Abstract
Spinal cord injury (SCI) is an insult to the spinal cord resulting in a change, either temporary or permanent, in its normal motor or sensory function, but the mechanism of neuron loss after spinal cord injury is still unclear. Long non-coding RNAs (lncRNAs) can play an important role in regulating cell physiological activities through competitively binding to miRNAs. However, there is still a lack of research on the effect of lncRNAs on SCI. In this study, we selected SCI gene expression data and miRNA expression data from the NCBI database for differential expression analysis, and predicted miRNA target genes. Subsequently, biological analysis of gene expression and miRNA changes was performed on a rat SCI model. The results showed that the expression level of lncRNA-MEG increased significantly in rat SCI model. Subsequently, we found that lncRNA-MEG can promote the expression level of PDCD4 by inhibiting miR-21-5p, which leads to neuronal cell apoptosis. Furthermore, knocking down of lncRNA-MEG with shRNA can reverse the effect of miR-21-5p and inhibit the effect of PDCD4 to reduce the expression of apoptosis-related proteins. Furthermore, we found lncRNA-MEG can regulate PDCD4 expression through miR-21-5p by targeting 3’UTR of PDCD4 in the OGD cell model. In summary, we first discovered lncRNA-MEG regulates neuronal cell apoptosis through miR-21-5p by targeting PDCD4 in SCI.
Keywords: Spinal cord injury, miR-21-5p, PDCD4, LncRNA-MEG
Introduction
Spinal cord injury (SCI) is mainly caused by direct damage to spinal cord tissue and cells caused by external forces. In addition, tumors, viral infections, vascular disease, spondylosis, secondary fractures caused by osteoporosis, and development-related diseases can also cause SCI [1-3]. When a spinal cord injury occurs, it can cause temporary or permanent damage to the spinal cord, resulting in loss of motor and sensory functions in the patient [4-6]. At present, the global incidence of spinal cord injury is about 10.4-83 cases/million/year [7]. In previous studies, researchers have found that the damage process of SCI is very complex, mainly including primary mechanical injury and secondary injury. Among them, mechanical damage caused directly by external forces will directly damage neuronal cells in the first time. Later, including vascular injury, edema, ischemia, changes in electrolytes, free radical generation, and delayed apoptosis of cells will cause the patient’s nervous system to be injured again [8-10]. Because the process of SCI injury is so complicated, the detailed mechanism of SCI is still undefined, which leads to the lack of appropriate methods for SCI treatment.
Some studies have shown that astrocytes in the nervous system will undergo a series of molecular, morphological and functional changes when SCI occurs [11-13]. The expression levels of glial fibrillary acidic protein and various cytokines in cells are significantly increased, thereby promoting the regeneration of nerve cells [14-17]. However, the target gene marker for SCI is ambiguous. Nowadays, researchers have found that non-coding RNA (ncRNA) can regulate gene expression in many biological processes [18]. MiR-137 attenuates spinal cord injury by modulating NEUROD4 through reducing inflammation and oxidative stress [19]. MiR-21-5p can participate in the repair process after neuronal cell injury. In this case, ncRNA may be a potential target for SCI. Therefore, more and more researchers have turned their attention to ncRNAs, hoping to discover the molecular mechanism of SCI progress.
In addition to miRNA, with the continuous development of genome sequencing technology, researchers have found that one type of ncRNA, lncRNA, also plays a very important role in the life process [20]. In previous research, researchers found that lncRNA can inhibit the transcription of genes encoding proteins by interfering with the binding of transcription factors to gene promoters or inducing histone modifications, thereby affecting the production of proteins and thus changing biological effects [21]. At the same time, lncRNA also has a regulatory mechanism for competing endogenous RNA [22]. That is, lncRNA can regulate the transcription and translation of coding genes through RNA-RNA interactions, and then affect biological processes. In 2015, researchers discovered that lncRNA uc.217 can promote the growth of dorsal neuron cells and promote the recovery of neural networks during peripheral nerve injury [23]. However, lncRNA related to the SCI process has not yet been found yet. Therefore, it is necessary to further study the role and mechanism of lncRNA in the SCI process.
In this study, we focused on the role of lncRNA in the SCI process. We first analyzed miRNAs in rats that may be related to the SCI process through biological information. Then we constructed a SCI rat model and performed qPCR on miRNAs predicted to be associated with SCI, with the hope to provide more insights for the researches of spinal cord injury.
Materials and methods
Cell culture
VSC4.1 cells were cultured in DMEM (Gibco, C11995500BT) and supplemented with 10% fetal calf serum (Lonsera, S711-001S), and 1% penicillin/streptomycin (Gibco) in a 37°C incubator containing 5% CO2.
For transfection, VSC4.1 cells in 24-well plates were transfected with the indicated plasmids (500 ng/well), or small interfering RNA (siRNA) at the indicated final concentrations in the culture medium using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After transfection for 72 h, cells were harvested for qPCR or immunoblotting.
Oxygen and glucose deprivation (OGD) model constructing
VSC4.1 were cultured using fetal bovine serum (FBS) and no-glucose Dulbecco’s modified Eagle’s medium (DMEM) with 5% CO2 and 1% O2 at 37°C for 6 hours after the cells having been washed with phosphate-buffered saline three times. After oxygen-glucose deprivation, the medium was changed with normal medium containing high-glucose DMEM, 10% FBS 1% penicillin/streptomycin in a 37°C incubator containing 5% CO2 for 24 hours.
