Summary
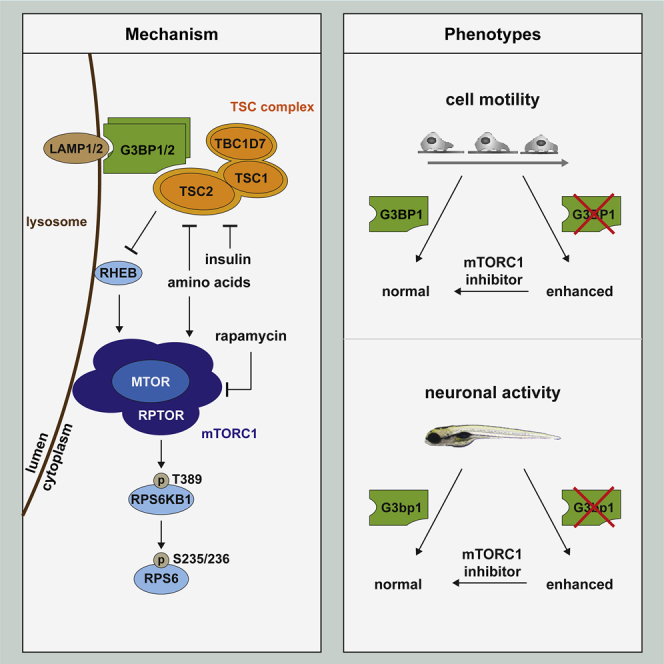
Ras GTPase-activating protein-binding proteins 1 and 2 (G3BP1 and G3BP2, respectively) are widely recognized as core components of stress granules (SGs). We report that G3BPs reside at the cytoplasmic surface of lysosomes. They act in a non-redundant manner to anchor the tuberous sclerosis complex (TSC) protein complex to lysosomes and suppress activation of the metabolic master regulator mechanistic target of rapamycin complex 1 (mTORC1) by amino acids and insulin. Like the TSC complex, G3BP1 deficiency elicits phenotypes related to mTORC1 hyperactivity. In the context of tumors, low G3BP1 levels enhance mTORC1-driven breast cancer cell motility and correlate with adverse outcomes in patients. Furthermore, G3bp1 inhibition in zebrafish disturbs neuronal development and function, leading to white matter heterotopia and neuronal hyperactivity. Thus, G3BPs are not only core components of SGs but also a key element of lysosomal TSC-mTORC1 signaling.
Keywords: TSC complex, mTORC1, G3BP1, G3BP2, lysosome, stress granule, metabolism, cancer, neuronal function
Graphical Abstract
Highlights
-
•
G3BPs act non-redundantly in the TSC-mTORC1 signaling axis
-
•
G3BPs reside at the lysosomal surface and inhibit mTORC1
-
•
The TSC complex requires G3BPs as its lysosomal tether
-
•
G3BP1 deficiency phenocopies TSC complex loss in cancer cells and neurons
Distinct from their contributions to stress granules, G3BPs regulate mTORC1 activity through spatial control of the TSC complex.
Introduction
The tuberous sclerosis complex (TSC) complex suppresses mechanistic target of rapamycin complex 1 (mTORC1) (Kim and Guan, 2019; Liu and Sabatini, 2020; Tee, 2018), a central driver of anabolism (Hoxhaj and Manning, 2019; Mossmann et al., 2018). mTORC1 hyperactivity causes diseases related to cellular overgrowth, migration, and neuronal excitability (Condon and Sabatini, 2019) and often arises from disturbances of the TSC complex, consisting of TSC complex subunit 1 (TSC1), TSC2, and TBC1 domain family member 7 (TBC1D7) (Dibble et al., 2012). In healthy cells, nutritional input such as insulin (Menon et al., 2014) and amino acids (Carroll et al., 2016; Demetriades et al., 2014) inhibits the TSC complex. The TSC complex acts as a GTPase-activating protein (GAP) that inhibits the small GTPase RHEB (Ras homolog-mTORC1 binding) (Garami et al., 2003; Inoki et al., 2003; Tee et al., 2003; Zhang et al., 2003), required for mTORC1 activation (Avruch et al., 2006; Long et al., 2005). Suppression of mTORC1 by the TSC complex takes place at mTORC1’s central signaling platform, the lysosomes (Demetriades et al., 2014; Menon et al., 2014). The molecular mechanisms anchoring RHEB and mTORC1 at lysosomes are understood in detail (Condon and Sabatini, 2019; Kim and Guan, 2019; Rabanal-Ruiz and Korolchuk, 2018). However, it is not yet clear how the TSC complex is recruited to lysosomes (Kim and Guan, 2019). We report that Ras GTPase-activating protein-binding proteins (G3BPs) act as a lysosomal tether of the TSC complex under nutrient sufficiency and starvation. G3BP1 and G3BP2 are primarily recognized as RNA-binding proteins that constitute core components of stress granules (SGs) (Alam and Kennedy, 2019; Reineke and Neilson, 2019; Riggs et al., 2020), and only a few SG-independent functions have been reported (Alam and Kennedy, 2019; Omer et al., 2020).
Results
G3BP1 inhibits mTORC1 in cells without SGs
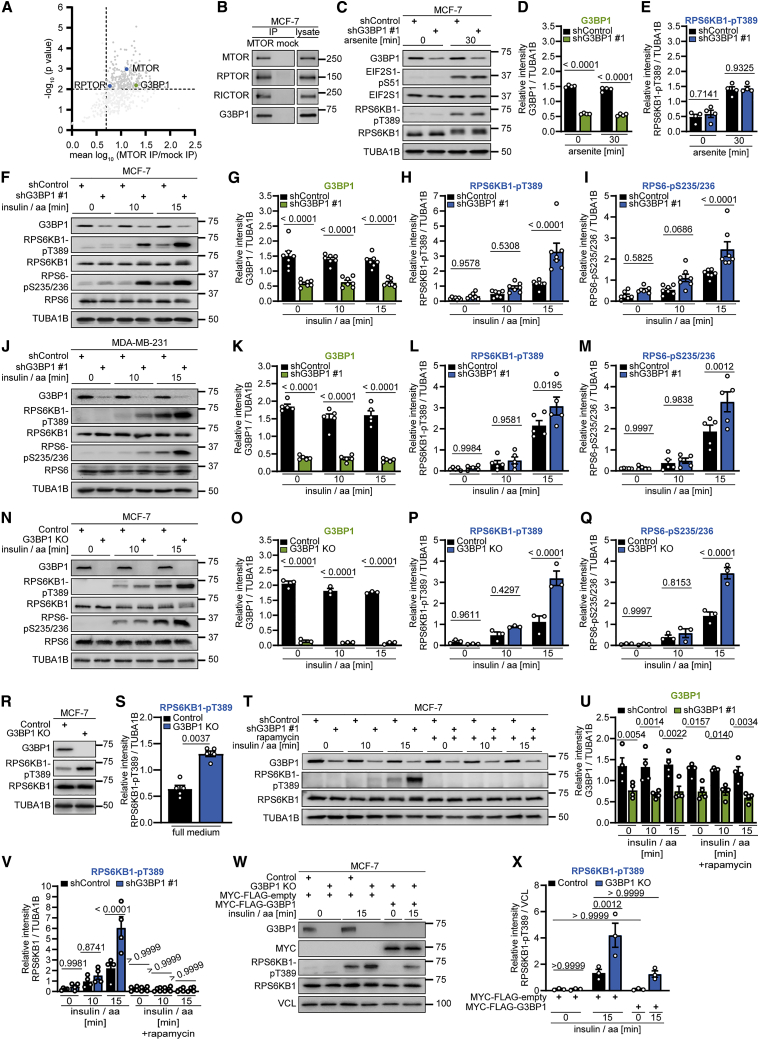
In an MTOR interactome (Schwarz et al., 2015), we observed enrichment of G3BP1 (Figures 1A and S1A). Co-immunoprecipitation (CoIP) in MCF-7 breast cancer cells corroborated that G3BP1 associates with MTOR, along with its interactors regulatory associated protein of MTOR complex 1 (RPTOR) and RPTOR independent companion of MTOR complex 2 (RICTOR) (Figures 1B and S1B). Inhibitors of mTORC1 and its upstream activator AKT1 (Kim and Guan, 2019; Liu and Sabatini, 2020; Tee, 2018) did not alter this association (Figures S1C and S1D). SGs inhibit mTORC1 (Thedieck et al., 2013; Wippich et al., 2013), and we tested involvement of the SG nucleator G3BP1 in this process. Arsenite, a frequently used inducer of SGs (Anderson et al., 2015), elicited a cytoplasmic punctate pattern of G3BP1 and eukaryotic translation initiation factor 3 subunit A (EIF3A) (Kedersha and Anderson, 2007; Figure S1E) and increased phosphorylation of the eukaryotic translation initiation factor 2 alpha (EIF2S1) at S51 (Figure 1C), a marker for conditions that induce SGs (Anderson and Kedersha, 2002). As reported earlier (Heberle et al., 2019; Thedieck et al., 2013; Wang and Proud, 1997), arsenite enhanced phosphorylation of the mTORC1 substrate ribosomal protein S6 kinase B1 (RPS6KB1) (Holz and Blenis, 2005) at T389 (RPS6KB1-pT389) (Figures 1C and 1E). G3BP1 knockdown did not alter RPS6KB1-pT389 levels (Figures 1C–1E and S1F–S1K; Table S1), indicating that, in cells with SGs, G3BP1 does not affect mTORC1 activity.
Figure 1.
G3BP1 suppresses mTORC1 activation by insulin and nutrients
(A) Re-analysis of the MTOR interactome (Schwarz et al., 2015). Shown are mean log10 ratios of proteins in MTOR versus mock IP.
(B) IP against MTOR or mock (rat immunoglobulin G [IgG]). n = 6.
(C) Arsenite-treated shG3BP1 #1 cells. n = 4.
(D) Quantitation of G3BP1 in (C). Shown are data points and mean ± SEM.
(E) Quantitation of RPS6KB1-pT389 in (C). Data are shown as in (D).
(F) Insulin and amino acid (insulin/aa)-stimulated shG3BP1 #1 cells. n = 7.
(G) Quantitation of G3BP1 in (F). Shown are data points and mean ± SEM.
(H) Quantitation of RPS6KB1-pT389 in (F). Data are shown as in (G).
(I) Quantitation of RPS6-pS235/236 in (F). Data are shown as in (G).
(J) Insulin/aa-stimulated shG3BP1 #1 cells. n = 5.
(K) Quantitation of G3BP1 in (J). Shown are data points and mean ± SEM.
(L) Quantitation of RPS6KB1-pT389 in (J). Data are shown as in (K).
(M) Quantitation of RPS6-pS235/236 in (J). Data are shown as in (K).
(N) Insulin/aa-stimulated G3BP1 KO cells. n = 3.
(O) Quantitation of G3BP1 in (N). Shown are data points and mean ± SEM.
(P) Quantitation of RPS6KB1-pT389 in (N). Data are shown as in (O).
(Q) Quantitation of RPS6-pS235/236 in (N). Data are shown as in (O).
(R) Full-medium-cultured G3BP1 KO cells. n = 5.
(S) Quantitation of RPS6KB1-pT389 in (R). Shown are data points and mean ± SEM.
(T) Rapamycin treatment of insulin/aa-stimulated shG3BP1 #1 cells. n = 4.
(U) Quantitation of G3BP1 in (T). Shown are data points and mean ± SEM.
(V) Quantitation of RPS6KB1-pT389 in (T). Data are shown as in (U).
(W) Insulin/aa-stimulated G3BP1 KO cells transfected with MYC-FLAG-G3BP1 (48 h). n = 3.
(X) Quantitation of RPS6KB1-pT389 in (W). Shown are data points and mean ± SEM.
See also Figures S1 and S2 and Table S1.
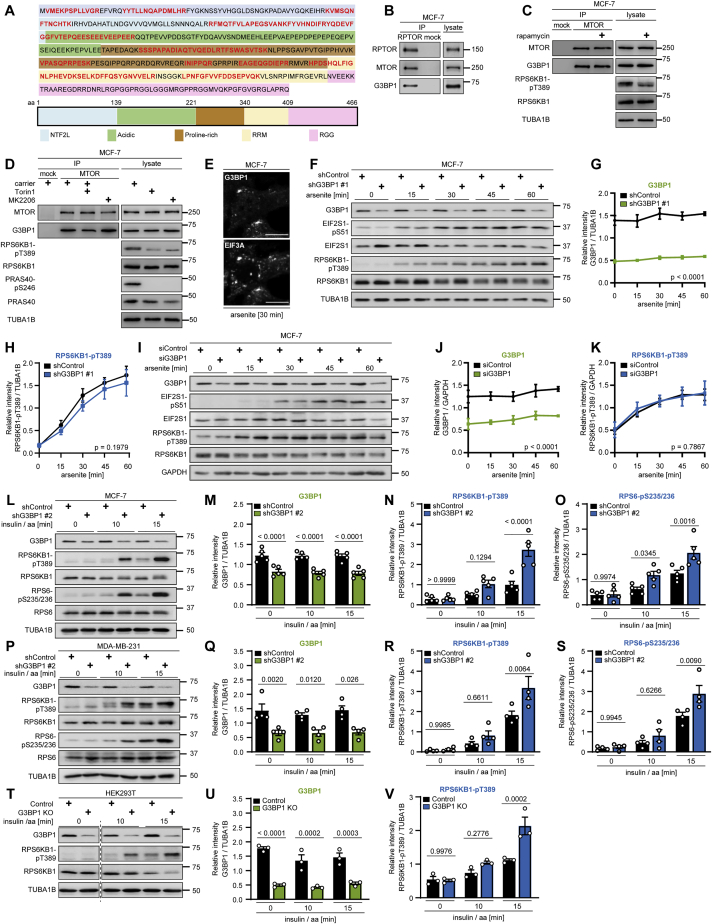
Figure S1.
G3BP1 does not alter mTORC1 activity upon arsenite stress but upon stimulation with insulin and nutrients, related to Figure 1
(A) Amino acid sequence of G3BP1. Protein domains indicated according to Reineke and Lloyd (2015) and highlighted in blue, green, brown, yellow and pink. G3BP1 peptides identified in MTOR IPs by mass spectrometry (Schwarz et al., 2015) shown in red. In total, 20 unique peptides were identified with a sequence coverage of 58.4%.
(B) IP against RPTOR (RPTOR#1 or #2) or mock (rat IgG). n = 3.
(C) IP against MTOR or mock (rat IgG) from rapamycin-treated cells. n = 3.
(D) IP against MTOR or mock (rat IgG) from Torin1 or MK2206treated cells. n = 3.
(E) IF analysis of G3BP1 and EIF3A in arsenite exposed cells. Scale bar, 10 μm. n = 3.
(F) Time course analysis of shG3BP1 #1 cells exposed to arsenite for up to 60 min. n = 3.
(G) Quantitation of G3BP1 in (F). Mean ± SEM.
(H) Quantitation of RPS6KB1-pT389 in (F). Data shown as in (G).
(I) Time course analysis of siG3BP1 cells exposed to arsenite for up to 60 min. n = 3.
(J) Quantitation of G3BP1 in (I). Mean ± SEM.
(K) Quantitation of RPS6KB1-pST389 in (I). Data shown as in (J).
(L) Insulin and amino acid (insulin/aa)-stimulated G3BP1 knockdown cells harboring a second shRNA sequence (shG3BP1 #2) targeting another exon than shG3BP1 #1 (Table S1). n = 5.
(M) Quantitation of G3BP1 in (L). Shown are data points and mean ± SEM.
(N) Quantitation of RPS6KB1-pT389 in (L). Data shown as in (M).
(O) Quantitation of RPS6-pS235/236 in (L). Data shown as in (M).
(P) Insulin/aa-stimulated shG3BP1 #2. n = 4.
(Q) Quantitation of G3BP1 in (P). Shown are data points and mean ± SEM.
(R) Quantitation of RPS6KB1-pT389 in (P). Data shown as in (Q).
(S) Quantitation of RPS6-pS235/236 in (P). Data shown as in (Q).
(T) Insulin/aa-stimulated G3BP1 KO cells generated with a second independent guide RNA against G3BP1 (sgRNA # 2, Table S1). Dashed line indicates cutting of immunoblot images to match the time points in (U) and (V). All time points were run on one gel. n = 3.
(U) Quantitation of G3BP1 in (T). Shown are data points and mean ± SEM.
(V) Quantitation of RPS6KB1-pT389 in (T). Data shown as in (U).
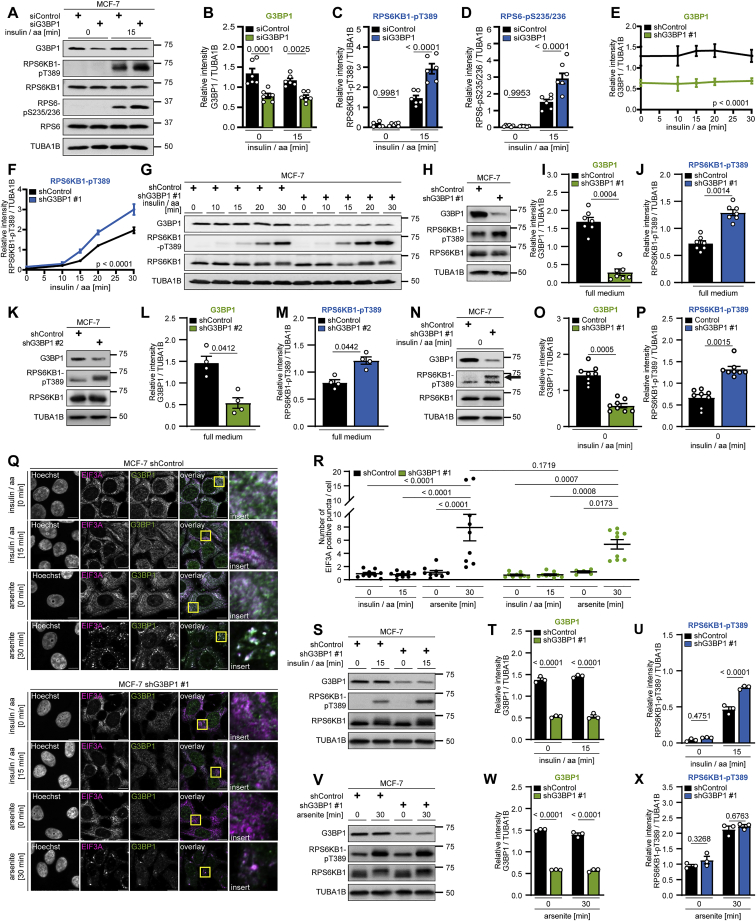
We tested whether G3BP1 influences mTORC1 activity under conditions not associated with SG formation. Insulin and amino acids activate metabolic signaling through mTORC1 (Menon et al., 2014; Shen et al., 2019; Wyant et al., 2017), and they enhanced phosphorylation of RPS6KB1-T389 and of its substrate ribosomal protein S6 (RPS6-pS235/236) 10 and 15 min after stimulation (Pende et al., 2004; Figures 1F, 1H, 1I, S1L, S1N, and S1O). G3BP1 knockdown by two different short hairpin RNA (shRNA) sequences (Table S1) further increased RPS6KB1-pT389 and RPS6-pS235/236 (Figures 1F–1I and S1L–S1O). In triple-negative MDA-MB-231 breast cancer cells, shG3BP1 knockdown also enhanced RPS6KB1-pT389 and RPS6-pS235/236 (Figures 1J–1M and S1P–S1S). Similar results were obtained when targeting G3BP1 by two different CRISPR-Cas9 single guide sequences (Table S1) in MCF-7 and HEK293T cells, respectively (Figures 1N–1Q and S1T–S1V), and by small interfering RNA (siRNA) knockdown in MCF-7 cells (Figures S2A–S2D). G3BP1 deficiency also increased RPS6KB1-T389 phosphorylation at later time points after stimulation (Figures S2E–S2G) and at steady state; i.e., in full (Figures 1R, 1S, and S2H–S2M) and in starvation medium (Figures S2N–S2P). Thus, RPS6KB1-T389 phosphorylation is enhanced in G3BP1-deficient cells. The mTORC1 inhibitor rapamycin prevented RPS6KB1-T389 hyperphosphorylation in G3BP1-deficient cells (Figures 1T–1V), showing it to be mediated by mTORC1. Re-expression of G3BP1 (Figures 1W and 1X) reversed RPS6KB1-T389 hyperphosphorylation in G3BP1 KO cells. We tested whether SGs were present in metabolically starved or stimulated cells (Figures S2Q and S2R). Arsenite served as a positive control. As expected, arsenite and amino acids + insulin enhanced mTORC1 activity (Figures S2S–S2X). Although arsenite induced SGs, no EIF3A puncta were visible in metabolically starved or stimulated cells (Figures S2Q and S2R). Thus, mTORC1 inhibition by G3BP1 under nutrient starvation and sufficiency occurs in the absence of SGs.
Figure S2.
G3BP1 inhibits mTORC1 in cells without SGs, related to Figure 1
(A) Insulin/aa-stimulated siG3BP1 cells. n = 6.
(B) Quantitation of G3BP1 in (A). Shown are data points and mean ± SEM.
(C) Quantitation of RPS6KB1-pT389 in (A). Data shown as in (B).
(D) Quantitation of RPS6-pS235/236 in (A). Data shown as in (B).
(E) Quantitation of G3BP1 in (G). Mean ± SEM.
(F) Quantitation of RPS6KB1-pT389 in (G). Data shown as in (E).
(G) Time course analysis of shG3BP1 #1 cells, insulin/aa-stimulated for up to 30 min. n = 3.
(H) shG3BP1 #1 cells cultured in full medium. n = 7.
(I) Quantitation of G3BP1 in (H). Shown are data points and mean ± SEM.
(J) Quantitation of RPS6KB1-pT389 in (H). Data shown as in (I).
(K) shG3BP1 #2 cells cultured in full medium. n = 4.
(L) Quantitation of G3BP1 in (K). Shown are data points and mean ± SEM.
(M) Quantitation of RPS6KB1-pT389 in (K). Data shown as in (L).
(N) Serum/aa-starved shG3BP1 #1 cells. Arrow, RPS6KB1-pT389 signal. n = 8, including re-analysis of improved contrast detections for data shown in Figures 1F–1H.
(O) Quantitation of G3BP1 in (N). Shown are data points and mean ± SEM. n = 8, including re-analysis of data shown in Figures 1F–1H.
(P) Quantitation of RPS6KB1-pT389 in (N). Data shown as in (O). n = 8, including re-analysis of improved contrast detections for data shown in Figures 1F–1H.
(Q) IF of shG3BP1 #1 cells. Cells were either serum/aa-starved and stimulated with insulin/aa for 15 min; or serum-starved and treated with arsenite for 30 min. Overlay: white, EIF3A and G3BP1 co-localization; magenta, EIF3A; green, G3BP1; inserts, magnifications of yellow square. Scale bar, 10 μm. n = 3, except shG3BP1 #1, arsenite [0 min], n = 2.
(R) Quantitation of data shown in (Q). Shown are data points and mean ± SEM.
(S) Immunoblot performed in parallel to IF data in (Q). Insulin/aa-stimulated shG3BP1 #1 cells. n = 3.
(T) Quantitation of G3BP1 in (S). Shown are data points and mean ± SEM.
(U) Quantitation of RPS6KB1-pT389 in (S). Data shown as in (T).
(V) Immunoblot performed in parallel to IF data in (Q). Arsenite-exposed shG3BP1 #1 cells. n = 3.
(W) Quantitation of G3BP1 in (V). Shown are data points and mean ± SEM.
(X) Quantitation of RPS6KB1-pT389 in (V). Data shown as in (W).
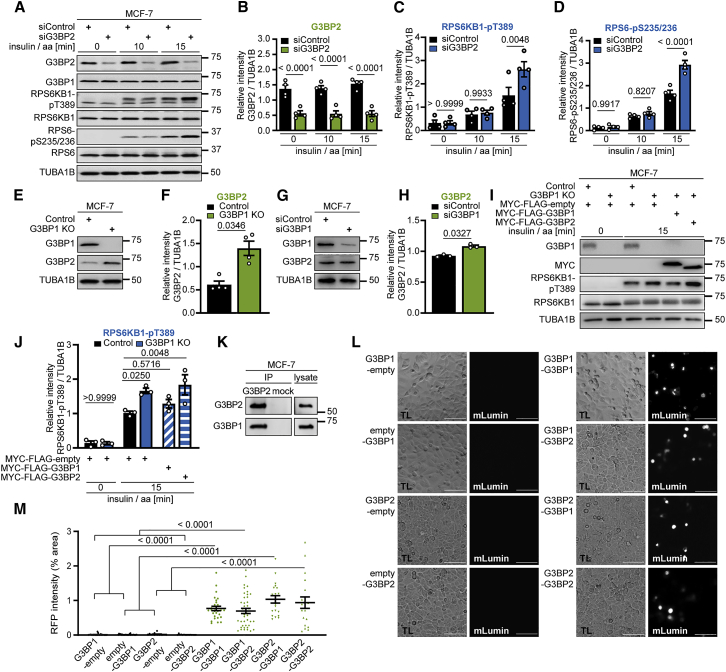
G3BP1 and G3BP2 suppress mTORC1 in a non-redundant manner
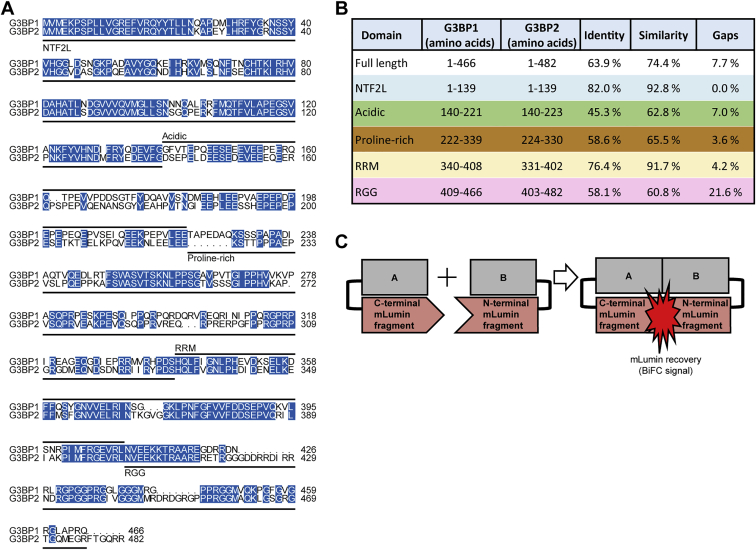
G3BP2 is highly similar to G3BP1 (Figures S3A and S3B; Kennedy et al., 2001) and can substitute for G3BP1 in SG assembly (Kedersha et al., 2016; Matsuki et al., 2013). Thus, we wanted to find out whether G3BP2 also compensates for G3BP1 in mTORC1 signaling. G3BP2 knockdown enhanced RPS6KB1-pT389 and RPS6-pS235/236 (Figures 2A–2D). In agreement with prior data (Kedersha et al., 2016), G3BP2 expression was enhanced ∼3-fold in G3BP1 knockout (KO) cells (Figures 2E and 2F) but less so upon G3BP1 knockdown (Figures 2G and 2H). If G3BP1 and G3BP2 were redundant, then an increase in G3BP2 levels would suppress the effect of G3BP1 KO. Contrary to this hypothesis, we observed a similar increase in RPS6KB1-pT389 in G3BP1 KO and knockdown cells (Figures 1P and 1H), in which the levels of G3BP2 differ substantially (Figures 2E–2H). To further test the redundancy, we performed a rescue experiment (Figures 2I and 2J). Only G3BP1, but not G3BP2, suppressed RPS6KB1-T389 hyperphosphorylation in G3BP1 KO cells. Thus, G3BP2 cannot compensate for G3BP1 loss. CoIP (Figure 2K) and bimolecular fluorescence complementation (BiFC) (Figures 2L and 2M) showed that G3BP1 and G3BP2 bind to each other. BiFC detects protein-protein interactions at a maximum distance of 10 nm (Hu et al., 2002; Figure S3C) and is indicative of close, likely direct contact. We conclude that G3BPs form a heterocomplex, which is in agreement with their non-redundancy in mTORC1 suppression.
