Abstract
Characterized by autophagy-associated protein disorders, autophagy participates in Taxol resistance in triple negative breast cancer (TNBC). As an evolutionarily conserved serine/threonine protein kinase with complex signaling pathway, mammalian target of rapamycin (mTOR) can regulate various cellular functions by phosphorylation of its downstream target proteins after activation. A large number of references have demonstrated that mTOR signaling pathway is related to autophagy and apoptosis. Formononetin (FMNT) has anticancer properties against breast, prostate and colon cancers. This study aimed to explore the regulatory effect of FMNT/miR-199a-3p/mTOR pathway on Taxol resistance and autophagy in breast cancer (BC). MiR-199a-3p, mTOR, LC3 and other autophagy related proteins were detected in Taxol sensitive and Taxol resistant TNBC cell lines, which were MDA-MB-231 and MDA-MB-231/Taxol, respectively. Cell viability and toxicity were determined by CCK-8 and MTT assay, respectively. The therapeutic effect of FMNT was evaluated in xenotransplantation model of nude mice. MiR-199a-3p was more highly expressed in MDA-MB-231/Taxol than in MDA-MB-231, while mTOR and p-mTOR decreased in MDA-MB-231/Taxol in comparison with MDA-MB-231, and autophagy activation and drug resistance were enhanced. In MDA-MB-231/Taxol cell line, the role of FMNT was verified to inhibit high miR-199a-3p expression. In addition, the combination therapy of FMNT and Taxol was found to be more effective in inhibiting autophagy and drug resistance. Moreover, mTOR was the target of miR-199a-3p, which was confirmed by dual luciferase reporter (DLR) gene assay. Oral administration of FMNT reduced tumor volume after MDA-MB-231/Taxol injection in vivo. Moreover, oral administration of FMNT and Taxol suppressed autophagy and Taxol resistance by restoring mTOR protein level to that of the parent MDA-MB-231, suggesting that miR-199a-3p can severe as a new target to overcome Taxol resistance in TNBC.
Keywords: Formononetin, Taxol, triple negative breast cancer, autophagy, miR-199a-3p
Introduction
Breast cancer (BC), as a common clinical female tumor, is one of the primary causes of female tumor death [1]. Triple negative breast cancer, or TNBC, which bears the responsibility for approximately 10%-20% of all breast carcinoma, is a type of BC with high diffusion, high histological grade, and poor prognosis [2,3]. Due to the fact that the tests of ER, PR and Her2 were all negative, it is difficult to carry out targeted therapy to patients [4]. Although chemotherapy can improve the condition of TNBC to a certain extent, the overall survival rate of patients is overwhelmingly poor as it has the characteristics of high recurrence and high metastasis [5]. Apart from that, long-term chemotherapy will lead to patients’ resistance to chemotherapy drugs, which is also one of the reasons for clinical treatment failure [6]. Therefore, solving TNBC drug resistance is the key to improving the prognosis of patients.
Formononetin (FMNT) is an active ingredient isolated from Astragalus membranaceus, which has many pharmacological effects [7]. Studies have revealed that FMNT plays a favorable role in tumor growth inhibition, wound healing, estrogen-like effect, antioxidant activity and anti-inflammation [8,9]. Moreover, it can induce apoptosis, inhibit cell cycle, angiogenesis and reverse multidrug resistance [10,12]. Zhou et al. [12] have proposed in their study that FMNT could enhance the antitumor effect of everolimus in BC MDA-MB-468 cells by inhibiting mTOR pathway. However, one recent study has found that FMNT can regulate the transcription of microRNA (miR) and participate in tumorigenesis [13]. It is not found that there is a miR betweenFMNT and mTOR pathway and exerts regulation at present. MiR-199a-3p, as a classic upstream miR of mTOR pathway, has been reported in many papers [14]. Therefore, we suspect that FMNT may participate in the development of mTOR pathway by mediating miR-199a-3p. However, researches on FMNT and chemotherapy resistance of TNBC are still scanty.
Autophagy, a kind of self-adaptation capacity, is a double-edged sword in tumor metastasis [15]. Generally, malignant cells have apoptosis defects, while autophagy refers to a form of cell death, which is the same as apoptosis [16,17]. Therefore, autophagy can be another mechanism of cell death when the tumor is deficient in apoptosis [18,19]. In the study of Huang et al. [20], it was found that FMNT may protect the heart of the elderly from ischemia/reperfusion injury by enhancing autophagy degradation. However, whether FMNT is related to autophagy in TNBC drug-resistant tumor cells is still under investigation.
Therefore, we aimed to explore the related mechanism of FMNT in taxol-resistant TNBC MDA-MB-231, with the goal of providing reference for the development of clinical novel drugs.
Methods and materials
Cell culture
In a humidity incubator with 5% CO2 at 37°C, human TNBC MDA-MB-231 and its Taxol-resistant cell lines were cultured in 1640 high glucose medium (BasalMedia Technologies, Shanghai, China) supplemented with fetal bovine serum (10%; Thermo Fisher-USA). The medium was replaced every two days, and the logarithmic growth phase cells were selected for experimental study. According to the grouping, the cells were added with the drug or reagent to be treated respectively, and treated 24 h later.
Cell transfection
Overexpression and inhibitor of microRNA-199a-3p (miR-199a-3p mimic and miR-199a-3p inhibitor respectively) and blank control (miR-NC) were purchased from RiboBio (Guangzhou, China), and the Lipofectamine 2000 kit (Invitrogen) was used for transfection of cytoplasms. The operation procedure was strictly in accordance with the manufacturer’s instructions.
qRT-PCR
Trizol (Invitrogen) was used for the extraction of the total RNA from cells or serum, and UV spectrophotometer and agarose gel electrophoresis were responsible for the determination of its purity, concentration and integrity. Subsequently, TaqMan™ Reverse Transcription Kit (Invitrogen, USA) was used for reverse transcription. Then, PCR amplification was carried out by a PrimeScript RT Master Mix kit (Takara Bio, Japan) and ABI7500 (ABI, USA), and the reaction system and conditions were operated according to the kit guidelines. Three parallel replicate wells were designed in the experiment, and all the specimens were determined three times. With U6 and β-actin as the internal reference for miR (microRNA) and GENE, respectively, 2-ΔΔct was used for data processing [21]. Primers are shown in Table 1.
Table 1.
