amplification of viral-specific RNA sequence. | Based on principles of rRT-PCR technology. Separation of reporter and quencher dye occurs through exonuclease activity of taq polymerase resulting in the production of fluorescence intensity detected through PCR software. |
Different viral targets are adopted by different countries. USA targets three regions within the N gene. Germany targets the E gene and RdRP genes of subgenus Sarbecovirus. Hong Kong undergoes screening assay of N gene followed by confirmation using ORF1b assay. Various molecular Diagnostic POC kits have been approved. |
|
| Serological | Antibody testing | It involves the identification of antibodies in the host or viral-specific proteins through assays such as neutralization assay (ELISA), chemiluminescent immunoassay and rapid diagnostic tests (lateral flow assay). In this process, the antigen-antibody complex is formed which is recognized by a secondary antibody that produces a detectable color change or shows light-emitting properties. |
Various immunodiagnostic POC kits have been approved for commercialization. These works on the principle of ELISA or lateral flow assay. |
|
| Laboratory and point of care devices | Identification of viral RNA sequence or viral-specific antigens/host antibodies. | Molecular devices work by identifying the specific sequence of the RNA genome and then amplifies it to the point it becomes easily detectable. Similarly, serology-based devices depend upon the ELISA technique for the identification of host antibodies and viral proteins. |
Various molecular devices have been approved for emergency use authorization. One such is Abbott ID that runs on Abbott’s ID NOWTM platform- a lightweight box that can be fit and adjusted in different locations. It gives positive results in 5 minutes and negative results in 13 minutes. |
|
| Radiological | Identification of clinical changes in the patient’s respiratory system | It is a medical imaging technique that uses CT scan and CXR for giving a greater view of internal organs, bones and soft tissues. | CT scan is appropriate for studying the symptoms of pneumonia. Different clinical stages were observed in the patients as the disease progressed. First GGO lesion to multifocal GGO lesion then septal thickening of inter and intralobular followed by the formation of the consolidated lesion was observed. |
1.Varied representation of clinical changes among patients. 2. Cannot be used alone, must be used with molecular diagnostic approaches for confirmation of results. |
| Cell culture | Isolation of SARS-CoV-2 on cell lines promoting growth and replication of the virus. | Vero E6 cell lines are very sensitive to the viral replication of SARS-CoV. Upon virus replication, cell death occurred due to apoptosis, and the cells which survived the replication process showed morphology similar to the uninfected cells supporting the production of these infectious agents. | Vero E6 cells expressing TMPRSS2 showed more production of viral mRNA transcripts suggesting a probable role of TMPRSS2 in virus replication. A ~10-fold higher activity was observed as compared to other cell lines. |
|
Abbreviations: r-RT-PCR, real-time reverse transcriptase-polymerase chain reaction; N, nucleocapsid; E, envelope; RdRP, RNA dependent RNA polymerase; ELISA, enzyme-linked immunosorbent assay; POC, point of care; CXR, chest X-ray; CT, computed tomography; GGO, ground-glass opacities; TMPRSS2, transmembrane protease serine 2; BSL, biosafety level.