Prediction of miRNA changes in rat SCI
From the NCBI database, gene expression data (GSE45006) and miRNA expression data (GSE19890) of rats with a spinal cord injury time of 7 days and their control group were selected. After downloading the above-mentioned original sequencing files from NCBI, we analyzed them through R software. The differential expression analysis was performed using the limma package, the marl package and the limma package to analyze the heat map of the expression differences during SCI, and the target genes of the selected miRNA were predicted using the three databases of miRDB, miRTarBase and TargetScan to obtain candidate miRNAs that were up-regulated or down-regulated and their corresponding potential target genes. The genes in the other set of data were matched. Network maps of successfully aligned genes and their miRNAs.
Construction of rat SCI model
All animal experiments were approved by the ethics committee and all animals were maintained and used in accordance with the guidelines of the Institutional Animal Care and Use Committee of Shanghai Changzheng Hospital. Ten 7-week-old male rats were randomly divided into a control group and an injury group, with 5 in each group. T10 thoracic segment was exposed and damaged with a heavy hammer by surgery, resulting in moderate spinal cord injury [24-26]. The BBB (Basso, Beattie, and Bresnahan) score was used to assess motor function changes of the hind limbs [27], and a sample of injured spinal cord tissue was collected 1 week later by cervical vertebra luxation.
ShRNA vector construction
Individual shRNA constructs targeting lncRNA-MEG were cloned into pLKO.1-EGFP-Puro. The primer sequence is as follows: Forword: 5’-CCGGTCATAGAGTCAAAGTATGAGCTTTTCAAGAGAAAGCTCATACTTTGACTCTATTTTTTG-3’; Reverse: 5’-AATTCAAAAAATAGAGTCAAAGTATGAGCTTTCTCTTGAAAAGCTCATACTTTGACTCTATGA.
Spinal cord in situ lentivirus injection
After the rat SCI model was constructed, we randomly divided SCI rats into a control group and a lentivirus injection group (5 rats in each group), and we used a 10 uL syringe to suck the lentiviral suspension (1 × 10/ml), fix it on the stereo injection rack, and connect the electric microinjection drive pump. We chose the injection point at the edge of the injury site, and control the injection speed at 30 ul/min through the electric microinjection drive pump. After the injection, slowly pull out the syringe and suture the wound. The random control group was injected with PBS.
HE staining
Seven days after the rat model was constructed, the rats were sacrificed, and the spinal cord tissue was removed, fixed and embedded in wax. Place the paraffin block on a microtome and cut the paraffin into 4-10 um thick slices. We used the adhesive to place the slice on a thin sheet, placed it on a baking sheet, absorb excess moisture with filter paper, and put it in an oven at 35 degrees overnight. Treat with xylene twice for 15 minutes and 5 minutes, wash it with 50% xylene + 50% ethanol, and then use 100%, 95%, 85%, 70%, 50% ethanol and water in order. The sections were stained in hematoxylin for about 20 minutes and rinsed with tap water for about 15 minutes to make the sections blue. The sections were discolored in a 1% hydrochloric acid ethanol solution, and rinsed in water. The sections were placed in 50%, 70%, and 80% ethanol for 5 minutes each, and then stained with 0.5% mono-red ethanol solution for 3 minutes. The sections were washed in 95% ethanol, and then placed in absolute ethanol for 5 minutes, and the filter paper was used to absorb excess ethanol. Cover the slide and place it under a microscope to observe and take pictures.
RNA extraction and qPCR
Total RNA was isolated from VSC4.1 cells or rat spinal cord tissue by Trizol method A reverse transcription reaction was performed at a final volume of 20 μL containing 1 μg total RNA and 4 μL 5 × PrimeScript RT Master Mix (Yeasen, China) with random primer or specific primers for miRNA. Afterwards, quantitative PCR was performed on a 10 μL mixture of 2 μL of reverse transcription product that was diluted by 20 times, 0.8 μL forward/reverse primer mixture for a final concentration of 0.4 μM, 5 μL 2 × SYBR Premix Ex TaqII (Yeasen, China), and DNase-free water added to reach the final volume. Reactions were performed using the following protocol: 95°C for 30 seconds and cycles of 95°C for 5 seconds and 60°C for 30 seconds for 40 cycles, followed by a dissociation stage. The primers used are listed in Table S1. Gene expression was normalized to the level of the house-keeping gene β-actin. Relative expression was calculated using the formula of 2-ΔΔCt.
Cell proliferation by CCK-8 assay
Five thousand cells were added to each well of 96-well plates, and each group was repeated three times. It was measured five more times every 24 hours. To measure the OD value, 10% CCK8 solution was added to fresh culture medium. The OD 450 nanometer value was measured after standing in an incubator at 37°C for 1 hour.
Tunel staining
The paraffin-embedded tissue samples were deparaffinized with xylene and different gradients of ethanol. Then 100 μL of proteinase K solution with a concentration of 20 n incuwas added into the tissue sample and the sample was incubated for 20 min at room temperature. Then we washed the tissue sample with PBS two times and placed the tissue sample in a staining jar containing FITC--12-dUTP Labling Mix (1 ug/ml), incubated for 5 min at room temperature in the dark, then washed with PBS three times. Finally, we analyzed the sample under a fluorescence microscope and took pictures.
Luciferase reporter assay
To examine the targeting sequence of miR-21-5p on 3’UTR of PDCD4, 500 ng WT or mutant pMIR-CMV vector or control pMIR-CMV was transfected to VSC4.1 cells using Hieff Trans™ Liposomal Transfection Reagent (Yeasen, Shanghai, China) according to the manufacturer’s instructions. Seventy two hours post transfection; the cells were harvested and lysed. Then the collected supernatant was subjected to detect luciferase activity using Dual-Luciferase Reporter Assay system (Promega). Each experiment was performed in triplicate.