Figure S3.
Sequence similarity between G3BP1 and G3BP2, related to Figure 2
(A) Sequence alignment of human G3BP1 (UniProt: Q13283) and G3BP2 (UniProt: Q9UN86). Protein domains are indicated according to Reineke and Lloyd (2015). Blue, identical residues.
(B) Sequence similarities of human G3BP1 and G3BP2. Sequence alignments done based on the domain regions defined for G3BP1 in Reineke and Lloyd (2015). Colors correspond to the domains marked in Figure S1A.
(C) Scheme of plasmids used for BiFC.
Figure 2.
G3BP1 and G3BP2 suppress mTORC1 in a non-redundant manner and form a heterocomplex
(A) Insulin/aa-stimulated siG3BP2 cells. n = 4.
(B) Quantitation of G3BP2 in (A). Shown are data points and mean ± SEM.
(C) Quantitation of RPS6KB1-pT389 in (A). Data are shown as in (B).
(D) Quantitation of RPS6-pS235/236 in (A). Data are shown as in (B).
(E) Serum/aa-starved G3BP1 KO cells. n = 4.
(F) Quantitation of G3BP2 in (E). Shown are data points and mean ± SEM.
(G) Serum/aa-starved siG3BP1 cells. n = 3.
(H) Quantitation of G3BP2 in (G). Shown are data points and mean ± SEM.
(I) Insulin/aa-stimulated G3BP1 KO cells transfected with MYC-FLAG-G3BP1 or MYC-FLAG-G3BP2 (48 h). n = 3.
(J) Quantitation of RPS6KB1-pT389 in (I). Shown are data points and mean ± SEM.
(K) IP against G3BP2 or mock (rabbit IgG). n = 2.
(L) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 3.
(M) Quantitation of data in (L). Shown are data points and mean ± SEM.
See also Figure S3.
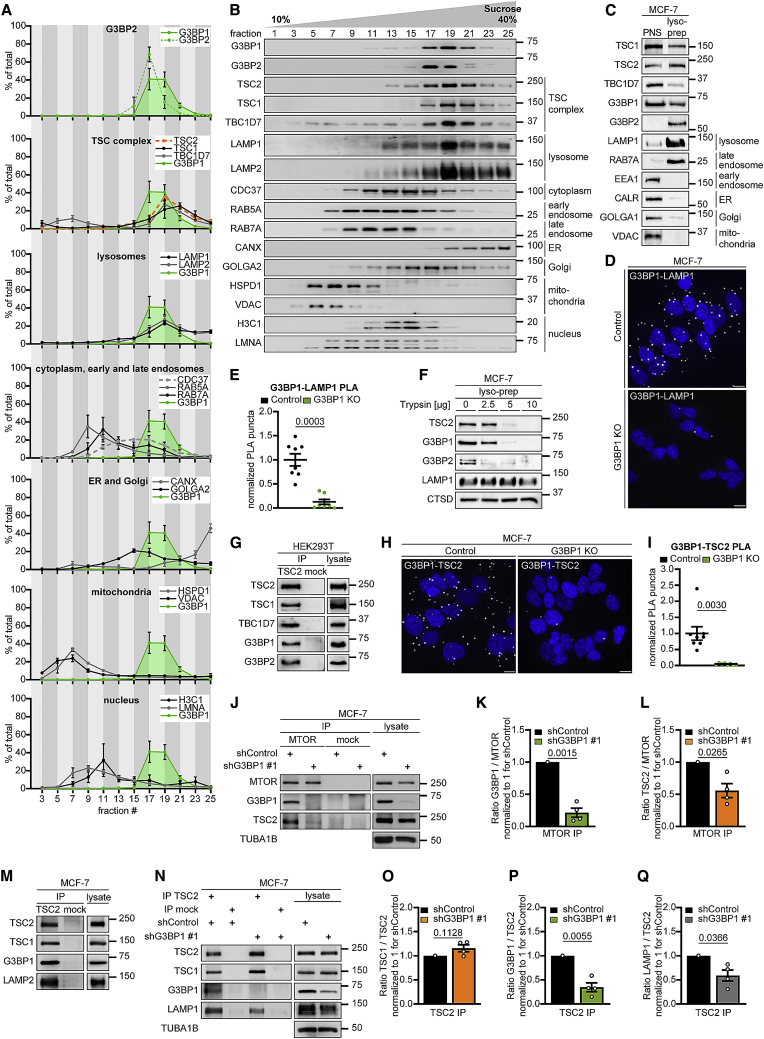
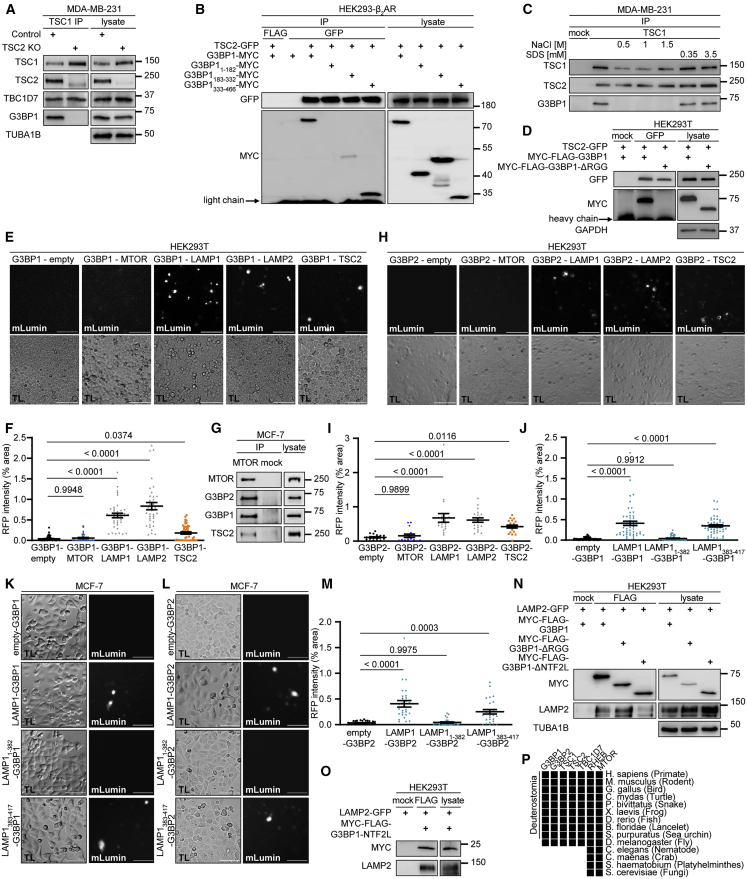
G3BPs reside at the lysosomal surface
To identify the subcellular compartment where G3BP1 and 2 act to inhibit mTORC1, we separated endosomal fractions of starved cells by sucrose density gradient centrifugation (Figures 3A and 3B). In line with earlier biochemical and immunofluorescence (IF) studies (Carroll et al., 2016; Demetriades et al., 2014; Menon et al., 2014), TSC1, TSC2, and TBC1D7 resided in the lysosomal fractions. In the absence of SGs, G3BP1 exhibits a ubiquitous cytoplasmic localization (Figure S2Q; Irvine et al., 2004), but so far no specific subcellular enrichment has been identified. We found that G3BP1 and G3BP2 reside in the same fractions as the TSC complex (Figures 3A and 3B), predominantly distributing to fractions containing lysosomal markers. Golgi apparatus, endoplasmic reticulum (ER), and cytoplasmic markers partially localized into the same fractions, suggesting that G3BPs reside at different subcellular locations. We further assessed their lysosomal localization by lysosome preparations (lyso-preps) (Figure 3C). Their purity was confirmed by enrichment of lysosomal and late endosomal markers and depletion of early endosomes and other organelles compared with the post-nuclear supernatant. We detected the G3BPs along with the TSC complex in the lyso-prep, indicative of their localization at late endosomes and/or lysosomes. In conjunction with sucrose density gradient fractionation (in which late endosomes segregate from lysosomes, the TSC complex and G3BP1/2) (Figures 3A and 3B), the lyso-prep allowed us to conclude that G3BPs localize to lysosomes. Proximity ligation assays (PLAs) (Figures 3D and 3E) confirmed in situ that G3BP1 resides close to the lysosomal protein lysosomal associated membrane protein 1 (LAMP1), at a distance of less than 40 nm (Debaize et al., 2017). Trypsin protease treatment of the lyso-preps (Figure 3F) showed that the TSC complex and the G3BPs were degraded, whereas the luminal protein cathepsin D (CTSD) and LAMP1, whose largest portion is luminal with a short cytoplasmic stretch (Eskelinen, 2006), were protected. Thus, G3BPs reside at the lysosomal surface along with the TSC complex.
Figure 3.
G3BP1 and G3BP2 reside at lysosomes
(A) Quantitation of data in (B). G3BP1, green area. Mean ± SEM.
(B) Sucrose density gradient separation of serum/aa-starved MCF-7 cells. n = 3.
(C) Lyso-prep with ferromagnetic nanoparticles. PNS, postnuclear supernatant. n = 3.
(D) PLA of G3BP1-LAMP1 in serum/aa-starved G3BP1 KO cells. PLA puncta, white dots; nuclei, blue (DAPI). Scale bar, 10 μm. n = 3.
(E) Quantitation of data in (D). Shown are data points and mean ± SEM. n = 8 technical replicates.
(F) Trypsin digest of lyso-preps prepared as in (C). n = 3 except for TSC2 (n = 2).
(G) IP against TSC2 (TSC2 #1) or mock (mouse IgG). n = 3.
(H) PLA of G3BP1-TSC2 in serum/aa-starved G3BP1 KO cells. PLA puncta, white dots; nuclei, blue (DAPI). Scale bar, 10 μm. n = 4.
(I) Quantitation of data in (H). Shown are data points and mean ± SEM. n = 8 technical replicates.
(J) IP against MTOR or mock (rat IgG); insulin/aa-stimulated shG3BP1 #1 cells (15 min). n = 4.
(K) Quantitation of G3BP1 in (J). Shown are data points and mean ± SEM.
(L) Quantitation of TSC2 in (J). Data are shown as in (K).
(M) IP against TSC2 (TSC2 #2 or #3) or mock (rabbit IgG). n = 3.
(N) IP against TSC2 (TSC2 #2) or mock (rabbit IgG); insulin/aa-stimulated shG3BP1 #1 cells (15 min). n = 4.
(O) Quantitation of TSC1 in (N). Shown are data points and mean ± SEM.
(P) Quantitation of G3BP1 in (N). Data are shown as in (O).
(Q) Quantitation of LAMP1 in (N). Data are shown as in (O).
See also Figure S4.
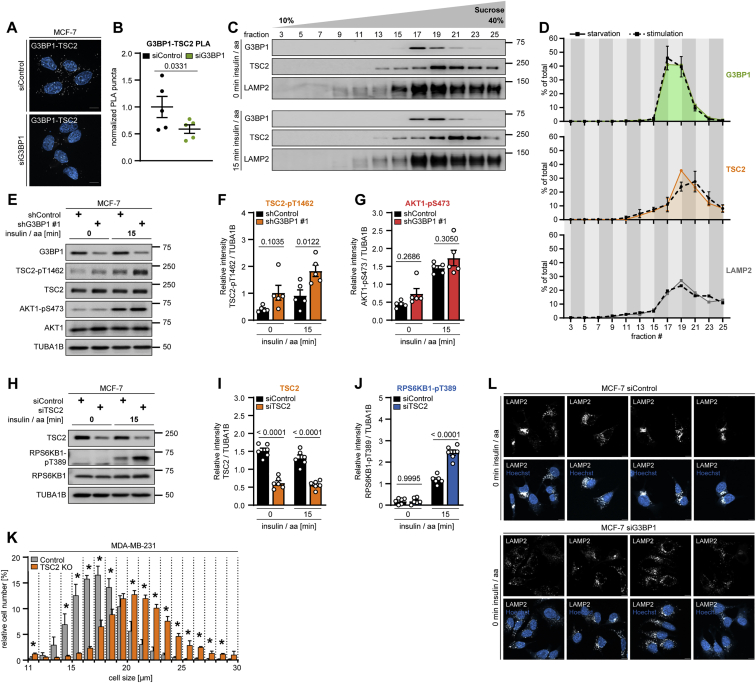
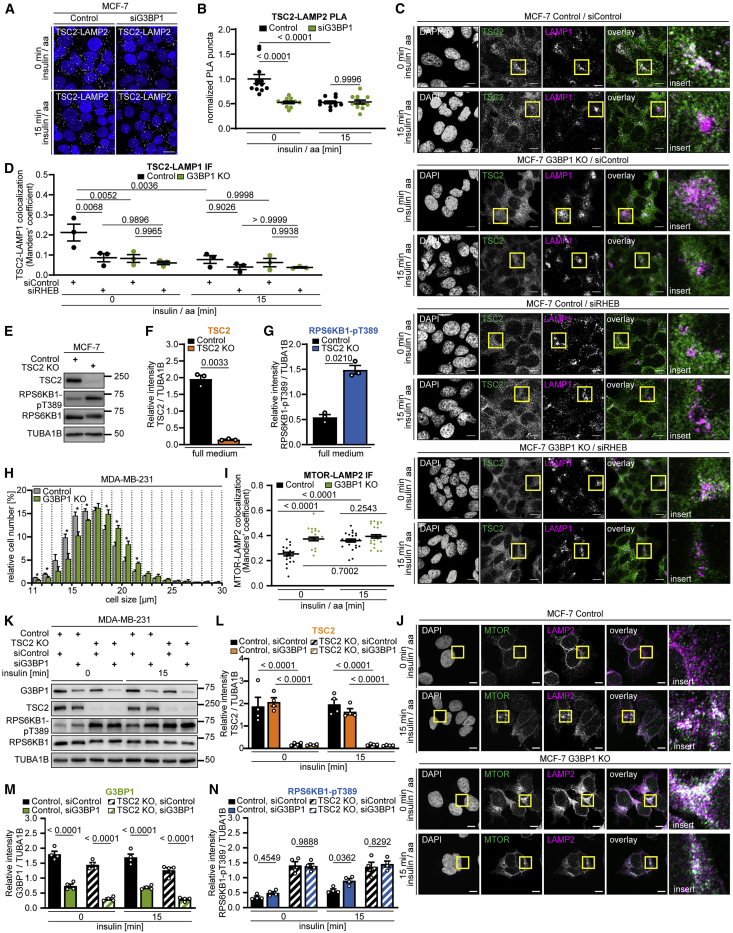
G3BP1 tethers the TSC complex to lysosomes and phenocopies lysosomal TSC functions
Like TSC1 and TBC1D7, G3BPs co-immunoprecipitated with TSC2 (Figure 3G). Thus, the TSC complex physically interacts with G3BPs. PLAs supported the association of G3BP1 with TSC2 in situ (Figures 3H, 3I, S4A, and S4B). G3BP1 was necessary for the TSC complex to act on MTOR as G3BP1 deficiency reduced the remaining TSC2-MTOR association (Figures 3J–3L) in nutrient-stimulated cells (Huang et al., 2008; Yang et al., 2020b). As a likely scenario, G3BP1 may inhibit mTORC1 by mediating the lysosomal localization of the TSC complex. We tested this in IPs of TSC2, which co-immunoprecipitated LAMP1 and LAMP2 (Figures 3M and 3N). Indeed, G3BP1 deficiency reduced TSC2-LAMP1 interaction (Figures 3N–3Q). We next wanted to find out whether G3BP1 is required for lysosomal re-localization of the TSC complex in nutrient-starved versus -stimulated cells (Carroll et al., 2016; Demetriades et al., 2014, 2016; Menon et al., 2014). Endosomal sucrose gradient fractionation was not suitable for this purpose because the nutrient-induced shift of the TSC complex away from lysosomes was not detectable (Figures S4C and S4D). PLAs showed that TSC2-LAMP2 association was highest in starved cells and decreased upon stimulation with amino acids and insulin (Figures 4A and 4B). In starved cells, G3BP1 knockdown reduced TSC2-LAMP2 association to a similar level as observed upon insulin and amino acid stimulation. In agreement, G3BP1 KO reduced TSC2-LAMP1 co-localization in starved cells to the same extent as metabolic stimulation (Figures 4C and 4D). We propose that, in G3BP1-deficient cells, impaired lysosomal recruitment of the TSC complex under starvation enhances mTORC1 activity, which results in faster mTORC1 phosphorylation dynamics upon metabolic stimuli and higher overall activity at steady state. We also observed increased TSC2 phosphorylation at the AKT target site T1462 (Figures S4E and S4F), known to be involved in its dissociation from the lysosome (Menon et al., 2014). As phosphorylation of AKT1 itself was not altered by G3BP1 deficiency (Figure S4G), lysosomal detachment may render the TSC complex more accessible to phosphorylation by AKT.
Figure S4.
G3BP1 phenocopies lysosomal TSC functions, related to Figures 3 and 4
(A) PLA of G3BP1-TSC2 in serum/aa-starved siG3BP1 cells. PLA puncta, white dots; nuclei, blue (DAPI). Scale bar, 10 μm. n = 3.
(B) Quantitation of data in (A). Shown are data points and mean ± SEM. n = 5 technical replicates.
(C) Sucrose density gradient of serum/aa-starved or insulin/aa-stimulated MCF-7 cells. n = 3.
(D) Quantitation of data in (C). Area under the curve highlighted in green (G3BP1), orange (TSC2), and gray (LAMP2), starved condition; dashed lines, stimulated condition. Mean ± SEM.
(E) Insulin/aa-stimulated shG3BP1 #1 cells. n = 5.
(F) Quantitation of TSC2-pT1462 in (E). Shown are data points and mean ± SEM.
(G) Quantitation of AKT1-pS473 in (E). Data shown as in (F).
(H) Insulin/aa-stimulated siTSC2 cells. n = 6.
(I) Quantitation of TSC2 in (H). Shown are data points and mean ± SEM.
(J) Quantitation of RPS6KB1-pT389 in (H). Data shown as in (I).
(K) Cell size of TSC2 KO cells. Mean ± SEM. *p < 0.05. n = 3.
(L) IF of LAMP2 positioning in serum/aa-starved siG3BP1 cells. White, LAMP2; blue (Hoechst), nuclei. Scale bar, 10 μm. n = 3.
Figure 4.
G3BP1 tethers the TSC complex to lysosomes
(A) PLA of TSC2-LAMP2 in insulin/aa-stimulated siG3BP1 cells (15 min, 1 μM insulin). PLA puncta, white dots; nuclei, blue (DAPI). Scale bar, 100 μm. n = 4.
(B) Quantitation of data in (A). Shown are data points and mean ± SEM. Control (0 min) normalized to 1. n = 12 technical replicates.
(C) IF of LAMP1-TSC2 co-localization in G3BP1 KO cells transfected with siRHEB; insulin/aa stimulation (1 μM insulin). Overlay: white, LAMP1-TSC2 co-localization; green, TSC2; magenta, LAMP1; insert, magnification of the yellow square. Scale bar, 10 μm. n = 3.
(D) Quantitation of data in (C). Shown are data points and mean ± SEM.
(E) TSC2 KO cells in full medium. n = 3.
(F) Quantitation of TSC2 in (E). Shown are data points and mean ± SEM.
(G) Quantitation of RPS6KB1-pT389 in (E). Data are shown as in (F).
(H) Size of G3BP1 KO cells. Mean ± SEM. *p < 0.05. n = 3.
(I) Quantitation of data in (J). Shown are data points and mean ± SEM.
(J) IF of MTOR-LAMP2 co-localization in G3BP1 KO cells. Overlay: white, MTOR-LAMP2 co-localization; green, MTOR; magenta, LAMP2; insert: magnification of the yellow square. Scale bar, 10 μm. n = 3.
(K) Insulin-stimulated TSC2 KO cells transfected with siG3BP1. n = 4.
(L) Quantitation of TSC2 in (K). Shown are data points and mean ± SEM. TSC2 was compared between control and TSC2 KO cells.
(M) Quantitation of G3BP1 in (K). Shown are data points and mean ± SEM. G3BP1 was compared between siControl and siG3BP1 in control or TSC2 KO cells.
(N) Quantitation of RPS6KB1-pT389 in (K). Data are shown as in (M).
See also Figure S4.
The TSC complex acts as a GAP on RHEB, and their interaction contributes to the lysosomal localization of the TSC complex (Carroll et al., 2016; Menon et al., 2014). Are the mechanisms by which G3BP1 and RHEB target the TSC complex to lysosomes interdependent? G3BP1 KO and RHEB knockdown reduced TSC2-LAMP1 co-localization to a similar extent, and they did not have an additive effect (Figures 4C and 4D), showing that G3BP1 and RHEB are both necessary for lysosomal recruitment of the TSC complex. Thus, the association with its GTPase is not sufficient for lysosomal localization of the TSC complex and it requires G3BP1 as an additional tether.
Like the components of the TSC complex, we propose that G3BP1 and G3BP2 act non-redundantly on mTORC1. In further support of this, inhibition of G3BP1 alone was sufficient to phenocopy TSC2 deficiency because the effect sizes of G3BP1 (Figures 1R and 1S) and TSC2 (Figures 4E–4G) KO on RPS6KB1-pT389 were similar. Also, knockdowns with similar efficiencies for G3BP1 (Figure 1F-1H) and G3BP2 (Figures 2A–2C) had similar effect sizes on RPS6KB1-pT389 as a TSC2 knockdown (Figures S4H–S4J). Loss of the TSC complex increases cell size (Figure S4K; Gao and Pan, 2001; Potter et al., 2001; Tapon et al., 2001). Cells were also enlarged upon G3BP1 KO (Figure 4H), and the increase was similar to that observed for interference with TBC1D7 (Dibble et al., 2012) or TSC1 (Potter et al., 2001; Rosner et al., 2003). G3BP1 KO also phenocopied the effects of TSC2 deficiency (Demetriades et al., 2014) in that lysosomal localization of MTOR was enhanced in starved cells (Figures 4I and 4J). Furthermore, G3BP1-deficient cells exhibited a more dispersed distribution of LAMP2 foci (Figures S4L and 4J), mimicking the dispersed lysosomal pattern in TSC2-deficient cells (Menon et al., 2014). Hence, G3BP1 inhibition is sufficient to phenocopy loss of the TSC complex.
G3BP1 suppresses mTORC1 via the TSC complex
In an epistasis experiment, we analyzed the effect of G3BP1 inhibition on mTORC1 activity in the presence or absence of TSC2 (Figures 4K–4N). We had previously stimulated cells with insulin and amino acids because they both signal through the TSC complex (Carroll et al., 2016; Demetriades et al., 2014, 2016). Amino acids also signal to mTORC1 via TSC complex-independent routes (Liu and Sabatini, 2020; Rabanal-Ruiz and Korolchuk, 2018). To exclusively assess mTORC1 inactivation via the TSC complex, we stimulated cells with insulin only. RPS6KB1-T389 was hyperphosphorylated to a similar extent in serum-starved or insulin-stimulated TSC2 KO cells because the TSC complex was absent. G3BP1 inhibition induced RPS6KB1-pT389 in control cells but not in TSC2 KO cells (Figures 4K and 4N). Thus, G3BP1 and the TSC complex act in the same pathway to suppress mTORC1.
TSC2 mediates formation of the G3BP1-TSC complex
Which TSC complex subunit mediates G3BP1 binding? TSC2 KO resulted in loss of G3BP1 from the TSC1-TBC1D7 complex (Figure 5A), indicating that G3BP1 binds TSC2. A C-terminal fragment of G3BP1 co-immunoprecipitated with TSC2-GFP to a similar extent as full-length G3BP1, whereas the middle part (with the proline-rich domain) and the N-terminal region (with the NTF2-like [NTF2L] domain) exhibited faint or no interaction (Figure 5B). Thus, the G3BP1 C terminus harboring the RNA recognition motif (RRM) and the arginine-glycine rich (RGG) repeats (Tourrière et al., 2003; Figure S1A) mediates binding to TSC2.
Figure 5.
G3BPs bridge TSC2 to LAMP proteins
(A) IP against TSC1 (TSC1 #1) or mock (rabbit IgG) in TSC2 KO cells. n = 3.
(B) IP against GFP or FLAG; transfection with the indicated plasmids. n = 5.
(C) IP against TSC1 (TSC1 #2) or mock (mouse IgG) incubated with NaCl or SDS. n = 3.
(D) IP against GFP or mock (mouse IgG); transfection with the indicated plasmids. n = 3.
(E) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 3.
(F) Quantitation of data in (E). Shown are data points and mean ± SEM.
(G) IP against MTOR or mock (rat IgG). n = 3.
(H) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 4.
(I) Quantitation of data in (H). Shown are data points and mean ± SEM.
(J) Quantitation of data in (K). Shown are data points and mean ± SEM.
(K) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 5.
(L) BiFC. Protein+C-terminal mLumin is indicated first; protein+N-terminal mLumin is indicated second. TL, transmitted light. Scale bar, 100 μm. n = 3.
(M) Quantitation of data in (L). Shown are data points and mean ± SEM.
(N) IP against FLAG or mock (mouse IgG); transfection with the indicated plasmids. n = 3.
(O) IP against FLAG or mock (mouse IgG); transfection with the indicated plasmids. n = 3.
(P) Phylogenetic analysis. Black square, protein present in species.
See also Figure S5.