Sequences of primers used in this study
| Gene | Primer sequences |
|---|---|
| miR-199a-3p | Sense sequence: 5’-ACACCAGCTGGGTACAGTAGTCTGCACA-3’ |
| Antisense sequence: 5’-GTGTCGTGGAGTCGGCAATTC-3’ | |
| ULK1 | Sense sequence: 5’-ACCGCATTCACAGCATCACT-3’ |
| Antisense sequence: 5’-CCGATCTACAGACCGACGCT-3’ | |
| Beclin1 | Sense sequence: 5’-ATGGAAGGGTCTAAGACGT-3’ |
| Antisense sequence: 5’-TCATTTGTTATAAAATTGTGA-3’ | |
| Vps34 | Sense sequence: 5’-GGACCTTCTGACCACGAT-3’ |
| Antisense sequence: 5’-GCAACAGCATAACGCCTC-3’ | |
| LC3-II | Sense sequence: 5’-AAACGCATTTGCCATCACA-3’ |
| Antisense sequence: 5’-GGACCTTCAGCAGTTTACAGTCAG-3’ | |
| m-TOR | Sense sequence: 5’-AGAAACTGCACGTCAGCACCA-3’ |
| Antisense sequence: 5’-CCATTCCAGCCAGTCATCTTTG-3’ | |
| β-actin | Sense sequence: 5’-GCTTCTAGGCGGACTGTTAC-3’ |
| Antisense sequence: 5’-CCATGCCAATGTTGTCTCTT-3’ |
Cell viability experiment
CCK-8 test was used for cell viability determination. The cells were resuspended to 5×103, seeded in plates (96-well), and pre-cultured in an incubator at 37°C with 5% CO2 and saturation humidity. The resistant strain was treated with different concentrations of Taxol (0, 1.5, 5, 7.5, 10, 50, 15 μM/L) for 24 h, and then 10 microliters of CCK-8 reagent was added into each well. The absorbance at 450 nm was measured after 2 h of culture.
Cytotoxicity test
Cytotoxicity was measured by 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) assay in this study. Cells were seeded into 96-well plates with 5×103 cells/well. After one overnight incubation, they were intervened by different concentrations of FMNT (0, 5, 15, 20, 40, 60, 80 μM/L) for 48 h. After the removal of the culture medium, 100 μl MTT with a final concentration of 500 μg/ml was added into each well for a 4-hour culture in an incubator at 37°C. Thereafter, 20% SDS solution (100 μl) was placed into the wells and the culture was continued for 20 h. Finally, the absorbance was measured at 570 nm by a microplate reader, and the cell viability value was expressed as the percentage of experimental group and control group.
FACS detection
The apoptosis rate and cell cycle were detected by FACS, which were briefly described as follows. The cells were adjusted to 1×106, seeded into plates (24-well), and immobilized with 70% chill ethanol 48 h later. Two hours after immobilization, the immobilized cells with 100 μl RNase (BD Biosciences, USA) were processed for 30 min of incubation at 37°C. After that, they were mixed with 400 μl propidium iodide and left in the dark at 4°C for 0.5 h. Likewise, the cells were adjusted to 1×105 and seeded into plates (24-well). The apoptotic cells were determined by fluorescein isothiocyanate (FITC)-Annexin V apoptosis detection kit (BD Biosciences) 48 h later. Flow cytometry (FACScan; BD Biosciences) was used to determine cell cycle and apoptosis. The experiments were repeated three times.
WB detection
Lysis of the cultured cells and the determination of protein concentration were conducted by RIPA buffer and BCA kit (both from Thermo Scientific Company, USA) respectively. The protein concentration was then regulated to 4 μg/μL, isolated by SDS-PAGE (12%), and transferred to a 0.22 μm PVDF membrane. Subsequently, it was blocked with 5% skim milk for 2 h, and added with primary antibody (Bcl2: 1:1000; Bax: 1:1000; Beclin1: 1:1000; ULK1: 1:2000; VPS34: 1:2000; mTOR: 1:2000; p-mTOR: 1:2000; LC3-II: 1:3000; α-Tublin: 1:3000, Abcam Company, USA) for overnight sealing at 4°C. After that, the primary antibody was washed away, and the goat anti-rabbit-HRP secondary antibody (1:5000; Abcam, USA) was added for one-hour incubation at 37°C, followed by three times of membrane washing with PBS, 5 min each. After blotting the extra liquid with filter paper, the membrane was developed by ECL. With α-Tublin as the internal reference, the protein bands were scanned and the grayscale values were analyzed in Quantity One software.
Immunofluorescence
Immunofluorescence staining was used for detecting the changes of autophagy-related proteins in cells. After rinsing with PBS three times (5 min each time), the cells were given 10 min of immobilization and then 10 min of infiltration using Triton X-100 (0.1%) in PBS. Thereafter, they were blocked at indoor temperature for 1 h, and processed for overnight incubation with anti-LC3A/B antibody at 4°C. On the second day, after being thoroughly washed with PBS, the cells were cultured with Cy3-labeled goat anti-rabbit IgG (Beyotime, China) at indoor temperature for 3 h. After staining the chromatin with DAPI for 10 min, the cells were evaluated under a fluorescence microscope (OLYMPAS IX71, OLYMPAS, Japan). The Alexa Fluor 488 fluorescence intensity was quantified using Image J software.
DLR assay
Through starBase 3.0 [22], Targetscan7.2 [23], miRDB [24], miRTarBase [25] and TarBase [26] prediction, it was found that miR-199a-3p had a targeted relationship with mTOR. Therefore, pmirGLO-mTOR9-3’UTR wild type (Wt) and mutant (Mut) were constructed and transferred to the luciferase reporter gene’s downstream for sequencing and identification of the constructed plasmids. The correctly sequenced plasmids were respectively co-transfected with miR-199a-3p mimic or miR-NC into 293T cells. Lipofectamine™ 2000 kit was used for transfection. The luciferase reporter vector was detected by following the guidelines of DLR assay system (Promega).