Notes: Data from Asrani et al.1515
Specimen Collection and Handling
For the effective diagnosis of an infection, the right specimen must be collected at the right time from the suspected patients.47 Nasopharyngeal (NP) swabs and oropharyngeal (OP) swabs are widely collected from the suspected individuals of SARS-CoV-2 infection.48 China was initially using OP swabs which showed 32% of active infections contrary to the 63% confirmed infections through NP swabs.49 US had highlighted the collection of NP swabs from the upper respiratory tract for detecting the viral RNA.50 NP swabs are more accurate for the diagnosis in comparison to the OP swabs but sometimes even they miss the detection of an early infection.51 A deeper sample must be obtained in such cases through bronchoscopy.52 Repeated testing for the patients.53 After isolation, a careful positioning of the swabs should be done on the viral transport medium.54
Besides OP and NP swabs, the most sensitive detection depends upon the use of samples collected from the lower and upper respiratory tract.51 These samples are in the form of BAL fluid and sputum. However, exposure to both of these samples increases the risk of transmission to the laboratory staff or healthcare personnel because they tend to generate aerosols.46 Hence, a highly trained staff with personal protective equipment (PPE) and higher BSL are recommended.55 Bronchoscopy is itself a difficult procedure requiring well-trained professionals for its implementation.52 The collection of samples from the upper respiratory tract is, however, easier and can be done in the areas where lesser facilities or resources are available. Reports say that virus was initially identified in the upper respiratory tract at 7 to 10 days after appearing of symptoms which then significantly decreased and later it peaked in the lower respiratory tract during the 2nd and 3rd weeks after the onset of illness.56 Earlier, feces, stools, and urine samples were used for the detection of other coronaviruses such as SARS-CoV and MERS-CoV.56–58
Other specimens that are currently being used in the detection of SARS-CoV-2 are saliva, serum, and rectal specimens.46 Diagnostic tests were performed on saliva samples collected from 12 patients of COVID-19 and 11 were found to be positive suggesting that saliva is a non-invasive specimen for close monitoring of this infection.59 Serum samples are used to check their performance in conducting the diagnosis. However, only 15% of pneumonia patients showed the presence of viral RNA in the serum.22 Rectal samples were found to be positive for SARS-CoV-2 infection in the patients of COVID-19.60
Besides specimen collection, their handling, transport, and storage are equally important for bringing out accurate diagnostic analysis. WHO has recommended certain steps to carry out this protocol in an efficient manner.61 This includes the immediate collection of samples as soon as a potential patient is identified. The collection of swabs should be done only through synthetic tips made up of nylon or Dacron with an aluminum or a plastic shaft. Right away the samples should be kept in viral transport media of no more than 2–3 mL dispensed in sterile tubes. Packaging and transport of specimen are equally important as their collection protocol. This includes sealing the neck of the sample vial by parafilm and covering its entire body with the absorbent material. The sample vials then must be placed in secondary containers like centrifuge tubes and finally should be locked in zip-pouch bags. These bags can be directly placed in thermocol boxes with a layer of frozen bags around them to maintain the required temperature of 2°C to 4°C. (https://www.mohfw.gov.in/pdf/5Sample%20collection_packaging%20%202019-nCoV.pdf)
Packaging, overnight shipping, and transport of the specimens must be followed the dangerous goods regulation of the International Air Transport Association (IATA) and UN 3373 Biological Substance for shipping regulations. Guidelines on transport planning and other essential documentation formalities required during the transport of specimens through airmail and railways may be assessed at (https://www.who.int/csr/resources/publications/biosafety/WHO_EMC_97_3_EN/en/). The storing of the samples must be done at −70°C or even lower if the delay is expected. The important considerations include the correct labeling of the sample with a patient’s ID number, medical record number, specimen type, and the date on which the sample was collected (https://www.fda.gov/media/134922/download).61
Safety Considerations During Diagnosis
To reduce the possibility of catching an infection among health professionals, there are certain guidelines on limiting the risk of transmission that needs to be strictly obeyed during the conduction of diagnostic tests (https://www.cdc.gov/coronavirus/2019-nCoV/lab/lab-biosafety-guidelines.html).47 Proper decontamination of the hospital wards and laboratories should be done regularly by using a 1% sodium hypochlorite solution. Protocols for disinfection of laboratories are described by the American Society for Microbiology (https://www.asm.org/Articles/Policy/Laboratory-Response-Network-LRN-Sentinel-Level-C). All microbiological-related specimen handling like smear preparation, staining procedures, viral culture, and isolation must be performed in at least BSL-3 (https://www.nuhospitals.com/blog/covid-19-specimen-management-and-laboratory-safety/). PPE like a mask, gloves, apron, face shield, and goggles should be worn during working hours.62 Workbench and machines such as centrifuge, laminar airflow after each use must be disinfected by the list of approved chemicals effective against SARS-CoV-2 as indicated by Pesticide Registration of United States Environment Protection Agency (https://www.epa.gov/pesticide-registration/list-n-disinfectants-use-against-sars-cov-2). After processing of the samples, the tubes and PPE must be autoclaved accompanied by incineration.63