Western blot
A total of 1 × 106 cells were preseeded in 10-cm dish and cultured 24 h. Then cells were harvested, lysed, and subject to SDS PAGE, then transferred on PVDF membrane, followed by the incubation with anti-Bax antibody (abcam, ab32503, 1:1000) anti-Bcl-2 antibody (abcam, ab19495, 1:1000), anti-Caspase-3 antibody (abcam, ab13847, 1:1000) and goat anti-Rabbit IgG (HRP) (abcam, ab6721, 1:5000). Membranes were visualized using the Immun-Star WesternC Chemiluminescence Kit (Bio-Rad) and images were captured using a ChemiDoc XRS + System and processed using ImageLab software (Bio-Rad).
Statistical analysis
All the experiments were repeated at least three times independently. Data are expressed as means ± SD. The difference between 2 groups was analyzed using Student’s T-test and the comparison among multiple groups was analyzed using one-way ANOVA test followed by post hoc test. Statistical significance was indicated at *P < 0.05, **P < 0.01 or ***P < 0.001.
Results
Biological analysis of gene expression level and miRNA expression level in rat SCI
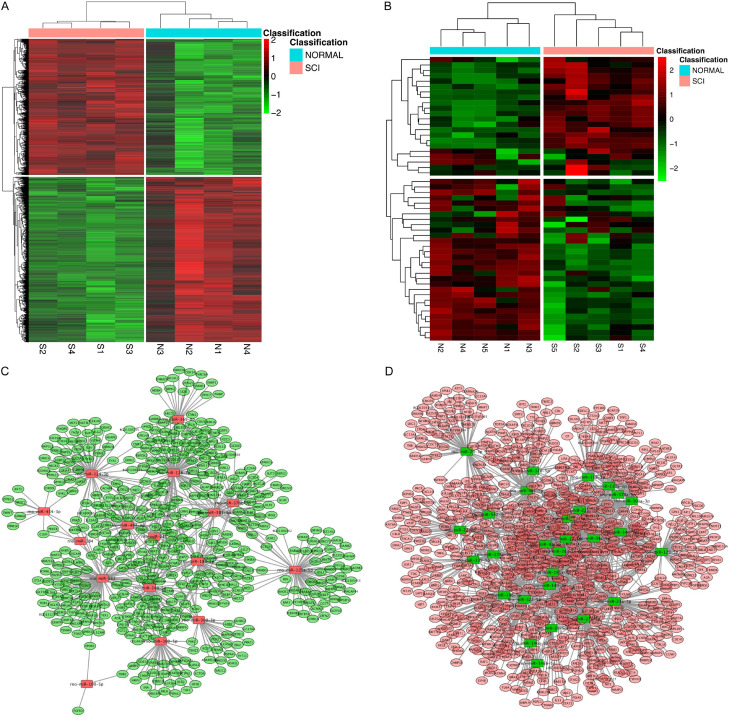
In order to investigate the possible changes of miRNA during SCI in rats, we used the limma package for differential expression analysis, 2189 up-regulated genes and 2656 down-regulated genes were obtained (Figure 1A) from the NCBI database the gene expression data (GSE45006) and miRNA expression data (GSE19890) of rats with a spinal cord injury time of 7 days. At the same time, we also performed differential expression analysis using the array package and limma package to obtain 20 up-regulated miRNAs and 30 down-regulated miRNAs (Figure 1B). After that, we combined the data of differentially expressed genes and differentially expressed miRNAs to perform screening and comparison in the three databases, miRDB, miRTarBase, and TargetScan, and found that there were 17 up-regulated miRNAs and 29 down-regulated miRNAs with a consistent change in miRNA and target genes (Figure 1C, 1D).
Figure 1.
Bioinformatics analysis of miRNA expression level in SCI rats. Gene expression data (GSE45006) and miRNA expression data (GSE19890) of rats and their control group were obtained from the NCBI database. (A, B) Heat map illustrating the expression level of protein coding gene (left) and miRNA (right) by the limma package (A), the array package and the limma package (B) in SCI. Red represents upwards and green represents downwards. (C, D) Screening comparisons were performed in three databases, miRDB, miRTarBase, and TargetScan, to analyze the candidate miRNAs that were up-regulated (C) and down-regulated (D) consistent with their potential target genes.
Validation of miRNA expression level in rat SCI model
In order to verify the data analyzed from high throughput sequencing, we constructed a SCI rat model as mentioned earlier. We first verified the success of the rat SCI model by BBB and HE staining (Figure 2A, 2B) and we found that the SCI rat model we constructed was successful.
Figure 2.