Animal experiment
Acquired from Shanghai Bikai Biotechnology, 4-5 weeks old Balb/c-nude nude mice were stochastically assigned to 7 groups (4 mice in each group): MDA-MB-231, MDA-MB-231/Taxol resistance, MDA-MB-231 Taxol resistance + Taxol, MDA-MB-231 Taxol resistance + FMNT, MDA-MB-231 Taxol resistance + 3-Methyladenine (3-MA), MDA-MB-231 Taxol resistance + FMNT + autophagy inhibitor 3-MA, and MDA-MB-231 Taxol resistance + FMNT + Taxol groups. Subcutaneous injection of MDA-MB-231 and MDA-MB-231/Taxol (5×107 cells/0.2 mL) was carried out on each mouse, and 3-MA was injected intraperitoneally (15 mg/kg). About two weeks later when the tumor diameter of nude mice increased to 5 mm, FMNT (30 mg/kg) or Taxol (50 mg/mouse) was given orally every three days, and the tumor size was measured every 4 days. The mice were killed 44 days later, and mTOR, P-mTOR, LC3-I/LC3-II, Beclin 1 and VPS34 protein expression in tumor tissues were detected by Western blot (WB). The research was approved by the Animal Ethics Committee of Shanghai Shuguang Hospital, and the experiment was conducted according to the Laboratory animal-Guideline for ethical review of animal welfare issued by China in 2018. Animals were all executed by making them inhale carbon dioxide (Concentration of CO2: 100%; duration: 2-3 min).
Statistical analysis
SPSS 17.0 software was used for statistical analysis. Differences between groups were verified by the t test. Univariate analysis was adopted for comparison among multiple groups, and Bonferroni post hoc test for comparison between two groups. All experiments were conducted 3 times or more, and all the data were described as the mean ± standard deviation (SD). Statistical significance was set at a p-value of < 0.05.
Results
FMNT inhibits MDA-MB-231/Taxol cell growth
At first, to verify the Taxol resistance of MDA-MB-231/Taxol, CCK-8 experiment was used to detect the effect of Taxol at different concentrations on MDA-MB-231/Taxol cell viability. Through test, we observed that MDA-MB-231/Taxol showed relatively high drug resistance at a concentration of 0-7.5 μm/L, and Taxol gradually decreased drug resistance when the concentration was 10-15 μm/L. For the safety of drug-resistant strains, we used 1 μM/L Taxol concentration to treat drug-resistant strains in subsequent experiments (Figure 1A). In addition, through the MTT experiment, we found that the concentration of FMNT had low toxicity to MDA-MB-231/Taxol in the range of 5-20 μM/L (< 10%), while the FMNT of 40-80 μM/L had obvious cytotoxicity to the drug-resistant strain, so we used the concentration of 15 μM/L to treat the drug-resistant strain in the follow-up experiment (Figure 1B). Finally, in order to determine whether FMNT affected MDA-MB-231/Taxol cell growth via autophagy, we used autophagy inhibitor 3-MA to treat the cells, and found that 3-MA (5 mM) reduced MDA-MB-231/Taxol viability, and FMNT also effectively suppressed MDA-MB-231/Taxol cell growth (inhibition rate: 60.2%). The results indicated that the inhibitory effect of FMNT was probably exerted through autophagy. Moreover, when FMNT and Taxol were combined to treat MDA-MB-231/Taxol, the inhibitory effect on the viability of drug-resistant cells was more significant (Figure 1C). The results indicated that FMNT may suppress MDA-MB-231/Taxol cell growth by inhibiting autophagy.