Limitations of Diagnostic Tests
Various limitations are associated with the use of each diagnostic procedure which should be well known before performing any test. The potential of molecular diagnostic tools is only restricted to the samples obtained from the respiratory tracts of the suspected individuals. Sputum, nasopharyngeal aspirates, BAL fluid, nasal aspirates, OP and NP swabs can only be tested through this approach. Also, the chances of false-negative results are high when the lab reagents are contaminated, used past their expiry date, samples are not timely collected from the right region, improper storage and transport of specimen, presence of amplification inhibitors in samples, and if the mutation rate of the virus is high during PCR cycle.64 Moreover, detection of virus in the specimens does not guarantee the infection caused by SARS-CoV-2. Additionally, less information available on the optimum specimen, their time of collection, the time when the viral RNA is present in the specimen is likely to hinder the process of sample collection followed by its detection through the molecular approach. In this scenario, multiple samples at different intervals of time from the patients are required to confirm the positive results (https://www.fda.gov/media/134922/download).61
Serological-based detection is not a regular and routinely used diagnostic method because of the insufficiency of commercial reagents.65,66 Proper specimen handling and maintenance is required before the performance of serological tests. The sensitivity and specificity of the test are also a big question mark on the efficacy of the diagnostic tool.47 Since the test measures the antibody response of the host towards an infection, the production of antibodies may vary among individuals.67 Usually, it takes 1 to 3 weeks for the antibodies to develop but some patients may take longer than that.68,69 The antibody response depends upon various factors including age, comorbid state, nutritional health, and severity of infection.70 It cannot detect the early phase of infection when the immune response of the patient is building and when the antibody titer is below a detectable level (https://www.fda.gov/medical-devices/letters-health-care-providers/important-information-use-serological-antibody-tests-covid-19-letter-health-care-providers). Many studies are conducted where confirmed patients through molecular approaches showed negative results when tests were performed through serology.71 This shows, the antibody response is slower and by the time we detect the pathogen it might have infected multiple hosts.72 Therefore, detection through this approach occurs mainly during the recovery phase of the patient, and hence, repeated testing at different intervals much be done for obtaining accurate results.73 One of the major shortcomings associated with the use of an immuno-diagnostic POC kit that detects the viral antigen in the samples show false-positive results.74 This may happen when the patient instead of being infected with SARS-CoV-2 has any other CoV infection that causes normal common cold-like symptoms and hence, generating a concern over its sensitivity that lies between 34% and 80%.75
A well-trained professional is required for the result analysis. Any misinterpretation of data can misguide the diagnosis which may increase the chance of further transmission.76
Since the varied representation of pneumonia is obtained by CXR and CT scan in different patients; therefore, it becomes difficult in concluding the actual pathogenic mechanism through radiology-based detection of the virus.77–79 Also, certain asymptomatic patients showed early changes under CT scan which is an excellent approach to identify the patients without symptoms; however, this was not always the case.80 Some people whose sample has been tested positive with SARS-CoV-2 through molecular approaches have shown no clinical changes under radiology-based detection, raising doubt among false-negative-RT-PCR tests or a limitation of CT scan for the viral detection.81 There also exists the possibility of false-positive results through CT tests. This happens when a patient suffering from pneumonia is not caused by SARS-CoV-2.82 Additionally, exposure of radiation to certain individuals like pregnant women and children might not be an appropriate method for conducting a diagnosis; however,; a low dose scanning may be recommended in these cases. There might also be chances of cross-infection during the scanning process indicating the risk in the use of CT scan as a potential diagnostic tool.82 Since CT scan provides better follow-up of the health emergency and also keeping in mind its limitations, it is reported that the best way of diagnosis is to combine the laboratory and clinical findings83 for facilitating the diagnosis of COVID-19.22,84
The lack of permissive cell lines in laboratories limits the use of viral culture procedures as a routine diagnosticmethods. Only trained professionals can properly handle the specimen. The main limiting factor is BSL-3 facility which controls the high risk of transmission.85
Conclusion and Future Direction
Diagnosis of COVID-19 starts with the specimen collection of the suspected patients by sterile nylon swabs with aluminum shafts from sputum, BAL fluid, aspirates, OP, and/or NP swabs. Since SARS-CoV-2 causes acute lung injuries, specimens from upper and lower respiratory tracts must be preferred. The collected samples should be immediately transferred to a suitable viral culture media, packaged, and transported overnight either through ships or airplanes following the standard rules and regulations issued by their respective authorities. The storage of the samples must be done at 2–4°C or −70°C if delayed.