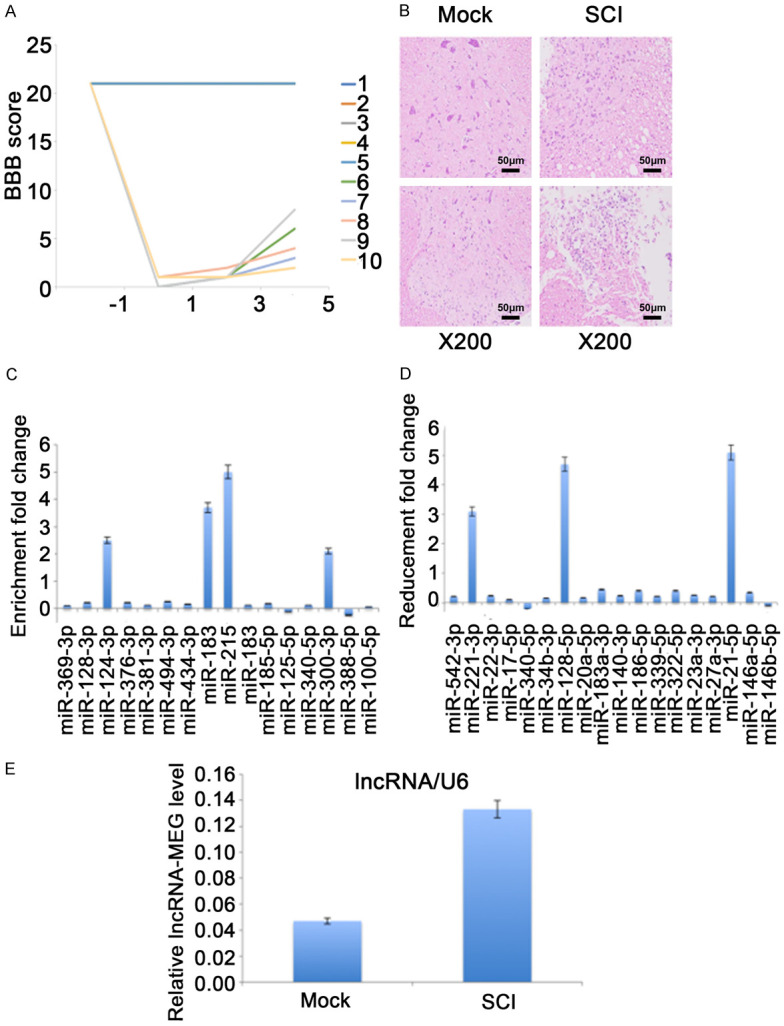
Verification of candidate ncRNA in SCI rat model. (A, B) Evaluation of rat SCI model construction process. (A) Before and after the rat model was established, the BBB score was used to evaluate the exercise ability of the rats. No. 1 to No. 5 are control group and No. 6 to No. 10 are SCI model group. (B) 7 days after the rat SCI model was established, rat spinal cord tissue was collected and analyzed by HE staining. All HE stained pictures were magnified 200 times. (C-E) Seven days after the rat SCI model was established, total RNA from rat spinal cord tissue was reverse transcripted with specific primers, and analyzed by qPCR. The expression level of predicted up-regulated miRNA (C), down-regulated miRNA (D) and lncRNA-MEG (E) are normalized to U6. Each data represented the mean ± SD of three independent experiments and were analyzed with T-test. *P < 0.05, **P < 0.01, ***P < 0.001.
On the seventh day of model construction, we sacrificed the rats and collected rat spinal cord tissue. We selected 10 up- and down-regulated candidate miRNAs for verification by qPCR. The results showed that the expression level of miR-124-3p, miR184, miR215, and miR-300-3 increased significantly, whilemiR-211-3p, miR-129-5p and miR-21-5p were significantly reduced (Figure 2C, 2D). Among them, the change of miR-21-5p was the most significant, so we focused our attention on miR-21-5p. Through bioinformatics prediction, we found that lncRNA-MEG may be an effector of miR-21-5p. Therefore, we examined the expression level of lncRNA-MEG in SCI rat models and control rats. The results showed that the expression of lncRNA-MEG was significantly increased in the SCI rat model (Figure 2E).
lncRNA-MEG inhibit neuronal apoptosis through miR-21-5p in SCI rats
To analyzing the role of lncRNA-MEG for SCI, we first screened siRNAs that could knockdown of lncRNA-MEG in VSC4.1 cells. After transiently transfecting siRNAs to VSC4.1 cells for 72 h, we detected the expression level of lncRNA-MEG by qPCR. The results indicated that siRNA-2 showed the best effect (Figure 3A). Therefore, we used the siRNA-2 sequence to construct shRNA lentiviral vectors for subsequent experiments.
Figure 3.
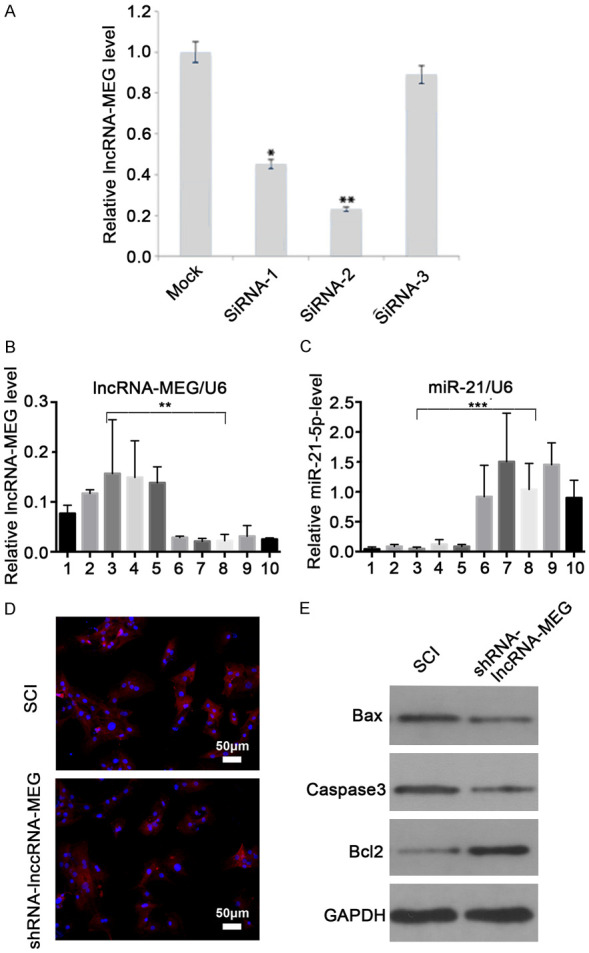
lncRNA-MEG inhibit neuronal apoptosis through miR-21-5p in SCI rats. (A) Screen for siRNAs that effectively inhibit lncRNA-MEG in VSC4.1 cells. VSC4.1 cells were transiently transfected with different siRNAs, and the relative knockdown of lncRNA-MEG was detected by qPCR. The relative changes were normalized for the mock group. Each data point represents the mean ± SD of three independent experiments and was analyzed with T-test. *P < 0.05, **P < 0.01. (B, C) In the SCI rat model, the spinal cord tissues of rats were collected for detection after 7 days. No. 1 to No. 5 are control group and No. 6 to No. 10 are SCI model group. The expression of lncRNA-MEG (B) and miR-21-5p (C) in different groups of rats were analyzed by qPCR. (D) Apoptosis in rat spinal cord group was identified by Tunnel staining. Blue represents DAPI and red represents apoptotic cells. (E) The levels of indicated proteins were measured with western Blot after lncRNA-MEG knocking down with shRNA lentivirus in SCI rat spinal cord tissues.