Figure 1.
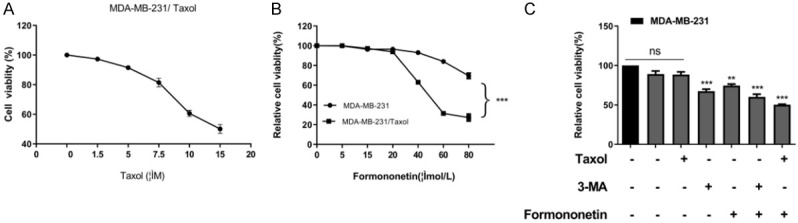
Effects of FMNT on the growth of MDA-MB-231/Taxol cells. A. Drug resistance of MDA-MB-231/Taxol cells treated with Taxol at different concentrations by CCK-8 assay. B. Survival of MDA-MB-231/Taxol cells treated with different concentrations of FMNT by MTT assay. *** indicated P < 0.001 compared between the two groups. C. Survival of MDA-MB-231/Taxol cells under different drug intervention by CCK-8 assay. ** indicated P < 0.01 vs. MDA-MB-231/Taxol; *** indicated P < 0.001 vs. MDA-MB-231/Taxol.
FMNT regulates cell cycle and induces apoptosis of MDA-MB-231/Taxol
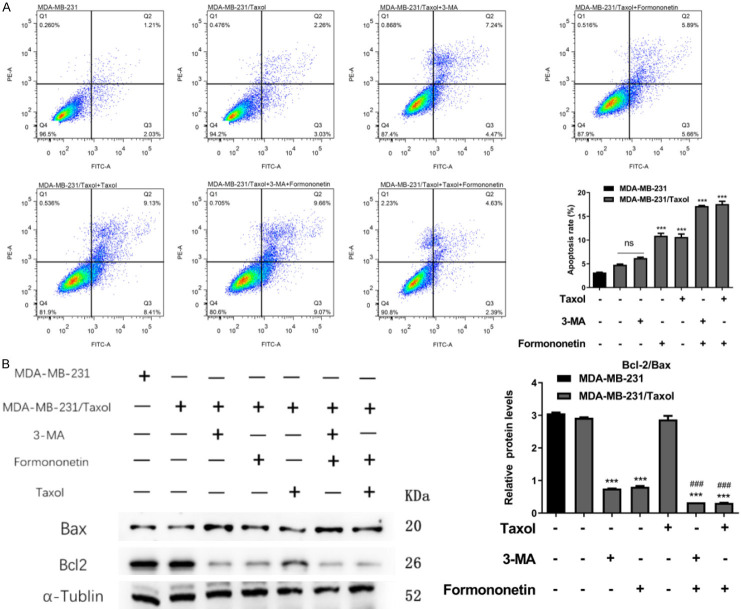
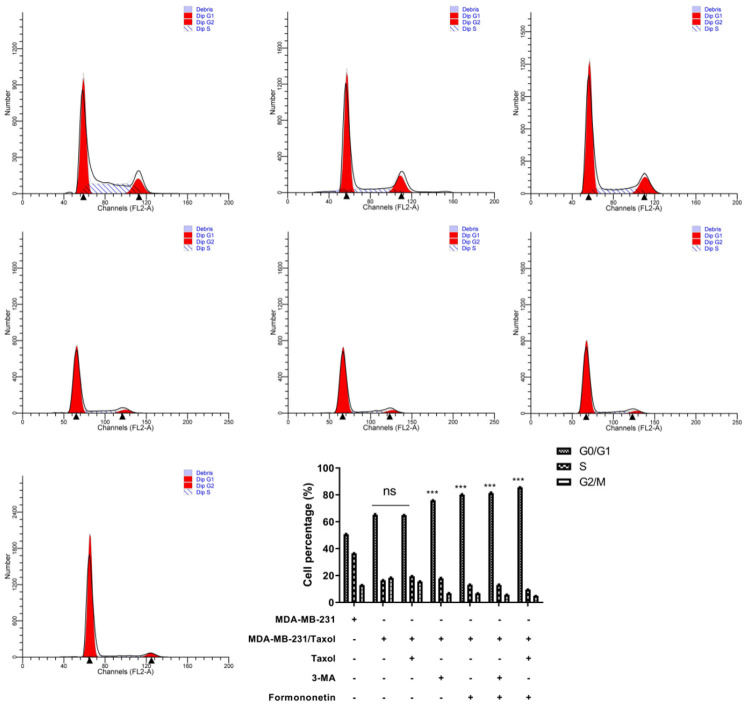
To further explore the effect of FMNT on MDA-MB-231/Taxol cell apoptosis, we examined MDA-MB-231/Taxol apoptosis after FMNT intervention. It was observed that both FMNT and 3-MA enhanced MDA-MB-231/Taxol cell apoptosis, and the apoptosis rate was even higher after the combined treatment with FMNT and Taxol (Figure 2A). Also, WB results demonstrated that the trend of apoptosis-related proteins was consistent with that of apoptosis (Figure 2B). Then, we detected the cell cycle of each group by flow cytometry (FC). It was found that both FMNT and 3-MA could obviously block the cell cycle of MDA-MB-231/Taxol in G1 phase, while the combined intervention of FMNT and Taxol had a more significant effect (Figure 3). It suggested that FMNT can promote MDA-MB-231/Taxol apoptosis by blocking G2/M phase.
Figure 2.
FMNT induces apoptosis of MDA-MB-231/Taxol cells. A. FACS detection of the effect of FMNT on apoptosis of MDA-MB-231/Taxol cells. B. WB detection of the effect of FMNT on the expression of MDA-MB-231/Taxol apoptosis-related proteins Bcl-2 and Bax; *** indicated P < 0.001 vs. MDA-MB-231/Taxol; ### indicated P < 0.001 vs. MDA-MB-231/Taxol or MDA-MB-231/Taxol+FMNT.
Figure 3.
Effects of FMNT on MDA-MB-231/Taxol cycle. *** indicated in intra-group comparison, P < 0.001.
FMNT inhibits autophagy of MDA-MB-231/Taxol cells
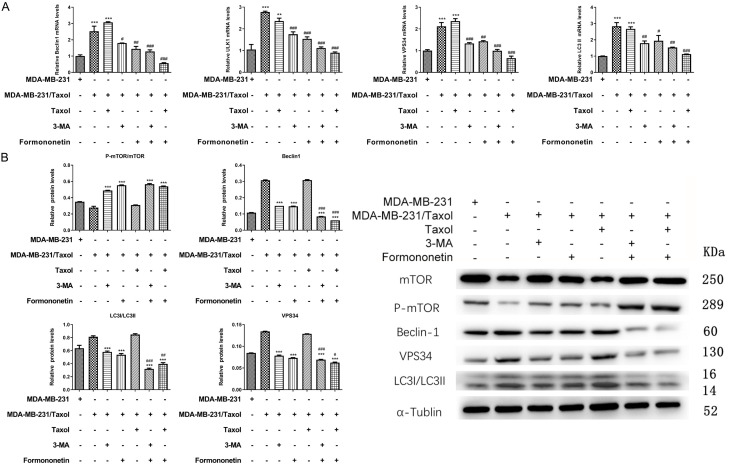
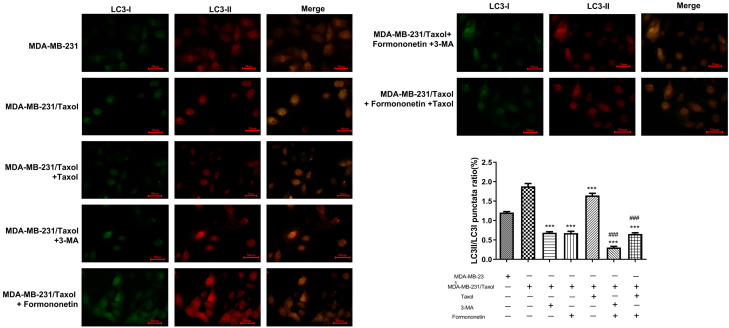
The above research has confirmed that FMNT may induce apoptosis of MDA-MB-231/Taxol cells by inhibiting autophagy, and thus inhibit cell growth. To verify our hypothesis, mRNA levels of autophagy-related genes, Beclin1, ULK1, VPS34 and LC3-II were examined by qRT-PCR. The results indicated that the autophagy level statistically elevated in MDA-MB-231/Taxol compared with in MDA-MB-231. FMNT or 3-MA evidently inhibited MDA-MB-231/Taxol cell autophagy, and the inhibitory effect was more profound when the FMNT and Taxol were in a combination therapy (Figure 4A). Besides, the results of protein detection were consistent with qRT-PCR results (Figure 4B). In addition, immunofluorescence staining exhibited less expressed autophagososomes in the MDA-MB-231 group. Conversely, more autophagolysosomes were presented in MDA-MB-231/Taxol and MDA-MB-231/Taxol+Taxol groups. FMNT or 3-MA notably suppressed MDA-MB-231/Taxol autophagy, and when FMNT was combined with Taxol, MDA-MB-231/Taxol cell autophagy can be more effectively inhibited (Figure 5). The results indicated that FMNT was capable of inhibiting the autophagy of MDA-MB-231/Taxol, thus killing tumor cells.
Figure 4.