The TSC complex is resistant to high salt (1.5 M NaCl) and detergent (3.5 mM SDS), indicative of the high affinity between TSC1, TSC2, and TBC1D7 (Dibble et al., 2012; Nellist et al., 1999). In contrast, G3BP1 was lost at 0.5 M NaCl (Figure 5C), suggesting that its binding to the TSC complex requires electrostatic interactions. The G3BP1 C terminus harbors an intrinsically disordered region (IDR) (Guillén-Boixet et al., 2020; Panas et al., 2019; Yang et al., 2020a), which, as is typical for IDRs (Forman-Kay and Mittag, 2013), contains a high density of positively charged arginine residues that mediate electrostatic interactions. G3BP1 binding was stable against denaturation by 3.5 mM SDS, a concentration that preferentially disrupts hydrophobic interactions (Højgaard et al., 2018). Thus, upon exposure to SDS, G3BP1 retains high affinity to the TSC complex, in a range similar to the affinity between TSC1 and TSC2 (Dibble et al., 2012). Because the TSC complex and G3BP1 form a highly stable complex that requires electrostatic interactions, we deleted the RGG domain, which encompasses the C-terminal IDR of G3BP1 (Guillén-Boixet et al., 2020; Yang et al., 2020a). TSC2 lost binding to G3BP1-ΔRGG (Figure 5D), demonstrating that the C-terminal IDR/RGG domain of G3BP1 interacts with TSC2.
G3BPs bridge TSC2 to LAMP proteins
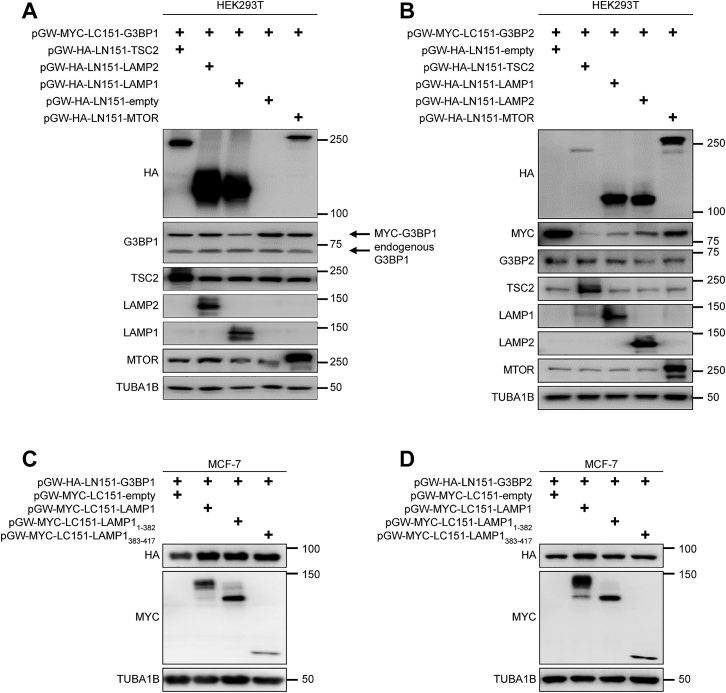
We next assessed the proximity of the G3BP1 association with TSC2, the LAMP1/2 proteins, and MTOR by BiFC (Figures 5E, 5F, and S5A). Cells that co-expressed G3BP1 with MTOR did not exhibit a BiFC signal. Thus, their interaction in IPs (Figure 1B) may not be direct but is possibly mediated by their common association with lysosomes. In contrast, BiFC signals for G3BP1 with LAMP1, LAMP2, and TSC2 were indicative of their close interaction. Similar results were obtained for G3BP2 (Figures 5G–5I and S5B). Because G3BPs are at the lysosomal surface (Figure 3), whereas LAMP proteins are mainly luminal with a short transmembrane and cytoplasmic portion, we wondered whether the latter is sufficient for binding. In line with this, G3BP1 and G3BP2 interacted with the transmembrane and cytoplasmic domains (LAMP1383–417) but not with the luminal part of LAMP1 (LAMP11–382) (Figures 5J–5M, S5C, and S5D). Loss of its N-terminal NTF2L domain (G3BP1-ΔNTF2L) prevented G3BP1 from binding to LAMP2 (Figure 5N). Conversely, the NTF2L domain was sufficient to co-immunoprecipitate LAMP2 (Figure 5O). LAMP2 remained bound to a G3BP1 fragment devoid of the RGG domain (Figure 5N) that mediates TSC2 interaction. Thus, G3BP1 binds to TSC2 via its C-terminal RGG domain and to LAMP2 via its N-terminal NTF2L domain, bridging TSC2 to the LAMP proteins.
Figure S5.
Expression of BiFC constructs, related to Figure 5
(A) Expression of BiFC fusion proteins used in Figures 5E and 5F. Cells transfected with the indicated plasmids. n = 3.
(B) Expression of BiFC fusion proteins used in Figures 5H and 5I. Cells transfected with the indicated plasmids. n = 3.
(C) Expression of BiFC fusion proteins used in Figures 5J and 5K. Cells transfected with the indicated plasmids. n = 3.
(D) Expression of BiFC fusion proteins used in Figures 5L and 5M. Cells transfected with the indicated plasmids. n = 3.
The G3BPs co-appeared with the TSC complex during evolution
In view of the key function of G3BP1/2 in TSC-mTORC1 signaling, we analyzed their phylogenetic distribution (Figure 5P). While MTOR and RHEB are present in the yeast S. cerevisiae, G3BPs appeared together with the TSC complex in D. melanogaster and in the clade of Deuterostomia. G3BP1 orthologs have been proposed in S. cerevisiae (Yang et al., 2014) and in C. elegans (Jedrusik-Bode et al., 2013). Sequence analyses (Database: NCBI BLASTP nr database, BLOSUM45 matrix, 19.02.2020) showed that the human protein UNC80 (Genbank: XP_016859383.1) has the highest similarity to the proposed S. cerevisiae G3BP1 ortholog BRE5 (UniProt: P53741). Although the C. elegans protein GTBP-1 (UniProt: Q21351) has the highest similarities to human G3BP1/2, they are low (e values 4e−7 and 0.12) and restricted to the NTF2L and RRM domains, of which they cover 23%, not reaching the threshold for our phylogenetic analysis. Thus, G3BP1 and G3BP2 orthologs emerged together with the TSC complex.
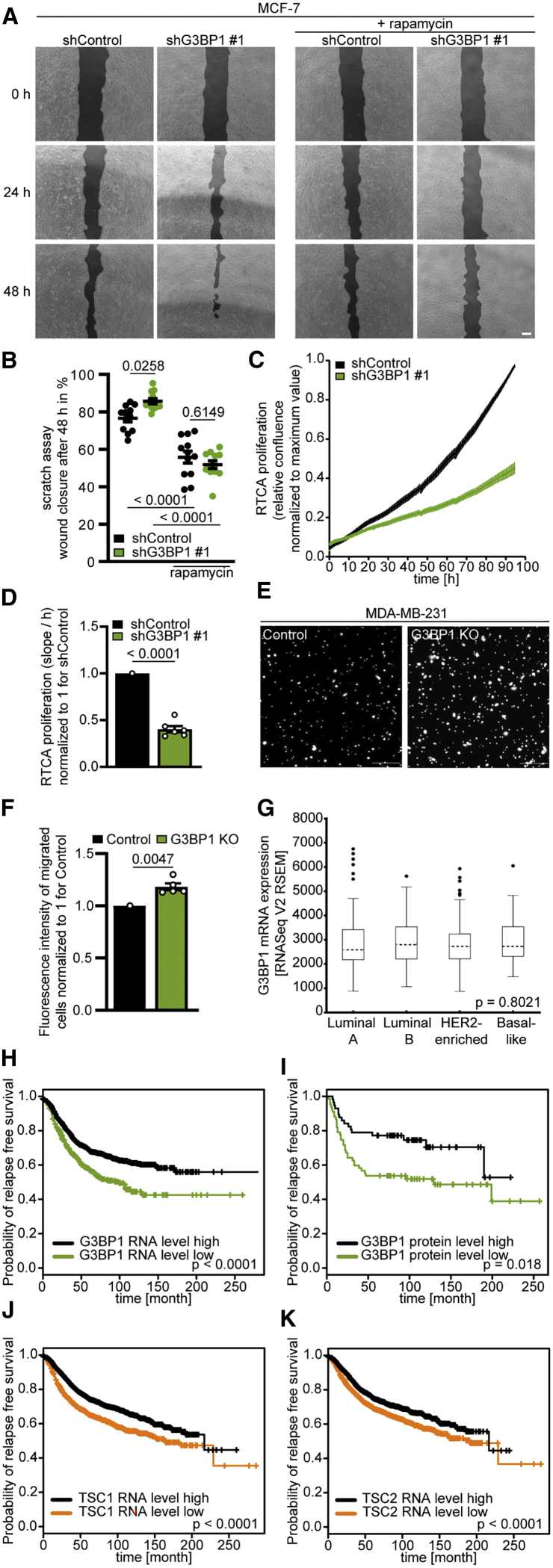
G3BP1 suppresses mTORC1-driven migration in breast cancer cells
Ablation of the TSC1 or TSC2 genes increases cancer cell motility and metastasis (Astrinidis et al., 2002; Goncharova et al., 2006). G3BP1 deficiency also enhanced cell motility in a scratch assay, which was abrogated by rapamycin (Figures 6A and 6B). In line with prior reports (Alam and Kennedy, 2019; Dou et al., 2016; Wang et al., 2018; Winslow et al., 2013), G3BP1 deficiency reduced proliferation (Figures 6C and 6D), confirming that the enhanced motility did not result from enhanced proliferation. Also in a Transwell migration assay (Figures 6E and 6F), G3BP1 KO cells exhibited enhanced migration. G3BP1 mRNA levels were similar in the four breast cancer subtypes (Koboldt et al., 2012; Figure 6G). Patients with G3BP1 mRNA or protein levels below the median exhibited shorter relapse-free survival (RFS) (Figure 6H, I), reminiscent of the shorter RFS in patients with low TSC1 or TSC2 (Figures 6J and 6K). Thus, G3BP1 and the TSC complex could be subtype-independent indicators of mTORC1 activity and cancer cell motility.
Figure 6.
G3BP1 suppresses mTORC1-driven migration in breast cancer cells
(A) Scratch assay with shG3BP1 #1 cells. Scale bar, 150 μm. n = 3.
(B) Quantitation of data in (A). Shown are data points and mean ± SEM.
(C) Real-time cell analysis (RTCA) of proliferation of shG3BP1 #1 MCF-7 cells. Mean ± SEM. n = 6.
(D) Quantitation of data in (C). Shown are data points and mean ± SEM.
(E) Transwell migration of G3BP1 KO cells (6–8 h). Scale bar, 150 μm. n = 5.
(F) Quantitation of data in (E). Shown are data points and mean ± SEM.
(G) G3BP1 mRNA expression. Expression values from The Cancer Genome Atlas (TCGA) processed and normalized by RNA-Seq by Expectation Maximization (RSEM) are classified according to PAM50. Data are shown as boxplots, median with 25th+75th percentiles as boxes, and 5th+95th percentiles as whiskers.
(H) Relapse free survival (RFS) of individuals with breast cancer based on G3BP1 RNA levels.
(I) RFS of individuals with breast cancer based on G3BP1 protein levels.
(J) RFS of individuals with breast cancer based on TSC1 RNA levels.
(K) RFS of individuals with breast cancer based on TSC2 RNA levels.
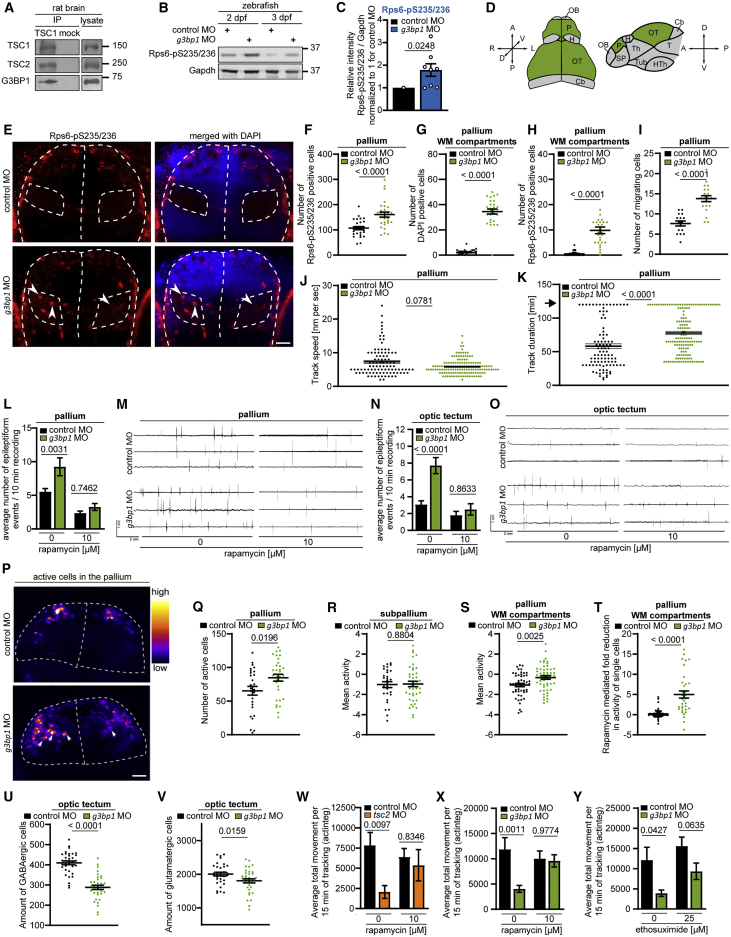
G3BP1 deficiency elicits mTORC1-driven neuronal phenotypes in vivo
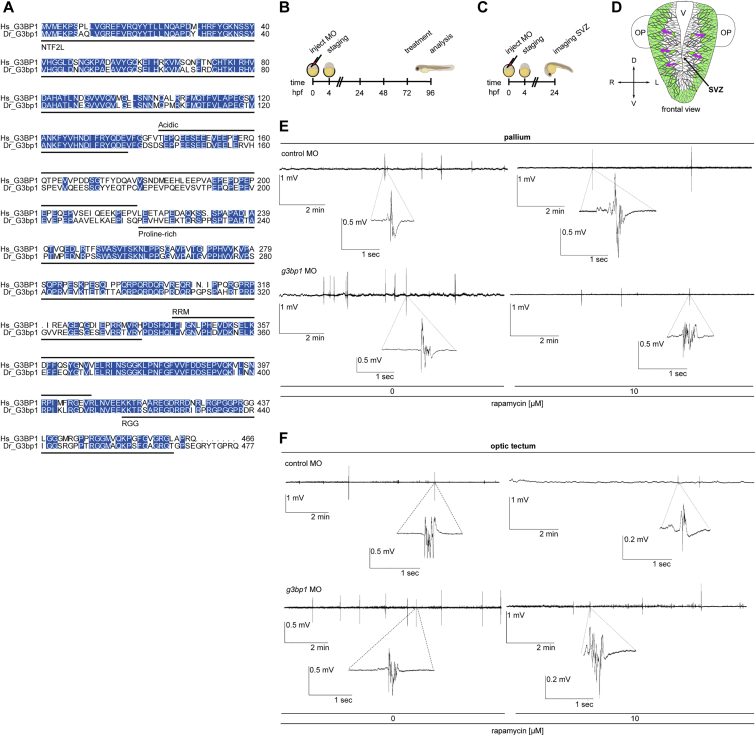
Loss of the TSC complex (Switon et al., 2017) and of G3BP1 (Martin et al., 2013; Zekri et al., 2005) elicits neuronal phenotypes. TSC1 IPs showed that G3BP1 binds the TSC complex in the rat brain (Figure 7A). We explored a possible similarity of neurodevelopmental G3BP1 and TSC2 phenotypes in zebrafish, where lack of Tsc2 elicits aberrant brain morphology, neuronal hyperexcitability, and seizures (Kedra et al., 2020; Kim et al., 2011; Scheldeman et al., 2017). The zebrafish G3bp1 and human G3BP1 orthologs exhibit 67.8% sequence identity (Figure S6A). g3bp1 inhibition by morpholino oligonucleotides (MO) enhanced mTORC1 activity, as determined by Rps6-pS235/236 (Figures 7B and 7C). We analyzed brain anatomy (Figure S6B) in the pallium (Figure 7D), the equivalent of the human cortex (Friedrich et al., 2010; Mueller and Wullimann, 2015; Parker et al., 2013), which is the main region involved in MTOR-related neurodevelopmental pathologies (Marsan and Baulac, 2018; Mühlebner et al., 2019). In Tsc2-deficient zebrafish, Rps6-pS235/236-positive cells mislocalize to the white matter (WM) of the pallium (Kedra et al., 2020; Kim et al., 2011). Similarly, g3bp1 morphants showed increased numbers of Rps6-pS235/236-positive cells (Figures 7E and 7F), many of which resided in the WM (Figures 7G and 7H). Twice as many neuronal progenitors migrated from the subventricular zone (SVZ) to outer brain layers (Figures S6C, S6D, and 7I). Although they exhibited similar velocity (Figure 7J), movement duration was prolonged (Figure 7K). Thus, aberrant migration dynamics may underlie neuron mislocalization to the WM in g3bp1 morphants. Non-invasive recordings of local field potentials (LFP) detect aberrant neuronal activity in epilepsy-related zebrafish models (Baraban et al., 2013; Hunyadi et al., 2017; Siekierska et al., 2019; Sourbron et al., 2016; Zhang et al., 2015b). LFP recordings from the pallia (Figure 7L, 7M, and S6E) and optic tecta (Figures 7N, 7O, and S6F) revealed neuronal hyperactivity in g3bp1 morphants, which was suppressed by rapamycin. At the single-cell level, increased numbers of active cells resided in the pallium (Figure S6B, 7P, and 7Q; Videos S1 and S2). While the mean neuronal activity in the subpallium was unchanged (Figure 7R), it was increased significantly in the WM of the pallium (Figure 7S). At the single-cell level, rapamycin also suppressed neuronal hyperactivity (Figure 7T). Neuronal network hyperactivity can result from imbalanced glutamatergic and GABAergic networks (Bozzi et al., 2018; Brenet et al., 2019). g3bp1 morphants showed a severe reduction of GABAergic neurons and a lesser reduction of glutamatergic neurons (Figures 7U, 7V, and S6B). Thus, an imbalance of GABAergic and glutamatergic networks may contribute to neuronal hyperactivity. In Tsc2-deficient zebrafish, anatomical changes and neuronal hyperexcitability are associated with non-motor seizures manifesting as decreased locomotor activity (Kedra et al., 2020; Scheldeman et al., 2017; Figure 7W). g3bp1 morphants recapitulated this behavior (Figure 7X and S6B), which was rescued by rapamycin (Figure 7X). Thus, mTORC1 accounts for their reduced locomotor activity. Similar to Tsc2-deficient zebrafish (Kedra et al., 2020), the antiepileptic drug ethosuximide reversed hypoactivity of the g3bp1 morphants (Figure 7Y). This is reminiscent of ethosuximide suppressing abnormal spike-and-wave discharges in mice with generalized non-motor absence seizures because of impaired cortico-striatal excitatory transmission (Miyamoto et al., 2019), suggesting that the hypoactivity of g3bp1 morphants may be caused by non-motor seizures.
Figure 7.
G3BP1 deficiency elicits mTORC1-driven neuronal phenotypes in vivo
(A) IP against TSC1 (TSC1 #3) or mock (rabbit IgG). n = 2.
(B) Zebrafish larvae injected with g3bp1 MO. dpf, days post fertilization. n = 4/day.
(C) Quantitation of Rps6-pS235/236 in (B), pooled for 2+3 dpf. Shown are data points and mean ± SEM.
(D) Dorsal and lateral view of a zebrafish larva brain. P, pallium; OT, optic tectum; H, habenula; Cb, cerebellum; OB, olfactory bulb; SP, subpallium; Th, thalamus; Tub, tuberculum; T, tegmentum; HTh, hypothalamus.
(E) IF of Rps6-pS235/236 in g3bp1 MO-injected zebrafish larvae. Nuclei, blue (DAPI); dashed white lines, white matter (WM) compartments of the pallium; arrows, Rps6-pS235/236-positive cells in the WM. Scale bar, 25 μm. n ≥ 29 larvae/condition.
(F) Quantitation of Rps6-pS235/236-positive cells in the pallium in (E). Shown are data points and mean ± SEM.
(G) Quantitation of cells in the WM in (E). Data are shown as in (F).
(H) Quantitation of Rps6-pS235/236-positive cells in the WM in (E). Data are shown as in (F).
(I) Quantitation of HuC-positive cells in g3bp1 MO zebrafish larvae (24 hpf [hours post fertilization]). Shown are data points and mean ± SEM. n ≥ 10 larvae/condition.
(J) Movement speed of single HuC-positive cells. Data are shown as in (I).
(K) Track duration of single HuC-positive cells. Data are shown as in (I). Arrow, maximum track duration.
(L) Quantitation of epileptiform events in LFP recordings from the pallia of g3bp1 MO zebrafish larvae (4 dpf). Mean ± SEM. n ≥ 34 larvae/condition.
(M) Representative LFP recordings for (L).
(N) Quantitation of epileptiform events in LFP recordings from optic tecta of g3bp1 MO zebrafish larvae (4 dpf). Mean ± SEM. n ≥ 20 larvae/condition.
(O) Representative LFP recordings for (N).
(P) Neuronal activity in pallia of Tg(HuC:GCaMP5G) zebrafish larvae injected with g3bp1 MO (4 dpf). Dashed white lines, pallium; arrows, ectopic cells with high neuronal activity in the WM; yellow/orange, high neuronal activity. Scale bar, 25 μm. n ≥ 27 larvae/condition.
(Q) Quantitation of active neuronal cells in (P). Shown are data points and mean ± SEM.
(R) Quantitation of mean neuronal activity in the subpallia of Tg(HuC:GCaMP5G) zebrafish larvae injected with g3bp1 MO (4 dpf). Shown are data points and mean ± SEM. n ≥ 15 larvae/condition.
(S) Quantitation of mean neuronal activity in the WM of Tg(HuC:GCaMP5G) zebrafish larvae injected with g3bp1 MO (4 dpf). Shown are data points and mean ± SEM. n ≥ 14 larvae/condition.
(T) Quantitation of rapamycin-mediated fold reduction in the activity of single cells in the WM of Tg(HuC:GCaMP5G) zebrafish larvae injected with g3bp1 MO (4 dpf). The number of active cells in rapamycin-treated larvae was normalized to those in untreated larvae. Shown are data points and mean ± SEM. n ≥ 14 larvae/condition.
(U) Quantitation of GABAergic cells in optic tecta of Tg(dlx5a/dlx6a-EGFP) x Tg(vglut2a:loxP-RFP-loxP-GFP) zebrafish larvae injected with g3bp1 MO (4 dpf). Shown are data points and mean ± SEM. n ≥ 34 larvae/condition.
(V) Quantitation of glutamatergic cells in optic tecta of Tg(dlx5a/dlx6a-EGFP) x Tg(vglut2a:loxP-RFP-loxP-GFP) zebrafish larvae injected with g3bp1 MO (4 dpf). Data are shown as in (U).
(W) Locomotor activity of tsc2 MO zebrafish larvae (4 dpf). Mean ± SEM. n ≥ 26 larvae/condition.
(X) Locomotor activity of g3bp1 MO zebrafish larvae (4 dpf). Mean ± SEM. n ≥ 36 larvae/condition.
(Y) Locomotor activity of g3bp1 MO zebrafish larvae (4 dpf). Mean ± SEM. n = 24, untreated, n = 36 ethosuximide-treated larvae/condition.
Figure S6.
Background information for the zebrafish experiments, related to Figure 7
(A) Sequence alignment of human (Hs) G3BP1 (UniProt: Q13283) and zebrafish (Dr) G3bp1 (UniProt: Q6P124). Protein domains indicated according to Reineke and Lloyd (2015). Blue, identical residues. The sequences share 67.8% identity and 77.4% similarity.
(B) Treatment scheme of the g3bp1 MO and control MO injected zebrafish larvae for the analyses at 4 dpf (96 hpf): IF analysis, non-invasive local field potential (LFP) recordings, cell activity measurements, GABAergic and glutamatergic network analysis, locomotor tracking; treatment with rapamycin or ethosuximide at 3 dpf (72 hpf).
(C) Treatment scheme for the neuronal migration experiments with the Tg(HuC:GCaMP5G) transgenic line. Migration of HuC positive cells from the subventricular zone (SVZ) toward outer layers was analyzed at 24 hpf.
(D) Schematic frontal view of the zebrafish front brain at 24 hpf. Green, HuC expressing cells. Magenta arrows, direction of migration from the SVZ to outer layers. V, ventricle; OP, olfactory placodes.
(E) LFPs from larval pallia. Representative 10 min recordings of non-invasive LFPs from g3bp1 MO or control MO injected zebrafish larvae. Treatment as indicated in (B). Magnification of a polyspiking event is shown for each condition. n > 34 larvae per condition.
(F) LFPs from larval optic tecta. Representative 10 min recordings of non-invasive LFPs from g3bp1 MO or control MO injected zebrafish larvae. Treatment as indicated in (B). Magnification of a polyspiking event is shown for each condition. n > 20 larvae per condition.
Red, zebrafish pallium; blue, WM compartments; yellow, active single cells in WM compartments.
Red, zebrafish pallium; blue, WM compartments; yellow, active single cells in WM compartments.
In summary, in vivo G3bp1 inhibition phenocopies the mTORC1-dependent effects of Tsc2 loss on brain function (Kedra et al., 2020; Scheldeman et al., 2017), highlighting the importance of this mechanism for nervous system development and function.
Discussion
G3BP1 was originally identified as a RasGAP-binding protein (Gallouzi et al., 1998; Kennedy et al., 2001; Parker et al., 1996). A role in the RAS pathway was proposed but later questioned (Annibaldi et al., 2011). We demonstrate that G3BP1’s assignment as a GAP-binding protein was correct, although for a different GAP, because it exerts this role by binding TSC2. It may be rewarding to revisit whether G3BPs bind to other RAS-related GAPs. In the insulin-mTORC1 axis, G3BP1 exerts its suppressor function through the TSC complex, but other GAPs may mediate the G3BPs’ roles in RAS (Parker et al., 1996), NFKB1 (Prigent et al., 2000), WNT (Bikkavilli and Malbon, 2011), and TGFB (Zhang et al., 2015a) signaling. Yet, these pathways crosstalk with the TSC complex (Ghosh et al., 2006; Inoki et al., 2006; Ma et al., 2005; Thien et al., 2015), which may also underlie a common role of the G3BPs in them.
