Graphical abstract
Keywords: Traumatic brain injury, Hydroxysafflor yellow A, Metabolomics, Network pharmacology, Mechanisms
Abstract
Traumatic brain injury (TBI) has become a leading cause of mortality, morbidity and disability worldwide. Hydroxysafflor yellow A (HSYA) is effective in treating TBI, but the potential mechanisms require further exploration. We aimed to reveal the mechanisms of HSYA against acute TBI by an integrated strategy combining metabolomics with network pharmacology. A controlled cortical impact (CCI) rat model was established, and neurological functions were evaluated. Metabolomics of brain tissues was used to identify differential metabolites, and the metabolic pathways were enriched by MetaboAnalyst. Then, network pharmacology was applied to dig out the potential targets against TBI induced by HSYA. The integrated network of metabolomics and network pharmacology was constructed based on Cytoscape. Finally, the obtained key targets were verified by molecular docking. HSYA alleviated the neurological deficits of TBI. Fifteen potentially significant metabolites were found to be involved in the therapeutic effects of HSYA against acute TBI. Most of these metabolites were regulated to recover after HSYA treatment. We found 10 hub genes according to network pharmacology, which was partly consistent with the metabolomics findings. Further integrated analysis focused on 4 key targets, including NOS1, ACHE, PTGS2 and XDH, as well as their related core metabolites and pathways. Molecular docking showed high affinities between key targets and HSYA. Region-specific metabolic alterations in the cortex and hippocampus were illuminated. This study reveals the complicated mechanisms of HSYA against acute TBI. Our work provides a novel paradigm to identify the potential mechanisms of pharmacological effects derived from a natural compound.
1. Introduction
Traumatic brain injury (TBI) is a leading cause of mortality, morbidity and disability worldwide [1]. The disease triggers a cascade of pathophysiological events, such as disrupting biochemical, metabolic, and molecular functions, disturbing brain cell homeostasis and impairing cognitive, motor, or neuropsychological health [2]. Despite large efforts to develop neuroprotective therapies for TBI, no drugs have been approved by the Food and Drug Administration (FDA).
Natural bioactive compounds tend to be promising agents against brain injury [3], [4]. Safflower (Carthamus tinctorius L.), a well-known traditional Chinese medicine, is widely used to treat cerebrovascular diseases. Hydroxysafflor yellow A (HSYA, C27H32O16, 612.500 g/mol, Fig. S1) is the main active ingredient of safflower and exerts antiinflammatory, antiapoptotic, antioxidative and neuroprotective effects [5], [6]. Our previous research demonstrated that HSYA could across the injured blood–brain barrier of TBI patients to exert a neuroprotective effect [7]. Further investigation suggested that HSYA prevents oxidative stress post TBI by increasing the activity of antioxidant enzymes [8]. However, the mechanisms and targets of HSYA in treating TBI have not been fully elucidated.
Given that TBI reflects perturbations in complex metabolic physiologies, metabolomics is powerful for monitoring the dynamic changes in pathological metabolites [9]. However, traditional metabolomics could only reflect the terminal variation of disease and treatment [10], [11], [12]. It is unclear about the endogenous mechanisms of metabolites’ changes, including how these metabolites are produced, what their upstream pathways and proteins are, and which proteins HSYA exerts effects through. Thus, metabolomics alone may limit the application of HSYA.
Currently, the paradigm of developing single target-based drugs as therapeutics has been challenged mainly due to lack of efficacy and emerging resistance [13]. Thus, natural compounds that selectively act on two or more targets of interest in theory should be more efficacious than single-target agents [14]. Network pharmacology appears in this setting to construct an alternative systems-level approach to find new drug candidates. Instead of looking for a single disease-causing gene and drugs which act solely on an individual target, the whole drug-disease network is considered with the aim to find multi-targets drugs to reduce side effects [15]. Nonetheless, network pharmacology is limited by the single computational methods that rely on public databases. Network pharmacology alone could only predict the possibility of compound-target combination and pathway analysis [16]. It is uncertain whether HSYA binds to targets in vivo and which effect HSYA exerts on targets: inhibition, activation or ineffective combination.
Therefore, we integrated metabolomics with network pharmacology. Untargeted metabolomics was applied to determine the influences of HSYA on TBI and to identify the essential metabolites. Subsequently, network pharmacology was performed to analyze the proteins and reactions that modulated the metabolites, as well as the targets that HSYA acted on. Collectively, this strategy compensates network pharmacology for lacking experimental validation and metabolomics for lacking upstream molecular mechanisms and drug-binding targets. This strategy will hopefully contribute to a better understanding of the therapeutic principle of natural compounds for TBI treatment.
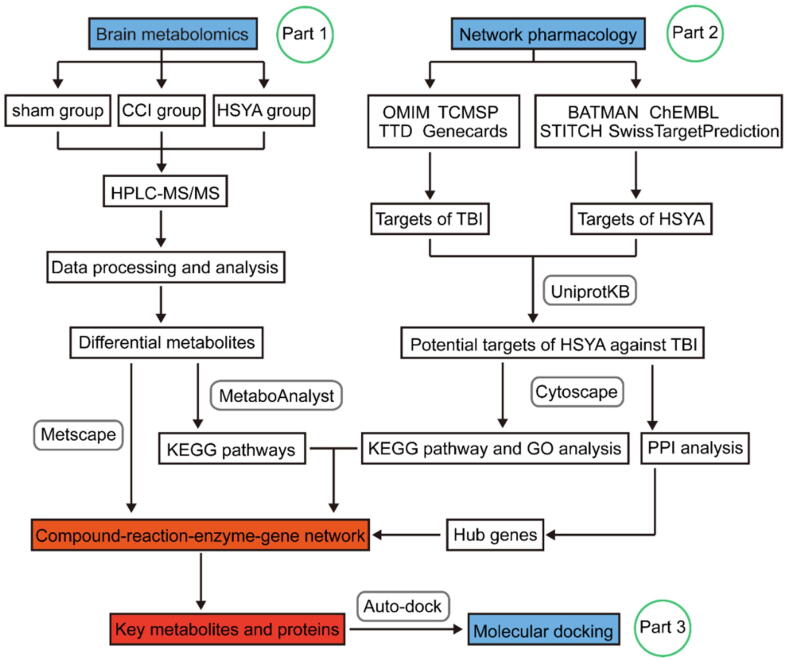
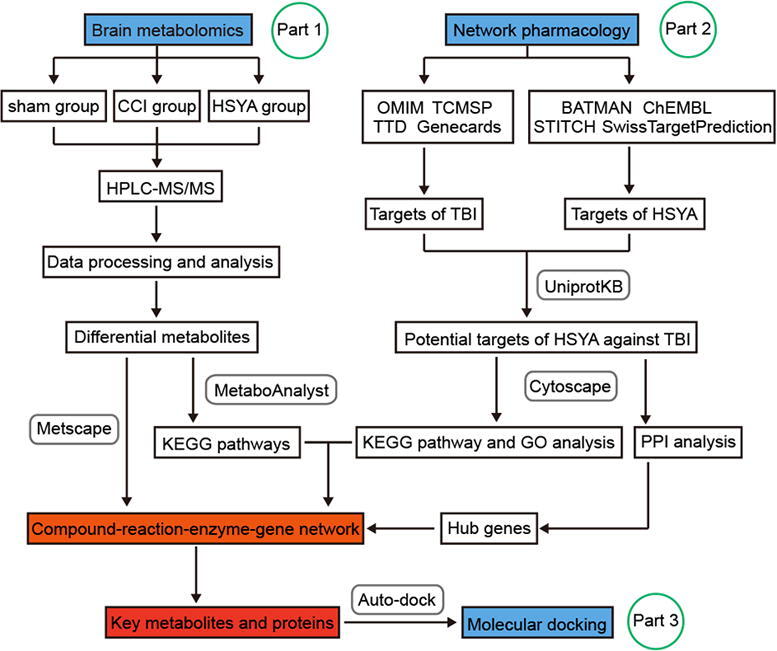
In the present study, we first developed a novel integrated strategy to explore the key targets and mechanisms of HSYA in treating acute TBI based on metabolomics and network pharmacology. Furthermore, we identified region-specific metabolic responses (cortex and hippocampus) in the HSYA treated rat model of TBI. This study provides new insight into the neuroprotective effects of HSYA in treating TBI. The research flowchart is shown in Fig. 1.
Fig. 1.
The schematic flowchart of the integrated strategy. The mechanisms of HSYA against TBI were analyzed by metabolomics of brain tissues (Part 1). Hub genes were extracted by network pharmacology (Part 2). Key metabolites and targets were identified and linked based on Part 1 and 2. These key targets were further verified by molecular docking (Part 3).