Effects of different intervention methods on autophagy-related genes and proteins in MDA-MB-231/Taxol cells by qRT-PCR and WB. A. qRT-PCR detection of the effects of different intervention methods on the mRNA levels of Beclin1, ULK1, VPS34 and LC3-II in MDA-MB-231/Taxol cells. *** indicated P < 0.001 vs. MDA-MB-231/Taxol; # indicated P < 0.05 vs. MDA-MB-231/Taxol+3-MA or MDA-MB-231/Taxol+FMNT; ### indicated P < 0.001 vs. MDA-MB-231/Taxol+3-MA or MDA-MB-231/Taxol+FMNT. B. WB detection of the effects of different intervention methods on protein levels of mTOR, p-mTOR, Beclin1, ULK1, VPS34 and LC3-II in MDA-MB-231/Taxol cells. *** indicated P < 0.001 vs. MDA-MB-231/Taxol; # indicated P < 0.05 vs. MDA-MB-231/Taxol+3-MA or MDA-MB-231/Taxol+FMNT; ### indicated P < 0.001 vs. MDA-MB-231/Taxol+3-MA or MDA-MB-231/Taxol+FMNT.
Figure 5.
Detection of LC3-I and LC3-II staining in cells after different interventions by immunofluorescence staining. * vs. MDA-MB-231/Taxol, # vs. MDA-MB-231/Taxol+3-MA orMDA-MB-231/Taxol+Formononetin.
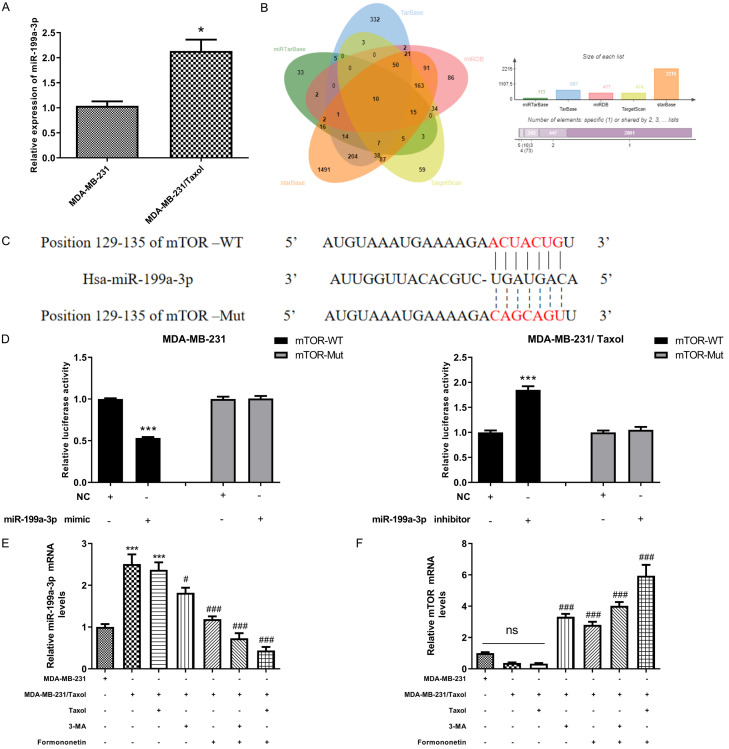
miR-199a-3p regulates mTOR in a targeted manner
References have revealed that miR-199a-3p is implicated in cancer and cardiomyocyte differentiation [27,28]. MiR-199a-3p expression as detected by qRT-PCR was more highly expressed in MDA-MB-231/Taxol than in MDA-MB-231 (Figure 6A). Moreover, we predicted the targeted binding between miR-199A-3P and mTOR through miRTarBase, TarBase, miRDB, TargetScan, starBase respectively (Figure 6B), which was subsequently verified by DLR. It was found that the fluorescence activity of mTOR-WT was decreased by miR-199a-3p mimic, while elevated by miR-199a-3p inhibitor (Figure 6C, 6D), indicating that miR-199a-3p regulated mTOR in a targeted manner. Based on the above results, we hypothesized that by inhibiting miR-199a-3P, FMNT may induce mTOR to the level of parent MDA-MB-231, thus inhibiting MDA-MB-231/Taxol autophagy and drug resistance. Subsequently, we detected miR-199a-3p and mTOR by qRT-PCR. It showed that FMNT could inhibit miR-199a-3p while restoring mTOR to the same level as that of the parent MDA-MB-231(Figure 6E, 6F), which validated the feasibility of our ideas indirectly.
Figure 6.
miR-199a-3p has a targeted relationship with mTOR. A. qRT-PCR detection of the relative expression of miR-199a-3p in MDA-MB-231 and MDA-MB-231/Taxol cells. * indicated P < 0.05 in comparison between the two groups. B. Multi-website prediction of targeted binding between miR-199a-3p and mTOR. C. Targeted binding sites and mutation sites of miR-199a-3p and mTOR. D. Dual luciferase reporter confirmed that there was a targeting relationship between miR-199a-3p and mTOR. *** indicated P < 0.001 in comparison between the two groups. E, F. Relative expression levels of miR-199a-3p and mTOR mRNA by qRT-PCR. *** indicated P < 0.001 vs. MDA-MB-231/Taxol; # indicated P < 0.05 vs. MDA-MB-231/Taxol+FMNT; ### indicated P < 0.001 vs. MDA-MB-231/Taxol+FMNT.
FMNT regulates MDA-MB-231/Taxol autophagy via miR-199a-3p/mTOR axis
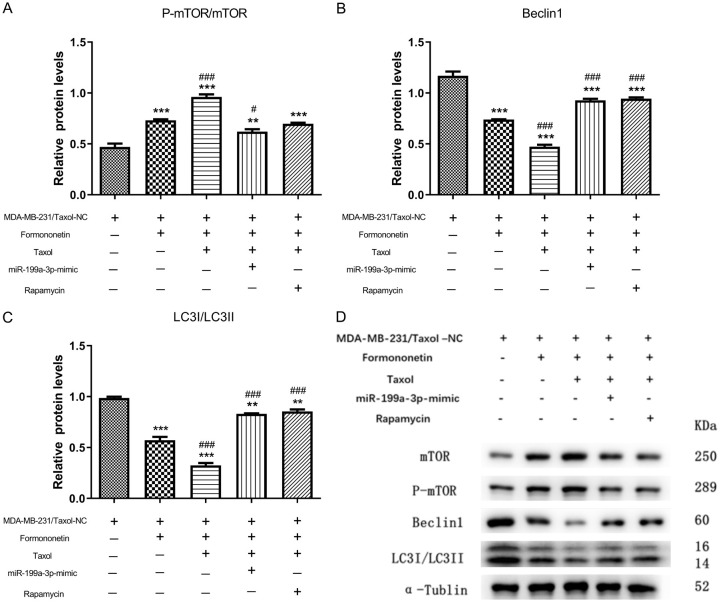
Further, for the purpose of verifying whether FMNT regulated MDA-MB-231/Taxol autophagy through miR-199a-3p/mTOR axis, we examined autophagy-related proteins in MDA-MB-231/Taxol after FMNT and miR-199a-3p mimic intervention. We found that FMNT alone could significantly suppress the autophagy of MDA-MB-231/Taxol, and FMNT combined with Taxol could further inhibit the autophagy level of it. When we treated miR-199a-3p mimic transfected drug-resistant cell lines with drugs, we found that the combined effect of FMNT and Taxol was weakened. Similarly, the combination of FMNT and Taxol was weakened by the addition of rapamycin (100 nM), an inhibitor of mTOR (Figure 7A-D). Thus, it was suggested that FMNT regulated mTOR level by inhibiting miR-199a-3P, thereby inhibiting MDA-MB-231/Taxol cell autophagy.