We have shown earlier that, in the presence of G3BP1-containing SGs, the sperm associated antigen 5 (SPAG5)-RPTOR complex decreases mTORC1 activity (Thedieck et al., 2013). Here we report that, in the absence of SGs, G3BP1 tethers the TSC complex to lysosomes. Why does G3BP1 inhibit mTORC1 upon metabolic starvation and sufficiency but not upon SG formation? Upon stress, activating (Heberle et al., 2019; Sfakianos et al., 2018; Wang and Proud, 1997; White et al., 2007; Wu et al., 2011) and inhibitory (Thedieck et al., 2013; Wippich et al., 2013) cues balance mTORC1 activity. Although it is tempting to speculate that G3BP1, as a SG nucleator, contributes to SG-mediated mTORC1 inhibition (Thedieck et al., 2013; Wippich et al., 2013), previous studies (Bley et al., 2015; Kedersha et al., 2016; Matsuki et al., 2013) and our own results (Figures S2Q and S2R) show that G3BP1 inhibition alone does not prevent SG formation. SG-inducing agents enhance TSC2 degradation (Heberle et al., 2019; Thedieck et al., 2013). Without TSC2, G3BP1 cannot bind to the TSC complex (Figure 5A) and, thus, cannot inhibit mTORC1. To conclude, upon SG formation, the TSC complex is reduced, and SG are not affected by G3BP1 deficiency; thus, neither mechanism can affect mTORC1 in a G3BP1-dependent manner.
Can the lysosomal localization of G3BPs be reconciled with functions in SGs and other subcellular compartments? SGs hitchhike on lysosomes (Liao et al., 2019), which may enable G3BPs to switch between their SG and lysosomal functions. G3BP1’s ubiquitous cytoplasmic distribution (Figure S2Q; Irvine et al., 2004) is reminiscent of the IF patterns for the TSC complex (Carroll et al., 2016; Demetriades et al., 2014) and MTOR (Betz and Hall, 2013), which also localize to multiple subcellular sites (Betz and Hall, 2013; Zhang et al., 2013). Thus, G3BPs may be relevant for TSC complex and mTORC1 function beyond lysosomes.
What can we conclude regarding the relevance of G3BP1 in cancer and neuronal disease? Its dual roles in oncogenic mTORC1 signaling and SG formation argue against G3BP1 as an anti-tumor drug target, as proposed by others (Alam and Kennedy, 2019; Anisimov et al., 2019; Zhang et al., 2012, 2019). G3BP1 inhibition is not sufficient to inhibit SG (Figures S2Q and S2R; Kedersha et al., 2016) but results in mTORC1 hyperactivation, known to drive cancer cell growth and motility (Condon and Sabatini, 2019; Crino, 2016; LiCausi and Hartman, 2018; Tee et al., 2016). G3BP1 may, however, be a promising marker associated with mTORC1 hyperactivity, which correlates with tumor sensitivity to mTORC1 inhibitors (Grabiner et al., 2014; Kwiatkowski and Wagle, 2014; Meric-Bernstam et al., 2012; Wagle et al., 2014). Whether disturbed function of G3BP1/2 in the TSC-mTORC1 axis contributes to the etiology of neuronal diseases also deserves evaluation. We scrutinized Genomics England (GEL) 100,000 Genomes Project data for mutations in G3BP1/2. 100 individuals had a clinical diagnosis of TSC disease with no pathogenic TSC1 or TSC2 variant, which is the most frequent cause of the disease (Borkowska et al., 2011; Curatolo et al., 2008; Jozwiak et al., 2020; Marcotte and Crino, 2006; Orlova and Crino, 2010). However, none of the 100 individuals showed likely pathogenic changes in G3BP1/2. Extending the analysis to all variants at the G3BP1 or G3BP2 loci in the GEL rare disease data (64,185 whole-genome sequences) identified nine individuals with unexplained, mostly neurological phenotypes and heterozygous variants in G3BP1 or G3BP2 strongly predicted to alter protein function (Table S2). A further G3BP1 loss-of-function variant was noted in the Epi4K cohort of individuals with epilepsy (Table S1 in Appenzeller et al., 2014). Six of the variants were apparently unique, being absent from gnomAD (https://gnomad.broadinstitute.org/), and four were present at extremely low allele frequencies. We conclude that G3BP1 and G3BP2 are unlikely to represent further genes determining the TSC disease phenotype. The numbers of observations in other neurological diseases were too small to statistically confirm or refute associations that will need to be addressed in larger and more specific cohorts. Interestingly, certain mutations of the TSC2 GAP domain that result in only partial loss of function and mutations of TBC1D7 also lead to neurological phenotypes that are clinically distinct to definite TSC disease (Alfaiz et al., 2014; Capo-Chichi et al., 2013; Hansmann et al., 2020). Mutations of these and further genes resulting in mTORC1 hyperactivation are linked with neuronal phenotypes, collectively referred to as “mTORopathies” (Crino, 2015; Wong and Crino, 2012). Future studies will shed light on whether G3BP1 and G3BP2 belong to this family. We advocate in-depth evaluation of the etiological and therapeutic relevance of G3BPs to cancer and neuronal disorders.
Another important question concerns the role of the G3BPs in the lysosomal dissociation of the TSC complex in response to insulin (Menon et al., 2014) or amino acids (Carroll et al., 2016; Demetriades et al., 2014). G3BP1 deficiency hyperactivates mTORC1 upon amino acids and insulin as well as insulin alone. It will be intriguing to explore whether posttranslational modifications in TSC2 (Huang and Manning, 2008) or G3BPs (Alam and Kennedy, 2019) differentially control their binding and regulate lysosomal TSC complex localization in response to different agonists.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CALR | Cell Signaling | Cat# 12238; RRID: AB_2688013 |
| CANX | Cell Signaling | Cat# 2679; RRID: AB_2228381 |
| CTSD | Cell Signaling | Cat# 2284; RRID: AB_10694258 |
| EEA1 | Cell Signaling | Cat# 3288; RRID: AB_2096811 |
| EIF2S1 | Cell Signaling | Cat# 9722; RRID: AB_2230924 |
| EIF2S1-pS51 | Cell Signaling | Cat# 9721; RRID: AB_330951 |
| EIF3A | Cell Signaling | Cat# 3411; RRID: AB_2096523 |
| FLAG | Sigma-Aldrich | Cat# F3165; RRID: AB_259529 |
| G3BP1 | Santa Cruz | Cat# sc-365338; RRID: AB_10846950 |
| G3BP1 | Santa Cruz | Cat# sc-81940; RRID: AB_1123055 |
| G3BP2 | Bethyl | Cat# A302-040A; RRID: AB_1576545 |
| GAPDH | Abcam | Cat# ab37187; RRID: AB_732651 |
| GAPDH (zebrafish) | Sigma-Aldrich | Cat# SAB2701826 |
| GFP | Roche | Cat# 11814460001; RRID: AB_390913 |
| Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Invitrogen | Cat# A-11001; RRID: AB_2534069 |
| Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Invitrogen | Cat# A-11011; RRID: AB_143157 |
| Goat anti-Mouse IgG (H+L) cross-adsorbed secondary, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21422; RRID: AB_2535844 |
| Goat anti-Rabbit IgG (H+L) cross-adsorbed secondary, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11008; RRID: AB_143165 |
| Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP-coupled | Thermo Fisher Scientific | Cat# 31430; RRID: AB_228307 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP-coupled | Thermo Fisher Scientific | Cat# 31460; RRID: AB_228341 |
| Goat anti-Rabbit IgG (H+L) Secondary Antibody, Dylight 800 (zebrafish) | Thermo Fisher Scientific | Cat# SA5-35571; RRID: AB_2556775 |
| Goat anti-Rat IgG (H+L) Secondary Antibody, HRP-coupled | Thermo Fisher Scientific | Cat# 31470; RRID: AB_228356 |
| GOLGA1 | Cell Signaling | Cat# 13192; RRID: AB_2798144 |
| GOLGA2 | Cell Signaling | Cat# 12480; RRID: AB_2797933 |
| HA | Roche | Cat# 11867423001; RRID: AB_390918 |
| Histone H3 (H3C1) | Bethyl | Cat# A300-822A; RRID: AB_597872 |
| HSP60 (HSPD1) | Cell Signaling | Cat# 12165; RRID: AB_2636980 |
| HSP90 (CDC37) | Cell Signaling | Cat# 4877; RRID: AB_2233307 |
| LMNA A/C | Cell Signaling | Cat# 2032; RRID: AB_2136278 |
| LAMP1 | Cell Signaling | Cat# 9091; RRID: AB_2687579 |
| LAMP1 | Developmental Studies Hybridoma Bank | Cat# H4A3; RRID: AB_2296838 |
| LAMP2 | Cell Signaling | Cat# 49067;RRID: AB_2799349 |
| LAMP2 | Santa Cruz | Cat# sc-18822; RRID: AB_626858 |
| LAMP2 | Developmental Studies Hybridoma Bank | Cat# H4B4; RRID: AB_2134755 |
| MTOR | Cell Signaling | Cat# 2983; RRID: AB_2105622 |
| MTOR epitope maps to residues 221 and 261 of human mTOR | Monoclonal Antibody Core Unit. Helmholtz Center Munich, Germany |
TQREP-3G6 |
| Mock antibody mouse | Santa Cruz | Cat# sc-2025; RRID: AB_737182 |
| Mock antibody rabbit | Bethyl | Cat# P120-101; RRID: AB_479829 |
| Sigma-Aldrich | Cat# I5006; RRID: AB_1163659 | |
| Mock antibody rat | Monoclonal Antibody Core Unit. Helmholtz Center Munich, Germany |
RmC3-7H8 |
| Mouse IgG HRP Linked Whole Ab | Merck | Cat# GENA931 |
| MYC-tag | Cell Signaling | Cat# 2276; RRID: AB_331783 |
| Rabbit IgG HRP Linked Whole Ab | Merck | Cat# GENA934; RRID: AB_2722659 |
| RPS6KB1 | Cell Signaling | Cat# 2708; RRID: AB_390722 |
| RPS6KB1-pT389 | Cell Signaling | Cat# 9206; RRID: AB_2285392 |
| RPS6KB1-pT389 | Cell Signaling | Cat# 9205; RRID: AB_330944 |
| RAB5A | Cell Signaling | Cat# 3547; RRID: AB_2300649 |
| RAB7A | Cell Signaling | Cat# 9367; RRID: AB_1904103 |
| RPTOR | Cell Signaling | Cat# 2280; RRID: AB_561245 |
| RPTOR #1 epitope maps to residues 686 and 704 of human Raptor | Monoclonal Antibody Core Unit. Helmholtz Center Munich, Germany |
RAP1-20C4 |
| RPTOR #2 | Bethyl | Cat# A300-553A; RRID: AB_2130793 |
| RPS6 | Cell Signaling | Cat# 2317; RRID: AB_2238583 |
| RPS6-pS235/236 | Cell Signaling | Cat# 4856; RRID: AB_2181037 |
| RPS6-pS235/236 (zebrafish) | Cell Signaling | Cat# 2211; RRID: AB_331679 |
| RPS6-pS235/236 (zebrafish) | Cell Signaling | Cat# 4858; RRID: AB_916156 |
| TSC1 | Cell Signaling | Cat# 4906; RRID: AB_2209790 |
| TSC1 #1 | Gift from Michael N. Hall, Basel, Switzerland (Molle, 2006). Generated according to van Slegtenhorst et al. (1998). | N/A |
| TSC1 #2 | Thermo Fisher Scientific (Invitrogen) | Cat# 37-0400; RRID: AB_2533292 |
| TSC1 #3 | Cell Signaling | Cat# 6935; RRID: AB_10860420 |
| TSC2 | Cell Signaling | Cat# 4308; RRID: AB_10547134 |
| TSC2 #1 | Thermo Fisher Scientific (Invitrogen) | Cat# 37-0500; RRID: AB_2533293 |
| TSC2 #2 epitope maps to residues 1535 and 1784 of human TSC2 | Gift from Michael N. Hall, Basel, Switzerland (Molle, 2006). Generated according to van Slegtenhorst et al. (1998). | N/A |
| TSC2 #3 | Abcam | Cat# ab52936; RRID: AB_883283 |
| TUBA1B | Abcam | Cat# ab108629; RRID: AB_10866252 |
| VDAC | Cell Signaling | Cat# 4661; RRID: AB_10557420 |
| For antibody dilutions, see Table S3 | ||
| Bacterial and Virus Strains | ||
| DB3.1 | Thermo Fisher Scientific | Cat# 11782018 (discontinued) |
| DH5-alpha | New England Biolabs | Cat# C2987H |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AEBSF (4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride) | Sigma-Aldrich | Cat# A8456 |
| Aprotinin | Sigma-Aldrich | Cat# A1153 |
| Benzamidine | Sigma-Aldrich | Cat# B6506 |
| Beta-Mercaptoethanol | GIBCO | Cat# 21-985-023 |
| Bromophenol Blue | Sigma-Aldrich | Cat# B5525 |
| BSA (bovine serum albumin) | Carl Roth | Cat# 8076.5 |
| Calcein AM Fluorescent Dye | Corning | Cat# 354216 |
| CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate) | Sigma-Aldrich | Cat# 3023 |
| CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate) (for IPs in rat brain tissue) |
Roth | Cat# 1479.3 |
| Complete Protease Inhibitor Cocktail | Sigma-Aldrich | Cat# D27802 |
| DABCO (1,4-diazabicyclo[2.2.2]octane) | Merck | Cat# 11836153001 |
| DexoMAG® | Liquids Research | N/A |
| DMEM (Dulbecco’s Modified Eagle’s Medium) w: 4.5 g/L Glucose, w/o: L-Glutamine, w: Sodium pyruvate, w: 3.7 g/L NaHCO3 | PAN | Cat# P04-03600 |
| DMEM (Dulbecco’s Modified Eagle’s Medium) used for PLA experiments in Figure 4A | Thermo Fisher Scientific | Cat# 41965-039 |
| DMEM (Dulbecco’s Modified Eagle’s Medium) used for lysosomal isolations and trypsin digests | Thermo Fisher Scientific | Cat# 41965-062 |
| DMSO (dimethyl sulfoxide) | Sigma-Aldrich | Cat# D2650 |
| Doxycycline | Sigma-Aldrich | Cat# D3447 |
| Duolink In Situ Mounting Medium with DAPI | Sigma-Aldrich | Cat# DUO82040 |
| Dynabeads Protein G for Immunoprecipitation | Thermo Fisher Scientific | Cat# 10009D |
| E64 | Sigma-Aldrich | Cat# E3132 |
| Ethosuximide | Sigma-Aldrich | Cat# 1001096708 |
| FBS (fetal bovine serum) | GIBCO | Cat# 10270106 |
| FBS (fetal bovine serum) | Sigma-Aldrich | Cat# F9665 |
| Glycerol | Sigma-Aldrich | Cat# G5516 |
| Glycine | Sigma-Aldrich | Cat# G7126 |
| HEPES ((4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) | Life technologies | Cat# 15630080 |
| HEPES ((4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) | Life technologies | Cat# 15630106 |
| HBSS (Hank’s Balanced Salt Solution) w/o: Phenol red, w: Ca and Mg, w: 0.35 g/L NaHCO3 | PAN | Cat# P04-32505 |
| Hoechst 33342 (dilution in IF: 1:100,000) | Invitrogen | Cat# H3570 |
| Insulin | Sigma-Aldrich | Cat# I1882 |
| IGEPAL CA-630 (NP40) | Sigma-Aldrich | Cat# I8896 |
| Imidazole | Sigma-Aldrich | Cat# I0250 |
| KCl (potassium chloride) | Sigma-Aldrich | Cat# P9541 |
| Leupeptin | Sigma-Aldrich | Cat# 103476-89-7 |
| L-glutamine | GIBCO | Cat# 25030024 |
| L-glutamine | GIBCO | Cat# 25030081 |
| Methanol | Klinipath | Cat# 4063-9005 |
| MgCl2 (magnesium chloride) | Sigma-Aldrich | Cat# M2670 |
| MK2206 | Axon Medchem | Cat# 1684 |
| Mowiol 4-88 | Carl Roth | Cat# 07131 |
| NaCl (sodium chloride) | Sigma-Aldrich | Cat# S7653 |
| NPG (n-propyl-gallate) | VWR | Cat# EM8.20599.0500 |
| PBS (phosphate-buffered saline) | PAN | Cat# P04-36500 |
| PBS (phosphate-buffered saline) for non-sterile washing | Biochrom | Cat# L182-50 |
| Penicillin/Streptomycin | GIBCO | Cat# 15140122 |
| Pepstatin A | Sigma-Aldrich | Cat# 26305-03-3 |
| PhosSTOP for IPs in brain tissue | Merck | Cat# 4906837001 |
| Phosphatase Inhibitor Cocktail 2 | Sigma-Aldrich | Cat# P5726 |
| Phosphatase Inhibitor Cocktail 3 | Sigma-Aldrich | Cat# P0044 |
| Phusion HF DNA Polymerase | Roche | Cat# M0530S |
| Pierce 16% Formaldehyde (w/v), Methanol-free | Thermo Fisher Scientific | Cat# 28908 |
| Polyacrylamide | Bio-Rad | Cat# 161-0159 |
| Polybrene | Sigma-Aldrich | Cat# H9268 |
| Prolong Gold antifade reagent with 4′,6-Diamidin-2-phenylindol (DAPI) | Thermo Fisher Scientific | Cat# P36935 |
| Protein G Sepharose beads | GE | Cat# 17061801 |
| Puromycin | Sigma-Aldrich | Cat# P8833 |
| PVDF (polyvinylidene difluoride) membrane | Millipore | Cat# IPVH00010 |
| Rapamycin | Calbiochem | Cat# 553210 |
| SDS (sodium dodecyl sulfate) | Sigma-Aldrich | Cat# 71725 |
| Sodium deoxycholate | Sigma-Aldrich | Cat# 30970 |
| Sodium fluoride | Sigma-Aldrich | Cat# 7681-49-4 |
| Sodium glycerophosphate | Sigma-Aldrich | Cat# G9322 |
| Sodium orthovanadate | Sigma-Aldrich | Cat# 13721-39-6 |
| Sodium pyrophosphate | Sigma-Aldrich | Cat# 13472-36-1 |
| Sucrose | Sigma-Aldrich | Cat# S2395 |
| T4 DNA Ligase | New England Biolabs | Cat# M0202S |
| Torin1 | Axon Medchem | Cat# 1833 |
| Transfectin | Biorad | Cat# 1703350 |
| Tricaine (MS-222) | Sigma-Aldrich | Cat# E10521 |
| TRIS base (tris(hydroxymethyl)aminomethane) | VWR | Cat# A1086.5000 |
| Triton X-100 | Sigma-Aldrich | Cat# 93443 |
| Trypan Blue | GIBCO | Cat# 15250061 |
| Trypsin | GIBCO | Cat# 15400054 |
| Tween-20 | MP Biomedicals | Cat# 11TWEEN201 |
| Critical Commercial Assays | ||
| Bio-Rad Protein Assay Dye Reagent Concentrate | Bio-Rad | Cat# 500-0006 |
| Duolink In Situ Red Starter Kit Mouse/Rabbit | Sigma-Aldrich | Cat# DUO92008 |
| Duolink® In Situ PLA® Probe Anti-Rabbit PLUS Affinity purified Donkey anti-Rabbit IgG (H+L) | Sigma-Aldrich | Cat# DUO92002; RRID: AB281940 |
| Duolink® In Situ PLA® Probe Anti-Mouse MINUS Affinity purified Donkey anti-Mouse IgG (H+L) | Sigma-Aldrich | Cat# DUO92004; RRID: AB_2713942 |
| JetPEI | Poly-Plus | Cat# 101-40N |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# L3000015 |
| Lipofectamine RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat# 13778150 |
| MidiPrepKit NUCLEOBOND XTRA MIDI | Macherey-Nagel | Cat# 740410.50 |
| NuPage MES SDS running buffer | Thermo Fisher Scientific | Cat# NP0002 |
| NuPage Novex 10% Bis-Tris gel | Thermo Fisher Scientific | Cat# NP0302BOX |
| Odyssey blocking buffer | Li-Cor | Cat# 927-40000 |
| Pierce BCA protein assay kit | Thermo Fisher Scientific | Cat# 23225 |
| Pierce ECL Western Blotting Substrate | Thermo Fisher Scientific | Cat# 32209 |
| RIPA buffer (zebrafish lysis) | Merck | Cat# R0278 |
| SuperSignal West FEMTO Maximum Sensitivity Substrate | Thermo Fisher Scientific | Cat# 34095 |
| Trans-Lentiviral shRNA Packaging Mix | Dharmacon | Cat# TLP5912 |
| Deposited Data | ||
| Invasive breast cancer (The Cancer Genome Atlas, TCGA, provisional) | http://www.cbioportal.org | RRID: SCR_014555 |
| TSC1 RNA expression data | http://www.kmplot.com | probeID: 209390_at |
| TSC2 RNA expression data | http://www.kmplot.com | probeID: 215735_s_at |
| G3BP1 RNA expression data | http://www.kmplot.com | probeID: 225007_at |
| G3BP1 protein expression data | http://www.kmplot.com | probeID: Q13283 |
| Experimental Models: Cell Lines | ||
| HEK293T | Thien et al. (2015) | N/A |
| HEK293T Control | This paper | N/A |
| HEK293T G3BP1 KO | This paper | N/A |
| HEK293-β2AR | Lavoie et al. (2002) | N/A |
| HeLa alpha Kyoto | Thedieck et al. (2007) | N/A |
| HeLa S3 | ATCC | Cat# CCL-2.2; RRID: CVCL_0058 |
| MCF-7 ACC115 | DSMZ | Cat# ACC115; RRID: CVCL_0031 |
| MCF-7 Control (for G3BP1) | This paper | N/A |
| MCF-7 Control (for TSC2) | This paper | N/A |
| MCF-7 G3BP1 KO | This paper | N/A |
| MCF-7 GFP-LC3 | Gift from Joern Dengjel, Fribourg, Switzerland | N/A |
| MCF-7 shControl | This paper | N/A |
| MCF-7 shG3BP1 #1 | This paper | N/A |
| MCF-7 shG3BP1 #2 | This paper | N/A |
| MCF-7 TSC2 KO | This paper | N/A |
| MDA-MB-231 | ATCC | Cat# HTB-26; RRID: CVCL_0062 |
| MDA-MB-231 TSC Control | This paper | N/A |
| MDA-MB-231 TSC2 KO | This paper | N/A |
| MDA-MB-231 shControl | This paper | N/A |
| MDA-MB-231 shG3BP1 #1 | This paper | N/A |
| MDA-MB-231 shG3BP1 #2 | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| AB Danio rerio | Zebrafish International Resource Center | Cat# ZL1; RRID: ZIRC_ZL1 |
| Tg(dlx5a/dlx6a-EGFP) x Tg(vglut2a:loxP-RFP-loxP-GFP) |
Noble et al. (2015)Tg(dlx5a/dlx6a-EGFP); Satou et al. (2012)Tg(vglut2a:loxP-RFP-loxP-GFP) |
N/A |
| Tg(HuC:GCaMP5G) | Ahrens et al. (2013) | N/A |
| Wistar Cmd:(WI)WU rats | Mossakowski Medical Research Centre Polish Academy of Sciences | N/A |
| Oligonucleotides | ||
| control MO: 5′-CCTCTTACCTCAGTTACAATTTATA-3′ | GeneTools | N/A |
| g3bp1 MO: 5′-TAACAAAGGGCAAGTCACCTGTGCA-3′ | GeneTools | N/A |
| tsc2 MO: 5′-CTGTGTGTTTTCTTACTGGACCATA-3′ | GeneTools | N/A |
| For primers, see Table S4 | N/A | |
| For RNA, see Table S5 | N/A | |
| Recombinant DNA | ||
| bFos-MYC-LC151 | Gift from Qingming Luo, Wuhan, China (Chu et al., 2009) | N/A |
| bJun-HA-LN151 | Gift from Qingming Luo, Wuhan, China (Chu et al., 2009) | N/A |
| lentiGuide-Puro | Sanjana et al. (2014) | RRID: Addgene_52963 |
| pCMV6-AN-MYC-DDK (MYC-FLAG-empty) | Origene | Cat# PS100016 |
| MYC-FLAG-G3BP1 | This paper | N/A |
| MYC-FLAG-G3BP1-ΔRGG (aa 410-466) | This paper | N/A |
| MYC-FLAG-G3BP1-ΔNTF2L (aa 1-139) | This paper | N/A |
| MYC-FLAG-G3BP1-NTF2L 1-139 | This paper | N/A |
| MYC-FLAG-G3BP2 | This paper | N/A |
| pCW-Cas9-Blast | Sanjana et al. (2014) | RRID: Addgene_83481 |
| pGW-MYC-LC151 | Stefan Pusch (Weiler et al., 2014) | N/A |
| pGW-HA-LN151 | Stefan Pusch (Weiler et al., 2014) | N/A |
| pGW-HA-LN151-G3BP1 | This paper | N/A |
| pGW-MYC-LC151-G3BP1 | This paper | N/A |
| pGW-MYC-LC151-G3BP1 1-182 | This paper | N/A |
| pGW-MYC-LC151-G3BP1 183-332 | This paper | N/A |
| pGW-MYC-LC151-G3BP1 333-466 | This paper | N/A |
| pGW-HA-LN151-G3BP2 | This paper | N/A |
| pGW-MYC-LC151-G3BP2 | This paper | N/A |
| pGW-HA-LN151-LAMP1 | This paper | N/A |
| pGW-MYC-LC151-LAMP1 | This paper | N/A |
| pGW-MYC-LC151-LAMP1 1-382 | This paper | N/A |
| pGW-MYC-LC151-LAMP1 383-417 | This paper | N/A |
| pGW-HA-LN151-LAMP2 | This paper | N/A |
| pGW-HA-LN151-mTOR | This paper | N/A |
| pGW-HA-LN151-TSC2 | This paper | N/A |
| pEGFP-C-LAMP2 | This paper | N/A |
| pEGFP-C-TSC2 | This paper | N/A |
| pEGFP-C (derivate of pDEST with a C-terminal EGFP tag) | Stefan Pusch | N/A |
| pDEST | Stefan Pusch; Clone repository of the DKFZ Genomics and Proteomics Core Facility (GPCF) |
N/A |
| pDONR201-tGFP | This paper | N/A |
| pDONR201-tGFP-LAMP1 383-417 | This paper | N/A |
| pENTR221-G3BP1 | Clone repository of the DKFZ Genomics and Proteomics Core Facility (GPCF) |
CloneId: 182373397 |
| pENTR223-G3BP2 | Clone repository of the DKFZ Genomics and Proteomics Core Facility (GPCF) |
CloneId: 192451551 |
| pENTR221-LAMP1 | Clone repository of the DKFZ Genomics and Proteomics Core Facility (GPCF) |
CloneId: 193137117 |
| pENTR221-LAMP2 | Clone repository of the DKFZ Genomics and Proteomics Core Facility (GPCF) |
CloneId: 115072391 |
| pSpCas9(BB)-2A-Puro (PX459) V2.0 | Addgene | RRID: Addgene_62988 |
| psPAX2 | Shalem et al. (2014) | RRID: Addgene_12260 |
| pMD2.G | Shalem et al. (2014) | RRID: Addgene_12259 |
| R777-E138 Hs.MTOR-nostop | Gift from Dominic Esposito, Addgene | Cat# 70422; RRID: Addgene_70422 |
| R777-E356 Hs.TSC2-nostop | Gift from Dominic Esposito, Addgene | Cat# 70640; RRID: Addgene_70640 |
| Software and Algorithms | ||
| Adobe Photoshop version CS5.1 | Adobe Systems Incorporated | RRID: SCR_014199;https://www.adobe.com/de/products/photoshop.html# |
| Bash script to identify the single nucleotide variants (SNV) within G3BP1 and G3BP2 | This paper | Document S3 |
| Cell Profiler version 3.1.5 | McQuin et al. (2018) | RRID: SCR_007358; https://www.cellprofiler.org/ |
| CGDS-R package version 1.2.6 | Gao et al. (2013) | https://github.com/cBioPortal/cgdsr |
| Dell Statistica version 13 | Dell Inc. | https://statistica.software.informer.com/13.0/ |
| EMBOSS Needle (Protein sequence analysis) | Madeira et al. (2019) | https://www.ebi.ac.uk/Tools/psa/emboss_needle/ |
| Fiji version 1.49v, 1.52p, 1.53c | Schindelin et al. (2012) | RRID: SCR_002285; https://imagej.net/Fiji.html#Downloads |
| GraphPad Prism version 7.04 and 8.0 | GraphPad Software | RRID: SCR_002798; https://www.graphpad.com/scientific-software/prism/ |
| ImageJ version 1.50b, 1.51r | Schneider et al. (2012) | RRID: SCR_003070; https://imagej.nih.gov/ij/ |
| Image Lab version 5.2.1 and 6.0.1 | Bio-Rad | RRID: SCR_014210; http://www.bio-rad.com/en-us/product/image-lab-software?ID=KRE6P5E8Z |
| ImageQuant TL version 8.1 | Cytiva | RRID: SCR_014246; https://us.vwr.com/store/product/25990402/imagequant-tl-8-1-cytiva-formerly-ge-healthcare-life-sciences |
| Image Studio Lite Version 5.2 | Li-Cor | RRID: SCR_013715; https://www.licor.com/bio/image-studio-lite/download |
| Imaris 9.1 | Bitplane AG | RRID: SCR_00730; https://imaris.oxinst.com/packages |
| NIS Elements version 4.13.04 | Nikon | RRID: SCR_014329; https://www.microscope.healthcare.nikon.com/de_EU/products/software/nis-elements |
| Phylogenetic analysis | Bockwoldt et al. (2019) | https://github.com/MolecularBioinformatics/Phylogenetic-analysis |
| RStudio | RStudio | RRID: SCR_000432; https://rstudio.com/products/rstudio/download/ |
| Tecan i-control software version 1.10.4.0 | Tecan | https://lifesciences.tecan.de/plate_readers/infinite_200_pro?p=tab-3 |
| TrackMate | Tinevez et al. (2017) | https://github.com/fiji/TrackMate/releases |
| TScratch | Gebäck et al. (2009) | RRID: SCR_014282; https://github.com/cselab/TScratch |
| ZEN2012 blue edition | Zeiss | https://www.zeiss.de/mikroskopie/produkte/mikroskopsoftware/zen.html#downloads |
| Other | ||
| 24 well plates | TPP | Cat# 92424 |
| 6 cm cell culture dish | Greiner bio-one | Cat# 628160 |
| 10 cm cell culture dish | TPP | Cat# 93100 |
| 15 cm cell culture dish | TPP | Cat# 93150 |
| 70 Ti Rotor for ultracentrifuge | Beckman Coulter | Cat# 337922 |
| AxioObserver Z1 | Zeiss | N/A |
| Beckman Optima L-70K Ultracentrifuge | Beckman Coulter | Cat# 8043-30-1187 |
| ChemiDoc XRS+ | Bio Rad | Cat# 1708265 |
| Cover Glass | VWR international | Cat# 631-0130 |
| E-plate 16 for RTCA | ACEA Biosciences, Inc. | Cat# 05469813001 |
| Falcon® 24-well TC-treated Cell Polystyrene Permeable Support Companion Plate, with Lid | Corning | Cat# 353504 |
| FluoroBlok 24-well Plate Permeable Support with 8.0 μm Colored PET Membrane | Corning | Cat# 351152 |
| FUSION FX7 with the DarQ-9 camera | Vilber | N/A |
| ibidi culture-insert 2 well | ibidi | Cat# 80209 |
| iBlot gel transfer stacks nitrocellulose membrane | Thermo Fisher Scientific | Cat# IB301002 |
| LAS-4000 mini camera system | GE Healthcare | N/A |
| LAS-4000 camera system | GE Healthcare | N/A |
| Lightsheet Z.1 | Zeiss | N/A |
| LS Column | Miltenyi Biotec | Cat# 130-042-401 |
| LSM 780 confocal microscope | Zeiss | N/A |
| Microscope slides | Thermo Fisher Scientific | Cat# 4951PLUS4 |
| Mini-PROTEAN® Tetra Vertical Electrophoresis Cell system | Bio Rad | Cat# 1658029FC |
| Nikon ECLIPSE Ti-E/B | Nikon | N/A |
| Odyssey 2.1 imaging system | Li-Cor, USA | N/A |
| QuadroMACS Separator | Miltenyi Biotec | Cat# 130-090-976 |
| RTCA Control Unit with RTCA Software | ACEA Biosciences, Inc. | Cat# 05454417001 |
| RTCA DP Analyzer | ACEA Biosciences, Inc. | Cat# 05469759001 |
| Tecan Infinite 200 PRO plate reader | Tecan | N/A |
| ZebraboxTM | ViewPoint | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead contact, Kathrin Thedieck (kathrin.thedieck@uibk.ac.at).