2. Material and methods
2.1. Reagents and materials
Hydroxysafflor yellow A (HSYA) was purchased from Shanghai Yuanye Bio-Technology Co., Ltd (Shanghai, China; purity: 90%, lot number: S26799). Ammonium acetate, ammonium hydroxide and methanol (grade: for HPLC) were provided by Sigma-Aldrich (St. Louis, MO, USA), and acetonitrile and H2O (grade: for HPLC) were obtained from J.T.Baker (PA, USA). All of the remaining reagents were of analytical grade.
2.2. Animals and the controlled cortical impact (CCI) model
7-week-old male specific-pathogen-free Sprague Dawley rats were obtained from the Laboratory Animal Centre of Central South University (Changsha, China). All rats were housed in a well-ventilated room at 25°C, with a 12 h dark-light cycle and free access to food and water. Animal care was performed under the guidelines of Central South University for the care and use of animals and the protocol was approved by the Medical Ethics Committee of Central South University. Rats were randomly assigned to 3 groups (n = 10 per day per group): sham group, CCI group and HSYA group. Rats in the HSYA group were orally administrated HSYA (0.87 mg/ml, dissolved in 0.9% saline) once a day at a dose of 13.88 mg/kg. Rats in the sham and CCI groups were treated with an equal volume of saline solution.
Replication of the CCI rat model was performed according to a previous study [17]. The parameters were set as follows: impact depth, 5.0 mm; striking speed, 6.0 m/s; dwell time, 50 ms. Rats in the sham group were operated identically to those in the CCI and HSYA groups, except for cortical impact. Rats were anaesthetized by intraperitoneal injection of sodium pentobarbital (60 mg/kg) and sacrificed at day 1 (n = 10 per group) and day 3 (n = 10 per group) after CCI. The ipsilateral cortex and hippocampus were collected from all of the rats after perfusion with ice-cold saline and stored at −80 °C for further use.
2.3. Neurological function testing
All animals were assessed by the modified neurologic severity score (mNSS) test and weight change. The 18-point mNSS comprises motor, sensory, reflex abilities and balance tests [18]. Higher scores indicate more serious damage. Rats were evaluated before and after injury to verify the neuroprotective effect of HSYA in the CCI model. The body weight of rats was recorded before and after injury, and the percentage of weight change was calculated.
2.4. Sample preparation
50 mg of tissue was homogenized with 400 µL of H2O. A BCA protein assay was performed to measure the total protein concentration on each of the individual homogenates. 100 µL aliquots of homogenates were precipitated by adding methanol and acetonitrile as a ratio of 1:1 (v/v). After vortexing for 30 s and sonicating for 10 min, the samples were incubated for 1 h at 20 °C and centrifuged at 20,000 g at 4 °C. Then, the supernatants were collected and dried in a vacuum concentrator. Finally, the dry extracts were reconstituted with 40 µL/mg acetonitrile and H2O (1:1, v/v) for HPLC/MS analysis. The pooled quality control (QC) samples were made by mixing 10 µL aliquots from each sample (one per six samples).
2.5. HPLC-MS/MS analysis
Metabolomics was applied using 1260 infinity high-performance liquid chromatography (Agilent, CA, USA) coupled with Q-Exactive MS/MS (Thermo, MA, USA). Chromatographic separations were performed on an amide column at 25 °C. The mobile phase consisted of water mixing with 25 mM ammonium acetate, 25 mM ammonium hydroxide (solvent A) and acetonitrile (solvent B). The gradient program was as follows: 90% B (0–1.0 min), 90 to 87% B (1.0–11.0 min), 87–80% B (11.0–14.0 min), 80–70% B (14.0–16.5 min), 70–50% B (16.5–18.5 min), 50–20% B (18.5–20.5 min), 20% B (20.5–25.0 min), 20–90% (25.0–25.1 min) and maintained at 90% B until 34 min. The injection volume was 4 µL and the flow rate was 0.4 mL/min. MS analysis was carried out on the Q-Exactive MS/MS in both positive and negative ion modes. Setting the relevant tuning parameters for the probe: aux gas heater temperature, 400 °C; spray voltage, 3.5 kV; sheath gas, 40 psi; auxiliary gas, 13 psi; capillary temperature, 350 °C. Building a DDA method as follows: full scan range was 60–900 m/z; maximum injection time for MS1 and ddMS2: 100 ms and 45 ms; resolution for MS1 and ddMS2: 70,000 and 17,500 respectively; automatic gain control for MS1 and ddMS2: 3e6 and 2e5; isolation window: 1.6 m/z; normalized collision energies: 10, 17, 25 or 30, 40, 50. Building a full scan method as follows: full scan range: 60 to 900 m/z; resolution: 140,000; maximum injection time: 100 ms; automatic gain control: 3e6 ions.
2.6. Data processing and analysis
The acquired raw files were preprocessed using Thermo Compound Discover 2.1 (Thermo, MA, USA) software. Intensities were corrected for signal drift and batch effect by fitting a locally quadratic (loess) regression model to the median intensity of pooled QC samples. The alpha parameter controlling the smoothing was set to 2 to avoid overfitting. After correction, the median area of all pooled QC samples was the same. The data were pretreated using the “80% rule” [19] to reduce the missing value input. HPLC-MS/MS analysis and data processing were conducted by KangChen Bio-tech (China). The features with relative standard deviations (RSDs) > 30% were removed from all the QC samples. The pretreated data were calibrated with median, transformed with log, and scaled with Pareto, then analyzed using principal component analysis (PCA), supervised partial least squares discrimination analysis (PLS-DA), and orthogonal partial least squared discriminant analysis (OPLS-DA) in R software (version 3.6.0) by the ropls R package. 7-round cross-validation and 200 permutation test were performed to evaluate the accuracy of the models. Features were further subjected to one-way analysis of variance (ANOVA) with a false discovery rate (FDR) at the univariate level to measure the significance of each metabolite (q-value). The features with variable importance in the projection (VIP) > 1 and q-value < 0.05 were considered to be differential compounds. These features were identified by performing retention time alignment, unknown compound detection, and compound grouping across all samples. For retention time alignment, the max time shift was 2 mins, and a tolerance of 0.5 min was used for grouping unknown compounds. Mass tolerance for feature detection and compound annotation was set as 10 ppm and 5 ppm respectively. The formula and accurate mass of each feature were submitted to ChemSpider (http://www.chemspider.com/) with 4 databases selected (BioCyc; Human Metabolome Database; Kyoto Encyclopedia of Genes and Genomes [KEGG]; LipidMAPS). The metabolite with the most references was considered as the terminal matching result. MS1 and ddMS2 were compared to the standard spectrum of the mzCloud database (https://www.mzcloud.org/), and the substance with the highest comparison rate was selected as the final identification result. Heat maps were displayed using the pheatmap package in R. Metabolic pathway analysis was performed by MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/).
2.7. Network pharmacology construction
To visualize the metabolite-protein-pathway network and to reveal the key metabolites and related proteins, network construction was applied via Cytoscape 3.7.2 (Cytoscape Consortium, CA, USA). The procedure was as follows (Fig. 1): (1) The candidate targets of TBI were screened by searching the keywords of “traumatic brain injury” in the gene map of the Online Mendelian Inheritance in Man (OMIM, https://omim.org/), therapeutic target database (TTD, http://db.idrblab.net/ttd/), Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://tcmspw.com/tcmsp.php) and genecards (https://www.genecards.org/). (2) The molecular targets of HSYA were filtered by searching the keywords “hydroxysafflor yellow A” from STITCH 5.0 (http://stitch.embl.de/), SwissTargetPrediction (http://www.swisstargetprediction.ch/), ChEMBL (https://www.ebi.ac.uk/chembl/) and A Bioinformatics Analysis Tool for Molecular Mechanism of Traditional Chinese Medicine (BATMAN-TCM, http://bionet.ncpsb.org/batman-tcm/). The PubChem CID (6443665) of HSYA was imported into BATMAN-TCM to obtain the compound-target-pathway network. SMILES or the name of HSYA was imported into SwissTargetPrediction or STITCH to acquire the related targets in Rattus norvegicus, respectively. (3) The intersection of (1) and (2) was considered the predicted target of HSYA against TBI. These targets were imported into UniProtKB (http://www.uniprot.org/) to standardize the gene and protein names. (4) A protein–protein interaction (PPI) network was established by STRING 11.0 (https://string-db.org/) and Cytoscape 3.7.2. Hub genes were obtained using CytoHubba in Cytoscape. (5) The pathway and Gene Ontology (GO) enrichments of potential targets were analyzed by ClueGO in Cytoscape. The KEGG pathway analysis was set as p-value < 0.05. (6) The identified differential metabolites in metabolomics were imported into Cytoscape equipped with MetScape to obtain the compound-reaction-enzyme-gene network. This construction was performed to visualize the interactions among the metabolites, pathways, enzymes and genes. (7) The key metabolites and proteins were recognized by combining the compound-reaction-enzyme-gene network with hub genes and metabolic pathways.