Triaging is done to ensure the critical patients receive diagnostic treatment and care on a priority basis followed by patients showing severe, moderate, and mild symptoms. Different technology-based diagnostic approaches are currently being employed. Health professionals need to follow the safety protocols issued by the WHO while diagnosing and being close to the patients. Disinfection of the laboratories and equipment, use of PPE and BSL-3 for viral cultures, proper disposal of waste tubes through autoclaving, and incineration must be done to ensure the risk-free status of the working environment. For devising effective therapeutic and diagnostic strategies in the future, understanding of behavior and activities of cells at the molecular level is required.86,87 For therapeutic advancements, indigenous flora of native areas of different countries should be explored for the screening of natural anti-viral compounds existing in the environment, and for diagnostic advancements better understanding of medical and healthcare systems is required to improvise the existing diagnostic and therapeutic strategies.38,86,88,89 Advancements in diagnosis and therapeutic strategies go hand in hand and thus, it is believed that improvisations in diagnosis would also help in identifying the target regions of SARS-CoV-2 providing avenues for devising vaccines and screening of anti-viral compounds.
Acknowledgments
MIH thanks the Council of Scientific and Industrial Research for the financial support (Grant No. 27 (0368)/20EMR-II). MFA, and AH acknowledge the generous support from Research Supporting Project (No. RSP-2020-122) by King Saud University, Riyadh, Kingdom of Saudi Arabia. SSS is very thankful to the Clifford Craig Foundation Launceston General Hospital (Tasmania, Australia) for Research Funding. We acknowledge Natural Science Foundation of Sichuan Province of China (Grant No. 21GJHZ0266).
Funding Statement
Indian Council of Medical Research (ISRM/12(22)/2020).
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman A, Sarkar A. Risk factors for fatal Middle East respiratory syndrome coronavirus infections in Saudi Arabia: analysis of the WHO line list, 2013–2018. Am J Public Health. 2019;109(9):1288–1293. doi: 10.2105/AJPH.2019.305186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020;91–98. doi: 10.1016/j.jare.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui DS, Azhar EE, Madani TA, et al. The continuing epidemic threat of novel coronaviruses to global health-the latest novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gralinski LE, Menachery VD. Return of the Coronavirus: 2019-nCoV. Viruses. 2020;12(2):135. doi: 10.3390/v12020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asrani P, Hassan MI. SARS-CoV-2 mediated lung inflammatory responses in host: targeting the cytokine storm for therapeutic interventions. Mol Cell Biochem. 2020:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surveillances V. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19)—China, 2020. China CDC Weekly. 2020;2(8):113–122. doi: 10.46234/ccdcw2020.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naqvi AAT, Fatima K, Mohammad T, et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol Basis Dis. 2020;1866(10):165878. doi: 10.1016/j.bbadis.2020.165878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asrani P, Hasan GM, Sohal SS, Hassan MI. Molecular basis of pathogenesis of coronaviruses: a comparative genomics approach to planetary health to prevent zoonotic outbreaks in the 21st century. OMICS. J Integr Biol. 2020. [DOI] [PubMed] [Google Scholar]
- 13.Dhama K, Sharun K, Tiwari R, et al. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohapatra RK, Pintilie L, Kandi V, et al. The recent challenges of highly contagious COVID‐19, causing respiratory infections: symptoms, diagnosis, transmission, possible vaccines, animal models, and immunotherapy. Chem Biol Drug Des. 2020;96(5):1187–1208. doi: 10.1111/cbdd.13761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asrani P, Eapen MS, Chia C, et al. Diagnostic approaches in COVID-19: clinical updates. Expert Rev Respir Med. 2020;1–16. doi: 10.1080/17476348.2021.1823833 [DOI] [PubMed] [Google Scholar]