Materials availability
All unique materials and reagents generated in this study are available from the Lead contact with a completed material transfer agreement.
Data and code availability
The custom Python scripts for the phylogenetic analysis are accessible via https://github.com/MolecularBioinformatics/Phylogenetic-analysis and were manually curated as described earlier (Bockwoldt et al., 2019).
The bash script to identify the single nucleotide variants (SNV) within G3BP1 and G3BP2 is provided in Document S3. The script was adapted from the ‘extract_variants_by_coordinate.sh’ script for germline variants (https://research-help.genomicsengland.co.uk/display/GERE/Extract+variants+by+coordinate) and was run on the command line within the Genomics England Research environment using bcftools (https://samtools.github.io/bcftools/).
Experimental model and subject details
Cell culture
Experiments were performed in HeLa alpha Kyoto cells (Thedieck et al., 2007), MCF-7 cells (ACC115), MCF-7 cells expressing GFP-LC3 (MCF-7-LC3), MDA-MB-231, HEK293T, and HEK293-β2AR cells. All cells, except for HEK293-β2AR (Lavoie et al., 2002), were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) with 4.5 g / L glucose, supplemented with 10% fetal bovine serum (FBS) and 3 mM L-glutamine (termed full medium) if not indicated otherwise. HEK293-β2AR were cultured in DMEM with 4.5 g / L glucose and 0.584 mM L-glutamine, supplemented with 10% FBS and 1% penicillin and streptomycin. All cell lines were maintained at 37°C in a 5% CO2 incubator and regularly tested for mycoplasma infection.
RNA knockdown experiments
siRNA knockdown of G3BP1, G3BP2 and RHEB was induced for two days using ON-TARGET plus SMARTpool siRNA at a final concentration of 40 nM. siRNA knockdown of TSC2 was induced for two days using ON-TARGET plus SMARTpool siRNA at a final concentration of 5 nM. As a negative control, a non-targeting scrambled siRNA pool (siControl) was used at the same concentration. siRNA transfection was performed using Lipofectamine 3000 or RNAiMAX transfection reagents according to the manufacturer’s protocols. The medium containing the transfection mix was replaced 6 hours after transfection. For PLA analysis in Figure 4A, siRNA knockdown of G3BP1 was induced for five days using siGENOME SMARTpool siRNA at a final concentration of 15 nM. Here siRNA against Renilla luciferase (Control) was used as a control.
Doxycyclin-inducible shRNA knockdown cell lines for G3BP1 were generated using the pTRIPZ system with the Trans-Lentiviral shRNA Packaging Mix (Horizon Discovery). Viral particles were produced using shRNA constructs targeting G3BP1 (shG3BP1 #1 or shG3BP1 #2) or a non-targeting scrambled control sequence (shControl) according to the manufacturer’s protocol. MCF-7-LC3 and MDA-MB-231 cells were transduced in three rounds. The cells were incubated with the viral supernatant containing 8 μg / mL polybrene for 16 hours, followed by 6 hours of fresh full medium. Antibiotic selection was carried out 48 hours post-transduction with 2 μg / mL puromycin for 7 days. Expression of the shRNA was induced with 2 μg / mL doxycycline for 4 days. Monoclonal cell populations were obtained by limiting dilutions. Knockdown efficiency was tested at protein level by immunoblotting.
Knockout cell lines
CRISPR/Cas9 knockout MCF-7 and MDA-MB-231 cell lines for G3BP1 and TSC2 were generated using a two-vector system as previously described (Sanjana et al., 2014). First, doxycyclin-inducible Cas9 expressing MDA-MB-231 and MCF-7 cell lines were generated by lentiviral transduction using the pCW-Cas9-Blast vector (Addgene plasmid # 83481) and thereafter selected with 5 μg / mL blasticidin for 48 hours, as described previously (Shalem et al., 2014). Next, the Cas9 expressing cells were transduced with the lentiGuide-Puro vector (Addgene plasmid # 52963) containing either no sgRNA (control), or sgRNA (sgRNA #1) targeting G3BP1 (G3BP1 KO) or TSC2 (TSC2 KO). These cells were selected with 2 μg / mL puromycin for 48 hours. Monoclonal cell populations were obtained by limiting dilutions. Cas9 expression was induced with 2 μg / mL doxycycline for 48 hours. Knockout efficiency was tested at protein level by immunoblotting.
Using a second independent sgRNA (sgRNA #2) G3BP1 was knocked out by CRISPR/Cas9 in HEK293T cells (Figures S1T–S1V) with the pSpCas9(BB)-2A-Puro (PX459) V2.0 vector (Addgene plasmid # 62988). The sgRNA #2 was cloned as previously described (Ran et al., 2013). The cells were transfected using Lipofectamine 3000 according to the manufacturer’s protocols and selected with 2 μg / mL puromycin for 48 hours. Monoclonal cell populations were obtained by limiting dilutions. Knockout efficiency was tested at protein level by immunoblotting.
If not indicated otherwise, G3BP1 knockout was performed using sgRNA #1. All different siRNA, shRNA and sgRNA sequences targeting G3BP1 are shown in Table S1. Table S5 lists all commercially available siRNAs and shRNAs used in this study.
Rat model
Adult Wistar Cmd:(WI)WU rats were obtained from the Mossakowski Medical Research Center of the Polish Academy of Sciences (Breeder no: 004) and kept in standard housing conditions with ad libitum access to food and water. To obtain brain tissue for immunoprecipitation, the rats were sacrificed according to protocol, which complied with the Republic of Poland Act of 15 December 2015 on the protection of animals used for scientific or educational purposes and the European Community Council Directive 2010/63/EU.
Zebrafish maintenance and breeding
Adult zebrafish of the AB (Zebrafish International Resource Center), Tg(dlx5a/dlx6a-EGFP) (Noble et al., 2015) x Tg(vglut2a:loxP-RFP-loxP-GFP) (Satou et al., 2012) and Tg(HuC:GCaMP5G) (Ahrens et al., 2013) strain were maintained under standard aquaculture conditions in UV-sterilized water at 28.5°C on a 14 hour light / 10 hour dark cycle.
Fertilized eggs were collected via natural spawning. Embryos and larvae were raised in embryo medium, containing 1.5 mM HEPES, pH 7.6, 17.4 mM NaCl, 0.21 mM KCl, 0.12 mM MgSO4 and 0.18 mM Ca(NO3)2 in an incubator on a 14 hour light / 10 hour dark cycle at 28.5°C. For all experiments described, larvae at 0-4 days post fertilization (dpf) were used. All experiments performed at the University of Leuven were approved by the Ethics Committee of the University of Leuven (Ethische Commissie van de KU Leuven, approval number 150/2015) and by the Belgian Federal Department of Public Health, Food Safety and Environment (Federale Overheidsdienst Volksgezondheid, Veiligheid van de Voedselketen en Leefmilieu, approval number LA1210199). All experiments performed at the International Institute of Molecular and Cellular Biology in which zebrafish embryos and larvae were used were conducted in accordance with the Act of 15 January 2015 on the protection of animals used for scientific and educational purposes, Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes and were approved by Institutional Animal Welfare Advisory Team at the International Institute of Molecular and Cellular Biology in Poland.
Antisense morpholino knockdown
To achieve knockdown of g3bp1 in zebrafish embryos, we used morpholino antisense oligonucleotides designed to target the Exon 2 – Intron 2 boundary of the g3bp1 mRNA (g3bp1 MO). The morpholino sequence, as synthesized by GeneTools was: 5′-TAACAAAGGGCAAGTCACCTGTGCA-3′. A fluoresceinated standard control (5′-CCTCTTACCTCAGTTACAATTTATA-3′) with a 3′ fluorescein group was used as a negative control (control MO). Embryos were microinjected at the one- or two-cell stage with 1 nL of either g3bp1 or control MO, corresponding to 8 ng of morpholino per injection. For the knockdown of tsc2 the morpholino sequence 5′-CTGTGTGTTTTCTTACTGGACCATA-3′ was used targeting the Exon 1 – Intron 1 boundary of the tsc2 mRNA (tsc2 MO). Embryos were microinjected at the one- or two-cell stage with 2.5 ng of tsc2 splice MO per injection. The same amount of control MO was used. The morpholino concentration used was defined by titration as the highest at which the larvae displayed no morphological abnormalities. At 3-4 hours after injections, staging was performed to assure the same age of the eggs in all groups.
Method details
Cell treatments
SG formation was induced with arsenite at a final concentration of 500 μM for the indicated time periods. Prior to arsenite stress, cells were washed with phosphate-buffered saline (PBS) and serum-starved for 16 hours.
Metabolic stimulation experiments: for serum and amino acid starvation, cells were washed in PBS and cultured for 16 hours in Hank’s balanced salt solution (HBSS). For stimulation with insulin and amino acids (insulin / aa), the medium was exchanged to DMEM supplemented with 3 mM L-glutamine and 100 nM insulin. 1 μM insulin was used for stimulation in Figures 4A and 4C.
For serum starvation, cells were washed in PBS and cultured for 16 hours in DMEM with 4.5 g / L glucose, supplemented with 3 mM L-glutamine. For stimulation with insulin alone (Figure 4K), insulin was directly added to the serum-free media for the time periods indicated.
Lyophilized rapamycin was dissolved in methanol to a concentration of 1 nmol / μL and aliquoted to 5 μL per tube. 5 μL aliquots were dried with open lids under a sterile cell culture hood and deep frozen at –80°C. Aliquots were thawed immediately before an experiment and methanol-dried rapamycin was directly dissolved in HBSS or DMEM to a final concentration of 20 or 100 nM, as indicated. Hence, no carrier was used in experiments with rapamycin. Rapamycin pretreatments in Figures 1T and S1C started 30 min prior to stimulation or lysis, respectively.
Torin1 and MK2206 were dissolved in dimethyl sulfoxide (DMSO) and added to full DMEM medium to a final concentration of 250 nM and 10 nM, respectively. Torin1 or MK2206 were added 30 min prior to lysis (Figure S1D).
G3BP1 or G3BP2 re-expression in G3BP1 KO cells
450,000 MCF-7 control and 650,000 G3BP1 KO cells were seeded in 60 mm dishes 24 hours prior to transfection. Cells were transiently transfected with 4 μg of either a MYC-FLAG-empty vector, MYC-FLAG-G3BP1 or MYC-FLAG-G3BP2 using JetPEI following the manufacturer’s protocol. On the next day, the cells were washed once with PBS and cultured for 16 hours in HBSS. For metabolic stimulation, the medium was exchanged to DMEM supplemented with 3 mM L-glutamine and 100 nM insulin for 15 min.
Cell size measurements
Cell size was measured and analyzed with a Casy electronic cell counter, following the manufacturer’s protocol. In short, 2.5 × 105 cells were seeded in 6-well plates. After 24 hours of culture, cells were trypsinized and taken up in full DMEM. Each cell line was measured twice; each measurement comprised three cycles of cell counts with intervals of 0.05 μm ranging from 0 to 50 μm. The sum of counts of viable cells in the range of 12 to 30 μm was plotted and quantified. Three biological replicates were performed per MDA-MB-231 cell line (KO and corresponding Control). Control and corresponding KO were compared with multiple unpaired t tests. p values are presented as stars above the corresponding bar graphs. ∗, p < 0.05.
Cloning
The coding sequences (CDS) of G3BP1, G3BP2, LAMP1 and LAMP2 were obtained from the clone repository of the DKFZ Genomics and Proteomics Core Facility (GPCF) as Gateway® compatible clones in pENTR221 or pENTR223. The CDS of MTOR and TSC2 were gifts from Dominic Esposito (Addgene plasmids # 70422 and # 70640) and obtained as Gateway® compatible clones in pDonor-255. All CDS were generated with and without STOP-codon in the Gateway-frame. If only one CDS cloning was successful in first instance, the cloning of the second was performed on the successful one and pDONR223 was used as ENTRY clone to reduce background by changing E.coli resistance from kanamycin to spectinomycin. After sequence verification, the CDS without STOP-codon were cloned into the BiFC destination vectors pGW-MYC-LC151 for G3BP1, G3BP2, and LAMP1, and pGW-HA-LN151 for G3BP1, G3BP2, LAMP1, LAMP2, MTOR and TSC2 by Gateway®-specific LR-reaction following the manufacturer’s protocol (Invitrogen). Previously, the vectors bFos-MYC-LC151 and bJun-HA-LN151 (Chu et al., 2009) were adapted for Gateway cloning. MYC-LC151 and HA-LN151 PCR-fragments were generated and cloned into modified pDEST26 vectors resulting in pGW-MYC-LC151 and pGW-HA-LN151, as previously described (Weiler et al., 2014). Using the Gateway®-specific LR reaction, TSC2 and LAMP2 were also cloned into pEGFP-C (TSC2-GFP and LAMP2-GFP). Three G3BP1 truncation constructs in pGW-MYC-LC151 were generated with primers placed at the end or start positions of each construct, respectively: G3BP11-182-MYC, G3BP1183-332-MYC and G3BP1333-466-MYC. AttB sites were added to the CDS by a two-step PCR. The first PCR was performed with hybrid primers, consisting of half of the AttB sites and the other half being gene specific. The second PCR was done with primers covering the complete AttB sites (see Table S4 for primer details). Following the same principle also a LAMP11-382 lysosomal truncation construct was generated resulting in pGW-MYC-LC151-LAMP11-382. Since the cytoplasmic domain of LAMP1 is very small, we fused it to turboGFP (tGFP). We therefore added the restriction sites for BamHl and Xhol in pDONR201-tGFP. LAMP1383-417 was amplified with primers adding the restriction sites of BglII and SalI, and subsequently cloned into pDONR201-tGFP via one tube ligation. After sequence verification tGFP-LAMP1383-417 was cloned into the BiFC destination vector resulting in pGW-MYC-LC151-LAMP1383-417 by Gateway®-specific LR-reaction. The respective primers can be found in Table S4.
G3BP1 was amplified by PCR using cDNA from HeLa S3 cells as template, and inserted into pCMV6-AN-MYC-DDK (MYC-FLAG-empty) by restriction cloning using AsiSI and MluI. MYC-FLAG-G3BP1-ΔRGG (lacking amino acids 410-466), MYC-FLAG-G3BP1-ΔNTF2L (lacking amino acids 1-139) and MYC-FLAG-G3BP1-NTF2L (amino acids 1-139) constructs were generated by Site-Directed Mutagenesis using Phusion HF DNA Polymerase and the vector pCMV6-AN-MYC-DDK containing G3BP1 (MYC-FLAG-G3BP1) as template. G3BP2 was amplified by PCR using pGW-MYC-LC151-G3BP2 (see above) as template, and inserted into pCMV6-AN-Myc-DDK (MYC-FLAG-G3BP2) by restriction cloning using AsiSI and MluI.
All plasmids were verified with restriction digest and Sanger sequencing.
Cell lysis and immunoblotting
For lysis, cells were washed with PBS and lysed with radio immunoprecipitation assay (RIPA) buffer (1% IGEPAL CA-630, 0.1% SDS, and 0.5% sodium deoxycholate in PBS) supplemented with Complete Protease Inhibitor Cocktail, Phosphatase Inhibitor Cocktail 2 and Cocktail 3. The protein concentration was measured using Protein Assay Dye Reagent Concentrate and adjusted to the lowest value. Cell lysates were mixed with sample buffer (10% glycerol, 1% beta-mercaptoethanol, 1.7% SDS, 62.5 mM TRIS base [pH 6.8], and bromophenol blue), and heated for 5 min at 95°C. Cell lysates were then loaded on SDS polyacrylamide gel electrophoresis (PAGE) gels with a concentration of 8%, 10%, 12% or 14% polyacrylamide. Polyacrylamide gels were prepared consisting of two distinct layers: a stacking and a separation gel. For the lower separation gel, polyacrylamide was diluted to the respective percentage with 375 mM TRIS base [pH 8.8]. For the upper stacking gel, polyacrylamide was mixed with 0.125 M TRIS base [pH 6.8] to a final concentration of 13%. Electrophoresis was carried out with a Mini-PROTEAN Tetra Vertical Electrophoresis Cell system that was filled with electrophoresis buffer (0.2 M glycine, 25 mM TRIS base, and 0.1% SDS), and an applied voltage of 90 to 150 V. Subsequently, proteins were transferred to polyvinylidene difluoride (PVDF) membranes using a Mini-PROTEAN Tetra Vertical Electrophoresis Cell system filled with blotting buffer (0.1 M glycine, 50 mM TRIS base, 0.01% SDS, [pH 8.3], and 10% methanol) and an applied voltage of 45 V for 2 hours. Afterward, membranes were blocked in 5% bovine serum albumin (BSA) – TRIS-buffered saline tween (TBST) buffer (0.15 M NaCl, 60 mM TRIS base, 3 mM KCl, and 0.1% Tween-20, [pH 7.4]). Membranes were incubated overnight with primary antibodies at 4°C, following the manufacturer’s instructions for the respective antibodies (antibody dilutions can be found in Table S3). The next day, membranes were washed in TBST buffer and incubated for at least one hour with the corresponding horseradish peroxidase (HRP) coupled secondary antibodies. For detection, Pierce ECL western blotting substrate or SuperSignal West FEMTO were used to detect chemiluminescence using a LAS-4000 camera system, a ChemiDoc XRS+ camera or a Fusion Fx camera. For graphical presentation, raw images taken with the LAS-4000 or Fusion camera were exported as RGB color TIFF files using ImageJ version 1.50b (Schneider et al., 2012), and further processed with Adobe Photoshop version CS5.1. Raw images taken with a ChemiDoc XRS+ camera were processed with Image Lab version 5.2.1 and exported for publication as TIFF files with 600 dpi resolution. Numbers on the right of each single immunoblot indicate the kilodalton (kDa) size of the nearest protein ladder signal.
Re-analysis of the MTOR interactome
Re-analysis of the MTOR interactome data reported by Schwarz et al. (2015) (Figure 1A). Volcano plot showing the mean log10 ratios of proteins detected by tandem mass spectrometry in MTOR versus mock immunoprecipitation (IP) experiments. Proteins quantified in at least two out of three biological replicates were plotted against the negative log10 p value (Student′s t test). Proteins with a mean ratio > 5 and a p value < 0.01 (sector highlighted in dark gray) were considered significantly enriched. G3BP1 is marked in green, the mTORC1 core components MTOR and RPTOR are marked in blue.
Immunoprecipitation (IP)
For IP experiments, cells were washed three times in ice-cold PBS and then harvested in CHAPS based IP lysis buffer (40 mM HEPES, 120 mM NaCl, [pH 7.5] and 0.3% CHAPS) supplemented with Complete Protease Inhibitor Cocktail, Phosphatase Inhibitor Cocktail 2 and Cocktail 3. The lysate volume was adjusted to 1 - 2.5 mL per 15 cm cell culture plate, depending on the cell density. The lysate was incubated under gentle agitation for 20 min at 4°C, centrifuged for 3 min at 600 g at 4°C, the pellet was discarded and the supernatant was transferred to fresh tubes. In case of multiple samples, the protein concentration was measured using Protein Assay Dye Reagent Concentrate and all samples were adjusted to the lowest value. The lysates were pre-incubated with 10 μL pre-washed Protein G covered Dynabeads per mL of lysate for 30 min at 4°C under gentle agitation. A fraction of each lysate was mixed with 5 x sample buffer, referred to as ‘lysate’ input in the figure panels. For IP, the pre-cleaned lysates were subdivided, and specific antibodies or isotype control IgG antibodies (mock condition) were added using 7.5 μg antibody per mL of pre-cleaned lysate. Isotype control IgG antibodies (mock antibodies) were used in the same concentration as the protein-specific antibodies. After 30 min at 4°C under gentle agitation, 37.5 μL pre-washed Protein G covered Dynabeads / mL lysate were added, and the incubation was continued for 90 min at 4°C under gentle agitation. Finally, beads were washed with CHAPS lysis buffer three times shortly and three times for 10 min at 4°C under gentle agitation, and taken up in 1 x sample buffer. Samples were heated for 5 min at 95°C and separated by SDS-PAGE. For IP experiments with TSC2 and respective mock antibodies, the samples were heated for 10 min at 70°C.
For TSC1-IPs with NaCl and SDS washes (Figure 5C), the IP was performed as detailed above but with a CHAPS-based IP lysis buffer without NaCl (40 mM HEPES, [pH 7.5] and 0.3% CHAPS). Before the final washing steps, the TSC1-IP was subdivided into six tubes. Each IP was washed with CHAPS-based lysis buffer supplemented with the indicated NaCl or SDS concentrations three times shortly and three times for 10 min at 4°C under gentle agitation, and taken up in 1 x sample buffer. Samples were heated for 10 min at 70°C and separated by SDS-PAGE. The TSC1 antibody (TSC1 #1) was a gift from Michael N. Hall and generated as previously described (Molle, 2006; van Slegtenhorst et al., 1998).