2.8. Molecular docking
The 3D structure of HSYA was obtained from PubChem Compound (https://www.ncbi.nlm.nih.gov/pccompound, PubChem CID: 6443665). The crystal structures of targets were acquired from the RCSB Protein Data Bank (https://www.rcsb.org/). Three protein targets were studied: acetylcholinesterase (ACHE, PDB ID: 1E66), neuronal nitric oxide synthase (NOS1, PDB ID: 4IMS), and prostaglandin-endoperoxide synthase 2 (PTGS2, PDB ID: 4RS0). HSYA and targets were converted from their native formats into pdbqt formats with AutoDockTools 1.5.6 [20]. The structures were optimized by deleting water molecules and adding hydrogen atoms. Then, the molecular docking study was performed using Autodock Vina. The coordinates of the target active pocket are listed in Table S1. Size_x = 60, size_y = 60, size_z = 60 in each target. The docking process was calculated by the Genetic Algorithm. All docking run options were default values. Finally, the docking results with the highest scores were visualized by PyMoL.
2.9. Statistical analysis
The data were expressed as the mean ± SD. Statistical analysis was performed by two-way ANOVA followed by Dunnett’s multiple comparisons using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA). A p-value of < 0.05 was considered to be statistically significant.
3. Results
3.1. HSYA treatment ameliorates neurological deficits in CCI rats
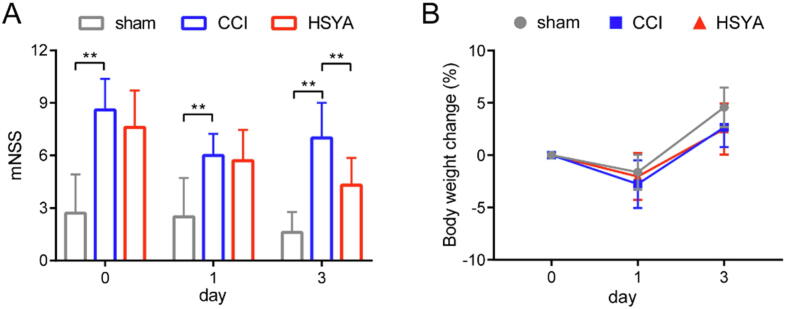
Day 0 means the day having the CCI operation. As shown in Fig. 2A, compared with the sham group, the mNSS scores in the CCI group were increased significantly on day 0, indicating the successful model we induced. On day 3, compared with the CCI group, HSYA treatment markedly decreased the mNSS scores of CCI rats. The initial weights were not significantly different among the three groups (Fig. S2). The change in weight did not differ significantly too.
Fig. 2.
Neurobehavioral scores (A) and body weight changes (B) on day 0, 1 and 3 after injury. All data are expressed as mean ± SD, n = 10, **p < 0.01.
3.2. Metabolomics profiling
A total of 1599 features in the cortex and 1687 in the hippocampus were determined after the data preprocessing. 87 metabolites in the cortex and 74 in the hippocampus were identified respectively. The stability and repeatability of metabolomics were evaluated by QC samples. As shown in Fig. S3A, 96.5% and 96.4% of metabolites had an RSD% < 30% in the cortex and hippocampus, respectively. Furthermore, unsupervised PCA and the representative total ion chromatograms (TICs) showed that QC samples behaved stably during the process (Fig. S3B–E). These data suggested the high stability of the instrument and the repeatability of the method.
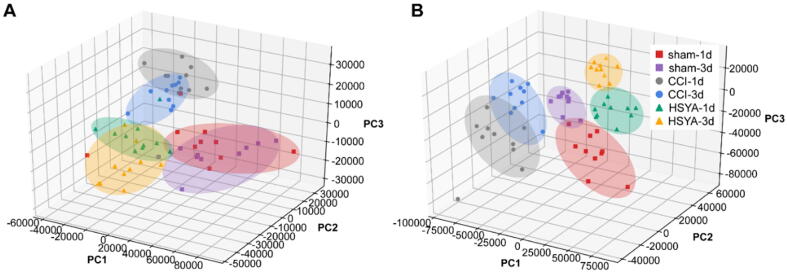
Fig. S4 shows the TICs of cortex samples on days 1 and 3. To investigate the separation among the sham, CCI and HSYA groups, we performed PCA and PLS-DA analysis. As shown in Fig. 3, PLS-DA displayed that the samples from the same group clustered together and samples from different groups distinguished well. The parameters of R2X, R2Y and Q2 in PLS-DA of cortex samples were 0.8, 0.712 and 0.528; The parameters of R2X, R2Y and Q2 in PLS-DA of hippocampus samples were 0.789, 0.736 and 0.62, respectively. These results indicated that the CCI operation and HSYA treatment caused obvious metabolic variations.
Fig. 3.
PLS-DA score plots of HSYA on CCI rats in the cortex (A) and hippocampus (B) on day 1 and 3 after injury.
3.3. Differential metabolite identification and pathway analysis
To identify the potential metabolites that contributed to the metabolic distinction, we performed OPLS-DA and ANOVA followed by FDR. Each OPLS-DA model showed good separation with high R2Y and Q2, indicating good explanative ability of sample classification information and cross-validated predictive capability (Table S2). Moreover, the permutation test showed the models were non-overfitting and reliable. Besides, new successful OPLS-DA models were built using random 80% of raw data as the training set and the rest 20% as the independent test set for validation. Fig. S5 showed good separation within the new models, R2Y and Q2 were all higher than 0.91 and 0.78, respectively (Table S2). The validation results with a good predictive accuracy further proved the reliability of models.
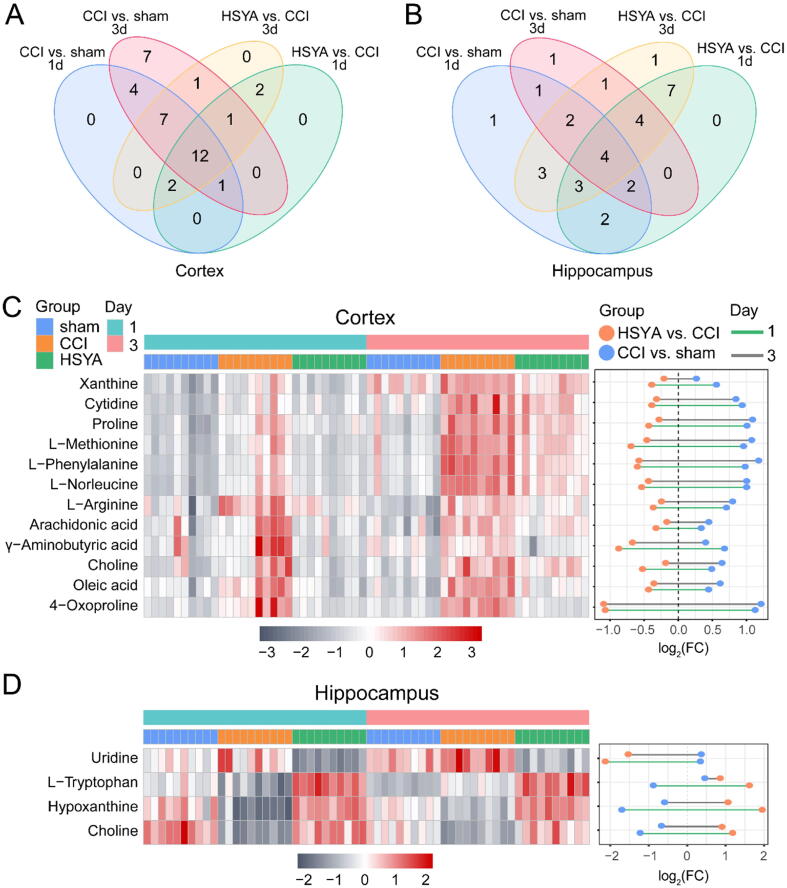
Based on VIP > 1 and q < 0.05, in the cortex, 26 and 33 metabolites were differentially expressed between the sham and CCI group on day 1 and 3, respectively; 18 and 25 metabolites were differentially expressed between the HSYA and CCI group on day 1 and 3, respectively. In the hippocampus, 18 and 15 metabolites were differentially expressed between the sham and CCI group on day 1 and 3, respectively; 22 and 25 metabolites were differentially expressed between the HSYA and CCI group on day 1 and 3, respectively. The information of these metabolites was shown in Supplementary File S1. Twelve and four metabolites were identified as differential metabolites that HSYA affected CCI rats in the cortex and hippocampus, respectively (Fig. 4, Table 1). The MS/MS spectra of differential metabolites are presented in Fig. S6. We calculated the accuracy of the metabolites by multivariate receiver operating characteristic (ROC) curves (Fig. S7). The ROC curves and predictive accuracy plots showed high diagnostic accuracies among the three groups.