Figure 7.
FMNT inhibits MDA-MB-231/Taxol autophagy by regulating miR-199a-3p/mTOR axis. A-C. WB detection of the effects of different intervention methods on p-mTOR/mTOR, Beclin1, and LC3-I/LC3-II in MDA-MB-231/Taxol cells. ** indicated P < 0.01 vs. MDA-MB-231/Taxol; *** indicated P < 0.001 vs. MDA-MB-231/Taxol; # indicated P < 0.05 vs. MDA-MB-231/Taxol+FMNT; ### indicated P < 0.001 vs. MDA-MB-231/Taxol+FMNT. D. WB detection of p-mTOR/mTOR, Beclin1, and LC3-I/LC3-II autophagy-related protein expression in cells.
FMNT inhibits tumor growth in MDA-MB-231/Taxol nude mice
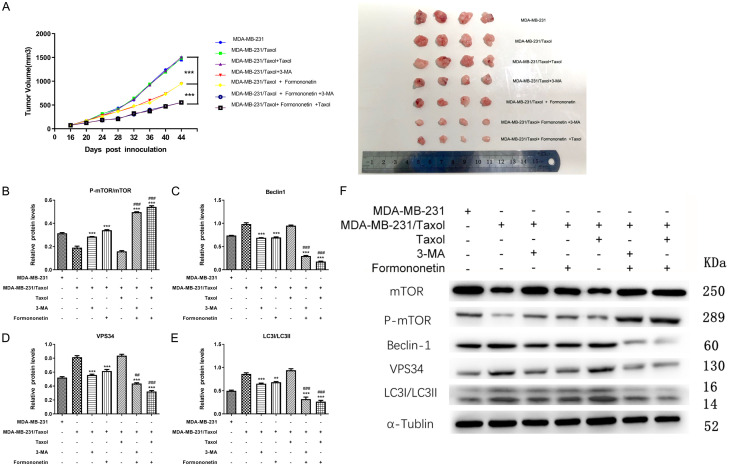
At last, an in vivo tumor model was established by subcutaneously injecting MDA-MB-231 and MDA-MB-231/Taxol into nude mice, so as to observe the cell growth. In comparison with the control group, FMNT statistically decreased mice tumor volume in the Taxol drug-resistant group, and the inhibitory effect was more significant in the combined treatment group (Figure 8A). In addition, WB detection verified that FMNT reduced Beclin1, VPS34 and LC3-I/LC3-II in the Taxol resistant group by regulating p-mTOR/mTOR (Figure 8B-F), suggesting that FMNT reduced autophagy through miR-199a-3p/mTOR axis, thus overcoming Taxol resistance.
Figure 8.
FMNT inhibits tumor growth in MDA-MB-231/Taxol nude mice. A. Changes of tumor volume in nude mice during 44 days of feeding. *** indicated P < 0.001 vs. MDA-MB-231/Taxol+FMNT. B-F. WB detection of the relative mRNA expression of Beclin1, VPS34, LC3-I/LC3-II, and p-mTOR/mTOR in tumor tissues of nude mice. ** indicated P < 0.01 vs. MDA-MB-231/Taxol; *** indicated P < 0.001 vs. MDA-MB-231/Taxol; ## indicated P < 0.01 vs. MDA-MB-231/Taxol+FMNT; ### indicated P < 0.001 vs. MDA-MB-231/Taxol+FMNT.
Discussion
TNBC is still a serious health problem. The lack of effective targeted therapy has resulted in unfavorable prognosis of patients with TNBC, and long-term chemotherapy triggers drug resistance in patients, which also lead to clinical treatment failure [29]. We observed by inhibiting miR-199a-3p that FMNT can restore mTOR to its parental level, and inhibit MDA-MB-231/Taxol cell autophagy, which has profound implications in developing potential therapeutic strategy for TNBC.
As the key component of Isoflavone Astragalus, FMNT has been proposed to have potential anticancer effect, which can inhibit the growth of various cancers such as liver cancer and BC [30,31]. However, whether FMNT plays the same role in TNBC has not been reported. This is the first study added in literature, to our knowledge, that has identified FMNT can inhibit the viability of MDA-MB-231/Taxol, indicating that FMNT may be a feasible drug for treating TNBC. Autophagy, as a hot research in the field of cancer, is considered as an essential way of anti-tumorigenesis [32,33]. For example, one study by Wang and others has revealed that Rasal2 inhibits the proliferation of breast cancer cells regulated by secretory autophagy [34]. One other study has showed that MTMR3 can regulate the proliferation of breast cancer, and cell cycle progression and autophagy are potential targets of breast cancer [35]. In addition, one study has pointed out that FMNT can protect the heart of the elderly from ischemia/reperfusion injury by enhancing autophagy degradation [36], which suggests that FMNT can regulate autophagy, but it is still unclear whether FMNT has the same effect in TNBC. Therefore, we analyzed the autophagy of FMNT in TNBC. In our research, MDA-MB-231/Taxol cells intervened by 3-MA were found to be able to inhibit the cell viability, and FMNT can effectively inhibit MDA-MB-231/Taxol cell growth, so we speculated that FMNT may play an anti-cancer role by inhibiting autophagy. Furthermore, by detecting the changes of apoptosis and cell cycle, we found that both FMNT and 3-MA promoted MDA-MB-231/Taxol cell apoptosis and blocked cell cycle in G1 phase. Moreover, it was observed that in the combined treatment group with FMNT and Taxol, MDA-MB-231/Taxol presented a higher apoptosis rate, and the cell cycle was more effectively blocked in the G1 phase. What’s more, in the combined treatment group intervened by FMNT and Taxol, MDA-MB-231/Taxol apoptosis as assessed by WB was noticeably increased, which was consistent with the FACS results. Further, the changes of autophagy-related genes and proteins in the cells were verified by immunofluorescence, qRT-PCR and WB experiments. The results identified that MDA-MB-231/Taxol cell autophagy was remarkably inhibited by FMNT or 3-MA alone, while the inhibitory effect was more profound after the combined treatment of FMNT and Taxol, suggesting that FMNT can promote apoptosis of MDA-MB-231/Taxol cells by inhibiting autophagy, thus inhibiting cell growth.