We injected the shRNA-lncRNA-MEG and control lentivirus into SCI rat spinal cord tissue in situ. After 7 days, the rats were sacrificed and the spinal cord tissue was collected. We first detected the expression levels of lncRNA-MEG and miR-21-5p in the SCI rats by qPCR. The results showed that compared with the control group, the expression of lncRNA-MEG after shRNA-lncRNA-MEG lentivirus injection decreased significantly, while the expression of miR-21-5p increased significantly (Figure 3B, 3C). After that, we performed tunnel staining to the spinal cord tissues, and the results showed that the number of apoptotic cells after lncRNA-MEG konckdown was significantly reduced compared with the control group. This indicates that interfering with lncRNA-MEG can reduce the apoptosis of rat neuronal cells (Figure 3D). In order to further verify the apoptosis of rat spinal cord tissue cells, we detected the expression level of apoptosis-related proteins BCL2, BAX, and cleaved-caspase3 by western blot. The results showed that after knockdown of lncRNA-MEG, the expression level of apoptosis-related protein BCL2 significantly increased, while the expression of BAX, and cleaved-caspase3 significantly decreased in the spinal cord neural tissue cells in SCI rats (Figure 3E). These results indicate that knockdown lncRNA-MEG can indeed inhibit neuronal apoptosis during SCI.
lncRNA-MEG affects the expression level of PDCD4 by inhibiting miR-21-5p
In previous studies, researchers have found that miR-21-5p can inhibit cell apoptosis by inhibiting PDCD4 expression in tongue squamous cell carcinoma [28]. However, it is unknown whether there are similar effects in the SCI process. Therefore, we decided to investigate the expression of PDCD4 in the spinal cord in SCI rats. We first detected PDCD4 expression in spinal cord tissue in control rats and SCI rats by qPCR. The results showed that compared with the control group, the mRNA expression level of PDCD4 in SCI rats increased significantly, matching with the expression level of miR-21-5p was significantly decreased (Figures 2C and 4A). After that, we further detected the mRNA expression level of PDCD4 in SCI rats after lncRNA-MEG knockdown. The results showed that, miR-21-5p significantly increased in SCI rats after knockdown of lncRNA-MEG, while mRNA and protein levels of PDCD4 decreased significantly (Figures 3C and 4B). These results suggested that during the process of SCI in rats, the expression level of lncRNA-MEG in injured rat neuronal cells increased, leading to a decrease in the expression level of miR-21-5p, which in turn led to an increase in the expression level of PDCD4, thereby promoting apoptosis of neuronal cells.
Figure 4.
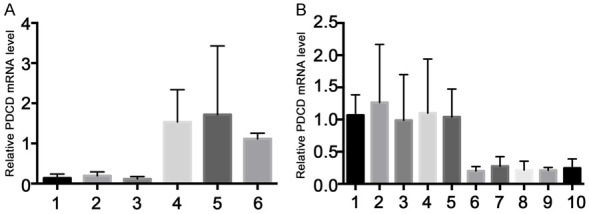
lncRNA-MEG affects the expression level of PDCD4. A. The expression level of PDCD4 in SCI rats and control rats was detected by qPCR. No. 1 to No. 3 are control group and No. 4 to No. 6 are SCI model group. B. The expression level of PDCD4 in SCI rats after knocking down of lncRNA-MEG with shRNA lentivirus was detected by qPCR. No. 1 to No. 5 are control group and No. 6 to No. 10 are SCI model group. Each data represented the mean ± SD of three independent experiments and were analyzed with one-way ANOVA-test. *P < 0.05, **P < 0.01, ***P < 0.001.
Effect of lncRNA-MEG on SCI process of rat neuron cells
In addition to animal models, we further explored the role of lncRNA-MEG in SCI models of neuron cell. To simulate the SCI process, we used rat neuron VSC4.1 cells to construct an oxygen and glucose deprivation (OGD) model. After constructing the cell model, we further explored the effect of lncRNA-MEG through infection with shRNA-lncRNA-MEG lentivirus. Just like the rat model, we first examined the expression of lncRNA-MEG, miR-21-5p, and PDCD4 mRNA in different groups by qCPR. The results showed that compared with the control group, the expression levels of lncRNA-MEG and PDCD4 increased significantly in the OGD model group, while the expression level of miR-21-5p decreased significantly. In the lncRNA-MEG knockdown OGD model group, the mRNA levels of lncRNA and PDCD4 decreased significantly compared with the OGD model group alone, but were still higher than the control group. The expression level of miR-21-5p was higher than that of the control group and the OGD group alone (Figure 5A-C). These results indicate that the lncRNA-MEG, miR-21-5p, and PDCD4 axes are an important factor in the SCI process of OGD simulation.
Figure 5.