Fig. 4.
The differential metabolites in CCI rats treated by HSYA. (A and B) Venn diagrams of the potential metabolites associated with CCI and HSYA treatment on day 1 and 3. (C and D) The heat maps and fold change dumbbell charts of potential metabolites. Data were calculated by the Pearson correlation method after mean centering and unit variance scaling.
Table 1.
The differential metabolites in HSYA treated CCI rats.
| Metabolites | Formula | Molecular Weight | RT (min) | MS/MS | Adduct | KEGG ID | Part | CCI vs. sham 1d |
HSYA vs. CCI 1d |
CCI vs. sham 3d |
HSYA vs. CCI 3d |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VIP | q-value | VIP | q-value | VIP | q-value | VIP | q-value | ||||||||
| L-Phenylalanine | C9H11NO2 | 165.078 | 5.991 | 53.040;77.039;119.073 | [M+H]+ | C00079 | Cortex | 5.181 | 0.000 | 4.321 | 0.005 | 6.829 | 0.000 | 6.070 | 0.000 |
| Choline | C5H13NO | 103.100 | 6.742 | 60.082;67.546;87.045 | [M+H]+ | C00114 | Cortex | 5.137 | 0.006 | 6.844 | 0.005 | 6.578 | 0.000 | 3.885 | 0.049 |
| Arachidonic acid | C20H32O2 | 304.241 | 0.929 | 97.066;205.195;303.233 | [M−H]- | C00219 | Cortex | 4.424 | 0.011 | 5.643 | 0.013 | 5.540 | 0.000 | 4.156 | 0.039 |
| L-Norleucine | C6H13NO2 | 131.094 | 6.840 | 56.050;69.071;86.097 | [M+H]+ | C01933 | Cortex | 4.286 | 0.000 | 3.516 | 0.000 | 4.917 | 0.000 | 4.150 | 0.000 |
| Oleic acid | C18H34O2 | 282.256 | 0.935 | 71.013;127.076;282.252 | [M−H]- | C00712 | Cortex | 4.024 | 0.000 | 4.630 | 0.000 | 4.481 | 0.000 | 4.158 | 0.000 |
| Cytidine | C9H13N3O5 | 243.085 | 4.208 | 266.075;134.033;91.752 | [M+H]+ | C00475 | Cortex | 3.717 | 0.000 | 2.094 | 0.040 | 3.878 | 0.000 | 2.554 | 0.025 |
| L-Proline | C5H9NO2 | 115.063 | 9.102 | 68.050;98.060;116.017 | [M+H]+ | C00148 | Cortex | 3.260 | 0.000 | 2.296 | 0.016 | 3.575 | 0.000 | 2.124 | 0.013 |
| L-Methionine | C5H11NO2S | 149.050 | 7.945 | 61.011;102.055;122.032 | [M+H]+ | C00073 | Cortex | 2.722 | 0.000 | 2.417 | 0.005 | 3.231 | 0.000 | 2.626 | 0.000 |
| 4-Oxoproline | C5H7NO3 | 129.041 | 7.498 | 82.028;84.044;127.896 | [M−H]- | C01877 | Cortex | 3.089 | 0.000 | 3.374 | 0.000 | 3.155 | 0.000 | 3.710 | 0.000 |
| L-Arginine | C6H14N4O2 | 174.111 | 21.974 | 70.066;116.071;175.119 | [M+H]+ | C00062 | Cortex | 3.516 | 0.000 | 2.700 | 0.010 | 3.076 | 0.000 | 1.919 | 0.08 |
| Xanthine | C5H4N4O2 | 152.032 | 2.984 | 65.997;108.019;151.025 | [M−H]- | C00385 | Cortex | 4.316 | 0.000 | 3.705 | 0.005 | 2.791 | 0.003 | 3.317 | 0.019 |
| γ-Aminobutyric acid | C4H9NO2 | 103.064 | 17.100 | 69.034;86.061;104.071 | [M+H]+ | C00334 | Cortex | 3.599 | 0.016 | 5.266 | 0.008 | 2.498 | 0.012 | 4.590 | 0.000 |
| Hypoxanthine | C5H4N4O | 136.038 | 2.295 | 82.041;104.108;106.065 | [M+H]+ | C00262 | Hippocampus | 10.170 | 0.000 | 12.954 | 0.000 | 7.231 | 0.000 | 11.798 | 0.000 |
| Choline | C5H13NO | 103.100 | 7.120 | 74.876;67.546;87.045 | [M+H]+ | C00114 | Hippocampus | 7.301 | 0.000 | 7.650 | 0.000 | 5.720 | 0.000 | 7.038 | 0.000 |
| Uridine | C9H12N2O6 | 244.069 | 2.273 | 82.028;140.034;200.056 | [M+Cl]- | C00299 | Hippocampus | 2.048 | 0.044 | 5.681 | 0.000 | 3.404 | 0.006 | 6.675 | 0.000 |
| L-Tryptophan | C11H12N2O2 | 204.089 | 6.620 | 74.023;116.049;203.082 | [M−H]- | C00078 | Hippocampus | 1.218 | 0.000 | 2.286 | 0.000 | 1.106 | 0.000 | 2.015 | 0.000 |
To visualize the variation in metabolites among the three groups, we plotted heat maps and dumbbell charts. Fig. 4C and D show that all of the candidate metabolites were changed in the CCI group, and most of them were reversed in the HSYA group, indicating that HSYA treatment could reduce metabolic perturbation.
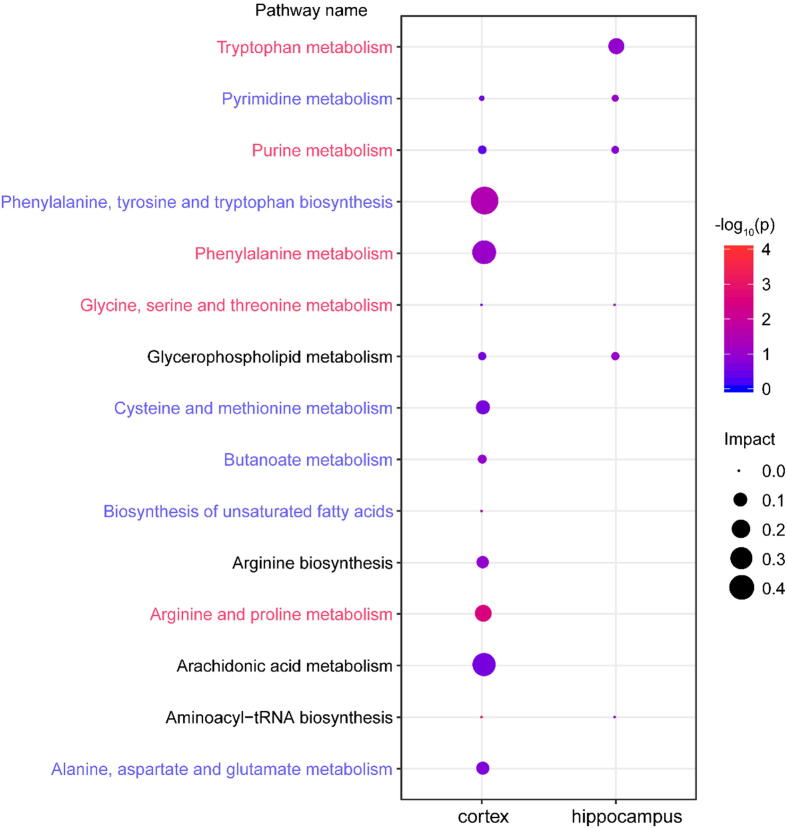
To explore the metabolic pathways of HSYA in CCI rats, we imported these differential metabolites to MetaboAnalyst 4.0. As shown in Fig. 5, based on pathway impact > 0.1, 4 pathways were affected significantly in the cortex, including arginine and proline metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, phenylalanine metabolism and arachidonic acid metabolism. The metabolites related to these pathways were L-arginine, L-proline, γ-aminobutyric acid, l-phenylalanine and arachidonic acid. Tryptophan metabolism was notably affected in the hippocampus with l-tryptophan as the related metabolite.
Fig. 5.
The metabolic pathways of significant metabolites in the cortex and hippocampus. Node size is based on impact values, node color is based on -log10(p) values. The pathways enriched in BATMAN-TCM are colored by names. The pathways marked in red are statistically different with a p-value < 0.05 in the BATMAN-TCM analysis. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Network pharmacology
To further explore the mechanisms of HSYA against TBI, we conducted network pharmacology. First, BATMAN-TCM was used for preliminary analysis. As shown in Fig. S8, brain injury was predicted as one of the related diseases of HSYA therapy. Furthermore, 11 pathways in the metabolomic analysis were also enriched in the BATMAN-TCM prediction, and 5 of them were significantly affected with adjusted p-values < 0.05 (Fig. 5). Such a high consistency validated the accuracy of pathway analysis in the metabolomics.