- 16.Huang G, Gong T, Wang G, et al. Timely diagnosis and treatment shortens the time to resolution of coronavirus disease (COVID-19) pneumonia and lowers the highest and last CT scores from sequential chest CT. Am J Roentgenol. 2020:1–7. [DOI] [PubMed] [Google Scholar]
- 17.Ruan S. Likelihood of survival of coronavirus disease 2019. Lancet Infect Dis. 2020;20(6):630–631. doi: 10.1016/S1473-3099(20)30257-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke RM. Active monitoring of persons exposed to patients with confirmed COVID-19—United States, January–February 2020. MMWR Morb Mortal Wkly Rep. 2020;69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. The laboratory diagnosis of COVID-19 infection: current issues and challenges. J Clin Microbiol. 2020;58(6). doi: 10.1128/JCM.00512-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X-W, Wu -X-X, Jiang X-G, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. BMJ. 2020;368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forman MS, Valsamakis A. Specimen collection, transport, and processing: virology. Manual of Clinical Microbiology, 10th Edition. American Society of Microbiology; 2011:1276–1288. [Google Scholar]
- 22.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorbalenya, A.E., Baker, S.C. et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sohrabi C, Alsafi Z, O’Neill N, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance. 2020;25(10):2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binnicker MJ. Emergence of a novel coronavirus disease (covid-19) and the importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin Chem. 2020;66(5):664–666. doi: 10.1093/clinchem/hvaa071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adhikari SP, Meng S, Wu Y-J, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):1–12. doi: 10.1186/s40249-020-00646-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Control, C.f.D. and Prevention. Information for Laboratories 2019 Novel Coronavirus. Wuhan, China; 2020. [Google Scholar]
- 33.Ayebare RR, Flick R, Okware S, Bodo B, Lamorde M. Adoption of COVID-19 triage strategies for low-income settings. Lancet Respir Med. 2020;8(4):e22. doi: 10.1016/S2213-2600(20)30114-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Zhou L, Yang Y, Peng W, Wang W, Chen X. Therapeutic and triage strategies for 2019 novel coronavirus disease in fever clinics. The Lancet Respiratory Medicine 2020;8(3):e11–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajwah S, Wilcock A, Towers R, et al. Managing the supportive care needs of those affected by COVID-19. Eur Respir Soc. 2020;55:2000815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharfstein JM, Becker SJ, Mello MM. Diagnostic testing for the novel coronavirus. JAMA. 2020;323(15):1437–1438. doi: 10.1001/jama.2020.3864 [DOI] [PubMed] [Google Scholar]
- 37.Control, C.f.D. and Prevention. Evaluating and Testing Persons for Coronavirus Disease 2019 (COVID-19). Centers for Disease Control and Prevention; 2020. Updated. [Google Scholar]
- 38.Kumari P, Singh A, Ngasainao MR, et al. Potential diagnostics and therapeutic approaches in COVID-19. Clin Chim Acta. 2020;510:488–497. doi: 10.1016/j.cca.2020.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suarez DL, Das A, Ellis E. Review of rapid molecular diagnostic tools for avian influenza virus. Avian Dis. 2007;51(s1):201–208. doi: 10.1637/7732-101006-REGR.1 [DOI] [PubMed] [Google Scholar]
- 40.Wynants L, Van Calster B, Bonten MM, et al. Prediction models for diagnosis and prognosis of covid-19 infection: systematic review and critical appraisal. BMJ. 2020;369. doi: 10.1136/bmj.m1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engvall E, Perlmann P.Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 1971:8(9):871–874. [DOI] [PubMed] [Google Scholar]
- 42.Runfeng L, Yunlong H, Jicheng H, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol Res. 2020;104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu J, Liu J, Zhao X, et al. Clinical characteristics of imported cases of COVID-19 in Jiangsu Province: a multicenter descriptive study. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang W, Cao Q, Qin L, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19): A multi-center study in Wenzhou city, Zhejiang, China. J Infect. 2020;80(4):388–393. doi: 10.1016/j.jinf.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH Radiological Society of North America; 2020.