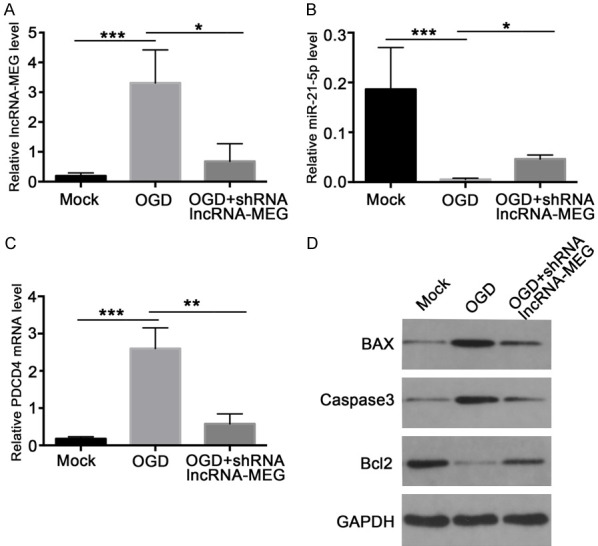
lncRNA-MEG affects the expression level of PDCD4 by inhibiting miR-21-5p in OGD Cell Model. The expression levels of lncRNA-MEG (A), miR-21-5p (B), and PDCD4 (C) in control group, OGD model group, and OGD model injection with shRNA-lncRNA-MEG lentiviral group were analyzed by qPCR in the OGD cell model. (D) The indicated protein expression levels were measured with western Blot in control group, OGD model group, and OGD model injection shRNA-lncRNA-MEG lentivirus group. Each data represented the mean ± SD of three independent experiments and were analyzed with T-test. *P < 0.05, **P < 0.01, ***P < 0.001.
In addition to changes in RNA levels, we also detected PDCD4 and the expression of apoptosis-related proteins during SCI simulations in OGD, such as BCL2, BAX, and cleaved-caspase3. The results showed that at the protein level, compared with the control group, the expression levels of PDCD4, BAX, and cleaved-caspase3 in the cells of the OGD model group increased significantly, while the expression levels of Bcl-2 decreased significantly. In addition, the expression level of PDCD4, BAX, and cleaved-caspase3 was significantly lower in the OGD model of lncRNA-MEG knockdown than in the OGD model group, but still higher than the control group, and the expression level of Bcl2 was also significantly lower than OGD group (Figure 5D).
lncRNA-MEG affects the expression level of PDCD4 by inhibiting miR-21-5p
MiRNAs are usually able to interact with the 3’UTR region of the target mRNA, thereby affecting the expression of the candidate gene. In previous studies, miR-21-5p was able to target with PTEN and Smad7 in the physiological process of cancer, and promote the proliferation of cancer cells [29,30]. Through bioinformatics prediction, we found that 3’UTR of PDCD4 may be a potential target of miR-21-5p (Figure 6A). Previous results have also shown that the expression level ofmiR-21-5p can affect the expression level of PDCD4, but whether miR-21-5p directly affects PDCD4 in neuronal cells is unknown. Therefore, we analyzed the interaction between miR-21-5p and PDCD4 through a luciferase reporter assay. We transiently transfected a siRNA inhibitor targeting miR-21-5p and a plasmid with a 3’UTR region of PDCD4 to VSC4.1 cells and detected the luciferase activity. The results showed that the level of the luciferase reporter gene with the 3’UTR of WT-type PDCD4 was higher when miR-21-5p was blocked by siRNA inhibitor compared to the group without, and there are significant statistical differences. But there was no statistical difference compared with mutant 3’UTR region of PDCD4 group. This suggested that miR-21-5p could inhibit PDCD4 expression by targeting the 3’UTR region of PDCD4 (Figure 6B). Afterwards, we further verified the effect of lncRNA-MEG on the 3’UTR of PDCD4 in an OGD cell model. Before we construct the OGD cell model, we transfected the 3’UTR plasmid of wild-type PDCD4 into VSC4.1 cells. Then the expression levels of luciferase reporter gene in different treatment groups were detected. As a result, we found that compared with the untreated group, the expression level of luciferase in the OGD model group increased significantly, while in the lncRNA-MEG knockdown group, the expression level of luciferase was significantly reduced (Figure 6C). This is consistent with our hypothesis that lncRNA-MEG affects PDCD4 expression by regulating miR-21-5p.
Figure 6.
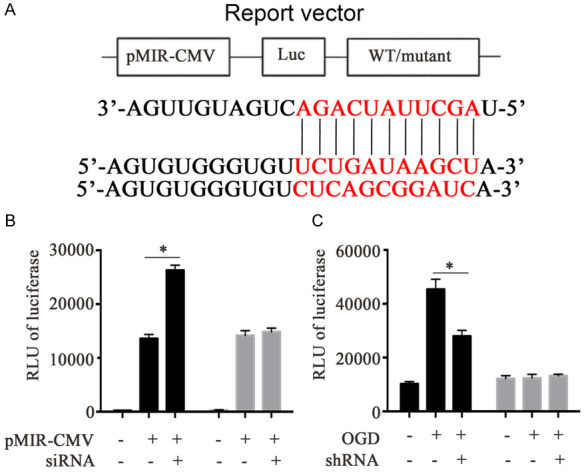
lncRNA-MEG affects the expression level of PDCD4 by inhibiting miR-21-5p. A. The predicted miR-21-5p binding sites on the 3’UTR of PDCD4. B. Luciferase activity in VSC4.1 cells co-transfected with siRNA targeting miR-21-5p and luciferase reporters containing wild-type or mutant type 3’-UTR of PDCD4. C. In the OGD model, the effect of shRNA-lncRNA-MEG on 3’UTR of PDCD4. Each data represented the mean ± SD of three independent experiments and were analyzed with T-test. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In recent years, with the continuous improvement of SCI animal models, the research on SCI has made great progress. Researchers have conducted in-depth research on molecular mechanisms such as neuronal apoptosis and proliferation during SCI [21,31,32]. Nevertheless, the various treatment strategies proposed based on the molecular mechanism of SCI still cannot solve the dilemma of SCI treatment well. Therefore, it is necessary to further study the molecular mechanism of SCI.