Subsequently, we collected the targets of TBI from the OMIM, TCMSP, TTD and Genecards databases and gathered the targets of HSYA from the BATMAN-TCM, ChEMBL, STITCH and SwissTargetPrediction databases. After matching the 86 HSYA-related targets with the TBI-related targets, 41 targets were identified as potential targets for HSYA to treat TBI (Table S3). All the intersected targets were normalized to their official symbols by the UniProt database.
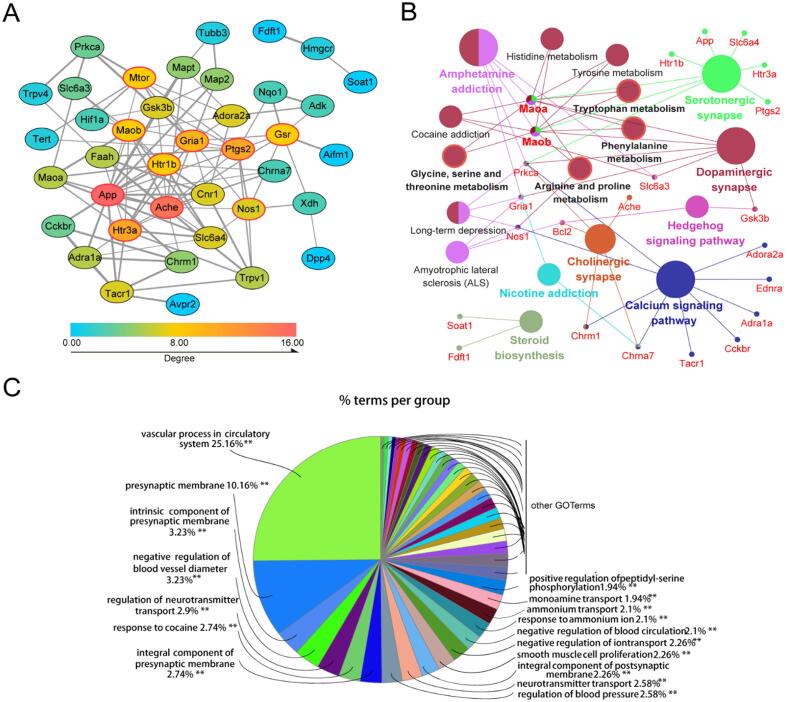
To identify the hub genes of HSYA against TBI, we constructed PPI network by Cytoscape. Fig. 6A gives a whole view of the relationships within 39 targets (the other two genes were disconnected). The hub genes were calculated by CytoHubba. Combining the scores of 10 computational methods, the top 10 genes were considered hub genes (ache, app, ptgs2, gria1, htr3a, nos1, mtor, gsr, maob, htr1b). The details are presented in Fig. 6A and Table S4.
Fig. 6.
Network pharmacology analysis of HSYA treating TBI. (A) The PPI network of HSYA treatment on TBI. Node color reflects its degree. The nodes with red borders represent the hub genes. (B) The KEGG pathways enrichment analysis by ClueGO. All pathways have a p-value of < 0.05. (C) The GO enrichment analysis of potential targets by ClueGO. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To decipher the neuroprotective function of the potential targets, we performed GO and KEGG pathway enrichment analyses by ClueGO (Fig. 6B and C). The top terms in GO analysis were vascular process in circulatory system (GO: 0003018), presynaptic membrane (GO: 0042734), intrinsic component of presynaptic membrane (GO: 0098889), negative regulation of blood vessel diameter (GO: 0097756), and regulation of neurotransmitter transport (GO: 0051588). According to the KEGG enrichment analysis, the pathways affected significantly were serotonergic synapse, dopaminergic synapse, cholinergic synapse and amino acid metabolism.
3.5. Integrated analysis of metabolomics and network pharmacology
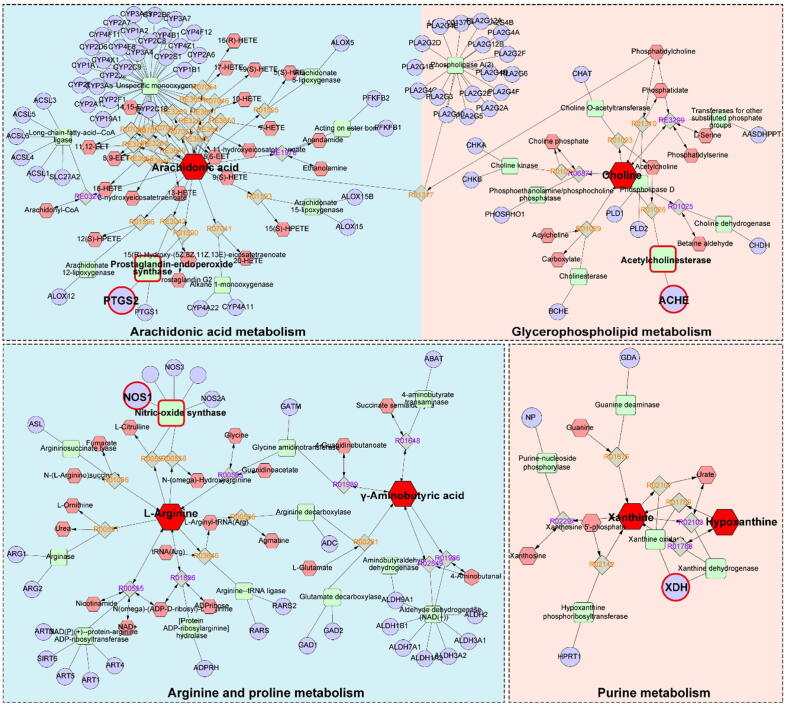
To obtain a comprehensive view of the mechanisms of HSYA against TBI, we constructed an interaction network based on metabolomics and network pharmacology (Fig. 7). Differential metabolites were imported into the MetScape plugin in Cytoscape to collect the compound-reaction-enzyme-gene networks. By matching the potential targets identified in network pharmacology with the genes in MetScape analysis, we found 4 key targets, including NOS1, ACHE, PTGS2 and xanthine oxidase (XDH) (Table 2). The related key metabolites were L-arginine, γ-aminobutyric acid, choline, arachidonic acid, xanthine and hypoxanthine. The affected pathways were arginine and proline metabolism, glycerophospholipid metabolism, arachidonic acid metabolism, and purine metabolism. They may play essential roles in the therapeutic effect of HSYA on TBI. Among these genes, NOS1, ACHE and PTGS2 are hub genes.
Fig. 7.
The compound-reaction-enzyme-gene networks of the key metabolites and targets. The red hexagons, grey diamonds, green round rectangle and purple circles represent the active compounds, reactions, proteins and genes, respectively. The key metabolites, proteins and genes were magnified. The pathways with a blue background are significantly regulated in the cortex. The pathways with a red background are significantly regulated in both the cortex and hippocampus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 2.
The information of key targets, metabolites and pathways.
| Related pathway | Key target | Key metabolite |
|---|---|---|
| Arginine and proline metabolisma | NOS1 | l-Arginine, γ-aminobutyric acid, l-proline |
| Arachidonic acid metabolisma | PTGS2 | Arachidonic acid |
| Glycerophospholipid metabolisma, b | ACHE | Choline |
| Purine metabolisma, b | XDH | Xanthine, hypoxanthine |
Pathways affected significantly in the cortex.
Pathways affected significantly in the hippocampus.