For FLAG-IP experiments (Figures 5N and 5O), 2 × 106 HEK293T cells per dish were seeded in 10 cm dishes (2 dishes per condition). 24 hours after seeding, the cells were co-transfected with 2 μg LAMP2-GFP (full length) and 1 μg pCMV6-MYC-FLAG-G3BP1 constructs (full-length or truncated versions) using Lipofectamine 3000 following the manufacturer’s protocol. After 24 hours, the cells were lysed and IP was performed as described above adjusting the samples to 1 – 1.5 mL per 10 cm cell culture plate, depending on the cell density. Protein G covered Dynabeads were used for the IP and samples were heated for 5 min at 95°C and separated by SDS-PAGE.
For GFP-IP experiments in HEK293T cells (Figure 5D), 1.7 × 106 cells per 10 cm dish were seeded (1 dish per condition). 24 hours after seeding, the cells were co-transfected with 2 μg TSC2-GFP (full length) and 1 μg pCMV6-MYC-FLAG-G3BP1 constructs (full-length or truncated versions) using Lipofectamine 3000 following the manufacturer’s protocol. After 48 hours of transient overexpression, the cells were lysed and IP was performed as described above adjusting the samples to 1 – 1.5 mL per 10 cm cell culture plate, depending on the cell density. Protein G covered Dynabeads were used for the IP and samples were heated for 10 min at 95°C and separated by SDS-PAGE.
For GFP-IP experiments in HEK293-β2AR cells (Figure 5B), 1.7 × 106 cells per dish were seeded in 10 cm dishes (2 dishes per condition). 24 hours after seeding, the cells were co-transfected with 2 μg TSC2-GFP (full length) and 1 μg G3BP1-myc constructs (full-length or truncated versions) using Transfectin (ratio 2:1) in FBS-free DMEM, following the manufacturer’s protocol. After 48 hours of transient overexpression, cells were washed once in ice-cold PBS and pooled into one tube per condition. Cells were centrifuged at 16,000 g for 1 minute at room temperature and resuspended in 1 mL of CHAPS-based IP lysis buffer, supplemented with protease inhibitors (100 μM Leupeptin, 100 μM Aprotinin, 1 μg / mL Pepstatin A) and phosphatase inhibitors (1 mM Sodium orthovanadate, 1 mM Sodium pyrophosphate, 1 mM Sodium fluoride). The cells were disrupted and the DNA was sheared through the repeated use of a syringe with a 21G x 0.80 mm needle. Afterward, the lysate was incubated on ice for 15 min at 4°C, centrifuged for 45 min at 16,000 g at 4°C, the pellet was discarded and the supernatant was transferred to fresh tubes. In case of viscous supernatants, the DNA shearing was repeated. Otherwise, the lysates were pre-incubated with 12 μL Protein G Sepharose beads per mL of lysate for 60 min at 4°C under gentle agitation. A fraction of each lysate was mixed with 5 x sample buffer (25 mM Tris-HCl [pH 6.8]; 4% (w/v) SDS; 3% (w/v) DTT; 0.02% (v/v) bromophenol blue), referred to as ‘lysate’ in the figure panels. For IP, the pre-cleared lysates were subdivided, and 1 μg/mL of anti-GFP antibody or anti-Flag antibody were added. After 3 hours at 4°C under gentle agitation, 12 μL Protein G Sepharose beads per mL lysate were added, and the incubation was continued for 60 min at 4°C under gentle agitation. Finally, beads were washed with CHAPS-based lysis buffer five times shortly and once for 5 min at 4°C under gentle agitation. In between the samples were centrifuged for 1 minute at 9,600 g to remove the supernatant. Finally, the IP samples were dissolved in 30 μL 1 x sample buffer. Samples were heated for 5 min at 95°C and separated by SDS-PAGE.
The Wistar Cmd:(WI)WU rats that were used to obtain brain tissue for IP of endogenous TSC1 (Figure 7A) were sacrificed according to protocol, which complied with European Community Council Directive 2010/63/EU. The cerebral cortex from one hemisphere of a rat brain was homogenized in 4 mL lysis buffer (40 mM Tris-HCl [pH 7.5], 120 mM NaCl) containing 0.3% CHAPS, supplemented with protease and phosphatase inhibitors, using a glass teflon homogenizer. The homogenate was diluted 1:1 with lysis buffer containing 0.1% CHAPS and incubated under gentle agitation for 90 min at room temperature. The brain lysate was centrifuged at 1,000 g, 4°C for 10 min, the pellet was discarded and the supernatant was transferred to fresh tubes. A fraction of each lysate was mixed with 4 x sample buffer, referred to as ‘lysate’ input in the figure panels. 30 μL of Protein G covered Dynabeads were pre-conjugated in lysis buffer containing 0.1% CHAPS with 4 μg of TSC1 antibody or isotype control rabbit IgG (mock condition) for 2 hours at 4°C. For IP, the pre-conjugated beads were incubated with the lysate at 4°C overnight under gentle agitation. Finally, beads were washed with lysis buffer containing 0.1% CHAPS four times for 3 min at 4°C under gentle agitation, and taken up in 1 x sample buffer. Samples were heated for 10 min at 95°C and separated by SDS-PAGE.
Sucrose gradients
Cells were lysed in homogenization buffer (50 mM Tris-HCl [pH 7.4], 250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 3 mM imidazole), supplemented with Complete Protease Inhibitor Cocktail and Phosphatase Inhibitor Cocktail 2 and Cocktail 3 on a rocking platform for 30 min at 4°C. Subsequently, cells were scraped and centrifuged at 12,000 g for 10 min at 4°C. The pellet was discarded, the supernatant was transferred to a fresh tube and the absolute protein concentration was determined with Protein Assay Dye Reagent Concentrate by calculating a BSA adjustment curve ranging from 0.5 mg / mL to 7.5 mg / mL BSA. 1.5 mg protein was loaded on 4 mL of a continuous sucrose gradient (10% to 40% sucrose) and centrifuged 194,000 x g for 16 hours. Each sample was divided into 26 fractions and 5 x sample buffer was added to a final concentration of 1 x. Every second fraction was analyzed by immunoblot. Quantitation of pixel intensities was performed as described below. Normalization was carried out against the average value of all lanes, and normalized values of each lane were plotted for n = 3 independent experiments. Quantitations in Figure 3A and S4D show pixel intensities of immunoblot detections for each fraction as % of the overall signal of all fractions.
Lysosome preparation (lyso-prep) with dextran coated nanoparticles
Lysosomes were loaded with ferromagnetic nanoparticles (DexoMAG) by incubating MCF-7 cells for 12 hours in DMEM with 10% FBS, 10% DexoMAG solution and 10 mM HEPES. Afterward, cells were washed twice with wash buffer (0.25 M sucrose, 10 mM triethanolamine, 10 mM acetic acid, [pH 7.8]), then scraped and pelleted by centrifugation at 150 g for 5 min. Cells were resuspended in homogenization buffer (wash buffer supplemented with 1 mM EDTA, and protease (5 mM benzaminidine, 5 µM E64, 1 mM AEBSF, 1 μM aprotinin, 1 μM leupeptin, 100 μM pepstatin A) and phosphatase (1 mM Sodium orthovanadate, 50 mM Sodium fluoride, 10 mM Sodium pyrophosphate, 10 mM Sodium glycerophosphate) inhibitors. Cells were homogenized by 10 strokes with a dounce homogenizer and nuclei were pelleted by centrifugation at 200 g for 10 min. An LS MACS column was attached to a QuadroMACS magnet, equilibrated with 1 mL PBS and 0.5% BSA, and then washed with 1 mL PBS. The postnuclear supernatant (PNS) was loaded onto the equilibrated column. The column was washed twice with 600 μL column wash buffer (PBS, 0.1 mM sucrose, and protease and phosphatase inhibitors). Finally, lysosomes (lyso-prep) were eluted by detaching the column from the magnet and plunging with 400 μL elution buffer (PBS, 0.5 mM sucrose, and protease and phosphatase inhibitors). All isolation steps were performed at 4°C. Protein amounts were calculated using a BCA assay and equal protein amounts of PNS and lyso-prep were separated by SDS-PAGE and analyzed by immunoblotting as detailed above, with nitrocellulose membranes and chemiluminescence detection using a ChemiDoc XRS+ camera.
Trypsin treatment of lyso-preps
10 μg of freshly isolated lyso-prep were incubated with 0, 2.5 μg, 5 μg or 10 μg trypsin, respectively at 37°C for 5 min and subsequently separated by SDS-PAGE and analyzed by immunoblotting with nitrocellulose membranes and chemiluminescence detection using a ChemiDoc XRS+ camera.
Immunofluorescence (IF)
In order to analyze SG assembly, cells were grown on coverslips and treated as indicated in the respective figures. Cells were washed with PBS and fixed with ice-cold methanol for 5 min on ice. After fixation, cells were washed three times with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 60 s. Cells were washed with PBS and blocked with 3% FBS in PBS for 30 min at room temperature, and incubated with primary antibodies against G3BP1 and EIF3A at 4°C overnight. The cells were washed three times with PBS and incubated with Alexa Fluor 568 and Alexa Fluor 488 labeled secondary antibodies and Hoechst 33342 at room temperature for 30 min in the dark. Afterward, cells were washed three times with PBS and twice in deionized water. The cells were mounted with Mowiol 4-88, including DABCO (1,4-diazabicyclo[2.2.2]octane) and supplemented with 10% NPG (n-propyl-gallate). Cells were analyzed by fluorescence microscopy. Images were taken using a wide-field AxioObserver Z1 microscope equipped with an Apotome, a 63x / 1.4 oil objective, and an AxioCamMRm CCD camera. For each experimental setup, the magnification and exposure times were adjusted to the condition with the brightest signal, and the settings were retained throughout for all conditions. For presentation in figures, regions of interest (ROI) were selected and maintained for all channels, and single layers of Z stacks were exported as TIFF with no compression using Zen2012 blue edition software, and brightness and contrast were adjusted for better visibility. For single channel images, channels were pseudo-colored white, for merge images, the Alexa 488 channel was left green and the Alexa 568 channel was left magenta. Pictures are representative of n = 3 independent experiments, except the 0 min arsenite condition in the shG3BP1 #1 cells (n = 2; Figure S2Q).
The number of EIF3A positive puncta / cell was analyzed on unprocessed image raw files without any adjustment using Fiji software version 1.49v (Schindelin et al., 2012), creating maximum intensity projections of all Z stacks. We used a background subtraction of 1, threshold adjustment with the intermodes function, and the ‘Analyze Particles’ function with a particle size from 0.2-infinity and a circularity from 0.5-1. SG were counted using the EIF3A channel. The number of EIF3A positive puncta / image was then normalized to the number of cells by counting the nuclei in the Hoechst channel and analyzed using a one-way ANOVA followed by a Sidak’s multiple comparisons test across n = 9 pictures from n = 3 independent experiments, except the 0 min arsenite condition in the shG3BP1 #1 cells for which only 6 pictures from n = 2 experiments were analyzed.
For analysis of lysosomal foci, LAMP2 was stained as described previously (Demetriades et al., 2016). Cells were grown on coverslips and treated as indicated. The medium was removed and cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After fixation, cells were permeabilized twice with 0.1% Tween-20 in PBS for 10 min. Cells were blocked with 0.1% Tween-20 and 0.1% BSA in PBS for 45 min at room temperature, and incubated with primary antibodies against LAMP2 diluted in 0.1% Tween-20 and 0.1% BSA in PBS at 4°C for 2 hours. Afterward, cells were washed four times for 15 min with 0.1% Tween-20 and 0.1% BSA in PBS and incubated with TRITC labeled secondary antibodies and Hoechst 33342 at room temperature for 1 hour in the dark. Afterward, cells were washed twice in 0.1% Tween-20 in PBS for 15 min and twice in deionized water and mounted with Mowiol 4-88, supplemented with DABCO and NPG, and analyzed by fluorescence microscopy. Images were taken using a wide-field AxioObserver Z1 microscope equipped with an Apotome, a 63x / 1.4 oil objective, and an AxioCamMRm CCD camera. For each experimental setup, the magnification and exposure times were adjusted to the condition with the brightest signal, and the settings were retained for all conditions. For presentation in figures, regions of interest (ROI) were selected and maintained for all channels, and single layers of Z stacks were exported as TIFF with no compression using Zen2012 blue edition software, and brightness and contrast were adjusted for better visibility. The TRITC channel of the LAMP2 staining was pseudo-colored white. Pictures are representative of n = 12 fields of view from n = 3 independent experiments.
TSC2-LAMP1 co-staining was performed as described previously (Carroll et al., 2016). Briefly, cells were grown on coverslips and treated as indicated in Figure 4C. The medium was removed and cells were fixed with 4% formaldehyde in PBS for 10 min at room temperature. After fixation, cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. Cells were blocked with 5% normal goat serum in PBS and 0.05% Tween-20 for 1 hour at room temperature, and incubated with primary antibodies against TSC2 and LAMP1 at 4°C overnight. The following day, cells were washed and incubated with the appropriate secondary antibodies for 1 hour at room temperature. Afterward, the cells were washed and coverslips were mounted on slides with Prolong Gold antifade reagent with 4′,6-Diamidin-2-phenylindol (DAPI). Cells were analyzed by fluorescence microscopy. Z stack images were taken using a Leica SP8 microscope, a 63x objective, 1.5x digital zoom and filters suitable for the used fluorophores. Identical settings were used to capture images across five to six separate fields (20 to 40 cells) of view. For presentation in figures, pictures were opened in Fiji (version 1.52p) and Z stacks were projected (max). Channels were split and brightness and contrast were adjusted for better visibility. Afterward channels were converted to RGB color. Regions of interest (ROI) were selected and coordinates were copied to maintain the same ROI in the different channels. For single channel images, channels were pseudo-colored white, for merge images, the Alexa 488 channel was left green and the Alexa 555 channel was pseudo-colored magenta. All images were exported as TIFF with no compression.
For TSC2-LAMP1 co-staining, the Manders’ coefficient was calculated using the Coloc2 plug-in of the ImageJ software (v1.51r). Prior to running the plug-in, a mask was made of the DAPI channel and subtracted from the other channels. A constant threshold was applied to all the images in the Z stack, and for every image within each experiment and the Manders’ colocalization coefficient was calculated. Differences in the tested conditions were analyzed using a one-way ANOVA followed by a Sidak’s multiple comparisons test across n = 5-6 fields of view from one dataset representative of at least n = 3 independent experiments.
For MTOR-LAMP2 co-staining cells were grown on coverslips and treated as indicated in Figure 4J. The medium was removed and cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. After fixation, cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min at room temperature. Cells were blocked with 3% FBS in PBS for 30 min at room temperature, and incubated with primary antibodies against MTOR and LAMP2 at 4°C overnight. The following day, cells were washed and incubated with the appropriate secondary antibodies for 1 hour at room temperature. Afterward, the cells were washed and coverslips were mounted on slides with Prolong Gold antifade reagent with DAPI.
Images were taken using a wide-field AxioObserver Z1 microscope equipped with an Apotome, a 40x / 1.4 oil objective, and an AxioCamMRm CCD camera. For each experimental setup, the magnification and exposure times were adjusted to the condition with the brightest signal, and the settings were retained for all conditions. For presentation in figures, single layers of Z stacks were exported as TIFF with no compression using Zen2012 blue edition software, and brightness and contrast were adjusted for better visibility. For presentation in figures, regions of interest (ROI) were selected. For single channel images, channels were pseudo-colored white, for merge images, the Alexa 488 channel was left green and the Alexa 555 channel was pseudo-colored magenta. All images were exported as TIFF with no compression.
For MTOR-LAMP2 co-staining, the Manders’ coefficient was calculated using the Coloc2 plug-in of the ImageJ software (v1.51r). Prior to running the plug-in, a mask was made of the DAPI channel and subtracted from the other channels. A constant threshold was applied to all the images in the Z stack, and for every image within each experiment and the Manders’ colocalization coefficient was calculated. Differences in the tested conditions were analyzed using a one-way ANOVA followed by a Sidak’s multiple comparisons test across n = 10-15 fields of view from one dataset representative of n = 3 independent experiments. In total n = 36 distinct fields of view were analyzed.
Bimolecular fluorescence complementation (BiFC)
For BiFC analysis, we made use of the red fluorophore mLumin (Chu et al., 2009; Weiler et al., 2014). For analysis of interaction between the G3BPs and TSC2, LAMP1, LAMP2 as well as mTOR, HEK293T cells were seeded in a 24 well plate at 100,000 cells / well in full medium 24 hours prior to transfection. The cells were transiently transfected with Lipofectamine 3000 following the manufacturer’s protocol in the following combinations: pGW-MYC-LC151-G3BP1 (G3BP1 fused to a C-terminal mLumin fragment) with empty pGW-HA-LN151 as a negative control (an N-terminal mLumin fragment only), and pGW-MYC-LC151-G3BP1 with either pGW-HA-LN151-LAMP1, pGW-HA-LN151-LAMP2, pGW-HA-LN151-MTOR, or pGW-HA-LN151-TSC2, respectively (an N-terminal mLumin fragment fused to LAMP1, LAMP2, MTOR or TSC2, respectively) (Figure 5E). For G3BP2, cells were transfected with pGW-MYC-LC151-G3BP2 and either empty pGW-HA-LN151 as a negative control, or pGW-HA-LN151-LAMP1, pGW-HA-LN151-LAMP2, pGW-HA-LN151-MTOR, pGW-HA-LN151-TSC2, respectively (Figure 5H). In order to achieve equal expression of all plasmids, 3 times the amount of DNA was used for the MTOR, TSC2 and empty control plasmids in comparison to the G3BP1, G3BP2, LAMP1, and LAMP2 plasmids. Cells were analyzed 48 hours after transfection using a wide-field AxioObserver Z1, equipped with a 10x / 0.3 Plan-NEO objective, an AxioCamMRm CCD camera and an mPlum (64 HE) filter. mLumin fluorescence was analyzed with Fiji version 1.49 using a background subtraction of 50, threshold adjustment from 20-max, a Gaussian Blur filter of 1 and the ‘Analyze Particles’ function with a particle size from 20-infinity. The mLumin fluorophore signal was measured in percent / image and compared between the different combinations by a one-way ANOVA followed by a Sidak’s multiple comparisons test across at least 3 independent fields of view from at least three independent datasets, respectively. In total at least 22 independent fields of view for G3BP1 and 15 independent fields of view for G3BP2 were analyzed. All pictures were taken from regions with a comparable cell density.
For analysis of G3BP1 and G3BP2 homo- and heterodimers, MCF-7 cells were seeded in a 24 well plate at a density of 100,000 cells / well in full medium. 24 hours after seeding, cells were transiently transfected with Lipofectamine 3000 following the manufacturer’s protocol in the following combinations: pGW-MYC-LC151-G3BP1 with empty pGW-HA-LN151 as a negative control, or pGW-MYC-LC151-G3BP1 with either pGW-HA-LN151-G3BP1 or pGW-HA-LN151-G3BP2, respectively. In addition, cells were also transfected with pGW-MYC-LC151-G3BP2 together with empty pGW-HA-LN151, or pGW-HA-LN151-G3BP1, or pGW-HA-LN151-G3BP2, respectively. Cells were also transfected with the respective BiFC constructs in the other orientation, namely empty pGW-MYC-LC151 with pGW-HA-LN151-G3BP1, as well as empty pGW-MYC-LC151 with pGW-HA-LN151-G3BP2 (Figure 2L). Cells were analyzed 48 hours after transfection using a wide-field AxioObserver Z1, equipped with a 10x / 0.3 Plan-NEO objective, an AxioCamMRm CCD camera and an mPlum (64 HE) filter. mLumin fluorescence was analyzed with Fiji version 1.49 using a background subtraction of 50, threshold adjustment from 40-max, a Gaussian Blur filter of 1 and the ‘Analyze Particles’ function with a particle size from 10-infinity. The mLumin fluorophore signal was measured in percent / image and compared between the different combinations by a one-way ANOVA followed by a Sidak’s multiple comparisons test across in total at least 14 independent fields of view from at least three independent datasets. All pictures were taken from regions with a comparable cell density.
For analysis of the interaction between the G3BPs and the cytoplasmic or luminal LAMP1 protein domains, MCF-7 cells were seeded in a 24 well plate at a density of 100,000 cells / well in full medium. 24 hours after seeding, cells were transiently transfected with Lipofectamine 3000 following the manufacturer’s protocol in the following combinations: pGW-HA-LN151-G3BP1 with either empty pGW-MYC-LC151, with pGW-myc-LC151-LAMP1, with pGW-myc-LC151-LAMP11-382, or with pGW-myc-LC151-LAMP1383-417, respectively (Figure 5K). Following the same principle, also pGW-HA-LN151-G3BP2 was transfected together with either empty pGW-MYC-LC151, pGW-myc-LC151-LAMP1, pGW-myc-LC151-LAMP11-382, or pGW-myc-LC151-LAMP1383-417, respectively (Figure 5L). Cells were analyzed 48 hours after transfection using a wide-field AxioObserver Z1, equipped with a 10x / 0.3 Plan-NEO objective, an AxioCamMRm CCD camera and an mPlum (64 HE) filter. mLumin fluorescence was analyzed with Fiji version 1.49 using a background subtraction of 50, threshold adjustment from 20-max, a Gaussian Blur filter of 1 and the ‘Analyze Particles’ function with a particle size from 20-infinity. The mLumin fluorophore signal was measured in percent / image and compared between the different combinations by a one-way ANOVA followed by a Sidak’s multiple comparisons test across in total at least 45 independent fields of view from five independent datasets for G3BP1, and at least 27 independent fields of view from three independent datasets for G3BP2. All pictures were taken from regions with a comparable cell density.
In all BiFC experiments shown, the protein fused to the C-terminal mLumin fragment is indicated first, and the protein fused to the N-terminal mLumin fragment is indicated second.
Proximity Ligation Assay (PLA)
For TSC2-LAMP2 PLAs (Figure 4A), MCF-7 cells were trypsinized and seeded in a 16-well chamber slide at a density of 4x104 cells per well 72 h after siRNA transfection with siG3BP1 or siRenilla luciferase. The following day, cells were washed twice with HBSS, starved in HBSS for 16 hours, and then stimulated for 15 min with high-glucose DMEM containing 4 mM glutamine and 1 μM insulin. Afterward, cells were washed once with PBS, fixed with 4% formaldehyde for 15 min and permeabilized with 0.1% Tween-20 in PBS for 5 min. The PLA was performed using the Duolink In Situ Red Starter Kit Mouse/ Rabbit according to the manufacturer’s instructions. Briefly, after permeabilization, the samples were blocked, and then incubated overnight with antibodies against LAMP2 and TSC2. The following day, the samples were incubated with the MINUS and PLUS PLA probes corresponding to the primary antibodies used, followed by ligation and rolling-circle amplification in the presence of Texas-Red labeled oligos to generate the PLA signal. Finally, the samples were mounted with DAPI-containing mounting medium. All incubations were performed in a humidity chamber using a volume of 40 μL per well. The experiment was imaged with a confocal microscope (SP8, Leica); twelve stacks (7-8 μm thick with 0.3 μm spacing between consecutive layers) per condition were acquired.
For G3BP1-TSC2 and G3BP1-LAMP1 PLAs, MCF-7 CRISPR control and G3BP1 KO cells were seeded in a 16-well chamber slide at a density of 2x104 cells per well. The following day, cells were washed twice with PBS, and incubated with HBSS for 16 hours. Afterward, cells were washed once with PBS and fixed with 4% formaldehyde for 5 min and permeabilized with 0.1% Triton X-100 in PBS for 5 min. The PLA was performed as described above with antibodies against G3BP1 and TSC2 or LAMP1. The slides were analyzed using an AxioObserver Z1 compound microscope equipped with an Apotome (6 pictures per slide), 63x objective, and Axiocam 702mono and Axiocam 298 color cameras. Six stacks (0.5 μm thick) per condition were acquired.
For quantitation of all PLAs, the number of PLA puncta was counted across maximum intensity projections of raw files of each stack using CellProfiler (McQuin et al., 2018) and then normalized to the number of DAPI-positive nuclei in that field of view. The mean of the respective control condition was set to 1. In Figure 4A the mean of the Control at 0 min insulin / aa stimulation was normalized to 1. For presentation in figures, maximum intensity projections were exported as TIFF, and brightness or contrast were adjusted for better visibility.
Migration assays
For analysis of cell migration into a cell-free space (scratch assays) (Figure 6A), 2-well ibidi culture-inserts were placed into 24-well plates, generating a cell-free gap of 500 μM. After knockdown induction for 4 days, 15,000 cells / well were seeded in 100 μL full DMEM medium. 4 replicates were seeded per condition and cell line (MCF-7 shControl and shG3BP1 #1) in the presence of 2 μg / mL doxycycline to induce shRNA expression. Where indicated, rapamycin was added during seeding to a final concentration of 20 nM (24 hours prior to the 0 h time point). After 24 hours, ibidi culture-inserts were removed and the medium was replaced with 1 mL full DMEM medium, supplemented with 20 nM rapamycin where indicated. Pictures were taken after 0, 24, and 48 hours with a Nikon ECLIPSE Ti-E/B inverted microscope, equipped with a 4x objective, using the NIS Elements version 4.13.04 software (settings: optimal frame size 1280 × 1024, no binning, 12 bit). Pictures were taken from two different regions in an automated manner by selecting the x- and y-coordinates of the 24-well plate, assuring that the same region of the scratch was monitored across all conditions. Pictures were exported as TIFF files converting the 12 bit to 16 bit and analyzed using the TScratch software (Gebäck et al., 2009) and a consistent threshold of 250. For quantitation, the width of the open wound area of the 48 hour time point was normalized to the initial scratch size at the 0 h time point and expressed as the percentage of wound closure. Data was compared using a one-way ANOVA followed by a Sidak’s multiple comparisons test across n = 12 scratches from 3 independent experiments.
For Transwell migration assays (Figure 6E), FluoroBlok cell culture inserts with a polyethylene terephthalate (PET) membrane which blocks the transmission of light between 400 and 700 nm were placed, one insert / well, into 24-well companion plates with 750 μL of full DMEM medium containing 10% FBS as chemoattractant. 100,000 MDA-MB-231 Control or G3BP1 KO cells were subsequently seeded in 500 μL DMEM supplemented with 3 mM L-glutamine and 2% FBS into the top chamber of the inserts. After 6-8 hours, medium was flipped out of the inserts, and inserts were placed into a second 24-well companion plate with 500 μL / well HBSS medium and a final concentration of 4 μg / mL Calcein fluorescent dye. Living cells were stained with Calcein for 1 hour. Finally, inserts were placed into a third 24-well plate with 500 μL HBSS / well, and fluorescence of migrated cells was measured with the Tecan i-control software (v1.10.4.0) and a Tecan Infinite 200 PRO plate reader equipped with 485 / 530 (excitation / emission) filter. Calcein fluorescence (494 / 517 (ex / em)) was measured from the bottom of the plate so that only those cells that migrated through the light-blocking PET membrane were measured. Three wells per cell line were measured in parallel as technical replicates. The average fluorescence value per well was calculated from nine reads / well (3 × 3) and the average fluorescence value from all three wells was normalized to 1 for the MDA-MB-231 Control cells and the fold change of the G3BP1 KO cells was compared using a paired Student’s t test across n = 5 independent experiments.