With the continuous development of next-generation sequencing and non-coding RNA research, more and more researches have focused on the role of non-coding RNA in the process of SCI. Several research groups have reported the correlation between multiple miRNAs and SCI [33-35]. However, the detailed molecular mechanism of lncRNA on SCI has not been reported.
In this study, we first analyzed the expression level of protein coding gene and miRNA in the SCI rat model from NCBI database through bioinformatics, and combined the two databse to predict target miRNAs that may be related to the SCI process. For the predicted candidate miRNAs, we performed single-point verification by constructing a SCI rat model. The results show that some of our predicted candidate miRNAs have consistent changes in the rat SCI model. For the most significant change of miR-21-5p, we further found that lncRNA-MEG has a competition with miR-21-5p. Therefore, we speculated that during the SCI process, lncRNA-MEG may promote the apoptosis of rat nerve cells by inhibiting miR-21-5p. Therefore, we explored the role of lncRNA-MEG in SCI rat model. As a result, we found that knocking down lncRNA-MEG can increase the expression of miR-21-5p, and by reducing Bax and cleaved Caspase-3, increasing Bcl-2 expression and inhibiting neuronal apoptosis. In further cell model experiments, we simulated the process of SCI through OGD models. It was found that lncRNA-MEG can also inhibit PDCD4 expression through miR-21-5p, and after knocking down lncRNA-MEG, the expression level of Bax and cleaved caspase3 decreased significantly, while the expression of Bcl-2 increased significantly. Furthermore, we validated lncRNA-MEG regulated the expression level of miR-21-5p and inhibit cell apoptosis in the OGD model by knocking down lncRNA-MEG. Finally, in order to confirm the binding target of miR-21-5p and PDCD4 and the effect of knocking down lncRNA-MEG on PDCD4 during SCI, we performed a luciferase reporter experiment. The results showed that miR-21-5p can target directly to the 3’UTR region of PDCD4, and that the knockdown of lncRNA-MEG can also inhibit the transcriptional activity of PDCD4 during SCI.
Nevertheless, there are some problems in our experiments that need further investigation. First, although miR-21-5p inhibits PDCD4 while 3’UTR leading to the inhibition of neuronal cell apoptosis and cell proliferation promotion, it is not clear which signal pathway involved in. Secondly, the results we have obtained are based on rat models and rat nerve cells, it is not mentioned whether human lncRNA-MEG have similar effects, which is worthy of further investigation. Finally, among the lncRNA-MEG, miR-21-5p, and PDCD4 axes, which step is more suitable to become the target of SCI treatment also needs our further research.
In summary, we found for the first time that lncRNA-MEG can affect the expression of PDCD4 through miR-21-5p, and then affect the apoptosis of neurons during SCI in rats. Therefore, lncRNA-MEG, miR-21-5p, and PDCD4 axis may be a key target for SCI treatment.
Acknowledgements
Project supported by National Natural Science Foundation of China (Grant No. 81702138).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ahuja CS, Nori S, Tetreault L, Wilson J, Kwon B, Harrop J, Choi D, Fehlings MG. Traumatic spinal cord injury-repair and regeneration. Neurosurgery. 2017;80:S9–S22. doi: 10.1093/neuros/nyw080. [DOI] [PubMed] [Google Scholar]
- 2.Brouwers EMJR, Meent HV, Curt A, Maier DD, Abel RF, Weidner N, Rupp R, Kriz J, de Haan AFJ, Kramer JK, Hosman AJF, Bartels RHMA EMSCI participants and investigators. Recovery after traumatic thoracic- and lumbar spinal cord injury: the neurological level of injury matters. Spinal Cord. 2020;58:980–987. doi: 10.1038/s41393-020-0463-1. [DOI] [PubMed] [Google Scholar]
- 3.Vivodtzev I, Tan AQ, Hermann M, Jayaraman A, Stahl V, Rymer WZ, Mitchell GS, Hayes HB, Trumbower RD. Mild to moderate sleep apnea is linked to hypoxia-induced motor recovery after spinal cord injury. Am J Respir Crit Care Med. 2020;202:887–890. doi: 10.1164/rccm.202002-0245LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackery A, Tator C, Krassioukov A. A global perspective on spinal cord injury epidemiology. J Neurotrauma. 2004;21:1355–1370. doi: 10.1089/neu.2004.21.1355. [DOI] [PubMed] [Google Scholar]
- 5.Fang H, Lin J, Liang L, Long X, Zhu X, Cai W. A nonsurgical and nonpharmacological care bundle for preventing upper urinary tract damage in patients with spinal cord injury and neurogenic bladder. Int J Nurs Pract. 2020;26:e12761. doi: 10.1111/ijn.12761. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Yang Q, Ma X. Synergistic effect of ascorbic acid and taurine in the treatment of a spinal cord injury-induced model in rats. 3 Biotech. 2020;10:50. doi: 10.1007/s13205-019-2032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karsy M, Hawryluk G. Modern medical management of spinal cord injury. Curr Neurol Neurosci Rep. 2019;19:65. doi: 10.1007/s11910-019-0984-1. [DOI] [PubMed] [Google Scholar]
- 8.Rowland JW, Hawryluk GW, Kwon B, Fehlings MG. Current status of acute spinal cord injury pathophysiology and emerging therapies: promise on the horizon. Neurosurg Focus. 2008;25:E2. doi: 10.3171/FOC.2008.25.11.E2. [DOI] [PubMed] [Google Scholar]