3.6. Molecular docking
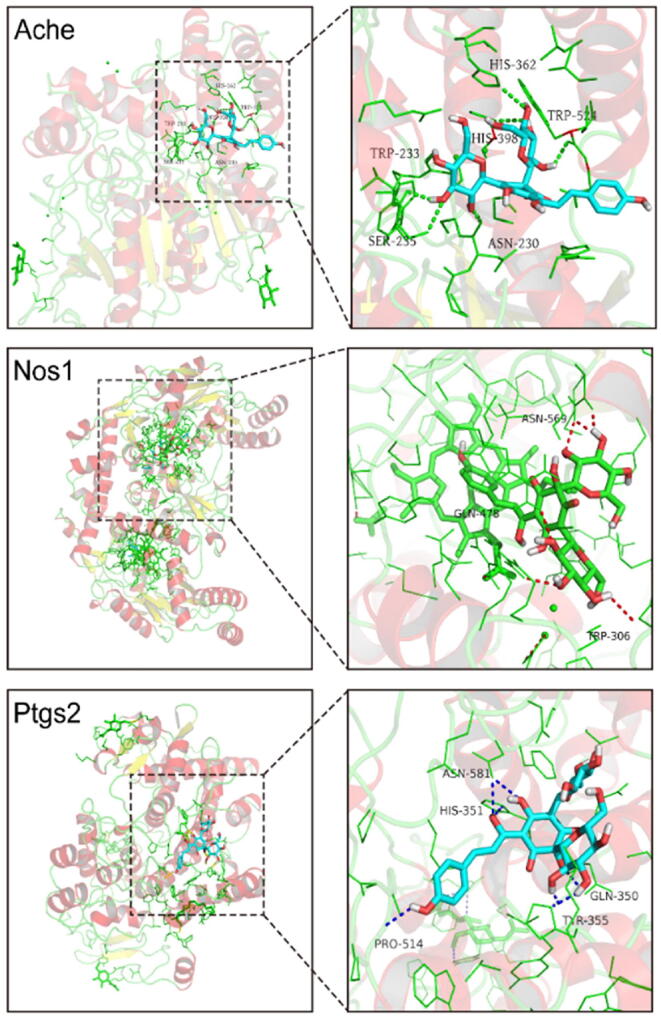
To further investigate the possibility of interaction between HSYA and the key targets, we applied molecular docking studies (Fig. 8). Three key targets could be analyzed by molecular docking after searching the RCSB Protein Data Bank database. The docking analysis of ACHE showed that HSYA made hydrogen-bonding interactions with PRO-232, SER-235, ASN-230, and TRP-524 at the active site. In the interaction with NOS1, HSYA made hydrogen-bonding interaction with TRP-306, GLN-478, and ASN-569. In addition, HSYA formed a pi-anion with GLU-592, a pi-alkyl with HEM-801, VAL-567, and a carbon-hydrogen bond with CL-806. In the interaction with PTGS2, HSYA formed hydrogen bonds with TYR-355, GLN-350, ASN-581, HIS-351, and PRO-514. HSYA also formed a carbon-hydrogen bond with ASP-584, ASP-347, and Pi-sigma with GLN-192. The binding energies of HSYA towards ACHE, NOS1 and PTGS2 were −7.9, −8.9 and −9.0 kcal/mol, respectively. These docking results indicated the high affinities between HSYA and the key targets, especially NOS1 and PTGS2.
Fig. 8.
The 3D interaction diagrams of HSYA and the key targets.
4. Discussion
Researchers are increasingly relying on metabolomics to study disease mechanisms and intervention strategies. We identified 12 significant metabolites of HSYA against TBI in the cortex and 4 in the hippocampus, as well as their related pathways. However, given the complexity and heterogeneity of metabolomics, data analysis and interpretation are collaborative efforts [21]. Network pharmacology greatly improves the screening of metabolites of HSYA against TBI and explicates the action mechanisms. By combining metabolomics with network pharmacology, we found 4 key targets (NOS1, ACHE, PTGS2, XDH), 7 key metabolites (l-arginine, γ-aminobutyric acid, l-proline, arachidonic acid, choline, xanthine, hypoxanthine) and 4 related pathways (arginine and proline metabolism, arachidonic acid metabolism, glycerophospholipid metabolism, purine metabolism). This strategy provides a suitable method to verify the results of the two approaches. It is also practicable to screen metabolites and targets in other natural compounds.
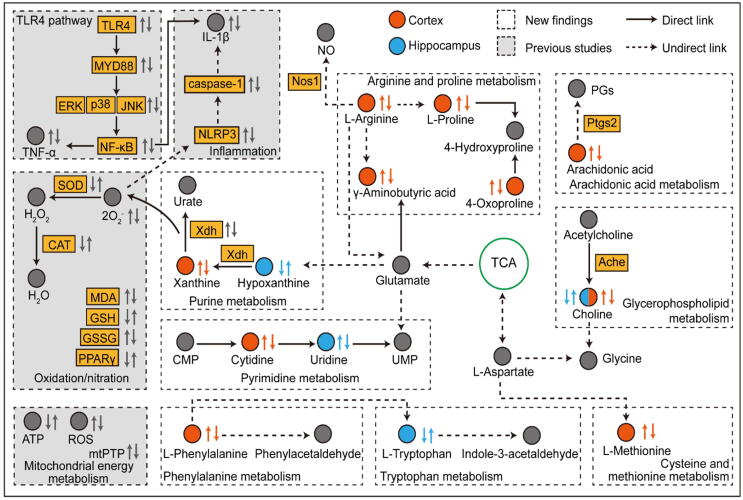
Previous studies have demonstrated the possible mechanisms of HSYA in treating TBI. We found that HSYA could effectively penetrate the injured blood–brain barrier of TBI patients [7]. HSYA exerts antioxidant effects by increasing superoxide dismutase (SOD), catalase (CAT) and glutathione (GSH), as well as decreasing malondialdehyde (MDA) and glutathione disulfide (GSSG) [8]. Sun et al. revealed that HSYA protects neurons from nitrosative stress by keeping PPARγ as a functional receptor [22]. Tian et al. reported that HSYA improves mitochondrial energy metabolism and inhibits the opening of mitochondrial permeability transition pores (mtPTPs) by scavenging free radicals in the brain [23]. Xu et al. proved that XDH may be a potential target of HSYA [24]. The direct binding of HSYA-XDH suppresses lipopolysaccharides (LPS) -induced reactive oxygen species (ROS), inhibits NLR family pyrin domain containing 3 (NLRP3) inflammasome and prevents the secretion of IL-1β in macrophages. Lv et al. suggested that HSYA exerts neurotrophic and anti-inflammatory functions by inhibiting toll-like receptor 4 (TLR4) pathway mediated signalling [25]. In our present study, we investigated the metabolic variations of the cortex and hippocampus, which are the most vulnerable regions post-TBI [26]. The cortex affected more metabolites and pathways with more complicated interaction networks than the hippocampus, which proved that metabolic disturbance is worse in the cortex than in the hippocampus at the acute stage [27]. Pathways affected significantly in the cortex are arginine and proline metabolism, arachidonic acid metabolism, glycerophospholipid metabolism and purine metabolism. Pathways affected markedly in the hippocampus are glycerophospholipid metabolism and purine metabolism. Fig. 9 shows the complex mechanisms of the interaction network, which contains the previous results and the new findings.
Fig. 9.
The interaction network based on metabolomics and network pharmacology. The first and the second arrow near metabolites from left to right denote changes in CCI vs. sham and HSYA vs. CCI groups, respectively.
Metabolomics studies are limited to a listing of potential metabolites and related pathways without further exploration of their direct relationships. Network pharmacology is a system biology-based methodology [16]. It evaluates drug polypharmacological effects at a molecular level to predict the interaction of natural products and proteins as well as to determine the major mechanisms [28]. Network pharmacology can further validate the therapeutic regulation of metabolic networks and facilitate the identification of key targets and biomarkers [29]. By combining metabolomics with network pharmacology, this integrated strategy finds the core targets and mechanisms and provides a more precise network of HSYA against TBI.
Arginine and proline metabolism represents a potential therapeutic pathway in acute brain injury [30]. l-arginine serves as a precursor to synthesize l-proline, nitric oxide (NO) and glutamate [31]. As the last step in the urea cycle, arginase cleaves l-arginine to form urea and l-ornithine. Then, l-ornithine synthesizes l-proline through the activity of ornithine aminotransferase [32]. l-proline is a promising biomarker of TBI that correlates positively with neurological deficits [33]. The increased level of l-proline in the CCI group implicates neurological impairment post-trauma. l-proline is required for extracellular matrix remodeling, especially for collagen synthesis. After treatment with HSYA, both l-arginine and l-proline were down-regulated, indicating a high demand for l-proline for tissue repair. Apart from the role in generating l-proline, arginine is also the substrate for NO synthase (NOS) to produce NO [30]. NO generated from endothelial NOS (NOS3) improves cerebral blood flow after brain injury [34]. However, the overproduction of NO from NOS1 may result in excitotoxicity and energy depletion of neurons [35], [36]. Molecular docking shows that HSYA binds tightly with NOS1, indicating that HSYA may reduce neurological impairment by inhibiting NOS1 and decreasing NO production. Glutamate is a primary excitatory neurotransmitter in the brain, while γ-aminobutyric acid is a major inhibitory neurotransmitter. Glutamate can be converted into γ-aminobutyric acid, which regulates homeostasis [37]. In the CCI group, the level of γ-aminobutyric acid was disturbed, which is consistent with a previous document [38]. After HSYA treatment, it was restored to normality.