It has been reported in many papers that miR participates in autophagy by regulating downstream target genes [37-39]. MiR-199a-3p, an early discovered miR, has been confirmed to participate in autophagy by regulating downstream target genes [40]. To verify miR-199a-3p expression in TNBC, we first measured its level in MDA-MB-231 and MDA-MB-231/Taxol after different interventions, and identified its high expression in MDA-MB-231/Taxol. Furthermore, with multiple online websites, we predicted that miR-199A-3p had a targeted relationship with mTOR. mTOR belongs to phosphatidylinositol kinase-related kinase family, which can modulate cell cycle arrest and immunosuppression. For the purpose of verifying the link between the two, we performed DLR analysis and found that miR-199a-3p exerted targeted regulation to mTOR. Based on the above results, we speculated that FMNT may inhibit MDA-MB-231/Taxol cell autophagy and drug resistance by mediating miR-199a-3p/mTOR axis. At last, through experiments conducted both in vivo and in vitro, we verified the role of FMNT in MDA-MB-231/Taxol. Experiments showed that FMNT up-regulated mTOR in cells and restored it to the parental level by inhibiting miR-199a-3p, thus inhibiting tumor cell growth and playing an anti-cancer role.
Through the above studies, we have identified the mechanism of FMNT in TNBC drug resistance, but the study still has some limitations. First of all, as a basic research, this study has not yet explored the clinical anti-tumor effect of FMNT. Secondly, early studies have found that FMNT can interfere with cell growth by regulating lncRNA, but whether FMNT can regulate the occurrence of MDA-MB-231/Taxol autophagy through lncRNA-miR-mRNA axis needs further study. Therefore, we hope to carry out more research in the future to supplement our findings.
Acknowledgements
1. Shanghai Municipal Health Commission Traditional Chinese Medicine Heritage and Technology Innovation Project (ZYCC2019020). 2. National Nature Science Nurturing Project of Baoshan Branch, Shuguang Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (GZRPYJJ-201702). 3. Zhang Weihong famous TCM inheritance studio (BSMZYGZS-201909). 4. Shanghai Key Specialty Training Project of Traditional Chinese Medicine.
Disclosure of conflict of interest
None.
References
- 1.Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg. 2017;69:313–317. doi: 10.1007/s13304-017-0424-1. [DOI] [PubMed] [Google Scholar]
- 2.Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, Crosetto N, Foukakis T, Navin NE. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173:879–893. e813. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marotti JD, de Abreu FB, Wells WA, Tsongalis GJ. Triple-negative breast cancer: next-generation sequencing for target identification. Am J Pathol. 2017;187:2133–2138. doi: 10.1016/j.ajpath.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Khosravi-Shahi P, Cabezon-Gutierrez L, Custodio-Cabello S. Metastatic triple negative breast cancer: optimizing treatment options, new and emerging targeted therapies. Asia Pac J Clin Oncol. 2018;14:32–39. doi: 10.1111/ajco.12748. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Yang J, Peng L, Sahin AA, Huo L, Ward KC, O’Regan R, Torres MA, Meisel JL. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat. 2017;161:279–287. doi: 10.1007/s10549-016-4059-6. [DOI] [PubMed] [Google Scholar]
- 6.Wein L, Loi S. Mechanisms of resistance of chemotherapy in early-stage triple negative breast cancer (TNBC) Breast. 2017;34(Suppl 1):S27–S30. doi: 10.1016/j.breast.2017.06.023. [DOI] [PubMed] [Google Scholar]
- 7.Jiang D, Rasul A, Batool R, Sarfraz I, Hussain G, Mateen Tahir M, Qin T, Selamoglu Z, Ali M, Li J, Li X. Potential anticancer properties and mechanisms of action of formononetin. Biomed Res Int. 2019;2019:5854315. doi: 10.1155/2019/5854315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Chen C, Zhang J. Effects and significance of formononetin on expression levels of HIF-1alpha and VEGF in mouse cervical cancer tissue. Oncol Lett. 2019;18:2248–2253. doi: 10.3892/ol.2019.10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Gong Y, Wang S, Gao F. Anti-colorectal cancer mechanisms of formononetin identified by network pharmacological approach. Med Sci Monit. 2019;25:7709–7714. doi: 10.12659/MSM.919935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Ni Q, Wang Y, Fan H, Li Y. Synergistic anticancer effects of formononetin and temozolomide on glioma C6 cells. Biol Pharm Bull. 2018;41:1194–1202. doi: 10.1248/bpb.b18-00002. [DOI] [PubMed] [Google Scholar]
- 11.Huang D, Wang C, Duan Y, Meng Q, Liu Z, Huo X, Sun H, Ma X, Liu K. Targeting Oct2 and P53: formononetin prevents cisplatin-induced acute kidney injury. Toxicol Appl Pharmacol. 2017;326:15–24. doi: 10.1016/j.taap.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Q, Zhang W, Li T, Tang R, Li C, Yuan S, Fan D. Formononetin enhances the tumoricidal effect of everolimus in breast cancer MDA-MB-468 cells by suppressing the mTOR pathway. Evid Based Complement Alternat Med. 2019;2019:9610629. doi: 10.1155/2019/9610629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang AL, Li Y, Zhao Q, Fan LQ. Formononetin inhibits colon carcinoma cell growth and invasion by microRNA149-mediated EphB3 downregulation and inhibition of PI3K/AKT and STAT3 signaling pathways. Mol Med Rep. 2018;17:7721–7729. doi: 10.3892/mmr.2018.