- 9.Ding Y, Song Z, Liu J. Aberrant lncRNA expression profile in a contusion spinal cord injury mouse model. Biomed Res Int. 2016;2016:9249401. doi: 10.1155/2016/9249401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Hu B, Cao F, Sun S, Zhang Y, Zhu Q. Down regulation of lncSCIR1 after spinal cord contusion injury in rat. Brain Res. 2015;1624:314–320. doi: 10.1016/j.brainres.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 11.Faulkner JR, Herrmann JE, Woo MJ, Tansey KE, Doan NB, Sofroniew MV. Reactive astrocytes protect tissue and preserve function after spinal cord injury. J Neurosci. 2004;24:2143–2155. doi: 10.1523/JNEUROSCI.3547-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu XY, Guo JW, Kou JQ, Sun YL, Zheng XJ. Repair mechanism of astrocytes and non-astrocytes in spinal cord injury. World J Clin Cases. 2020;8:854–863. doi: 10.12998/wjcc.v8.i5.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada S, Hara M, Kobayakawa K, Matsumoto Y, Nakashima Y. Astrocyte reactivity and astrogliosis after spinal cord injury. Neurosci Res. 2018;126:39–43. doi: 10.1016/j.neures.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Pekny M, Nilsson M. Astrocyte activation and reactive gliosis. Glia. 2005;50:427–434. doi: 10.1002/glia.20207. [DOI] [PubMed] [Google Scholar]
- 15.Pekny M, Wilhelmsson U, Pekna M. The dual role of astrocyte activation and reactive gliosis. Neurosci Lett. 2014;565:30–38. doi: 10.1016/j.neulet.2013.12.071. [DOI] [PubMed] [Google Scholar]
- 16.Wilhelmsson U, Bushong EA, Price DL, Smarr BL, Phung V, Terada M, Ellisman MH, Pekny M. Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proc Natl Acad Sci U S A. 2006;103:17513–17518. doi: 10.1073/pnas.0602841103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karimi-Abdolrezaee S, Billakanti R. Reactive astrogliosis after spinal cord injury-beneficial and detrimental effects. Mol Neurobiol. 2012;46:251–264. doi: 10.1007/s12035-012-8287-4. [DOI] [PubMed] [Google Scholar]
- 18.Yu B, Zhou S, Yi S, Gu X. The regulatory roles of non-coding RNAs in nerve injury and regeneration. Prog Neurobiol. 2015;134:122–139. doi: 10.1016/j.pneurobio.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Dai J, Xu LJ, Han GD, Sun HL, Zhu GT, Jiang HT, Yu GY, Tang XM. MiR-137 attenuates spinal cord injury by modulating NEUROD4 through reducing inflammation and oxidative stress. Eur Rev Med Pharmacol Sci. 2018;22:1884–1890. doi: 10.26355/eurrev_201804_14709. [DOI] [PubMed] [Google Scholar]
- 20.Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandran R, Mehta SL, Vemuganti R. Non-coding RNAs and neuroprotection after acute CNS injuries. Neurochem Int. 2017;111:12–22. doi: 10.1016/j.neuint.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao C, Wang J, Zhang H, Zhou S, Qian T, Ding F, Gu X, Yu B. Long non-coding RNA uc. 217 regulates neurite outgrowth in dorsal root ganglion neurons following peripheral nerve injury. Eur J Neurosci. 2015;42:1718–1725. doi: 10.1111/ejn.12966. [DOI] [PubMed] [Google Scholar]
- 24.Cao Y, Wu T, Yuan Z, Li D, Ni S, Hu J, Lu H. Three-dimensional imaging of microvasculature in the rat spinal cord following injury. Sci Rep. 2015;5:12643. doi: 10.1038/srep12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, Ren S, Fu K, Wu Q, Wu J, Hou L, Pan H, Sun L, Zhang J, Wang B, Miao Q, Sun G, Bonicalzi V, Canavero S, Ren X. Restoration of motor function after operative reconstruction of the acutely transected spinal cord in the canine model. Surgery. 2018;163:976–983. doi: 10.1016/j.surg.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Guo H, Lu Z, Sun K, Jin Q. Hyperglycemia aggravates spinal cord injury through endoplasmic reticulum stress mediated neuronal apoptosis, gliosis and activation. Biomed Pharmacother. 2019;112:108672. doi: 10.1016/j.biopha.2019.108672. [DOI] [PubMed] [Google Scholar]
- 27.Tian DS, Jing JH, Qian J, Chen L, Zhu B. Effect of oscillating electrical field stimulation on motor function recovery and myelin regeneration after spinal cord injury in rats. J Phys Ther Sci. 2016;28:1465–1471. doi: 10.1589/jpts.28.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennelli G, Galuppini F, Barollo S, Cavedon E, Bertazza L, Fassan M, Guzzardo V, Pelizzo MR, Rugge M, Mian C. The PDCD4/miR-21 pathway in medullary thyroid carcinoma. Hum Pathol. 2015;46:50–57. doi: 10.1016/j.humpath.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Yan X, Liu Y, Kong X, Ji J, Zhu H, Zhang Z, Fu T, Yang J, Zhang Z, Liu F, Gu Z. MicroRNA-21-5p are involved in apoptosis and invasion of fibroblast-like synoviocytes through PTEN/PI3K/AKT signal. Cytotechnology. 2019;71:317–328. doi: 10.1007/s10616-018-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui H, He Y, Chen S, Zhang D, Yu Y, Fan C. Macrophage-derived miRNA-containing exosomes induce peritendinous fibrosis after tendon Injury through the miR-21-5p/Smad7 pathway. Mol Ther Nucleic Acids. 2019;14:114–130. doi: 10.1016/j.omtn.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serviss JT, Johnsson P, Grandér D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. 2014;5:234. doi: 10.3389/fgene.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rinn JL. lncRNAs: linking RNA to chromatin. Cold Spring Harb Perspect Biol. 2014;6:a018614. doi: 10.1101/cshperspect.a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.