Neuroinflammation is a key pathological response to brain injury. Proinflammatory molecules disperse throughout the brain and cause neuronal damage [39]. PTGS2, also known as cyclooxygenase-2, plays a central role in the acute inflammatory cascade by converting arachidonic acid into bioactive prostanoids. PTGS2 is the best target for anti-inflammatory drugs [40]. We observed an elevated level of arachidonic acid in the CCI group, which is consistent with the previous study [41]. HSYA downregulates arachidonic acid, which exerts an anti-inflammatory effect on TBI. Further molecular docking analysis indicated that the mechanism may be involved in inhibiting PTGS2 by HSYA.
In glycerophospholipid metabolism, choline is a major precursor of membrane phospholipids and acetylcholine, which plays an essential role in cognitive function [42]. ACHE hydrolyses acetylcholine to choline and thereby terminates synaptic transmission. TBI causes neuronal depolarizations that deplete acetylcholine. When there is no available choline to generate acetylcholine, additional choline is utilized from membrane phospholipids, which results in neuronal damage [43]. In the hippocampus, the activity of ACHE is decreased post-trauma [44] with a low level of choline. HSYA upregulates choline to protect the cell membrane from disruption. In the cortex, low activity of acetylcholine triggers the excessive release of choline postinjury and HSYA reverses it to normal conditions.
Oxidative stress occurs shortly after TBI by releasing ROS [45]. At the end of purine metabolism, hypoxanthine catabolizes to xanthine by XDH, which is one of the major sources of ROS [46]. The balance of hypoxanthine and xanthine is disrupted post-TBI and restored after HSYA treatment, suggesting that HSYA protects organisms from oxidative stress. This is consistent with our previous work [8]. The mechanism may involve suppressing XDH according to the network pharmacology analysis, which is consistent with the results of Xu et al. [24].
Tryptophan metabolism is also implicated in the hippocampus. l-tryptophan, an essential amino acid required for protein synthesis, is the obligatory substrate of serotonin and kynurenine [47]. The hippocampus is a notable region with a general change in l-tryptophan and its metabolites [48]. Consistent with the previous study, we observed a decreased level of l-tryptophan in the CCI group on day 1, which may negatively impact cognitive function, brain structure and mood [49], HSYA reversed this alteration. On day 3, the increased l-tryptophan in the CCI group demonstrates that the organism tries to repair homeostasis. Moreover, HSYA accelerates this recovery process.
5. Conclusion
In this study, we first developed a novel integrated strategy to explore the key targets and mechanisms of HSYA in treating acute TBI based on metabolomics and network pharmacology. We identified the region-specific metabolic responses towards HSYA therapy in the cortex and hippocampus. The integrated analysis revealed 4 key targets as well as related metabolites and pathways. These targets were further validated by molecular docking. This research offers data and theoretical support for an in-depth study of mechanisms and lays a foundation for clinical application. Further systematic molecular biology experiments are needed to verify the accurate mechanisms. It also provides a novel paradigm to identify the potential mechanisms of pharmacological effects derived from a natural compound.
CRediT authorship contribution statement
Teng Li: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Wei Zhang: Data curation, Investigation, Funding acquisition. En Hu: Conceptualization, Methodology, Software, Validation, Formal analysis. Zhengji Sun: Methodology, Software, Visualization. Pengfei Li: Methodology, Validation, Resources. Zhe Yu: Methodology, Software, Validation. Xiaofei Zhu: Formal analysis, Visualization. Fei Zheng: Methodology, Software, Visualization. Zhihua Xing: Supervision, Project administration, Funding acquisition. Zian Xia: Resources, Supervision, Funding acquisition. Feng He: Methodology, Validation. Jiekun Luo: Resources, Project administration. Tao Tang: Resources, Writing - review & editing, Supervision. Yang Wang: Conceptualization, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was funded by grants from the National Natural Science Foundation of China (No. 81973665, 81673719, 81874409 and 81803948), Hunan Provincial Natural Science Foundation of China (No. 2019JJ30042), The Outstanding Youth Fund of Hunan Natural Science (No. 2020JJ2024), and Innovation-Driven Project of Central South University (No. 2020CX047).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.01.033.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Maas A.I.R., Menon D.K., Adelson P.D., Andelic N., Bell M.J., Belli A. Traumatic brain injury: integrated approaches to improve prevention, clinical care, and research. Lancet Neurol. 2017;16:987–1048. doi: 10.1016/S1474-4422(17)30371-X. [DOI] [PubMed] [Google Scholar]
- 2.Di Pietro V., Yakoub K.M., Caruso G., Lazzarino G., Signoretti S., Barbey A.K. Antioxidant Therapies in Traumatic Brain Injury. Antioxidants (Basel, Switzerland) 2020;9(3):260. doi: 10.3390/antiox9030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheff S.W., Ansari M.A. Natural compounds as a therapeutic intervention following traumatic brain injury: the role of phytochemicals. J Neurotrauma. 2017;34(8):1491–1510. doi: 10.1089/neu.2016.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H.-S., Qi S.-H., Shen J.-G. One-compound-multi-target: combination prospect of natural compounds with thrombolytic therapy in acute ischemic stroke. Curr Neuropharmacol. 2016;15(1):134–156. doi: 10.2174/1570159X14666160620102055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen L., Xiang Y., Kong L., Zhang X., Sun B., Wei X. Hydroxysafflor yellow A protects against cerebral ischemia-reperfusion injury by anti-apoptotic effect through PI3K/Akt/GSK3β pathway in rat. Neurochem Res. 2013;38(11):2268–2275. doi: 10.1007/s11064-013-1135-8. [DOI] [PubMed] [Google Scholar]
- 6.Yu L.u., Liu Z., He W., Chen H., Lai Z., Duan Y. Hydroxysafflor yellow a confers neuroprotection from focal cerebral ischemia by modulating the crosstalk between JAK2/STAT3 and SOCS3 signaling pathways. Cell Mol Neurobiol. 2020;40(8):1271–1281. doi: 10.1007/s10571-020-00812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng C, Peng W, Xia Z, Wang Y. Plasma and cerebrospinal fluid pharmacokinetics of hydroxysafflor yellow A in patients with traumatic brain injury after intravenous administration of Xuebijing using LC-MS/MS method. Xenobiotica; the fate of foreign compounds in biological systems 2020; 50: 545-51. [DOI] [PubMed]
- 8.Wang Y, Zhang C, Peng W, Xia Z, Gan P, Huang W, et al. Hydroxysafflor yellow A exerts antioxidant effects in a rat model of traumatic brain injury. Molecular medicine reports 2016; 14: 3690-6. [DOI] [PMC free article] [PubMed]
- 9.Nicholson J.K., Wilson I.D. Opinion: understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discovery. 2003;2(8):668–676. doi: 10.1038/nrd1157. [DOI] [PubMed] [Google Scholar]
- 10.Simón-Manso Y., Lowenthal M.S., Kilpatrick L.E., Sampson M.L., Telu K.H., Rudnick P.A. Metabolite profiling of a NIST Standard Reference Material for human plasma (SRM 1950): GC-MS, LC-MS, NMR, and clinical laboratory analyses, libraries, and web-based resources. Anal Chem. 2013;85(24):11725–11731. doi: 10.1021/ac402503m. [DOI] [PubMed] [Google Scholar]
- 11.Singh K., Trivedi R., Verma A., D'souza M.M., Koundal S., Rana P. Altered metabolites of the rat hippocampus after mild and moderate traumatic brain injury - a combined in vivo and in vitro (1) H-MRS study. NMR Biomed. 2017;30(10):e3764. doi: 10.1002/nbm.3764. [DOI] [PubMed] [Google Scholar]
- 12.Fu C., Wu Q., Zhang Z., Xia Z., Ji H., Lu H. UPLC-ESI-IT-TOF-MS metabolomic study of the therapeutic effect of Xuefu Zhuyu decoction on rats with traumatic brain injury. J Ethnopharmacol. 2019;245:112149. doi: 10.1016/j.jep.2019.112149. [DOI] [PubMed] [Google Scholar]
- 13.Kibble M., Saarinen N., Tang J., Wennerberg K., Mäkelä S., Aittokallio T. Network pharmacology applications to map the unexplored target space and therapeutic potential of natural products. Nat Prod Rep. 2015;32(8):1249–1266. doi: 10.1039/c5np00005j. [DOI] [PubMed] [Google Scholar]
- 14.Hopkins A.L. Network pharmacology. Nat Biotechnol. 2007;25(10):1110–1111. doi: 10.1038/nbt1007-1110. [DOI] [PubMed] [Google Scholar]
- 15.Gertsch J. Botanical drugs, synergy, and network pharmacology: forth and back to intelligent mixtures. Planta Med. 2011;77(11):1086–1098. doi: 10.1055/s-0030-1270904. [DOI] [PubMed] [Google Scholar]
