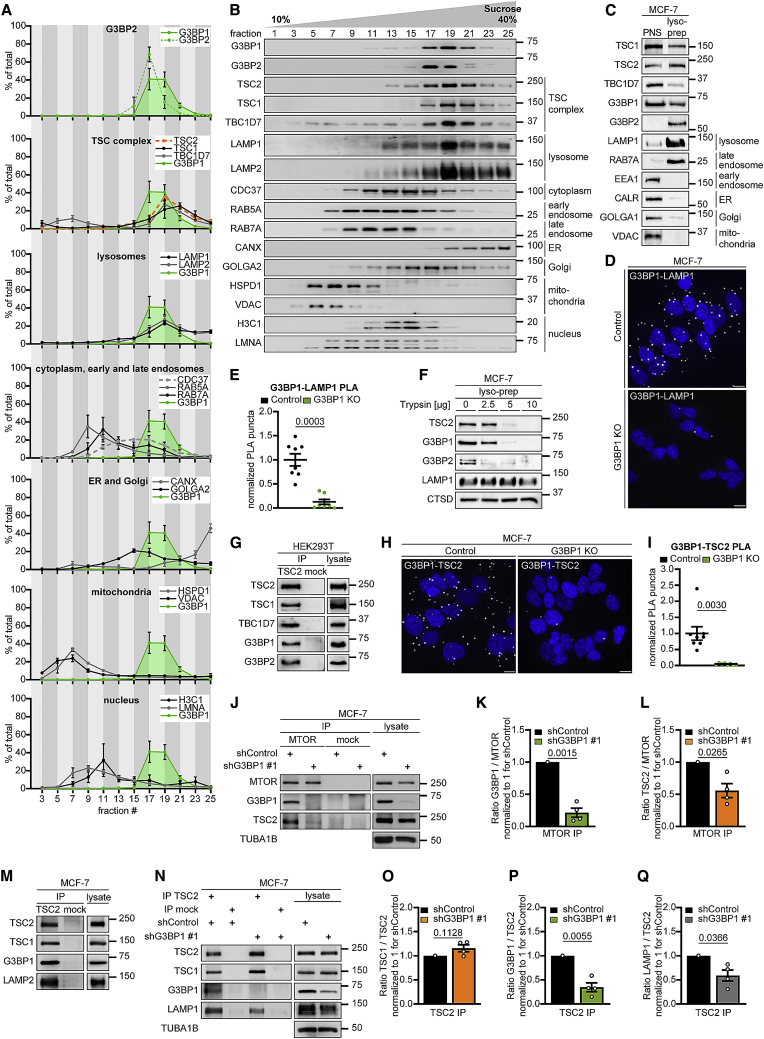
Figure 3.
G3BP1 and G3BP2 reside at lysosomes
(A) Quantitation of data in (B). G3BP1, green area. Mean ± SEM.
(B) Sucrose density gradient separation of serum/aa-starved MCF-7 cells. n = 3.
(C) Lyso-prep with ferromagnetic nanoparticles. PNS, postnuclear supernatant. n = 3.
(D) PLA of G3BP1-LAMP1 in serum/aa-starved G3BP1 KO cells. PLA puncta, white dots; nuclei, blue (DAPI). Scale bar, 10 μm. n = 3.
(E) Quantitation of data in (D). Shown are data points and mean ± SEM. n = 8 technical replicates.
(F) Trypsin digest of lyso-preps prepared as in (C). n = 3 except for TSC2 (n = 2).
(G) IP against TSC2 (TSC2 #1) or mock (mouse IgG). n = 3.
(H) PLA of G3BP1-TSC2 in serum/aa-starved G3BP1 KO cells. PLA puncta, white dots; nuclei, blue (DAPI). Scale bar, 10 μm. n = 4.
(I) Quantitation of data in (H). Shown are data points and mean ± SEM. n = 8 technical replicates.
(J) IP against MTOR or mock (rat IgG); insulin/aa-stimulated shG3BP1 #1 cells (15 min). n = 4.
(K) Quantitation of G3BP1 in (J). Shown are data points and mean ± SEM.
(L) Quantitation of TSC2 in (J). Data are shown as in (K).
(M) IP against TSC2 (TSC2 #2 or #3) or mock (rabbit IgG). n = 3.
(N) IP against TSC2 (TSC2 #2) or mock (rabbit IgG); insulin/aa-stimulated shG3BP1 #1 cells (15 min). n = 4.
(O) Quantitation of TSC1 in (N). Shown are data points and mean ± SEM.
(P) Quantitation of G3BP1 in (N). Data are shown as in (O).
(Q) Quantitation of LAMP1 in (N). Data are shown as in (O).
See also Figure S4.