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callegari E, D’Abundo L, Guerriero P, Simioni C, Elamin BK, Russo M, Cani A, Bassi C, Zagatti B, Giacomelli L, Blandamura S, Moshiri F, Ultimo S, Frassoldati A, Altavilla G, Gramantieri L, Neri LM, Sabbioni S, Negrini M. miR-199a-3p modulates MTOR and PAK4 pathways and inhibits tumor growth in a hepatocellular carcinoma transgenic mouse model. Mol Ther Nucleic Acids. 2018;11:485–493. doi: 10.1016/j.omtn.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619–1628. doi: 10.1080/15548627.2017.1343770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, Zhang DM, Chen ZS. Autophagy and multidrug resistance in cancer. Chin J Cancer. 2017;36:52. doi: 10.1186/s40880-017-0219-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das CK, Linder B, Bonn F, Rothweiler F, Dikic I, Michaelis M, Cinatl J, Mandal M, Kogel D. BAG3 overexpression and cytoprotective autophagy mediate apoptosis resistance in chemoresistant breast cancer cells. Neoplasia. 2018;20:263–279. doi: 10.1016/j.neo.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang LH, Yang AJ, Wang M, Liu W, Wang CY, Xie XF, Chen X, Dong JF, Li M. Enhanced autophagy reveals vulnerability of P-gp mediated epirubicin resistance in triple negative breast cancer cells. Apoptosis. 2016;21:473–488. doi: 10.1007/s10495-016-1214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z, Liu Y, Huang X. Formononetin may protect aged hearts from ischemia/reperfusion damage by enhancing autophagic degradation. Mol Med Rep. 2018;18:4821–4830. doi: 10.3892/mmr.2018.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, Chiew MY, Tai CS, Wei TY, Tsai TR, Huang HT, Wang CY, Wu HY, Ho SY, Chen PR, Chuang CH, Hsieh PJ, Wu YS, Chen WL, Li MJ, Wu YC, Huang XY, Ng FL, Buddhakosai W, Huang PC, Lan KC, Huang CY, Weng SL, Cheng YN, Liang C, Hsu WL, Huang HD. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou D, Kavakiotis I, Maniou S, Skoufos G, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qu F, Zheng J, Gan W, Lian H, He H, Li W, Yuan T, Yang Y, Li X, Ji C, Yan X, Xu L, Guo H. MiR-199a-3p suppresses proliferation and invasion of prostate cancer cells by targeting Smad1. Oncotarget. 2017;8:52465–52473. doi: 10.18632/oncotarget.17191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park KM, Teoh JP, Wang Y, Broskova Z, Bayoumi AS, Tang Y, Su H, Weintraub NL, Kim IM. Carvedilol-responsive microRNAs, miR-199a-3p and -214 protect cardiomyocytes from simulated ischemia-reperfusion injury. Am J Physiol Heart Circ Physiol. 2016;311:H371–83. doi: 10.1152/ajpheart.00807.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jhan JR, Andrechek ER. Triple-negative breast cancer and the potential for targeted therapy. Pharmacogenomics. 2017;18:1595–1609. doi: 10.2217/pgs-2017-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meng J, Ai X, Lei Y, Zhong W, Qian B, Qiao K, Wang X, Zhou B, Wang H, Huai L, Zhang X, Han J, Xue Y, Liang Y, Zhou H, Chen S, Sun T, Yang C. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics. 2019;9:573–587. doi: 10.7150/thno.27654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J, Zhang X, Wang Y, Ye Y, Huang Z. Differential ability of formononetin to stimulate proliferation of endothelial cells and breast cancer cells via a feedback loop involving MicroRNA-375, RASD1, and ERalpha. Mol Carcinog. 2018;57:817–830. doi: 10.1002/mc.22531. [DOI] [PubMed] [Google Scholar]
- 32.Liang G, Ling Y, Mehrpour M, Saw PE, Liu Z, Tan W, Tian Z, Zhong W, Lin W, Luo Q, Lin Q, Li Q, Zhou Y, Hamai A, Codogno P, Li J, Song E, Gong C. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19:65. doi: 10.1186/s12943-020-01152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Zhang M, Shan R, Wang YJ, Chen J, Huang J, Sun LQ, Zhou WB. MTMR3 is upregulated in patients with breast cancer and regulates proliferation, cell cycle progression and autophagy in breast cancer cells. Oncol Rep. 2019;42:1915–1923. doi: 10.3892/or.2019.7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Yin X, Yang Y. Rasal2 suppresses breast cancer cell proliferation modulated by secretory autophagy. Mol Cell Biochem. 2019;462:115–122. doi: 10.1007/s11010-019-03615-7. [DOI] [PubMed] [Google Scholar]
- 35.Huang Z, Liu Y, Huang X. Formononetin may protect aged hearts from ischemia/reperfusion damage by enhancing autophagic degradation. Mol Med Rep. 2018;18:4821–4830. doi: 10.3892/mmr.2018.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin QG, Liu W, Mo YZ, Han J, Guo ZX, Zheng W, Wang JW, Zou XB, Li AH, Han F. Development of prognostic index based on autophagy-related genes analysis in breast cancer. Aging (Albany NY) 2020;12:1366–1376. doi: 10.18632/aging.102687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Wang Y, Xiao X, Cheng W, Hu L, Yao W, Qian Z, Wu W. MiR-204 inhibits hepatocellular cancer drug resistance and metastasis through targeting NUAK1. Biochem Cell Biol. 2019;97:563–570. doi: 10.1139/bcb-2018-0354. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Zhong Y, Li L. miR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC) J Transl Med. 2020;18:69. doi: 10.1186/s12967-020-02242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Wang P, Wu LL, Yan J, Pang XY, Liu SJ. miR-26a-5p inhibit gastric cancer cell proliferation and invasion through mediated Wnt5a. Onco Targets Ther. 2020;13:2537–2550. doi: 10.2147/OTT.S241199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y, Zhang G, Wu B, Yang W, Liu Z. miR-199a-5p represses protective autophagy and overcomes chemoresistance by directly targeting DRAM1 in acute myeloid leukemia. J Oncol. 2019;2019:5613417. doi: 10.1155/2019/5613417. [DOI] [PMC free article] [PubMed] [Google Scholar]