Microscope pictures of the migrated cells were taken with a Nikon ECLIPSE Ti-E/B inverted microscope, equipped with a 4x objective and a GFP filter, using the NIS Elements version 4.13.04 software (settings: optimal frame size 1280 × 1024, no binning, 12 bit). Pictures were exported as TIFF files converting the 12 bit to 16 bit. The representative pictures were exported as TIFF with no compression and a pseudo-colored white Calcein signal using Zen2012 blue edition software.
Proliferation assays
Cell proliferation was monitored using an xCELLigence real-time cell analysis (RTCA) system, allowing real-time, label free cellular analysis. After knockdown induction for 4 days, MCF-7 cells (MCF-7 shControl and shG3BP1 #1) were seeded in duplicates at a total of 2,000 cells per E-plate 16 chamber following the manufacturer’s protocol, in the presence of 2 μg / mL doxycycline. Proliferation was measured as the relative change in electrical impedance every 30 min for 5 days until the cells reached the stationary growth phase. Proliferation was analyzed using the RTCA software 1.2. For presentation of the growth curves in Figure 6C, the measured impedance is displayed as relative confluence, normalized to 1 for the maximum value. In Figure 6D RTCA proliferation is shown as slope/h; shControl (normalized to 1) and shG3BP1 #1 cells were compared using a paired two-tailed Student’s t test across n = 6 independent experiments.
G3BP1 expression analyses
G3BP1 mRNA expression analysis (Figure 6G). Clinical and RNaseq data of invasive breast cancer (TCGA, provisional) were downloaded from cBio Cancer Genomics Portal (http://www.cbioportal.org) using the CGDS-R package (Gao et al., 2013), and classified according to PAM50. For 522 patients, information on the breast cancer subtype was available, of which 514 had RNaseq V2 data for G3BP1. A Kruskal-Wallis ANOVA by ranks was applied to evaluate subtype-dependent differences in G3BP1 transcription. n = 231 for luminal A, n = 127 for luminal B, n = 58 for HER2-enriched and n = 97 for basal-like. Data are shown as boxplots, representing the median with 25th and 75th percentiles as boxes and 5th and 95th percentiles as whiskers. The p value of the Kruskal-Wallis ANOVA by ranks is shown.
Survival analyses
The Kaplan Meier Plotter database (http://www.kmplot.com; Györffy et al., 2010; Szász et al., 2016) was used for survival analysis. Relapse free survival (RFS) was assessed in breast cancer patients based on gene expression of G3BP1 (probeID: 225007_at) (Figure 6H), TSC1 (probeID: 209390_at) (Figure 6J), and TSC2 (probeID: 215735_at) (Figure 6K). Outlier gene arrays were excluded leaving 1764 patients for analysis of G3BP1 and 3571 patients for analyses of TSC1/TSC2. n = 1224 with high G3BP1 mRNA expression; n = 409 with low G3BP1 mRNA expression; n = 2541 with high TSC1 mRNA expression; n = 1030 with low TSC1 mRNA expression; n = 1712 with high TSC2 mRNA expression; n = 1859 with low TSC2 mRNA expression.
RFS analysis in relation to G3BP1 protein expression (Figure 6I) also was based on data available in the Kaplan-Meier Plotter database, which included 126 patients. n = 57 with high G3BP1 protein expression; n = 67 with low G3BP1 protein expression.
For all calculations, patients were split based on the best performing expression threshold and log-rank p values were calculated.
Zebrafish treatments
For pharmacological assessment, 3 dpf larvae were individually placed into the wells of a 24-well or 96-well-plate, with each well containing 400 μL or 100 μL, respectively, of a freshly prepared 10 μM rapamycin or 25 μM ethosuximide solution in embryo medium. The untreated larvae were treated similarly with 400 μL or 100 μL of embryo medium. Larvae were incubated for 24 hours, and non-invasive local field potential recordings, locomotor tracking and in vivo imaging experiments were carried out as described below.
Zebrafish larvae lysis and immunoblotting
For Rps6-pS235/236 analysis 10 zebrafish larvae (2-3 dpf) were pooled per condition and independent experiment (n = 4) and homogenized in RIPA buffer supplemented with Complete Mini Protease Inhibitor cocktail. A Pierce BCA protein assay kit was used to determine the protein concentration of the lysates. 40 μg of protein were separated on a NuPage Novex 10% Bis-Tris gel, using SDS-PAGE with NuPage MES SDS running buffer, followed by dry transfer to an iBlot gel transfer stacks nitrocellulose membrane with an iBlot Dry Blotting System, which was then blocked for 1 hour at room temperature in Odyssey blocking buffer. Overnight incubation at 4°C with a primary antibody against RPS6-pS235/236 was followed by incubation with Dylight secondary goat antibody to rabbit IgG for 1 hour at room temperature. A rabbit antibody against GAPDH was used as a loading control. For detection, fluorescence signal was detected using an Odyssey 2.1 imaging system (Li-Cor, USA). For graphical presentation, raw images were further processed with Adobe Photoshop version CS5.1.
IF analysis of the zebrafish pallium
For IF analysis of the zebrafish pallium, 4 dpf zebrafish larvae were fixed with 4% paraformaldehyde and 0.1 M sodium fluoride for whole-mount immunostaining with DAPI and an antibody against RPS6-pS235/236. The whole front brains were imaged using a Z.1 Light sheet microscope, equipped with a 40x objective (NA 1.0). Images were analyzed in Fiji.
In vivo imaging of migrating neuronal progenitors from the subventricular zone (SVZ)
For analysis of cell migration in vivo, we utilized the Tg(HuC:GCaMP5G) stable transgenic zebrafish line in the mitfa−/−;royorbisson−/− genetic background (Ahrens et al., 2013) to follow the migration of newly-born HuC-positive neuronal progenitors (Kim et al., 1996) from the subventricular zone (SVZ) toward the outer layers. Tg(HuC:GCaMP5G) zebrafish larvae were injected with g3bp1 MO or control MO. 24 hours post fertilization (hpf), embryos were anesthetized with 0.02% tricaine (MS-222) and embedded in 2% agarose for imaging. The front brains were imaged using a Z.1 Light sheet microscope, equipped with a 40x objective (NA 1.0). Images were taken with 0.8 zoom in a time-lapse mode every 5 min for 2 hours in 32 degrees (approximately 2.5 hours of zebrafish development).
Images were processed and analyzed in Fiji (version 1.53c). Each hemisphere was analyzed separately. Briefly, spatial drift was corrected across time points, then 3D projections were generated for quantification of the number of HuC-positive cells migrating from the SVZ to the telencephalon. The single cell tracking of movement dynamics was performed with TrackMate (Tinevez et al., 2017).
Non-invasive local field potential (LFP) recordings
Brain activity of 4 dpf zebrafish larvae was assessed by performing non-invasive local field potential (LFP) recordings, reading the electrical signal from the skin of the larvae’s head (Zdebik et al., 2013). g3bp1 MO or control MO injected zebrafish larvae were treated on 3 dpf for 24 hours with rapamycin or left untreated. A glass pipet (containing the recording electrode), filled with artificial cerebrospinal fluid (124 mM NaCl, 2 mM KCl, 2 mM MgSO4, 2 mM CaCl2, 1.25 mM KH2PO4, 26 mM NaHCO3 and 10 mM glucose), was positioned on the skin above the optic tectum or the pallium using a stereomicroscope. The differential signal between the recording electrode and the reference electrode was amplified 10,000 times by DAGAN 2400 amplifier (Minnesota, USA), band pass filtered at 0.3-300 Hz and digitized at 2 kHz via a PCI-6251 interface (National Instruments, UK) with WinEDR (John Dempster, University of Strathclyde, UK). Recordings lasted for 10 min and were analyzed with Clampfit 10.2 software (Molecular Devices Corporation, USA). A polyspiking discharge was scored positive when its amplitude exceeded three times the amplitude of the baseline and it had a duration of at least 50 ms. At least n = 34 larvae for LFPs of zebrafish pallia (Figure 7L) and at least n = 20 larvae for LFPs of zebrafish optic tecta (Figure 7N) were analyzed.
Three representative 10 minute LFP recordings are shown for control and g3bp1 MO without and with rapamycin, respectively in Figure 7M (pallia) and Figure 7O (optic tecta).
In vivo imaging of pan-neuronal activity
The Tg(HuC:GCaMP5G) (Ahrens et al., 2013) stable zebrafish line in the mitfa−/−;royorbisson−/− genetic background was used for pan-neuronal activity imaging. g3bp1 MO or control MO injected zebrafish larvae were treated on 3 dpf for 24 hours with rapamycin or left untreated. 4 dpf zebrafish larvae were embedded in 2% agarose for imaging and the tails were cleared of agarose to allow movement. The front brains were imaged using lightsheet microscopy (Z.1 Lightsheet microscope, Zeiss) and 40x objective (NA 1.0) at zoom 0.8x. Images were taken in a time-lapse mode every 7 s for 3 min. Images were processed and analyzed in Fiji2 similarly to Kedra et al. (2020). Each hemisphere was analyzed separately. Briefly, spatial drift was corrected across time points, then changes in the fluorescence intensity were calculated using the measurement tool in Fiji and Rstudio (rstudio.com) in a semi-automated manner. The linear model was fitted to empirical data using the LOESS regression and smoothing function in RStudio (least-squares regression in localized subsets).
In vivo imaging of glutamatergic and GABAergic networks
Tg(dlx5a/dlx6a-EGFP) x Tg(vglut2a:loxP-RFP-loxP-GFP) zebrafish (Noble et al., 2015; Satou et al., 2012) larvae, injected with control or g3bp1 MO, were anesthetized at 4 dpf with tricaine (MS-222), immobilized in 2% low melting point agarose on a cover glass. For imaging of the glutamatergic and GABAergic networks in the optic tectum a two-photon LSM 780 confocal microscope equipped with an LD LCI Plan Apo 25x/0.8 objective was used. The EGFP and RFP markers were respectively excited at 488 nm and 561 nm and emission was recorded at 493 / 548 nm and 593 / 656 nm, respectively. All stacks were visualized and analyzed using Imaris 9.1. GABAergic and glutamatergic cells were counted as 5.5 μm and 3.5 μm diameter spheres respectively, and sorted by their fluorescence. The threshold was adapted in order to count all cells.
Locomotor activity recordings
The locomotor behavior of 4 dpf zebrafish larvae was evaluated by means of an automated tracking device (ZebraboxTM). tsc2 MO or g3bp1 MO and their respective control MO injected larvae were treated on 3 dpf for 24 hours with rapamycin (Figures 7W and 7X) or ethosuximide (Figure 7Y) or left untreated. Zebrafish larvae were arrayed individually into a 24-well plate that contained 400 μL of embryo medium per well. After an adaptation phase of 5 min in the Zebrabox, the locomotor activity was tracked for 15 min in the dark. The total movement was expressed in “actinteg” units, which are defined as the sum of all image pixel changes detected during the time of the tracking experiment.
Human Genomic Analysis
The 100,000 Genomes Project data (GEL, The National Genomics Research and Healthcare Knowledgebase v5, Genomics England. https://doi.org/10.6084/m9.figshare.4530893.v5. 2019.) was accessed and analyzed through the Genomics England Research Environment via https://re.extge.co.uk/ovd/. A bash script (Document S3) was written to identify all single nucleotide variants (SNV) within G3BP1 at chr5:151771954-151812785 (GRCh38/hg38, Genbank: NM_198395) and G3BP2 at chr4:75642769-75673483 (GRCh38/hg38, Genbank: NM_203505). Genomes of 64,185 participants with rare diseases and matched family members were analyzed. Variants were filtered for protein altering SNVs. Depth of coverage at variant loci was determined in probands and their available relatives. Minor allele frequencies (MAFs) were assessed using GnomAD (https://gnomad.broadinstitute.org/). In silico prediction scores predicting pathogenicity were determined using SIFT, PolyPhen, CADD and exomiser.
Quantification and statistical analysis
Immunoblot quantitation
Quantitation of raw images taken with a LAS-4000 camera system or FUSION FX7 with DarQ-9 camera was performed using ImageQuant TL Version 8.1. Background subtraction was performed using the rolling ball method with a defined radius of 200 for all images. Quantitation of raw images taken with a ChemiDoc XRS+ camera system was performed using Image Lab version 5.2.1. For all images, pixel values of a single lane were normalized to the average value of all lanes, and then normalized to the loading control indicated in the respective immunoblot figure. Quantitation of raw images taken with an Odyssey 2.1 imaging system (Li-Cor) was performed using Image Studio Lite Version 5.2. For images from immunoblot analysis of zebrafish samples (Figure 7B), pixel values of a single lane were normalized to the single value of the loading control GAPDH and then to the Control MO within each experiment.
Protein sequence analysis
To analyze the sequence similarities between human G3BP1 (UniProt: Q13283) and human G3BP2 (UniProt: Q9UN86) and their domains, or between human and zebrafish G3BP1 (UniProt: Q6P124) EMBOSS Needle (Madeira et al., 2019) with the Blosum62 matrix was used. Visualization of sequence alignments was done using Texshade based on a ClustalW alignment of the whole protein sequences. The domains indicated for G3BP1 were based on Reineke and Lloyd (2015).
Phylogenetic analysis
To identify possible orthologs in other species, the human proteins G3BP1 (UniProt: Q13283), G3BP2 (UniProt: Q9UN86), TSC1 (UniProt: Q92574), TSC2 (UniProt: P49815), TBC1D7 (UniProt: Q9P0N9), RHEB (UniProt: Q15382), and MTOR (UniProt: P42345) were used as query proteins for a blastp+ search (Camacho et al., 2009) against the NCBI non-redundant protein sequence database (nr; version 2017-11). The e value cut-off for identified proteins was 1e-30 with a maximum of 20,000 target sequences, disabled low-complexity filtering, using the BLOSUM62 matrix, a word size of 6 and gap opening and extension costs of 11 and 1, respectively. The results were parsed and filtered using custom Python scripts (https://github.com/MolecularBioinformatics/Phylogenetic-analysis) and manually curated as described earlier (Bockwoldt et al., 2019).
Statistical analysis
GraphPad Prism version 7.04 or 8.03 was used for statistical analysis and statistical presentation of quantitations. p values are presented in the figures above or below the compared conditions.
Where two conditions were compared, either a paired two-tailed Student’s t test (Figures 1S, 2F, 2H, 3E, 3I, 3K, 3L, 3O–3Q, 4F, 4G, 6D, 6F, 7C, S2I, S2J, S2L, S2M, S2O, S2P, and S4B) or unpaired two-tailed Student’s t test (Figures 7F, 7I, 7Q–7S, 7U, and 7V), or an unpaired two-tailed Mann-Whitney test (Figures 7G, 7H, 7J, 7K, and 7T) was performed. Cell size was compared using multiple unpaired t tests (Figures 4H and S4K).
If more than two conditions were compared, a one-way ANOVA followed by a Sidak’s multiple comparisons test was applied (Figures 1D, 1E, 1G–1I, 1K–1M, 1O–1Q, 1U–1X, 2B–2D, 2J, 2M, 4B, 4D, 4I, 4L–4N, 5F, 5I, 5J, 5M, 6B, 7L, 7N, 7W–7Y, S1M–S1O, S1Q–S1S, S1U, S1V, S2B–S2D, S2R, S2T, S2U, S2W, S2X, S4F, S4G, S4I, and S4J). In the case of immunoblot time courses more than two conditions were compared using a two-way ANOVA (Figures S1G, S1H, S1J, S1K, S2E, and S2F).
For G3BP1 expression analysis (Figure 6G) a Kruskal-Wallis ANOVA by ranks was performed using Dell Statistica version 13. For the analysis of relapse-free survival (Figures 6H–6K) the Kaplan-Meier Plotter was used and a log-rank test was applied.
For each experiment, the number of replicates is indicated in the figure legend with “n” designating the number of biological replicates if not indicated otherwise. For bar graphs, the corresponding dot plots were overlaid.
For each experiment the appropriate controls were used as described above: Control KO cells for G3BP1 KO; Control KO cells for TSC2 KO; shControl cells for shG3BP1 #1 and shG3BP1 #2; siControl transfected cells for siG3BP1, siG3BP2, siRHEB and siTSC2; siRenilla transfected cells (Control) for siG3BP1 (Figure 4A); control MO injected zebrafish larvae for g3bp1 MO; control MO injected zebrafish larvae for tsc2 MO.
Acknowledgments
We thank the DKFZ Light Microscopy Facility; M.N. Hall for TSC1 and TSC2 antibodies (Molle, 2006); D. Esposito for Addgene plasmids 70422 and 70640; Q. Luo for the bFos-myc-LC151 and bJun-HA-LN-151 plasmids; M. Orger for Tg(HuC:GCaMP5G); M. Ekker for Tg(dlx5a/dlx6a-EGFP); S. Higashijima for Tg(vglut2a:loxP-RFP-loxP-GFP); J. Maes for zebrafish microinjections; the Cell and Tissue Imaging Cluster (CIC) for microscopy supported by Hercules AKUL/11/37 and FWO G.0929.15 (to P. Vanden Berghe); and J. Utikal and his lab, K. Breuker, S.A. Fernandes, L.F. Somarribas Patterson, M. Rodriguez Peiris, and A. Sadik for support and helpful discussions. We acknowledge support from research awards from the German Tuberous Sclerosis Foundation 2019 (to M.T.P.) and 2017 (to K.T.); the German Research Foundation (Excellence Initiative GSC-4, Spemann Graduate School to M.T.P.; TH 1358/3-1 to K.T.; SFB 430 1389–UNITE Glioblastoma to A.v.D., S.P., and C.A.O.; and Germany’s Excellence Strategy EXC 294 and EXC-2189-Projektnummer 390939984, CRC850, and CRC1381 to R.B.); the Graduate School of Medical Sciences of the University of Groningen (to M.C.S.); the German TS Foundation (to K.T.); Stichting TSC Fonds (to K.T. and M.N.); TS Alliance and TS Association UK (to M.N.); BMBF e:Med initiatives MAPTor-NET (031A426B to K.T.), GlioPATH (01ZX1402 to C.A.O., K.T., and S.T.); a Rosalind Franklin Fellowship of the University of Groningen (to K.T.); the PoLiMeR Innovative Training Network (Marie Skłodowska-Curie grant agreement 812616 to K.T.) and the MESI-STRAT project (grant agreement 754688 to C.A.O., I.H., and K.T.), which received funding from the European Union Horizon 2020 Research and Innovation Program; the European Research Council (ERC) under the European Union Horizon 2020 Research and Innovation Program (grant agreement 757729) and the Max Planck Society (to C.D.); the Austrian Science Fund (FWF DK W11 and P32608 to T.Y. and L.A.H.); the Molecular Cell Biology and Oncology PhD Program at the Medical University of Innsbruck (MCBO; to T.Y. and L.A.H.); the Fund for O6260 Research Foundation–Flanders (FWO; 11F2919N to A.-S.d.M.), the British Skin Foundation and Vice-Chancellor’s Fellowship, University of Bristol (to B.C.); a long-term EMBO postdoctoral fellowship (ALT-755-2018 to G.F.); a TEAM grant from the Foundation for Polish Science (POIR.04.04.00-00-5CBE/17-00 to J.J. and A.K.); a Polish National Science Centre Etiuda grant (2020/36/T/NZ3/00132 to M.K.); the University of Leuven (grant C32/18/067 to A.S.); a Fellowship for Extraordinary Young Scientists from the Polish Ministry of Science and Higher Education (to J.Z.); and a Sêr Cymru II Precision Medicine Fellowship (to H.W.). This research was made possible by access to the data and findings generated by the 100,000 Genomes Project, managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care) and funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK, and the Medical Research Council have also funded research infrastructure. The 100,000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. The Genomics England Research Consortium members and affiliations can be found online with this article in the Document S2.
Author contributions
M.T.P., U.R., and M.C.S. planned/conducted/analyzed experiments and wrote the manuscript. A.M.H., B.B., B.H., K.K., I.v.t.L.-K., M.R., A.R., F.R., and S.W. supported the experiments. A.M.H., A.K., L.B., and C.D. supported in-depth data interpretation. R.B. supported project initiation. J.M.R.P. and S.P. performed cloning. S.P. and A.v.D supported BiFC. A.-S.d.M., M.K., C.S., A.S., P.d.W., J.Z., and J.J. performed zebrafish analyses. S.R.C. and W.P. performed lyso-preps. M.B. and I.H. performed phylogenetic analyses. B.C. and V.I.K. performed IF. G.F. and A.A.T. performed TSC2-LAMP2 PLAs. M.E.G.d.A. supported cell size analyses. M.N., L.A.H., and T.Y. supported CRISPR experiments. M.M. and J.J. performed rat brain IP. O.T.-Q. and E.S. performed IP for Figure 5B. S.T. analyzed expression data. L.E.T., J.R.S., and H.D.W. analyzed GEL data. C.A.O. and K.T. planned/guided the project and wrote the manuscript. All authors participated in in-depth discussions and revised the manuscript. Apart from first/last authors, all authors are listed alphabetically.
Declaration of interests
The authors declare no competing interests.
Published: January 25, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.12.024.
Contributor Information
Christiane A. Opitz, Email: c.opitz@dkfz.de.
Kathrin Thedieck, Email: kathrin.thedieck@uibk.ac.at.