- 16.Zhong Y., Luo J., Tang T., Li P., Liu T., Cui H. Exploring pharmacological mechanisms of xuefu zhuyu decoction in the treatment of traumatic brain injury via a network pharmacology approach. Evidence-based complementary and alternative medicine: eCAM. 2018;2018:1–20. doi: 10.1155/2018/8916938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y., Fan X., Tang T., Fan R., Zhang C., Huang Z. Rhein and rhubarb similarly protect the blood-brain barrier after experimental traumatic brain injury via gp91(phox) subunit of NADPH oxidase/ROS/ERK/MMP-9 signaling pathway. Sci Rep. 2016;6(1) doi: 10.1038/srep37098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P., Tang T., Liu T., Zhou J., Cui H., He Z. Systematic analysis of tRNA-derived small RNAs reveals novel potential therapeutic targets of traditional chinese medicine (buyang-huanwu-decoction) on intracerebral hemorrhage. Int J Biol Sci. 2019;15(4):895–908. doi: 10.7150/ijbs.29744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn W.B., Broadhurst D., Begley P., Zelena E., Francis-McIntyre S., Anderson N. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc. 2011;6(7):1060–1083. doi: 10.1038/nprot.2011.335. [DOI] [PubMed] [Google Scholar]
- 20.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eicher T., Kinnebrew G., Patt A., Spencer K., Ying K., Ma Q. Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites. 2020;10(5):202. doi: 10.3390/metabo10050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun L.i., Xu Y.-W., Han J., Xiao C., Cao S.-S., Liang H. Hydroxysafflor yellow a shows protection against PPARγ inactivation in nitrosative neurons. Oxid Med Cell Longevity. 2018;2018:1–13. doi: 10.1155/2018/9101740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian J., Li G., Liu Z., Fu F. Hydroxysafflor yellow A inhibits rat brain mitochondrial permeability transition pores by a free radical scavenging action. Pharmacology. 2008;82(2):121–126. doi: 10.1159/000141653. [DOI] [PubMed] [Google Scholar]
- 24.Xu X., Guo Y., Zhao J., Wang N., Ding J., Liu Q. Hydroxysafflor yellow A inhibits LPS-induced NLRP3 inflammasome activation via binding to xanthine oxidase in mouse RAW264.7 macrophages. Mediators Inflamm. 2016;2016:8172706. doi: 10.1155/2016/8172706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lv Y., Qian Y., Ou-yang A., Fu L. Hydroxysafflor yellow A attenuates neuron damage by suppressing the lipopolysaccharide-induced TLR4 pathway in activated microglial cells. Cell Mol Neurobiol. 2016;36(8):1241–1256. doi: 10.1007/s10571-015-0322-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carron S.F., Alwis D.S., Rajan R. Traumatic brain injury and neuronal functionality changes in sensory cortex. Front Syst Neurosci. 2016;10:47. doi: 10.3389/fnsys.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall E.D., Sullivan P.G., Gibson T.R., Pavel K.M., Thompson B.M., Scheff S.W. Spatial and temporal characteristics of neurodegeneration after controlled cortical impact in mice: more than a focal brain injury. J Neurotrauma. 2005;22(2):252–265. doi: 10.1089/neu.2005.22.252. [DOI] [PubMed] [Google Scholar]
- 28.Sheng S., Wang J., Wang L., Liu H., Li P., Liu M. Network pharmacology analyses of the antithrombotic pharmacological mechanism of Fufang Xueshuantong Capsule with experimental support using disseminated intravascular coagulation rats. J Ethnopharmacol. 2014;154(3):735–744. doi: 10.1016/j.jep.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 29.Yu H., Chen J., Xu X., Li Y., Zhao H., Fang Y. A systematic prediction of multiple drug-target interactions from chemical, genomic, and pharmacological data. PLoS ONE. 2012;7(5):e37608. doi: 10.1371/journal.pone.0037608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fouda A.Y., Eldahshan W., Narayanan S.P., Caldwell R.W., Caldwell R.B. Arginase pathway in acute retina and brain injury: therapeutic opportunities and unexplored avenues. Front Pharmacol. 2020;11:277. doi: 10.3389/fphar.2020.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu G., Morris S.M., Jr. Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336(Pt 1):1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caldwell R.W., Rodriguez P.C., Toque H.A., Narayanan S.P., Caldwell R.B. Arginase: a multifaceted enzyme important in health and disease. Physiol Rev. 2018;98(2):641–665. doi: 10.1152/physrev.00037.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louin G., Neveux N., Cynober L., Plotkine M., Marchand-Leroux C., Jafarian-Tehrani M. Plasma concentrations of arginine and related amino acids following traumatic brain injury: Proline as a promising biomarker of brain damage severity. Nitric Oxide Boil Chem. 2007;17(2):91–97. doi: 10.1016/j.niox.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Garry P.S., Ezra M., Rowland M.J., Westbrook J., Pattinson K.T.S. The role of the nitric oxide pathway in brain injury and its treatment–from bench to bedside. Exp Neurol. 2015;263:235–243. doi: 10.1016/j.expneurol.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 35.Förstermann U., Sessa W.C. Nitric oxide synthases: regulation and function. Eur Heart J. 2012;33(7):829–837. doi: 10.1093/eurheartj/ehr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown G.C. Nitric oxide and neuronal death. Nitric Oxide Biol Chem. 2010;23(3):153–165. doi: 10.1016/j.niox.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Guerriero R.M., Giza C.C., Rotenberg A. Glutamate and GABA imbalance following traumatic brain injury. Curr Neurol Neurosci Rep. 2015;15:27. doi: 10.1007/s11910-015-0545-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amorini A.M., Lazzarino G., Di Pietro V., Signoretti S., Lazzarino G., Belli A. Severity of experimental traumatic brain injury modulates changes in concentrations of cerebral free amino acids. J Cell Mol Med. 2017;21(3):530–542. doi: 10.1111/jcmm.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonsack B., Heyck M., Kingsbury C., Cozene B., Sadanandan N., Lee J.-Y. Fast-tracking regenerative medicine for traumatic brain injury. Neural Regener Res. 2020;15(7):1179. doi: 10.4103/1673-5374.270294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aïd S., Bosetti F. Targeting cyclooxygenases-1 and -2 in neuroinflammation: Therapeutic implications. Biochimie. 2011;93(1):46–51. doi: 10.1016/j.biochi.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang S., Ma Y., Liu Y., Que H., Zhu C., Liu S. Arachidonic acid: a bridge between traumatic brain injury and fracture healing. J Neurotrauma. 2012;29(17):2696–2705. doi: 10.1089/neu.2012.2442. [DOI] [PubMed] [Google Scholar]
- 42.Blusztajn J.K., Slack B.E., Mellott T.J. Neuroprotective actions of dietary choline. Nutrients. 2017:9. doi: 10.3390/nu9080815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wurtman R.J. Choline metabolism as a basis for the selective vulnerability of cholinergic neurons. Trends Neurosci. 1992;15(4):117–122. doi: 10.1016/0166-2236(92)90351-8. [DOI] [PubMed] [Google Scholar]
- 44.Donat C.K., Schuhmann M.U., Voigt C., Nieber K., Schliebs R., Brust P. Alterations of acetylcholinesterase activity after traumatic brain injury in rats. Brain Inj. 2007;21(10):1031–1037. doi: 10.1080/02699050701630359. [DOI] [PubMed] [Google Scholar]
- 45.Dasuri K., Zhang L.e., Keller J.N. Oxidative stress, neurodegeneration, and the balance of protein degradation and protein synthesis. Free Radical Biol Med. 2013;62:170–185. doi: 10.1016/j.freeradbiomed.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 46.Ozturk E., Demirbilek S., Kadir But A., Saricicek V., Gulec M., Akyol O. Antioxidant properties of propofol and erythropoietin after closed head injury in rats. Prog Neuro-Psychopharmacol Biol Psychiatry. 2005;29(6):922–927. doi: 10.1016/j.pnpbp.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 47.Höglund E., Øverli Ø., Winberg S. Tryptophan metabolic pathways and brain serotonergic activity: a comparative review. Front Endocrinol. 2019;10:158. doi: 10.3389/fendo.2019.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li C.-C., Jiang N., Gan L., Zhao M.-J., Chang Q.i., Liu X.-M. Peripheral and cerebral abnormalities of the tryptophan metabolism in the depression-like rats induced by chronic unpredicted mild stress. Neurochem Int. 2020;138:104771. doi: 10.1016/j.neuint.2020.104771. [DOI] [PubMed] [Google Scholar]
- 49.Durham W.J., Foreman J.P., Randolph K.M., Danesi C.P., Spratt H., Masel B.D. Hypoaminoacidemia characterizes chronic traumatic brain injury. J Neurotrauma. 2017;34(2):385–390. doi: 10.1089/neu.2015.4350. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.