- 46.Loeffelholz MJ, Tang Y-W. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerg Microbes Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. Available from: https://apps.who.int/iris/handle/10665/331329. Accessed January 6, 2021.
- 48.Guan W-J, Ni Z-Y, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020. doi: 10.1001/jama.2020.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Center for Disease Control and Prevention. Interim Guidelines for Collecting, Handling, and Testing Clinical Specimens from Persons Under Investigation (PUIs) for Coronavirus Disease 2019 (COVID-19). Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed January 6, 2021.
- 51.Cheng PK, Wong DA, Tong LK, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363(9422):1699–1700. doi: 10.1016/S0140-6736(04)16255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pritchett MA, Oberg CL, Belanger A, et al. Society for Advanced Bronchoscopy Consensus Statement and Guidelines for bronchoscopy and airway management amid the COVID-19 pandemic. J Thorac Dis. 2020;12(5):1781–1798. doi: 10.21037/jtd.2020.04.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charlton CL, Babady E, Ginocchio CC, et al. Practical guidance for clinical microbiology laboratories: viruses causing acute respiratory tract infections. Clin Microbiol Rev. 2018;32(1):e00042–00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falsey AR, Formica MA, Walsh EE. Simple method for combining sputum and nasal samples for virus detection by reverse transcriptase PCR. J Clin Microbiol. 2012;50(8):2835. doi: 10.1128/JCM.01473-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balog-Way DH, McComas KA. COVID-19: reflections on trust, tradeoffs, and preparedness. J Risk Res. 2020:1–11. [Google Scholar]
- 56.Al-Abdely HM, Midgley CM, Alkhamis AM, et al. Middle East respiratory syndrome coronavirus infection dynamics and antibody responses among clinically diverse patients, Saudi Arabia. Emerg Infect Dis. 2019;25(4):753. doi: 10.3201/eid2504.181595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poissy J, Goffard A, Parmentier-Decrucq E, et al. Kinetics and pattern of viral excretion in biological specimens of two MERS-CoV cases. J Clin Virol. 2014;61(2):275–278. doi: 10.1016/j.jcv.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu D, Zhang Z, Jin L, et al. Persistent shedding of viable SARS-CoV in urine and stool of SARS patients during the convalescent phase. Eur J Clin Microbiol Infect Dis. 2005;24(3):165–171. doi: 10.1007/s10096-005-1299-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.To KK-W, Tsang OT-Y, Yip CC-Y, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71(15):841–843. doi: 10.1093/cid/ciaa149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Organization, W.H. Coronavirus Disease (COVID-19) Technical Guidance: Laboratory Testing for 2019-nCoV in Humans. Geneva, Switzerland; 2020. [Google Scholar]
- 62.Iwen PC, Stiles KL, Pentella MA. Safety considerations in the laboratory testing of specimens suspected or known to contain the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Am J Clin Pathol. 2020;153(5):567–570. doi: 10.1093/ajcp/aqaa047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan SS, Yan B, Saw S, et al. Practical laboratory considerations amidst the COVID-19 outbreak: early experience from Singapore. J Clin Pathol. 2020:jclinpath-2020-206563. doi: 10.1136/jclinpath-2020-206563 [DOI] [PubMed] [Google Scholar]
- 64.Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624 [DOI] [PubMed] [Google Scholar]
- 65.Deiris J, Lai S, Poon L. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shao X, Guo X, Esper F, Weibel C, Kahn JS. Seroepidemiology of group I human coronaviruses in children. J Clin Virol. 2007;40(3):207–213. doi: 10.1016/j.jcv.2007.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Okba NM, Muller MA, Li W, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv. 2020. [Google Scholar]