Supplemental information
References
- Ahrens M.B., Orger M.B., Robson D.N., Li J.M., Keller P.J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- Alam U., Kennedy D. Rasputin a decade on and more promiscuous than ever? A review of G3BPs. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:360–370. doi: 10.1016/j.bbamcr.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaiz A.A., Micale L., Mandriani B., Augello B., Pellico M.T., Chrast J., Xenarios I., Zelante L., Merla G., Reymond A. TBC1D7 mutations are associated with intellectual disability, macrocrania, patellar dislocation, and celiac disease. Hum. Mutat. 2014;35:447–451. doi: 10.1002/humu.22529. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N. Stressful initiations. J. Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- Anderson P., Kedersha N., Ivanov P. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta. 2015;1849:861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov S., Takahashi M., Kakihana T., Katsuragi Y., Kitaura H., Zhang L., Kakita A., Fujii M. G3BP1 inhibits ubiquitinated protein aggregations induced by p62 and USP10. Sci. Rep. 2019;9:12896. doi: 10.1038/s41598-019-46237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annibaldi A., Dousse A., Martin S., Tazi J., Widmann C. Revisiting G3BP1 as a RasGAP binding protein: sensitization of tumor cells to chemotherapy by the RasGAP 317-326 sequence does not involve G3BP1. PLoS ONE. 2011;6:e29024. doi: 10.1371/journal.pone.0029024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller S., Balling R., Barisic N., Baulac S., Caglayan H., Craiu D., De Jonghe P., Depienne C., Dimova P., Djémié T., EuroEPINOMICS-RES Consortium. Epilepsy Phenome/Genome Project. Epi4K Consortium De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrinidis A., Cash T.P., Hunter D.S., Walker C.L., Chernoff J., Henske E.P. Tuberin, the tuberous sclerosis complex 2 tumor suppressor gene product, regulates Rho activation, cell adhesion and migration. Oncogene. 2002;21:8470–8476. doi: 10.1038/sj.onc.1205962. [DOI] [PubMed] [Google Scholar]
- Avruch J., Hara K., Lin Y., Liu M., Long X., Ortiz-Vega S., Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- Baraban S.C., Dinday M.T., Hortopan G.A. Drug screening in Scn1a zebrafish mutant identifies clemizole as a potential Dravet syndrome treatment. Nat. Commun. 2013;4:2410. doi: 10.1038/ncomms3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C., Hall M.N. Where is mTOR and what is it doing there? J. Cell Biol. 2013;203:563–574. doi: 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikkavilli R.K., Malbon C.C. Arginine methylation of G3BP1 in response to Wnt3a regulates β-catenin mRNA. J. Cell Sci. 2011;124:2310–2320. doi: 10.1242/jcs.084046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N., Lederer M., Pfalz B., Reinke C., Fuchs T., Glaß M., Möller B., Hüttelmaier S. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 2015;43:e26. doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockwoldt M., Houry D., Niere M., Gossmann T.I., Reinartz I., Schug A., Ziegler M., Heiland I. Identification of evolutionary and kinetic drivers of NAD-dependent signaling. Proc. Natl. Acad. Sci. USA. 2019;116:15957–15966. doi: 10.1073/pnas.1902346116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkowska J., Schwartz R.A., Kotulska K., Jozwiak S. Tuberous sclerosis complex: tumors and tumorigenesis. Int. J. Dermatol. 2011;50:13–20. doi: 10.1111/j.1365-4632.2010.04727.x. [DOI] [PubMed] [Google Scholar]
- Bozzi Y., Provenzano G., Casarosa S. Neurobiological bases of autism-epilepsy comorbidity: a focus on excitation/inhibition imbalance. Eur. J. Neurosci. 2018;47:534–548. doi: 10.1111/ejn.13595. [DOI] [PubMed] [Google Scholar]
- Brenet A., Hassan-Abdi R., Somkhit J., Yanicostas C., Soussi-Yanicostas N. Defective Excitatory/Inhibitory Synaptic Balance and Increased Neuron Apoptosis in a Zebrafish Model of Dravet Syndrome. Cells. 2019;8:1199. doi: 10.3390/cells8101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capo-Chichi J.M., Tcherkezian J., Hamdan F.F., Décarie J.C., Dobrzeniecka S., Patry L., Nadon M.A., Mucha B.E., Major P., Shevell M. Disruption of TBC1D7, a subunit of the TSC1-TSC2 protein complex, in intellectual disability and megalencephaly. J. Med. Genet. 2013;50:740–744. doi: 10.1136/jmedgenet-2013-101680. [DOI] [PubMed] [Google Scholar]
- Carroll B., Maetzel D., Maddocks O.D., Otten G., Ratcliff M., Smith G.R., Dunlop E.A., Passos J.F., Davies O.R., Jaenisch R. Control of TSC2-Rheb signaling axis by arginine regulates mTORC1 activity. eLife. 2016;5:e11058. doi: 10.7554/eLife.11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Zhang Z., Zheng Y., Yang J., Qin L., Lu J., Huang Z.L., Zeng S., Luo Q. A novel far-red bimolecular fluorescence complementation system that allows for efficient visualization of protein interactions under physiological conditions. Biosens. Bioelectron. 2009;25:234–239. doi: 10.1016/j.bios.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Condon K.J., Sabatini D.M. Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 2019;132:jcs222570. doi: 10.1242/jcs.222570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino P.B. mTOR signaling in epilepsy: insights from malformations of cortical development. Cold Spring Harb. Perspect. Med. 2015;5:a022442. doi: 10.1101/cshperspect.a022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino P.B. The mTOR signalling cascade: paving new roads to cure neurological disease. Nat. Rev. Neurol. 2016;12:379–392. doi: 10.1038/nrneurol.2016.81. [DOI] [PubMed] [Google Scholar]
- Curatolo P., D’Argenzio L., Cerminara C., Bombardieri R. Management of epilepsy in tuberous sclerosis complex. Expert Rev. Neurother. 2008;8:457–467. doi: 10.1586/14737175.8.3.457. [DOI] [PubMed] [Google Scholar]
- Debaize L., Jakobczyk H., Rio A.G., Gandemer V., Troadec M.B. Optimization of proximity ligation assay (PLA) for detection of protein interactions and fusion proteins in non-adherent cells: application to pre-B lymphocytes. Mol. Cytogenet. 2017;10:27. doi: 10.1186/s13039-017-0328-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C., Doumpas N., Teleman A.A. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell. 2014;156:786–799. doi: 10.1016/j.cell.2014.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C., Plescher M., Teleman A.A. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat. Commun. 2016;7:10662. doi: 10.1038/ncomms10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C.C., Elis W., Menon S., Qin W., Klekota J., Asara J.M., Finan P.M., Kwiatkowski D.J., Murphy L.O., Manning B.D. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou N., Chen J., Yu S., Gao Y., Li Y. G3BP1 contributes to tumor metastasis via upregulation of Slug expression in hepatocellular carcinoma. Am. J. Cancer Res. 2016;6:2641–2650. [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Forman-Kay J.D., Mittag T. From sequence and forces to structure, function, and evolution of intrinsically disordered proteins. Structure. 2013;21:1492–1499. doi: 10.1016/j.str.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich R.W., Jacobson G.A., Zhu P. Circuit neuroscience in zebrafish. Curr. Biol. 2010;20:R371–R381. doi: 10.1016/j.cub.2010.02.039. [DOI] [PubMed] [Google Scholar]
- Gallouzi I.E., Parker F., Chebli K., Maurier F., Labourier E., Barlat I., Capony J.P., Tocque B., Tazi J. A novel phosphorylation-dependent RNase activity of GAP-SH3 binding protein: a potential link between signal transduction and RNA stability. Mol. Cell. Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A., Zwartkruis F.J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S.C., Hafen E., Bos J.L., Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Gebäck T., Schulz M.M., Koumoutsakos P., Detmar M. TScratch: a novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques. 2009;46:265–274. doi: 10.2144/000113083. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Tergaonkar V., Rothlin C.V., Correa R.G., Bottero V., Bist P., Verma I.M., Hunter T. Essential role of tuberous sclerosis genes TSC1 and TSC2 in NF-kappaB activation and cell survival. Cancer Cell. 2006;10:215–226. doi: 10.1016/j.ccr.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Goncharova E.A., Goncharov D.A., Lim P.N., Noonan D., Krymskaya V.P. Modulation of cell migration and invasiveness by tumor suppressor TSC2 in lymphangioleiomyomatosis. Am. J. Respir. Cell Mol. Biol. 2006;34:473–480. doi: 10.1165/rcmb.2005-0374OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabiner B.C., Nardi V., Birsoy K., Possemato R., Shen K., Sinha S., Jordan A., Beck A.H., Sabatini D.M. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. 2014;4:554–563. doi: 10.1158/2159-8290.CD-13-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Boixet J., Kopach A., Holehouse A.S., Wittmann S., Jahnel M., Schlüßler R., Kim K., Trussina I.R.E.A., Wang J., Mateju D. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell. 2020;181:346–361.e17. doi: 10.1016/j.cell.2020.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- Hansmann P., Brückner A., Kiontke S., Berkenfeld B., Seebohm G., Brouillard P., Vikkula M., Jansen F.E., Nellist M., Oeckinghaus A., Kümmel D. Structure of the TSC2 GAP Domain: Mechanistic Insight into Catalysis and Pathogenic Mutations. Structure. 2020;28:933–942.e4. doi: 10.1016/j.str.2020.05.008. [DOI] [PubMed] [Google Scholar]
- Heberle A.M., Razquin Navas P., Langelaar-Makkinje M., Kasack K., Sadik A., Faessler E., Hahn U., Marx-Stoelting P., Opitz C.A., Sers C. The PI3K and MAPK/p38 pathways control stress granule assembly in a hierarchical manner. Life Sci. Alliance. 2019;2 doi: 10.26508/lsa.201800257. e201800257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højgaard C., Sørensen H.V., Pedersen J.S., Winther J.R., Otzen D.E. Can a Charged Surfactant Unfold an Uncharged Protein? Biophys. J. 2018;115:2081–2086. doi: 10.1016/j.bpj.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz M.K., Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J. Biol. Chem. 2005;280:26089–26093. doi: 10.1074/jbc.M504045200. [DOI] [PubMed] [Google Scholar]
- Hoxhaj G., Manning B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 2019;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.D., Chinenov Y., Kerppola T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Huang J., Manning B.D. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem. J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Dibble C.C., Matsuzaki M., Manning B.D. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol. Cell. Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunyadi B., Siekierska A., Sourbron J., Copmans D., de Witte P.A.M. Automated analysis of brain activity for seizure detection in zebrafish models of epilepsy. J. Neurosci. Methods. 2017;287:13–24. doi: 10.1016/j.jneumeth.2017.05.024. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., Guan K.L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Ouyang H., Zhu T., Lindvall C., Wang Y., Zhang X., Yang Q., Bennett C., Harada Y., Stankunas K. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- Irvine K., Stirling R., Hume D., Kennedy D. Rasputin, more promiscuous than ever: a review of G3BP. Int. J. Dev. Biol. 2004;48:1065–1077. doi: 10.1387/ijdb.041893ki. [DOI] [PubMed] [Google Scholar]
- Jedrusik-Bode M., Studencka M., Smolka C., Baumann T., Schmidt H., Kampf J., Paap F., Martin S., Tazi J., Müller K.M. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J. Cell Sci. 2013;126:5166–5177. doi: 10.1242/jcs.130708. [DOI] [PubMed] [Google Scholar]
- Jozwiak S., Kotulska K., Wong M., Bebin M. Modifying genetic epilepsies - Results from studies on tuberous sclerosis complex. Neuropharmacology. 2020;166:107908. doi: 10.1016/j.neuropharm.2019.107908. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Kedersha N., Panas M.D., Achorn C.A., Lyons S., Tisdale S., Hickman T., Thomas M., Lieberman J., McInerney G.M., Ivanov P., Anderson P. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol. 2016;212:845–860. doi: 10.1083/jcb.201508028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedra M., Banasiak K., Kisielewska K., Wolinska-Niziol L., Jaworski J., Zmorzynska J. TrkB hyperactivity contributes to brain dysconnectivity, epileptogenesis, and anxiety in zebrafish model of Tuberous Sclerosis Complex. Proc. Natl. Acad. Sci. USA. 2020;117:2170–2179. doi: 10.1073/pnas.1910834117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy D., French J., Guitard E., Ru K., Tocque B., Mattick J. Characterization of G3BPs: tissue specific expression, chromosomal localisation and rasGAP(120) binding studies. J. Cell. Biochem. 2001;84:173–187. doi: 10.1002/jcb.1277. [DOI] [PubMed] [Google Scholar]
- Kim J., Guan K.L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019;21:63–71. doi: 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- Kim C.H., Ueshima E., Muraoka O., Tanaka H., Yeo S.Y., Huh T.L., Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 1996;216:109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Speirs C.K., Solnica-Krezel L., Ess K.C. Zebrafish model of tuberous sclerosis complex reveals cell-autonomous and non-cell-autonomous functions of mutant tuberin. Dis. Model. Mech. 2011;4:255–267. doi: 10.1242/dmm.005587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D.C., Fulton R.S., McLellan M.D., Schmidt H., Kalicki-Veizer J., McMichael J.F., Fulton L.L., Dooling D.J., Ding L., Mardis E.R., Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski D.J., Wagle N. mTOR Inhibitors in Cancer: What Can We Learn from Exceptional Responses? EBioMedicine. 2014;2:2–4. doi: 10.1016/j.ebiom.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C., Mercier J.F., Salahpour A., Umapathy D., Breit A., Villeneuve L.R., Zhu W.Z., Xiao R.P., Lakatta E.G., Bouvier M., Hébert T.E. Beta 1/beta 2-adrenergic receptor heterodimerization regulates beta 2-adrenergic receptor internalization and ERK signaling efficacy. J. Biol. Chem. 2002;277:35402–35410. doi: 10.1074/jbc.M204163200. [DOI] [PubMed] [Google Scholar]
- Liao Y.C., Fernandopulle M.S., Wang G., Choi H., Hao L., Drerup C.M., Patel R., Qamar S., Nixon-Abell J., Shen Y. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell. 2019;179:147–164.e20. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiCausi F., Hartman N.W. Role of mTOR Complexes in Neurogenesis. Int. J. Mol. Sci. 2018;19:1544. doi: 10.3390/ijms19051544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X., Lin Y., Ortiz-Vega S., Yonezawa K., Avruch J. Rheb binds and regulates the mTOR kinase. Curr. Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Ma L., Chen Z., Erdjument-Bromage H., Tempst P., Pandolfi P.P. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte L., Crino P.B. The neurobiology of the tuberous sclerosis complex. Neuromolecular Med. 2006;8:531–546. doi: 10.1385/NMM:8:4:531. [DOI] [PubMed] [Google Scholar]
- Marsan E., Baulac S. Review: Mechanistic target of rapamycin (mTOR) pathway, focal cortical dysplasia and epilepsy. Neuropathol. Appl. Neurobiol. 2018;44:6–17. doi: 10.1111/nan.12463. [DOI] [PubMed] [Google Scholar]
- Martin S., Zekri L., Metz A., Maurice T., Chebli K., Vignes M., Tazi J. Deficiency of G3BP1, the stress granules assembly factor, results in abnormal synaptic plasticity and calcium homeostasis in neurons. J. Neurochem. 2013;125:175–184. doi: 10.1111/jnc.12189. [DOI] [PubMed] [Google Scholar]
- Matsuki H., Takahashi M., Higuchi M., Makokha G.N., Oie M., Fujii M. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 2013;18:135–146. doi: 10.1111/gtc.12023. [DOI] [PubMed] [Google Scholar]
- McQuin C., Goodman A., Chernyshev V., Kamentsky L., Cimini B.A., Karhohs K.W., Doan M., Ding L., Rafelski S.M., Thirstrup D. CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol. 2018;16:e2005970. doi: 10.1371/journal.pbio.2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S., Dibble C.C., Talbott G., Hoxhaj G., Valvezan A.J., Takahashi H., Cantley L.C., Manning B.D. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156:771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F., Akcakanat A., Chen H., Do K.A., Sangai T., Adkins F., Gonzalez-Angulo A.M., Rashid A., Crosby K., Dong M. PIK3CA/PTEN mutations and Akt activation as markers of sensitivity to allosteric mTOR inhibitors. Clin. Cancer Res. 2012;18:1777–1789. doi: 10.1158/1078-0432.CCR-11-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto H., Tatsukawa T., Shimohata A., Yamagata T., Suzuki T., Amano K., Mazaki E., Raveau M., Ogiwara I., Oba-Asaka A. Impaired cortico-striatal excitatory transmission triggers epilepsy. Nat. Commun. 2019;10:1917. doi: 10.1038/s41467-019-09954-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle K.-D. University of Basel; 2006. Regulation of the mammalian target of rapamycin complex 2 (mTORC2). PhD thesis. [Google Scholar]
- Mossmann D., Park S., Hall M.N. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer. 2018;18:744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- Mueller T., Wullimann M. Elsevier Science; 2015. Atlas of Early Zebrafish Brain Development: A Tool for Molecular Neurogenetics. [Google Scholar]
- Mühlebner A., Bongaarts A., Sarnat H.B., Scholl T., Aronica E. New insights into a spectrum of developmental malformations related to mTOR dysregulations: challenges and perspectives. J. Anat. 2019;235:521–542. doi: 10.1111/joa.12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellist M., van Slegtenhorst M.A., Goedbloed M., van den Ouweland A.M., Halley D.J., van der Sluijs P. Characterization of the cytosolic tuberin-hamartin complex. Tuberin is a cytosolic chaperone for hamartin. J. Biol. Chem. 1999;274:35647–35652. doi: 10.1074/jbc.274.50.35647. [DOI] [PubMed] [Google Scholar]
- Noble S., Godoy R., Affaticati P., Ekker M. Transgenic Zebrafish Expressing mCherry in the Mitochondria of Dopaminergic Neurons. Zebrafish. 2015;12:349–356. doi: 10.1089/zeb.2015.1085. [DOI] [PubMed] [Google Scholar]
- Omer A., Barrera M.C., Moran J.L., Lian X.J., Di Marco S., Beausejour C., Gallouzi I.E. G3BP1 controls the senescence-associated secretome and its impact on cancer progression. Nat. Commun. 2020;11:4979. doi: 10.1038/s41467-020-18734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova K.A., Crino P.B. The tuberous sclerosis complex. Ann. N Y Acad. Sci. 2010;1184:87–105. doi: 10.1111/j.1749-6632.2009.05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Kedersha N., Schulte T., Branca R.M., Ivanov P., Anderson P. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J. Cell Biol. 2019;218:2425–2432. doi: 10.1083/jcb.201801214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker F., Maurier F., Delumeau I., Duchesne M., Faucher D., Debussche L., Dugue A., Schweighoffer F., Tocque B. A Ras-GTPase-activating protein SH3-domain-binding protein. Mol. Cell. Biol. 1996;16:2561–2569. doi: 10.1128/mcb.16.6.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.O., Brock A.J., Walton R.T., Brennan C.H. The role of zebrafish (Danio rerio) in dissecting the genetics and neural circuits of executive function. Front. Neural Circuits. 2013;7:63. doi: 10.3389/fncir.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M., Um S.H., Mieulet V., Sticker M., Goss V.L., Mestan J., Mueller M., Fumagalli S., Kozma S.C., Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C.J., Huang H., Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Prigent M., Barlat I., Langen H., Dargemont C. IkappaBalpha and IkappaBalpha /NF-kappa B complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J. Biol. Chem. 2000;275:36441–36449. doi: 10.1074/jbc.M004751200. [DOI] [PubMed] [Google Scholar]
- Rabanal-Ruiz Y., Korolchuk V.I. mTORC1 and Nutrient Homeostasis: The Central Role of the Lysosome. Int. J. Mol. Sci. 2018;19:818. doi: 10.3390/ijms19030818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L.C., Lloyd R.E. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J. Virol. 2015;89:2575–2589. doi: 10.1128/JVI.02791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L.C., Neilson J.R. Differences between acute and chronic stress granules, and how these differences may impact function in human disease. Biochem. Pharmacol. 2019;162:123–131. doi: 10.1016/j.bcp.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs C.L., Kedersha N., Ivanov P., Anderson P. Mammalian stress granules and P bodies at a glance. J. Cell Sci. 2020;133 doi: 10.1242/jcs.242487. jcs242487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M., Hofer K., Kubista M., Hengstschläger M. Cell size regulation by the human TSC tumor suppressor proteins depends on PI3K and FKBP38. Oncogene. 2003;22:4786–4798. doi: 10.1038/sj.onc.1206776. [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C., Kimura Y., Higashijima S. Generation of multiple classes of V0 neurons in zebrafish spinal cord: progenitor heterogeneity and temporal control of neuronal diversity. J. Neurosci. 2012;32:1771–1783. doi: 10.1523/JNEUROSCI.5500-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheldeman C., Mills J.D., Siekierska A., Serra I., Copmans D., Iyer A.M., Whalley B.J., Maes J., Jansen A.C., Lagae L. mTOR-related neuropathology in mutant tsc2 zebrafish: Phenotypic, transcriptomic and pharmacological analysis. Neurobiol. Dis. 2017;108:225–237. doi: 10.1016/j.nbd.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J.J., Wiese H., Tölle R.C., Zarei M., Dengjel J., Warscheid B., Thedieck K. Functional Proteomics Identifies Acinus L as a Direct Insulin- and Amino Acid-Dependent Mammalian Target of Rapamycin Complex 1 (mTORC1) Substrate. Mol. Cell. Proteomics. 2015;14:2042–2055. doi: 10.1074/mcp.M114.045807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfakianos A.P., Mellor L.E., Pang Y.F., Kritsiligkou P., Needs H., Abou-Hamdan H., Désaubry L., Poulin G.B., Ashe M.P., Whitmarsh A.J. The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ. 2018;25:1766–1780. doi: 10.1038/s41418-018-0076-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Valenstein M.L., Gu X., Sabatini D.M. Arg-78 of Nprl2 catalyzes GATOR1-stimulated GTP hydrolysis by the Rag GTPases. J. Biol. Chem. 2019;294:2970–2975. doi: 10.1074/jbc.AC119.007382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekierska A., Stamberger H., Deconinck T., Oprescu S.N., Partoens M., Zhang Y., Sourbron J., Adriaenssens E., Mullen P., Wiencek P., C4RCD Research Group. AR working group of the EuroEPINOMICS RES Consortium Biallelic VARS variants cause developmental encephalopathy with microcephaly that is recapitulated in vars knockout zebrafish. Nat. Commun. 2019;10:708. doi: 10.1038/s41467-018-07953-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourbron J., Schneider H., Kecskés A., Liu Y., Buening E.M., Lagae L., Smolders I., de Witte P. Serotonergic Modulation as Effective Treatment for Dravet Syndrome in a Zebrafish Mutant Model. ACS Chem. Neurosci. 2016;7:588–598. doi: 10.1021/acschemneuro.5b00342. [DOI] [PubMed] [Google Scholar]
- Switon K., Kotulska K., Janusz-Kaminska A., Zmorzynska J., Jaworski J. Molecular neurobiology of mTOR. Neuroscience. 2017;341:112–153. doi: 10.1016/j.neuroscience.2016.11.017. [DOI] [PubMed] [Google Scholar]
- Szász A.M., Lánczky A., Nagy Á., Förster S., Hark K., Green J.E., Boussioutas A., Busuttil R., Szabó A., Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Ito N., Dickson B.J., Treisman J.E., Hariharan I.K. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Tee A.R. The Target of Rapamycin and Mechanisms of Cell Growth. Int. J. Mol. Sci. 2018;19:880. doi: 10.3390/ijms19030880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A.R., Manning B.D., Roux P.P., Cantley L.C., Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Tee A.R., Sampson J.R., Pal D.K., Bateman J.M. The role of mTOR signalling in neurogenesis, insights from tuberous sclerosis complex. Semin. Cell Dev. Biol. 2016;52:12–20. doi: 10.1016/j.semcdb.2016.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K., Polak P., Kim M.L., Molle K.D., Cohen A., Jenö P., Arrieumerlou C., Hall M.N. PRAS40 and PRR5-like protein are new mTOR interactors that regulate apoptosis. PLoS ONE. 2007;2:e1217. doi: 10.1371/journal.pone.0001217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K., Holzwarth B., Prentzell M.T., Boehlke C., Kläsener K., Ruf S., Sonntag A.G., Maerz L., Grellscheid S.N., Kremmer E. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 2013;154:859–874. doi: 10.1016/j.cell.2013.07.031. [DOI] [PubMed] [Google Scholar]
- Thien A., Prentzell M.T., Holzwarth B., Kläsener K., Kuper I., Boehlke C., Sonntag A.G., Ruf S., Maerz L., Nitschke R. TSC1 activates TGF-β-Smad2/3 signaling in growth arrest and epithelial-to-mesenchymal transition. Dev. Cell. 2015;32:617–630. doi: 10.1016/j.devcel.2015.01.026. [DOI] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- van Slegtenhorst M., Nellist M., Nagelkerken B., Cheadle J., Snell R., van den Ouweland A., Reuser A., Sampson J., Halley D., van der Sluijs P. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum. Mol. Genet. 1998;7:1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- Wagle N., Grabiner B.C., Van Allen E.M., Hodis E., Jacobus S., Supko J.G., Stewart M., Choueiri T.K., Gandhi L., Cleary J.M. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov. 2014;4:546–553. doi: 10.1158/2159-8290.CD-13-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Proud C.G. p70 S6 kinase is activated by sodium arsenite in adult rat cardiomyocytes: roles for phosphatidylinositol 3-kinase and p38 MAP kinase. Biochem. Biophys. Res. Commun. 1997;238:207–212. doi: 10.1006/bbrc.1997.7273. [DOI] [PubMed] [Google Scholar]
- Wang Y., Fu D., Chen Y., Su J., Wang Y., Li X., Zhai W., Niu Y., Yue D., Geng H. G3BP1 promotes tumor progression and metastasis through IL-6/G3BP1/STAT3 signaling axis in renal cell carcinomas. Cell Death Dis. 2018;9:501. doi: 10.1038/s41419-018-0504-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler M., Blaes J., Pusch S., Sahm F., Czabanka M., Luger S., Bunse L., Solecki G., Eichwald V., Jugold M. mTOR target NDRG1 confers MGMT-dependent resistance to alkylating chemotherapy. Proc. Natl. Acad. Sci. USA. 2014;111:409–414. doi: 10.1073/pnas.1314469111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Cardenas A.M., Marissen W.E., Lloyd R.E. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2007;2:295–305. doi: 10.1016/j.chom.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Winslow S., Leandersson K., Larsson C. Regulation of PMP22 mRNA by G3BP1 affects cell proliferation in breast cancer cells. Mol. Cancer. 2013;12:156. doi: 10.1186/1476-4598-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F., Bodenmiller B., Trajkovska M.G., Wanka S., Aebersold R., Pelkmans L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell. 2013;152:791–805. doi: 10.1016/j.cell.2013.01.033. [DOI] [PubMed] [Google Scholar]
- Wong M., Crino P.B. mTOR and Epileptogenesis in Developmental Brain Malformations. In: Noebels J.L., Avoli M., Rogawski M.A., Olsen R.W., Delgado-Escueta A.V., editors. Jasper’s Basic Mechanisms of the Epilepsies. Oxford University Press; 2012. pp. 1241–1254. [Google Scholar]
- Wu X.N., Wang X.K., Wu S.Q., Lu J., Zheng M., Wang Y.H., Zhou H., Zhang H., Han J. Phosphorylation of Raptor by p38beta participates in arsenite-induced mammalian target of rapamycin complex 1 (mTORC1) activation. J. Biol. Chem. 2011;286:31501–31511. doi: 10.1074/jbc.M111.233122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant G.A., Abu-Remaileh M., Wolfson R.L., Chen W.W., Freinkman E., Danai L.V., Vander Heiden M.G., Sabatini D.M. mTORC1 Activator SLC38A9 Is Required to Efflux Essential Amino Acids from Lysosomes and Use Protein as a Nutrient. Cell. 2017;171:642–654.e12. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Shen Y., Garre E., Hao X., Krumlinde D., Cvijović M., Arens C., Nyström T., Liu B., Sunnerhagen P. Stress granule-defective mutants deregulate stress responsive transcripts. PLoS Genet. 2014;10:e1004763. doi: 10.1371/journal.pgen.1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Mathieu C., Kolaitis R.M., Zhang P., Messing J., Yurtsever U., Yang Z., Wu J., Li Y., Pan Q. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell. 2020;181:325–345.e28. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Zhang Y., Ting C.Y., Bettedi L., Kim K., Ghaniam E., Lilly M.A. The Rag GTPase Regulates the Dynamic Behavior of TSC Downstream of Both Amino Acid and Growth Factor Restriction. Dev. Cell. 2020;55:272–288.e5. doi: 10.1016/j.devcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdebik A.A., Mahmood F., Stanescu H.C., Kleta R., Bockenhauer D., Russell C. Epilepsy in kcnj10 morphant zebrafish assessed with a novel method for long-term EEG recordings. PLoS ONE. 2013;8:e79765. doi: 10.1371/journal.pone.0079765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zekri L., Chebli K., Tourrière H., Nielsen F.C., Hansen T.V., Rami A., Tazi J. Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol. Cell. Biol. 2005;25:8703–8716. doi: 10.1128/MCB.25.19.8703-8716.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao X., Saucedo L.J., Ru B., Edgar B.A., Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhang S., He H., Zhao W., Chen J., Shao R.G. GAP161 targets and downregulates G3BP to suppress cell growth and potentiate cisplaitin-mediated cytotoxicity to colon carcinoma HCT116 cells. Cancer Sci. 2012;103:1848–1856. doi: 10.1111/j.1349-7006.2012.02361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kim J., Alexander A., Cai S., Tripathi D.N., Dere R., Tee A.R., Tait-Mulder J., Di Nardo A., Han J.M. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Ma Y., Zhang S., Liu H., He H., Li N., Gong Y., Zhao S., Jiang J.D., Shao R.G. Involvement of Ras GTPase-activating protein SH3 domain-binding protein 1 in the epithelial-to-mesenchymal transition-induced metastasis of breast cancer cells via the Smad signaling pathway. Oncotarget. 2015;6:17039–17053. doi: 10.18632/oncotarget.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kecskés A., Copmans D., Langlois M., Crawford A.D., Ceulemans B., Lagae L., de Witte P.A., Esguerra C.V. Pharmacological characterization of an antisense knockdown zebrafish model of Dravet syndrome: inhibition of epileptic seizures by the serotonin agonist fenfluramine. PLoS ONE. 2015;10:e0125898. doi: 10.1371/journal.pone.0125898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.H., Wang J.X., Cai M.L., Shao R., Liu H., Zhao W.L. The roles and mechanisms of G3BP1 in tumour promotion. J. Drug Target. 2019;27:300–305. doi: 10.1080/1061186X.2018.1523415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Red, zebrafish pallium; blue, WM compartments; yellow, active single cells in WM compartments.
Red, zebrafish pallium; blue, WM compartments; yellow, active single cells in WM compartments.
Data Availability Statement
The custom Python scripts for the phylogenetic analysis are accessible via https://github.com/MolecularBioinformatics/Phylogenetic-analysis and were manually curated as described earlier (Bockwoldt et al., 2019).
The bash script to identify the single nucleotide variants (SNV) within G3BP1 and G3BP2 is provided in Document S3. The script was adapted from the ‘extract_variants_by_coordinate.sh’ script for germline variants (https://research-help.genomicsengland.co.uk/display/GERE/Extract+variants+by+coordinate) and was run on the command line within the Genomics England Research environment using bcftools (https://samtools.github.io/bcftools/).