- 68.Liu Y, Diao B, Ren F, Wang Y, Ding J, Huang Q. Diagnostic indexes of a rapid IgG. IgM combined antibody test for SARS-CoV-2. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027–2034. doi: 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gorse GJ, Donovan MM, Patel GB. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus‐associated illnesses. J Med Virol. 2020;92(5):512–517. doi: 10.1002/jmv.25715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020:1–5. [DOI] [PubMed] [Google Scholar]
- 72.Liu W, Liu L, Kou G, et al. Evaluation of Nucleocapsid and Spike Protein-based ELISAs for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lou B, Li T, Zheng S, et al. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang N, Li S-Y, Yang X-L, et al. Serological evidence of bat SARS-related coronavirus infection in humans, China. Virol Sin. 2018;33(1):104–107. doi: 10.1007/s12250-018-0012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Che X-Y, Qiu L-W, Liao Z-Y, et al. Antigenic cross-reactivity between severe acute respiratory syndrome—associated coronavirus and human coronaviruses 229E and OC43. J Infect Dis. 2005;191(12):2033–2037. doi: 10.1086/430355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hyman WA. Risks associated with diagnostic devices. J Clin Eng. 1986;11(4):273–278. doi: 10.1097/00004669-198607000-00003 [DOI] [PubMed] [Google Scholar]
- 77.Jeffrey P Chest CT findings in 2019 novel Coronavirus (2019-nCoV) infections from Wuhan. China: key Points for the Radiologist: 8 2019–2020; 2020. [DOI] [PMC free article] [PubMed]
- 78.Kim H. Outbreak of novel coronavirus (COVID-19): What is the role of radiologists? European Radiology. 2020;30:3266–3267. [DOI] [PMC free article] [PubMed]
- 79.Lee KS. Pneumonia associated with 2019 novel coronavirus: can computed tomographic findings help predict the prognosis of the disease? Korean J Radiol. 2020;21(3):257–258. doi: 10.3348/kjr.2020.0096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425–434. doi: 10.1016/S1473-3099(20)30086-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lan L, Xu D, Ye G, et al. Positive RT-PCR test results in patients recovered from COVID-19. JAMA. 2020;323(15):1502–1503. doi: 10.1001/jama.2020.2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li M. Chest CT features and their role in COVID-19. Radiol Infect Dis. 2020;7(2):51–54. doi: 10.1016/j.jrid.2020.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J Med Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) from a symptomatic patient. Jama. 2020;323(16):1610–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Anas Shamsi TM, Anwar S, AlAjmi MF, et al. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci Rep. 2020;40(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mohammad T, Shamsi A, Anwar S, et al. Identification of high-affinity inhibitors of SARS-CoV-2 main protease: towards the development of effective COVID-19 therapy. Virus Res. 2020;288:198102. doi: 10.1016/j.virusres.2020.198102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fatima U, Rizvi SSA, Fatima S, Hassan MI. Impact of hydroxychloroquine/chloroquine in COVID-19 therapy: two sides of the coin. J Interferon Cytokine Res. 2020;40(10):469–471. doi: 10.1089/jir.2020.0105 [DOI] [PubMed] [Google Scholar]
- 89.Khan S, Fakhar Z, Hussain A, et al. Structure-based identification of potential SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2020:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tu YF, Chien CS, Yarmishyn AA, Lin YY, Luo YH, Lin YT, Lai WY, Yang DM, Chou SJ, Yang YP, Wang ML. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. 2020:Jan;21(7):2657. [DOI] [PMC free article] [PubMed] [Google Scholar]